Significance
Pneumococcal meningitis, the most frequent cause of bacterial meningitis in adults, is associated with substantial morbidity and mortality. In a prospective, nationwide cohort of patients with pneumococcal meningitis, macrophage migration inhibitory factor (MIF), a proinflammatory mediator, was identified as a previously unidentified genetic marker of patient’s outcome. High-expression MIF alleles were associated with disease severity and death, a finding consistent with the harmful consequences of robust proinflammatory cytokine responses on brain edema and neuronal damage in the course of bacterial meningitis. These results provide strong evidence that functional MIF polymorphisms are genetic predictors of morbidity and mortality of pneumococcal meningitis and suggest that MIF is a potential target for immune-modulating adjunctive therapies.
Keywords: macrophage migration inhibitory factor, polymorphism, meningitis, sepsis, innate immunity
Abstract
Pneumococcal meningitis is the most frequent and critical type of bacterial meningitis. Because cytokines play an important role in the pathogenesis of bacterial meningitis, we examined whether functional polymorphisms of the proinflammatory cytokine macrophage migration inhibitory factor (MIF) were associated with morbidity and mortality of pneumococcal meningitis. Two functional MIF promoter polymorphisms, a microsatellite (−794 CATT5–8; rs5844572) and a single-nucleotide polymorphism (−173 G/C; rs755622) were genotyped in a prospective, nationwide cohort of 405 patients with pneumococcal meningitis and in 329 controls matched for age, gender, and ethnicity. Carriages of the CATT7 and −173 C high-expression MIF alleles were associated with unfavorable outcome (P = 0.005 and 0.003) and death (P = 0.03 and 0.01). In a multivariate logistic regression model, shock [odds ratio (OR) 26.0, P = 0.02] and carriage of the CATT7 allele (OR 5.12, P = 0.04) were the main predictors of mortality. MIF levels in the cerebrospinal fluid were associated with systemic complications and death (P = 0.0002). Streptococcus pneumoniae strongly up-regulated MIF production in whole blood and transcription activity of high-expression MIF promoter Luciferase reporter constructs in THP-1 monocytes. Consistent with these findings, treatment with anti-MIF immunoglogulin G (IgG) antibodies reduced bacterial loads and improved survival in a mouse model of pneumococcal pneumonia and sepsis. The present study provides strong evidence that carriage of high-expression MIF alleles is a genetic marker of morbidity and mortality of pneumococcal meningitis and also suggests a potential role for MIF as a target of immune-modulating adjunctive therapy.
Acute community-acquired bacterial meningitis is a life-threatening disease associated with substantial morbidity and mortality and ranks among the top 10 infectious causes of death (1). Streptococcus pneumoniae is the most common cause of bacterial meningitis in adults of all age groups, accounting for 50–70% of cases in developed countries (2). Pneumococcal meningitis is associated with a mortality ranging from 19% to 37% (3, 4). Neurological sequelae such as hearing loss, focal deficits, and motor and cognitive impairments significantly affect the quality of life of survivors (5–7). Predisposing factors for pneumococcal meningitis include pneumonia, otitis, sinusitis, cerebrospinal fluid (CSF) leaks, splenectomy or asplenic states, debilitating conditions (i.e., alcoholism, cirrhosis, diabetes, and cancer), and primary or acquired immune deficiencies (i.e., multiple myeloma, hypogammaglobulinemia, sickle cell anemia, HIV/AIDS, and the use of immunosuppressive agents). Genetic studies of extreme phenotypes have revealed that patients with single-gene inborn errors in MyD88, IRAK4, and NEMO affecting the activation of the canonical TLR and IL-1R signaling pathways or in complement factor two are prone to pneumococcal diseases (8, 9). In addition, case-control candidate gene studies identified polymorphisms of genes associated either with increased susceptibility to (MBL2 and PTPN22) or with protection from (TIRAP, NFKBIA, and NFKBIE) pneumococcal disease (2, 8, 9). The increased susceptibility was related to reduced concentrations of the mannose binding lectin (variants of MBL2) or to an increased activity of the PTPN22 phosphatase (variants of PTPN22) (10, 11). The protection afforded by the polymorphic TIRAP variant is mediated by an attenuation of TLR2 signal transduction due to a defective recruitment of the TIRAP variant to TLR2 (12). The functional effects of the polymorphisms of the NFKBIA and NFKBIE genes coding for the inhibitors of NF-κB (IκB) are unknown (13).
Cytokines are critical effector molecules of the immune system and play a central role in the orchestration of host defenses against infection. Until now, no polymorphism of cytokine genes (including TNF, IL6, IL10, and LTA) has been associated with susceptibility to and outcome of invasive pneumococcal infection (2). Within this large family of mediators, macrophage migration inhibitory factor (MIF) occupies a special place (14, 15). Unrelated to classical cytokine families (tumor necrosis factor, chemokines, interleukins, or interferons), MIF is a constitutively expressed proinflammatory cytokine acting at the interface of the immune and endocrine systems. Within the innate immune system, MIF positively regulates TLR4 expression, inhibits activation-induced and p53-dependent apoptosis of macrophages, and counter-regulates the anti-inflammatory and immunosuppressive effects of glucocorticoids in part by a down-regulation of mitogen-activated protein kinase phosphatase-1 (16–19). MIF is up-regulated in inflammatory, infectious, and autoimmune diseases functioning as a modulator of innate and adaptive immunity (20–22).
Functional polymorphisms of the MIF gene locus include a microsatellite repeat of five to eight CATT tetranucleotide (CATT5–8) at position −794 (rs5844572) and a single-nucleotide polymorphism (SNP) of a G-to-C transition at position −173 (−173G/C; rs755622) (23, 24). Genetic studies have revealed a complex picture of the role of polymorphic MIF alleles in the pathogenesis of autoimmune diseases (20, 25). Few studies have been performed in patients with infectious diseases, especially in patients with bacterial sepsis (20, 25). We therefore examined the impact of the MIF gene locus on the susceptibility to, severity of, and outcome of pneumococcal meningitis in a large, nationwide cohort of patients. Functional studies of polymorphic MIF promoters were conducted in human monocytic cells stimulated with S. pneumoniae and analyzed by Luciferase reporter assays. Lastly, the effect of an anti-MIF treatment strategy was evaluated in a mouse model of S. pneumoniae pneumonia and sepsis.
Results
Pneumococcal Meningitis Cohort.
A total of 461 patients with culture-proven, community-acquired S. pneumoniae meningitis and 343 controls matched for age (median, 59.4 y vs. 60.1 y), gender (female: 53% vs. 50.8%), and ethnicity (Caucasian: 94.2% vs. 96.0%) were enrolled in a prospective, nationwide cohort study. DNA samples were available from 434 patients and 329 controls, who were all Caucasians. The baseline characteristics of patients are shown in Table 1. Briefly, 80.4% of the patients were bacteremic, and 43.7% required intensive care unit (ICU) admission for shock or respiratory failure. During hospitalization, 79.6% developed neurological complications, and 38.1% developed systemic complications. Outcome was unfavorable [defined as a Glasgow Outcome Score (GOS) 1–4] in 133 patients (32.8%). A total of 30 patients (7.5%) died (GOS of 1).
Table 1.
Characteristics of controls and patients with pneumococcal meningitis
| Characteristics | Controls (n = 343) | Patients (n = 461) | Caucasian patients with DNA and outcome (n = 405) |
| Age (years) | 59.4 ± 18.1 | 60.1 ± 20.8 | |
| Female gender | 146 (49.2) | 220 (47.0) | |
| Caucasian ethnicity | 329 (96.0) | 434 (94.2) | |
| Predisposing factors (n = 405) | |||
| Otitis/sinusitis | 191 (47.3) | ||
| Immunocompromised status | 100 (24.7) | ||
| Body temperature in °C (n = 401) | 39.0 ± 1.5 | ||
| Glasgow coma scale <8 (indicating coma) (n = 402) | 53 (13.2) | ||
| Bacteremia (n = 357) | 287 (80.4) | ||
| Neurologic complications (n = 404) | 257 (79.6) | ||
| Systemic complications (n = 396) | 151 (38.1) | ||
| Complications (n = 405) | |||
| Shock | 25 (6.2) | ||
| Respiratory failure | 93 (23.0) | ||
| ICU admission | 177 (43.7) | ||
| Mechanical ventilation | 140 (34.6) | ||
| Outcome | |||
| GOS (n = 405) | |||
| 1: Death | 30 (7.5) | ||
| 2: Persistent vegetative state | 1 (0.2) | ||
| 3: Severe disability | 18 (4.4) | ||
| 4: Moderate disability | 84 (20.7) | ||
| 5: Good recovery | 272 (67.2) | ||
| Hearing loss at discharge (n = 368) | 41 (11.1) |
Data are mean ± interquartile range or number (percent).
Association Between MIF Polymorphisms and Susceptibility to, Severity of, and Outcome of Pneumococcal Meningitis.
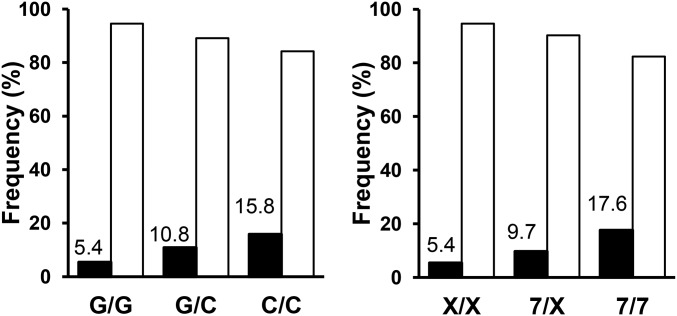
Allelic frequencies and genotypes for the −173 G/C (rs755622) and CATT5–8 (rs5844572) polymorphisms of patients and controls are presented in Table S1. No deviation from the Hardy–Weinberg equilibrium (HWE) was observed for the −173 G/C SNP and CATT5–8 microsatellite. The allele frequencies, genotypes, and haplotypes of the −173 G/C and of the CATT5–8 polymorphisms were similar in patients and controls (P > 0.5), indicating that there was no association between MIF polymorphisms and susceptibility to pneumococcal meningitis. Carriage of the −173 C or of the CATT7 high MIF expression alleles were associated with unfavorable outcome [−173 C: odds ratio (OR) 1.9, P = 0.003; CATT7: OR 1.89, P = 0.005], respiratory failure (−173 C: OR 1.71, P = 0.03), and death (−173 C: OR: 2.6, P = 0.01; CATT7: OR 2.27, P = 0.03) (Table 2). The association between the −173 C or CATT7 high MIF expression allelic variants and death was also significant by using an additive mode of inheritance (Fig. 1). Carriage of the CATT7 allele was associated with markers of inflammation such as C-reactive protein (P = 0.02) and erythrocyte sedimentation rate (P = 0.02) and with indicators of disease severity (i.e., a CSF leukocyte count of <1,000 cells per mm3; P = 0.05) (22). The association between the CATT7 and death remained significant in a multivariate logistic regression model (OR 5.12, P = 0.04) and tended to be associated with unfavorable outcome (OR 2.61, P = 0.07) after adjustment for relevant covariables (Table 3).
Table S1.
MIF allelic frequencies and genotypes in patients with pneumococcal meningitis and in controls
| MIF gene polymorphism | Allele | Patients, n (%) | HWE P | Controls, n (%) | HWE P | Genotype | Patients, n (%) | Controls, n (%) | P |
| −173 G/C (rs755622) | G | 697 (80.3) | 542 (82.4) | GG | 283 (65.2) | 223 (67.8) | |||
| C | 171 (19.7) | 0.3 | 116 (17.6) | 0.9 | GC | 131 (30.2) | 96 (29.2) | ||
| CC | 20 (4.6) | 10 (3.0) | 0.5 | ||||||
| CATT5–8 (rs5844572) | CATT 5 | 213 (24.6) | 164 (25.0) | CATT5–5 | 26 (6.0) | 24 (7.3) | |||
| CATT 6 | 513 (59.2) | 398 (60.7) | CATT5–6 | 129 (29.8) | 99 (30.2) | ||||
| CATT 7 | 139 (16.1) | 94 (14.3) | CATT6–6 | 153 (35.3) | 117 (35.7) | ||||
| CATT 8 | 1 (0.1) | 0.9 | 0 (0) | 0.7 | CATT6–7 | 77 (17.8) | 65 (19.8) | ||
| CATT5–7 | 32 (7.4) | 17 (5.2) | |||||||
| CATT7–7 | 15 (3.5) | 6 (1.8) | |||||||
| CATT5–8 | 0 (0) | 0 (0) | |||||||
| CATT6–8 | 1 (0.2) | 0 (0) | 0.6 | ||||||
| CATT/−173 haplotype | 5G | 212 (24.5) | 163 (24.9) | ||||||
| 6G | 479 (55.3) | 375 (57.3) | |||||||
| 6C | 36 (4.2) | 22 (3.4) | |||||||
| 7C | 132 (15.2) | 92 (14.1) | |||||||
| 8C | 1 (0.1) | 0 (0) | |||||||
| 5C | 1 (0.1) | 0 (0) | |||||||
| 7G | 5 (0.6) | 2 (0.3) | 0.8 |
Calculation of allele frequencies based on 2xN patients or controls. Genotypes were obtained from 434 (−173 G/C) or 433 (CATT5–8) patients and 329 (−173 G/C) or 328 (CATT5–8) controls.
Table 2.
Association between MIF gene polymorphisms and patient’s outcome
| Polymorphisms | Unfavorable outcome (GOS ≤ 4; n = 133) | Respiratory failure (n = 93) | Death (n = 30) | |||
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| GC/CC vs. GG | 1.9 (1.24–2.92) | 0.003 | 1.71 (1.06–2.74) | 0.03 | 2.6 (1.01–3.78) | 0.01 |
| 55/5X vs. XX | 0.88 (0.58–1.33) | 0.54 | 0.87 (0.54–1.39) | 0.56 | 0.66 (0.31–1.43) | 0.30 |
| 66/6X vs. XX | 0.98 (0.57–1.68) | 0.93 | 1.05 (0.57–1.93) | 0.89 | 1.48 (0.50–4.38) | 0.48 |
| 77/7X vs. XX | 1.89 (1.21–2.96) | 0.005 | 1.41 (0.98–2.63) | 0.06 | 2.27 (1.07–4.83) | 0.03 |
The analyses were performed by using a univariate logistic regression model assuming a dominant mode of inheritance.
Fig. 1.
Carriage of −173 C or CATT7 high-expression MIF allelic variants is associated with death from pneumococcal meningitis. Proportions of survivors (open bars) and nonsurvivors (filled bars) for the −173 (G/G, G/C, and C/C) and CATT7 (X/X, 7/X, and 7/7) genotypes. Logistic regression analysis was performed using dominant and additive modes of inheritance (P = 0.01 and 0.01 for −173 G/C and P = 0.03 and 0.01 for CATT7, respectively).
Table 3.
Logistic regression analysis of factors associated with death and unfavorable outcome
| Prediction | OR (95% CI) | P |
| Death (n = 30/398) | ||
| Shock | 26.06 (5.36–48.09) | 0.02 |
| Carriage of CATT7 allele | 5.12 (1.11–23.67) | 0.04 |
| Immunocompromised state | 2.85 (1.16–6.98) | 0.09 |
| Male gender | 2.48 (0.99–6.17) | 0.05 |
| Age (per year increase) | 1.04 (1.01–1.08) | 0.02 |
| Unfavorable outcome (n = 133/405) | ||
| Systemic complications on admission | 15.8 (1.80–137.97) | 0.01 |
| Otitis or sinusitis | 3.79 (2.21–6.49) | <0.001 |
| Carriage of CATT7 allele | 2.61 (0.91–7.50) | 0.07 |
| Male gender | 1.67 (0.96–2.91) | 0.07 |
| Age (per year increase) | 1.01 (0.99–1.03) | 0.18 |
| GCS > 12 | 0.54 (0.29–1.01) | 0.06 |
The analyses were performed by using multivariate logistic regression models. GCS, Glasgow coma score.
MIF Levels in the CSF and Morbidity and Mortality of Pneumococcal Meningitis.
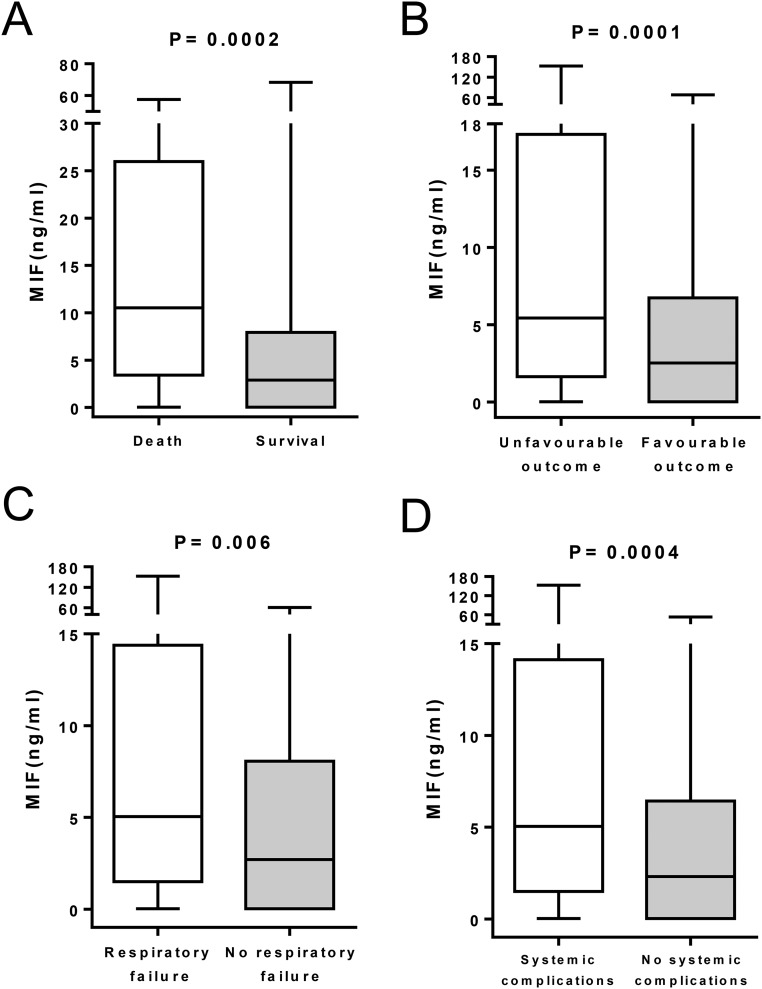
MIF CSF levels were measured in 242 patients (52% of the patients included in the study). The median concentration was 2.65 ng/mL (range: 0.025–323 ng/mL). The MIF CSF levels were similar in patients in whom antibiotic treatment was started before or after the lumbar puncture (median [range]: 2.49 [0.02–323.22] vs. 3.49 [0.02–68.34] ng/mL, P = 0.22). MIF CSF levels were markedly higher in nonsurvivors than in survivors (10.80 [3.29–89.11] vs. 3.19 [0.02–323.22] ng/mL, P = 0.0002) and also higher in patients with than in patients without unfavorable outcome (P = 0.0001), respiratory failure (P = 0.006), or systemic complications (P = 0.0004) (Fig. S1).
Fig. S1.
Associations between CSF MIF levels with death, unfavorable outcome (GOS ≤ 4), respiratory failure and systemic complications (n = 242).
S. pneumoniae Induces MIF Production in Whole Blood and MIF Promoter Activity in THP-1 Monocytes.
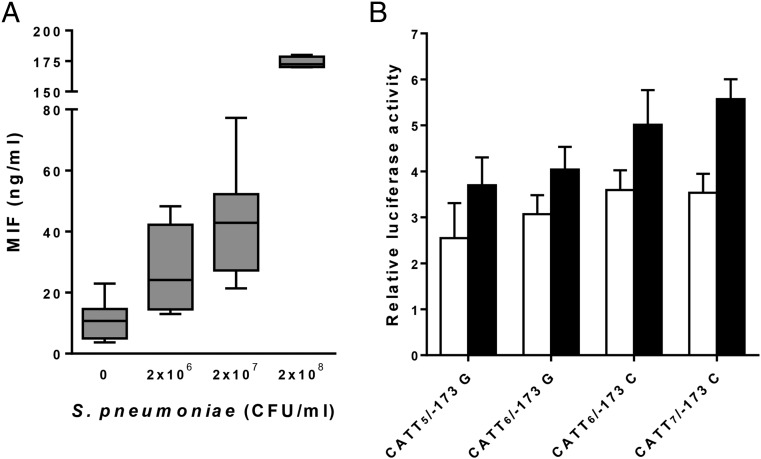
Given that MIF polymorphisms and MIF levels in the CSF were associated with morbidity and mortality of pneumococcal meningitis, we examined whether S. pneumoniae up-regulated blood levels of MIF and whether polymorphic MIF alleles affected the transcriptional activity of the MIF promoter. S. pneumoniae induced a strong and dose-dependent up-regulation of MIF production in human whole blood (Fig. 2A). After 24 h, the median concentrations of MIF in cell culture supernatants increased 2.4-, 3.8-, and 15.5-fold upon stimulation with 2 × 106, 2 × 107, and 2 × 108 CFU/mL S. pneumoniae (P = 0.005 and 0.001, and P < 10−8 compared with unstimulated control cells). We then evaluated the transcriptional activity of the four most common polymorphic MIF promoters (CATT5/−173 G, CATT6/−173 G, CATT6/−173 C, and CATT7/−173 C) in human THP-1 monocytes (Fig. 2B). The transcriptional activity was up-regulated after stimulation with S. pneumoniae and was highest with the CATT6/−173 C and CATT7/−173 C constructs (P = 0.03 and 0.003 compared with CATT5/−173 G).
Fig. 2.
S. pneumoniae induces MIF production in whole blood and polymorphic MIF alleles affect the transcriptional activity of the MIF promoter. (A) Whole blood from five healthy volunteers was stimulated for 24 h with S. pneumoniae. MIF concentrations were measured by ELISA. Bottom, median, and top lines of the box mark the 25th, 50th, and 75th percentiles, respectively. Vertical lines with whiskers show the range of values. P = 0.005 (2 × 106), P = 0.001 (2 × 107), and P < 10−8 (2 × 108) compared with control. (B) THP-1 monocytes were transiently transfected with CATT5/−173 G, CATT6/−173 G, CATT6/−173 C, and CATT7/−173 C MIF promoter pGL3 and Renilla luciferase vectors. Cells were stimulated for 24 h with (filled bars) or without (open bars) S. pneumoniae (MOI 10). Data are means + SD of three independent experiments performed in triplicate. P = 0.05 (CATT6/−173 G), P = 0.04 (CATT6/−173 C), and P = 0.007 (CATT7/−173 C) for S. pneumoniae vs. control. After S. pneumoniae stimulation, P = 0.03 (CATT6/−173 C) and P = 0.003 (CATT7/−173 C) compared with CATT5/−173 G.
Anti-MIF Antibody Protects Against Lethal Pneumococcal Pneumonia and Sepsis.
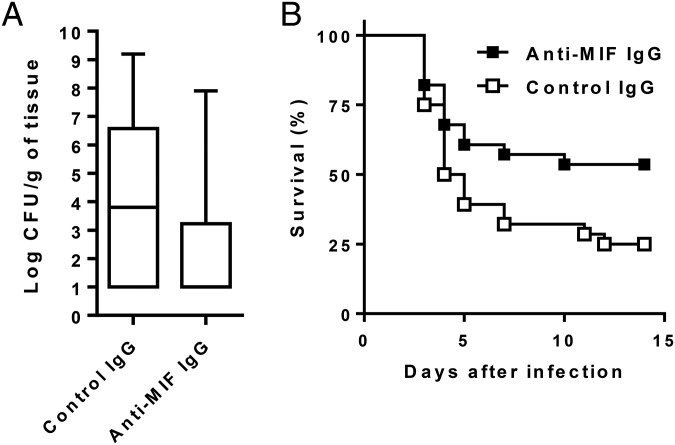
Given that high-expression MIF alleles and high MIF CSF levels were associated with poor patient outcome, we tested the impact of an anti-MIF treatment strategy in a mouse model of pneumococcal pneumonia and sepsis (Fig. 3). Compared with mice treated with control antibody, mice treated with anti-MIF antibody had lower bacterial counts in the lung (median [range] log CFU per gram of tissue: 3.8 [1–9.2] vs. 1 [1–7.9], P = 0.008) and higher survival (25% vs. 53.6%, P = 0.04), indicating that neutralization of MIF was associated with enhanced bacterial clearance and substantial survival benefit in this lethal model of pneumococcal sepsis.
Fig. 3.
Anti-MIF antibody protects against lethal pneumococcal pneumonia and sepsis. Mice were treated with anti-MIF or nonimmune (control) IgG (2 mg injected intraperitoneally) given 2 h before an intranasal inoculation of 104 CFU of S. pneumoniae. (A) Box plots of bacterial counts in the lung 48 h after infection. Data are from five independent experiments with a total of 34 mice per treatment group. Bottom, median, and top lines of the box mark the 25th, 50th, and 75th percentiles, respectively. Vertical lines with whiskers show the range of values. P = 0.008 (Mann–Whitney u test). (B) Kaplan–Meier survival plot. Data points are from three independent experiments with a total of 28 mice per treatment group. P = 0.04 (log-rank test).
Discussion
In this large, nationwide, and well-characterized cohort of patients with community-acquired pneumococcal meningitis, high-expression MIF alleles and elevated levels of MIF in the CSF were identified as predictors of morbidity and mortality. These results suggest that a vigorous MIF response is detrimental in patients with pneumococcal meningitis, a finding consistent with the harmful consequences of robust proinflammatory cytokine responses on brain edema and neuronal damage in the course of bacterial meningitis (26).
Up to now, studies of polymorphisms of cytokine genes have failed to demonstrate association between TNF, IL6, IL10, or LTA and the susceptibility to or the outcome of bacterial meningitis (2). MIF is therefore, to our knowledge, the first cytokine whose gene polymorphisms are associated with morbidity and mortality of bacterial meningitis. MIF may exert its effects by promoting the migration, tissue seeding, and survival of myeloid cells (such as neutrophils and monocytes) that are important players in the pathogenesis of bacterial meningitis (26, 27). At a cellular level, MIF up-regulates proinflammatory responses of immune cells via the activation the MAPK and PI3K/Akt signaling pathways downstream of the CD74–CD44 and CXCR2–CXCR4 MIF receptor complex, which is likely to be detrimental in patients with bacterial meningitis (26, 27).
Few and fairly small studies have looked at MIF in patients with pneumococcal diseases (28, 29). In one study, elevated levels of MIF were detected in the CSF of 31 patients with bacterial meningitis (29). Albeit higher in 11 patients with pneumococcal meningitis than in 15 patients with meningococcal meningitis, MIF CSF levels were not associated with patient outcome. In a study that included 15 patients with pneumococcal meningitis, the frequency of the CATT7 allele, but not that of the −173 C allele, was higher in patients with meningitis than in those with non-CNS pneumococcal infections (28). Associations between MIF genotypes and other clinical variables were not reported. Moreover, given the absence of uninfected control subjects, it was not possible to determine whether the carriage of a CATT7 allele was a risk factor for the development of pneumococcal meningitis. The present study included a large number of patients and had sufficient power to examine these questions. Neither the CATT5–8 nor the −173 G/C MIF polymorphisms was a risk factor for susceptibility to pneumococcal meningitis. However, high-expression MIF alleles and high levels of MIF levels in the CSF were associated with markers of inflammation, indicators of disease severity, complications, and mortality.
Studies exploring the relationship between functional polymorphisms of the MIF gene locus and susceptibility to, severity of, and outcome of infectious and autoimmune diseases have yielded a complex global picture (20, 25). In clinical studies of infectious diseases, carriage of high-expression MIF alleles (CATT7/8) or haplotypes (CATT6/7/−173 C) was associated with severe complications of malaria (30), with mortality in patients with severe sepsis (31), and with survival in patients with community-acquired pneumonia (32). Carriage of the low-expression MIF allele (CATT5) protected children from meningococcemia (33), but it predisposed older adults to Gram-negative bacteremia (34). Notwithstanding the possibility that the impact of MIF polymorphisms may vary according to the age of the host, the site of infections, and the type of microorganism, it also is possible that confounding factors (such as selection biases, ambiguous phenotypes, patient and pathogen heterogeneity, and lack of power) account for these discrepant findings (2, 35, 36). To minimize the influence of potential confounders, we elected to conduct our study in a large and homogeneous cohort of patients with community-acquired bacterial meningitis caused by a single pathogen (S. pneumoniae). One of the strengths of the study was the use of a well-defined phenotype. Working with a nationwide cohort also protected against the potential risk of single-center biases. The choice of controls (patient’s partners or proxies) limited the risk of socioeconomic and environmental mismatching. Power calculations indicated that the study had 80% power to detect ORs in the range of 1.5–3.0 for the main study endpoints.
The study also has some limitations. It included only Caucasians. Given that MIF polymorphisms are unequally distributed in different populations (20), our findings therefore might not be extrapolated to other races or ethnic groups. Blood samples were not available to measure MIF in the systemic circulation. However, one study looking at MIF levels in patients with CNS infections did not find an association between MIF levels in the CSF and inflammation biomarkers in the systemic circulation (37–40). Attempts to correlate MIF polymorphisms with MIF CSF levels were unsuccessful. This result may be due to the relatively limited number of CSF and DNA specimen pairs. Additionally, the range of MIF CSF levels was rather broad, likely because lumbar punctures were performed in patients at different stages of the disease. It is also conceivable that carriage of a high-expression MIF allele plays a detrimental role at the very early stage of disease, promoting inflammation, blood–brain barrier disruption, and bacterial neuro-invasion, as shown both for MIF or IFN lambda in West Nile encephalitis (41, 42). MIF-driven early pathogenic effects may indeed not be correlated with CSF parameters measured just once at a later stage of the disease. In that scenario, both MIF polymorphisms and MIF expression levels might be independent markers of severity at different stages of the disease. Finally, given that MIF is constitutively expressed by innate immune cells and markedly up-regulated upon infection, the release of intracellular MIF pools from dying cells may have increased MIF levels in the CSF, compromising pairwise analyses of MIF gene polymorphisms and MIF protein levels.
A large body of experimental and clinical evidence indicates that MIF is an upstream mediator of the host antimicrobial defense response and a powerful immunomodulating cytokine. It has therefore been investigated as a target for therapeutic interventions in infectious, inflammatory, and autoimmune diseases (19, 32–34, 37–40, 43–45). The observation that MIF is a genetic marker of morbidity and mortality of pneumococcal meningitis is in line with the results obtained in animal models of sepsis and in clinical studies that have linked high levels of MIF with life-threatening bacterial infections (21, 22, 46–49). In a murine model of pneumococcal colonization, the clearance of pneumococci from the nasopharynx, a critical step in the pathogenesis of invasive pneumococcal diseases, was more rapid in wild-type mice than in MIF-deficient mice (45). This finding suggested that MIF might be a protective factor against pneumococcal diseases. However, in the present study, there was no relationship between MIF expression and susceptibility to pneumococcal meningitis. In a model of pneumococcal pneumonia, wild-type mice had higher bacterial load, higher innate immune cell counts in the lung parenchyma, more severe lung pathology, and a higher mortality rate than MIF-deficient mice (50), indicating that MIF played a detrimental role in severe pneumococcal infection. Consistent with this observation, high-expression MIF alleles and MIF CSF levels were predictors of disease severity and poor outcome in patients with pneumococcal meningitis. Furthermore, anti-MIF therapy using either neutralizing antibodies (present study) or a small-molecule inhibitor of MIF (50) promoted bacterial clearance and increased survival in a lethal pneumococcal sepsis. Likewise, humanized anti-MIF antibodies were shown to be protective in a lethal mouse Escherichia coli peritonitis model (51). A first-in-class anti-MIF monoclonal antibody (BAX69/imalumab) is currently under evaluation in phase 1/2a clinical trials in patients with metastatic colorectal cancer or with ovarian cancer and malignant ascites.
All together, the present study provides strong evidence that carriage of high-expression MIF alleles is a genetic marker of morbidity and mortality of pneumococcal meningitis and also suggests a potential role for MIF as a target of immune-modulating adjunctive therapies for bacterial meningitis and sepsis.
Materials and Methods
Bacteria Preparation and Whole-Blood Stimulation Assay.
A serotype 19 S. pneumoniae strain (kind gift of Gilbert Greub, Institute of Microbiology, Centre Hospitalier Universitaire Vaudois and University of Lausanne) isolated from the CSF of a patient with acute community-acquired meningitis was used in all stimulation experiments. Bacteria were grown to log phase in brain–heart infusion medium, washed, and resuspended in PBS at an OD620 of 1.0. Whole blood from five healthy Caucasian subjects was drawn into S-monovette-tubes (Sarstedt) containing 16 U of heparin per mL, diluted fivefold in phenol red-free RPMI medium 1640 containing 2 mM l-glutamine (Life Technologies) (52). Blood was incubated at 37 °C with S. pneumoniae (53). Penicillin and streptomycin (100 µg/mL) were added to the cultures 1 h after stimulation. Cells were pelleted (500 × g for 5 min at room temperature) 24 h after stimulation and cell-free supernatants were used to measure MIF concentrations by ELISA (R&D Systems). The lower limit of detection of the assay was 15.6 pg/mL lactate dehydrogenase (LDH) was measured by using the International Federation of Clinical Chemistry method to evaluate cell death. LDH concentration in samples (9.5–66 units/L) remained in the normal range (i.e., <250 units/L).
Patient Cohort and Controls.
After identification by the Netherlands Reference Laboratory for Bacterial Meningitis (Academic Medical Center, Amsterdam) patients >16 y of age and with a positive CSF culture were enrolled between March 2006 and June 2011 in a nationwide prospective cohort study (54). The study was approved by the medical ethics committee of the University of Amsterdam. Treating physicians obtained informed written consent from either the patient or their legal representatives. Patients with culture-negative community-acquired or with hospital-acquired bacterial meningitis were excluded. Controls consisted of co-dwelling nonrelated patient proxies who shared environment and socioeconomic milieu and provided written informed consent (55). Demographic data including age, gender, and ethnicity for both controls and patients were collected. A detailed medical history and extensive clinical and laboratory data were obtained from all patients. Outcome was evaluated at discharge according to the GOS, with the following scoring system: score of 1 for death; score of 2 for persistent vegetative state; score of 3 for severe disability, defined as a conscious patient who is dependent for daily activities; score of 4 for moderate disability, defined as a patient with some deficits, capable of living independently, but unable to return to work; and score of 5 for a good recovery (56). A favorable outcome was defined as a score of 5 and an unfavorable outcome as a score of 1–4. A CSF leukocyte count <1,000 per mm3 was used as an indicator of disease’s severity (57).
Blood Collection and Genotyping.
Blood from patients and controls was collected in sodium EDTA. DNA extraction was performed by the Gentra Puregene isolation kit (Qiagen) with quality control evaluation for DNA yield and purity. The −794 CATT5–8 microsatellite (rs5844572) and the −173 G/C SNP (rs755622) were genotyped as reported (33). Given the frequencies of the −173 C minor allele and of the CATT7 allele in the Caucasian population, the study had 80% power to detect ORs of at least 1.5 for the susceptibility to infection, 1.9 for an unfavorable outcome, and 3.0 for mortality for these two alleles. These numbers were calculated by using the Quanto 1.2.3 software (biostats.usc.edu/software).
CSF Collection and MIF Measurement.
CSF was obtained from the first lumbar puncture and centrifuged, and supernatant was aliquoted and stored at −80 °C until analysis. Human MIF levels in the CSF were measured by the Luminex technology using a Milliplex assay (Millipore). The lower limit of detection of the assay was 12 pg/mL.
MIF Promoter Activity.
A fragment ranging from −1,073 to −129 bp of the MIF gene of the −794 CATT5/−173 G, −794 CATT6/−173 G, −794 CATT6/−173 C, or −794 CATT7/-173 C genotype was cloned in the pGL3-basic luciferase vector (Promega) (58). The human monocytic THP-1 (TIB-202; American Type Culture Collection) cell line was cultured in RPMI medium 1640 containing 2 mM l-glutamine, 50 μM 2-mercaptoethanol, and 10% (vol/vol) heat-inactivated FCS (Sigma-Aldrich). A total of 40,000 THP-1 cells in 96-well plates were transfected with 350 ng of the luciferase reporter vector together with 50 ng of the Renilla pRL-TK vector (Promega) (59). After 7 h, cells were incubated with S. pneumoniae [multiplicity of infection (MOI) 10]. Penicillin and streptomycin (100 μg/mL) were added to the cultures 1 h after stimulation, and incubation was continued for an additional 23 h. Luciferase and Renilla luciferase activities were measured by using the Dual-Luciferase Reporter Assay System (Promega). Results were expressed as the ratio of luciferase activity to Renilla luciferase activity. An empty vector served as a negative control (18).
Mouse Model of Pneumococcal Pneumonia and Sepsis.
Animal experimentations were approved by the Office Vétérinaire du Canton de Vaud (authorization no. 877.8) and performed according to the institution and Animal Research: Reporting of In Vivo Experiments guidelines (www.nc3rs.org.uk/arrive-guidelines). Female BALB/cByJ mice (10 wk old; Charles River Laboratories) were housed under specific pathogen-free conditions. Mice (8–10 per treatment group) were injected intraperitoneally with 2 mg of rabbit anti-MIF or nonimmune (control) IgG (obtained as described in ref. 37) administered 2 h before an intranasal inoculation of 104 CFU of S. pneumoniae in anesthetized mice. Survival was monitored at least twice daily until the end of the experiment. In selected experiments, mice were killed 48 h after infection to obtain blood and lung tissue for the measurements of bacterial counts.
Statistical Analyses.
Graphs were generated with GraphPad Prism (Version 6.0; GraphPad Software). Data were analyzed by using STATA (Version 11.1; StataCorp LP) and IBM SPSS Statistics (Version 21.0; IBM). Genotype frequencies among patients and controls and HWE were calculated by using the χ2 test. Tests for associations between MIF genotypes and susceptibility to and severity of pneumococcal meningitis were performed by using logistic regression models. Stepwise multivariate selection was performed by using a P value > 0.2 for removal of variables in the model. Sex, age, and polymorphism carriage were forced into the model. Polymorphisms were analyzed by using dominant (comparison of heterozygotes plus homozygotes vs. wild-type carriers) and additive modes of inheritance (assessment of the effect of each additional copy of the minor allele). Strength of relationships between CSF levels and continuous variables was evaluated by using Spearman’s correlation tests. Dichotomous variables were compared with the χ2 test. Continuous variable for patient’s characteristics and disease parameters were assessed by using the Mann–Whitney U or Kruskall–Wallis tests. MIF promoter activity and concentrations of MIF released in whole blood after stimulation with S. pneumoniae were analyzed by using one-way ANOVA for multiple comparisons. The log-rank test was used to compare the Kaplan–Meier survival curves. All tests were two-tailed, and P values < 0.05 were considered to indicate statistical significance.
Acknowledgments
We thank Agnieszka Wόjtowicz for power calculations. This work was supported by Swiss National Science Foundation Grants 138488 (to T.C.), 145014 and 149511 (to T.R.), and 144054 (to P.-Y.B.); European Research Council Starting Grant 281156 (to D.v.d.B.); Netherlands Organization for Health Research and Development Vidi Grant 016.116.358 (to D.v.d.B.); and Netherlands Organisation for Scientific Research Veni Grant 916.13.078 (to M.C.B.). A.S. was supported by the Porphyrogenis Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520727113/-/DCSupplemental.
References
- 1.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brouwer MC, et al. Host genetic susceptibility to pneumococcal and meningococcal disease: A systematic review and meta-analysis. Lancet Infect Dis. 2009;9(1):31–44. doi: 10.1016/S1473-3099(08)70261-5. [DOI] [PubMed] [Google Scholar]
- 3.Brouwer MC, Tunkel AR, van de Beek D. Epidemiology, diagnosis, and antimicrobial treatment of acute bacterial meningitis. Clin Microbiol Rev. 2010;23(3):467–492. doi: 10.1128/CMR.00070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thigpen MC, et al. Emerging Infections Programs Network Bacterial meningitis in the United States, 1998-2007. N Engl J Med. 2011;364(21):2016–2025. doi: 10.1056/NEJMoa1005384. [DOI] [PubMed] [Google Scholar]
- 5.Edmond K, et al. Global and regional risk of disabling sequelae from bacterial meningitis: A systematic review and meta-analysis. Lancet Infect Dis. 2010;10(5):317–328. doi: 10.1016/S1473-3099(10)70048-7. [DOI] [PubMed] [Google Scholar]
- 6.Heckenberg SG, Brouwer MC, van der Ende A, Hensen EF, van de Beek D. Hearing loss in adults surviving pneumococcal meningitis is associated with otitis and pneumococcal serotype. Clin Microbiol Infect. 2012;18(9):849–855. doi: 10.1111/j.1469-0691.2011.03668.x. [DOI] [PubMed] [Google Scholar]
- 7.Jit M. The risk of sequelae due to pneumococcal meningitis in high-income countries: A systematic review and meta-analysis. J Infect. 2010;61(2):114–124. doi: 10.1016/j.jinf.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Casanova JL, Abel L, Quintana-Murci L. Human TLRs and IL-1Rs in host defense: Natural insights from evolutionary, epidemiological, and clinical genetics. Annu Rev Immunol. 2011;29:447–491. doi: 10.1146/annurev-immunol-030409-101335. [DOI] [PubMed] [Google Scholar]
- 9.Kasanmoentalib ES, Brouwer MC, van de Beek D. Update on bacterial meningitis: Epidemiology, trials and genetic association studies. Curr Opin Neurol. 2013;26(3):282–288. doi: 10.1097/WCO.0b013e328360415c. [DOI] [PubMed] [Google Scholar]
- 10.Chapman SJ, et al. PTPN22 and invasive bacterial disease. Nat Genet. 2006;38(5):499–500. doi: 10.1038/ng0506-499. [DOI] [PubMed] [Google Scholar]
- 11.Roy S, et al. Oxford Pneumoccocal Surveillance Group MBL genotype and risk of invasive pneumococcal disease: A case-control study. Lancet. 2002;359(9317):1569–1573. doi: 10.1016/S0140-6736(02)08516-1. [DOI] [PubMed] [Google Scholar]
- 12.Khor CC, et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet. 2007;39(4):523–528. doi: 10.1038/ng1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman SJ, et al. IkappaB genetic polymorphisms and invasive pneumococcal disease. Am J Respir Crit Care Med. 2007;176(2):181–187. doi: 10.1164/rccm.200702-169OC. [DOI] [PubMed] [Google Scholar]
- 14.Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153(3731):80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- 15.David JR. Delayed hypersensitivity in vitro: Its mediation by cell-free substances formed by lymphoid cell-antigen interaction. Proc Natl Acad Sci USA. 1966;56(1):72–77. doi: 10.1073/pnas.56.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calandra T, et al. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377(6544):68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell RA, et al. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: Regulatory role in the innate immune response. Proc Natl Acad Sci USA. 2002;99(1):345–350. doi: 10.1073/pnas.012511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roger T, Chanson AL, Knaup-Reymond M, Calandra T. Macrophage migration inhibitory factor promotes innate immune responses by suppressing glucocorticoid-induced expression of mitogen-activated protein kinase phosphatase-1. Eur J Immunol. 2005;35(12):3405–3413. doi: 10.1002/eji.200535413. [DOI] [PubMed] [Google Scholar]
- 19.Roger T, David J, Glauser MP, Calandra T. MIF regulates innate immune responses through modulation of Toll-like receptor 4. Nature. 2001;414(6866):920–924. doi: 10.1038/414920a. [DOI] [PubMed] [Google Scholar]
- 20.Bucala R. MIF, MIF alleles, and prospects for therapeutic intervention in autoimmunity. J Clin Immunol. 2013;33(Suppl 1):S72–S78. doi: 10.1007/s10875-012-9781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calandra T, Roger T. Macrophage migration inhibitory factor: A regulator of innate immunity. Nat Rev Immunol. 2003;3(10):791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lue H, Kleemann R, Calandra T, Roger T, Bernhagen J. Macrophage migration inhibitory factor (MIF): Mechanisms of action and role in disease. Microbes Infect. 2002;4(4):449–460. doi: 10.1016/s1286-4579(02)01560-5. [DOI] [PubMed] [Google Scholar]
- 23.Baugh JA, et al. A functional promoter polymorphism in the macrophage migration inhibitory factor (MIF) gene associated with disease severity in rheumatoid arthritis. Genes Immun. 2002;3(3):170–176. doi: 10.1038/sj.gene.6363867. [DOI] [PubMed] [Google Scholar]
- 24.Donn RP, Shelley E, Ollier WE, Thomson W. British Paediatric Rheumatology Study Group A novel 5′-flanking region polymorphism of macrophage migration inhibitory factor is associated with systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2001;44(8):1782–1785. doi: 10.1002/1529-0131(200108)44:8<1782::AID-ART314>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 25.Renner P, Roger T, Calandra T. Macrophage migration inhibitory factor: Gene polymorphisms and susceptibility to inflammatory diseases. Clin Infect Dis. 2005;41(Suppl 7):S513–S519. doi: 10.1086/432009. [DOI] [PubMed] [Google Scholar]
- 26.Mook-Kanamori BB, Geldhoff M, van der Poll T, van de Beek D. Pathogenesis and pathophysiology of pneumococcal meningitis. Clin Microbiol Rev. 2011;24(3):557–591. doi: 10.1128/CMR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernhagen J, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13(5):587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 28.Doernberg S, et al. Association of macrophage migration inhibitory factor (MIF) polymorphisms with risk of meningitis from Streptococcus pneumoniae. Cytokine. 2011;53(3):292–294. doi: 10.1016/j.cyto.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Østergaard C, Benfield T. Macrophage migration inhibitory factor in cerebrospinal fluid from patients with central nervous system infection. Crit Care. 2009;13(3):R101. doi: 10.1186/cc7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Awandare GA, et al. MIF (macrophage migration inhibitory factor) promoter polymorphisms and susceptibility to severe malarial anemia. J Infect Dis. 2009;200(4):629–637. doi: 10.1086/600894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehmann LE, et al. A MIF haplotype is associated with the outcome of patients with severe sepsis: A case control study. J Transl Med. 2009;7:100. doi: 10.1186/1479-5876-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yende S, et al. The influence of macrophage migration inhibitory factor gene polymorphisms on outcome from community-acquired pneumonia. FASEB J. 2009;23(8):2403–2411. doi: 10.1096/fj.09-129445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renner P, et al. A functional microsatellite of the macrophage migration inhibitory factor gene associated with meningococcal disease. FASEB J. 2012;26(2):907–916. doi: 10.1096/fj.11-195065. [DOI] [PubMed] [Google Scholar]
- 34.Das R, et al. Functional polymorphisms in the gene encoding macrophage migration inhibitory factor are associated with Gram-negative bacteremia in older adults. J Infect Dis. 2014;209(5):764–768. doi: 10.1093/infdis/jit571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bochud PY, Bochud M, Telenti A, Calandra T. Innate immunogenetics: A tool for exploring new frontiers of host defence. Lancet Infect Dis. 2007;7(8):531–542. doi: 10.1016/S1473-3099(07)70185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet. 2012;13(3):175–188. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- 37.Calandra T, et al. Protection from septic shock by neutralization of macrophage migration inhibitory factor. Nat Med. 2000;6(2):164–170. doi: 10.1038/72262. [DOI] [PubMed] [Google Scholar]
- 38.Beishuizen A, Thijs LG, Haanen C, Vermes I. Macrophage migration inhibitory factor and hypothalamo-pituitary-adrenal function during critical illness. J Clin Endocrinol Metab. 2001;86(6):2811–2816. doi: 10.1210/jcem.86.6.7570. [DOI] [PubMed] [Google Scholar]
- 39.Emonts M, et al. Association between high levels of blood macrophage migration inhibitory factor, inappropriate adrenal response, and early death in patients with severe sepsis. Clin Infect Dis. 2007;44(10):1321–1328. doi: 10.1086/514344. [DOI] [PubMed] [Google Scholar]
- 40.Sprong T, et al. Macrophage migration inhibitory factor (MIF) in meningococcal septic shock and experimental human endotoxemia. Shock. 2007;27(5):482–487. doi: 10.1097/01.shk.0000246898.65692.34. [DOI] [PubMed] [Google Scholar]
- 41.Arjona A, et al. Abrogation of macrophage migration inhibitory factor decreases West Nile virus lethality by limiting viral neuroinvasion. J Clin Invest. 2007;117(10):3059–3066. doi: 10.1172/JCI32218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lazear HM, et al. Interferon-λ restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci Transl Med. 2015;7(284):284ra59. doi: 10.1126/scitranslmed.aaa4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calandra T, Spiegel LA, Metz CN, Bucala R. Macrophage migration inhibitory factor is a critical mediator of the activation of immune cells by exotoxins of Gram-positive bacteria. Proc Natl Acad Sci USA. 1998;95(19):11383–11388. doi: 10.1073/pnas.95.19.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roger T, et al. Macrophage migration inhibitory factor deficiency is associated with impaired killing of gram-negative bacteria by macrophages and increased susceptibility to Klebsiella pneumoniae sepsis. J Infect Dis. 2013;207(2):331–339. doi: 10.1093/infdis/jis673. [DOI] [PubMed] [Google Scholar]
- 45.Das R, et al. Macrophage migration inhibitory factor promotes clearance of pneumococcal colonization. J Immunol. 2014;193(2):764–772. doi: 10.4049/jimmunol.1400133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernhagen J, et al. MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature. 1993;365(6448):756–759. doi: 10.1038/365756a0. [DOI] [PubMed] [Google Scholar]
- 47.Lehmann LE, et al. Plasma levels of macrophage migration inhibitory factor are elevated in patients with severe sepsis. Intensive Care Med. 2001;27(8):1412–1415. doi: 10.1007/s001340101022. [DOI] [PubMed] [Google Scholar]
- 48.Roger T, Glauser MP, Calandra T. Macrophage migration inhibitory factor (MIF) modulates innate immune responses induced by endotoxin and Gram-negative bacteria. J Endotoxin Res. 2001;7(6):456–460. [PubMed] [Google Scholar]
- 49.Calandra T, Froidevaux C, Martin C, Roger T. Macrophage migration inhibitory factor and host innate immune defenses against bacterial sepsis. J Infect Dis. 2003;187(Suppl 2):S385–S390. doi: 10.1086/374752. [DOI] [PubMed] [Google Scholar]
- 50.Weiser JN, et al. Macrophage migration Inhibitory factor is detrimental in pneumococcal pneumonia and a target for therapeutic immunomodulation. J Infect Dis. 2015;212(10):1677–1682. doi: 10.1093/infdis/jiv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kerschbaumer RJ, et al. Neutralization of macrophage migration inhibitory factor (MIF) by fully human antibodies correlates with their specificity for the β-sheet structure of MIF. J Biol Chem. 2012;287(10):7446–7455. doi: 10.1074/jbc.M111.329664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roger T, et al. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood. 2011;117(4):1205–1217. doi: 10.1182/blood-2010-05-284711. [DOI] [PubMed] [Google Scholar]
- 53.Lugrin J, et al. Histone deacetylase inhibitors repress macrophage migration inhibitory factor (MIF) expression by targeting MIF gene transcription through a local chromatin deacetylation. Biochim Biophys Acta. 2009;1793(11):1749–1758. doi: 10.1016/j.bbamcr.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Woehrl B, et al. Complement component 5 contributes to poor disease outcome in humans and mice with pneumococcal meningitis. J Clin Invest. 2011;121(10):3943–3953. doi: 10.1172/JCI57522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Little J, et al. Reporting, appraising, and integrating data on genotype prevalence and gene-disease associations. Am J Epidemiol. 2002;156(4):300–310. doi: 10.1093/oxfordjournals.aje.a000179. [DOI] [PubMed] [Google Scholar]
- 56.Jennett B, Bond M. Assessment of outcome after severe brain damage. Lancet. 1975;1(7905):480–484. doi: 10.1016/s0140-6736(75)92830-5. [DOI] [PubMed] [Google Scholar]
- 57.van de Beek D, et al. Clinical features and prognostic factors in adults with bacterial meningitis. N Engl J Med. 2004;351(18):1849–1859. doi: 10.1056/NEJMoa040845. [DOI] [PubMed] [Google Scholar]
- 58.Roger T, Ding X, Chanson AL, Renner P, Calandra T. Regulation of constitutive and microbial pathogen-induced human macrophage migration inhibitory factor (MIF) gene expression. Eur J Immunol. 2007;37(12):3509–3521. doi: 10.1002/eji.200737357. [DOI] [PubMed] [Google Scholar]
- 59.Delaloye J, et al. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 2009;5(6):e1000480. doi: 10.1371/journal.ppat.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]