Significance
The recognition of foreign peptide-MHCs by T cells is a central event in adaptive immunity that triggers antigen-specific immune responses against infections and cancer. To study antigen-specific T cells, we devised a peptide-MHC dodecamer that can sensitively detect and specifically stain these T cells, especially low-affinity and rare ones. This dodecamer technology is superior to most current peptide-MHC multimers, compatible with existing reagents, inexpensive to make, and easy to use. It has been successfully applied to studies of human and murine antigen-specific αβ and γδ T cells by flow cytometry and mass cytometry. Thus, this dodecamer constitutes an important tool for the investigation of antigen-specific T cells in basic and clinical research.
Keywords: peptide-MHC, dodecamer, avidity, antigen-specific T cell
Abstract
Here we report a peptide-MHC (pMHC) dodecamer as a “next generation” technology that is a significantly more sensitive and versatile alternative to pMHC tetramers for the detection, isolation, and phenotypic analysis of antigen-specific T cells. In particular, dodecamers are able to detect two- to fivefold more antigen-specific T cells in both human and murine CD4+ and CD8+ αβ T-cell compartments compared with the equivalent tetramers. The low-affinity, tetramer-negative, dodecamer-positive T cells showed comparable effector cytokine responses as those of high-affinity, tetramer-positive T cells. Dodecamers are able to detect early stage CD4+CD8+ double-positive thymocytes on which T-cell receptors are 10- to 30-fold less dense than mature T cells. Dodecamers also show utility in the analysis of γδ T cells and in cytometry by time-of-flight applications. This construct has a simple structure with a central scaffold protein linked to four streptavidin molecules, each having three pMHC ligands or other molecules. The dodecamer is straightforward and inexpensive to produce and is compatible with current tetramer technology and commercially available streptavidin conjugates.
T-cell receptors (TCRs) detect antigens in the form of peptides bound to peptide-major histocompatibility complex (pMHC) molecules on the surface of antigen-presenting cells. TCR–pMHC interactions determine the selection, development, differentiation, fate, and function of a T cell. However, TCRs bind monomeric pMHCs with very low binding affinities (Kd, ∼1–200 μM, 1,000- to 200,000-fold weaker than a typical antibody–antigen interaction) and with fast dissociation rates (koff, ∼0.05 s−1) in solution (1). To increase the binding avidity and circumvent the problem of fast dissociation, we previously engineered a pMHC tetramer to detect antigen-specific T cells by conjugating four biotinylated pMHC monomers to a single fluorescent-labeled streptavidin (2). This fulfilled a critical need in both basic and clinical immunology to be able to identify and characterize often very rare specific T cells in a population. Subsequent improvements in sensitivity, manufacture, and combinatorial labeling have made this methodology even more useful (3–7). However, there is a sharp drop-off in tetramer binding in the lower affinity range (∼150 μM) (8) so there have been a number of higher valency alternatives including pentamers (ProImmune), lipid vesicles (9), octamers (10), dextramers (11), and quantum dot (QD)-based multimers (12). Some of these clearly have improved sensitivity, and this is important in detecting T cells with especially low affinities (13, 14). For example, naive T cells and thymocytes, which express low-level and/or low-affinity TCRs, show little-to-no tetramer staining. Furthermore, αβ T cells and γδ T cells that do not bind antigen-specific tetramers can still produce significant antigen-specific cytokine responses (15, 16). MHC class II tetramers are also problematic in cytometry by time-of-flight mass spectrometry (CyTOF), which is an advanced version of flow cytometry that can simultaneously measure more than 40 parameters on single cells (17). The extensive washing steps, harsh fixation conditions, complicated sample injection process, and sensitivity of the time-of-flight mass spectrometry make the low-avidity MHC class II tetramers unsuitable for CyTOF studies.
To augment the avidity of the tetramer, we have engineered a biotinylated scaffold protein linked to four streptavidin tetramers, each capable of binding three biotinylated pMHC monomers. We then used the resulting dodecamer (Greek for “12”) to detect low-affinity αβ and γδ T cells in blood and tissue samples from humans and mice. Compared with tetramers and at least some multimers, this construct has better sensitivity, stronger signal strength, higher binding avidity, and a much slower dissociation rate. For the various specificities that we have analyzed, it can identify two- to fivefold more specific T cells than an equivalent tetrameric reagent.
Results
Dodecamer Construction.
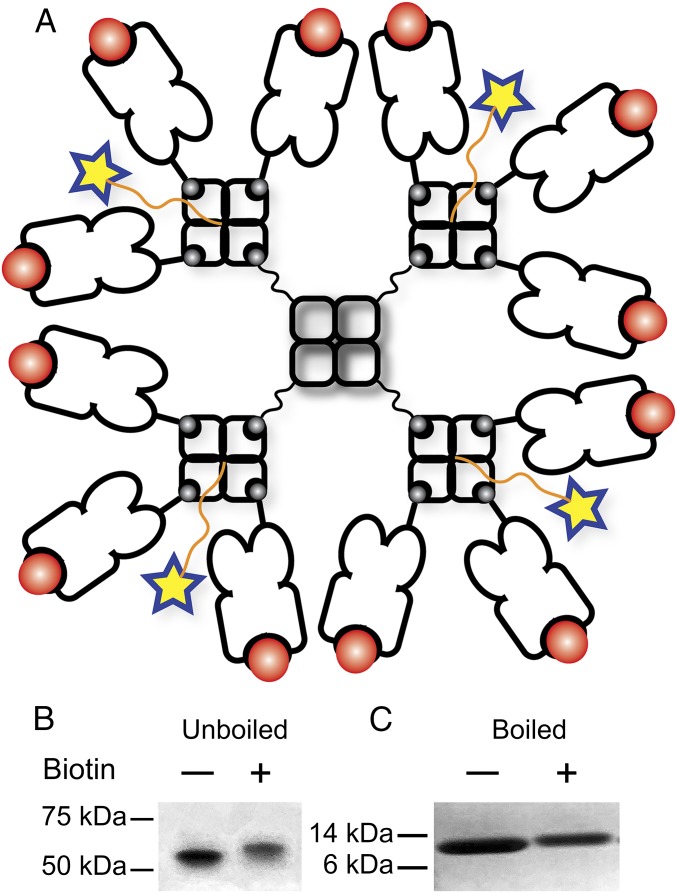
To overcome the limitations of current tetramer technology, we aimed to engineer a dodecamer with higher avidity to detect and quantify antigen-specific T cells, especially rare and low-affinity ones. To accomplish this, we added a cysteine residue at the C terminus of an inactive streptavidin subunit that has no biotin binding sites (18). After expression, refolding, and purification, a tetrameric scaffold protein was assembled bearing four terminal cysteines, which were subsequently biotinylated by 1-Biotinamido-4-[4′-(maleimidomethyl)cyclohexanecarboxamido]butane (BMCC-biotin) (molecular weight: 534). The scaffold protein with four biotin sites is the centerpiece of the dodecamer. As illustrated in Fig. 1A, a biotinylated scaffold protein associates with four commercial or homemade fluorescent/metal-labeled streptavidin molecules, each of which further associates with three biotinylated pMHC monomers. Thus, a biotinylated scaffold protein allows the formation a pMHC dodecamer. The expression and biotinylation of the scaffold protein were confirmed by SDS/PAGE. This tetrameric scaffold protein has a molecular weight of ∼53.4 and ∼55.5 kDa before and after biotinylation (Fig. 1B). Consistently, a monomeric subunit has a molecular weight of ∼13.3 and ∼13.9 kDa before and after biotinylation (Fig. 1C). The increase in the molecular weight of the scaffold protein in both the tetrameric and the monomeric forms is consistent with the expected results (Fig. 1 B and C).
Fig. 1.
Generation of a pMHC dodecamer. (A) Molecular structure of a pMHC dodecamer. A tetrameric scaffold protein was site-specifically conjugated to four biotins that bind to four fluorescent/metal-labeled streptavidin molecules. These four streptavidins further bind to 12 biotinylated pMHCs and form a pMHC dodecamer. (B) SDS/PAGE of the unbiotinylated and the biotinylated tetrameric scaffold protein under nondenaturing conditions. (C) SDS/PAGE of the unbiotinylated and the biotinylated monomeric scaffold protein under denaturing conditions to break the tetramer into monomers.
Comparison of Dodecamers to Different Multimer Formats.
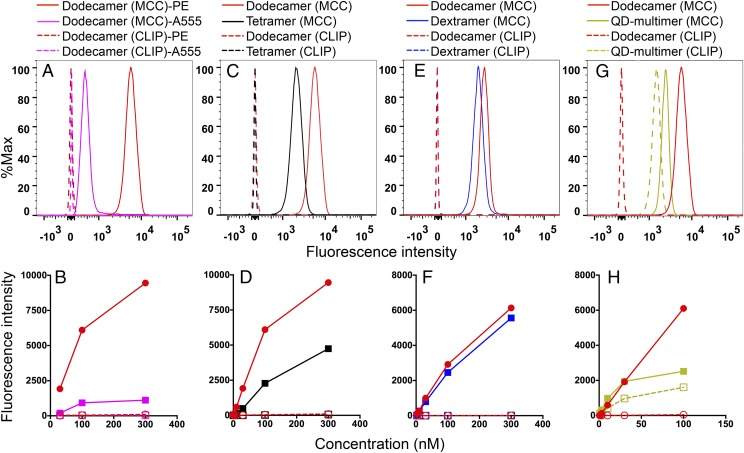
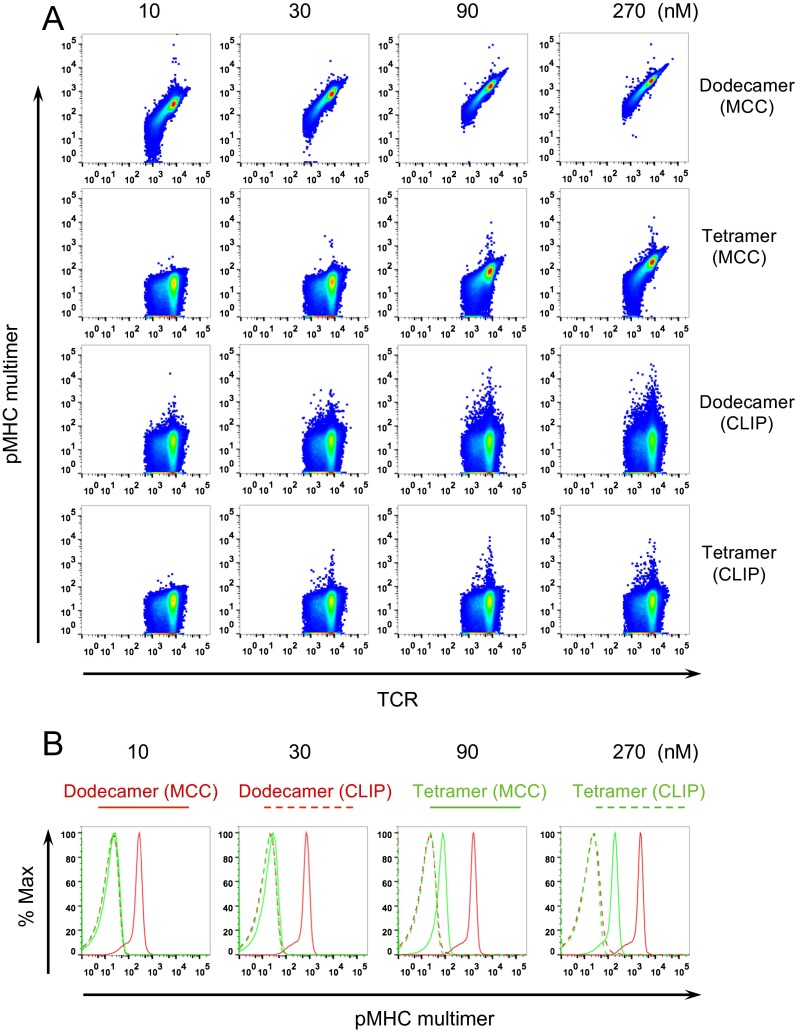
We first tested the functionality of our newly engineered dodecamers labeled with different fluorophores. The purified scaffold molecule was incubated with commercially available phycoerythrin (PE)-labeled streptavidin or Alexa555 (A555)-labeled streptavidin, and then incubated with biotinylated pMHC monomers containing specific moth cytochrome c (MCC) peptide (19) or control human class II-associated invariant chain peptide (CLIP) to form pMHC dodecamers. Dodecamers labeled with either the large fluorescent protein PE (∼250 kDa) or the small fluorescent dye A555 (∼1 kDa) specifically stained antigen-specific naive T cells in preparations of transgenic 5C.C7 splenocytes. The PE-labeled dodecamer gave ∼10 times better signal than the A555-labeled dodecamer as illustrated in Fig. 2A and Fig. S1A. This is expected because PE is a large fluorescent protein consisting of multiple fluorescent units whereas A555 is a small, singly fluorescent molecule. Both control dodecamers produced negligible nonspecific staining (Fig. 2 A and B). These data show that the dodecamer is compatible with different types of fluorophores for detecting antigen-specific T cells. We next compared dodecamers with tetramers. As shown in Fig. 2C, dodecamers gave a fivefold better signal than tetramers. Dodecamers yielded minimal background staining at 4 °C, comparable to tetramers over a broad range of concentrations (Fig. 2 C and D and Fig. S1B). We next compared dodecamers with dextramers (Fig. 2 E and F). Dextramers are hetereogeneous mixtures of polymers of different sizes conjugated to pMHC molecules. Unlike dextramers, dodecamers have a defined structure that allows precise calculation of their molar concentration to quantitatively study antigen-specific T cells. To directly compare dodecamer and dextramer staining, we had to rely on the molar concentration of the fluorescent streptavidin that was conjugated to these two multimers. Here we found a slight advantage in staining intensity (∼20%) versus dextramers over a range of concentrations (Fig. 2 E and F and Fig. S1C). Finally, we compared dodecamers with QD-based multimers. We generated QD multimers by incubating QD–streptavidin conjugates with biotinylated pMHC monomers. We found that dodecamers stained significantly better than QD multimers. In addition, we observed that QD multimers produced high levels of nonspecific staining, whereas the level of nonspecific staining with dodecamers was negligible (Fig. 2 G and H and Fig. S1D). Overall, pMHC dodecamers gave excellent results.
Fig. 2.
Antigen-specific stains of transgenic 5C.C7 naive T cells using different pMHC multimers. Dodecamers, tetramers, dextramers, and QD multimers were made using IEk monomer loaded with the specific MCC peptide recognized by the 5C.C7 TCR (solid dots and lines) or an irrelevant CLIP peptide (dashed dots and lines). Splenocytes were stained with an antibody mixture (Materials and Methods) and specific (MCC) or control (CLIP) dodecamers, tetramers, dextramers, and QD multimers. Histograms are gated on naive T cells and show representative stains at 100 nM (A, C, E, and G), and the curves (B, D, F, and H) summarize the mean fluorescence intensities at different concentrations. (A and B) T cells stained by PE-labeled dodecamers (red) and A555-labeled dodecamers (purple). (C and D) Staining comparison between PE-labeled dodecamers (red) and tetramers (black). (E and F) Staining comparison between PE-labeled dodecamers (red) and dextramers (blue). (G and H) Staining comparison between PE-labeled dodecamers (red) and QD605-labeled multimers (gold).
Fig. S1.
Dose-dependent staining of 5C.C7 naive T cells by different pMHC multimers. (A–C) Splenocytes were stained with an antibody mixture (Materials and Methods) and specific (MCC) dodecamers, tetramers, and dextramers. Histograms are gated on naive T cells. (A) Naive T cells stained with PE-labeled dodecamers and A555-labeled dodecamers at 30 nM (red), 100 nM (blue), and 300 nM (orange). (B) Staining comparison between PE-labeled dodecamers and tetramers at 0.3 nM (red), 1 nM (blue), 3 nM (orange), 10 nM (light green), 30 nM (dark green), 100 nM (brown), and 300 nM (purple). (C) Staining comparison between PE-labeled dodecamers and dextramers at 3 nM (red), 10 nM (blue), 30 nM (orange), 100 nM (light green), and 300 nM (dark green). (D) Nonspecific binding of QD multimers. Splenocytes were stained with an antibody mixture (Materials and Methods) and specific (MCC) or control (CLIP) QD multimers. Histograms are gated on naive T cells and show staining by MCC or CLIP QD multimers at 1 nM (red), 3 nM (blue), 10 nM (orange), 30 nM (light green), and 100 nM (dark green).
Binding Properties of Dodecamers.
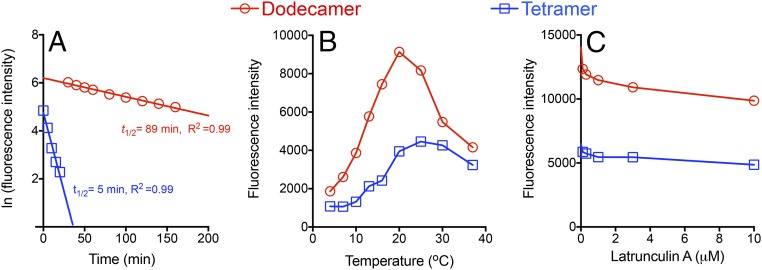
To evaluate the binding stability of pMHC dodecamers, we measured the apparent dissociation rates of a tetramer and a dodecamer. For 5C.C7 naive T cells, a tetramer binds TCRs with a half-life of 5 min, whereas a dodecamer binds TCRs with a half-life of 90 min (Fig. 3A), which equates to a 16-fold slower dissociation rate for the dodecamer. This much slower dissociation rate is most likely responsible for much of the improved binding avidity of the dodecamer because most tetramer-dependent assays require much more than 5 min to execute. TCR–pMHC interactions are also highly dependent on temperature (20), so we also tested the effect of temperature on pMHC multimer staining. We found that temperature influences the binding of both the dodecamer and the tetramer and that the temperature dependence positively correlates with the binding valency of the multimer. The temperature dependence of the dodecamer showed an inverted V shape. The dodecamer staining rapidly increases from 4 °C to 20 °C, peaks at 20 °C, and then quickly decreases from 20 °C to 37 °C (Fig. 3B). In comparison, tetramer binding is much less dependent on temperature. Tetramer staining slowly increases from 4 °C to 25 °C, peaks at 25 °C, and then slightly decreases from 25 °C to 37 °C. The binding of both multimers decreases at temperatures exceeding 25 °C, possibly due to TCR internalization, which occurs after T-cell activation dependent on the cytoskeleton. Because the cytoskeleton actively regulates TCR diffusion, binding, and internalization (21), we investigated the effect of latrunculin A, which disrupts actin filaments, on pMHC multimer staining. We found that latrunculin A significantly reduced the binding of both the dodecamer and the tetramer in a dose-dependent manner (Fig. 3C).
Fig. 3.
Biophysical properties of dodecamers and tetramers binding to TCRs. (A) Dissociation kinetics of pMHC multimers binding to 5C.C7 naive T cells at 4 °C. The half-lives (t1/2) of a dodecamer and a tetramer were determined using real-time flow cytometry in the presence of saturating amounts of anti-IEk blocking antibody. (B) Temperature dependence of MHC multimer staining. (C) Disruption of cytoskeleton using latrunculin A reduces pMHC multimer binding.
Staining of Human αβ T Cells with Dodecamers.
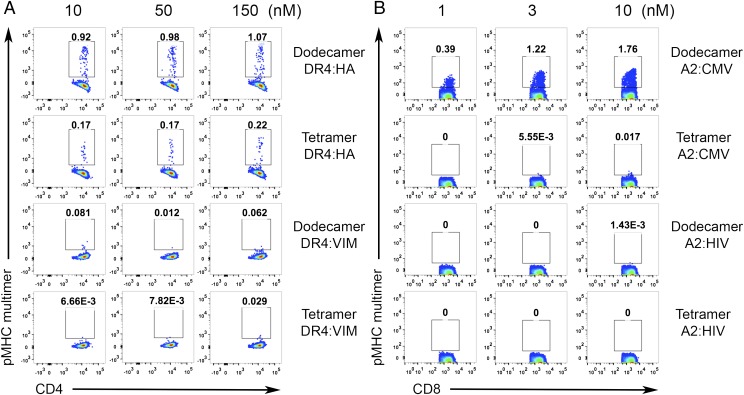
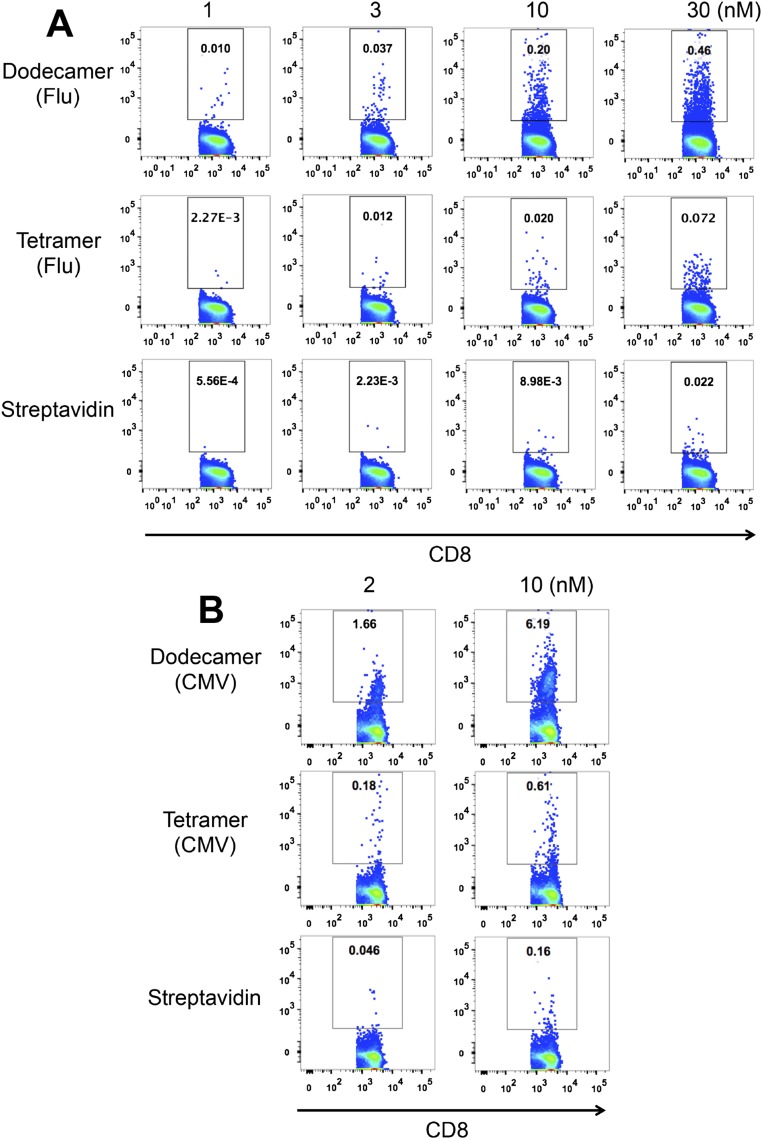
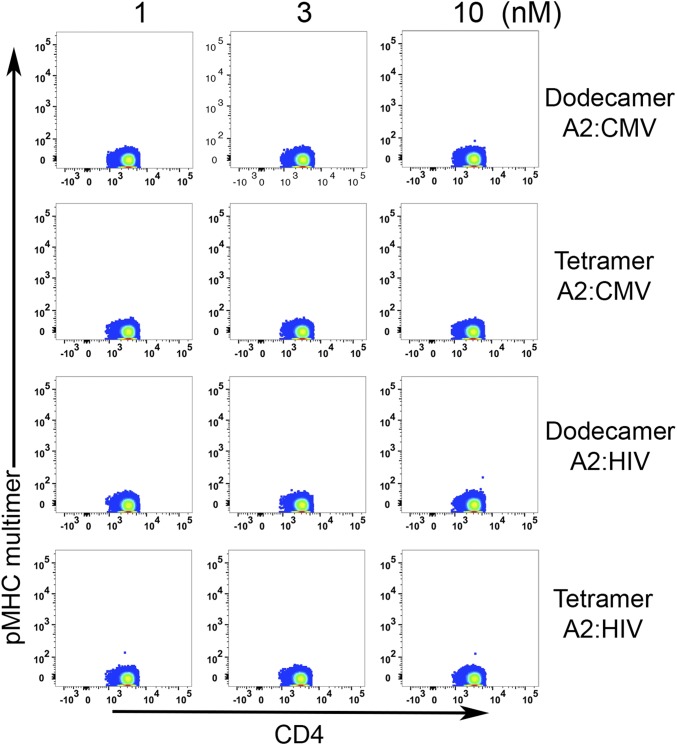
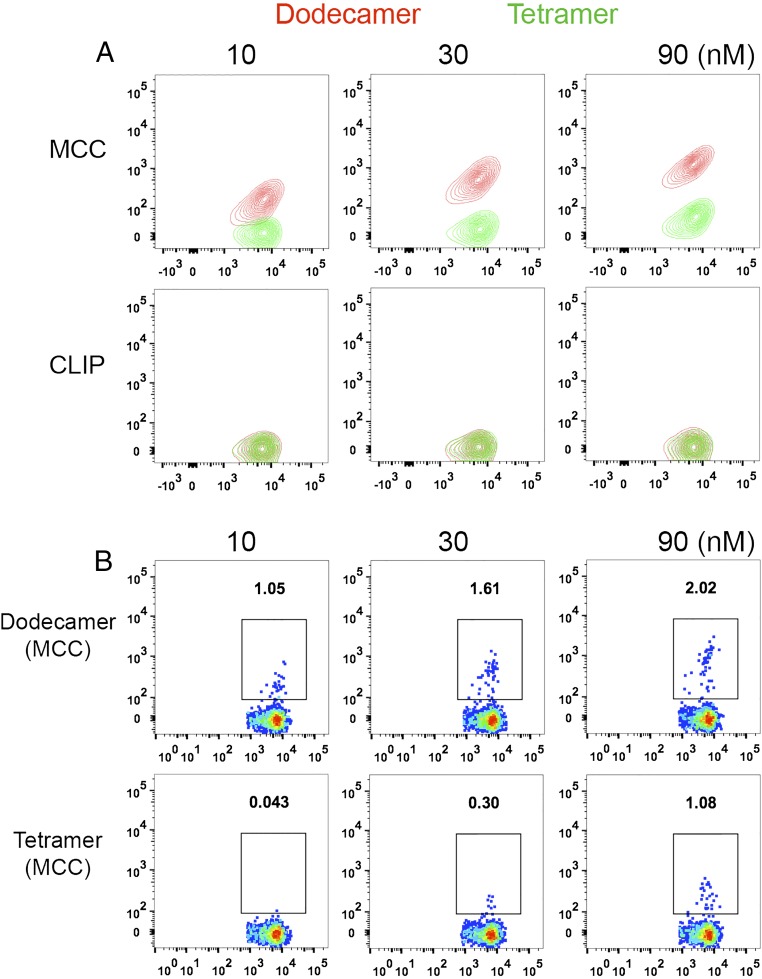
One important use of pMHC multimers involves the detection of antigen-specific T cells from human blood samples. Tetramer staining has been the primary method for this over the past decade. To increase the efficiency of detection, T cells are usually purified from peripheral blood mononuclear cells (PBMCs), and the rare antigen-specific T cells are further enriched after tetramer staining (6, 7, 22). Dodecamers can also be used for T-cell enrichment, especially for rare and low-affinity T cells, such as CD4+ T cells specific for influenza hemagglutinin (HA). Compared with tetramers, dodecamers significantly augmented the enrichment efficiency. For example, at 10 nM, dodecamers already could identify 0.92% antigen-specific T cells, but at 150 nM, tetramers could detect only 0.22% antigen-specific T cells. Dodecamers also had comparable nonspecific staining (as tetramers) for T-cell enrichment (Fig. 4A). For high-frequency antigen-specific T cells, dodecamers may even eliminate the need for enrichment in some cases, allowing direct staining and detection of antigen-specific T cells from human PBMCs at 4 °C (we have found that multimer staining generates higher backgrounds for human cells at room temperature) (Fig. S2). As shown in Fig. 4B, cytomegalovirus (CMV)-specific CD8+ T cells were easily detected by a cognate HLA-A2:CMV dodecamer from human PBMCs without enrichment. However, these CD8+ T cells were undetectable by a cognate HLA-A2:CMV tetramer at low concentrations (1–10 nM) at 4 °C (Fig. 4B) [Note that HLA-A2:CMV tetramer detected antigen-specific CD8+ T cells at room temperature (Fig. S2B)]. Meanwhile, a control HLA-A2:HIV dodecamer produced negligible nonspecific staining similar to that of a control HLA-A2:HIV tetramer (Fig. 4B). Furthermore, the staining was specific to CD8+ T cells, and we detected negligible nonspecific bindings to CD4+ T cells in the PBMCs (Fig. S3). Overall, these results suggest that traditional tetramers may significantly underestimate the actual frequency of antigen-specific T cells in the repertoire, which might be physiologically important in maintaining and mediating the effector function and homeostasis of the adaptive immune system.
Fig. 4.
Antigen-specific staining of human influenza-specific CD4+ T cells (A) or CMV-specific CD8+ T cells (B) by dodecamers and tetramers. Dodecamers and tetramers were made using HLA-DR4 loaded with either a hemagglutinin peptide HA306 or an irrelevant VIM peptide (A) or with HLA-A2 loaded with either a CMV peptide or an irrelevant HIV peptide (B) (6). The influenza-specific CD4+ T cells were enriched before FACS analyses using a protocol developed by Jenkins et al. (7) (A). The pseudocolor plots show representative staining of influenza-specific CD4+ T cells (A) or CMV-specific CD8+ T cells (B) in human PBMCs by dodecamers and tetramers at different concentrations. The percentages of T cells staining positive are labeled in each panel.
Fig. S2.
Antigen-specific staining of Flu-specific (A) and CMV-specific (B) CD8+ T cells by dodecamers and tetramers at different concentrations at room temperature (22 °C). Total human PBMCs were stained with an antibody mixture (Materials and Methods) and specific (HLA-A2:Flu or HLA-A2:CMV) dodecamers and tetramers or control (PE–streptavidin). Plots are gated on CD8+ T cells.
Fig. S3.
Background staining analysis. Dodecamers and tetramers were made using HLA-A2 loaded with either a CMV peptide or an irrelevant HIV peptide. The pseudocolor plots show representative background staining of CD4+ T cells in total human PBMCs by dodecamers and tetramers at different concentrations.
Detection of Rare and Low-Affinity T Cells by Dodecamers.
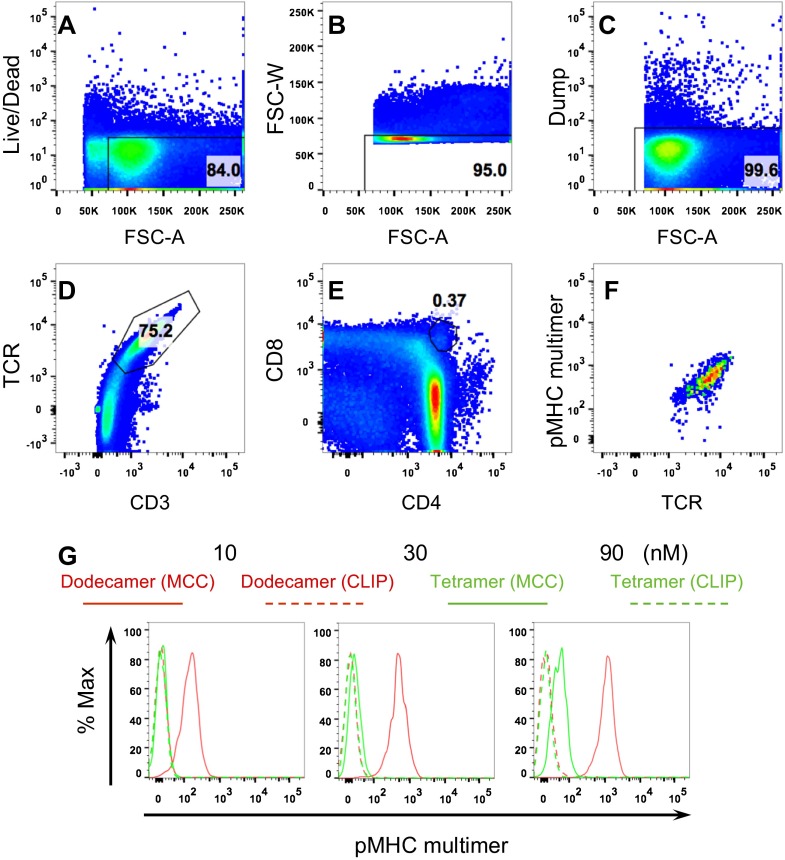
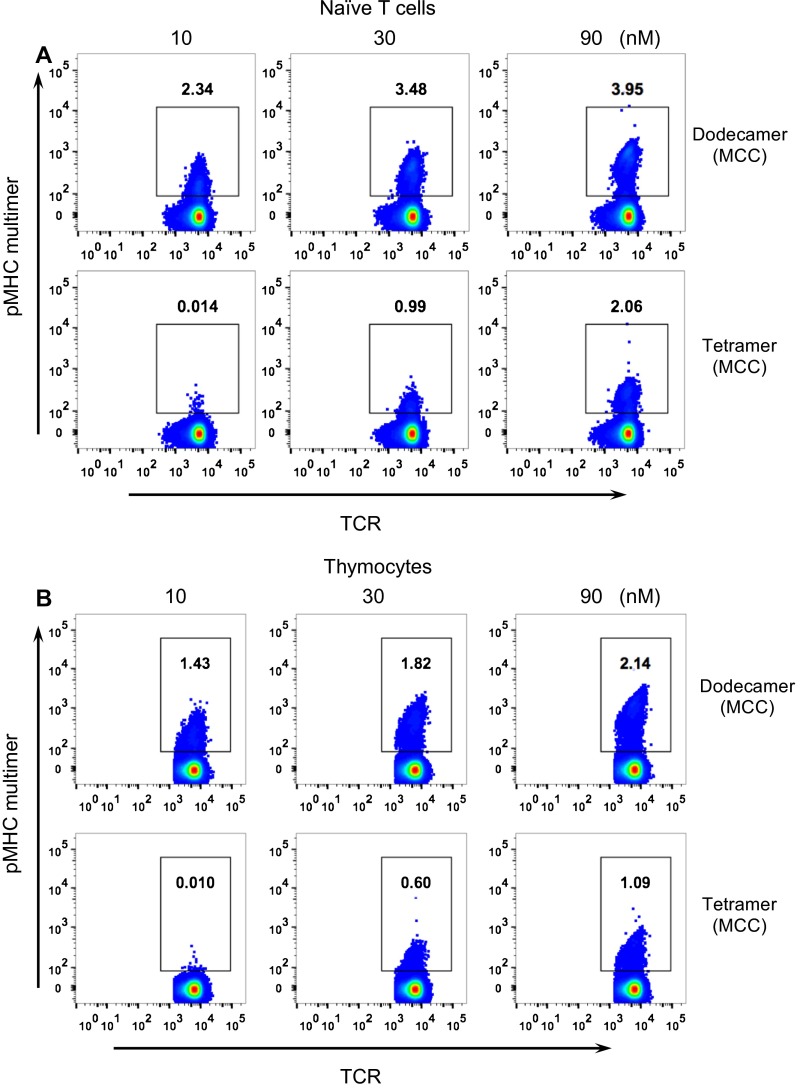
Because a dodecamer has a much higher pMHC valency compared with a tetramer, and is also much larger, we tested whether a dodecamer is able to stain rare and low-affinity αβ T cells that are usually difficult to detect with tetramers, such as thymocytes. The binding affinity of thymocytes has been generally recognized as a key determinant for T-cell selection (23). However, immature, “double-positive” CD4+CD8+ thymocytes express 10- to 30-fold lower levels of TCR than mature T cells and consequently are refractory to tetramer staining (24). As expected, cytochrome c/I-Ek tetramers showed minimal staining of 5C.C7 TCR+CD3+ thymocytes even when used at high concentrations (∼90 nM) (Fig. S4). In contrast, the dodecamer specifically stained 5C.C7 TCR+CD3+ thymocytes even at low concentrations (∼10 nM) (Fig. S4). We further analyzed the CD4+CD8+ double-positive thymocyte population (Fig. S5). The dodecamer readily stained the rare and low-affinity CD4+CD8+ thymocytes, which were undetectable using the tetramer reagent (Fig. 5A and Fig. S5G). We further used this dodecamer to detect antigen-specific 5C.C7 β–chain-transgenic splenic T cells and thymocytes, which all express the same TCRβ chain together with different TCRα chains (24), leading to differences in TCR affinity. Compared with the tetramer, the dodecamer detected significantly more specific cells in 5C.C7 β-chain naive T cells (Fig. S6A), TCR+CD3+ thymocytes (Fig. S6B), and CD4+CD8+ thymocytes (Fig. 5B). These data further indicate the utility of dodecamers in identifying populations of specific T cells that have been difficult to study because of low TCR densities or avidities.
Fig. S4.
Staining of TCR+ CD3+ thymocytes of 5C.C7 αβ transgenic mice by dodecamers and tetramers. Total 5C.C7 thymocytes were collected from transgenic mouse thymuses and stained with an antibody mixture (Materials and Methods) and specific (MCC) or control (CLIP) dodecamers and tetramers. TCR+ CD3+ thymocytes were gated from other cells. The pseudocolor plots (A) and histograms (B) show the staining of TCR+ CD3+ thymocytes at 10, 30, 90, and 270 nM.
Fig. S5.
Representative gating of CD4+ CD8+ double-positive thymocytes of 5C.C7 αβ transgenic mice. (A) Thymocytes were stained by live/dead aqua stain to identify live cells. (B–E) Plots are gated on CD4+ CD8+ double-positive thymocytes. (F) Representative staining of CD4+ CD8+ double-positive thymocytes by 30 nM MCC-IEk dodecamer. (G) Staining of CD4+ CD8+ double-positive thymocytes of 5C.C7 αβ transgenic mice by dodecamers and tetramers. Total 5C.C7 thymocytes were collected from transgenic mouse thymuses and stained with an antibody mixture (Materials and Methods) and specific (MCC) or control (CLIP) dodecamers and tetramers. Plots are gated on CD4+ CD8+ thymocytes. The histograms show the staining of CD4+ CD8+ thymocytes at 10, 30, and 90 nM.
Fig. 5.
Staining of rare and low-affinity T cells by dodecamers. (A) 5C.C7 αβ transgenic CD4+CD8+ double-positive thymocytes were stained by specific (MCC) and control (CLIP) dodecamers (red) and tetramers (green) at different concentrations. (B) 5C.C7 β-chain CD4+CD8+ double-positive thymocytes were stained by specific (MCC) dodecamers and tetramers at different concentrations. The percentages of pMHC multimer positive T cells are indicated in each panel.
Fig. S6.
(A) Staining of naive 5C.C7 β-chain T cells by dodecamers and tetramers. 5C.C7 β-chain splenocytes were stained with an antibody mixture (Materials and Methods) and specific (MCC) dodecamers and tetramers. Plots are gated on naive 5C.C7 β-chain T cells. The pseudocolor plots show the staining of naive 5CC.7 β-chain T cells at 10, 30, and 90 nM. (B) Staining of TCR+ CD3+ 5C.C7 β-chain thymocytes by dodecamers and tetramers. Total 5C.C7 β-chain thymocytes were stained with an antibody mixture (Materials and Methods) and specific (MCC) dodecamers and tetramers. Plots are gated on TCR+ CD3+ 5C.C7 β-chain thymocytes. The pseudocolor plots show the staining of TCR+ CD3+ 5C.C7 β-chain thymocytes at 10, 30, and 90 nM.
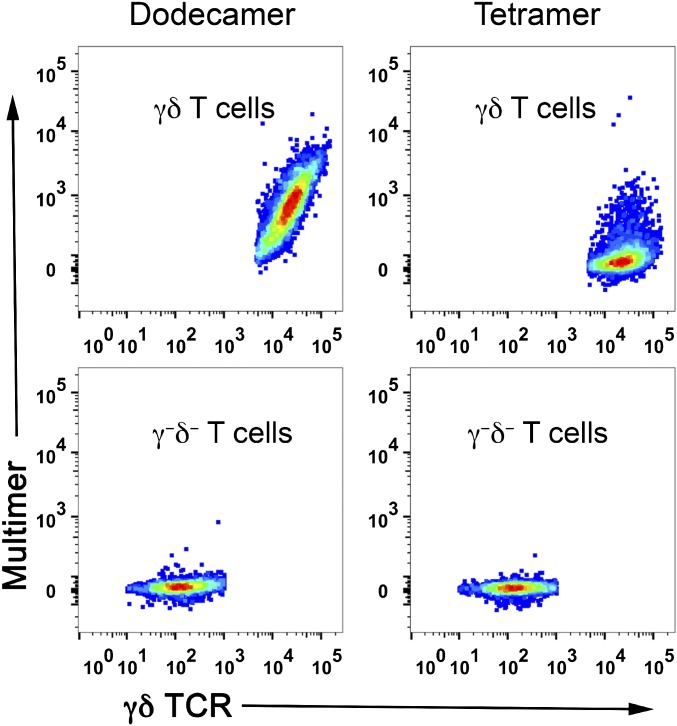
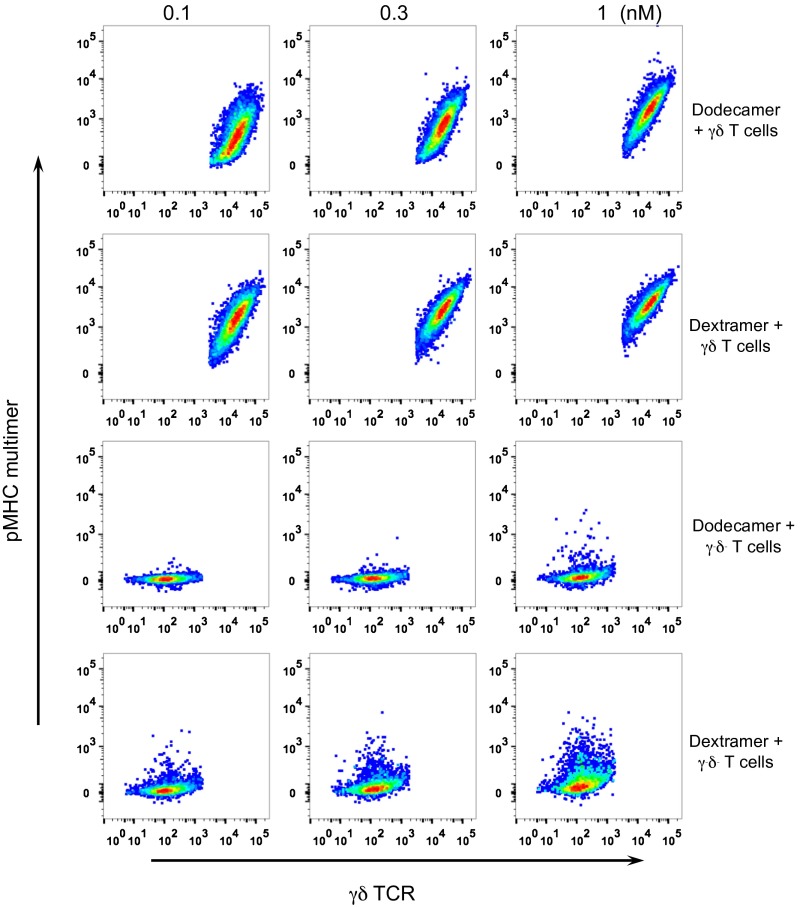
γδ T cells are a small subset of T cells that express γδ TCRs on their surface. We have also used dodecamers to successfully stain a T-cell line expressing δγ TCRs. We first compared staining with dodecamers to staining with tetramers. As expected, dodecamers stained γδ TCR-expressing T cells much better than tetramers (Fig. S7), which is consistent with our previous results for αβ T cells (Figs. 2–5). We further compared staining by dodecamers versus dextramers. Both dodecamers and dextramers showed comparable staining intensities (Fig. S8: compare top two rows), although dodecamers had the added benefit of less nonspecific binding (Fig. S8: compare bottom two rows).
Fig. S7.
Antigen-specific staining of γδ TCR-expressing cells (Top row) and γδ-negative cells (Bottom row) by dodecamers and tetramers. The pseudocolor plots show representative staining at 1 nM.
Fig. S8.
Antigen-specific staining of γδ TCR-expressing cells by dodecamers and dextramers. γδ TCR-expressing cells (Top two rows) and γδ-negative cells (Bottom two rows) were stained with dodecamers and dextramers at 0.1, 0.3, and 1 nM.
Application of Dodecamers to Single-Cell Mass Cytometry.
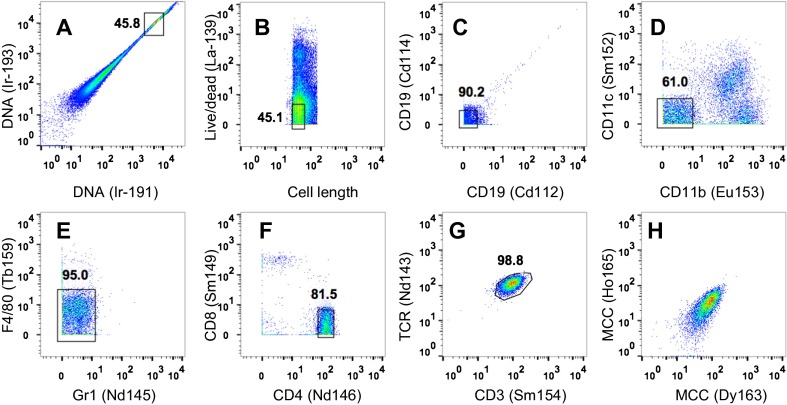
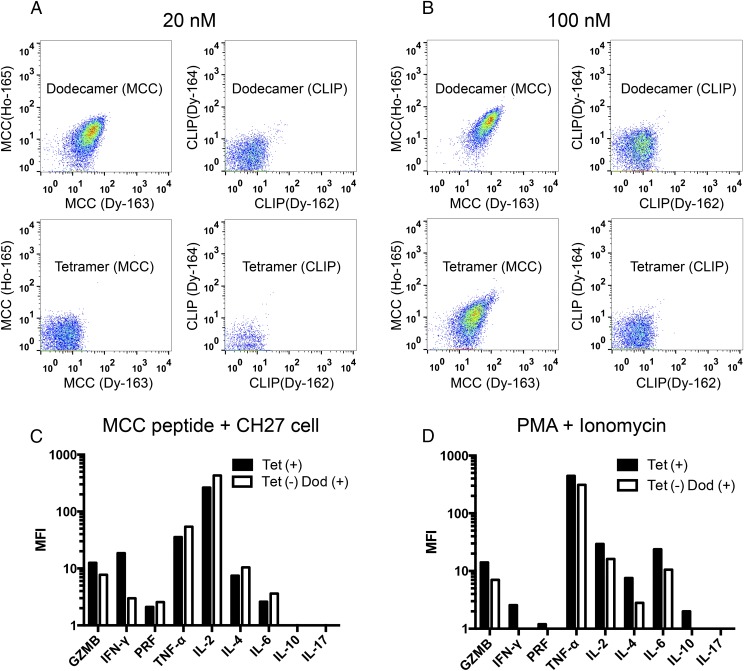
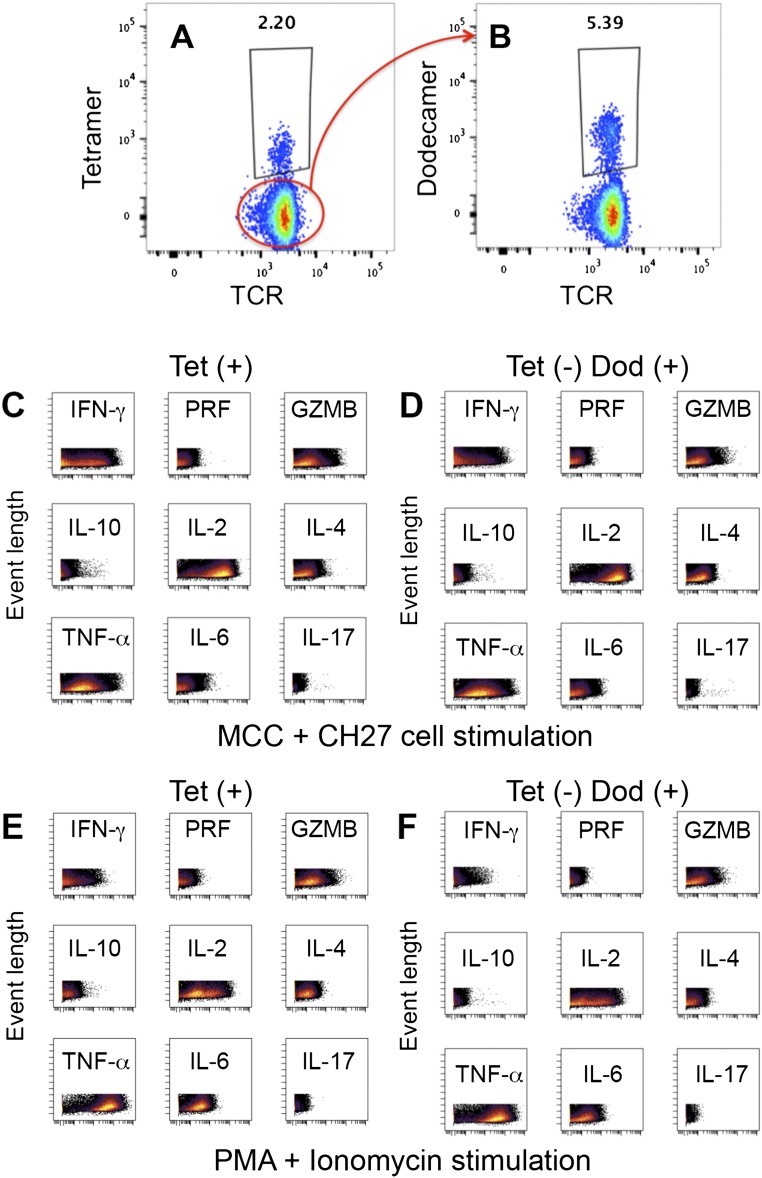
Single-cell mass cytometry, also known as CyTOF, is a new type of flow cytometry in which antibodies coupled to heavy metal isotopes are used to stain molecules of interest on cells (17, 25). CyTOF can simultaneously measure more than 40 parameters on a single cell without the problem of overlapping excitation and emission spectra that is inherent to conventional fluorescence-based flow cytometry (17, 25). Although we had some success in using MHC class I tetramers to analyze CD8+ T cells by CyTOF (6, 17), the detection of antigen-specific CD4+ T cells using MHC class II tetramers has been variable, possibly due to the harsh experimental conditions needed for CyTOF measurements, such as extensive washing, cell fixation, and the complex sample introduction system of the machine. The high binding avidity of dodecamers may overcome these barriers, so we tested the application of dodecamers in CyTOF. 5C.C7 splenocytes were incubated with metal-labeled MHC class II tetramers or dodecamers and then analyzed by high-throughput single-cell CyTOF (Fig. S9). Dodecamers can readily stain antigen-specific 5C.C7 naive T cells with high specificity (Fig. 6 A and B). Consistent with the results obtained by conventional flow cytometry, dodecamers stained significantly better than tetramers in CyTOF. The staining of the 20-nM dodecamer is better than that of the 100-nM tetramer in CyTOF (Fig. 6 A and B). Furthermore, to evaluate the function of low-affinity T cells detected by dodecamers but missed by tetramers, we sorted tetramer (+) T cells (Fig. S10A) and tetramer (−) dodecamer (+) T cells (Fig. S10B) from the splenocytes of a 5C.C7 β-chain mouse using flow cytometry. Sorted T cells were stimulated by either MCC peptide-pulsed CH27 cells (Fig. 6C) or phorbol 12-myristate 13-acetate (PMA)/Ionomycin (Fig. 6D) and then stained by ten metal-labeled antibodies to detect the effector cytokines. Tetramer (−) dodecamer (+) T cells showed a comparable cytokine expression profile to that of tetramer (+) T cells (Fig. 6 C and D and Fig. S10 C–F). The data highlighted the functional importance of low-affinity T cells missed by tetramers. Thus, dodecamers should be useful in CyTOF-based studies for detecting, phenotyping, quantifying, and analyzing antigen-specific T cells.
Fig. S9.
Representative analysis of antibody and pMHC dodecamer-stained cells using CyTOF. (A) DNA content stained by an iridium-191/193 interchelator is used to identify individual cells. (B) The cells are also gated for uniformity in the length of signal received by the ion detector and by exclusion of a live-dead viability stain. (C–G) Plots are gated on naive 5C.C7 T cells. (H) Representative staining of naive 5C.C7 T cells by a 100-nM metal-labeled specific MCC-IEk dodecamer.
Fig. 6.
The application of dodecamers in single-cell mass cytometry. (A and B) Naive 5C.C7 T cells were stained with 20 nM (A) or 100 nM (B) metal-labeled dodecamers and tetramers and analyzed by CyTOF. Dodecamers and tetramers were made using IEk MHC loaded with the specific MCC peptide recognized by the 5C.C7 TCR or an irrelevant CLIP peptide. (C and D). Cytokine analysis by CyTOF. Tetramer (+) T cells and tetramer (−) dodecamer (+) T cells were stimulated by either 1 μM MCC peptide and CH27 cells (C) or PMA + Ionomycin (D). Stimulated cells were intracellularly stained with a metal-labeled antibody mixture and analyzed by CyTOF. Data were shown as mean fluorescence intensity (MFI) after subtracting background staining.
Fig. S10.
(A and B) Isolation of tetramer (+) T cells and tetramer (−) dodecamer (+) T cells by fluorescence-activated cell sorting. Splenocytes isolated from a 5C.C7 single β-chain transgenic mouse were stained with APC-Cy7–labeled anti-CD19, anti-CD11b, anti-Gr-1, and anti-F4/80 antibodies and followed by anti-APC magnetic beads for cell selection. (A) Negatively selected T cells were stained with live/dead aqua stain, pacific blue-labeled anti-TCRβ antibody, FITC-labeled anti-CD4 antibody, and PE-labeled tetramer (90 nM). Aqua (−) APC-Cy7 (−) pacific blue (+) FITC (+) PE (+) cells were sorted as tetramer (+) cells. (B) Tetramer (−) T cells were further stained with PE-labeled dodecamer (90 nM). Aqua (−) APC-Cy7 (−) pacific blue (+) FITC (+) PE (+) cells were sorted as tetramer (−) dodecamer (+) cells. (C–F) Cytokine analysis by CyTOF. Tetramer (+) T cells and tetramer (−) dodecamer (+) T cells were stimulated by either 1 μM MCC peptide and CH27 cells (C and D) or PMA + Ionomycin (E and F). Stimulated cells were stained with a metal-labeled antibody mixture (Materials and Methods) and analyzed by CyTOF. The expression of IFN-γ, PRF, GZMB, IL-10, IL-2, IL-4, TNF-α, IL-6, and IL-17 in the same T cells are shown in each panel.
Discussion
Peptide-MHC tetramers have had a major impact on the analysis of T cells in a wide variety of applications (26). Although various pMHC multimers have been developed, such as dimers, pentamers, octamers, dextramers, and polymers (13, 14), tetramers remain the most common reagents for the profiling of antigen-specific T cells, mainly due to their simple structure, relatively high binding avidity, and low nonspecific staining. The development of T-cell enrichment methods in conjunction with advances in flow cytometry has further enabled the study of antigen-specific T cells using pMHC tetramers (13). However, tetramer technology is not perfect. The performance of tetramers has been less than adequate in the detection of antigen-specific CD4+ T cells, low-affinity αβ T cells, and rare γδ T cells, especially when analyzed by CyTOF (13, 14). In these cases, the binding avidity of tetramers is probably insufficient. The maximum valency of a tetramer is four, but it is likely that only three would be able to bind TCRs on the cell surface at any one time because of the fourfold symmetry of the streptavidin molecule (27). To increase this sensitivity, we have engineered a pMHC dodecamer, which has a maximum valency of 12 peptide-MHCs. This construct shows excellent sensitivity, high binding avidity, slow dissociation kinetics, and low background staining. It has a relatively simple structure and retains important features of a tetramer. Recent advances in MHC tetramer technology, such as photocleavable peptide exchange (28), tetramer-guided epitope mapping (29), T-cell enrichment (7, 22), antibody stabilization staining (30), and combinatorial staining (5, 6), can be easily used with dodecamer reagents. We can also make dodecamers for nonclassical MHC molecules and non-MHC molecules (13). The dodecamer is compatible with commercially available fluorescent/metal-labeled streptavidin, is easy to manufacture, and has advantages over standard tetramers and other multimers for many applications.
MHC class I tetramers have been successfully applied to the study of CD8+ T cells. However, it has been difficult to apply MHC class II tetramers to the study of CD4+ T cells because it is more difficult to produce MHC class II molecules and the binding of the coreceptor CD4 to MHC class II is very weak (31, 32). To quantify rare naive antigen-specific CD4+ T cells in the T-cell repertoire, Jenkins and colleagues used an enrichment technique (7) initially developed by Wucherpfennig and colleagues (22). This gave important insights into a critical part of the repertoire (7). For cells from all sources, the greatly enhanced stability of dodecamers will make the enrichment protocol more efficient (Fig. 4A) in capturing low-frequency and low-affinity CD4+ T cells. For cells from transgenic mice, our dodecamer technology can even bypass these very demanding cell enrichment steps and directly stain both monoclonal and polyclonal antigen-specific CD4+ naive T cells (Figs. 2 and 3), CD3+TCR+ thymocytes (Fig. S4 and Fig. S6B), and even CD4+ CD8+ double-positive thymocytes (Fig. 5) from a large pool of different cells without any T-cell–enriching steps. Clearly, the high-binding valency of dodecamers compensates for the low affinity of TCRs and the weak binding of the CD4 coreceptor. Dodecamers also have significant advantages over most other multimers such as dextramers and QD multimers. Compared with dextramers, dodecamers have only a modest staining advantage (Fig. 2 E and F) but less nonspecific binding (Fig. S8). Compared with QD multimers, dodecamers are superior in both staining intensity and background (Fig. 2 G and H).
We have successfully used dodecamers to stain antigen-specific γδ T cells and human CD4+ and CD8+ αβ T cells with significantly improved sensitivity over tetramers in flow cytometry (Fig. 4 and Figs. S2, S7, and S8). Dodecamers also showed significant advantages over tetramers in CyTOF applications (Fig. 6 A and B). Furthermore, using FACS we have purified low-affinity T cells by dodecamers that were missed by tetramers (Fig. S10 A and B). The low-affinity tetramer (−) dodecamer (+) T cells showed comparably strong cytokine expression as the high-affinity tetramer (+) T cells in a CyTOF-based multiplex cytokine analysis (Fig. 6 C and D and Fig. S10 C–F). These data highlight the functional importance of lower-affinity T cells that escape detection by tetramers. These experiments demonstrate the great potential of dodecamers in the analysis of antigen-specific T cells in both flow cytometry and mass cytometry.
Because pMHC dodecamers can effectively identify functionally important, low-affinity T cells missed by tetramers (Fig. 6 C and D), we also think that pMHC dodecamers will be applied to the studies of other low-affinity T cells, such as those that typically predominate in tumor-specific responses and autoimmune diseases, which escape detection by traditional tetramer staining (13, 14). Recently, a similar construct (33) was reported that was able to stimulate T-cell activation, similar to what has been described previously for tetrameric reagents (13, 14). Therefore, we expect that the dodecamers described here can also be used to manipulate T-cell responses in vivo and in vitro and to develop dodecamer-based targeted immunotherapy. Other variations along these lines might involve putting two different pMHCs on one construct or mixing biotinylated costimulatory molecules together with specific pMHCs.
Materials and Methods
Engineering of a Scaffold Protein.
A single cysteine was added at the C terminus of an inactive streptavidin subunit that has no biotin-binding site using a site-directed mutagenesis kit (Agilent) to create a scaffold protein monomer. Scaffold protein monomer was expressed in BL21 Escherichia coli as described (18) and refolded to form a tetrameric scaffold protein tagged with four cysteines, which was purified by fast protein liquid chromatography (FPLC). The four cysteines on a tetrameric scaffold protein were biotinylated using maleimide chemistry with EZ-Link BMCC-Biotin (Thermo Scientific). Briefly, cysteine-tagged scaffold protein was treated with 10 mM Tris(2-carboxyethyl)phosphine for 10 min and followed by incubation with BMCC-Biotin at a 1:100 molar ratio overnight at room temperature. Excess BMCC-Biotin was removed using 7K MWCO Zeba Spin Desalting Columns twice (Thermo Scientific). The biotinylation efficiency was tested using a Pierce Fluorescence Biotin Quantitation Kit (Thermo Scientific), and it was found that the biotin/scaffold protein ratio was ∼3 in a typical experiment. The biotinylation efficiency can be further improved by optimizing experimental conditions. Biotinylated scaffold protein was further purified by FPLC and then analyzed by SDS/PAGE.
Generation of pMHC Dodecamers and Other Multimers.
In most cases, to perform simple cell-staining experiments, biotinylated scaffold protein was mixed and incubated with fluorescently labeled streptavidin at a molar ratio of 1:4 for 1 h at room temperature, followed by incubation with biotinylated pMHC at a molar ratio of 1:12 for an additional hour at room temperature. Such a process is straightforward and easy to use; however, it led to the formation of a mixture of multimers with different binding valencies due to the uncompleted biotinylation of scaffold proteins and the binding kinetics of biotin–streptavidin interactions. In some experiments [such as dissociation assays (Fig. 3A)], to make dodecamers, biotinylated scaffold protein was mixed with fluorescently labeled streptavidin at a molar ratio of 1:20, and the pentameric molecular complex was purified using FPLC before the addition of biotinylated pMHC. To generate QD multimers, streptavidin-conjugated QD605 was incubated with biotinylated pMHC at a 1:28 molar ratio for 1 h at room temperature. To generate pMHC dextramers, fluorescently labeled streptavidin was incubated with biotinylated pMHC monomers at a molar ratio of 3.5:1. After 1 h at room temperature, biotinylated dextramer was added at a molar ratio of 60:1, and the mixture was incubated for 1 h or longer at room temperature before use. PE-labeled streptavidin was purchased from eBioscience and Alexa-labeled streptavidin was purchased from Thermo Fisher Scientific.
Cells.
TCRα/TCRβ transgenic 5C.C7 mice (B10.A/Rag2 KO background) and TCRβ transgenic 5C.C7 (B10.BR background) mice were bred and maintained in the Research Animal Facility at Stanford University’s Department of Comparative Medicine Animal Facility (protocol 3540) in accordance with guidelines of the National Institutes of Health. All mouse studies were approved by the Stanford Administrative Panel of Laboratory Animal Care (Stanford University, Stanford, CA). Mouse splenocytes and thymocytes were obtained and used for experiments. γδ TCR-expressing 58α−β− cells and γδ negative 58α−β− cells have been described (34). PBMCs from nonidentifiable healthy blood donors were isolated by density centrifugation of leukocyte reduction shuttles from the Stanford Blood Center according to Institutional Review Board (IRB) (“Minimal Risk Research Related Activities at Stanford Blood Center,” Protocol 13942) protocol. Informed consent was obtained. For the purposes of pMHC multimer staining experiments, HLA-A2–positive donor samples were typed and identified by the Stanford Blood Center. These samples were also serotyped for CMV infection status. HLA-DR4–positive donors samples were typed by the Children’s Hospital Oakland Research Institute. PBMCs were cryopreserved and stored in liquid nitrogen before use.
Flow Cytometry.
Mouse splenocytes and thymocytes were first incubated with 1% anti-CD16/CD32 Fc blocker, 10% (vol/vol) rat serum, and 10% (vol/vol) syrian hamster serum on ice for 30 min. Mouse cells were further stained with pMHC dodecamers, tetramers, dextramers, or QD605-based multimers and costained with an antibody mixture containing FITC-labeled anti-CD8, APC-Cy7–labeled anti-CD4, Alexa700-labeled anti-CD3, APC-labeled anti-TCRβ, pacific blue-labeled anti-CD19, anti-CD11b, anti-CD11c, anti-Gr1, anti-F4/80, and aqua live/dead cell stain. I-Ek molecules loaded with MCC (ANERADLIAYLKQATK) or CLIP (PVSKMRMATPLLMQA) peptides were used for the staining of mouse 5C.C7 transgenic CD4+ T cells. Frozen human PBMCs were thawed and rested overnight (5). To stain human CD8+ T cells, human PBMCs were incubated with pMHC dodecamers or tetramers and costained with an antibody mixture including FITC-labeled anti-CD8α, Alexa700-labeled anti-CD3, QD605-labeled anti-CD45RA, brilliant violet-labeled anti-CD62L, PerCP/Cy5.5-labeled anti-CD4, anti-CD19, anti-CD33, anti-CD14, and aqua live/dead cell stain. HLA-A2 molecules loaded with CMV (NLVPMVATV), Flu (GILGFVFTL), and HIV (SLYNTVATL) peptides were used for the staining of human antigen-specific CD8+ T cells. To profile rare influenza-specific CD4+ T cells, cells stained with pMHC dodecamers or tetramers were magnetically enriched (7) and costained with a mixture of antibodies containing PE/Texas Red-labeled anti-CD4, PE/Cy5 non-CD4 exclusion markers, and near-IR live/dead cell stain. HLA-DR4 molecules loaded with HA 306–318 (GGPKYVKQNTLKLAT) and VIM 61–80 with arginine to citrulline substitution (YAT[Cit]SSAV[Cit]L[Cit]SSVPGV[Cit]LL; Cit: Citrulline) peptides were used for the staining of human antigen-specific CD4+ T cells. Most cells were stained at 4 °C although some were stained at other temperatures. To test the effect of temperature on pMHC multimer staining, mouse cells were incubated for 1 h in a thermocycler at the indicated temperatures. In experiments with latrunculin A, mouse cells were pretreated with latrunculin A (0.1–10 μM) for 1 h and then stained with pMHC tetramers or dodecamers and the mouse antibody mixture for 1 additional hour at room temperature (22 °C). Cells were washed and analyzed on a BD LSR II flow cytometer. To measure the dissociation rates of tetramers and dodecamers, 5C.C7 splenocytes were incubated with a 100-nM Alexa488-labeled tetramer or dodecamer for 1 h at 22 °C in the presence of PE-labeled anti-CD8, APC-Cy7–labeled anti-CD4, Alexa700-labeled anti-CD3, pacific blue-labeled anti-CD19, anti-CD11b, anti-CD11c, anti-Gr1, anti-F4/80, and aqua live/dead cell stain. Cells were pelleted and resuspended in 200 μL FACS buffer in the presence of 100 μg/mL 14.4.1 anti-I-Ek MHC antibody on ice (35). The fluorescence intensities of 10,000 cells were measured at different time points after adding the anti–I-Ek MHC-blocking antibody. Flow cytometry data were analyzed by FlowJo software. A first-order decay kinetic model was used to fit the mean fluorescence intensities at different time points and to obtain the off-rate, koff, and the half-life, t1/2 (34).
Mass Cytometry.
Biotinylated I-Ek was refolded with either MCC peptide or CLIP peptide. For dodecamerization, biotinylated scaffold protein was incubated with metal-labeled streptavidin (1:4 molar ratio) for 1 h followed by incubation with pMHC monomers (1:12 molar ratio) for 1 more hour at room temperature. Tetramers were prepared as described (6). Each pMHC multimer was barcoded with two different metal labels. 5C.C7 mouse splenocytes were incubated with 1% anti-CD16/CD32 Fc blocker, 10% (vol/vol) rat serum, and 10% (vol/vol) syrian hamster serum on ice for 30 min; stained by dodecamers or tetramers for 1 h at room temperature; and then incubated with a metal-labeled antibody mixture including anti-TCRβ (Nd143), anti-CD3 (Sm154), anti-CD4 (Nd146), anti-CD8 (Sm149), anti-CD11b (Eu153), anti-CD11c (Sm152), anti-CD19 (Cd112 and Cd114), anti-Gr1 (Nd145), and anti-F4/80 (Tb159). Splenocytes were washed with CyFACS buffer and stained with metal-labeled live/dead (La139) medium for 20 min on ice (6). Splenocytes were washed and fixed in 2% paraformaldehyde CyPBS for 1 h on ice. After treatment with permeabilization buffer (eBioscience), cells were washed, resuspended in milliQ water, and analyzed by CyTOF.
Multiplex Cytokine Analysis.
Splenocytes were isolated from 5C.C7 single β-chain transgenic mice, and cells were stained with APC-Cy7–labeled anti-CD19, anti-CD11b, anti–Gr-1, and anti-F4/80 antibodies followed by anti-APC magnetic beads for cell selection. Positively selected cells were eluted and stained with aqua live/dead cell stain and pacific blue-labeled anti-TCRβ antibody. Aqua (−) pacific blue (−) APC-Cy7 (+) cells were sorted as antigen-presenting cells. Negatively selected cells were stained with aqua live/dead cell stain, pacific blue-labeled anti-TCRβ antibody, FITC-labeled anti-CD4 antibody, and PE-labeled tetramer (90 nM). Aqua (−) APC-Cy7 (−) pacific blue (+) FITC (+) PE (+) cells were sorted as tetramer (+) T cells (Fig. S10A). The rest of the T cells were further stained with PE-labeled dodecamer (90 nM). Aqua (−) APC-Cy7(−) pacific blue (+) FITC (+) PE (+) cells were sorted as tetramer (−) dodecamer (+) T cells (Fig. S10B). Sorted T cells were expanded in vitro by coculturing with antigen-presenting cells (1:1 ratio), 1 μM MCC peptides, and 100 U/mL IL-2 for 4 d. Expanded T cells were washed and further cultured with 100 U/mL IL-2 for 24 h, followed by cell ficoll and wash. To evaluate the cytokine expression, T cells were stimulated with either 1 μM MCC peptide-pulsed CH27 cells (1:1 ratio) or PMA/Ionomycin in the presence of protein transport inhibitor mixture (eBioscience) for 4 h. Stimulated cells were intracellularly stained with a metal-labeled antibody mixture including TCRβ (Pr141), IL-10 (Nd150), IL-2 (Gd155), IL-4 (Gd157), TNF-α (Dy162), IL-6 (Dy163), IFN-γ (Ho165), IL-17 (Er166), Granzyme B (GZMB) (Yb171), and Perforin (PRF) (Yb172) antibodies and then analyzed by CyTOF (Fig. S10 C–F).
Acknowledgments
We thank the NIH Tetramer Core Facility at Emory University for providing I-Ek MHC monomers; Allison Nicole Nau, Yuling Wei, and Naresha Saligrama for helping with flow cytometry and mouse experiments; and Nelida Prado for animal care. This work was supported by NIH Grants U19 AI090019 and U19 AI057229 (to M.M.D.); the Howard Hughes Medical Institute (M.M.D.); and NIH Grant K99AI106941 (to J.H.).
Footnotes
Conflict of interest statement: J.H. and M.M.D. are inventors on a patent application (US Provisional Application No. 62/194,726) that applies to detection, phenotyping, and quantification of cells with multimeric binding reagent.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602488113/-/DCSupplemental.
References
- 1.Davis MM, et al. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 2.Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274(5284):94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 3.Toebes M, et al. Design and use of conditional MHC class I ligands. Nat Med. 2006;12(2):246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 4.Hadrup SR, et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat Methods. 2009;6(7):520–526. doi: 10.1038/nmeth.1345. [DOI] [PubMed] [Google Scholar]
- 5.Newell EW, Klein LO, Yu W, Davis MM. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat Methods. 2009;6(7):497–499. doi: 10.1038/nmeth.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newell EW, et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol. 2013;31(7):623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moon JJ, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27(2):203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolton G, et al. Comparison of peptide-major histocompatibility complex tetramers and dextramers for the identification of antigen-specific T cells. Clin Exp Immunol. 2014;177(1):47–63. doi: 10.1111/cei.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallet-Designe VI, et al. Detection of low-avidity CD4+ T cells using recombinant artificial APC: Following the antiovalbumin immune response. J Immunol. 2003;170(1):123–131. doi: 10.4049/jimmunol.170.1.123. [DOI] [PubMed] [Google Scholar]
- 10.Guillaume P, et al. Soluble major histocompatibility complex-peptide octamers with impaired CD8 binding selectively induce Fas-dependent apoptosis. J Biol Chem. 2003;278(7):4500–4509. doi: 10.1074/jbc.M208863200. [DOI] [PubMed] [Google Scholar]
- 11.Batard P, et al. Dextramers: New generation of fluorescent MHC class I/peptide multimers for visualization of antigen-specific CD8+ T cells. J Immunol Methods. 2006;310(1-2):136–148. doi: 10.1016/j.jim.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Chattopadhyay PK, et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12(8):972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 13.Davis MM, Altman JD, Newell EW. Interrogating the repertoire: Broadening the scope of peptide-MHC multimer analysis. Nat Rev Immunol. 2011;11(8):551–558. doi: 10.1038/nri3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wooldridge L, et al. Tricks with tetramers: How to get the most from multimeric peptide-MHC. Immunology. 2009;126(2):147–164. doi: 10.1111/j.1365-2567.2008.02848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabatino JJ, Jr, Huang J, Zhu C, Evavold BD. High prevalence of low affinity peptide-MHC II tetramer-negative effectors during polyclonal CD4+ T cell responses. J Exp Med. 2011;208(1):81–90. doi: 10.1084/jem.20101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schild H, et al. The nature of major histocompatibility complex recognition by gamma delta T cells. Cell. 1994;76(1):29–37. doi: 10.1016/0092-8674(94)90170-8. [DOI] [PubMed] [Google Scholar]
- 17.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36(1):142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howarth M, et al. A monovalent streptavidin with a single femtomolar biotin binding site. Nat Methods. 2006;3(4):267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newell EW, et al. Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k) J Immunol. 2011;186(10):5823–5832. doi: 10.4049/jimmunol.1100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whelan JA, et al. Specificity of CTL interactions with peptide-MHC class I tetrameric complexes is temperature dependent. J Immunol. 1999;163(8):4342–4348. [PubMed] [Google Scholar]
- 21.Huppa JB, Davis MM. T-cell-antigen recognition and the immunological synapse. Nat Rev Immunol. 2003;3(12):973–983. doi: 10.1038/nri1245. [DOI] [PubMed] [Google Scholar]
- 22.Day CL, et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest. 2003;112(6):831–842. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janeway CA, Travers P, Walport M, Capra JD. Immunobiology: The Immune System in Health and Disease. 4th Ed. Elsevier Science Ltd/Garland Publishing; New York: 1999. pp. 241–257. [Google Scholar]
- 24.Baldwin KK, Trenchak BP, Altman JD, Davis MM. Negative selection of T cells occurs throughout thymic development. J Immunol. 1999;163(2):689–698. [PubMed] [Google Scholar]
- 25.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doherty PC. The tetramer transformation. J Immunol. 2011;187(1):5–6. doi: 10.4049/jimmunol.1101297. [DOI] [PubMed] [Google Scholar]
- 27.Boniface JJ, et al. Initiation of signal transduction through the T cell receptor requires the multivalent engagement of peptide/MHC ligands [corrected] Immunity. 1998;9(4):459–466. doi: 10.1016/s1074-7613(00)80629-9. [DOI] [PubMed] [Google Scholar]
- 28.Grotenbreg GM, et al. Discovery of CD8+ T cell epitopes in Chlamydia trachomatis infection through use of caged class I MHC tetramers. Proc Natl Acad Sci USA. 2008;105(10):3831–3836. doi: 10.1073/pnas.0711504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reijonen H, Kwok WW. Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes. Methods. 2003;29(3):282–288. doi: 10.1016/s1046-2023(02)00350-x. [DOI] [PubMed] [Google Scholar]
- 30.Tungatt K, et al. Antibody stabilization of peptide-MHC multimers reveals functional T cells bearing extremely low-affinity TCRs. J Immunol. 2015;194(1):463–474. doi: 10.4049/jimmunol.1401785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong Y, Kern P, Chang H, Reinherz E. T cell receptor binding to a pMHCII ligand is kinetically distinct from and independent of CD4. J Biol Chem. 2001;276(8):5659–5667. doi: 10.1074/jbc.M009580200. [DOI] [PubMed] [Google Scholar]
- 32.Huppa JB, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463(7283):963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fairhead M, et al. SpyAvidin hubs enable precise and ultrastable orthogonal nanoassembly. J Am Chem Soc. 2014;136(35):12355–12363. doi: 10.1021/ja505584f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng X, et al. γδ T cells recognize a microbial encoded B cell antigen to initiate a rapid antigen-specific interleukin-17 response. Immunity. 2012;37(3):524–534. doi: 10.1016/j.immuni.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huppa JB, Gleimer M, Sumen C, Davis MM. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat Immunol. 2003;4(8):749–755. doi: 10.1038/ni951. [DOI] [PubMed] [Google Scholar]