Significance
Thiopeptides are a subclass of ribosomally synthesized natural products with complex structures and potent antimicrobial activities. Here we describe a general strategy that allows the incorporation of noncanonical amino acids into thiopeptides by introducing orthogonal amber suppressor aminoacyl-tRNA synthetase/tRNA pairs into a thiocillin-producing strain of Bacillus cereus. We show that thiocillin variants harboring a noncanonical amino acid with bioorthogonal chemical reactivity can be further modified to create probes for biological studies. This work should significantly enhance our ability to manipulate the structures and properties of ribosomally produced natural products by recombinant methods.
Keywords: noncanonical amino acid, biosynthesis, natural products, thiopeptides, antibiotic
Abstract
Thiopeptides are a subclass of ribosomally synthesized and posttranslationally modified peptides (RiPPs) with complex molecular architectures and an array of biological activities, including potent antimicrobial activity. Here we report the generation of thiopeptides containing noncanonical amino acids (ncAAs) by introducing orthogonal amber suppressor aminoacyl-tRNA synthetase/tRNA pairs into a thiocillin producer strain of Bacillus cereus. We demonstrate that thiopeptide variants containing ncAAs with bioorthogonal chemical reactivity can be further postbiosynthetically modified with biophysical probes, including fluorophores and photo-cross-linkers. This work allows the site-specific incorporation of ncAAs into thiopeptides to increase their structural diversity and probe their biological activity; similar approaches can likely be applied to other classes of RiPPs.
Thiopeptides are a subclass of ribosomally synthesized and posttranslationally modified peptides (RiPPs) that are characterized by a posttranslationally modified 12- to 14-amino acid macrocycle constrained by a central nitrogen-containing heterocycle (1, 2). Members of the thiopeptide family have potent activities against gram-positive bacterial pathogens, such as methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE), and penicillin-resistant Streptococcus pneumoniae (PRSP) (3). Recent studies have shown that thiopeptide-encoding sequences are widely distributed in the genomes of the human microbiota and contribute significantly to microbe–host interactions (4). In addition, it has been shown that thiopeptides have antimalarial (5) and antiproliferative activity in human cancers (6). Thus, there is considerable interest in the synthesis of thiopeptide variants to improve the pharmacological properties of naturally occurring thiopeptides for use in medicine, as well as to study their mechanism of action.
Several methods have been used to produce thiopeptide variants, including biosynthetic pathway engineering (7, 8), semisynthesis (9), and total chemical synthesis (10), with the latter providing the greatest control over chemical structure. However, the total synthesis of variants in sufficient quantities for mechanistic or therapeutic purposes is often time-consuming and costly. The ability to genetically encode noncanonical amino acids (ncAAs) in bacteria has provided an alternative approach to incorporating building blocks with novel structures and chemistries into ribosomally biosynthesized proteins (11), cyclic peptides (12, 13), lanthipeptides (14, 15), and lasso peptides (16, 17). However, methods to heterologously express thiopeptides in Escherichia coli, where the genetic incorporation of ncAAs with orthogonal nonsense or frameshift suppressor aminoacyl-tRNA synthetase/tRNA (aaRS/tRNA) pairs is well-established (11), have not yet been reported. Moreover, current techniques do not allow the efficient genetic encoding of ncAAs in thiopeptide-producing strains, most of which are gram-positive bacteria (18).
Here we report the genetic incorporation of ncAAs into polypeptides in the gram-positive host Bacillus cereus 14579 using orthogonal amber suppressor aaRS/tRNA pairs. Using this method, we demonstrate the production of variants of the mature full-length thiopeptide thiocillin containing ncAAs at defined sites. Thiocillins are naturally derived from a ribosomally synthesized 52-residue precursor peptide that undergoes 13 posttranslational modifications to yield the mature thiopeptide antibiotic in B. cereus 14579 (19). To illustrate the utility of the expression of thiopeptides with an expanded genetic repertoire, ncAAs with bioorthogonal chemical reactivity were incorporated into thiocillin and subsequently modified with fluorescent and photoaffinity probes. The latter chemistry was used to demonstrate direct binding of thiocillin to the ribosomal protein L11.
Results
Genetic Incorporation of ncAAs in B. cereus.
To genetically encode ncAAs in B. cereus, we used the orthogonal pyrrolysyl-tRNA synthetase (PylRS) from Methanosarcina barkeri and the amber suppressor tRNAPylCUA from Methanosarcina mazei. Variants of this pair have demonstrated high efficiency and fidelity for the incorporation of a wide range of ncAAs in response to the nonsense codons in E. coli (20). Because Bacillus is phylogenetically similar to Escherichia and shares tRNA identity elements with E. coli tRNAs (21), we expected that this tRNA/aaRS pair would similarly be orthogonal (i.e., not cross-react with host tRNAs and aaRSs) in B. cereus. Therefore, we sought to establish an expression system that allows amber suppression with the PylRS/tRNAPylCUA pair in the B. cereus host.
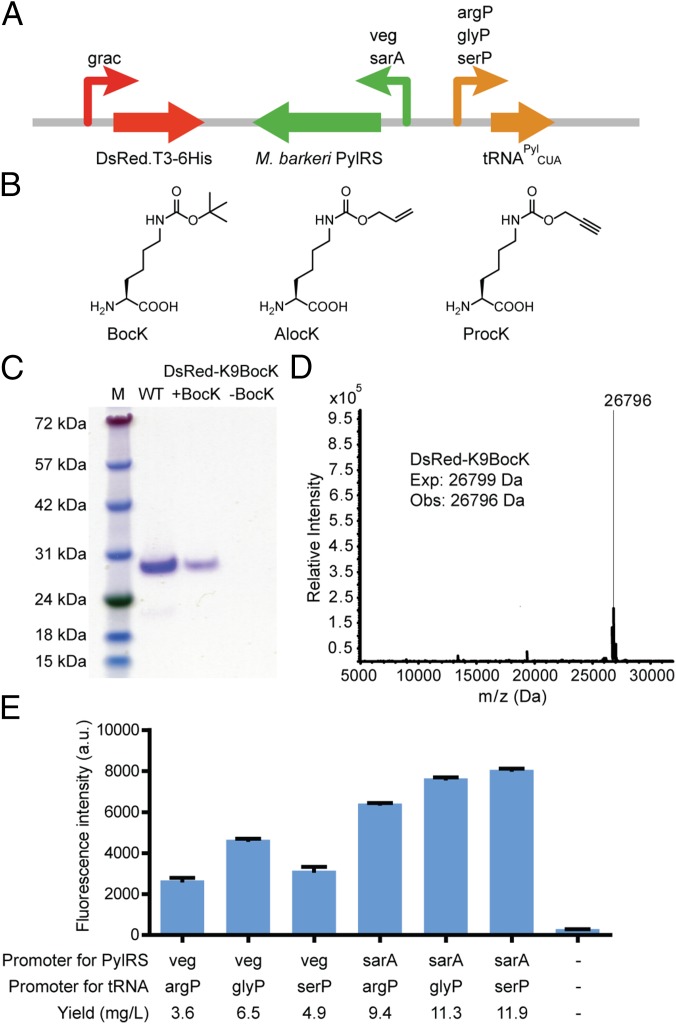
To transcribe the tRNAPylCUA in B. cereus, we tested the endogenous promoters (from −200 to −1 bp upstream) and terminators (200 bp downstream) of Bacillus subtilis glycyl-tRNA, seryl-tRNA, and arginyl-tRNA (referred to as glyP, serP, and argP, respectively). To express PylRS, a codon-optimized gene (PylRS-OptBC) was used to enable efficient transcription in the low-GC-content Bacillus host (22). PylRS-OptBC was expressed under the transcriptional control of the sarA promoter [a global regulator of virulence in S. aureus (23)] or of the veg promoter (which is constitutively expressed during vegetative growth in B. subtilis). Both of these promoters have been used to express heterologous proteins in many Bacillus strains (24). A previously described optimized ribosomal binding site for gram-positive bacteria, the translation initiation region, was used to start translation (25). The PylRS-OptBC and tRNAPylCUA genes and their regulatory elements were then inserted into the B. cereus–E. coli shuttle vector (26), pHT01, to create plasmid pRIPP (Fig. 1A).
Fig. 1.
Overview of genetic incorporation of ncAAs in B. cereus. (A) Schematic of the gene clusters for ncAA incorporation in B. cereus depicting promoters tested for PylRS and tRNA expression. (B) Structures of ncAAs: Nε-(tert-butoxycarbonyl)-l-lysine (BocK), Nε-allyloxycarbonyl-l-lysine (AlocK), and Nε-prop-2-ynyloxycarbonyl-l-lysine (ProcK). (C) SDS/PAGE analysis showing Ni-NTA–purified wild-type DsRed (WT) and DsRed-K9BocK proteins produced by suppression with the PylRS/tRNAPylCUA pair under sarA/serP promoters in B. cereus. Expression of DsRed-K9BocK was carried out in the presence (+BocK) or absence (−BocK) of 1 mM BocK. M, molecular weight markers in kDa. (D) ESI-MS analysis of DsRed-K9BocK revealed a mass of 26,796 Da (calculated 26,799 Da). (E) Comparison of suppression efficiencies of different promoters for the PylRS-OptBC/tRNAPylCUA pair as measured by DsRed-K9BocK fluorescence, with yields of protein after Ni-NTA purification listed below. B. cereus was transformed with plasmids harboring different promoter pairs, and the assays were performed in LB media in the presence of 1 mM BocK and induced with 1 mM IPTG. a.u., arbitrary units. Assays were performed in triplicate, and error bars represent the SD.
To test the amber-suppression efficiency of the different promoter pairs in B. cereus, we used a quantitative assay based on the expression of the fluorescent protein DsRed. The ncAA Nε-(tert-butoxycarbonyl)-l-lysine (BocK) (Fig. 1B) is efficiently incorporated by the PylRS in response to the TAG codon (27) in E. coli and was used as a prototypical ncAA at the permissive site K9 of DsRed. We inserted the gene for the DsRed-K9TAG mutant with a His6 tag at its C terminus into plasmid pRIPP under the control of an IPTG (isopropyl β-d-1-thiogalactopyranoside)-inducible Pgrac promoter to afford pRIPP-DsRed(K9TAG) (Fig. 1A). As expected, substitution of K9 with BocK did not significantly affect the fluorescence of the DsRed protein when expressed in E. coli (SI Appendix, Fig. S1). pRIPP-DsRed(K9TAG) plasmids with different promoter pairs for PylRS and tRNAPylCUA were next tested in B. cereus, and the expression of DsRed with the K9BocK mutation was induced by addition of IPTG. Purified protein was observed in cultures supplemented with 1 mM BocK, whereas no protein was detected for cultures in the absence of the ncAA (Fig. 1C). Electrospray ionization (ESI)-MS analysis of the purified DsRed-K9BocK further confirmed homogeneous incorporation of BocK with no detectable misincorporation of other amino acids (Fig. 1D). The highest fluorescence was observed when PylRS and tRNAPylCUA were driven by sarA and serP promoters, respectively (Fig. 1E). This fluorescence corresponded to a yield of 12 mg/L purified mutant DsRed-K9BocK protein, compared with 42 mg/L for wild-type protein. This promoter pair was therefore used for all subsequent experiments. The above results confirmed that the PylRS/tRNAPylCUA pair is functional and orthogonal in B. cereus, and supported the investigation of the incorporation of ncAAs into thiopeptides.
Production of Thiocillin Variants Containing ncAAs.
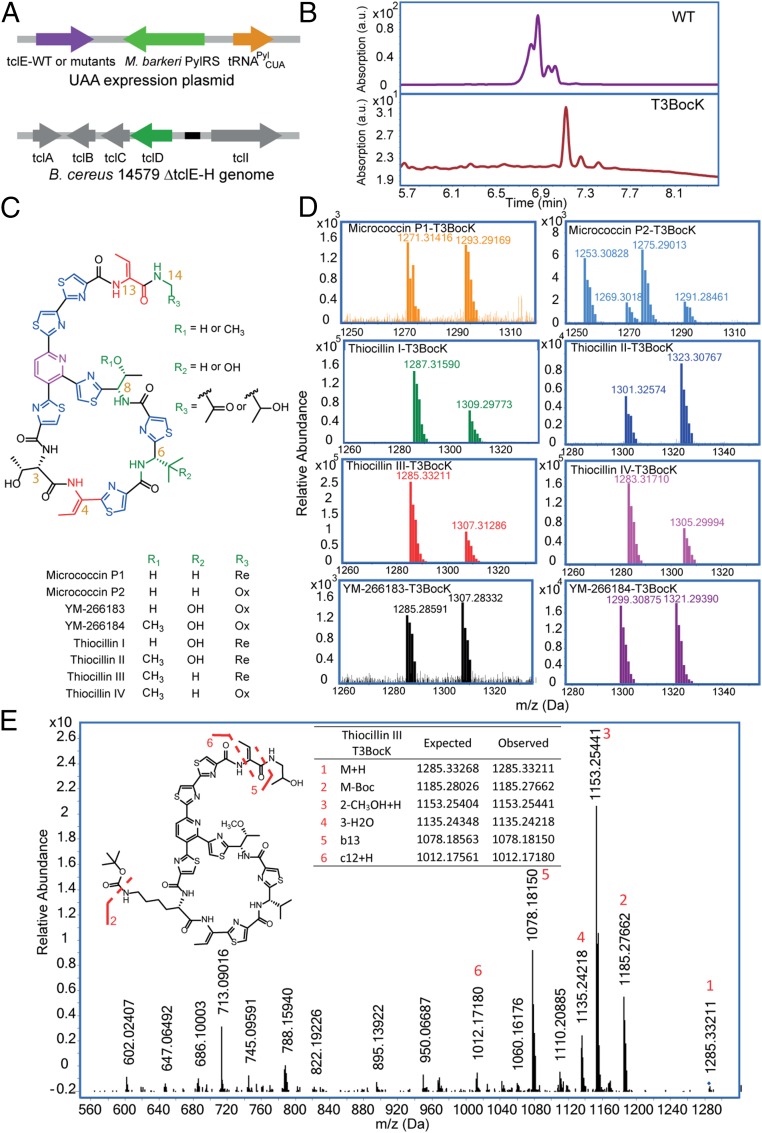
To facilitate the purification and characterization of thiocillin variants containing ncAAs, we used a previously reported strain (B. cereus 14579 ΔtclE-H) in which the genes for the precursor peptide for thiocillin and its variants had been deleted by homologous recombination (7). To test complementation of the knockout with pRIPP, a single copy of wild-type tclE was inserted behind the Pgrac promoter in pRIPP to afford pRIPP-tclE(wt). This plasmid was then electroporated into B. cereus ΔtclE-H (Fig. 2A). To test for expression of wild-type thiocillin, transformed B. cereus ΔtclE-H + pRIPP-tclE(wt) cultures were grown in LB media with IPTG induction for 60 h and pelleted cell material was extracted with methanol as previously described (7, 19, 28). Wild-type thiocillin and congeners were validated in methanolic extracts by HPLC purification and LC-MS analysis (SI Appendix, Fig. S2).
Fig. 2.
Expression and characterization of thiocillin variants with ncAAs and their characterization. (A) Schematic of the complementation of the B. cereus 14579 ΔtclE-H (knockout) strain with a plasmid containing one copy of tclE wild-type gene or tclE amber nonsense mutants and the optimized PylRS-OptBC/tRNAPylCUA pair. (B) HPLC UV traces (350 nm) of products from wild-type and T3BocK thiocillin variant expression by B. cereus transformed with pRIPP-tclE(wt) and pRIPP-tclE(T3TAG), respectively. RiPP expression was performed in LB media supplemented with 1 mM IPTG for 60 h. Thiopeptides were isolated by methanol extraction of pelleted cell material; 1 mM BocK was added for expression of T3BocK thiocillin. The y axis is adjusted to the largest peak in each trace. (C) Structure of thiocillin with the three positions of naturally occurring stochastic posttranslational modifications (R1, R2, R3) in green. Nomenclature follows ref. 18. (D) High-resolution mass for each of the T3BocK thiocillin congeners obtained from a single T3BocK thiocillin expression. The presence of [M+H]+ and [M+Na]+ ions confirmed the successful incorporation of BocK into all variants. For micrococcin P2-T3BocK, masses 1269.3018 and 1291.28461 correspond to the [M+H]+ and [M+Na]+ for the expected variant, respectively. Other masses in the spectra are unidentified products that copurify with this congener. (E) Representative MS/MS fragmentation pathways observed for T3BocK variant shown for Thiocillin III-T3BocK confirmed site-specific incorporation of BocK. Six unique fragments are labeled in the spectrum that harbor the BocK ncAA.
We next examined the incorporation of ncAAs into thiocillin by suppression of a TAG codon placed at specific sites in the tclE gene. We initially focused on substitutions at the T3 site, which is not involved in forming the trithiazolylpyridine core structure and has been previously substituted with other canonical amino acids (7). The TAG codon was introduced at the T3 site by site-directed mutagenesis to afford plasmid pRIPP-tclE(T3TAG). Expression of thiocillin T3BocK variants from B. cereus in shake flasks in the presence of 1 mM BocK was monitored by reverse-phase HPLC and LC-MS of B. cereus methanol extracts. After 2 d, HPLC peaks with longer retention times were observed, likely due to the increased hydrophobicity of thiocillin variants containing the aliphatic BocK versus threonine. No peak was observed for a control culture in which BocK was not added (Fig. 2B and SI Appendix, Fig. S3). LC-MS analysis of these new peaks confirmed that they had the exact masses for all eight thiocillin T3BocK congeners (Fig. 2 C and D). The total yield for the four most abundant T3BocK variants after HPLC purification was 686 μg/L, compared with 11 mg/L for wild-type thiocillin variants (Table 1). MS/MS analysis of the most abundant variant, Thiocillin III-T3BocK (which has an O-methylation on Thr8), confirmed the site-specific incorporation of BocK at the T3 position in thiocillin on the basis of its unique fragmentation pattern (Fig. 2E).
Table 1.
Summary of thiocillin variants produced in this study and their antibiotic efficacy against B. subtilis 168
| Mutant | Compounds observed* | Yield, μg/L† | MIC, μg/mL‡ |
| T3BocK | 8/8 | 686 | 1.5 |
| T3ProcK | 8/8 | 943 | 3 |
| T3AlocK | 4/8 | 713 | 1.5 |
| T4BocK | 0/8 | n.d. | n.d. |
| T4ProcK | 2/8 | n.d. | n.d. |
| T4AlocK | 2/8 | n.d. | n.d. |
| V6BocK | 1/4 | n.d. | n.d. |
| V6ProcK | 2/4 | n.d. | n.d. |
| V6AlocK | 0/4 | n.d. | n.d. |
| T8BocK | 2/4 | 180 | 0.75 |
| T8ProcK | 2/4 | 436 | 1.5 |
| T8AlocK | 4/4 | 667 | 1.5 |
| T13BocK | 2/8 | 125 | >4 |
| T13ProcK | 3/8 | 623 | >4 |
| T13AlocK | 5/8 | 889 | >4 |
| Wild type | 8/8 | 11,200 | 0.38 |
n.d., not determined.
Details of expression and analysis can be found in SI Appendix, Table S1.
Representative yield after purification of the most abundant compounds.
Minimum inhibitory concentrations were determined by standard dilution assay; experiments were performed in triplicate.
Next, we explored the ability to substitute other ncAAs at distinct sites in thiocillin and its variants using Nε-allyloxycarbonyl-l-lysine (AlocK) and Nε-prop-2-ynyloxycarbonyl-l-lysine (ProcK) (Fig. 1B), which provide bioorthogonal alkene and alkyne moieties for postbiosynthetic modifications, respectively (29, 30). In addition, to test the tolerance of ncAAs at different sites, we also examined substitutions at positions T4, V6, T8, and T13 with all three ncAAs. These positions were chosen based on previously demonstrated tolerance to mutation with natural amino acids (7). In total, 15 separate expressions were carried out with the potential for production of 96 novel compounds [including all possible stochastic modifications found for wild-type thiocillin (19)]. The incorporation of at least one ncAA was tolerated at every site, as demonstrated by the detection of at least one variant for each site by LC-MS. Only 2 of the 15 expressions failed to yield any detectable thiocillins: T4BocK and V6AlocK (Table 1 and SI Appendix, Figs. S4–S8 and Table S1). T3, T8, and T13 variants were purified by HPLC; yields varied from 125 μg/L (T13BocK) to nearly 1 mg/L (T3ProcK) (Table 1). The minimal inhibitory concentrations (MICs) for the purified thiopeptides against B. subtilis were determined by serial dilution liquid culture assay. The T3 and T8 ncAA variants retained activity, albeit at markedly lower potency than the wild-type thiocillin, whereas ncAA variants of T13 were inactive, even at the highest concentration tested (4 μg/mL) (Table 1).
Postbiosynthetic Modifications of Thiocillin Variants.
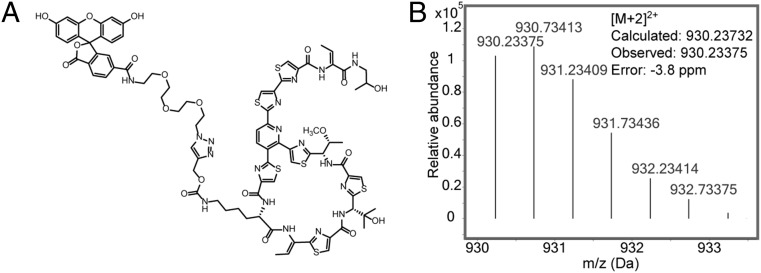
The postbiosynthetic modification of polypeptides containing natural or nonnaturally occurring amino acids with bioorthogonal chemical reactivity can be used to install biophysical probes such as fluorescence labels or affinity tags (31, 32) (Fig. 3A). To determine whether ncAAs can be used as chemical handles for the subsequent modification of thiocillin with a fluorophore, the T3ProcK variant Thiocillin II, with two reactive hydroxyl groups and a propargyl side chain, was expressed and purified by HPLC. Thiocillin II-T3ProcK was then subjected to a copper-catalyzed azide-alkyne cycloaddition reaction (CuAAC) with fluorescein-PEG2-azide in the presence of sodium ascorbate, Tris(benzyltriazolylmethyl)amine (TBTA), and CuSO4. The Thiocillin II-T3ProcK–fluorescein conjugate was isolated in >95% yield and confirmed by LC-MS analysis (Fig. 3B).
Fig. 3.
Postbiosynthetic modifications of Thiocillin II variant. (A) Chemical structure of the Thiocillin II-T3ProcK–fluorescein conjugate. (B) High-resolution MS for the Thiocillin II-T3ProcK–fluorescein conjugate revealed the [M+2H]2+ ion.
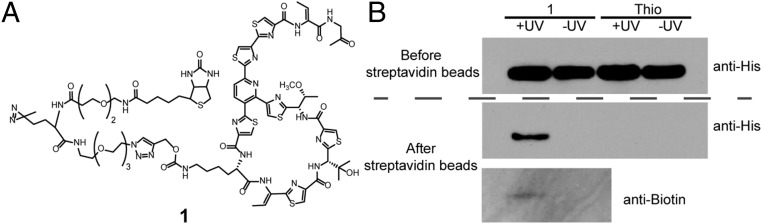
Next, we introduced a photo-cross-linker into thiocillin to covalently cross-link it to its putative intracellular target, the L11 ribosomal protein (33, 34). Thiocillin is hypothesized to exert its antimicrobial activity by binding to the L11 protein of the 50S ribosome, as has been shown for other members of this class of thiopeptides. However, direct evidence for thiocillin binding has not been demonstrated. To this end, we conjugated the ProcK variant YM-266183-T3ProcK to a probe containing both biotin as an affinity tag and a photoreactive diazirine cross-linker (SI Appendix, Fig. S9). To conjugate the functionalized linker to YM-266183-T3ProcK, both the linker and YM-266183-T3ProcK were mixed under CuAAC conditions to afford the final product 1 in >95% yield. 1 was characterized by HPLC and LC-MS, which showed a single peak (Fig. 4A and SI Appendix, Fig. S10). The MIC for the probe was determined to be 3 μg/mL, the same as the unconjugated YM-266183-T3ProcK, which indicated that conjugation did not interfere with antibiotic activity. To facilitate detection of the cross-linked L11 protein by Western analysis, we transformed a plasmid bearing the L11 protein with a His6 tag under the control of the Pgrac promoter into B. subtilis. Cell lysate from an overnight culture that contained the His6-tagged L11 protein was supplemented with 1 μM 1 or wild-type thiocillin and incubated at 25 °C for 30 min. The samples were irradiated with 365-nm light and the cross-linked material was pulled down with streptavidin-conjugated magnetic beads. A band corresponding to L11 protein was observed only in the lysate supplemented with 1 and UV-irradiated, indicating covalent linkage of 1 and L11 protein (Fig. 4B, Middle). The cross-link was further confirmed by Western blot analysis with an anti-biotin antibody (Fig. 4B, Bottom). The above results demonstrate that ncAAs can serve as useful probes of RiPP function.
Fig. 4.
Thiocillin variant with a photoaffinity probe. (A) Chemical structure of YM-266183-T3ProcK probe 1. (B) Western blot analysis of B. subtilis lysate with anti-His antibody before (Top) and after (Middle) streptavidin affinity purification and with anti-biotin antibody after (Bottom) purification confirmed the cross-linking of thiocillin to ribosomal L11 protein. B. subtilis lysate was treated with the probe (Thio-Biotin) or wild-type thiocillin (Thio) at 25 °C for 30 min before being subjected to 365-nm UV irradiation at 4 °C for 10 min. Controls were incubated at the same temperature without UV irradiation.
Discussion
We have extended the genetic encoding of ncAAs to the gram-positive bacterium B. cereus. This advance allows the recombinant introduction of ncAAs with diverse structures and properties into ribosomally produced natural products. Using the thiocillin gene cluster, we have shown that the posttranslational biosynthetic machinery responsible for thiocillin maturation is permissive to substitutions with ncAAs. At sites T3, T4, V6, V8, and T13, which have been previously substituted with canonical amino acids (7), we have demonstrated that ncAAs do not inhibit the core posttranslational modifications that lead to the mature natural product. In addition, stochastic hydroxylation, methylation, and oxidation modifications proceed to varying degrees, in similar fashion to the previously reported natural amino acid variants (7, 28). In total, 45 novel ncAA-modified compounds were identified. ncAA substitution at sites T3, T8, and T13 afforded higher yields of mature product than at sites T4 and V6. This discrepancy in permissibility to ncAA mutation may be due to low suppression efficiency of the TAG codon at these sites, or to intolerance of the posttranslational modification machinery to a change in chemical structure at these sites. Similar intolerance to mutation has been previously reported with natural amino acids at the V6 position, supporting the latter hypothesis (7, 28). Because methanolic extraction from the cell material was used to isolate variants as previously described (7, 19, 28), it is unknown whether these new ncAA-modified variants are substrates for the ABC transporter tclW or the efflux pump tclX of the endogenous thiocillin gene cluster. No overt toxicity in the producing host was observed, suggesting the copies of ribosomal L11 proteins in the thiocillin cluster encoded by tclQ and tclT were still able to protect the host against new variants that retained antimicrobial activity.
Although a T3K mutant was previously shown to abrogate activity, substitution with ncAAs, to afford T3BocK, T3ProcK, and T3AlocK mutants, retained activity on B. subtilis, albeit with 10-fold decreased potency compared with wild type. This may be a result of lysine N-succinylation in the T3K mutant, which may inhibit activity, and was not found to occur on the ncAAs (7). This method may be generalized by using one of several “photo-caged” unnatural amino acids that have been developed to incorporate natural amino acids that are protected from posttranslational modification in similar fashion and subsequently postbiosynthetically deprotected to yield novel thiopeptides not otherwise expressible in vivo. Although no mutants were found with enhanced activity in this study, new chemical functionalities provided by ncAAs to thiopeptide biosynthesis offer the potential to screen a diverse chemical space for mutants with increased activity in the future (8, 13).
The ability to introduce ncAAs into RiPPs also provides easy access to thiopeptides modified with biological probes. For example, ProcK was used as a bioorthogonal chemical handle to attach a fluorophore via a copper-catalyzed Huisgen 1,3-dipolar cycloaddition (“click” chemistry). This reaction is highly specific for the alkyne group of ProcK, and did not produce side products from modification of other chemical functionalities in thiocillin. This is advantageous compared with previous methods using thiol-Michael addition to dehydroamino acids (35), as these residues occur frequently in thiopeptides (36). A similar approach was used to create an activity probe containing a biotin-diazirine linker that allows cross-linking of thiocillin to its cellular target. This experiment conclusively demonstrated a direct interaction between thiocillin and ribosomal protein L11. This approach may be particularly useful to study microbe–microbe and microbe–host interactions in human microbiota and anticancer activities, where targets are less well defined.
In addition to their identification in soil-dwelling bacteria, it has recently become appreciated that RiPPs are among the most widely distributed and diverse biosynthetic gene clusters encoded by the human microbiota. Among these, thiopeptide gene clusters, including thiocillin, are prolific (4). This is underscored by the discovery of putative thiopeptide biosynthetic gene clusters in 4.5% of the Bacillus genomes and 39% of the Streptomyces genomes sequenced to date (37). Thus, we expect that the combination of RiPP biosynthetic gene cluster manipulation and unnatural amino acid methodology will provide a powerful tool to both study and enhance the biological activity of this important class of natural products.
Materials and Methods
Expression and Purification of DsRed.T3 Mutants from B. cereus.
An overnight culture of B. cereus transformed with pRIPP-DsRed(K9X) in LB media with chloramphenicol (3 μg/mL) was diluted 100-fold into 50 mL LB media supplemented with chloramphenicol (3 μg/mL) and ncAAs (1 mM) at 37 °C. The cells were allowed to grow for ∼2 h, when the OD600 reached 0.6, and IPTG was added to a final concentration of 1 mM to induce protein expression. The cells were grown for an additional 16 h at 30 °C before harvesting by centrifugation at 4,750 × g for 10 min. The cell pellet was lysed by sonication and the resulting cell lysate was clarified by centrifugation at 20,000 × g for 20 min at 4 °C. DsRed.T3 mutants were purified on Ni-NTA resin (Qiagen) following the manufacturer’s instructions.
Expression and Purification of Thiocillin Variants from B. cereus.
An overnight culture of B. cereus transformed with pRIPP-tclE(T3TAG) in LB media with chloramphenicol (3 μg/mL) was diluted 100-fold into 500 mL LB media supplemented with chloramphenicol (3 μg/mL) and appropriate ncAAs (1 mM) in a 2.5-L Ultra Yield Flask (Thomson Instrument) at 37 °C. Cells were allowed to grow for ∼2 h, when the OD600 reached 0.6, and IPTG was added to a final concentration of 1 mM to induce expression. The cells were grown for an additional 60 h at 30 °C with shaking at 200 rpm before harvesting by centrifugation at 9,000 × g for 10 min. Fifty milliliters methanol together with 15 g sodium sulfate were added to the pellet, and the mixture was vortexed vigorously and allowed to sit for at least 30 min. The mixture was then clarified by centrifugation at 6,000 × g for 5 min and filtered through Whatman filter paper. Methanol was removed by vacuum and the solid was solubilized in 10 mL methanol for HPLC and LC-MS analysis. Further purification of each congener was done by preparative RP-HPLC. Compounds in methanol were eluted in a gradient of buffer A (0.1% TFA in water) and B (0.1% TFA in acetonitrile): 10–30% for the first 3 min and increasing to 100% B over 17 min, then isocratic at 100% B for 10 min before switching back to 10% B. The elution of congeners was monitored by absorption at 350 nm, and the fractions were pooled and further characterized by LC-MS.
High-Resolution Mass Spectrometry and MS/MS Analysis.
An Agilent 6520 accurate-mass quadrupole-time-of-flight (QTOF) instrument was used to carry out the high-resolution mass spectrometry, which was equipped with reverse-phase liquid chromatography and an electrospray ionization source. For proteins, samples in PBS were injected (10 μL) at a concentration of 0.2 mg/mL and separated on a 150-mm reverse-phase C8 wide-pore column heated to 70 °C to improve peak resolution. Proteins were eluted in a gradient of H2O + 0.1% formic acid (solvent A) and acetonitrile + 0.1% formic acid (solvent B) using the following method: 5% B for 2 min, 5–60% B for 10 min, 60–80% B for 1 min, followed by a wash (95% B) and reequilibration (5% A) phase. ESI source settings were 350 °C, 10 L/min, 40 psig (pounds per square inch gage) nebulizer nitrogen gas, 200 V fragmentor, and 4,500 V capillary. Figures were created by extraction of the total ion count across the entire area of protein elution (roughly 8–10 min) and deconvolution of charge envelopes using Agilent Qualitative Analysis software with BioConfirm.
For thiocillin variants, purified or crude samples in methanol were injected (10 μL) and separated on a 150-mm reverse-phase C8 wide-pore column heated to 70 °C to improve peak resolution. Thiocillin variants were eluted in a gradient of H2O + 0.1% formic acid (solvent A) and acetonitrile + 0.1% formic acid (solvent B) using the following method: 5% B for 2 min, 5–95% B for 7 min, 95% B for 11 min, followed by a reequilibration (5% A) phase. ESI source settings were 350 °C, 10 L/min, 40 psig nebulizer nitrogen gas, 200 V fragmentor, and 4,500 V capillary. Figures were created by extraction of the total ion count across the entire area of thiocillin variant elution with absorption at 350 nm (roughly 6–8 min) using Agilent Qualitative Analysis software with BioConfirm.
Photo-Cross-Linking with Thiocillin-T3ProcK Derivatives.
Probe 1 was prepared by standard peptide-coupling techniques, followed by CuAAC with T3ProcK. L11 protein with a C-terminal His tag was inserted into pHT01 vector and the resulting plasmid was transformed into B. subtilis 168 to facilitate detection by Western blot. The supernatant of cell lysate that contained the ribosome complex was incubated with 1 mM 1 or wild-type thiocillin at 25 °C for 30 min before 365-nm light irradiation at 4 °C for 10 min. The mixture was purified with streptavidin-conjugated magnetic beads and the eluants were subjected to Western blot analysis.
Supplementary Material
Acknowledgments
We acknowledge Kristen Williams for her assistance in manuscript preparation. This work was supported by NIH Grant GM62159-14 (to P.G.S.). This is Paper 29273 of The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602733113/-/DCSupplemental.
References
- 1.Just-Baringo X, Albericio F, Álvarez M. Thiopeptide engineering: A multidisciplinary effort towards future drugs. Angew Chem Int Ed Engl. 2014;53(26):6602–6616. doi: 10.1002/anie.201307288. [DOI] [PubMed] [Google Scholar]
- 2.Walsh CT, Malcolmson SJ, Young TS. Three ring posttranslational circuses: Insertion of oxazoles, thiazoles, and pyridines into protein-derived frameworks. ACS Chem Biol. 2012;7(3):429–442. doi: 10.1021/cb200518n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q, Liu W. Biosynthesis of thiopeptide antibiotics and their pathway engineering. Nat Prod Rep. 2013;30(2):218–226. doi: 10.1039/c2np20107k. [DOI] [PubMed] [Google Scholar]
- 4.Donia MS, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158(6):1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoof S, et al. Antiplasmodial thiostrepton derivatives: Proteasome inhibitors with a dual mode of action. Angew Chem Int Ed Engl. 2010;49(19):3317–3321. doi: 10.1002/anie.200906988. [DOI] [PubMed] [Google Scholar]
- 6.Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat Chem. 2011;3(9):725–731. doi: 10.1038/nchem.1114. [DOI] [PubMed] [Google Scholar]
- 7.Acker MG, Bowers AA, Walsh CT. Generation of thiocillin variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J Am Chem Soc. 2009;131(48):17563–17565. doi: 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young TS, Dorrestein PC, Walsh CT. Codon randomization for rapid exploration of chemical space in thiopeptide antibiotic variants. Chem Biol. 2012;19(12):1600–1610. doi: 10.1016/j.chembiol.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, et al. Radical-mediated enzymatic carbon chain fragmentation-recombination. Nat Chem Biol. 2011;7(3):154–160. doi: 10.1038/nchembio.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aulakh VS, Ciufolini MA. Total synthesis and complete structural assignment of thiocillin I. J Am Chem Soc. 2011;133(15):5900–5904. doi: 10.1021/ja110166x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 12.Tianero MDB, Donia MS, Young TS, Schultz PG, Schmidt EW. Ribosomal route to small-molecule diversity. J Am Chem Soc. 2012;134(1):418–425. doi: 10.1021/ja208278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young TS, et al. Evolution of cyclic peptide protease inhibitors. Proc Natl Acad Sci USA. 2011;108(27):11052–11056. doi: 10.1073/pnas.1108045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, Yang X, Garg N, van der Donk WA. Production of lantipeptides in Escherichia coli. J Am Chem Soc. 2011;133(8):2338–2341. doi: 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bindman NA, Bobeica SC, Liu WR, van der Donk WA. Facile removal of leader peptides from lanthipeptides by incorporation of a hydroxy acid. J Am Chem Soc. 2015;137(22):6975–6978. doi: 10.1021/jacs.5b04681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al Toma RS, et al. Site-directed and global incorporation of orthogonal and isostructural noncanonical amino acids into the ribosomal lasso peptide capistruin. ChemBioChem. 2015;16(3):503–509. doi: 10.1002/cbic.201402558. [DOI] [PubMed] [Google Scholar]
- 17.Piscotta FJ, Tharp JM, Liu WR, Link AJ. Expanding the chemical diversity of lasso peptide MccJ25 with genetically encoded noncanonical amino acids. Chem Commun (Camb) 2015;51(2):409–412. doi: 10.1039/c4cc07778d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem Rev. 2005;105(2):685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 19.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc Natl Acad Sci USA. 2009;106(8):2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan W, Tharp JM, Liu WR. Pyrrolysyl-tRNA synthetase: An ordinary enzyme but an outstanding genetic code expansion tool. Biochim Biophys Acta. 2014;1844(6):1059–1070. doi: 10.1016/j.bbapap.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue H, Shen W, Giegé R, Wong JTF. Identity elements of tRNA(Trp). Identification and evolutionary conservation. J Biol Chem. 1993;268(13):9316–9322. [PubMed] [Google Scholar]
- 22.Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry. 2013;52(10):1828–1837. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manna AC, Bayer MG, Cheung AL. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J Bacteriol. 1998;180(15):3828–3836. doi: 10.1128/jb.180.15.3828-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukushima T, Ishikawa S, Yamamoto H, Ogasawara N, Sekiguchi J. Transcriptional, functional and cytochemical analyses of the veg gene in Bacillus subtilis. J Biochem. 2003;133(4):475–483. doi: 10.1093/jb/mvg062. [DOI] [PubMed] [Google Scholar]
- 25.Cheng X, Patterson TA. Construction and use of lambda PL promoter vectors for direct cloning and high level expression of PCR amplified DNA coding sequences. Nucleic Acids Res. 1992;20(17):4591–4598. doi: 10.1093/nar/20.17.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen HD, Phan TTP, Schumann W. Expression vectors for the rapid purification of recombinant proteins in Bacillus subtilis. Curr Microbiol. 2007;55(2):89–93. doi: 10.1007/s00284-006-0419-5. [DOI] [PubMed] [Google Scholar]
- 27.Mukai T, et al. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun. 2008;371(4):818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 28.Bowers AA, Acker MG, Koglin A, Walsh CT. Manipulation of thiocillin variants by prepeptide gene replacement: Structure, conformation, and activity of heterocycle substitution mutants. J Am Chem Soc. 2010;132(21):7519–7527. doi: 10.1021/ja102339q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen DP, et al. Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA synthetase/tRNA(CUA) pair and click chemistry. J Am Chem Soc. 2009;131(25):8720–8721. doi: 10.1021/ja900553w. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, et al. Genetically encoded alkenyl-pyrrolysine analogues for thiol-ene reaction mediated site-specific protein labeling. Chem Sci. 2012;3(9):2766–2770. [Google Scholar]
- 31.Zhang W, Curtin C, Franco C. Towards manipulation of post-biosynthetic events in secondary metabolism of plant cell cultures. Enzyme Microb Technol. 2002;30(6):688–696. [Google Scholar]
- 32.Bindman NA, van der Donk WA. A general method for fluorescent labeling of the N-termini of lanthipeptides and its application to visualize their cellular localization. J Am Chem Soc. 2013;135(28):10362–10371. doi: 10.1021/ja4010706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wienen B, et al. Ribosomal protein alterations in thiostrepton- and micrococcin-resistant mutants of Bacillus subtilis. J Biol Chem. 1979;254(16):8031–8041. [PubMed] [Google Scholar]
- 34.Walsh CT, Acker MG, Bowers AA. Thiazolyl peptide antibiotic biosynthesis: A cascade of post-translational modifications on ribosomal nascent proteins. J Biol Chem. 2010;285(36):27525–27531. doi: 10.1074/jbc.R110.135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoof S, Baumann S, Ellinger B, Arndt H-D. A fluorescent probe for the 70 S-ribosomal GTPase-associated center. ChemBioChem. 2009;10(2):242–245. doi: 10.1002/cbic.200800642. [DOI] [PubMed] [Google Scholar]
- 36.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30(1):108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blin K, et al. antiSMASH 2.0—A versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41(Web Server issue):W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.