Significance
Mechanisms of competition are not well-studied in the mammalian gut microbiota, especially among abundant species of this ecosystem. Theoretical models predict that antagonistic mechanisms should profoundly affect member fitness, yet identification and experimental analyses of such factors are few. Here we show that the abundant gut species Bacteroides fragilis produce T6SSs that deploy previously undescribed toxins able to antagonize numerous species of human gut Bacteroidales. We show that these systems are synthesized in the mammalian gut and allow a producing strain to outcompete a sensitive isogenic strain. Our data support that GA3 T6SSs likely allow the producing B. fragilis strain to create a protected niche in the human colon.
Keywords: Bacteroides, T6SS, gut microbiota, bacterial competition, type VI secretion
Abstract
Type VI secretion systems (T6SSs) are multiprotein complexes best studied in Gram-negative pathogens where they have been shown to inhibit or kill prokaryotic or eukaryotic cells and are often important for virulence. We recently showed that T6SS loci are also widespread in symbiotic human gut bacteria of the order Bacteroidales, and that these T6SS loci segregate into three distinct genetic architectures (GA). GA1 and GA2 loci are present on conserved integrative conjugative elements (ICE) and are transferred and shared among diverse human gut Bacteroidales species. GA3 loci are not contained on conserved ICE and are confined to Bacteroides fragilis. Unlike GA1 and GA2 T6SS loci, most GA3 loci do not encode identifiable effector and immunity proteins. Here, we studied GA3 T6SSs and show that they antagonize most human gut Bacteroidales strains analyzed, except for B. fragilis strains with the same T6SS locus. A combination of mutation analyses, trans-protection analyses, and in vitro competition assays, allowed us to identify novel effector and immunity proteins of GA3 loci. These proteins are not orthologous to known proteins, do not contain identified motifs, and most have numerous predicted transmembrane domains. Because the genes encoding effector and immunity proteins are contained in two variable regions of GA3 loci, GA3 T6SSs of the species B. fragilis are likely the source of numerous novel effector and immunity proteins. Importantly, we show that the GA3 T6SS of strain 638R is functional in the mammalian gut and provides a competitive advantage to this organism.
Bacteria that live in communities have numerous mechanisms to compete with other strains and species. The ability to acquire nutrients is a major factor dictating the success of a species in a community. In addition, the production of secreted factors, such as bacteriocins, that competitively interfere or antagonize other strains/species, also contributes to a member’s fitness in a community. In the microbe-dense human gut ecosystem, such factors and mechanisms of antagonism by predominant members are just beginning to be described, as are models predicting the relevance of these competitive interactions to the microbial community (1). Bacteroidales is the most abundant order of bacteria in the human colonic microbiota, and also the most temporally stable (2). The fact that numerous gut Bacteroidales species stably cocolonize the human gut at high density raises the question of how these related species and strains interact with each other to promote or limit each other’s growth. We previously showed that coresident Bacteroidales strains intimately interact with each other and exchange large amounts of DNA (3) and also cooperate in the utilization of dietary polysaccharides (4). To date, two types of antagonistic factors/systems have been shown to be produced by human gut Bacteroidales species: secreted antimicrobial proteins (5) and T6SSs (3, 6, 7). However, neither of these antagonistic processes has been analyzed to determine if they provide a competitive advantage in the mammalian intestine.
Type VI secretion systems (T6SSs) are contact-dependent antagonistic systems used by some Gram-negative bacteria to intoxicate other bacteria or eukaryotic cells. The T6 apparatus is a multiprotein, cell envelope spanning complex comprised of core Tss proteins. A key component of the machinery is a needle-like structure, similar to the T4 contractile bacteriophage tail, which is assembled in the cytoplasm where it is loaded with toxic effectors (8–10). Contraction of the sheath surrounding the needle apparatus drives expulsion of the needle from the cell, delivering the needle and associated effectors either into the supernatant of in vitro grown bacteria, or across the membrane of prey cells. Identified T6SS effectors include cell wall degrading enzymes (11), proteins that affect cell membranes such as phospholipases (12) and pore-forming toxins (13, 14), proteins that degrade NAD(P)+ (15), and nucleases (16). The effector protein is produced with a cognate immunity protein, typically encoded by the adjacent downstream gene (17), which protects the producing cell from the toxicity of the effector. Although both eukaryotic and bacterial cells are targeted by T6SS effectors (18), most described T6SSs target Gram-negative bacteria.
We previously performed a comprehensive analysis of all sequenced human gut Bacteroidales stains and found that more than half contain T6SS loci (7). These T6SSs are similar to the well-described T6SSs of Proteobacteria in that remote orthologs of many Proteobacterial Tss proteins are encoded by Bacteroidales T6SS regions, with the exception of proteins that likely comprise the transmembrane complex, which are distinct. The T6SS loci of human gut Bacteroidales species segregate into three distinct genetic architectures (GA), designated GA1, GA2, and GA3, each with highly identical segments within a GA comprising the core tss genes (7). GA1 and GA2 T6SS loci are present on large ∼80- to 120-kb integrative conjugative elements (ICE) that are extremely similar at the DNA level within a GA. Due to the ability of these T6SS regions to be transferred between strains via ICE, GA1 and GA2 T6SS loci are present in diverse human gut Bacteroidales species. GA3 T6SS loci are confined to Bacteroides fragilis and are not contained on conserved ICE (7).
Although T6SS loci of a particular GA are highly identical to each other, each GA has internal regions of variability where the genes differ between strains (7). The variable regions of GA1 and GA2 T6SS loci contain genes encoding the identifiable toxic effector and cognate immunity proteins found in these regions. Unlike the GA1 and GA2 T6SS loci, there are no identifiable genes encoding toxin or immunity proteins in the two variable regions or other areas of GA3 T6SS loci. The present study was designed to answer three fundamental questions regarding GA3 T6SS loci: (i) Because no known effectors/immunity proteins are encoded by these regions, are they involved in bacterial antagonism? And if so, what prey cells do they target? (ii) Do the variable regions contain genes encoding effector and immunity proteins? and (iii) If GA3 T6SSs mediate bacterial antagonism, do they provide a competitive advantage in the mammalian gut?
Results
Deletion of T6SS Genes and Analysis of Antagonism.
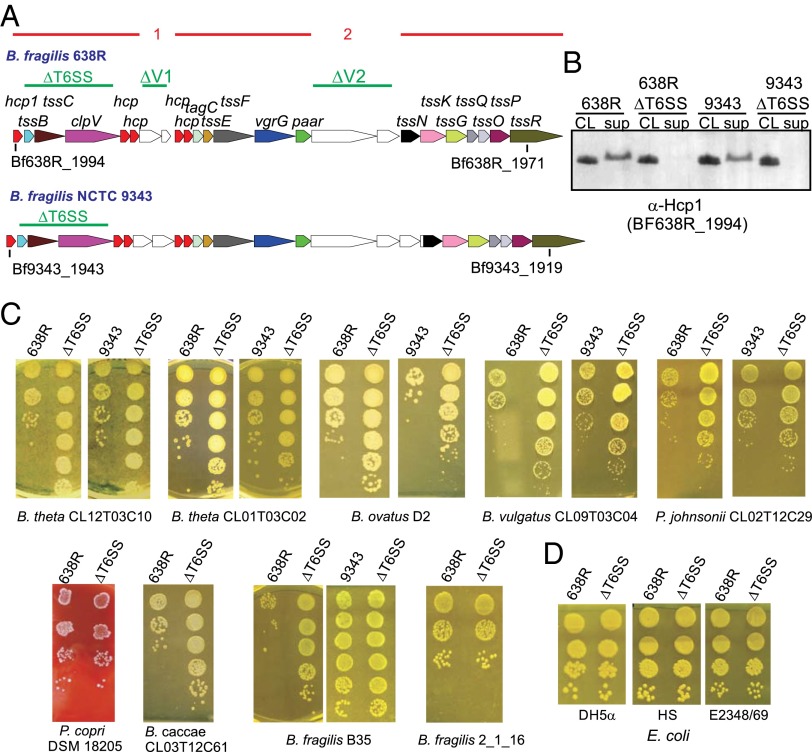
The two most studied B. fragilis strains, NCTC 9343 and 638R, each have a single T6SS locus, both of which are GA3, and are distinct from each other in both variable regions (Fig. 1A). Therefore, we studied the ability of the T6SSs of these two strains to antagonize other Bacteroidales strains/species. To abrogate firing of the T6SS apparatus, we made nonpolar unmarked deletion mutants removing three adjacent genes from these two strains (tssB-clpV; shown in Fig. 1A as ∆T6SS). A hallmark of functional T6 firing is the release of the major needle protein, Hcp, into the supernatant. Each GA3 T6SS locus typically encodes five distinct Hcp proteins with very little similarity to each other (<47% similarity) (7). We previously found that the Hcp encoded by the first gene of GA3 loci, labeled hcp1, is the closest ortholog to Hcp proteins encoded by GA1 and GA2 T6 regions of gut Bacteroidales, leading us to predict that this is the major structural Hcp protein. To confirm this prediction, we performed RNAseq analysis of B. fragilis 9343 grown to midlog. As predicted, the first hcp gene of this region, BF9343_1943, is expressed more than 40-fold the level of the next most expressed hcp (Table S1), and thus, Hcp1 is likely the major structural protein of the T6SS apparatus. This gene is conserved in all identified GA3 T6SS regions (7).
Fig. 1.
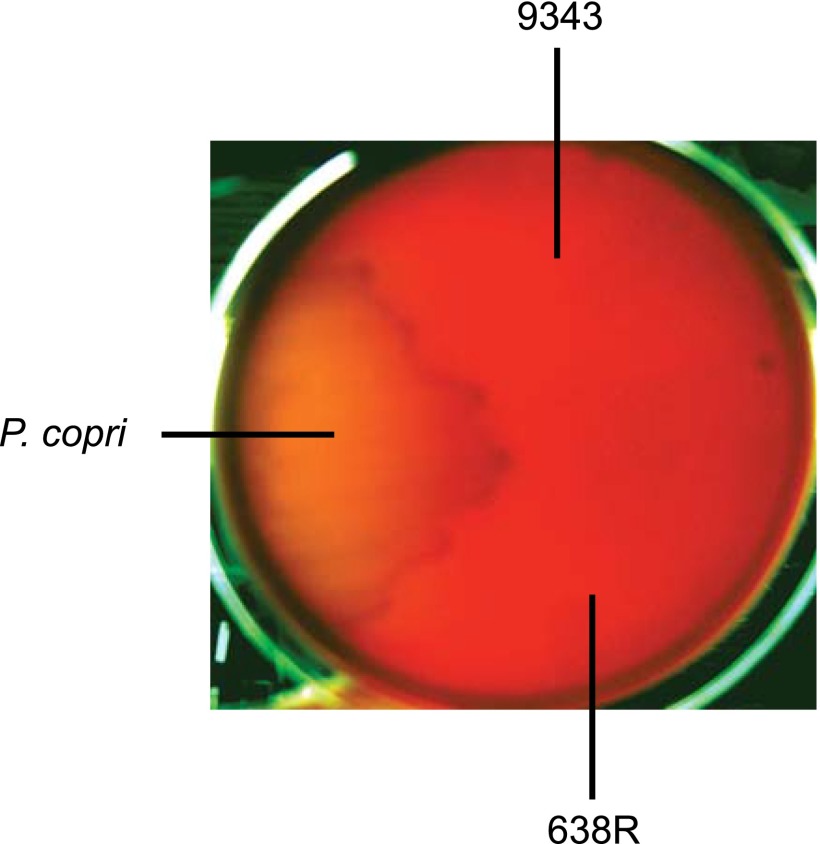
Phenotypic analysis of ΔT6SS mutants. (A) ORF maps of the GA3 T6SS loci of B. fragilis strains 9343 and 638R. Regions of 95% or greater DNA identity between GA3 T6SS loci are shown as red lines above the ORF maps, with the numbered breaks representing two variable regions between GA3 T6SS loci. Genes in the variable regions are white. The regions deleted in the various mutants are shown above the ORF maps with green lines. (B) Western immunoblots of 638R and 9343 probed with antiserum to Hcp1 showing that this protein is not secreted into the culture supernatant (sup) in the ∆T6SS mutants. CL, cell lysate. (C) Antagonism of Bacteroidales strains by the T6SSs of B. fragilis 638R or 9343 following overnight coculture with wild-type or ∆T6SS isogenic mutant. Bacteroides caccae CL03T12C61, B. fragilis 2_1_16, and P. copri were antagonized by the 9343 ΔT6SS mutant in a non-T6SS manner, so results are only shown for 638R. (D) Antagonism studies using E. coli strains as prey.
Table S1.
Transcriptional analysis of genes of the T6SS region of B. fragilis NCTC 9343
| Gene | Locus tag | Putative function | RNA-Seq values | |
| Expression | RPKM | |||
| tetR | BF9343_1944 | Transcriptional regulator | 74 | 102 |
| hcp | BF9343_1943 | Hcp needle protein | 10,547 | 13,976 |
| tssB | BF9343_1942 | Contractile sheath protein | 636 | 829 |
| tssC | BF9343_1941 | Contractile sheath protein | 666 | 871 |
| clpV | BF9343_1940 | AAA+ ATPase | 302 | 399 |
| hcp | BF9343_1939 | Hcp protein | 240 | 323 |
| hcp | BF9343_1938 | Hcp protein | 69 | 92 |
| unk | BF9343_1937 | 76 | 101 | |
| unk | BF9343_1936 | 143 | 195 | |
| hcp | BF9343_1935 | Hcp protein | 164 | 213 |
| hcp | BF9343_1934 | Hcp protein | 145 | 191 |
| tagC | BF9343_1933 | 67 | 88 | |
| tssE | BF9343_1932 | Baseplate protein | 182 | 239 |
| tssF | BF9343_1931 | TssK–TssFG membrane-associated subcomplex | 111 | 145 |
| vgrG | BF9343_1930 | Spike component | 133 | 175 |
| paar | BF9343_1929 | Spike component | 80 | 108 |
| unk | BF9343_1928 | 96 | 129 | |
| unk | BF9343_1927 | 69 | 94 | |
| unk | BF9343_1926 | 57 | 77 | |
| tssN | BF9343_1925 | Transmembrane protein | 84 | 109 |
| tssK | BF9343_1924 | TssK–TssFG membrane-associated subcomplex | 101 | 132 |
| tssG | BF9343_1923 | TssK–TssFG membrane-associated subcomplex | 28 | 36 |
| tssQ | BF9343_1922 | Transmembrane protein | 71 | 91 |
| tssO | BF9343_1921 | Transmembrane protein | 49 | 65 |
| tssP | BF9343_1920 | Transmembrane protein | 103 | 135 |
| tssR | BF9343_1919 | Lipoprotein | 60 | 79 |
RPKM, reads per kilobase of transcript per million mapped reads.
In the T6SS deletion mutants of both strains, Hcp1 is synthesized but not secreted into the supernatant (Fig. 1B), confirming the T6S defect. Next, the ability of these T6SSs to target other Bacteroidales strains was tested by coculture with wild-type 638R or 9343 and their isogenic ΔT6SS mutants. The strains selected were each resistant to either tetracycline or erythromycin to allow for selection of the prey strain. With only a few exceptions, coculture of Bacteroidales strains with wild-type 9343 brought about a 2-log reduction in cfu compared with when cocultured with its ΔT6SS mutant, and for strain 638R, a 3-log reduction resulted compared with its ΔT6SS mutant (Fig. 1C). This difference was not due to preferential transfer of the antibiotic resistance genes from the prey to the ΔT6SS strain (Fig. S1). These GA3 T6SSs targeted numerous Bacteroides species as well as Parabacteriodes, but not the one Prevotella copri strain tested. Of the Bacteroidales strains tested, B. fragilis strain B35 is not antagonized by the T6SS of strain 9343, and B. fragilis strain 2_1_16 is not antagonized by the T6SS of strain 638R. Analysis of the genome sequence of B. fragilis 2_1_16 revealed that it has the same T6SS locus as that of 638R (7), suggesting that this strain is protected because it encodes the cognate immunity protein(s). The genome of B. fragilis B35 has not been sequenced; however, we predicted that it has a GA3 T6SS locus similar to that of 9343 and therefore has the necessary immunity protein(s) to prevent antagonism by this strain. Using primers from conserved genes flanking the variable regions, we amplified and sequenced both variable regions of strain B35 and found that they are 100% identical to those of 9343. These data strongly suggest that immunity proteins, and likely the toxic effectors, are contained in one or both of the GA3 variable regions. In addition, these collective data demonstrate that B. fragilis GA3 T6SSs are able to target diverse human gut Bacteroidales strains, including B. fragilis.
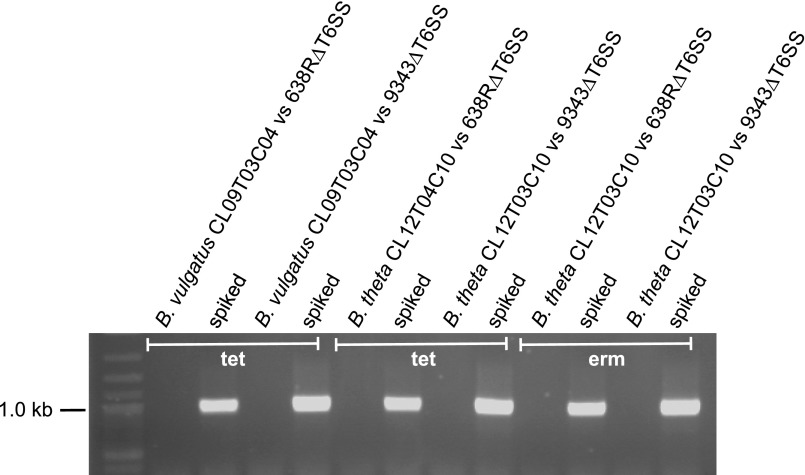
Fig. S1.
Analysis for the presence of predator strain (638R or 9343) on the antibiotic plate following coculture with antibiotic resistant prey strains B. vulgatus CL09T03C04 and B. thetaiotaomicron CL12T03C10. To ensure that the bacteria arising on the erythromycin and/or tetracycline plates following coculture assays were not from the transfer of the antibiotic-resistant gene to the predator strain, the bacteria growing on the third spot from the bottom of cocultures with the T6SS mutants were removed and grown overnight in the antibiotic to ensure dead bacteria were not analyzed. A total of 1 mL of this culture was pelleted, or 1 mL with the addition of 1 µL of 638R or 9343 (spiked). The bacteria were resuspended in 1 mL water, and 1 µL was analyzed by PCR with primers to GA3 conserved regions. No predator bacteria were detected in these samples. The spiked samples show that this method would identify predator bacteria that are present below 1/1,000 of the population if present. B. thetaiotaomicron CL12T03C10 was analyzed for transfer of both the tetracycline and erythromycin resistance genes during coculture.
To determine if these T6SSs are able to target Escherichia coli, we tested three different strains as prey; the laboratory strain DH5α, the symbiotic gut strain HS, and the enteropathogenic strain E2348/69. Our results show that the T6SSs of B. fragilis 9343 and 638R do not antagonize these E. coli strains (Fig. 1D; results shown for strain 638R).
Proteins Encoded in the 638R GA3 Variable Regions Are Necessary for Antagonism and Protection.
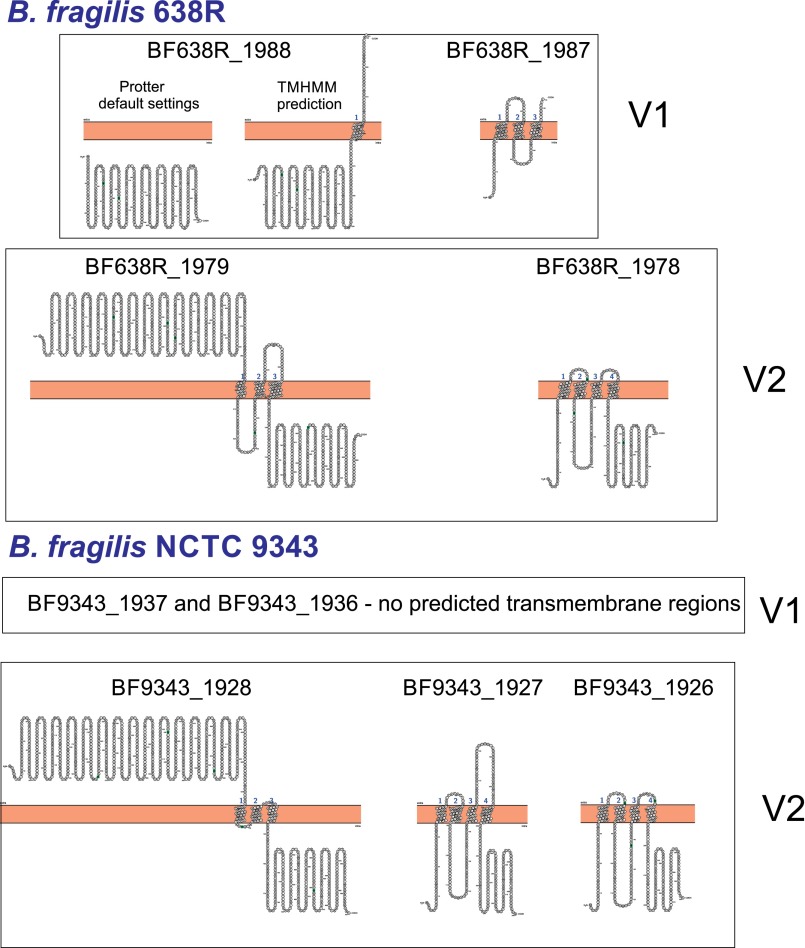
None of the four proteins encoded by the 638R variable regions or the five proteins encoded by 9343 variable regions (Fig. 1A) are similar to known proteins or contain any described motifs. Protter analysis (19) predicted that all genes of variable region 2 (V2) (Fig. 1A) of both strains encode transmembrane proteins, with either three or four predicted membrane spanning domains per protein (Fig. S2). In addition, proteins from V2 regions are orthologous between strains with the first gene of each region sharing 54% similarity and the second gene of the 638R region 48% and 51% similar to the two small proteins encoded in the 9343 V2 region. Proteins encoded by the variable region 1 (V1) are not orthologous between strains, and only the small protein encoded by the 638R V1 regions was predicted to have transmembrane spanning domains by Protter (Fig. S2).
Fig. S2.
Predicted transmembrane topology of proteins encoded by variable regions of GA3 T6SS loci of B. fragilis 638R and 9343. Predictions made using Protter (20) for all proteins with the addition of TMHMM analysis for BF638R_1987.
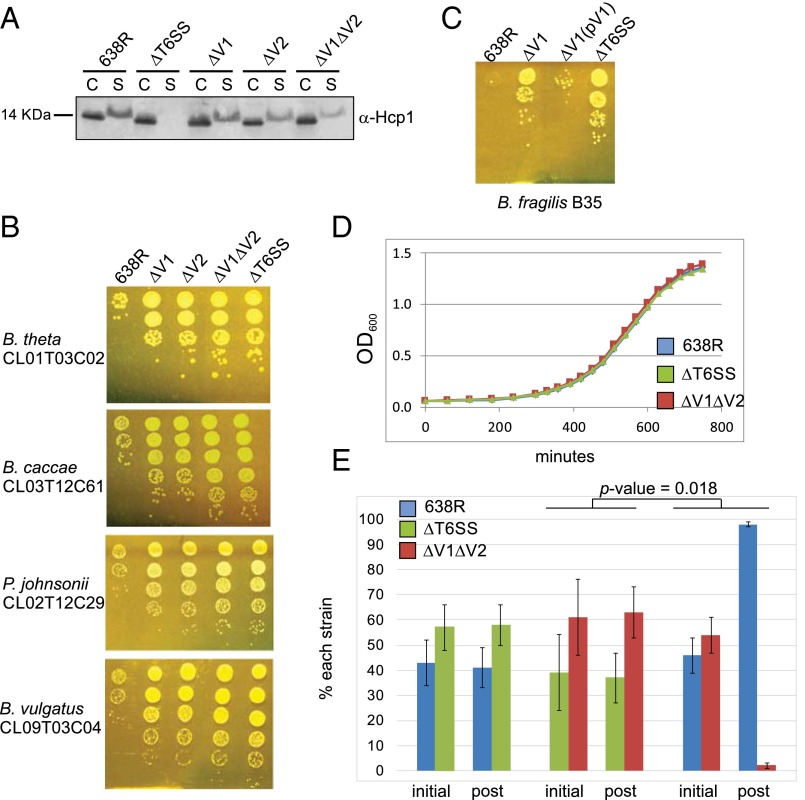
To determine if these variable region genes encode effector and immunity proteins, we made mutants deleted in each of the variable regions individually (ΔV1 and ΔV2) and together (ΔV1ΔV2) in strain 638R (Fig. 1A). Unlike the ΔT6SS mutant, we found that the T6SS machinery is operational in all three mutants, as evidenced by the secretion of Hcp1 into the supernatant (Fig. 2A). These data supported our prediction that genes contained in the variable regions encode T6SS proteins that are not part of the core T6SS machinery. However, T6 firing was not as robust in these mutants compared with wild type, especially in the double-deletion mutant (Fig. 2A).
Fig. 2.
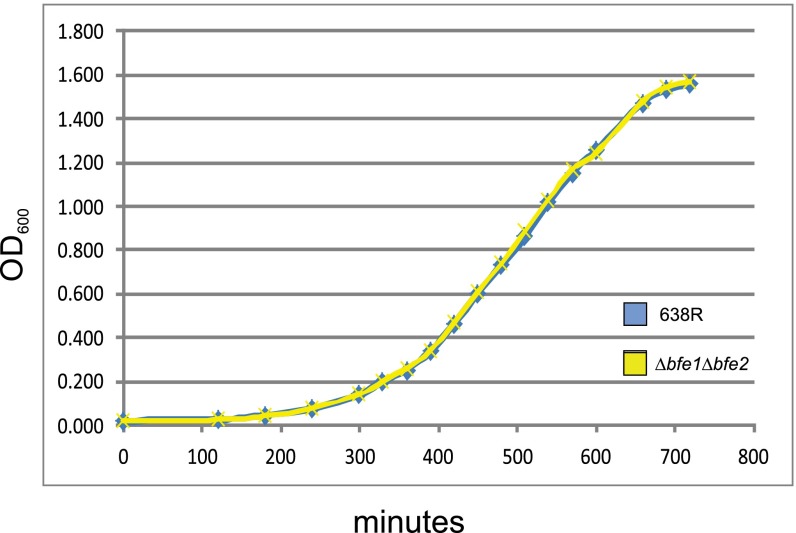
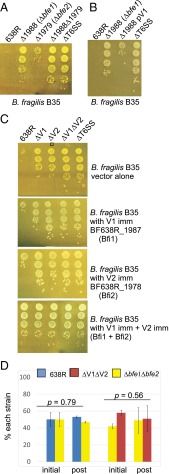
Genes in variable regions encode effector and immunity proteins. (A) Western immunoblots of 638R and its isogenic mutants probed with α-Hcp1 showing that Hcp1 is secreted into the supernatant of mutants in the variable regions but not in the T6SS mutant. C, cell lysate; S, supernatant. (B) Antagonism of Bacteroidales strains by the T6SSs of B. fragilis 638R and isogenic mutants following overnight coculture. (C) Demonstration of restoration of antagonism of the 638R∆V1 mutant when both genes of this region are present in trans (pV1). (D) In vitro growth curves of 638R and isogenic mutants (averages of biological triplicates). (E) In vitro competition assays between 638R and isogenic mutants. Percent of each strain in the inoculum at the start of the experiment (initial) is compared with the ratio after overnight coculture (post). Average of two experiments. Error bars represent SD. The change in the percentage of ∆V1∆V2 when cocultured with 638R is significantly different from when it is cocultured with ∆T6SS.
Because we predicted that each variable region would encode an effector and immunity protein pair, we expected that mutants deleted in genes of one variable region would still antagonize sensitive strains due to the products encoded by the other variable region. Although antagonism still occurred in each deletion mutant, deletion of either variable region resulted in a significant attenuation of antagonism, typically two orders of magnitude less than wild type (Fig. 2B). As predicted, the double-deletion mutant was completely attenuated similar to the ΔT6SS mutant with three orders of magnitude difference compared with wild type. These data demonstrate that factors necessary for antagonism are encoded in the variable regions, and that loss of the products of one variable region severely affects antagonism by the second region. Two possibilities may explain this second finding. Either the two effectors from the variable regions are both necessary for full antagonism, or the lack of one variable region leads to a T6 defect such as efficient loading of effectors onto the needle, or firing of the T6 apparatus. This second possibility is supported by the reduced amount of Hcp1 in the supernatant of variable region mutants (Fig. 2A). Previous analysis of 54 B. fragilis GA3 T6SS loci revealed 20 distinct regions based on the variable regions (7). We did not find a correlation of one V1 region with a particular V2 region; rather, some strains with the same V1 have different V2 regions and vice versa. Therefore, we predicted that V1 effectors do not require particular V2 products for antagonism.
To provide further evidence that genes in the variable regions encode immunity proteins necessary to protect the strain from its effectors, we performed coculture experiments using 638R, which is able to kill and protect itself; ∆T6SS, which is unable to kill but should produce immunity proteins; and ΔV1ΔV2, which we predict is unable to kill or protect itself. We first determined that all three strains grow with the same kinetics in monoculture in rich broth (Fig. 2D). Coculture studies clearly indicated that ∆T6SS is not antagonized by wild type, whereas ΔV1 ΔV2 is antagonized by wild type, but not by ∆T6SS as expected (P = 0.018; Fig. 2E). Therefore, immunity protein(s) are encoded in the 638R GA3 T6SS variable region(s).
Identification of Effector and Immunity Proteins.
In T6SS regions of Proteobacteria, immunity genes are typically present just downstream of the gene encoding its cognate effector and are generally smaller than the effector genes (17). Therefore, we predicted that the product encoded by the first gene of each variable region is the toxic effector and that of the second gene is the cognate immunity protein. We created deletion mutants of each putative effector gene individually and a mutant with deletions in both putative effector genes. Analysis of the double-deletion mutant showed that it had the same in vitro growth kinetics as the wild-type strain (Fig. S3). Coculture studies with B. fragilis B35 showed that both single gene deletions reduced the ability of this strain to antagonize; however, the deletion of the effector gene in V1 reduced antagonism more dramatically than deletion of the effector gene of V2 (Fig. 3A). Deletion of the effector genes of both regions resulted in a mutant unable to mediate T6 killing, similar to the ∆T6SS mutant (Fig. 3A). We were able to clone the V1 region genes into E. coli, but were unable to clone the V2 region genes. The 638R V1 genes placed in trans in the 638R Δ1988 mutant (V1 effector mutant) restored this mutant’s ability to antagonize (Fig. 3B).
Fig. S3.
In vitro growth curves of 638R and isogenic double-effector deletion mutant. Averages of biological triplicates.
Fig. 3.
Identification of effector and immunity proteins. B. fragilis B35 is used as the prey strain in all assays because it contains variable regions 1 and 2 that are distinct from those of 638R. (A) Analysis of antagonism of strain B35 with 638R mutants deleted for each putative effector gene or a mutant with both deletions. (B) Restoration of antagonism of the 638R∆1988 (∆bfe1) mutant when both genes of this region are present in trans (pV1). (C) Protection of B35 from antagonism by 638R T6SS when putative 638R immunity genes BF638R_1987 (bfi1) and 638R_1978 (bfi2) are present in trans. Each immunity protein confers protection against the corresponding 638R effector and both together provide full protection from antagonism by wild-type 638R. (D) In vitro competition assays showing that the double-effector deletion mutant is protected from killing by 638R and is also unable to antagonize ∆V1∆V2, because there are not significant differences in the ratios of the initial and post samples. Averages of two experiments. Error bars show SD.
To determine if the second genes of each variable region encode immunity proteins, we cloned 638R_1987 (V1) and BF639R_1978 (V2) singly or together and placed them in trans in strain B35 to see if these products could protect this strain from antagonism by 638R. Compared with vector alone, addition of each gene protected B35 from killing by the effector encoded in its cognate region, but not by the effector of the heterologous region, and the presence of both genes in B35 fully protected it from killing by wild-type 638R (3-log protection; Fig. 3C, Lower). Therefore, BF638R_1987 and BF638R_1978 encode immunity proteins that protect the strain from antagonism by their adjacent and upstream encoded effectors.
As expected, the double-effector deletion mutant, which encodes both immunity proteins, is not antagonized by wild-type 638R, and is also not able to antagonize the ∆V1∆V2 mutant in vitro (Fig. 3D). Based on these collective data, we designate the V1 region effector and immunity protein as Bfe1 and Bfi1, and the effector and immunity proteins encoded by the V2 region as Bfe2 and Bfi2, respectively (B. fragilis effector or immunity).
The GA3 T6SS Is Functional in the Mammalian Gut.
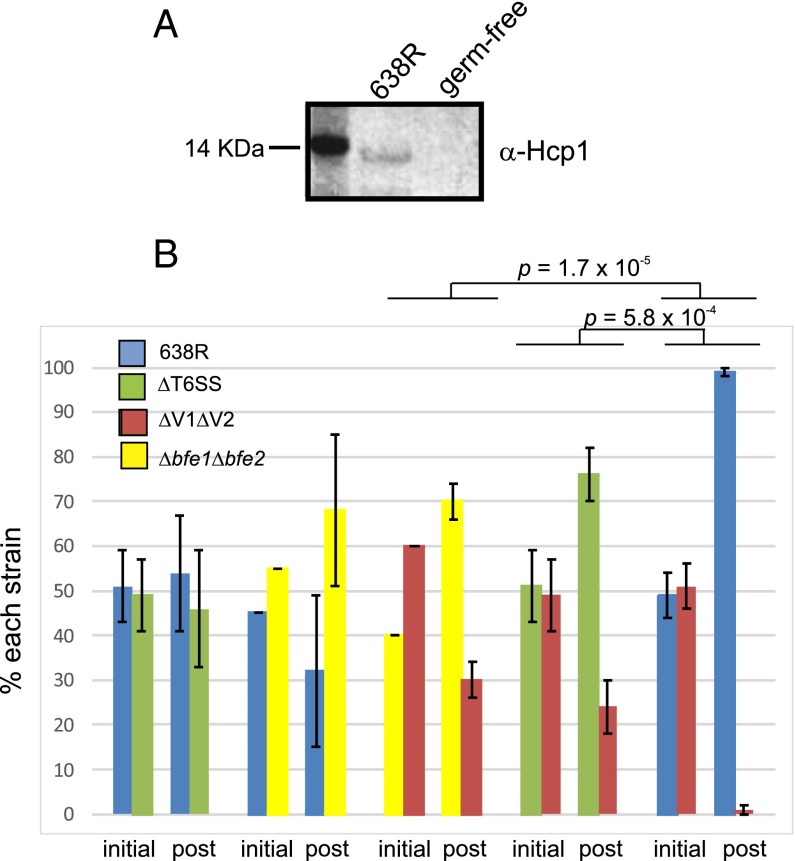
Despite our finding that more than half of human gut Bacteroidales species contain T6SS regions (7), there are still no data showing that these secretion systems are functional in the mammalian gut. To address this question, we first tested whether Hcp is present in the feces of gnotobiotic mice monocolonized with strain 638R. In comparison with germ-free mice, we could detect Hcp1 in the feces of mice colonized with 638R demonstrating that this important component of the T6SS machinery is synthesized in the mammalian gut (Fig. 4A).
Fig. 4.
Evidence that the 638R GA3 T6SS is operational in the mammalian gut. (A) Western immunoblot analysis showing that Hcp1 is present in the feces of mice monocolonized with B. fragilis 638R compared with germ-free mice. (B) In vivo competition studies showing that 638R antagonizes the ΔV1ΔV2 isogenic mutant in the mouse intestine. The percent of each strain in the inoculum (initial) and present in the stool after 7 d of cocolonization (post). Four mice were used for each group.
Next, we tested the ability of strain 638R or its ∆T6SS mutant to inhibit Bacteroides thetaiotaomicron strain CL01T03C02 when coinoculated into gnotobiotic mice. Seven days after introduction of the two strains into three mice per group, the percentage of B. thetaiotaomicron in the feces of mice when coinoculated with either 638R or ∆T6SS increased compared with its percentage in the initial inoculum, and the increases were not significantly different whether coinoculated with 638R or ∆T6SS (P = 0.18; Table S2). This same experiment was repeated except that a community of six Bacteroidales type strains (Fig. S4) was introduced into mice with either 638R or ∆T6SS. In both of these communities, the percentage of 638R or ∆T6SS in the inoculum was 14% (Table S2). Seven days after introduction of the community, <1% of the resulting bacteria in the feces were 638R or ∆T6SS. Therefore, under these experimental conditions, we could not detect an effect of the 638R T6SS in vivo. These results are likely due to two main factors. First, these human-derived Bacteroides strains have differing abilities to colonize the mouse gut. Second, because T6SSs require cell-to-cell contact for killing, sensitive bacteria must occupy the same niche to be antagonized.
Table S2.
In vivo colonization of 638R and ∆T6SS with Bacteroidales
| Sample | B. thetaiotaomicron | Bacteroidales community | ||
| CL01T03C02* | ||||
| 638R | ∆T6SS | 638R | ∆T6SS | |
| Inoculum percentage | 39 | 44 | 14 | 14 |
| Mouse 1 D7 | 21 | 20 | <1 | <1 |
| Mouse 2 D7 | 26 | 21 | <1 | <1 |
| Mouse 3 D7 | 29 | 23 | <1 | <1 |
| Average | 25 ± 4 | 21 ± 2 | <1 | <1 |
Numbers shown are initial or final percentages of 638R or ∆T6SS.
No significant difference between change in B. thetaiotaomicron percentages when cocolonized with 638R or ∆T6SS mutant. P = 0.18.
Fig. S4.
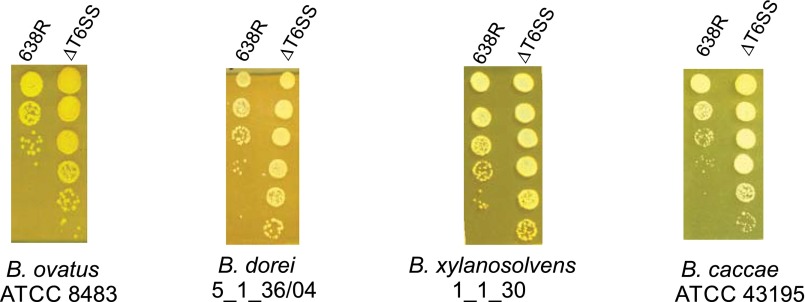
In vitro antagonism of Bacteroidales-type strains by the T6SS of B. fragilis 638R. Type strains used for in vivo competition include Bacteroides ovatus ATCC 8484, Bacteroides dorei 5_1_36/04, Bacteroides xylanisolvens 1_1_30, Bacteroides caccae ATCC 43195, Parabacteroides distasonis 8503, and Parabacteroides merdae 43184. Because these strains are not normally resistant to erythromycin, we mated plasmid pFD340 into these strains to allow for selection of the prey strain using erythromycin selection. We were able to transfer this plasmid into the four Bacteroides strains. In vitro cocultures were incubated overnight with a ratio of 1:1 predator:prey, and 10-fold dilutions were plated on erythromycin plates. All four Bacteroides-type strains are sensitive to killing by the T6SS of B. fragilis 638R.
To eliminate these inherent colonization differences and to ensure that the prey and predator strains have overlapping niches, we performed competition assays using wild type, ∆T6SS, ΔV1ΔV2, and ∆bfe1∆bfe2. The use of this isogenic set eliminated contributions of other factors that dictate variable colonization of different strains in the mouse intestine and allowed us to directly assess the contribution of the T6SS. After 7 d of cocolonization, we found that ΔV1ΔV2 decreased in abundance when cocolonized with ∆T6SS or ∆bfe1∆bfe2, but the decrease was significantly less than the decrease of ΔV1ΔV2 when cocolonized with 638R (P = 5.8 × 10−4 or 1.7 × 10−5, respectively; Fig. 4B). Therefore, the T6SS of this human gut Bacteroidales strain confers a competitive advantage in the mammalian gut.
Discussion
This study analyzed a specific subset of T6SSs of human gut Bacteroidales, termed genetic architecture 3 (GA3) T6SSs, which are specific to B. fragilis. These T6SS loci differ from GA1 and GA2 T6SS loci in that they are not contained on conserved ICE and therefore are not readily transferred between Bacteroidales species. In addition, GA3 T6SSs do not have effector and immunity proteins similar to those previously described. We showed that the effector and immunity proteins of GA3 T6SS regions are encoded by two variable regions within these loci. Our prior analysis of GA3 T6SS loci revealed that of the 54 GA3 regions analyzed, 20 distinct GA3 T6SS regions were detected, with differences almost exclusively confined to the two variable regions. Therefore, the GA3 T6SSs are likely a source of numerous novel effector and immunity proteins.
In vitro, these T6SSs are able to target most human gut Bacteroidales strains tested that do not contain the cognate immunity proteins. However, E. coli strains are not antagonized by these T6SSs, indicating either that the effectors are not active/toxic in E. coli, or the T6 machinery does not target E. coli. In future studies, it will be important to elucidate the mechanism of action of the Bfe1 and Bfe2 effectors and how they target sensitive strains, and why E. coli and P. copri DSM 18205 are not affected. Analyses are yet to be performed to determine if the GA1 and GA2 T6SSs of human gut Bacteroidales strains also target Bacteroidales species. Because GA1 and GA2 T6SS loci are contained on ICE and shared between coresident Bacteroidales strains in the human gut (3), it is possible that these antagonistic systems have different targets. Communal sharing of these antagonistic systems suggests they may function as a defense strategy against a common competitor or predator.
In this study, we show that GA3 T6SSs are synthesized and functional in the mammalian gut. Our data suggest that these contact-dependent killing systems likely function to create a locally protected niche in the human gut, and sensitive strains are likely not targeted unless they share an overlapping niche. In support of this prediction, we recently showed that Bacteroidales strains with distinct T6SSs, including GA3 T6SSs, are coresident in the human gut with numerous other Bacteroidales species at high density (7), indicating that these systems may function more for localized competition rather than strain elimination.
Materials and Methods
All primers used in this study are listed in Table S3.
Table S3.
Primers used in this study
| Purpose | Primer | Sequence* |
| Construction of His-BF638R_1994 (Hcp) | Forward | GTTTCATATGGCATTTAGAGCAACCTTAAGCTTTG |
| Reverse | ACTTGGATCCTTACTTCGGCCAATTCTGTTCA | |
| Construction of B. fragilis 638R ΔT6SS | Left flank forward | CGTAGGATCCCTCTTTGATTACTTCAAGGGATGC |
| Left flank reverse | ATACACGCGTCACTTCATTACCACCAATTCCATA | |
| Right flank forward | TGTAACGCGTAGGGAATACAGTTGTTGTCGATGT | |
| Right flank reverse | AGTAGGATCCAATTAAAATTCCCATTTCCAATGA | |
| Construction of B. fragilis 9343 ΔT6SS | Left flank forward | CACTGGATCCAGGGCGTATCAATCCTACTATCAC |
| Left flank reverse | ATACACGCGTCACTTCATTACCACCAATTCCATA | |
| Right flank forward | AGTGACGCGTATGGAGATATTGATTGGAATGTGA | |
| Right flank reverse | AAACGGATCCATCTTCTTTACGGAAGGTGTCAAA | |
| Construction of B. fragilis 638R ΔV1 | Left flank forward | ATCGGGATCCCCAAAGGAAATATAACGGTAATCG |
| Left flank reverse | GTTTACGCGTTGACATTAAATTCACTTTGCGTTT | |
| Right flank forward | TTGTACGCGTTTTTGTGGTGTTTTTCATAGTTGT | |
| Right flank reverse | CAATGGATCCAACAGGTCTGTAGGGGACACAAT | |
| Construction of B. fragilis 638R ΔV2 | Left flank forward | GAAGGAGCTCATATTCAAGCAACTGGGTGAGAAG |
| Left flank reverse | TTTTGGTACCAGTTTACCTCCCGTAGCTTCTTTT | |
| Right flank forward | AATAGGTACCACGACGGAAAGAAAGAGAAAAATA | |
| Right flank reverse | TCACGAGCTCGTATCCCTTCTCTGTCCACTTCAC | |
| Construction of B. fragilis 638R Δ1988 | Left flank forward | AGACGGATCCGATGCCGCTATTGATTTGATAGAC |
| (∆Bfe1638R) | Left flank reverse | GTTTACGCGTTGACATTAAATTCACTTTGCGTTT |
| Right flank forward | TGTGACGCGTTAGAGCTTCTGAATTGGAAATGTG | |
| Right flank reverse | CTATGGATCCAGCAGTCGGATAAGAGGGTCTAT | |
| Construction of B. fragilis 638R Δ1979 | Left flank forward | TCAAGGATCCGCAACTATTCCCAATTGTCTCAA |
| (∆Bfe2638R) | Left flank reverse | TTTTACGCGTAGTTTACCTCCCGTAGCTTCTTTT |
| Right flank forward | GGCAACGCGTAGCAGAAATAGAACCTAAATGGCA | |
| Right flank reverse | TCCCGGATCCTAGTTCTTCACTAATTCGCTTCCC | |
| Amplification of BF638R_1978 | Forward | ACGGGGATCCAGCAGAAATAGAACCTAAATGGCA |
| (Bfi2638R) | Reverse | GATGGGATCCTTGTCAAAATGGGATGTAACGTAG |
| Amplification of BF638R_1987 | Forward | GTGTGGATCCTAGAGCTTCTGAATTGGAAATGTG |
| (Bfi1638R) | Reverse | AAAAGGATCCCTGCATGATATGCACTAATCAAAA |
| Amplification of BF638R_1987-1978 for coexpression | Forward | GTGTGGATCCTAGAGCTTCTGAATTGGAAATGTG |
| Reverse | AAAAGAATTCCTGCATGATATGCACTAATCAAAA | |
| Forward | ACGGGAATTCAGCAGAAATAGAACCTAAATGGCA | |
| Reverse | GATGGGATCCTTGTCAAAATGGGATGTAACGTAG | |
| Amplification of BF638R_1988-87 (pV1) | Forward | GTTTGGATCCATTTTGATGAATGGAATTAGCTTTG |
| Reverse | AAAAGGATCCCTGCATGATATGCACTAATCAAAA | |
| Primers to differentiate 638R or ΔT6SS from ΔV1ΔV2 | ATTTTGATGAATGGAATTAGCTTTG | |
| TGTGACCTCATACCCATTTTGTAG | ||
| TTTCCGTATCTTCTCCCATATACC | ||
| TATCCATAATCTTCTCAGCCCAAT | ||
| Primers to differentiate 638R from ΔT6SS | CGAATTAAAGACCGAATAAAAGACA | |
| TCTGAACTGTTTCATTGATGTTCA | ||
| AGAATTGGCCGAAGTAATCATTTA | ||
| CTTTGATTTTACGTGCCTGTAATG | ||
| Amplify V1 of B. fragilis B35 for sequencing | Forward | ATGGAATTAGCTTTGACAATCACT |
| Reverse | TGCATCCAACGAATATTTTTATTT | |
| Amplify V2 of B. fragilis B35 for sequencing | Forward | CAATAACCATGTTACTGTCGAATG |
| Reverse | GTTTGTTCTCTCATTACTTTTCAGTCTT |
Restriction sites are underlined.
Construction of ΔT6SS, ΔV1, ΔV2, ΔV1ΔV2, Δ1988, Δ1979, and Δ1988 Δ1979.
Gene nomenclature is based on GenBank designations. A deletion removing tssB, tssC, and tssH was created in both B. fragilis strains 638R and 9343 (ΔT6SS). These genes were chosen because they are necessary for the T6 machinery to fire but do not affect synthesis of Hcp (Fig 2A). DNA segments upstream and downstream of the regions to be deleted were PCR amplified and digested with BamHI and MluI and cloned by three-way ligation into the BamHI site of the suicide vector pNJR6 (20). The resulting plasmids were conjugally transferred into B. fragilis 638R or 9343 using helper plasmid R751, and cointegrates were selected by erythromycin resistance. Cointegrates were passaged in nonselective medium to allow for second homologous recombination cross-out. Erythromycin-sensitive cross-outs were screened by PCR for the mutant genotype. B. fragilis 638R variable region mutants were created by deleting BF638R1988-87 (ΔV1), BF638R_1979-78 (ΔV2), and both of the gene pairs (ΔV1ΔV2). DNA segments were PCR amplified and digested with BamHI and MluI for the deletion of ΔV1 or SacI and KpnI for the deletion of ΔV2 and cloned by three way ligation into the BamHI or SacI site of pNJR6, respectively. B. fragilis 638R effector gene deletion mutants were created by deleting BF638R_1988, BF638R_1979 and both of the genes together (Δ1988Δ1979). DNA segments were PCR amplified and digested with BamHI and MluI and cloned by three-way ligation into the BamHI site of pNJR6. The cointegrate and cross-out steps for all deletion mutants were performed as described above.
Cloning 638R Variable Region Genes.
BF638R_1988-87, BF638R_1987, BF638R_1978 were PCR amplified and the products were cloned into the BamHI site of pFD340 (21). For coexpression of BF638R_1987 and BF638R_1978, a three-way ligation was performed ligating genes together using an EcoRI site engineered into the primers, and the products were cloned into the BamHI site of pFD340.
RNA Sequence Analysis.
RNA was extracted from midlog-phase bacteria grown in basal medium (22) using the Zymo Research RNA MiniPrep kit. Samples were treated with RQ1 DNase, then cleaned and concentrated using Zymo RNA Clean and Concentrator kit. For ribosomal depletion, Ribo-Zero magnetic kit (Epicentre) was used, adding 1 µg RiboGuard RNase inhibitor. RNA was cleaned using the Zymo RNA Clean and Concentrator kit. The concentration, purity, and integrity of RNA were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). RNA input to library construction was of high quality with RNA integrity number values >7. cDNA libraries were prepared and sequenced for RNA sequence (RNA-Seq) using Illumina HiSeq2500 machine to generate 50-bp paired-end reads. The results from two independent experiments performed on different days were combined. RNA-Seq data were analyzed using RockHopper open source software (23). A total of 108,176,502 from 130,538,906 reads (82.87%) were mapped to the NCTC 9343 genome.
Western Immunoblot Analyses and Preparation of Anti-BF638R_1994 (Hcp1) Antiserum.
An N-terminal His-tagged fusion of BF638R_1994 was constructed by cloning into the NdeI–BamHI sites of pET16b (Novagen), and the plasmid was transformed into E. coli BL21/DE3. Following induction, His-BF638R_1994 was purified using the ProBond Purification System (Life Technologies) and the eluted protein was dialyzed against PBS. Antiserum to purified His-BF638R_1994 was prepared in rabbits by Lampire Biologicals using the EXPRESS-LINE polyclonal antiserum protocol. For Western immunoblot analyses, bacteria or supernatants were boiled in LDS sample buffer and subjected to electrophoresis using NuPAGE 4–12% polyacrylamide gradient gels (Life Technologies). The contents of the gels were transferred to PVDF membranes, blocked, and probed with α-BF638R_1994 (α-Hcp1) followed by alkaline phosphatase-labeled α-rabbit IgG, and developed with BCIP/NBT (KPL).
Bacterial Growth Studies.
Overnight cultures of B. fragilis 638R, ∆T6SS, ∆V1∆V2, or ∆1988∆1979 were diluted 1:1,000 in supplemented basal medium and growth was monitored as increase in OD600. Biological triplicates were performed for each strain.
Antagonism (Coculture) Studies.
Log-phase cultures of B. fragilis 9343 or its isogenic ΔT6SS mutant and 638R wild type and its isogenic mutants, ∆T6SS, ∆V1∆V2, ∆1988∆1979 were mixed in a ratio of 10:1 with log-phase cultures of Bacteroidales strains. A total of 5 µL of the above mixtures were spotted on BHIS plates, or Brucella blood plates for P. copri, and incubated anaerobically at 37 °C overnight. The spots were excised, suspended in PBS and serial 10-fold dilutions were plated to erythromycin (5 µg/mL) or tetracycline (6 µg/mL) plates to quantify sensitivity of the strains to the T6SSs. For P. copri, before plating the 10-fold dilutions, 200 µL of 48 µg/mL tetracycline was spread on the Brucella blood plates, which we determined inhibited the growth of strains 638R and 9343, but not of P. copri (Fig. S5). E. coli strains DH5α, HS, and E2348/69 were cocultured similarly as prey Bacteroides strains; however, overnight coincubations were conducted under both anaerobic and aerobic conditions. After overnight coincubations, serial dilutions were plated onto BHIS plates and incubated aerobically to select for E. coli.
Fig. S5.
Brucella blood plate with 200 µL of 48 µg/mL tetracycline overlaid selectively grows P. copri DSM 18205 but not the two B. fragilis predator strains.
In Vitro Antagonism by B. fragilis 638R and Its Isogenic Mutants.
Log-phase cultures of B. fragilis 638R and its isogenic mutants were mixed at ratios of 1:1 (actual percentage shown in Figs. 2E and 3D). A total of 5 µL of the above mixtures were spotted on supplemented brain heart infusion (BHIS) plates and incubated anaerobically at 37 °C overnight. The spots were then excised, suspended in PBS, and plated on BHIS for single colonies. Multiplex PCRs were performed from a minimum of 90 individual colonies to differentiate strains. The results of two independent experiments are shown in Figs. 2E and 3D.
In Vivo Antagonism Assays.
Mouse studies were approved by the Harvard Medical Area Standing Committee on Animals. Swiss–Webster germ-free mice (4–8 wk old) were obtained from the Harvard Digestive Diseases Center gnotobiotic facility and housed in sterile OptiMice cages (Animal Care Systems); four were used for each experimental condition. Mice were colonized by gavage with B. fragilis 638R or ∆T6SS mixed 1:1 with B. thetaiotaomicron CL01T03C02 or with a Bacteroidales community (Fig. S4; Table S2). Fresh fecal samples from each mouse in each experimental condition were collected 7 d after cocolonization, diluted in PBS, and plated for single colonies. B. fragilis 638R or ∆T6SS were quantified by PCR from between 90 and 200 colonies per mouse. In separate experiments, mice were colonized by gavage with combinations of B. fragilis 638R, ∆T6SS, ∆V1∆V2, and/or Δ1988 Δ1979 (∆bfe1∆bfe2). Fresh fecal samples were collected 7 d after cocolonization, diluted in PBS, and plated for single colonies. Wild-type and mutant colonies were differentiated by PCR using primers shown in Table S3. At least 90 colonies per mouse were analyzed.
Statistical Analyses.
Comparisons were made between experiments using a metric that takes into account the change between initial and postratios between strains:
Here, θ denotes a proportion, P denotes the post, and I denotes the initial measurement; a and b denote the two strains in the experiment. This metric is analogous to the difference between log ratios of the post and initial measurements, except the arcsine transform was used instead of the log transform to allow for proportions of zero. Hypothesis testing was performed using a two-sample, two-tailed t test.
Acknowledgments
We thank M. Coyne for computational analyses, G. Gerber for statistical analyses, V. Yeliseyev for assistance with mouse experiments, and the Harvard Digestive Diseases Center gnotobiotic facility Grant P30DK034854 to the Brigham and Women’s Hospital Center for Clinical and Translational Metagenomics. This work was funded by Public Health Services Grants R01AI093771 and R01AI120633 from the NIH National Institute of Allergy and Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522510113/-/DCSupplemental.
References
- 1.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: Networks, competition, and stability. Science. 2015;350(6261):663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 2.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341(6141):1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coyne MJ, Zitomersky NL, McGuire AM, Earl AM, Comstock LE. Evidence of extensive DNA transfer between Bacteroidales species within the human gut. MBio. 2014;5(3):e01305–e01314. doi: 10.1128/mBio.01305-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. 2014;24(1):40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatzidaki-Livanis M, Coyne MJ, Comstock LE. An antimicrobial protein of the gut symbiont Bacteroides fragilis with a MACPF domain of host immune proteins. Mol Microbiol. 2014;94(6):1361–1374. doi: 10.1111/mmi.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell AB, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16(2):227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyne MJ, Roelofs KG, Comstock LE. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics. 2016;17(1):58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silverman JM, et al. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell. 2013;51(5):584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitney JC, et al. Genetically distinct pathways guide effector export through the type VI secretion system. Mol Microbiol. 2014;92(3):529–542. doi: 10.1111/mmi.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hachani A, Allsopp LP, Oduko Y, Filloux A. The VgrG proteins are “à la carte” delivery systems for bacterial type VI effectors. J Biol Chem. 2014;289(25):17872–17884. doi: 10.1074/jbc.M114.563429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell AB, et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475(7356):343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell AB, et al. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature. 2013;496(7446):508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng J, Ho B, Mekalanos JJ. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One. 2011;6(8):e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyata ST, Unterweger D, Rudko SP, Pukatzki S. Dual expression profile of type VI secretion system immunity genes protects pandemic Vibrio cholerae. PLoS Pathog. 2013;9(12):e1003752. doi: 10.1371/journal.ppat.1003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitney JC, et al. An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell. 2015;163(3):607–619. doi: 10.1016/j.cell.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma LS, Hachani A, Lin JS, Filloux A, Lai EM. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe. 2014;16(1):94–104. doi: 10.1016/j.chom.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durand E, Cambillau C, Cascales E, Journet L. VgrG, Tae, Tle, and beyond: The versatile arsenal of type VI secretion effectors. Trends Microbiol. 2014;22(9):498–507. doi: 10.1016/j.tim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30(6):884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- 20.Stevens AM, Shoemaker NB, Salyers AA. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J Bacteriol. 1990;172(8):4271–4279. doi: 10.1128/jb.172.8.4271-4279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith CJ, Rogers MB, McKee ML. Heterologous gene expression in Bacteroides fragilis. Plasmid. 1992;27(2):141–154. doi: 10.1016/0147-619x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 22.Pantosti A, Tzianabos AO, Onderdonk AB, Kasper DL. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect Immun. 1991;59(6):2075–2082. doi: 10.1128/iai.59.6.2075-2082.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClure R, et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41(14):e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]