Significance
Catechol oxidases and tyrosinases belong to the family of polyphenol oxidases (PPOs). In contrast to tyrosinases, catechol oxidases were so far defined to lack hydroxylase activity toward monophenols. Aurone synthase (AUS1) is a plant catechol oxidase that specializes in the conversion of chalcones to aurones (flower pigments). We evidence for the first time, to our knowledge, hydroxylase activity for a catechol oxidase (AUS1) toward its natural monophenolic substrate (chalcone). The presented first crystal structure of a plant pro-PPO provides insights into its activation mechanisms, and based on biochemical and structural studies of AUS1, we propose a novel catalytic reaction mechanism for plant PPOs. The proven hydroxylase functionality of AUS1 suggests that other catechol oxidases might also be involved in the plant’s secondary metabolism.
Keywords: crystal structure, tyrosinase, catechol oxidase, type III copper protein, mechanism
Abstract
Tyrosinases and catechol oxidases belong to the family of polyphenol oxidases (PPOs). Tyrosinases catalyze the o-hydroxylation and oxidation of phenolic compounds, whereas catechol oxidases were so far defined to lack the hydroxylation activity and catalyze solely the oxidation of o-diphenolic compounds. Aurone synthase from Coreopsis grandiflora (AUS1) is a specialized plant PPO involved in the anabolic pathway of aurones. We present, to our knowledge, the first crystal structures of a latent plant PPO, its mature active and inactive form, caused by a sulfation of a copper binding histidine. Analysis of the latent proenzyme’s interface between the shielding C-terminal domain and the main core provides insights into its activation mechanisms. As AUS1 did not accept common tyrosinase substrates (tyrosine and tyramine), the enzyme is classified as a catechol oxidase. However, AUS1 showed hydroxylase activity toward its natural substrate (isoliquiritigenin), revealing that the hydroxylase activity is not correlated with the acceptance of common tyrosinase substrates. Therefore, we propose that the hydroxylase reaction is a general functionality of PPOs. Molecular dynamics simulations of docked substrate–enzyme complexes were performed, and a key residue was identified that influences the plant PPO’s acceptance or rejection of tyramine. Based on the evidenced hydroxylase activity and the interactions of specific residues with the substrates during the molecular dynamics simulations, a novel catalytic reaction mechanism for plant PPOs is proposed. The presented results strongly suggest that the physiological role of plant catechol oxidases were previously underestimated, as they might hydroxylate their—so far unknown—natural substrates in vivo.
Tyrosinases and catechol oxidases are polyphenol oxidases (PPOs) and belong to the family of type III copper proteins. They are widespread among plants, fungi, and bacteria (1, 2). Plant PPOs display a high diversity, and often several PPO genes are present in one organism (2, 3). Tyrosinases catalyze the hydroxylation and oxidation of monophenols (e.g., tyrosine) to o-diphenols and the corresponding o-quinones. Due to the fact that catechol oxidases are unable to hydroxylate common tyrosinase substrates (e.g., tyrosine and tyramine), they were so far defined to lack the hydroxylation (monophenolase) activity and to catalyze solely the oxidation of o-diphenols to o-quinones (diphenolase activity). Plant PPOs are expressed as latent proenzymes (∼64 kDa), in which the active site of the enzyme is shielded by the C-terminal domain (∼19 kDa). Presumably, the C-terminal domain is proteolytically cleaved during the maturation process, resulting in a fully active enzyme (4, 5). The latent proenzymes can be activated in vitro by proteases, an acidic pH, fatty acids, or detergents (e.g., SDS) (6). PPOs are found in almost all plants and have been assigned primarily to enzymatic browning reactions and to the protection of organisms against biotic and abiotic stress (1, 7). However, in most cases, their individual physiological role and especially their natural substrates remain unknown. Beside aurone formation, their specific involvement in biosynthetic processes was shown only in a few cases (1, 7–11). Recently, Sullivan (7) wondered whether there exist more PPOs that are involved in the plant’s secondary metabolism and whether the few so far known specialized PPOs represent only “the tip of the iceberg.”
More than one decade ago, a vacuole-localized PPO, aureusidin synthase, was identified to be a key enzyme in the anabolic aurone (yellow pigments) formation in petals of Antirrhinum majus (12, 13). Recently, it has been reported that aurone biosynthesis in Asteraceae species (4-deoxyaurone formation) (14) differs in various aspects from the 4-hydroxyaurone biosynthesis in A. majus. A PPO homolog, termed aurone synthase (AUS1, Uniprot A0A075DN54), was assigned to be involved in 4-deoxyaurone biosynthesis in petals of Coreopsis grandiflora (tickseed, Asteraceae) (15, 16). As with most plant PPOs, this enzyme possesses an N-terminal chloroplast transit peptide and a thylakoid transfer domain and is hence predicted to be transported to the thylakoid lumen (16). AUS1 comprises the conserved motifs of common plant PPOs—for example, a thioether bridge between a cysteine and a CuA binding histidine and a phenylalanine (“blocker” or “gate” residue) above the active site. AUS1 exhibits an insertion in a loop region near the active site and is therefore classified as a member of the group 2 PPOs (15). So far uniquely, it possesses a disulfide linkage between the shielding C-terminal domain and the catalytically active main core. The latent proenzyme has been shown to undergo an allosteric activation mechanism, caused by in situ-formed, highly reactive o-quinones (15). Mechanistic considerations regarding the differences between tyrosinases and catechol oxidases were so far based on the comparison of structures of plant catechol oxidases (17, 18) with structures of several bacterial (19, 20) and fungal (21, 22) tyrosinases. However, the comparability of fungal and bacterial with plant PPOs is very limited due to the existence of the plant PPOs’ bulky gate residue, which will have a general high influence in substrate coordination. Recently, the first crystal structure of a plant tyrosinase from walnut leaves was reported, and it was revealed that the residues located at the entrance of the active site are more important for the acceptance of tyramine and tyrosine than the previously suggested restriction of the active site (23). However, the differences in the enzymatic activity and the mechanism of catechol oxidases and tyrosinases still remain striking questions.
In the following, to our knowledge, the first crystal structures of a latent plant pro-PPO as well as its mature activated and an inactive form, caused by a sulfation of a copper binding histidine, are presented. Insights into the in vitro activation and the allosteric activation of the latent enzyme were obtained by the analysis of the interdomain interactions of the proenzyme. A combination of molecular dynamics (MD) simulations of docked natural substrates to active AUS1 and its hydroxylase activity toward its natural substrate leads to the proposal of a novel catalytic reaction mechanism of plant PPOs.
Results
Overall Structure of Active AUS1.
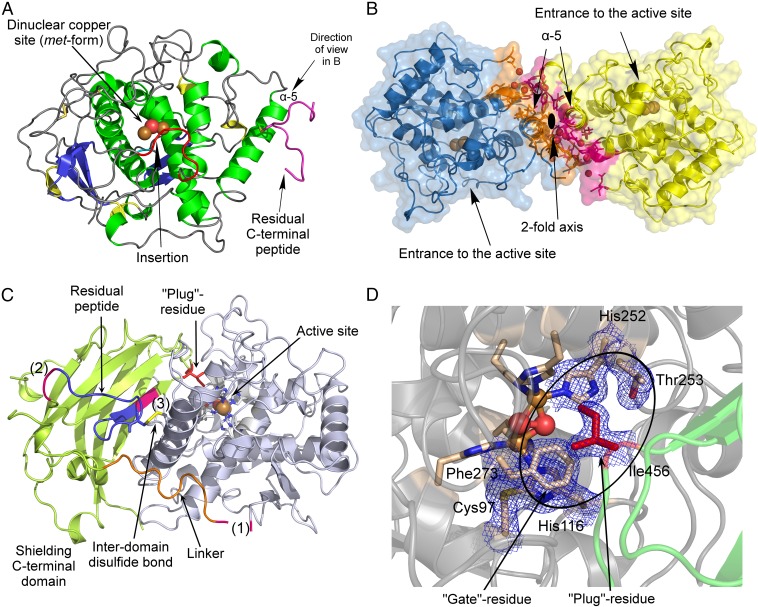
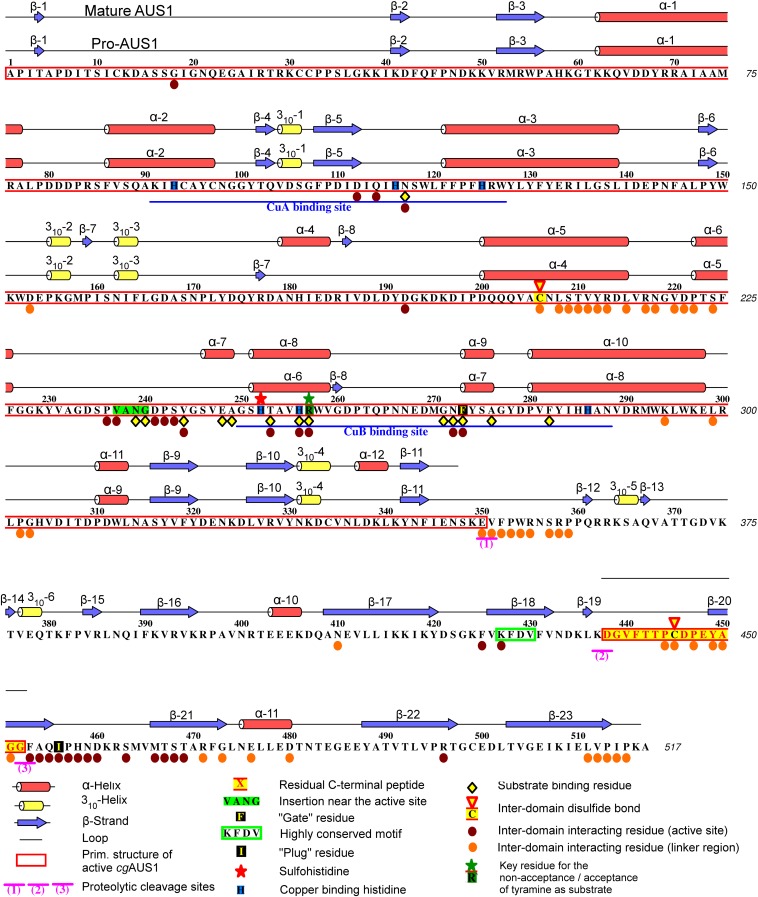
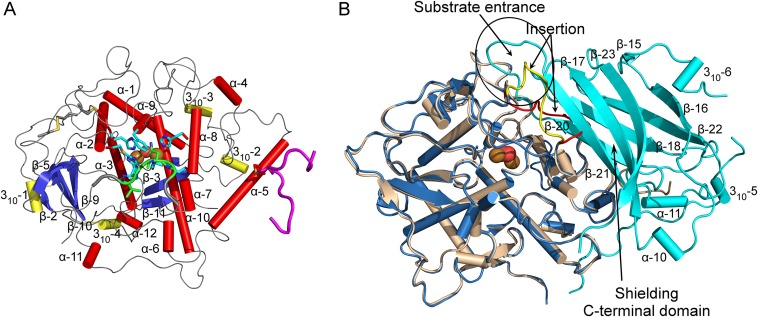
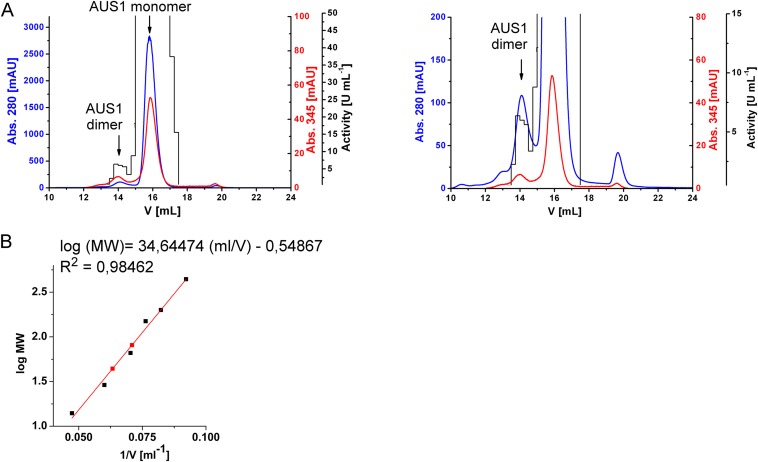
Mature AUS1 (41.6 kDa) crystallized in space groups P212121 and P1211, both with four monomers in the asymmetric unit, yielding crystals of comparable quality (24) (diffracting to ∼1.6 Å; Tables S1 and S2). The enzyme’s structure possesses a very high structural similarity to the plant catechol oxidases from Ipomoea batatas (18) [ibCO; Protein Data Bank (PDB) ID code 1BT3; rmsd of 0.81 Å between 296 atom pairs] and Vitis vinifera (17) (vvCO; PDB ID 2P3X; rmsd of 0.79 Å between 284 atom pairs) as well as to the plant tyrosinase from J. regia (23) (jrTYR; PDB ID code 5CE9; rmsd of 0.82 Å between 287 atom pairs). Mature AUS1 shares ∼47% sequence identity with each of the active catechol oxidases and ∼48% with the active tyrosinase. Structural divergences to these PPOs are the unique features (15) of AUS1 (Fig. 1A and Figs. S1 and S2A): the remaining C-terminal peptide (Asp338 to Gly452)—connected to the catalytically active main core by a disulfide bond (Cys206–Cys445)—and the insertion of four amino acids (V237ANG240) that form a cavity near the active site. Both crystal forms possess homodimeric assemblies of their monomers [analyzed with PISA (Proteins, Interfaces, Structures and Assemblies)] (25), and a portion of mature AUS1 was found in a dimeric state by size exclusion chromatography (Fig. S3), evidencing that the dimer exists in solution. The biological assembly reveals a so far unknown role of the residual C-terminal peptide, as it forms a main building block of the homodimeric interface (Fig. 1B). The quaternary structure is stabilized by four hydrogen bonds, 11 bridging water molecules, and several hydrophobic interactions between the residues of the C-terminal peptide of one monomer and the α-helix of the second monomer that carries the C-terminal peptide. These interactions result in a buried surface area ranging from 1,523 to 2,323 Å2 and a free energy of dissociation of the dimer ranging from 2.6 to 8.9 kcal/mol. The active sites of the dimeric monomers are facing opposite directions, due to their twofold rotational symmetry relation.
Table S1.
Data collection and refinement statistics (molecular replacement) for crystal structures of mature AUS1 (natural source)
| Data | Active AUS1—CuB:sulfohistidine, ∼1.4:1 | Inactive AUS1, sulfohistidine ∼90% |
| PDB entry | 4Z0Y | 4Z0Z |
| Data collection | ||
| Space group | P212121 | P1211 |
| Cell dimensions | ||
| a, b, c, Å | 88.72, 90.56, 182.59 | 51.53, 183.52, 78.09 |
| α, β, γ, ° | 90.0, 90.0, 90.0 | 90.0, 94.5, 90.0 |
| Resolution, Å | 45.65–1.60 (1.66–1.60)a | 48.10–1.60 (1.66–1.60) |
| Rmerge | 0.108 (0.842) | 0.107 (0.830) |
| I/σI | 11.3 (1.8) | 8.1 (1.6) |
| Number of measured reflections | 1,221,727 (83,125) | 647,241 (66,932) |
| Number of unique reflections | 192,560 (18,419) | 187,078 (18,928) |
| Completeness, % | 99.4 (96.1) | 98.7 (99.9) |
| Redundancy | 6.3 (4.5) | 3.5 (3.5) |
| CC1/2 | 0.998 (0.626) | 0.993 (0.508) |
| Refinement | ||
| Resolution, Å | 45.65–1.60 (1.66–1.60)a | 48.10–1.60 (1.66–1.60) |
| Rwork/Rfree | 0.157/0.194 | 0.165/0.200 |
| Number of atoms | 13,612 | 13,452 |
| Protein | 11,507 | 11,405 |
| Ligand/ion | 38 | 4 |
| Water | 2,067 | 2,043 |
| B factors, Å2 | 20.3 | 21.4 |
| Protein | 18.1 | 19.3 |
| Ligand/ion | 27.2 | 16.5 |
| Water | 32.3 | 33.1 |
| rmsd | ||
| Bond lengths, Å | 0.008 | 0.008 |
| Bond angles, ° | 1.17 | 1.09 |
| CC* | 1.000 (0.878) | 0.998 (0.821) |
| Ramachandran plot | ||
| Most favored, % | 97.2 | 96.7 |
| Allowed, % | 2.8 | 3.3 |
One crystal was used.
Values in parentheses are for the highest resolution shell.
Table S2.
Data collection and refinement statistics (molecular replacement) for crystal structures of latent AUS1 (natural source and recombinantly expressed)
| Data | Pro-AUS1, natural source | Pro-AUS1, cocrystallized with TEW, recombinantly expressed | Oxy-pro-AUS1, cocrystallized with TEW, recombinantly expressed |
| PDB entry | 4Z11 | 4Z12 | 4Z13 |
| Data collection | |||
| Space group | P1211 | P1211 | P1211 |
| Cell dimensions | |||
| a, b, c, Å | 62.57, 174.1, 102.54 | 52.99, 110.41, 94.99 | 52.88, 109.75, 94.76 |
| α, β, γ, ° | 90.0, 105.3, 90.0 | 90.0, 95.8, 90.0 | 90.0, 96.3, 90.0 |
| Resolution, Å | 46.38–2.50 (2.59–2.50)a | 48.15–1.85 (1.92–1.85) | 47.41–1.78 (1.84–1.78) |
| Rmerge | 0.145 (0.862) | 0.118 (2.729) | 0.112 (1.337) |
| I/σI | 7.6 (2.0) | 8.3 (0.6) | 8.6 (1.0) |
| Number of measured reflections | 242,803 (24,273) | 415,481 (42,647) | 388,959 (38,335) |
| Number of unique reflections | 71,372 (7,074) | 90,863 (8,954) | 101,113 (9,851) |
| Completeness, % | 97.8 (97.7) | 98.1 (97.0) | 98.4 (96.2) |
| Redundancy | 3.4 (3.4) | 4.6 (4.8) | 3.8 (3.9) |
| CC1/2 | 0.986 (0.559) | 0.996 (0.293) | 0.996 (0.453) |
| Refinement | |||
| Resolution, Å | 46.38–2.50 (2.59–2.50)a | 48.15–1.85 (1.92–1.85) | 47.41–1.78 (1.84–1.78) |
| Rwork/Rfree | 0.183/0.234 | 0.172/0.205 | 0.157/0.196 |
| Number of atoms | 16,266 | 8,544 | 8,974 |
| Protein | 15,738 | 7,976 | 8,020 |
| Ligand/ion | 8 | 98 | 96 |
| Water | 520 | 470 | 858 |
| B factors, Å2 | 55.1 | 51.5 | 37.1 |
| Protein | 55.4 | 50.3 | 35.4 |
| Ligand/ion | 49.8 | 153.0 | 114.5 |
| Water | 45.3 | 50.0 | 43.9 |
| rmsd | |||
| Bond lengths, Å | 0.003 | 0.007 | 0.007 |
| Bond angles, ° | 0.66 | 0.99 | 1.02 |
| CC* | 0.996 (0.847) | 0.999 (0.673) | 0.999 (0.790) |
| Ramachandran plot | |||
| Most favored, % | 97.7 | 97.0 | 97.4 |
| Allowed, % | 2.3 | 3.0 | 2.6 |
One crystal was used.
Values in parentheses are for highest resolution shell.
Fig. 1.
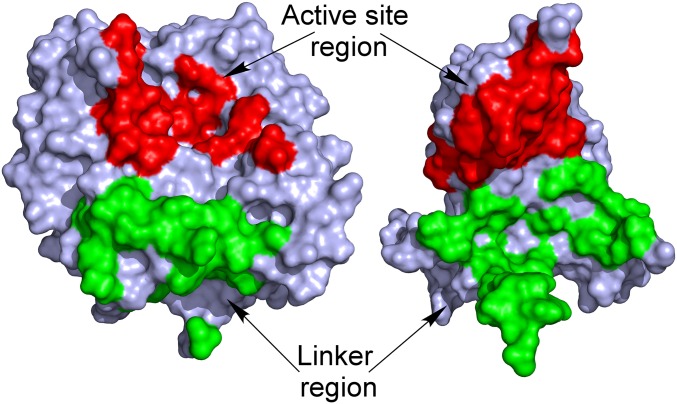
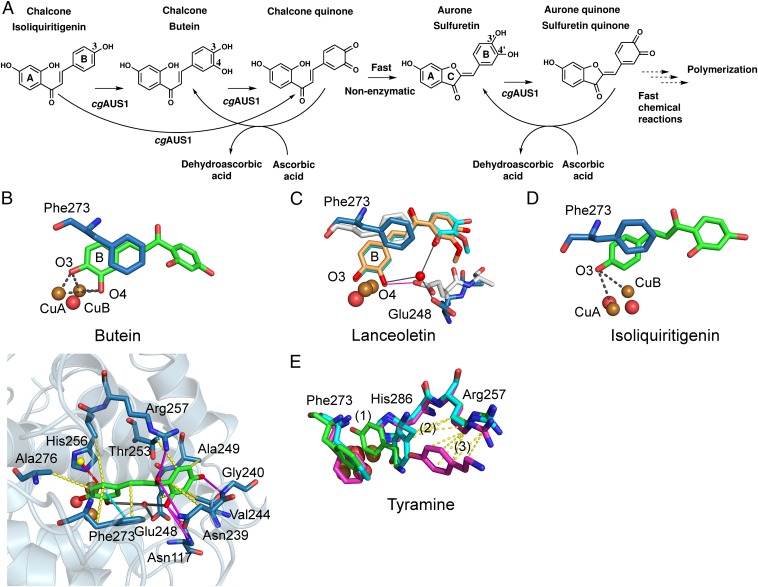
Overall structures of latent and mature aurone synthase. (A) Top view of mature aurone synthase. The coloring was performed according to the secondary structure features (α-helices, green; 310-helices, yellow; β-sheets, blue), and the characteristic features of AUS1 are colored red (loop carrying the insertion V237ANG240) and magenta (residual C-terminal peptide). (B) Side view of the dimeric biological assembly of mature AUS1. The interacting residues are colored orange (monomer chain B) and magenta (monomer chain D). (C) Overall structure of latent aurone synthase. The features of the C-terminal domain are as follows: the three proteolytic cleavage sites, colored magenta and labeled as (1), (2), and (3); the linker region (orange) that connects the catalytically active domain (gray) and the shielding C-terminal domain (green); and the residual peptide of the C-terminal domain (blue) and the active site shielding Ile456 (plug residue) (red). (D) The active site is shielded by the C-terminal residue Ile456, which is responsible for the latency of the proenzyme and functions like a plug [2Fo-Fc electron density map (blue mesh) contoured at 1.0 σ].
Fig. S1.
Primary structure, secondary structure, and features of latent and mature AUS1.
Fig. S2.
Secondary structure features [assigned using DSSP (64); compare with Fig. S1] and superimposition of mature and latent AUS1. (A) Secondary structure features of mature AUS1 (α-helices, red tubes; 310-helices, yellow tubes; β-strands, blue). (B) Superposition of active (colored beige) and latent (catalytically active domain, blue; shielding C-terminal domain, cyan) AUS1 (rmsd of 0.47 Å between 338 atom pairs). The location of the loop carrying the insertion is different in the latent proenzyme (latent, yellow; active, red).
Fig. S3.
Size exclusion chromatogram of mature AUS1 [41.6 kDa (15)] on Superdex 200 Increase. A buffer containing 50 mM sodium citrate and 1.5 M sodium chloride, pH 5.4, was used, and a flow rate of 0.5 mL/min was applied. Enzymatic assays for active AUS1 were performed by monitoring the oxidation of 50 µM fisetin in 125 mM sodium citrate pH 5.5 at 280 nm in a total volume of 1 mL (15). (A) Full (Left) and magnified (Right) chromatogram of mature AUS1. The dimeric AUS1 eluted at 14.10 mL (estimated mass, 81.0 kDa), and the monomeric AUS1 eluted at 15.80 mL (estimated mass, 44.1 kDa). (B) The calibration was performed using thyroglobulin (669 kDa), apoferritin (443 kDa), beta-amylase (200 kDa), alkoholdehydrogenase (150 kDa), albumin (66 kDa), carbonic anhydrase (29 kDa), and lysozyme (14 kDa).
Overall Structure of Latent AUS1.
Latent AUS1 was found to be monomeric, and the overall structure of the main core of the latent proenzyme is almost identical to the overall structure of the mature enzyme (rmsd of 0.47 Å between 338 atom pairs), except for the location of one loop, which gives way for the shielding C-terminal domain (Fig. S2B). Interestingly, this loop contains the characteristic insertion (V237ANG240) of the group 2 PPOs (15). The C-terminal domain begins with a short linker region of approximately 10 amino acids, located in the vicinity of the interdomain disulfide linkage (Fig. 1C). The C-terminal domain displays seven β-strands that form a jelly roll barrel motif with the four loops being arranged in a staggered manner above the active site. The latent enzyme displayed very low activity (toward butein) and an allosteric activation (in the presence of in situ-formed o-quinones) (15). Therefore, the active site of a small portion of latent enzyme molecules has to be accessible for substrates. A loop region of the jelly roll motif as well as the loop that carries the insertion show weak or even completely missing electron density. This demonstrates the flexibility of this region possibly being an entrance for the substrates (Fig. S2B). The interface between the catalytically active main core and the shielding C-terminal domain is fairly large and composed of several hydrogen bonds, hydrophobic interactions, and water bridges. However, the interdomain interactions are concentrated at two different locations (Fig. 2). One area is located near the linker region and the interdomain disulfide linkage, whereas the other is located in the vicinity of the active site. The residue Ile456 is of crucial importance, as it blocks the entrance to the active site and functions similar to a “plug” (Fig. 1D). This plug residue is located outside of the active site and on top of the gate residue (Phe273). It is stabilized by its hydrophobic interactions with the gate residue, one CuB binding histidine (His252), and Thr253.
Fig. 2.
Interactions between the shielding C-terminal domain and the catalytically active domain of latent AUS1. The latent structure is segmented by its domains (left, catalytically active domain; right, shielding C-terminal domain), and their solvent-accessible surface is presented. The interdomain interactions are located at two distinct areas: in the vicinity of the active site (colored red) and in the vicinity of the linker region (colored green).
Copper Binding Sites of met-, oxy-, and Inactive Forms of AUS1.
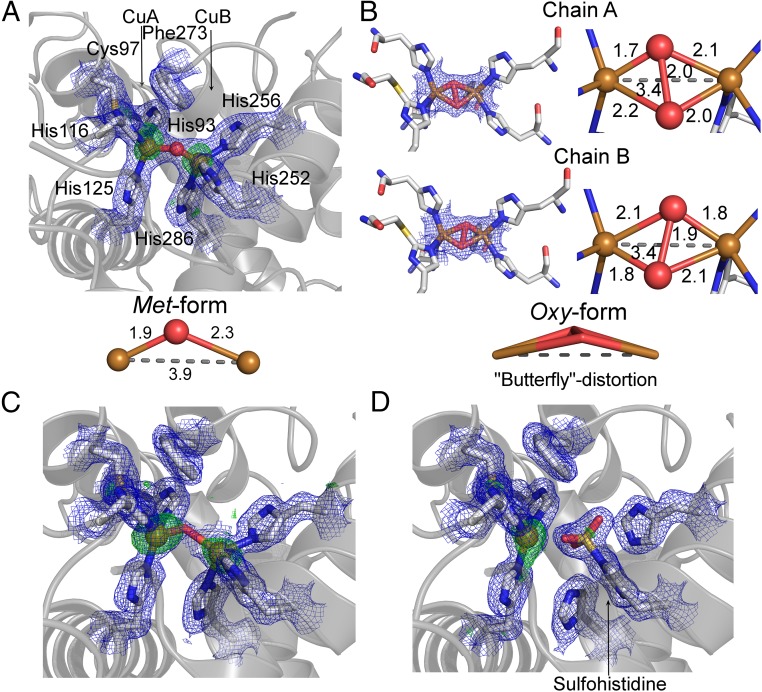
Active, latent, and recombinantly expressed latent AUS1 crystallized in the resting met-form, in which the copper ions (CuII–CuII distance, ∼4 Å) are bridged by a hydroxide ion or a water molecule (Fig. 3A). The geometry of the met-form of AUS1 is fully consistent with previously published structures (18, 23). To generate the oxy-form, crystals of recombinantly expressed AUS1 were soaked in cryoprotectant solution containing hydrogen peroxide (24). The copper ions (CuII) of the obtained crystal structure are bridged by a peroxide ion, and the Cu–Cu distance is reduced to ∼3.4 Å (Fig. 3B). The O–O distance of 1.9–2.0 Å (not restrained during refinements) indicates a Cu2O2 geometry that lies between the bis-μ-oxo and the μ-η2:η2-peroxo geometry, which exists in equilibrium. Interestingly, the complex exhibits a “butterfly” distortion (distortion of the Cu–O–O–Cu torsion angles). The Cu–O distances seem to slightly differ between chain A and chain B. However, the resolution (∼1.8 Å; Tables S1 and S2) of the obtained dataset is not high enough to provide an unambiguous statement about the precise Cu–O distance.
Fig. 3.
Copper binding site of AUS1. The 2Fo-Fc electron density map (blue mesh) is contoured at 1.0 σ, and the anomalous Fourier difference map in A, C, and D (green mesh) is contoured at 3.0 σ. (A) Native met-copper center of recombinantly expressed AUS1. (B) Oxy-copper center of recombinantly expressed AUS1 (μ-η2:η2-peroxo geometry; crystal soaked in H2O2). The oxygen atoms are slightly unsymmetrically bound to the copper atoms, and the μ-η2:η2 peroxo complex displays a butterfly distortion. (C) Copper binding site of active AUS1 (occupancy CuB, ∼0.55). For clarity, the sulfohistidine (occupancy, ∼0.45) is not shown. (D) Inactive AUS1 (occupancy sulfohistidine, 0.9).
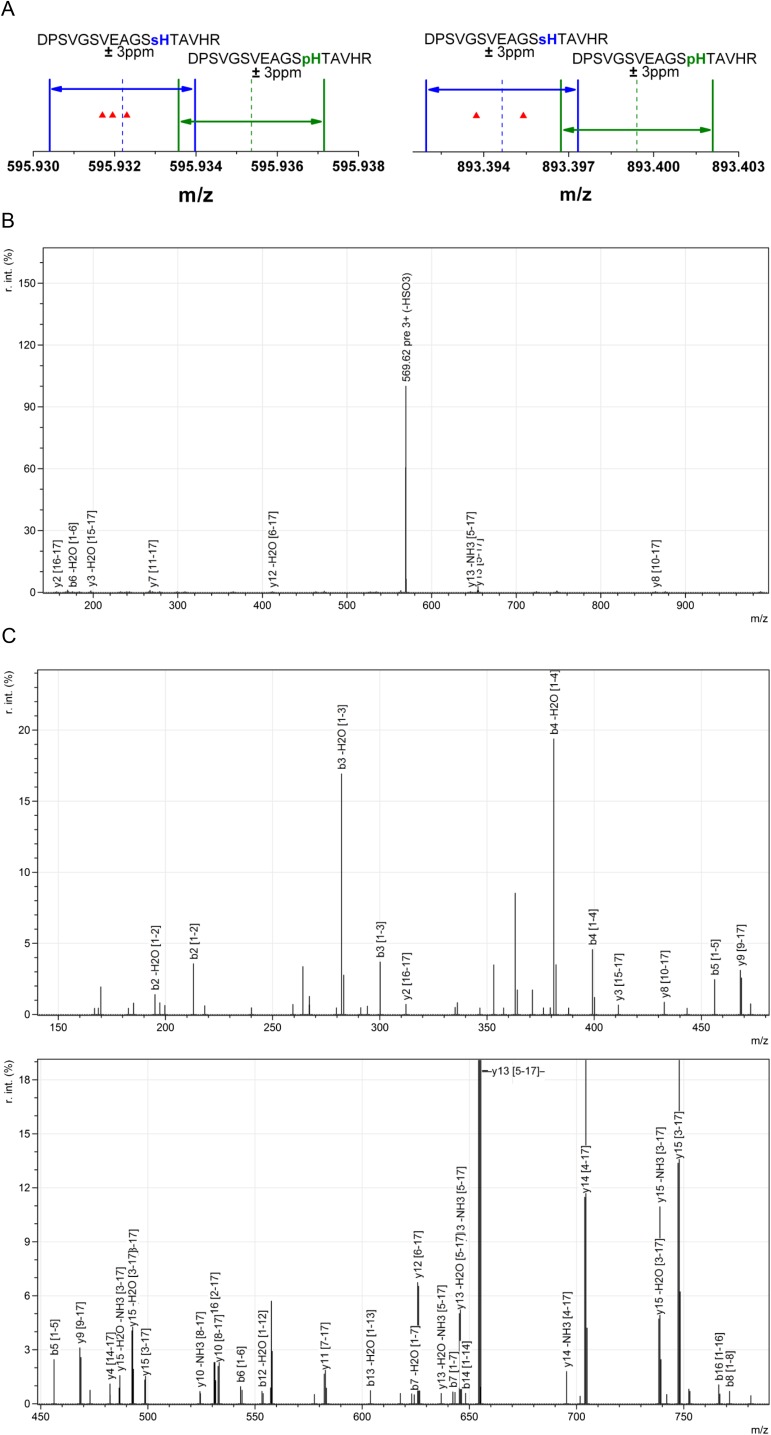
The crystal structures of mature AUS1 (Fig. 3 C and D), purified from the natural source, displayed an unexpected electron density at the CuB binding His252. This was particularly the case in crystal structures from enzyme samples that exhibited a high portion of sulfation or phosphorylation (24), making it impossible to place a histidine-coordinated copper atom into the density. However, the density matched very well to the density of a phosphohistidine or sulfohistidine. High-resolution mass spectrometric analysis revealed that the histidine residue was sulfated (Fig. S4). Due to the sulfation, the histidine loses its ability to participate in copper coordination. This leads to the loss of the CuB atom (Fig. 3D) and to the loss of enzymatic activity of the modified enzyme molecules.
Fig. S4.
HPLC–ESI–MS data of the peptide carrying the sulfohistidine. (A) MS1 data of the modified peptide possessing a neutral loss of 80 Da in MS2 spectra. The mass precision of the performed measurements is better than 3 ppm. The MS1 masses match to a sulfonation of the modified peptide, whereas a putative phosphorylation can be excluded. (B) Exemplary MS2 spectrum of 595.93 m/z (MS1) displaying a neutral loss of 80 Da. (C) MS3 spectrum of 569.6 m/z (MS2) originating from 595.93 m/z (MS1). (D) List of matched peptide fragments.
Reactivity of AUS1 and Insights into the Reaction Mechanism by MD Simulations.
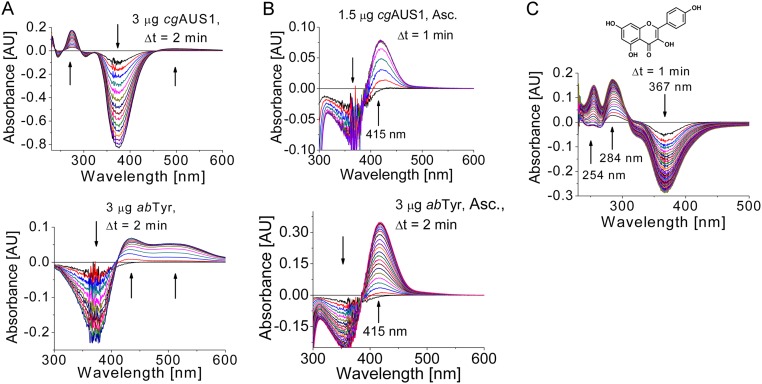
Recently, we reported that AUS1 lacks monophenolase activity (o-hydroxylation of monophenolic compounds) and displays exclusively diphenolase activity (oxidation of diphenolic compounds to the corresponding o-quinones) (15, 16). However, a modification of the activity assay by adding ascorbic acid as a reducing agent revealed that AUS1 in fact possesses hydroxylase activity toward isoliquiritigenin (Fig. S5 A and B) and that consecutive reactions were responsible for the previously reported false-negative statements. In the absence of a reducing agent, the highly reactive quinoid intermediates and products polymerize and are therefore undetectable by HPLC analysis. Furthermore, the reaction product sulfuretin has been reported to be a suicide substrate for AUS1 (15), for which the utilization of isoliquiritigenin strongly depended on the amount of enzymes used in the activity assay. Additionally, the difference spectra of isoliquiritigenin differed between AUS1 and abTYR (Fig. S5 A and B). However, in the presence of a reducing agent that traps the highly reactive quinoid intermediates, the difference spectra show identical absorption changes, which is evidence that the differences observed in the absence of a reducing agent are related not to the hydroxylation reaction but to the complex consecutive reactions. Furthermore, the enzyme showed hydroxylation activity toward kaempferol (Fig. S5C). In contrast to tyrosinases, AUS1 did not accept tyramine and tyrosine as substrates (10 µg active AUS1; 2 mM tyramine in 1 mL total volume; recordings up to 1 h; the experiments were performed with and without the addition of hydrogen peroxide and ascorbic acid).
Fig. S5.
Monophenolase activity of AUS1 and its comparison with mushroom tyrosinase (abTYR). The reaction medium (1 mL) contained 50 µM substrate (A and B, isoliquiritigenin; C, kaempferol) in 100 mM sodium phosphate pH 6.5. (A) Comparison of difference spectra of the reaction of isoliquiritigenin catalyzed by AUS1 (Top) and abTYR (Bottom) without a reducing agent. In contrast to the reaction catalyzed by mushroom tyrosinase, only an extremely weak and very broad absorption band at a higher wavelength than expected appears, although the disappearance of substrate is visible. The reason for the occurring different reaction products remains unclear. However, the comparison of the enzymes in the presence of a reducing agent (compare with B) indicates that the highly reactive quinoid intermediates and products are responsible for the observed differences. (B) Difference spectra of the hydroxylation reaction (indicated by an increasing and narrow absorption band at 415 nm) (15, 16) catalyzed by AUS1 (Top) and abTYR (Bottom) in the presence of 1 mM sodium ascorbate as the reducing agent to trap the highly reactive quinoid intermediates and products. (C) Difference spectra of the hydroxylation reaction of kaempferol catalyzed by AUS1 in the presence of 5 mM H2O2 (41).
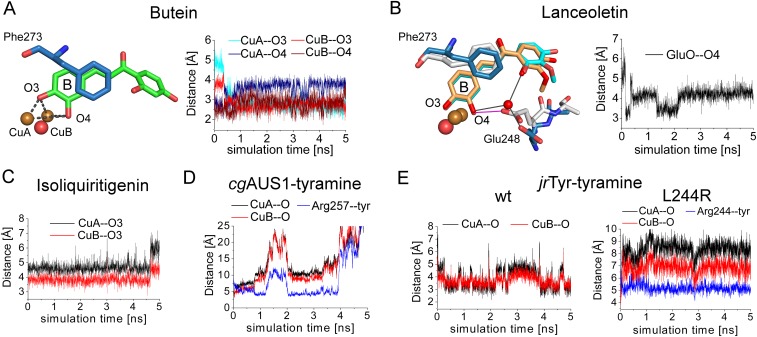
The chalcone isoliquiritigenin and the chalcone–aurone pairs of butein and lanceoletin (corresponding aurones, sulfuretin and leptosidin) naturally occur in Coreopsis species [summarized in Molitor et al. (15)]. To validate obtained molecular docking results of butein to AUS1, two different poses were subjected to MD simulations with the met-form of AUS1. During the simulation of one docking pose, the reactive B-ring of the chalcone remained in the active site, whereas the A-ring was very flexible and interacted with several different residues. The other docking pose resulted in a stable substrate enzyme complex, where the diphenolic substrate rapidly moved deeper into the cavity, which is formed by the extended loop region (V237ANG240) (compare with Fig. 1A), and remained there relatively stably during the simulation time. The substrate’s oxygen atoms are almost centered between the two copper atoms (Fig. 4B and Fig. S6A). To mimic the substrate position during the electron transfer, the simulation was repeated with restrained Cu–O–substrate bonds (26), resulting in a considerably reduced fluctuation of the unreactive A-ring due to its stronger interaction with protein residues of the cavity. Butein is stabilized by hydrogen bonds with the residues Asn117, Asn239, Arg257, and Gly240 (backbone); by hydrophobic interactions with the residues Asn239, Val244, Ala249, Thr253, His256, Phe273, and Ala276; and by residue Glu248 through a water bridge (Fig. 4B and the substrate binding residues are highlighted in Fig. S1).
Fig. 4.
Reactivity and in silico-obtained substrate–enzyme complexes of AUS1. (A) Reaction scheme of the hydroxylation of isoliquiritigenin in the presence of ascorbic acid to trap highly reactive quinoid intermediates and products. (B) MD simulation snapshot of AUS1 in complex with butein. The reactive oxygen atoms at the B-ring of butein (O3, O4) are almost symmetrically located between CuA and CuB. The enzyme–substrate interactions are visualized by magenta lines (hydrogen bonding), yellow dashes (hydrophobic interactions), cyan dashed lines (π–π stacking), red dashes (cation–π interactions), and gray lines (water bridges). (C) Superimposition of snapshots obtained from different MD simulations of AUS1 with lanceoletin as the substrate. Residue Glu248 coordinates an oxygen atom of the A-ring through the formation of a water bridge (blue sticks; water bridges, gray lines) or one reactive oxygen atom (O4, B-ring) directly (white sticks; hydrogen bonding, magenta lines). (D) MD simulation snapshot of AUS1 in complex with isoliquiritigenin. (E) Three snapshots [0 ps, green (1); 44 ps, cyan (2); and 344 ps, magenta (3)] of the MD simulation of AUS1 with tyramine. Hydrophobic interactions of tyramine with Arg257 are visualized by yellow dashes.
Fig. S6.
Distance plots of the performed MD simulations. (A) Distances of the reactive oxygen atoms of butein (O3 and O4) to both copper atoms of AUS1. (B) Distance of the carboxylic oxygen atom to the substrate’s copper-coordinating oxygen atom. (C) Distances of the reactive oxygen atom of isoliquiritigenin to the CuA and CuB atoms of AUS1. (D) Distances of the reactive oxygen atom of tyramine to the CuA and CuB atoms of AUS1. The centers of masses were used to determine the distance of tyramine to the side chain of Arg257 (blue line). (E) Distances of the reactive oxygen atom of tyramine to the CuA and CuB atoms of the wild type (Left) and the L244R mutant (Right) of jrTYR. The centers of masses were used to determine the distance of tyramine to the side chain of Arg244 (blue line).
The unrestraint but stable docking position of butein was then taken as a basis for the simulation of the diphenolic substrate lanceoletin and the monophenolic substrates isoliquiritigenin and tyramine. The interactions of lanceoletin with AUS1 were almost identical with those of butein, but with additional stabilization of the methoxyl group by hydrogen bonds with Gly240 and hydrophobic interactions arising from Val244, Ala249, and Glu248. Furthermore, the residues Gly271 and Asn271 form water bridges with the carbonyl oxygen of the chalcone. Interestingly, when the bridging water molecule is missing (water molecules in the simulation box were placed randomly), the highly conserved residue Glu248 is capable of directly coordinating one copper-coordinating hydroxyl group of the substrate (Fig. 4C and Fig. S6B). This residue has been proposed to participate in the water-mediated deprotonation of substrates (17, 20, 23). However, the MD simulations indicate that it might even be directly involved in substrate deprotonation (Fig. 4C).
For the simulation of the monophenols isoliquiritigenin and tyramine, the oxy-form of the enzyme was used. Even without a Cu–O–substrate distance restraint, isoliquiritigenin remained very stable within the cavity, with identical enzyme–substrate interactions as in the case of the diphenols butein and lanceoletin under restrained conditions. The distances of both copper atoms to the reactive hydroxyl group ranged mainly between 3.5 and 4.5 Å (Fig. 4D and Fig. S6C). In contrast, tyramine rapidly left the binuclear copper site of AUS1 (within 75 ps) due to attractive interactions with Arg257 (Fig. 4E and Fig. S6D), which is in accordance with the absence of monophenolase activity of AUS1 toward tyramine. The characteristic insertion (V237ANG240) of AUS1 might additionally interact with tyramine. To obtain more generalized information about the role of Arg257, simulations of tyramine in complex with jrTYR and with the corresponding L244R mutant of jrTYR were performed. Tyramine remained within the active site of jrTYR, whereas it left the active site of the L244R mutant rapidly (Fig. S6E). These results indicate a crucial role of the residues in this position (Fig. S1) for the acceptance or refusal of tyramie as a substrate for plant PPOs.
Exemplary video files of the simulation of AUS1 in complex with butein (Movies S1–S3) are available.
Discussion
Overall Structures of Mature AUS1 and of the First Latent Plant PPO.
The most relevant differences to the plant PPO homologs ibCO (17), vvCO (18), and jrTYR (23) are the characteristic four-amino-acid insertion of the group 2 PPOs and the short residual C-terminal peptide, connected to the main core by a disulfide bond. The insertion forms a binding pocket for the natural substrates (chalcones) of AUS1 (discussed below). The disulfide linkage was proposed to stabilize the C-terminal domain in the latent proenzyme (15). However, the crystal structures of mature AUS1 reveal that the main function of the interdomain linkage is the retention of the residual peptide, as it is a main building block of the homodimeric interface (Fig. 1B). The structure of the latent proenzyme exhibits two domains: the catalytically active domain and the C-terminal domain that is responsible for the latency of the proenzyme. The dominating structural motif of the C-terminal domain is a jelly roll barrel. Similar motifs are also present in the shielding domains of fungal (21, 27) (C-terminal), arthropod (28) (N-terminal), and insect (29) (N-terminal) PPOs. This taxa-spanning structural homology might indicate another, so far unknown, function of the shielding domains. Due to the interdomain disulfide linkage of the main core with the C-terminal domain, the proteolytic activation of the latent enzyme has to occur at three different sites (compare with Fig. S1) to result in the active enzyme (15). Two proteolytic cleavage sites are exposed on the surface (1 and 2 in Fig. 1C). In contrast, the third is covered by the C-terminal domain as well as by the main core but becomes accessible after the proteolytic cleavage at the first two cleavage sites. This suggests, and is supported by the purification of “semicleaved” latent forms from the natural source (15), that the proteolytic activation occurs stepwise and is targeted. The separation of the interdomain interactions into two distinct areas (Fig. 2) reflects specific roles of each area. The interactions close to the linker region will be responsible for the general structural integrity of the proenzyme. This is supported by the fact that the interdomain disulfide linkage is also part of this area. The strength of the interactions near the active site will affect the latency of the proenzyme. Furthermore, we propose that the in vitro activation (acidic activation and SDS activation) (6) as well as the allosteric activation (15) of latent plant PPOs are primarily caused by a structural rearrangement in this region. The transfer of the detailed knowledge of the interdomain interactions (Fig. S1) to other plant PPO homologs will enable the targeted modulation of the kinetic properties of recombinantly expressed proenzymes (30).
The active site of fungal (21, 27), bacterial (31), insect (29), and arthropod (28) pro-PPOs is blocked by a substrate mimicking the “placeholder” residue (tyrosine or phenylalanine, originating from the shielding domain) and located within the active site. However, due to the presence of the bulky gate residue above the active site (phenylalanine, originating from the main core), the shielding strategy in plant PPOs has to be different. The gate residue would have to swing away to make space for the placeholder residue (compare with Figs. 4 and 5). As this is energetically slightly unfavorable, other stabilizing interactions would be necessary for compensation. The crystal structures of the first latent plant pro-PPO reveal that plant PPOs overcome this issue by a smaller hydrophobic plug residue (Ile456 in AUS1; valine, leucine, and isoleucine in other plant PPOs) located above the gate residue and outside of the active site (Fig. 1D).
Fig. 5.
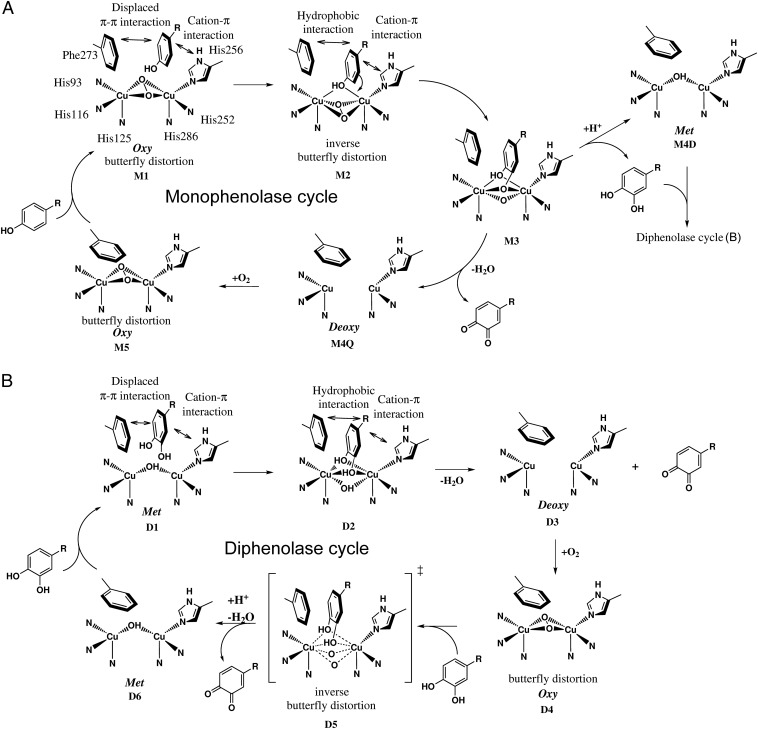
Proposed catalytic reaction mechanism of plant PPOs. (A) Monophenolase cycle: The monophenolic substrate is guided to the binuclear copper site [oxy-form, Cu(II)2O2, μ-η2:η2 peroxide complex] by displaced π–π interactions with the tilt out gate residue Phe273 and cation–π interaction with the CuB binding histidine His256 (M1). When the substrate binds to the copper atoms, hydrophobic interactions with the substrate’s para-substituent become important additionally. The reactive substrate oxygen atom binds equally to both copper ions (M2), and the peroxide ligand is transferred to an inverse butterfly distortion (ligand field molecular dynamical simulations, the Cu–O–O–Cu torsion angles of the peroxo ligand fluctuate by ±20°) (32). Nucleophilic attack of the Cu2O2 moiety by the substrate results either in an o-diphenolic product and the met-form [M4D, Cu(II)2OH] or in an o-quinone and the deoxy-form [M4Q, Cu(I)2]. During product release, the gate residue swings back to its preferred position. Finally, oxygen uptake closes the catalytic cycle (M5). (B) Diphenolase cycle: The principles of the diphenolase cycle are similar to the monophenolase cycle described in A. The reactive substrate oxygen atoms bind equally to both copper ions of the met-form (D1, D2), and the o-quinone is released after electron transfer to the binuclear copper site resulting in the deoxy-form (D3). Oxygen uptake results in the oxy-form (D4) and substrate binding results in an inverse butterfly distortion in the transition state (D5). The catalytic cycle is closed by the release of an o-quinone resulting in the met-form of the binuclear copper site.
Copper Binding Site of AUS1.
The reactive oxy-form of AUS1 exhibits a butterfly distortion (Fig. 3B) similar to the distortion found in the oxy-form of Streptomyces castaneoglobisporus tyrosinase (scTYR) (31). Because no indications of a distortion were found in ligand field molecular dynamical simulations of the oxy-form of tyrosinase (32), it has been suggested that the distortion in scTYR might be caused by a hydrogen bonding interaction of the placeholder residue (tyrosine) with the oxygen atoms. However, the oxy-form of AUS1 indicates that the butterfly distortion is a general characteristic of the oxy-form of PPOs, as AUS1 does not possess a placeholder residue that might cause this distortion.
Irreversible inactivation of tyrosinase from Agaricus bisporus (PPO3; abTYR) has been reported to occur by incubation with sulfite ions (33). The authors evidenced the sulfation of a CuB-coordinating histidine and speculated that the sulfation (accompanied by the loss of the CuB atom) presumably occurred at His263 (His256 in AUS1). The crystal structure of AUS1, however, reveals that the sulfuryl group is transferred to the Nε atom of His252 (His259 in PPO3; abTYR). The consistent discovery of this inactivation in both PPOs might indicate an infiltration of sulfite ions during the purification procedure of AUS1. However, there exist several matters that evoke doubts about this:
-
i)
The chemicals used to purify AUS1 are not only commonly used in protein purification, the identical chemicals were also used in the purification of tyrosinases from J. regia (34) and A. bisporus (35) and catechol oxidase from V. vinifera (24) in either similar or even identical methods. Mass spectrometric analyses [HPLC–ESI–MS (HPLC electrospray ionization mass spectrometry) of digested protein samples and ESI–Q-TOF–MS (electrospray ionization quadrupole time-of-flight mass spectrometry) of intact protein samples] were performed routinely, and none of the PPO samples exhibited any indication of such a modification (34, 35).
-
ii)
The latent form of AUS1 also did not display any modification.
-
iii)
The purification has been performed independently two times with approximately 1 y between them. The sulfohistidine occupancy varied from ∼0.45 (first batch, 6 kg petal tissue) to ∼0.9 (second batch, 9 kg petal tissue) and provides hints that the modification is either caused by sulfated compounds of petals of C. grandiflora or occurred in vivo.
Other sources of this modification might be a reaction of AUS1 with sulfated compounds within the petals of C. grandiflora during the isolation and purification procedure or an in vivo modification of AUS1 by a membrane-bound sulfotransferase. The latter possibility is very unlikely, as a controlled in vivo PPO inactivation has not been reported so far and we do not have any hints that this occurred in petals of C. grandiflora. In contrast, flavonoid sulfate esters are common in plants, especially in Asteraceae species (36). Four flavonol sulfotransferases were identified in Flaveria chloraefolia and Flaveria bidentis and catalyze sequential sulfation of quercetin to quercetin 3,7,3′,4′-tetrasulfate (37, 38). Interestingly, it has been crystallographically evidenced that a direct sulfuryl group transfer from p-nitrophenylsulfate to histidine, resulting in an identical sulfohistidine as observed in AUS1, represents an intermediate stage in the reaction mechanism of a bacterial sulfotransferase (39). This suggests that a direct sulfuryl transfer might also have taken place in the case of AUS1, as several flavonoids (e.g., quercetin and fisetin) (15) represent substrates for PPOs. So far, the preparation of samples of crude extracts (without the use of ammonium sulfate) from petals of C. grandiflora, suitable for analysis by HPLC–ESI–MS, failed due to the extremely high amount of pigments and polyphenols that interfere with performed SDS/PAGE analysis. However, the development of effective tyrosinase inhibitors has become increasingly important in the cosmetic, medicinal, and agricultural industries for application as antibrowning and depigmenting agents, and the inactivation of a PPO by a sulfonated substrate would have a high impact in this field of research.
Reactivity, Enzyme–Substrate Simulations, and Reaction Mechanism of Plant PPOs.
AUS1 is a catechol oxidase type of PPO, as no reactivity toward the common tyrosinase substrates tyramine and tyrosine was detectable. However, AUS1 showed monophenolase activity toward its natural substrate isoliquiritigenin (chalcone) and the flavonol kaempferol (Fig. S5). A similar reactivity has been reported for catechol oxidase from Aspergillus oryzae (aoCO4) (40). This fungal PPO showed activity toward aminophenol and guaiacol but not toward tyrosine. Historically, the definition that catechol oxidases lack the hydroxylase activity was primarily based on the acceptance of common tyrosinase substrates (e.g., p-cresol, tyrosine, tyramine) and not on the hydroxylation of their natural substrates (or at least a wider range of potential substrates). Due to the evidenced hydroxylase activity of AUS1 and aoCO4 (40), the question arises as to whether this definition is still valid. There is no doubt that catechol oxidases are incapable of hydroxylating tyramine or tyrosine, however AUS1 and aoCO4 reveal that they might hydroxylate various other monophenols (e.g., their potential natural substrates). Furthermore, the comparison of the crystal structures of plant catechol oxidases (PDB ID codes 2P3X and 1BT3) with the structure of a plant tyrosinase (PDB ID code 5CE9) did not reveal relevant structural differences except in the potential substrate binding residues (23). In this study, we identified a substrate binding residue (by performing MD simulations, discussed below) that is able to stabilize or destabilize the tyrosinase substrate tyramine within the active site of plant PPOs. This supports the presumption that no general structural limitation regarding the hydroxylase activity of plant PPOs exists, except the prerequisite of a substrate stabilization in a way that a hydroxylation reaction can occur. However, further investigations will be necessary to prove a putative general hydroxylase activity of PPOs. Therefore, we suggest using a substantially broader range of compounds (including the class of flavonoids—e.g., chalcones, flavonols, flavanols) in activity assays for plant PPOs. Furthermore, we suggest varying the conditions by adding reducing agents (to avoid polymerization or enzyme inactivation) or hydrogen peroxide. The latter is useful not only in omitting the lag phase of the hydroxylation reaction but also in detecting hydroxylase activity toward compounds that were previously reported to be tyrosinase inhibitors but in fact are hydroxylized and oxidized by tyrosinase (41) [e.g., arbutin and isoeugenol (42), phloridzin and phloretin (43)].
The very specific interactions observed during the MD simulations of AUS1 with the naturally occurring chalcones strongly support the hypothesis that the crystallized enzyme represents a specialized enzyme involved in aurone biosynthesis (15, 16). Furthermore, the proposed importance of the characteristic insertion of the group 2 PPOs in substrate binding (15) is confirmed (Fig. 4B and Movies S1–S3). However, although the substrates are strongly stabilized, it should be mentioned that the diphenols do not “have” to bind within this binding pocket. PPOs accept an enormous variety of diphenolic substrates (e.g., chlorogenic acid, 4-tert-butylcatechol, 3,5-di-tert-butylcatechol), suggesting that even unspecific binding to the copper center can result in product formation. Apart from the characteristic insertion of AUS1, the involvement of several residues at the copper binding sites (Fig. 4B and Fig. S1) in substrate coordination will be transferrable to other plant PPOs.
It has been shown that in a bacterial tyrosinase (from Bacillus megaterium; bmTYR) the substrates bind to the CuA atom, and therefore, it was concluded that the monophenols will perform a substrate rotation during the hydroxylation reaction (20). The authors proposed that the substrates will bind to the same copper atom in plant catechol oxidases. Based on these assumptions, it was further concluded that the hydroxylation reaction is impossible in plant catechol oxidases, as the bulky gate residue in combination with the rigidity of the CuA site (thioether bond) will hinder the substrate rotation. However, the proposed mechanism is incompatible with the hydroxylation activity of jrTYR (34, 44) as well as the observed hydroxylation activity of AUS1, as both possess a bulky gate residue and a thioether bond. Furthermore, no explanation for the lack of monophenolase activity of aoCO4 toward tyrosine as a substrate (40) was given.
Naturally, classical MD simulations are not suitable to obtain information about the geometry of the substrate–copper binding scenario, as they do not take into account quantum mechanical effects or the Jahn–Teller distortion. However, the strong hydrophobic interactions of the substrate with the gate residue (Phe273) and His256 direct the B-ring to the active site, where the substrate’s o-diphenolic oxygen atoms are almost equally positioned between both copper ions (Fig. 4 B and C and Fig. S6). The positions of the oxygen atoms are in a way comparable with the position of the butterfly distorted oxygen atoms of the oxy-form, demonstrating the general possibility of this coordination. Furthermore, the monophenol isoliquiritigenin is stabilized in a position that allows the hydroxylation in the ortho position to the copper-coordinating hydroxyl group (Fig. 4D). These observations in combination with the detected hydroxylation activity of AUS1 (Fig. S5) and jrTYR (23, 34, 45) led us to propose a novel mechanism for the hydroxylation and oxidation of phenolic substrates by plant PPOs (Fig. 5). In this mechanism, no substrate rotation is required, as the substrate is coordinated equally to both copper ions. This removes the contradiction of the previously proposed mechanisms with the experimentally proven monophenolase activities of plant PPOs. Furthermore, the MD simulations of tyramine in the active site of AUS1, jrTYR, and the L244R mutant of jrTYR strongly suggest that the residue at the position of Arg257 (compare with Fig. S1) is one of the key residues for the acceptance of tyramine as a substrate for plant PPOs. It should be noted that the interactions will be different in fungal and bacterial PPOs, as they do not possess a bulky gate residue and the substrates are bound to the CuA atom (20). However, during MD simulations the classical tyrosinase substrate tyramine is destabilized in the active center of plant PPOs by an arginine in this position (AUS1, L244R mutant of jrTYR), whereas it is stabilized by a leucine in this position (jrTYR). It has been reported recently that vvCO, possessing a lysine in this position, showed weak activity toward tyrosine and tyramine, and vvCO was therefore classified as a tyrosinase (46). However, a comparison of the kinetic data of vvCO and jrTYR indicates that tyramine will presumably not be the natural substrate of vvCO [Km(vvCO) = 7.7 mM (46); Km(jrTYR) = 0.274 mM (23)]. This example demonstrates that in the long term the classification and the nomenclature of plant PPOs will have to change to avoid misunderstandings.
As a consequence of the evidenced hydroxylation activity of AUS1 toward its natural substrate in combination with the consistent proposed novel reaction mechanism, another complexion is put on the function of plant PPOs. So far, the role of plant PPOs has primarily been assigned to enzymatic browning reactions, with only few exceptions [e.g., aureusidin synthase (12, 13) and aurone synthase (15, 16)]. However, we demonstrated that the monophenolase activity of plant PPOs seems to depend only on whether the monophenol can be stabilized within the active site or not. There is no doubt that the hydroxylase activity represents the primary functionality of tyrosinases (e.g., in the hydroxylation of tyrosine in the melanin biosynthetic pathway). If catechol oxidases also possess hydroxylase activity, it seems reasonable that the above statement is also true for this type of enzyme. As a consequence, it could be assumed that the catechol oxidases are involved in anabolic pathways by hydroxylating their—so far unknown—natural substrates. Hence, it can be concluded that the identification of the few plant PPOs involved in the secondary metabolism does indeed represent just the “tip of the iceberg” (7).
There is no doubt that there will be a long interdisciplinary way to go (including metabolomics and transcriptomic and proteomic analyses) until the role of plant PPOs is clarified, particularly with regard to their complex diversity. The substrate binding residues (compare Fig. 4 and Fig. S1) around the CuA and CuB binding sites differ considerably within the family of PPOs (2, 3), indicating the occurrence of individual natural substrates. Therefore, the upcoming challenges will be to cluster the PPOs according to their substrate binding residues, to identify the potential natural substrates (or substrate scaffolds), and to validate the involvement of the corresponding PPOs in the plants’ secondary metabolism.
Methods
Protein Purification, Crystallization, Data Collection, Structure Determination, and Refinement.
The protein purification of active and latent AUS1 from petals of C. grandiflora is described in Molitor et al. (15), and the purification of recombinantly expressed AUS1 is described in Kaintz et al. (16). The crystallization of the obtained enzyme samples and the data collection, using synchrotron radiation, of the obtained crystals have been described in Molitor et al. (24). The corresponding data collections of the presented crystal structures were performed using synchrotron radiation at 100 K (PDB ID code 4Z0Y—P14, PETRA III, EMBL, DESY; λ = 1.23953 Å; PDB ID code 4Z0Z—I04-1, DIAMOND; λ = 0.9173 Å; PDB 4Z11—ID23-1, ESRF; λ = 0.972499 Å; PDB 4Z12—P11, PETRA III, DESY; λ = 1.0247 Å; PDB 4Z13—I04-1, DIAMOND; λ = 0.9173 Å). Automated refinements of all models (obtained as described below) were performed by phenix.refine from the PHENIX suite (47) followed by manual structure completion and correction using COOT (48).
Initial phases for the active AUS1 form (space group P212121) were obtained by molecular replacement using the BALBES webserver (49), which used the crystal structure of catechol oxidase from I. batatas (PDB ID code 1BT3, sequence identity ∼47%) as a search model. The refined structure of AUS1 was then used as a search model for structure determination of the remaining obtained datasets by molecular replacement using PHASER (MR) from the PHENIX program suite. The structure of the latent AUS1 form was obtained by automated model building using AUTOBUILD from the PHENIX suite, after placing the refined catalytically active main core with PHASER (MR) in the asymmetric unit. Restraints for the polyoxometalate hexatungstotellurate (VI), used as a cocrystallization agent for recombinantly expressed AUS1 (24, 50), were obtained with phenix.reel and phenix.elbow as previously reported (27, 51, 52) using the crystal structure of hexatungstotellurate (VI) (53). The quality of the models was validated by MOLPROBITY (54). All models were refined to excellent stereochemistry without any Ramachandran outliers, and the respective refinement statistics are presented in Tables S1 and S2.
The models were deposited in the PDB (www.rcsb.org) as ID codes 4Z0Y, 4Z0Z, 4Z11, 4Z12, and 4Z13.
Protein Digestion and HPLC–ESI–MS Measurements of Inactive AUS1.
The experiments were performed by the company Proteome Factory. The protein sample was denatured by incubation in 8 M urea for 30 min at room temperature. For reduction, a final concentration of 5 mM Tris(2-carboxyethyl)phosphine was added, followed by incubation for 20 min at room temperature. Then iodoacetamide was added to a final concentration of 10 mM. After incubation for another 20 min in the dark, the samples were diluted to 0.8 M urea and subsequently digested by two endoproteases (trypsin and AspN) with an enzyme:protein ratio of 1:50 (wt/wt) according to the Proteome Factory’s protein digestion standard operating procedures. The acidified peptide populations were applied to nanoLC–ESI–MS (LTQ-FT, Thermo Finnigan) analyses using a 60-min nanoLC gradient (Agilent 1100 nanoLC system) with solvent A [0.1% (vol/vol) formic acid in 5% (vol/vol) acetonitrile] and solvent B [0.1% (vol/vol) formic acid in 99.9% acetonitrile].
The mass accuracy was better than 3 ppm for MS data and ±0.02 Da for MS2 data. MS2 data analyses were done using MASCOT (Matrix Science), whereas the MS data were analyzed using Qualbrowser (Thermo Finnigan). The induced modification carbamidomethylation (C) was allowed during MS2 data searches, along with possible modifications such as oxidation (M), deamidation (NQ), and phosphorylation/sulfation (H). The precursor showing the predicted modification and a strong neutral loss signal was subject to dedicated MS3 experiments.
Molecular Docking and MD Simulations.
The structures of active AUS1 (PDB ID code 4Z0Y) and jrTYR (PDB ID code 5CE9) were prepared for molecular docking studies and MD simulations by the addition of missing side chain atoms using COOT and the removal of the residual C-terminal peptide of AUS1. Molecular docking of butein was performed with AutoDock VINA (55) by the use of the deoxy-form of active AUS1 and setting the gate residue PHE273 as a flexible residue. The search exhaustiveness was set to the maximal value of 2,000 (default 8). MD simulations of obtained enzyme–substrate complexes were performed with GROMACS (56) version 4.6.7 package applying the AMBER99SB force field (57). The ligand topologies were generated using the SWISSPARAM webserver (58). RESP (restrained electrostatic potential) charges were obtained from the RED webserver (59) by applying the HF/6–31G* level of theory (Gaussian09) to calculate the electrostatic potential. For the simulation of o-diphenols in complex with AUS1, the met-form was used and new residues were defined for the copper atoms, the copper-coordinating histidines, and the thioether forming cysteine, according to Bochot et al. (60). For the simulation of monophenols in complex with AUS1, the oxy-form was used and parameters of the dinuclear copper site were adopted from Deeth and Diedrich (32). The protein–ligand complex was solvated in a dodecahedral box (10 Å edge distance) of TIP3PBOX water molecules. The charge of the system was neutralized by replacing water molecules by the respective number of sodium ions. Energy minimization was performed with the steepest step algorithm with a maximum of 3,000 steps and an energy tolerance of 1,000 kJ⋅mol−1⋅nm−1. After convergence, the system was equilibrated in two phases: NVT ensemble (isothermal-isochoric: constant number of particles, volume, and temperature) and NPT ensemble (isothermal-isobaric: constant number of particles, pressure, and temperature), each for 100 ps at 300 K. MD simulations were carried out by applying periodic boundary conditions. The LINCS (Linear Constraint Solver) algorithm was used to constrain all bond lengths, and the particle-mesh Ewald method was used to calculate long-range electrostatic interactions. The Berendsen coupling algorithm was used for temperature, and the Parrinello–Rahman coupling algorithm was used for pressure control. After the two equilibration runs, a 5-ns simulation was carried out with, unless otherwise stated, no position restraints.
Structural Analysis and Graphical Presentation.
The quaternary structure of the obtained crystal structures was analyzed with PISA (Proteins, Interfaces, Structures and Assemblies) (25). Protein interfaces and ligand interactions were analyzed and visualized with LigPlot+ (61) and PLIP (Protein–Ligand Interaction Profiler) (62). Molecular graphic images and movies were generated with PyMOL (www.pymol.org/) and VMD (Visual Molecular Dynamics) (63), respectively. The secondary structure features were assigned using DSSP (Dictionary of Secondary Structures of Proteins) (64).
Supplementary Material
Acknowledgments
We thank Kristina Djinovic-Carugo and Georg Mlynek [Max F. Perutz Laboratories (MFPL), Vienna Biocenter] for their kind support during the early stages of this research. We thank beamline scientists Elspeth Gordon (ESRF ID23-1, mx1450), Anja Burkhardt (DESY P11, I-20120633 EC), and Alice Douangamath (Diamond Light Source I04-1, MX8476) for their generous support during the allocated beam times. We give special thanks to Gleb Bourenkov and Victor S. Lamzin (DESY/EMBL) for the opportunity of data collection at beamline P14 during “European School for Macromolecular Crystallography (ESMAX) 2012.” For cultivating C. grandiflora and for taking care of the plant fields, we thank the Horticultural Department of Molecular Systems Biology, UZA1-Glashaus1, University of Vienna—in particular Thomas Joch and Andreas Schröfl—and the gardeners of the experimental garden Augarten—especially Miroslav Crep and Erich Wagner. We also thank Aleksandar Bijelic, Matthias Pretzler, and Ioannis Kampatsikas for valuable discussions concerning this work. The research was funded by the Austrian Science Fund (FWF): P25217 and the Deutsche Forschungsgemeinschaft (Ro 1084/8-1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4Z0Y, 4Z0Z, and 4Z11–4Z13).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523575113/-/DCSupplemental.
References
- 1.Mayer AM. Polyphenol oxidases in plants and fungi: Going places? A review. Phytochemistry. 2006;67(21):2318–2331. doi: 10.1016/j.phytochem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Tran LT, Taylor JS, Constabel CP. The polyphenol oxidase gene family in land plants: Lineage-specific duplication and expansion. BMC Genomics. 2012;13:395. doi: 10.1186/1471-2164-13-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dirks-Hofmeister ME, Singh R, Leufken CM, Inlow JK, Moerschbacher BM. Structural diversity in the dandelion (Taraxacum officinale) polyphenol oxidase family results in different responses to model substrates. PLoS One. 2014;9(6):e99759. doi: 10.1371/journal.pone.0099759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King RS, Flurkey WH. Effects of limited proteolysis on broad bean polyphenoloxidase. J Sci Food Agric. 1987;41(3):231–240. [Google Scholar]
- 5.Marusek CM, Trobaugh NM, Flurkey WH, Inlow JK. Comparative analysis of polyphenol oxidase from plant and fungal species. J Inorg Biochem. 2006;100(1):108–123. doi: 10.1016/j.jinorgbio.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Yoruk R, Marshall MR. Physicochemical properties and function of plant polyphenol oxidase: A review1. J Food Biochem. 2003;27(5):361–422. [Google Scholar]
- 7.Sullivan ML. Beyond brown: Polyphenol oxidases as enzymes of plant specialized metabolism. Front Plant Sci. 2014;5:783. doi: 10.3389/fpls.2014.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahler D, et al. Polyphenoloxidase silencing affects latex coagulation in Taraxacum species. Plant Physiol. 2009;151(1):334–346. doi: 10.1104/pp.109.138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richter C, Dirks ME, Gronover CS, Prüfer D, Moerschbacher BM. Silencing and heterologous expression of ppo-2 indicate a specific function of a single polyphenol oxidase isoform in resistance of dandelion (Taraxacum officinale) against Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact. 2012;25(2):200–210. doi: 10.1094/MPMI-04-11-0082. [DOI] [PubMed] [Google Scholar]
- 10.Araji S, et al. Novel roles for the polyphenol oxidase enzyme in secondary metabolism and the regulation of cell death in walnut. Plant Physiol. 2014;164(3):1191–1203. doi: 10.1104/pp.113.228593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho MH, et al. (+)-Larreatricin hydroxylase, an enantio-specific polyphenol oxidase from the creosote bush (Larrea tridentata) Proc Natl Acad Sci USA. 2003;100(19):10641–10646. doi: 10.1073/pnas.1934562100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama T, et al. Aureusidin synthase: A polyphenol oxidase homolog responsible for flower coloration. Science. 2000;290(5494):1163–1166. doi: 10.1126/science.290.5494.1163. [DOI] [PubMed] [Google Scholar]
- 13.Ono E, et al. Localization of a flavonoid biosynthetic polyphenol oxidase in vacuoles. Plant J. 2006;45(2):133–143. doi: 10.1111/j.1365-313X.2005.02625.x. [DOI] [PubMed] [Google Scholar]
- 14.Miosic S, et al. 4-Deoxyaurone formation in Bidens ferulifolia (Jacq.) DC. PLoS One. 2013;8(5):e61766. doi: 10.1371/journal.pone.0061766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molitor C, et al. Latent and active aurone synthase from petals of C. grandiflora: A polyphenol oxidase with unique characteristics. Planta. 2015;242(3):519–537. doi: 10.1007/s00425-015-2261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaintz C, et al. Cloning and functional expression in E. coli of a polyphenol oxidase transcript from Coreopsis grandiflora involved in aurone formation. FEBS Lett. 2014;588(18):3417–3426. doi: 10.1016/j.febslet.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klabunde T, Eicken C, Sacchettini JC, Krebs B. Crystal structure of a plant catechol oxidase containing a dicopper center. Nat Struct Biol. 1998;5(12):1084–1090. doi: 10.1038/4193. [DOI] [PubMed] [Google Scholar]
- 18.Virador VM, et al. Cloning, sequencing, purification, and crystal structure of Grenache (Vitis vinifera) polyphenol oxidase. J Agric Food Chem. 2010;58(2):1189–1201. doi: 10.1021/jf902939q. [DOI] [PubMed] [Google Scholar]
- 19.Sendovski M, Kanteev M, Ben-Yosef VS, Adir N, Fishman A. First structures of an active bacterial tyrosinase reveal copper plasticity. J Mol Biol. 2011;405(1):227–237. doi: 10.1016/j.jmb.2010.10.048. [DOI] [PubMed] [Google Scholar]
- 20.Goldfeder M, Kanteev M, Isaschar-Ovdat S, Adir N, Fishman A. Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins. Nat Commun. 2014;5:4505. doi: 10.1038/ncomms5505. [DOI] [PubMed] [Google Scholar]
- 21.Fujieda N, et al. Crystal structures of copper-depleted and copper-bound fungal pro-tyrosinase: Insights into endogenous cysteine-dependent copper incorporation. J Biol Chem. 2013;288(30):22128–22140. doi: 10.1074/jbc.M113.477612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ismaya WT, et al. Crystal structure of Agaricus bisporus mushroom tyrosinase: Identity of the tetramer subunits and interaction with tropolone. Biochemistry. 2011;50(24):5477–5486. doi: 10.1021/bi200395t. [DOI] [PubMed] [Google Scholar]
- 23.Bijelic A, Pretzler M, Molitor C, Zekiri F, Rompel A. The structure of a plant tyrosinase from walnut leaves reveals the importance of “substrate-guiding residues” for enzymatic specificity. Angew Chem Int Ed Engl. 2015;54(49):14677–14680. doi: 10.1002/anie.201506994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molitor C, Mauracher SG, Rompel A. Crystallization and preliminary crystallographic analysis of latent, active and recombinantly expressed aurone synthase, a polyphenol oxidase, from Coreopsis grandiflora. Acta Crystallogr F Struct Biol Commun. 2015;71(Pt 6):746–751. doi: 10.1107/S2053230X15007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Wise O, Coskuner O. New force field parameters for metalloproteins I: Divalent copper ion centers including three histidine residues and an oxygen-ligated amino acid residue. J Comput Chem. 2014;35(17):1278–1289. doi: 10.1002/jcc.23622. [DOI] [PubMed] [Google Scholar]
- 27.Mauracher SG, Molitor C, Al-Oweini R, Kortz U, Rompel A. Latent and active abPPO4 mushroom tyrosinase cocrystallized with hexatungstotellurate(VI) in a single crystal. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 9):2301–2315. doi: 10.1107/S1399004714013777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masuda T, Momoji K, Hirata T, Mikami B. The crystal structure of a crustacean prophenoloxidase provides a clue to understanding the functionality of the type 3 copper proteins. FEBS J. 2014;281(11):2659–2673. doi: 10.1111/febs.12812. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Wang Y, Jiang H, Deng J. Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes. Proc Natl Acad Sci USA. 2009;106(40):17002–17006. doi: 10.1073/pnas.0906095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leufken CM, Moerschbacher BM, Dirks-Hofmeister ME. Dandelion PPO-1/PPO-2 domain-swaps: The C-terminal domain modulates the pH optimum and the linker affects SDS-mediated activation and stability. Biochim Biophys Acta. 2015;1854(2):178–186. doi: 10.1016/j.bbapap.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Matoba Y, Kumagai T, Yamamoto A, Yoshitsu H, Sugiyama M. Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J Biol Chem. 2006;281(13):8981–8990. doi: 10.1074/jbc.M509785200. [DOI] [PubMed] [Google Scholar]
- 32.Deeth RJ, Diedrich C. Structural and mechanistic insights into the oxy form of tyrosinase from molecular dynamics simulations. J Biol Inorg Chem. 2010;15(2):117–129. doi: 10.1007/s00775-009-0577-6. [DOI] [PubMed] [Google Scholar]
- 33.Kuijpers TF, Gruppen H, Sforza S, van Berkel WJ, Vincken JP. The antibrowning agent sulfite inactivates Agaricus bisporus tyrosinase through covalent modification of the copper-B site. FEBS J. 2013;280(23):6184–6195. doi: 10.1111/febs.12539. [DOI] [PubMed] [Google Scholar]
- 34.Zekiri F, et al. Purification and characterization of tyrosinase from walnut leaves (Juglans regia) Phytochemistry. 2014;101:5–15. doi: 10.1016/j.phytochem.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mauracher SG, et al. High level protein-purification allows the unambiguous polypeptide determination of latent isoform PPO4 of mushroom tyrosinase. Phytochemistry. 2014;99:14–25. doi: 10.1016/j.phytochem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gidda SK, Varin L. Biochemical and molecular characterization of flavonoid 7-sulfotransferase from Arabidopsis thaliana. Plant Physiol Biochem. 2006;44(11-12):628–636. doi: 10.1016/j.plaphy.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Varin L, Ibrahim RK. Partial purification and characterization of three flavonol-specific sulfotransferases from Flaveria chloraefolia. Plant Physiol. 1989;90(3):977–981. doi: 10.1104/pp.90.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varin L, Ibrahim RK. Partial purification and some properties of flavonol 7-sulfotransferase from Flaveria bidentis. Plant Physiol. 1991;95(4):1254–1258. doi: 10.1104/pp.95.4.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malojcić G, et al. A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2008;105(49):19217–19222. doi: 10.1073/pnas.0806997105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hakulinen N, Gasparetti C, Kaljunen H, Kruus K, Rouvinen J. The crystal structure of an extracellular catechol oxidase from the ascomycete fungus Aspergillus oryzae. J Biol Inorg Chem. 2013;18(8):917–929. doi: 10.1007/s00775-013-1038-9. [DOI] [PubMed] [Google Scholar]
- 41.García-Molina Mo, et al. Hydrogen peroxide helps in the identification of monophenols as possible substrates of tyrosinase. Biosci Biotechnol Biochem. 2013;77(12):2383–2388. doi: 10.1271/bbb.130500. [DOI] [PubMed] [Google Scholar]
- 42.Ortiz-Ruiz CV, Garcia-Molina MdelM, Serrano JT, Tomas-Martinez V, Garcia-Canovas F. Discrimination between alternative substrates and inhibitors of tyrosinase. J Agric Food Chem. 2015;63(8):2162–2171. doi: 10.1021/jf5051816. [DOI] [PubMed] [Google Scholar]
- 43.Ortiz-Ruiz CV, et al. Identification of p-hydroxybenzyl alcohol, tyrosol, phloretin and its derivate phloridzin as tyrosinase substrates. Bioorg Med Chem. 2015;23(13):3738–3746. doi: 10.1016/j.bmc.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 44.Escobar MA, Shilling A, Higgins P, Uratsu SL, Dandekar AM. Characterization of polyphenol oxidase from walnut. J Am Soc Hortic Sci. 2008;133(6):852–858. [Google Scholar]
- 45.Zekiri F, Bijelic A, Molitor C, Rompel A. Crystallization and preliminary X-ray crystallographic analysis of polyphenol oxidase from Juglans regia (jrPPO1) Acta Crystallogr F Struct Biol Commun. 2014;70(Pt 6):832–834. doi: 10.1107/S2053230X1400884X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fronk P, et al. Polyphenoloxidase from Riesling and Dornfelder wine grapes (Vitis vinifera) is a tyrosinase. Food Chem. 2015;183:49–57. doi: 10.1016/j.foodchem.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Long F, Vagin AA, Young P, Murshudov GN. BALBES: A molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bijelic A, Rompel A. The use of polyoxometalates in protein crystallography—An attempt to widen a well-known bottleneck. Coord Chem Rev. 2015;299(0):22–38. doi: 10.1016/j.ccr.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bijelic A, et al. Hen egg-white lysozyme crystallisation: Protein stacking and structure stability enhanced by a Tellurium(VI)-centred polyoxotungstate. ChemBioChem. 2015;16(2):233–241. doi: 10.1002/cbic.201402597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mauracher SG, Molitor C, Al-Oweini R, Kortz U, Rompel A. Crystallization and preliminary X-ray crystallographic analysis of latent isoform PPO4 mushroom (Agaricus bisporus) tyrosinase. Acta Crystallogr F Struct Biol Commun. 2014;70(Pt 2):263–266. doi: 10.1107/S2053230X14000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt KJ, Schrobilgen GJ, Sawyer JF. Hexasodium hexatungstotellurate(VI) 22-hydrate. Acta Crystallogr C. 1986;42(9):1115–1118. [Google Scholar]
- 54.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pronk S, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorin EJ, Pande VS. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys J. 2005;88(4):2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zoete V, Cuendet MA, Grosdidier A, Michielin O. SwissParam: A fast force field generation tool for small organic molecules. J Comput Chem. 2011;32(11):2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 59.Vanquelef E, et al. R.E.D. Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011;39(Web Server issue):W511–W517. doi: 10.1093/nar/gkr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bochot C, et al. Unsymmetrical binding modes of the HOPNO inhibitor of tyrosinase: From model complexes to the enzyme. Chemistry. 2013;19(11):3655–3664. doi: 10.1002/chem.201202643. [DOI] [PubMed] [Google Scholar]
- 61.Laskowski RA, Swindells MB. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 62.Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43(W1):W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 64.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.