Significance
The results from the reported studies indicate that the human lagging strand polymerase, pol δ, maintains a loose association with the sliding clamp, proliferating cell nuclear antigen (PCNA), while replicating and rapidly dissociates on stalling, leaving PCNA behind on the DNA. This behavior has profound implications on lagging strand synthesis as it limits the extent of processive DNA synthesis on undamaged DNA and promotes the rapid dissociation of pol δ on stalling at a replication-blocking lesion. This challenges the accepted models for polymerase recycling and exchange on the lagging strand and instead suggests passive mechanisms for the human system. These studies provide valuable insight for future experiments in the fields DNA replication, DNA repair, and DNA damage tolerance.
Keywords: lagging strand, stability, PCNA, DNA polymerase delta, translesion DNA synthesis
Abstract
In eukaryotes, DNA polymerase δ (pol δ) is responsible for replicating the lagging strand template and anchors to the proliferating cell nuclear antigen (PCNA) sliding clamp to form a holoenzyme. The stability of this complex is integral to every aspect of lagging strand replication. Most of our understanding comes from Saccharomyces cerevisae where the extreme stability of the pol δ holoenzyme ensures that every nucleobase within an Okazaki fragment is faithfully duplicated before dissociation but also necessitates an active displacement mechanism for polymerase recycling and exchange. However, the stability of the human pol δ holoenzyme is unknown. We designed unique kinetic assays to analyze the processivity and stability of the pol δ holoenzyme. Surprisingly, the results indicate that human pol δ maintains a loose association with PCNA while replicating DNA. Such behavior has profound implications on Okazaki fragment synthesis in humans as it limits the processivity of pol δ on undamaged DNA and promotes the rapid dissociation of pol δ from PCNA on stalling at a DNA lesion.
During S-phase of the cell cycle, genomic DNA must be faithfully copied in a short period. Replicative DNA polymerases (pols) alone are distributive and must anchor to ring-shaped sliding clamps to achieve the high degree of processivity required for efficient DNA replication. The highly conserved toroidal structure of sliding clamps has a central cavity large enough to encircle double-stranded DNA (dsDNA) and slide freely along it. Thus, such an association effectively tethers the pol to DNA, substantially increasing the extent of continuous replication. The eukaryotic sliding clamp, proliferating cell nuclear antigen (PCNA), is trimer of identical subunits aligned head-to-tail, forming a ring with two structurally distinct faces. Each subunit consists of two independent domains connected by an interdomain connecting loop (IDCL). The “front” face of the homotrimeric PCNA ring contains all IDCLs and is a platform for interaction with the eukaryotic replicative pols, ε and δ, which are responsible for the faithful replication of the leading and lagging strands, respectively (1, 2). Specifically, the well-conserved PCNA-interacting peptide (PIP) box within replicative pols makes extensive contact with an IDCL of PCNA and displays conserved residues that “plug” into the proximal hydrophobic patches. The amino acid sequence of a canonical PIP box is QXXhXXaa, where X represents any amino acid, h is a hydrophobic residue (usually L, I, or M), and a is an aromatic residue (usually F or Y) (3).
Unlike the leading strand, the lagging strand is synthesized discontinuously in short Okazaki fragments that are processed and ligated together to form a continuous strand (4). In eukaryotes, each Okazaki fragment is initiated by the bifunctional DNA pol α/primase complex that lays down cRNA/DNA hybrid primers every 100–250 nucleotides (nt) on the exposed template for the lagging strand. The intermittent single-stranded DNA (ssDNA) is protected from cellular nucleases by replication protein A (RPA), a ssDNA binding protein that also prevents formation of alternative DNA structures. The clamp loader, replication factor C (RFC), recognizes these hybrid primers abutted by RPA and loads PCNA onto each such that the front face of the clamp is oriented toward the 3′ end of the nascent primer/template (P/T) junction where DNA synthesis will initiate. An incoming pol δ subsequently captures the loaded PCNA ring, forming a holoenzyme, and DNA synthesis initiates (2, 5).
The stability of the lagging strand holoenzyme is integral to various aspects of Okazaki fragment synthesis. For eukaryotes, most of our understanding comes from studies in Saccharomyces cerevisae, where the three-subunit pol δ is extremely stable with PCNA on DNA (koff < 2 × 10−3 s−1, t1/2 > 5 min). Once DNA synthesis is initiated from a nascent primer, the dramatically slow koff ensures every nucleotide within a given Okazaki fragment is faithfully duplicated before dissociation. On the other hand, such high stability necessitates an active mechanism for displacement of pol δ once DNA synthesis stops (6, 7). This situation arises when a pol δ holoenzyme encounters either the 5′ RNA end of a downstream Okazaki fragment (pol recycling) or distortions to the native sequence that it cannot accommodate (pol exchange), such as common byproducts of UV radiation exposure (8). Pol recycling allows the scarce pol δ to be reused during S-phase, whereas pol exchange permits a specialized pol to bind to PCNA and synthesize past the offending damage [translesion DNA synthesis (TLS)] so that pol δ may resume synthesis (9–12). However, studies on the human pol δ holoenzyme are lacking, and hence, the mechanisms by which polymerase recycling and exchange occur are unknown. To gain insight, we designed a unique kinetic assay to measure the stability of the pol δ holoenzyme. Surprisingly, the results indicate that human pol δ maintains a loose association with PCNA. Such behavior has profound implications on lagging strand synthesis as it limits the extent of processive DNA synthesis and promotes the rapid dissociation of pol δ from PCNA on stalling.
Results
Monitoring a Single Turnover of Primer Extension by Human pol δ.
Human pol δ is comprised of four subunits: three accessory subunits (p50/POLD2, p66/POLD3, and p12/POLD4) and a large catalytic subunit (p125/POLD1) that contains both DNA polymerase and exonuclease domains. The p125, p50, and p66 subunits are homologs of the S. cerevisae pol3, pol31, and pol32 subunits, respectively. A homolog of the human p12 subunit is absent in S. cerevisae. Three of the four human pol δ subunits can interact directly with PCNA. The p66 accessory subunit contains a canonical PIP box, whereas the p125 catalytic subunit and the p12 accessory subunit both contain noncanonical PIP domains where the conserved Q residue has been replaced with an alternative amino acid (13–21). The homotrimeric PCNA sliding clamp contains three identical binding sites for PIP box-containing proteins and, hence, human pol δ may simultaneously bind all three subunits within a given PCNA trimer, similar to that observed in S. cerevisae (22). Indeed, sequential removal of the p12 and p66 accessory subunits from the human pol δ assembly reduced the extent of PCNA-dependent DNA synthesis in a stepwise manner, suggesting that all PCNA-interacting subunits are required to form a holoenzyme with optimal DNA synthetic activity (23). In all experiments discussed herein, only the complete, four-subunit human pol δ complex was used. Furthermore, pol exchange in eukaryotes involves the conjugation of single ubiquitin moieties (i.e., monoubiquitination) to lysine residues (K164) of PCNA residing at a stalled P/T junction. This posttranslational modification (PTM) is essential for optimal TLS activity in mammalian cells (24), but its role in pol exchange is under intense debate. To gain insight into a potential role of this PTM involving the lagging strand pol δ, we synthesized monoubiquitinated PCNA [referred to herein as (Ub)3-PCNA] that contains a single ubiquitin moiety on K164 of each monomer within a homotrimeric clamp ring (25). The conjugation of ubiquitin to PCNA has no effect on the interaction of PCNA with RFC (Fig. S1B) or on the ability of RFC to load PCNA onto DNA (Fig. S2), in agreement with observations from similar studies in S. cerevisiae (26). Thus, any observed effect on the DNA synthetic activity of an assembled pol δ holoenzyme will not be attributable to the amount of (Ub)3-PCNA loaded onto DNA.
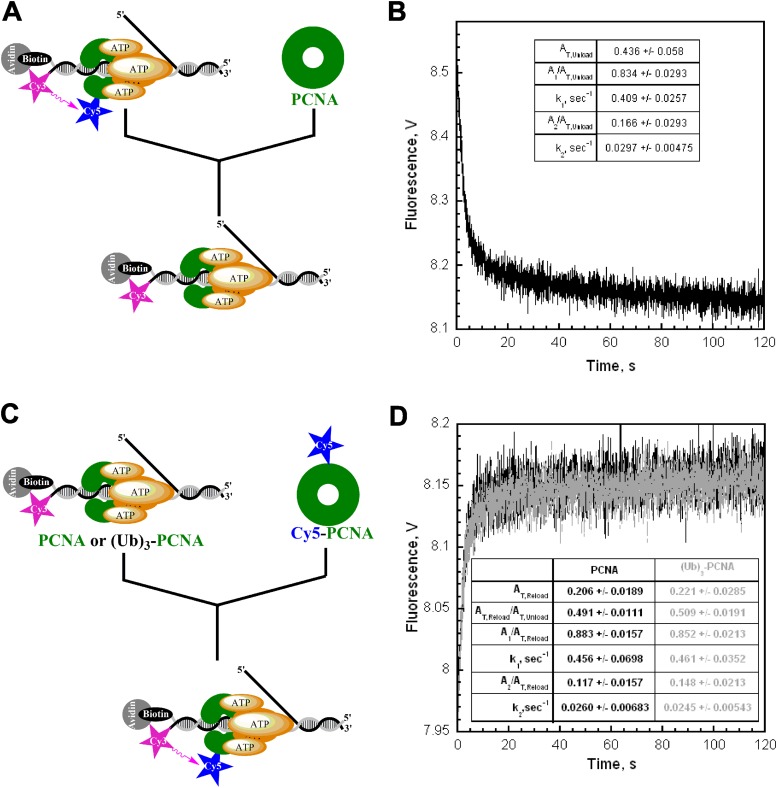
Fig. S1.
Characterization of human proteins used in this study. (A) Exonuclease activities of WT and exonuclease-deficient human pol δ. Assays were performed in duplicate. 5′-Labeled 29-mer primer alone (ssDNA) or annealed in the P29/Bio62 substrate was incubated with the indicated pol δ enzymes. Average activities and SDs from duplicate reactions are presented. *At the initial time point (1.5 min), all of the substrate was degraded, setting a lower limit for exonuclease activity of WT human pol δ at 200 nM/1.5 min = 133.33 nM/min. Thus, the D402A mutation reduces the exonuclease activity of human pol δ at least 14.4-fold on an optimal substrate for exonuclease activity. Exonuclease activity was not visible on the P29/Bio62 DNA substrate with the D402A mutant. (B) Activities of PCNA clamps in stimulating human RFC ATPase activity. The ATPase activity was determined at 25 °C in an assay solution containing 125 nM RFC, 125 nM PCNA (WT or monoubiquitinated, Ub), 125 nM P/T DNA (500 nMNeutravidin), 1 mM ATP, 3 mM phosphoenolpyruvate, 200 mM NADH, and 6–8 U phosphoenolpyruvate kinase-lactate dehydrogenase mix. For each condition, the initial rates of ATP hydrolysis are reported as an average of three independent experiments ± SD. (Ub)3-PCNA stimulated the DNA-dependent ATPase activity of RFC to the same extent as WT PCNA, indicating a fully functional sliding clamp ring.
Fig. S2.
Monitoring the loading and unloading of PCNA by FRET. (A) Schematic representation of experimental procedure for B. (B) Cy5-PCNA (100 nM) loaded onto Cy3-P/T DNA (200 nM) by RFC (100 nM) in the presence of 1 mM ATP was mixed with 2 µM unlabeled PCNA chase and the decrease in FRET signal was followed. The unloading traces were fit to a double-exponential equation. The rate constants and their relative amplitudes are reported. The overall amplitude (AT,Unload) represents the total amount of PCNA loaded onto and unloaded from DNA. (C) Schematic representation of experimental procedure for D. (D) Unlabeled PCNA (black trace, 100 nM) was loaded onto Cy3-P/T DNA (200 nM) by RFC (100 nM) in the presence of 1 mM ATP and mixed with Cy5-PCNA (100 nM) at a 1:1 ratio of labeled to unlabeled PCNA, and the increase in FRET signal was followed. The loading traces were fit to a double-exponential equation, and the rate constants and their relative amplitudes are reported. Under these conditions, the only source of RFC is from the preassembled RFC•ATP•PCNA•Cy3-P/T DNA complex. All unlabeled PCNA is actively unloaded from Cy3-P/T DNA by RFC and rapidly exchanges with Cy5-PCNA. Thus, the rate of Cy5-PCNA loading is entirely rate limited by dissociation of the RFC•PCNA complex from Cy3-P/T DNA and, hence, the rate of appearance of the FRET signal is the same as the rates observed for the disappearance of the FRET signal in B. Furthermore, the overall amplitude of the FRET increase (AT,Reload) is exactly one-half the overall amplitude for the FRET decrease in B (AT,Unload) due to the complete unloading of PCNA from Cy3-P/T DNA by RFC (5). The same behavior was observed with unlabeled (Ub)3-PCNA (gray trace), demonstrating that the conjugation of ubiquitin to PCNA has no effect on the dynamic loading/unloading mechanism of human RFC; RFC loaded (Ub)3-PCNA onto DNA and, in the absence of pol δ, dissociated back into solution taking all loaded (Ub)3-PCNA along with it. Together with Fig. S1B, this demonstrates that the conjugation of ubiquitin to PCNA has no effect on its interaction with RFC and, hence, the ability of RFC to load PCNA onto DNA. Thus, any observed effect on the DNA synthetic activity of the pol δ holoenzyme will not be attributable to the amount of (Ub)3-PCNA loaded onto DNA.
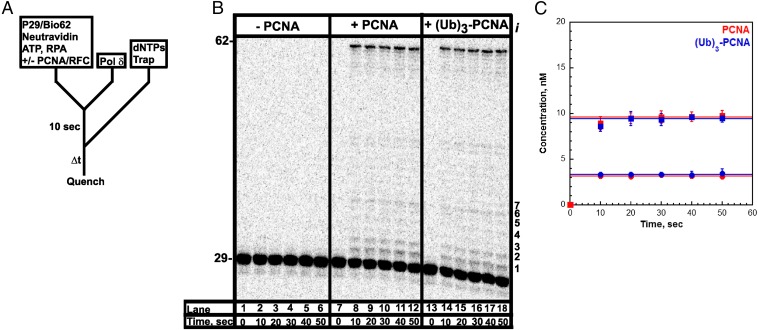
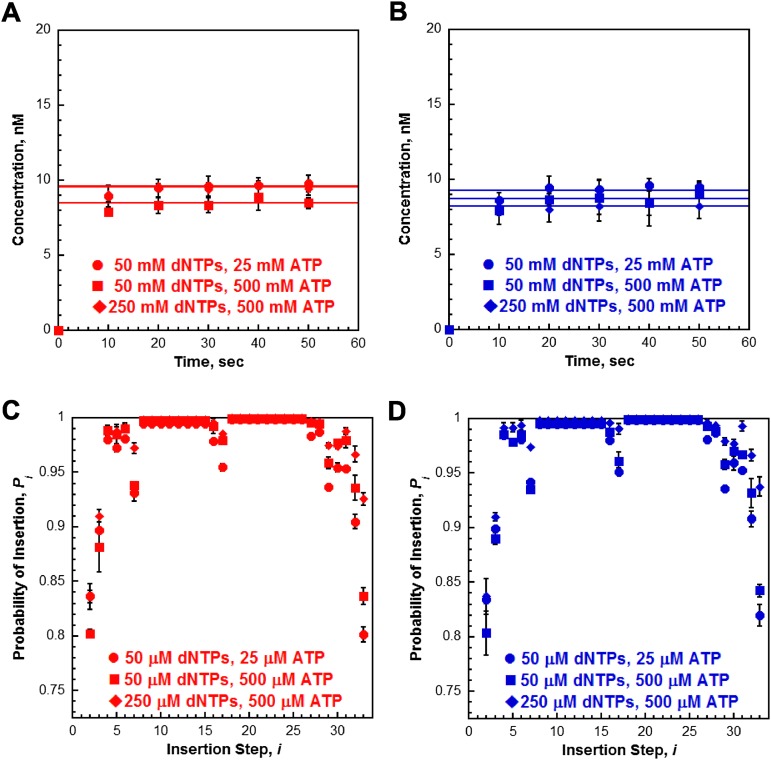
To probe the stability of the pol δ holoenzyme, a holoenzyme must be distinguished from pol δ alone. We used a DNA substrate (Fig. 1) that mimics a nascent P/T junction on the lagging strand to monitor primer extension by pol δ during a single DNA binding event (Fig. 2A). Under the conditions used (physiological pH, ionic strength, and dNTP concentration), pol δ alone did not extend the primer (Fig. 2B). Increasing the preincubation time did not promote primer extension, nor did increasing the concentration of dNTPs (Fig. S3). These observations are in agreement with the inability of pol δ alone to form a stable complex with a native P/T DNA substrate (27) and suggest that the DNA binding affinity of human pol δ alone is dramatically low such that dissociation from the DNA substrate is much faster than dNTP binding and/or insertion. Primer extension was only observed in the presence of RFC and a PCNA, either unmodified or monoubiquitinated PCNA. Thus, assembly of a pol δ holoenzyme is indicated by DNA synthesis. The activities of the assembled holoenzymes were identical (Fig. 2C and Table 1). Synthesis of the full-length product was complete within the first time point (10 s), yet primer extension continued up to at most 20 s. This behavior suggests there are at least two populations of holoenzymes that cannot be interconverted: a fast, processive population (kpol ∼ 100 s−1, discussed later) and a slow population (kpol ≤ 0.485 s−1; SI Text). It is highly unlikely the latter reflects holoenzymes lacking the smallest pol δ subunit (p12), as exclusion of p12 only decreases kpol 4.6-fold (28). After 20 s, all DNA synthesis by the assembled pol δ holoenzymes has ceased as the total amount of primer extension products remains constant, and the relative abundance of each primer extension product does not change (Fig. S6). The primer extension products plateau at a concentration (i.e., the amplitude) less than the concentration of the DNA substrate, indicating a single turnover of DNA synthesis has been monitored. Hence, the amplitude is equal to the concentration of assembled holoenzymes that are competent for DNA synthesis. Equivalent values were obtained for holoenzymes assembled with either PCNA or (Ub)3-PCNA, indicating that (Ub)3-PCNA has no effect on the assembly of the pol δ holoenzyme.
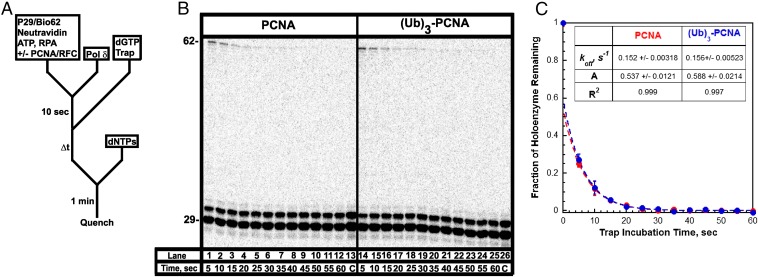
Fig. 1.
Sequence of the biotinylated primer/template P29/Bio62. The size of the double-stranded P/T region (29 bp) is in agreement with the size of an initiating hybrid P/T and the requirements for assembly of a human pol δ holoenzyme by RFC (5, 70). The biotin attached to the 3′-end of the template strand was prebound to neutravidin, preventing loaded PCNA from sliding off the 5′ end of the primer. The single-stranded DNA (ssDNA) adjacent to the 3′ end of the P/T junction was prebound with excess RPA. The size of this region (33 nt) is consistent with the size of ssDNA covered by a single RPA molecule (22–30 nt) (71). To monitor extension of the primer by pol δ within a single binding encounter, the 29-mer primer was 32P-end-labeled before annealing and reactions were carried out in the presence of a passive DNA trap. In all experiments, the trap is unlabeled substrate lacking a biotin tag and containing a 3′-dideoxy-terminated primer (referred to as 29ddC/62).
Fig. 2.
Assembly of a pol δ holoenzyme on the P29/Bio62 DNA substrate is characterized by primer extension. (A) Schematic representation of the experiment performed to monitor primer extension by a pol δ holoenzyme. The pol δ holoenzyme was assembled on the P29/Bio62 by the addition of RPA, PCNA, ATP, RFC, and pol δ in succession. PCNA was either unmodified (PCNA) or monoubiquitinated [(Ub)3-PCNA]. Synthesis by assembled holoenzymes was initiated by the simultaneous addition of dNTPs and a passive DNA trap. The experiment for pol δ alone was performed identically except for the omission of PCNA and RFC. (B) 16% denaturing sequencing gel of the primer extension products for pol δ alone (lanes 1–6) and pol δ holoenzymes assembled with either PCNA (lanes 7–12) or (Ub)3-PCNA (lanes 13–18). The sizes of the substrate and full-length product are indicated on the left and the insertion step (i) for each primer extension product up to 36 nt in length (i = 7) is indicated on the right. (C) Quantification of the primer extension products (total, ■) and the fully extended primer (●) for holoenzymes assembled with either PCNA or (Ub)3-PCNA. Each point represents the average ± SD of three independent experiments. For the full-length 62-mer product, all data points were fit to a flat line where the y-intercept reflects the amplitude. The total amount of primer extension products increased up to at most 20 s and plateaued thereafter. Such behavior was independent of the preincubation time (Fig. S3) and was not attributable to limiting concentrations of ATP and/or dNTPs (Fig. S4 A and B) or mis-incorporation of the first nucleotide (Fig. S5). For primer extension products, data points after t = 10 s were fit to a flat line where the y-intercept reflects the amplitude. Values are reported in Table 1.
Fig. S3.
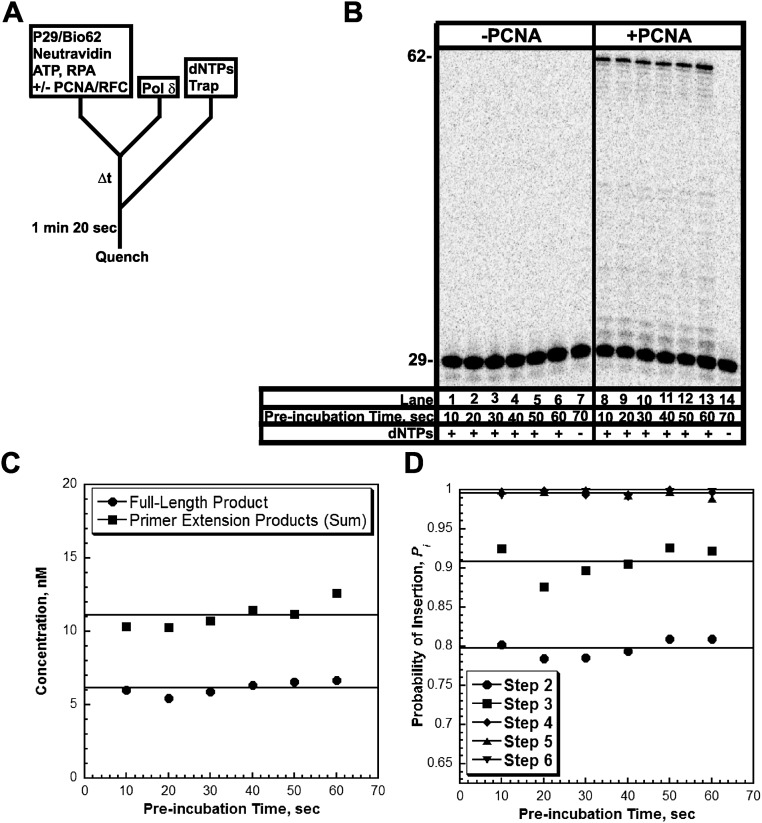
Formation of the pol δ holoenzyme is independent of preincubation time. (A) Schematic representation of experiment performed to monitor extent of holoenzyme formation with increasing preincubation time. (B) 16% denaturing sequencing gel of the primer extension products for pol δ alone (lanes 1–7) and a pol δ holoenzyme assembled with PCNA (lanes 8–14) in the presence of 250 µM of each dNTP and 0.5 mM ATP. The size of the substrate and full-length product are indicated on the left. As a control for each condition, an aliquot was removed after 70 s of preincubation and quenched before the addition of dNTPs and trap (lanes 7 and 14). (C) Quantification of the primer-extension products (total, ■) and the fully-extended primer (●) for holoenzymes assembled with PCNA. Data points were fit to a flat line. As can be seen, the ratio of full-length product to the total amount of primer extension products was maintained when the preincubation time was increased. This demonstrates that the two populations of pol δ holoenzymes are independent of the preincubation time and are not interconvertible. (D) Quantification of the probability of insertion for steps 2 through 6 for holoenzymes assembled with PCNA. Data points were fit to a flat line.
Table 1.
Amplitudes calculated from single turnover primer extension assays described in Fig. 2
| 50 µM dNTPs | 250 µM dNTPs | |||||
| 0.025 mM ATP | 0.5 mM ATP | 0.5 mM ATP | ||||
| WT-PCNA | (Ub)3-PCNA | WT-PCNA | (Ub)3-PCNA | WT-PCNA | (Ub)3-PCNA | |
| Primer extension products (total), nM | 9.63 ± 0.061 | 9.47 ± 0.062 | 8.50 ± 0.129 | 8.74 ± 0.109 | 9.54 ± 0.066 | 8.22 ± 0.081 |
| Full-length product, nM | 3.17 ± 0.027 | 3.36 ± 0.031 | 3.81 ± 0.074 | 3.71 ± 0.030 | 5.40 ± 0.0320 | 4.88 ± 0.064 |
Fig. S6.
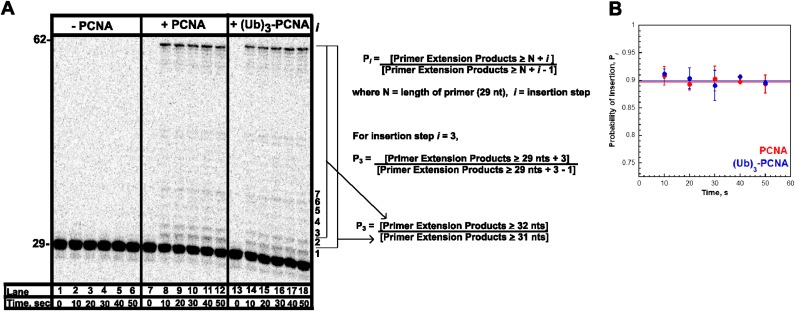
Calculation of the probability of insertion, Pi. (A) 16% denaturing sequencing gel from Fig. 2 of the primer extension products for pol δ alone (lanes 1–6) and pol δ holoenzymes assembled with either PCNA (lanes 7–12) or (Ub)3-PCNA (lanes 13–18). The size of the substrate and full-length product are indicated on the left, and the insertion step (i) for each primer extension product up to 36 nt in length (i = 7) is indicated on the right. At each dNTP insertion step, i, the probability that a pol δ holoenzyme will insert another dNTP, Pi, is equal to the concentration of all primer extension products up to at least length N + i (where N is the length of the primer) divided by the concentration of all primer extension products up to at least length N + i – 1. As an example, for insertion step i = 3, P3 is equal to the concentration of all primer extension products that are least 32 nt in length (N + i = 29 + 3 = 32) divided by the concentration of all primer extension products that are least 31 nt in length (N + i − 1 = 29 + 3 − 1 = 31). This was carried for each time point. As can be seen in B, Pi for i = 3 remains constant after 10 s, reaffirming that only a single turnover of DNA synthetic activity by the pol δ holoenzyme has been monitored. By fitting the data from 20 to 50 s to a flat line, the y-intercept yields an average value of Pi = 0.897 ± 0.0126 for i = 3. Such calculations were carried out for insertion steps i = 2 to i = 33, and the results are depicted in Fig. 3A.
Fig. S5.
Inclusion of only the first dNTP to be inserted does not affect primer extension by a pol δ holoenzyme. Primer extension was monitored as depicted in Fig. S4 except that only dGTP (250 µM) was introduced with the trap. Primer-extension products (total, ●) for holoenzyme assembled with PCNA. Note that primer extension occurs up to 20 s and plateaus thereafter, demonstrating that erroneous dNTP insertion is not responsible for slower primer extension.
Measuring the Processivity of the Human pol δ Holoenzyme.
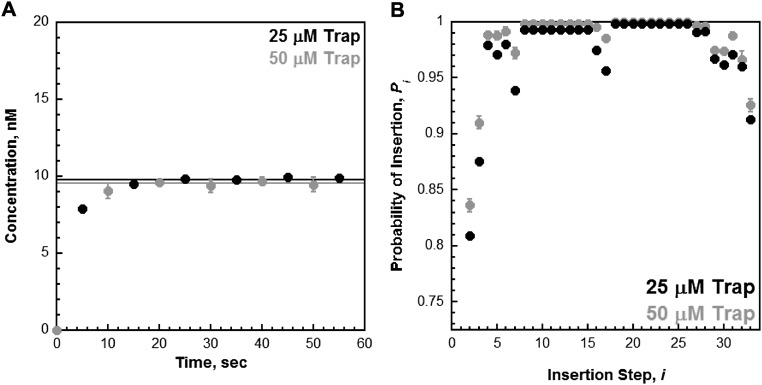
Under the conditions of the assay, the extent of processive DNA synthesis by assembled pol δ holoenzymes can be quantitatively analyzed at single nucleotide resolution. At each dNTP insertion step, i, the probability that a pol δ holoenzyme will insert another dNTP, Pi, is equivalent for the holoenzymes assembled with either PCNA or (Ub)3-PCNA (Fig. 3). Together, this demonstrates that assembly of the pol δ holoenzyme and its consequent processivity are unaffected by PCNA monoubiquitination. Interestingly, maximal Pi values are not observed until i = 18, where more than 40% of the initially assembled holoenzymes have already aborted. From i = 18 to i = 26, Pi remains constant and then drops off as the pol δ holoenzyme approaches the end of the DNA template. Kinetically, Pi is defined as Pi = kpol/(kpol + koff) where koff is the rate constant for dissociation of pol δ from the DNA substrate. Given the observance of at least two pol δ holoenzyme populations that insert dNTPs with vastly different rate constants (kpol), the aborted primer extension products observed at the onset of DNA synthesis (i ≤ 17) are from the slow holoenzyme population (low Pi), whereas longer products (i ≥ 18) are synthesized only by the fast, processive population (maximal Pi). Based on the concentration of all primer extension products that are least 47 nt in length (N + i = 29 + 18 = 47), this suggests that the faster, processive population may account for ∼60% of the pol δ holoenzymes assembled with either PCNA or (Ub)3-PCNA.
Fig. 3.
Measuring the processivity of pol δ holoenzymes at single nucleotide resolution. (A) The probability of insertion (Pi) for each step (i) beyond the first insertion was calculated as described in Materials and Methods for the experiments depicted in Fig. 2. The results for holoenzymes assembled with PCNA or (Ub)3-PCNA are plotted vs. the insertion step, and each data point represents the average ± SD of at least three independent experiments. The lower Pi values observed at the onset of primer extension are not due to limiting dNTP concentration (Fig. S4 C and D) or the trap actively assisting in the dissociation of holoenzymes as they traverse the DNA substrate (Fig. S9B). From i = 18 to i = 26, Pi plateaus and remains constant. Within this range (shaded gray), Pi is 0.999 ± 7.25 × 10−5 and 0.998 ± 2.99 × 10−4 for holoenzymes assembled with PCNA and (Ub)3-PCNA, respectively. (B) Fraction of holoenzymes remaining. Based on the maximal values for Pi (Pmax) obtained from the data in A, the fraction of holoenzymes (y) remaining at insertion step, i, was determined for pol δ holoenzymes assembled with either PCNA or (Ub)3-PCNA by the function y = Pmaxi. For comparison, this was carried out for the pol δ holoenzyme from S. cerevisae assembled with PCNA (yellow) at a comparable ionic strength (165 mM, data adapted from ref. 6). (Left) All calculated values (i = 1–4,000). (Right) Data within the size of eukaryotic Okazaki fragment (i = 100–250).
Once an assembled pol δ holoenzyme initiates DNA synthesis from a nascent primer, the probability of insertion, Pi, determines the fraction of pol δ holoenzymes that will reach the 5′ end of the downstream duplex region before dissociation. For instance, the pol δ holoenzyme from S. cerevisae inserts dNTPs very fast (kpol = 72–150 s−1) and is extremely stable when stalled on DNA (koff ≤ 2.31 × 10−3 s−1), yielding a Pi value of essentially 1.0 (Pi ≥ 0.9999). Thus, less than 1% of pol δ holoenzymes dissociate before the nascent primer is completely extended (0.9999250 = 0.9962). Indeed, a pol δ holoenzyme from S. cerevisiae can replicate an entire 7.4-kb ssDNA circular plasmid within a single binding encounter (6, 7, 29, 30). As observed in Fig. 3A, Pi for human pol δ plateaus at 0.999 ± 7.25 × 10−5and 0.998 ± 2.99 × 10−4 for holoenzymes assembled with PCNA and (Ub)3-PCNA, respectively. Based on these Pi values, ∼14% of human pol δ holoenzymes dissociate before inserting 100 dNTPs and ∼31% dissociate before inserting 250 dNTPs (Fig. 3B). Thus, at physiological pH, ionic strength, and dNTP concentration, a significant portion of human pol δ holoenzymes may dissociate into solution before finishing an Okazaki fragment, in stark contrast to S. cerevisae. Such contrast is not unforeseen as the highly limited processivity of the human pol δ holoenzyme has been well documented. Specifically, synthesis of the full-length product (7.4 kb) by a human pol δ holoenzyme on a singly primed M13 substrate required long incubation times, a large excess of pol δ, and the lack of a trap. Furthermore, a distribution of primer extension products appeared and persisted between the native primer and the full-length product. This behavior has been observed for the native four subunit human pol δ complex obtained directly from human cells (31), as well as recombinant four subunit complexes expressed and purified from either insect cells (21, 23, 31–33) or Escherichia coli (34, 35). All of these aforementioned properties are classic descriptions of a nonprocessive DNA pol and are in complete contrast to the behavior of S. cerevisae pol δ on the same DNA substrate (6, 7, 29, 30). Indeed, the experimentally measured Pi values presented in Fig. 3 suggest that more than half of all pol δ holoenzymes assembled with PCNA dissociate before synthesizing 695 nt (0.999695 = 0.499), necessitating multiple binding events for complete replication of a M13 template. Thus, the Pi values measured in these studies agree with the previous qualitative assessments on the limited processivity of the human pol δ holoenzyme compared with S. cerevisiae. As alluded to above, lower Pi values for the human pol δ holoenzyme reflect either a faster koff, a slower kpol, or a combination of both possibilities. Next, we designed an assay to directly measure koff for the human pol δ holoenzyme.
Stability of the pol δ Holoenzyme on Stalling.
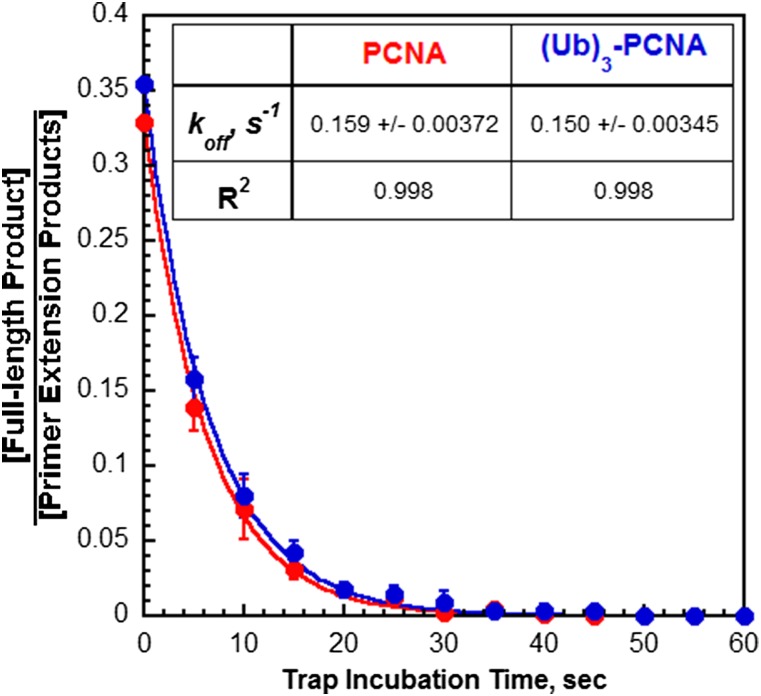
Assembly of a pol δ holoenzyme is indicated by primer extension (Fig. 2). We exploited this to probe the stability of pol δ holoenzymes under running start conditions that mimic stalling at a DNA lesion (Fig. 4A). Only long primer extension products (i ≥ 28) were observed above i = 1 (Fig. 4B), and analysis revealed that a substantial portion of assembled holoenzymes rapidly dissociated on stalling, whereas the remaining portion dissociated over the next 30 s or so (Fig. 4C). Data points from 5 to 60 s fit very well to single exponential decays and yielded identical results for the assembled holoenzymes. koff values of 0.152 ± 0.00318 s−1 and 0.156 ± 0.00523 s−1 were obtained for pol δ holoenzymes assembled with PCNA and (Ub)3-PCNA, respectively. Extrapolation of the fits back to time 0 yields amplitudes of 0.537 ± 0.0121 and 0.588 ± 0.0214 for each respective holoenzyme, suggesting that the observed population of pol δ holoenzymes accounts for ∼60% of the total population. This value bears a striking resemblance to the amount of the fast, processive population that was predicted based on the distribution of primer extension products in Fig. 2. Together with the aforementioned results, this analysis suggests that the slow holoenzyme population immediately disassembles on stalling and is not detected, whereas only the fast, processive population remains on DNA for some time. Only the latter population can synthesize the full-length product and the proportion of primer extension products that are completely extended decreases with trap incubation time (Fig. S7). Fitting these data yielded koff values equivalent to those reported in Fig. 4C. Thus, the assay and analysis only describe the fast, processive pol δ holoenzymes and indicate that koff is unaffected by PCNA monoubiquitination. Clearly, (Ub)3-PCNA does not promote disassembly of the human pol δ holoenzyme.
Fig. 4.
Dissociation of a stalled pol δ holoenzyme. (A) Schematic representation of experiment performed to monitor stability of the pol δ holoenzyme. First, the pol δ holoenzyme was assembled as in Fig. 2A. After a 10-s preincubation, trap and dGTP (the first dNTP to be incorporated) were added simultaneously to select for assembled pol δ holoenzymes that are competent for DNA synthesis. After varying incubation times, aliquots were removed, mixed with a dNTP chase containing all dNTPs, and then quenched after 1 min. Under these conditions, only pol δ holoenzymes that remain assembled on the P29/Bio62 substrate after the specified incubation time will be able to extend the primer to i = 2 and beyond. Hence, the probability of insertion for i = 2 is equal to the fraction of pol δ holoenzymes remaining. (B) 16% denaturing sequencing gel of the primer extension products for pol δ holoenzymes assembled with either PCNA (lanes 1–13) or (Ub)3-PCNA (lanes 14–26). The size of the substrate and full-length product is indicated on the left. As a control for each condition, an aliquot was removed after 2 min of preincubation and quenched before the addition of dNTPs (lanes 13 and 26). (C) The fraction of holoenzymes associated with the P29/Bio62 DNA substrate as a function of trap incubation time. Each point represents the average ± SD of three independent experiments for holoenzymes assembled with either PCNA or (Ub)3-PCNA. Data points from 5 to 60 s were fit to a single exponential decay. Extrapolation of the fit back to t = 0 yields the amplitude. The amplitudes, rate constants, and R2 values for each fit are reported.
Fig. S7.
Directly monitoring the stability of the fast, processive pol δ holoenzymes. From Fig. 4B, it can be seen that the proportion of primer extension products that are extended to the full-length 62-mer product on chasing with dNTPs decreases as a function of trap incubation time. For each trap incubation time, the concentration of the full-length product was divided by the total concentration of all primer extension products, and the ratios are plotted as a function of trap incubation time. For t = 0 s, the ratio was calculated from the amplitudes reported in Table 1. Each data point represents the average ± SD of three independent experiments for holoenzymes assembled with either PCNA or (Ub)3-PCNA. All points were fit to a single exponential decay, and the rate constants and R2 values are reported. Only the fast, processive population of pol δ holoenzyme can extend to the full-length 62-mer product. Thus, the reported rate constants only describe dissociation of the fast, processive pol δ holoenzymes from the P29/Bio62 DNA substrate (koff). These values exactly agree with those reported in Fig. 4C, demonstrating that the assay and analysis described in Fig. 4 only account for fast, processive pol δ holoenzymes.
The rate constant for dNTP insertion, kpol, may be calculated by rearranging the equation Pi = kpol/(kpol + koff) to kpol = (koff × Pi)/(1 − Pi). From the experimentally determined values of Pi (Fig. 3) and koff (Fig. 4) for the fast, processive population, kpol values of 108 and 102 s−1 are calculated for pol δ holoenzymes assembled with PCNA and (Ub)3-PCNA, respectively. These values agree with that previously reported for pol δ from S. cerevisiae (72–150 s−1) and with a recent measurement (87 s−1) obtained via rapid quench for the four-subunit human pol δ complex expressed and purified from insect cells (6, 28–30), indicating the fast kpol is conserved in humans and independent of PCNA monoubiquitination. Altogether, the results presented in this study demonstrate that monoubiquitination of PCNA has no effect on the assembly (Fig. S2, Fig. 2, and Table 1), activity (Figs. 2 and 3), or disassembly (Fig. 4 and Fig. S7) of the human pol δ holoenzyme. Given the drastic contrast in the behavior of the slow holoenzyme population (kpol ≤ 0.485 s−1, koff ≥ 0.970 s−1; SI Text), it is not physiologically relevant and will not be discussed further.
Discussion
The Instability of the Human pol δ Holoenzyme and Okazaki Fragment Synthesis.
The experiments in Figs. 2 and 3 demonstrate that anchoring to PCNA increases the Pi of human pol δ from 0.00 to 0.999 ± 7.25 × 10−5 and 0.998 ± 2.99 × 10−4 for holoenzymes assembled with PCNA and (Ub)3-PCNA, respectively. Presumably, this is due to the stabilization of pol δ on DNA (decrease in koff). However, kpol in the presence and absence of PCNA has yet to be directly compared, and thus, a simultaneous enhancement of kpol cannot be ruled out. By directly monitoring dissociation of the human pol δ holoenzyme on stalling (Fig. 4), koff values of 0.152 ± 0.00318 s−1 and 0.156 ± 0.00523 s−1 were obtained for holoenzymes assembled with PCNA or (Ub)3-PCNA, respectively. Under the conditions of the assay, pol δ may dissociate from the P29/Bio62 DNA substrate by one of three pathways; (i) pol δ first dissociates from DNA and then dissociates from PCNA encircling the P/T junction; (ii) pol δ first separates from PCNA and then “free” pol δ dissociates from DNA; or (iii) pol δ first dissociates from DNA and the “disengaged” pol δ•PCNA complex slides off the unblocked end of the template, which lacks biotin/neutravidin. Human PCNA slides along DNA extremely fast (i.e., diffusion coefficient ∼ 1.0 μM2/s), even when its hydrodynamic radius is dramatically increased by a bound protein (36). Hence, the third pathway cannot be measured by the assay and would not be observed in Fig. 4. The results from Fig. 2 demonstrate that pol δ will not extend the primer at physiological pH, ionic strength, and dNTP concentration, suggesting that the DNA binding affinity of human pol δ alone is dramatically low such that dissociation from the DNA substrate is much faster (i.e., >>100 s−1) than dNTP binding and/or insertion and cannot be measured/observed by the assay. This behavior agrees with the inability of S. cerevisae pol δ to form a stable complex with a native P/T DNA substrate in the absence of PCNA (27). Altogether, this suggests that, on stalling of the pol δ holoenzyme, dissociation of pol δ from DNA is entirely rate limited by the dissociation of pol δ from PCNA encircling the P/T junction (either pathway 1 or pathway 2). Thus, koff observed in Fig. 4 reports on the dissociation of pol δ from PCNA or (Ub)3-PCNA and reveals the surprising instability of this complex (t1/2 < 4.57 s) compared with S. cerevisiae (t1/2 > 300 s). From the equation Pi = kpol/(kpol + koff), we calculate kpol values of 108 and 102 s−1 for pol δ holoenzymes assembled with PCNA and (Ub)3-PCNA, respectively. These values agree with that previously reported for pol δ from human (87 s−1) and S. cerevisiae (72–150 s−1), indicating the fast kpol is conserved in humans (6, 28–30). Thus, complete extension of a nascent RNA/DNA hybrid primer to a downstream duplex 100–250 nt away requires ∼1.0–2.5 s. Altogether, the results of the current studies provide the first demonstration, to our knowledge, that the nonprocessive behavior (low Pi) of the human pol δ holoenzyme compared with S. cerevisiae is due to a faster koff and not a slower kpol. Furthermore, these studies clearly indicate that monoubiquitination of PCNA has no effect on the properties of the human pol δ holoenzyme, in agreement with that observed in S. cerevisiae (26).
As illustrated in Fig. 3B, a substantial portion (∼14–31%) of human pol δ may release from the lagging strand template before completing DNA synthesis of a given Okazaki fragment, in stark contrast to S. cerevisiae (<1%). Based on the concentration of pol δ in the cell (1.96 × 104 copies of p125 per cell), the size of the diploid genome (∼6 × 109 bp), and the length of Okazaki fragments in humans (100–250 nt), each pol δ must be reused ∼1,220–3,060 times per S-phase (4, 37). Thus, it appears that pol δ is most likely recycled back to a nascent RNA/DNA hybrid primer on releasing prematurely from a given Okazaki fragment. Under this assumption, ssDNA gaps between adjacent Okazaki fragments may arise and persist during unperturbed S-phase in humans. However, extensive efforts have revealed this is not the case in vivo. In mammalian cells, all replication origins do not fire simultaneously at the onset of S-phase. Rather, DNA synthesis progresses by the activation of replication origins at different times over the course of an extended S-phase (8–12 h), such that only a fraction of replication origins are active at any given time (ref. 38 and references cited therein). Thus, the need to recycle limiting pol δ during S-phase may not be so dire. Furthermore, the rate of replication fork progression in various human cell lines is only ∼1.6 kb/min (38–40), suggesting that an Okazaki fragment is exposed every 3.8–9.4 s on average, much longer than the time required (∼1.0–2.5 s) for one or more human pol δ holoenzymes to completely extend a nascent RNA/DNA hybrid primer to the downstream duplex. In the event that pol δ dissociates prematurely from the lagging strand template, PCNA is almost certainly left behind on the DNA for some time as spontaneous opening of the human PCNA ring is dramatically slower (t1/2 = 9.63 min) (5). Indeed, recent in vivo evidence suggests that enzyme-catalyzed recycling of scarce PCNA during unperturbed S-phase will not occur until the adjacent Okazaki fragments are ligated together (41). Thus, PCNA is retained on the Okazaki fragment on dissociation of pol δ, and hence, the pol δ holoenzyme does not need to be reassembled from scratch to complete DNA synthesis. Last, when replicated, simian virus 40 (SV40) DNA was extracted from infected human cells and visualized by electron microscopy, internal ssDNA gaps were not observed under nonperturbed conditions. For this model system, all aspects of replicating the double-stranded SV40 DNA except for helicase-assisted unwinding are carried out by the replication machinery of the human host cell. In the absence of DNA damage, small internal ssDNA gaps (50–300 nt) indicative of premature release and recycling of pol δ were not observed within the replicated regions of SV40 DNA molecules (42, 43). Altogether, this suggests that lagging strand synthesis within a human cell may not require maximum processivity/efficiency and that aborted Okazaki fragments are rebound in the event pol δ dissociates prematurely (Fig. 5A). Hence, more than one pol δ holoenzyme may contribute to the synthesis of a given Okazaki fragment, in contrast to S. cerevisiae.
Fig. 5.
Lagging strand synthesis in humans. (A) Native template. While replicating an undamaged lagging strand template, the human pol δ holoenzyme inserts dNTPs with a rate constant of 108 s−1, more than 710-fold faster than the rate constant for dissociation of pol δ from PCNA encircling DNA (koff). Thus, at each dNTP insertion step, a pol δ holoenzyme has a 99.9 ± 7.25 × 10−3% chance of inserting another dNTP rather than dissociating into solution. In the event pol δ dissociates into solution before completing a given Okazaki fragment, PCNA is left behind on the lagging strand template. Pol δ rebinds the residual PCNA and DNA synthesis resumes from the aborted P/T junction. Thus, ssDNA gaps in replicated regions of the lagging strand template, i.e., behind a progressing replication fork, are absent on native templates (B) Damaged template. During S-phase, the lagging strand template DNA may be compromised by modifications (X) that pol δ cannot accommodate (i.e., kpol = 0). Upon encountering such lesions, pol δ rapidly dissociates from DNA (t1/2 < 4.6 s), leaving PCNA behind. An incoming TLS pol only needs to bind PCNA residing at the stalled P/T junction to attempt TLS. Pol δ may rebind to the resident PCNA and stalled P/T junction as in A, but pol δ-mediated DNA synthesis cannot resume on the afflicted Okazaki fragment until the lesion is bypassed by one or more TLS pols. Distributive DNA synthesis by TLS pols allows Pol δ-mediated DNA synthesis to resume beyond the lesion and the nascent DNA is completely extended to the 5′ end of the downstream Okazaki fragment. These “on the fly” TLS events occur in the absence of PCNA monoubiquitination and do not result in the formation of ssDNA gaps opposite the offending lesion. In the absence of a successful on the fly TLS event, pol δ is recycled to the upstream Okazaki fragments, leaving behind a ssDNA gap extending from the DNA lesion to the 5′ terminus of the downstream Okazaki fragment. These persistent ssDNA gaps coated with RPA serve as the signal for monoubiquitination of PCNA on the lagging strand. In this scenario, TLS across the offending DNA lesion and subsequent “filling in” and sealing of the ssDNA gap occurs behind the replication fork, i.e., postreplicative gap filling TLS.
The Instability of the Human pol δ Holoenzyme and Translesion DNA Synthesis.
During S-phase, the template DNA may be compromised by modifications that replicative pols cannot accommodate (2). For instance, the lagging strand pol δ in humans cannot replicate past common byproducts of lipid peroxidation and synthesis on the afflicted Okazaki fragment abruptly stops (44, 45). Such arrests may be overcome by TLS where pol δ is exchanged for a TLS pol that also binds to the front face of PCNA, either through a noncanonical PIP domain or a BRCT (BRCA1 C terminus) domain (9). With a more open pol active site and the lack of an associated proofreading activity, TLS pols are able to support stable, yet potentially erroneous, nucleotide incorporation opposite damaged templates, allowing synthesis by pol δ to resume. In humans, TLS involves the monoubiquitination of PCNA and at least seven TLS pols with varying fidelities. However, remarkably low error rates are observed in vivo after exposure to various DNA-damaging agents, indicating a highly efficient process (2, 46). Currently, the mechanism for pol exchange, the role of monoubiquitinated PCNA in particular, is under scrutiny.
In addition to their PCNA binding domains, most eukaryotic TLS pols such as pol η contain ubiquitin binding domains (UBDs) that may selectively increase their affinity for monoubiquitinated PCNA (9). A popular model for pol exchange during TLS purports that ubiquitin moieties on PCNA recruit TLS pols to sites of DNA damage where they actively displace/exchange with a stalled replicative pol. In S. cerevisae, the extremely processive pol δ holoenzyme dissociates very slowly when stalled on DNA (koff ≤ 2 × 10−3 s−1) and an active pol exchange model would be fitting for TLS (6, 7). Indeed, a recent characterization of the exchange between S. cerevisae pol δ and pol η provided staunch supporting evidence (7). However, various experimental observations suggest this model may not be conserved in humans. As stated above, multiple independent reports have noted the highly limited processivity of the human pol δ holoenzyme compared with S. cerevisiae, suggesting that the human pol δ holoenzyme is relatively unstable, and, hence, an active pol exchange mechanism may be unnecessary for TLS on the lagging strand (21, 23, 31–35). Furthermore, the binding affinity of a noncanonical PIP domain of human pol η for PCNA (0.4 μM) is more than 190-fold tighter than the affinity of its UBD for ubiquitin (∼77 μM), arguing against a selectively enhanced affinity for monoubiquitinated PCNA through additive binding domains (47, 48). Indeed, three of the seven TLS pols in mammalian cells lack a UBD and the UBD of human pol η is dispensable for pol η-mediated TLS in vivo (46, 49). Altogether, these studies suggest that an active pol exchange mechanism proceeding through a transient (Ub)3-PCNA•pol δ•pol η intermediate is not conserved in humans.
The experiments depicted in Fig. 4A mimic the scenario in which a pol δ holoenzyme encounters a modest DNA lesion that it cannot accommodate, such as common byproducts of lipid peroxidation (1,N2-ethenoguanine and 1,N6-ethenoadenine) and the major UV-induced lesion, cis-syn cyclobutane pyrimidine dimers (CPDs). These relatively small modifications are not envisioned to significantly alter the structure/dynamics of the lagging strand template. However, human pol δ cannot replicate past them, suggesting that disassembly of the pol δ holoenzyme on encountering such lesions is predominantly driven by an abrupt decrease in kpol. Indeed, blockage of strand elongation by human pol δ was observed directly 3′ to these lesions rather than upstream (8, 44, 45, 50). The results presented in Fig. 4C demonstrate that human pol δ maintains a loose association with PCNA while replicating and rapidly dissociates from DNA on stalling (t1/2 < 4.6 s), leaving PCNA behind. Thus, an incoming TLS pol would only need to bind PCNA residing at the stalled P/T junction to attempt TLS (Fig. 5B), in agreement with the observation that only the PCNA binding domains of human pol η are essential and sufficient for pol η-mediated TLS in vivo (49). Indeed, when the pol δ holoenzyme stability assays were repeated using catalytically inactive human pol η as trap, koff values of 0.115 ± 0.0112 s−1 and 0.166 ± 0.0174 s−1 were obtained for holoenzymes assembled with PCNA or (Ub)3-PCNA, respectively (Table 2), in agreement with the values presented in Fig. 4. In vivo, this full-length, human pol η mutant retains all biological functions except DNA synthesis activity (51, 52). Thus, equivalent koff values for pol δ holoenzymes assembled with either PCNA or (Ub)3-PCNA suggests that the binding of TLS pols to PCNA is governed by the passive dissociation of pol δ. The ubiquitin moieties on PCNA do not serve to recruit TLS pols to sites of DNA damage where they actively displace stalled pol δ. Altogether, the results presented in this study demonstrate that monoubiquitination of PCNA has no effect on the stability of the pol δ holoenzyme in the absence or presence of a TLS pol. To our knowledge, this is the first direct evidence that pol exchange during TLS on the lagging strand is not an active process in humans proceeding through a (Ub)3-PCNA•pol δ•pol η complex.
Table 2.
Rate constants for the dissociation (koff) of pol δ from PCNA loaded onto DNA measured in the presence of various traps
| Trap | PCNA | (Ub)3-PCNA |
| 29ddC/62 primer template DNA, s−1 | 0.152 ± 0.00318 | 0.156 ± 0.00523 |
| Catalytically “dead” full-length pol η, s−1 | 0.115 ± 0.0112 | 0.166 ± 0.0174 |
Human pol η accurately replicates across thymine-thymine CPDs (TT-CPDs) in vitro and is responsible for the highly error-free (>90%) bypass of UV-induced CPDs in vivo (53, 54). Interestingly, the DNA binding affinity of human pol η alone is not enhanced by the presence of a TT-CPD, and it progressively weakens on extending a P/T junction such that the pol η•DNA complex is destabilized after incorporating only two dNTPs beyond the lesion (55). Human pol η does contain two noncanonical PIP domains, only one of which is essential and sufficient for pol η-mediated TLS in vivo (49). However, the affinity of these noncanonical PIP domains for PCNA is moderately compromised by the replacement of conserved residues in the canonical sequence with alternative amino acids (47). Thus, PCNA stimulates the processivity of human pol η very weakly such that only four to seven dNTPs are inserted within a single DNA binding encounter (56), in stark contrast to that observed for human pol δ in the current study (Figs. 2 and 3). The distributive behavior of human pol η is also observed in other human TLS pols such as pol κ and pol ι and it is befitting of TLS as it limits potentially erroneous DNA synthesis by TLS pols (57).
Together with the results presented in the current study, we suggest that TLS and the subsequent resumption of DNA synthesis by pol δ may occur in the absence of PCNA monoubiquitination on the lagging strand, as illustrated in the model in Fig. 5B. The results presented in Fig. 4 demonstrate that human pol δ rapidly dissociates from DNA on stalling at a DNA lesion it cannot accommodate, leaving PCNA behind. An incoming TLS pol(s) only needs to bind PCNA at the stalled P/T junction to attempt TLS. This process is dictated by the passive dissociation of pol δ (Table 2) and, hence, pol δ may rebind to the resident PCNA and stalled P/T junction. However, pol δ-mediated DNA synthesis cannot resume on the afflicted Okazaki fragment until the lesion is bypassed by one or more TLS pols. Once bypass occurs, the distributive behavior of the TLS pol(s) allows pol δ-mediated DNA synthesis to resume beyond the lesion and the nascent DNA is completely extended to the 5′ end of the downstream Okazaki fragment. These TLS events allow DNA synthesis to continue without formation of ssDNA gaps opposite DNA lesions. Such behavior (referred to as “on the fly” TLS) was first observed in DT40 avian cells irradiated with UV and, indeed, found to be independent of PCNA monoubiquitination (58). Recent cellular studies suggest that ubiquitination-independent TLS events occur in murine (24, 59) and human cells (60) irradiated with UV, perhaps in a similar manner. However, monoubiquitination of PCNA is required for optimal TLS in mammalian cells (24). This discrepancy raises two key questions: (i) what is the temporal correlation between monoubiquitination of PCNA and TLS on the lagging strand and (ii) what is the role(s) PCNA monoubiquitination during TLS? The signal for monoubiquitination of PCNA at a stalled P/T junction is the buildup and persistence of RPA-coated ssDNA downstream of the offending damage (2). Considering observations from previous in vivo studies (see below), we propose the following model (Fig. 5B). After a mounting number of failed on the fly TLS attempts, pol δ is eventually recycled to the upstream Okazaki fragments to keep up with ongoing “bulk” DNA synthesis. Such events would leave behind an ssDNA gap opposite the offending lesion. Indeed, when SV40-transformed human cells were irradiated with UV, ssDNA gaps were observed in the replicated portions distal to the replication forks within SV40 DNA molecules and the size of the gaps (50–300 nt) agreed with the expectation for incomplete synthesis of Okazaki fragments (42, 43). In similar fashion, on SV40-based plasmids containing a single unique site CPD in the lagging strand template, synthesis of the Okazaki fragment containing the CPD lesion was selectively inhibited compared with the flanking fragments. Furthermore, strand elongation past and beyond the lesion was observed in only a fraction of the analyzed molecules (50, 61). Altogether, these observations suggest that the lagging strand template is reprimed 5′ to the damage, and pol δ is recycled to nascent primers upstream of the damage, leaving behind a ssDNA gap extending from the CPD lesion to the 5′ terminus of the downstream Okazaki fragment. In this scenario, TLS across the offending DNA lesion and subsequent “filling in” and sealing of the ssDNA gap occurs behind the replication fork, i.e., postreplicative gap-filling. This model originated in the 1970s from studies on UV-irradiated mammalian cells and was the prevailing view for TLS before the discovery of PCNA monoubiquitination (42, 62, 63). Perhaps these persistent ssDNA gaps coated with RPA serve as the signal for monoubiquitination of PCNA on the lagging strand. Indeed, postreplicative gap filling in DT40 avian cells irradiated with UV is dependent on PCNA monoubiquitination (58).
The present study unveiled the passive exchange of human DNA pols at stalled P/T junctions, and such behavior is in tune with ubiquitin-independent (on the fly) TLS. However, such events have yet to be demonstrated in vitro and supporting evidence has mostly been inferred from cellular and genetic studies, as noted above. In addition, we demonstrate that PCNA monoubiquitination has no effect on the assembly, activity, or disassembly of the human pol δ holoenzyme. So, what is the role(s) monoubiquitinated PCNA during postreplicative gap filling in humans and how is it executed? The results from the current study provide the foundation to directly test each model in future biochemical experiments and delineate the molecular details of human TLS. This initiative is imperative to deciphering the elusive role(s) of monoubiquitinated PCNA in human TLS.
Materials and Methods
Materials.
[γ-32P]ATP was purchased from PerkinElmer, and all unlabeled dNTPs were obtained from Denville Scientific. T4 polynucleotide kinase was purchased from New England Biolabs. Neutravidin was purchased from Sigma-Aldrich.
Oligonucleotides and Recombinant Proteins.
DNA substrates were synthesized by Integrated DNA Technologies and purified on denaturing polyacrylamide gels. Concentrations were determined from the absorbance at 260 nm using the calculated extinction coefficients. For annealing the P/T DNA substrates, the primer strand was mixed with a 1.1-fold excess of template in 1× annealing buffer (10 mM Tris⋅HCl, pH 8.0, 100 mM NaCl, and 1 mM EDTA), heated to 95 °C for 5 min, and allowed to slowly cool to room temperature. For experiments in which DNA synthesis was measured, the primer was 5′-labeled with 32P using [γ-32P]ATP and T4 polynucleotide kinase before annealing.
A truncated form of RFC (hRFCp140ΔN555) described previously was used in all of the reported studies and is referred to as simply RFC throughout the text (5). The plasmids for expression of WT human RPA were a generous gift from Marc Wold (University of Iowa, Iowa City, IA). RPA was expressed and purified from E. coli, and the concentration was determined from the reported extinction coefficient as described (64). WT and exonuclease-deficient human pol δ (pol δ) were expressed and purified from E. coli as described previously (34). The exonuclease-deficient human pol δ mutant contains a single point mutation (D402A) within the exonuclease domain of the catalytic subunit (p125/POLD1). This conserved residue is critical for divalent metal binding and the D402A mutation completely abolishes exonuclease activity (45). Details of the exonuclease-deficient human pol δ expression vectors will be described elsewhere. The concentration of the p125 subunit was determined by slight modifications to a published protocol and the concentration of the four-subunit DNA pol δ complex was expressed as the concentration of the p125 subunit (SI Materials and Methods). The exonuclease-deficient DNA pol δ D402A mutant is completely devoid of exonuclease activity on the 29-mer primer when annealed in the P29/Bio62 DNA substrate (Fig. S1A), in agreement with a previous report (45). This pol was used in all of the reported primer extension and stability assays and is referred to simply as pol δ throughout the text. PCNA was expressed in E. coli and purified by a published protocol (65). PCNA concentrations were determined from the absorbance at 280 nm using the calculated extinction coefficients. The expression plasmid for WT human ubiquitin (pRSUB) was a generous gift from Keith D. Wilkinson (Emory University School of Medicine, Atlanta, GA). The expression plasmid for UbcH5c(S22R) was purchased from Addgene (66). The human cDNA clone of UBA1 was purchased from OriGene. Detailed protocols for the expression and purification of recombinant ubiquitin, Uba1, and UbcH5c(S22R) proteins can be found in SI Materials and Methods. Monoubiquitinated PCNA containing single ubiquitin moieties conjugated to K164 of each monomer within a PCNA ring was synthesized enzymatically and purified by slight modifications to a published protocol (25). Please refer to SI Materials and Methods for details.
Primer Extension Assays.
All primer extension and holoenzyme stability (see below) experiments were performed at 25 °C in an assay buffer consisting of 1× replication buffer [25 mM TrisOAc, pH 7.7, 10 mM Mg(OAc)2, 125 mM KOAc] supplemented with 0.1 mg/mL BSA and 1 mM DTT. For all experiments, the final ionic strength was adjusted to 200 mM by addition of appropriate amounts of KOAc. All reagents, substrate, and protein concentrations listed are final reaction concentrations. The pol δ holoenzyme was assembled by pre-equilibrating a mixture of 50 nM P29/Bio62 DNA and 200 nM neutravidin at 25 °C in a temperature-controlled water bath. RPA (125 nM) was added, and the mixture was allowed to equilibrate for 2 min. PCNA [62.5 nM of either PCNA or (Ub)3-PCNA trimer] was added, and the mixture was allowed to equilibrate for 3.5 min. ATP (25 μM) was added, and the mixture was allowed to equilibrate for 30 s. The ATP concentration was kept low unless otherwise indicated to minimize the erroneous insertion of ATP by human pol δ (67). However, the PCNA loading and unloading activities of RFC are maximal at this low concentration of ATP (5) and, thus, the extent of holoenzyme formation and subsequent extension of the primer is not limited by the amount of PCNA loaded onto DNA by RFC (Table 1 and Fig. S4 A and B). RFC (62.5 nM) was added, and the mixture was allowed to equilibrate for 1 min 50 s before pol δ (100 nM) was added. After 10–60 s, DNA synthesis by assembled holoenzymes was initiated by simultaneous addition of dNTPs (50 or 250 μM of each) and trap (50 μM). The concentration of trap (50 μM) was chosen so that it operates passively to sequester any pol δ that dissociates from the P29/Bio62 DNA substrate over an extended incubation (Figs. S8 and S9); 50 µM of each dNTP is within the physiological range of concentrations for each dNTP in a dividing human cell (68). At variable times, aliquots of the reaction were removed, quenched with 250 mM EDTA, pH 8.0, and diluted 3.8-fold into 95% formamide/25 mM EDTA/0.01% (wt/vol) tracking dye solution. Primer extension products were analyzed on 16% sequencing gels. Before loading onto gel, samples were heated at 95 °C for 5 min and immediately chilled in ice water for 5 min. Gel images were obtained on a Typhoon Model 9410 imager. The radioactivity in each band on a gel was quantified with ImageQuant (GE Healthcare), and the data were plotted with Kaleidagraph (Synergy). For the full-length 62-mer product, all data points were fit to a flat line where the y-intercept reflects the amplitude. For the total amount of primer extension products, data points after t = 10 s were fit to a flat line where the y-intercept reflects the amplitude. Values for the amplitudes are reported in Table 1.The amplitude for primer extension is equal to the concentration of assembled pol δ holoenzymes that are competent for DNA synthesis. It should be noted that the DNA synthetic activity of pol δ was not monitored in our previous report, and formation of the pol δ holoenzyme was indicated by pol δ capturing loaded PCNA from DNA-bound RFC. The results indicated that pol δ stabilized a stoichiometric amount of loaded PCNA on DNA, demonstrating the maximum efficiency of pol δ holoenzyme formation (5). Under all experimental conditions reported in the current study, the amplitude for primer extension was less than the concentration of pol δ when the accessory proteins (RPA, PCNA, RFC) were in excess of the P29/Bio62 DNA substrate. Such behavior was independent of dNTP and ATP concentrations, as well as preincubation time (Fig. 2). Altogether, this suggests that all assembled pol δ holoenzymes are not competent for primer extension.
Fig. S4.
Increasing ATP and/or dNTP concentration does not affect activity of the pol δ holoenzyme. (A and B) Primer extension was monitored as depicted in Fig. 2A except the concentration of ATP was increased to 0.5 mM or the concentrations of ATP and dNTPs were increased to 0.5 mM and 250 µM, respectively. Primer-extension products (total) for holoenzymes formed with either PCNA (A) or(Ub)3-PCNA (B). For comparison, the data from Fig. 2C are included in each,and the conditions are indicated in the figure legend. Each point represents the average ± SD of three independent experiments. As can be seen, primer extension was still not completed within the first time point (10 s) when the concentration of ATP or the concentration of both ATP and dNTPs were increased.This suggests that dNTPs are saturating and PCNA is loaded onto the DNA correctly. (C and D) Probability of insertion (Pi) for experiments in A and B. The results for holoenzymes assembled with PCNA (C) or (Ub)3-PCNA (D) are plotted vs. the insertion step, and each point represents the average ± SD of at least three independent experiments. For comparison, the data from Fig. 3A are included in each, and the conditions are indicated in the figure legend.
Fig. S8.
29ddC/62 is an efficient trap. Primer extension was monitored as depicted in Fig. 2. (A) 16% denaturing sequencing gel of the primer extension products for pol δ alone (lanes 1–13) and a pol δ holoenzyme (lanes 14–26) in the presence of 50 µM 29ddC/62 DNA trap. The size of the substrate and full-length product are indicated on the left. Primer extension is not observed for polδ alone (lanes 1–13) over the entire time course. (B) Quantification of the primer-extension products (total, ●) for holoenzyme formed with PCNA. The amount of primer extension products remains flat over the entire time course. Together, this demonstrates the efficiency of the 29ddC/62 DNA to trap any pol δ introduced into solution during an extended incubation.
Fig. S9.
29ddC/62 acts as a passive trap. Primer extension was monitored as depicted in Fig. 2 with 250 µM of each dNTP, 0.5 mM ATP, and 25 µM 29ddC/62 trap. (A) Primer-extension products (total, ●) for holoenzyme formed with PCNA. For comparison, the data (black) are overlaid on data from Fig. S4A obtained under the same conditions except with 50 µM 29ddC/62 trap (gray). The primer extension products build up to the same level and then plateau thereafter, demonstrating that only a single turnover of primer extension is observed in the presence of the 29ddC/62 trap. (B) Probability of insertion (Pi, ●) for holoenzyme formed with PCNA. For comparison, the data (black) are overlaid on data from Fig. S6C obtained under the same conditions except with 50 µM 29ddC/62 trap (gray). Decreasing the concentration of 29ddC/62 trap twofold does not increase the probability of insertion (Pi) for any insertion step, in particular those for which Pi is low. For instance, Pi = 0.836 ± 0.00579 for i = 2 at 50 μM trap indicating that kpol is 5.11-fold greater than koff. If trapping was active, i.e., a second-order kinetic process, the observed rate constant for dissociation should decrease twofold when the concentration of trap is decreased twofold. In this scenario, kpol would now be 10.22-fold greater than koff and Pi should increase to 0.911. However, Pi remains constant for i = 2 at 25 μM trap (0.810 ± 0.0126). Together with the data presented in A, this demonstrates that 29ddC/62 trap does not actively assist in the dissociation of holoenzymes from the P29/Bio62 substrate. Rather, 29ddC/62 serves as a passive trap.
Probability of Insertion, Pi.
The processivity of pol δ holoenzymes can be analyzed at single nucleotide resolution in the experiments depicted in Fig. 2. For each dNTP insertion step, i, the probability that a pol δ holoenzyme will insert another dNTP, Pi, is equal to the concentration of all primer extension products up to at least length N + i (where N is the length of the primer) divided by the concentration of all primer extension products up to at least length N + i − 1. These calculations were carried out for each time point within the plateau (20–50 s), and the average was taken. Please see Fig. S6 for a detailed example. Primer extension products between the size of 36 and 43 nt and between the size of 46 and 54 nt cannot be accurately quantified individually. In these regions, the primer extension products were quantified together as a whole, and we assume that the probability of insertion in these regions remains constant and is perpetuated. For example, the probability that a primer extension product 46 nt in length will be extended to a product of 55 nt in length, a total of nine insertion steps, is equal to (P18–26)9. To calculate (P18–26), the concentration of all primer extension products at least 55 nt in length was divided by the concentration of all primer extension products at least 46 nt in length, and the ninth root of this ratio reflects the average probability of insertion (P18–26) for steps i = 18 through i = 26.
Pol δ Holoenzyme Stability Assays.
The pol δ holoenzyme was assembled as described above with either PCNA or (Ub)3-PCNA. All reagents, substrate, and protein concentrations listed are final reaction concentrations. After a 10-s preincubation with pol δ, holoenzymes were selected for by the simultaneous addition of dGTP (50 μM) and 29ddC/62 DNA trap (50 μM). The holoenzymes were then incubated for 5–60 s. After the indicated time of incubation, an aliquot was removed and mixed with dNTPs (50 μM final concentration of each) to initiate DNA synthesis by holoenzymes still bound to the P29/Bio62 DNA substrate. After 1 min, the reactions were quenched and analyzed as described above. Only dGTP was included during the trap incubation to limit a potential bias for fast, processive pol δ holoenzymes. Inclusion of the first two or three dNTPs to be incorporated would have permitted primer extension up to i = 5 and i= 8, respectively (Figs. 1 and 3A), where 30–40% of the assembled pol δ holoenzymes will have already dissociated. Under either of these conditions, only stable, processive pol δ holoenzymes would survive the trap incubation and further extend the primer upon addition of the dNTP chase.
To ascertain the extent of erroneous primer extension beyond i = 1 during the trap incubation, these experiments were repeated except aliquots were quenched before the addition of the remaining dNTPs. The fraction of primer extension products beyond i = 1, referred to as c, remained constant over the entire trap incubation time and the average was taken. For holoenzymes assembled with either PCNA (c = 0.0167 ± 0.00237) or (Ub)3-PCNA (c = 0.0245 ± 0.000753), the observed values were minimal but nonetheless accounted for as follows. For each time point, the fraction of holoenzymes associated with the P29/Bio62 DNA substrate was determined by F = (At – c)/(P2 – c) where At is equal to the fraction of primer extension products that are greater than 30 nt in length at a trap incubation time of t and P2 is the probability of insertion for i = 2 from Fig. 3. Thus, the term P2 – c reflects the range for each holoenzyme and allows data to be normalized. Hence, at t = 0, At = P2 and F = 1 for each respective holoenzyme.
As a control, these experiments were repeated using catalytically inactive human pol η as trap. This mutant was generated by inactivating point mutations in conserved residues (D115A, E116A) within the full-length, human pol η protein that are necessary for catalytic activity (52, 53, 69). Site-directed mutagenesis, expression, and purification of this protein will be described elsewhere. The stability assays were carried out as described above with minor differences in the final concentrations of the substrate and proteins: 10 nM P29/Bio62 DNA, 40 nM neutravidin, 50 nM RPA, 50 nM PCNA trimer [either WT or (Ub)3-PCNA], 10 nM RFC, 50 nM pol δ, and 500 nM catalytically inactive pol η. The concentration of catalytically inactive pol η was chosen based on its ability to inhibit the activity of the human pol δ holoenzyme for at least 2 min.
SI Text
Estimated Rate Constants for the Slow pol δ Holoenzyme Population.
By 10 s, primer extension by the fast, processive holoenzyme has been completed for some time (Fig. 2). Therefore, the increase in the total amount of primer extension products after 10 s is only from a slower holoenzyme population (at least one). The total amount of primer extension products increases from 10 to 20 s and plateaus thereafter, signaling completion. A conservative estimate is that 10 s represents at least seven half-lives for kpol for the slow holoenzyme, i.e., more than 99% of the primer has been extended by at least one nucleotide. This estimate equates to a rate constant of 0.485 s−1 for the slow holoenzyme population [kpol = ln 2/t1/2 = ln 2/(10 s/7) = 0.485 s−1]. This value is more than 210-fold slower than the calculated value for the fast, processive population.
By 5 s, all of the slow pol δ holoenzyme has dissociated into solution (Fig. 4). A conservative estimate is that 5 s represents at least seven half-lives for koff for the slow holoenzyme, i.e., more than 99% of this population has disassembled. This equates to a rate constant of 0.970 s−1 for the slow holoenzyme population [kpol = ln 2/t1/2 = ln 2/(5 s/7) = 0.970 s−1]. This value is more than 6.4-fold faster than the experimentally measured value for the fast, processive population.
SI Materials and Methods
Recombinant Human Proteins.
Human ubiquitin was purified by modifications to a published protocol (55). The plasmid for expression of WT human ubiquitin (pRSUB) was transformed into the E. coli strain BL21(DE3) (Novagen) by electroporation, and transformants were selected on LB (Luria Broth) plates containing 50 μg/mL ampicillin. After a 24-h growth at 37 °C, a single colony was suspended in 100 mL LB media supplemented with 100 μg/mL ampicillin and grown at 37 °C overnight. This culture was diluted with 4 L LB media supplemented with 100 μg/mL ampicillin and grown at 37 °C to A600 = 1.0, at which point IPTG (isopropyl β-D-1-thiogalactopyranoside) was added to 0.5 mM and incubation was continued for 3 h at 37 °C. Cells were collected by centrifugation at 6,340 × g for 15 min and frozen at −80 °C. Cells were resuspended in lysis buffer (50 mM Tris⋅HCl, pH 6.8, 0.5 mM EDTA, and 1 mM DTT) and lysed by four cycles of 1-min sonication and cooling to <10 °C. The cell suspension was centrifuged at 9,000 × g for 20 min. Nonubiquitin proteins were precipitated from the supernatant by dropwise addition of perchloric acid at 4 °C to a final volume of 5% (vol/vol). After centrifugation at 9,000 × g for 20 min, the supernatant was dialyzed at 4 °C against 2 L 0.05 M ammonium acetate buffer, pH 4.5, for 9 h. This was repeated in 2 L fresh dialysis buffer for an additional 9 h. The dialyzed solution was very cloudy. Ubiquitin was resolved from the dialyzed supernatant by cation exchange chromatography on a 10-mL CM-Sepharose Fast Flow Column (Pharmacia Biotech) equilibrated with 0.05 M ammonium acetate buffer, pH 4.5. Unbound protein was removed by washing the column with five column volumes (CVs) of the same buffer, and ubiquitin was eluted with 10 CV of 0.1 M ammonium acetate, pH 5.5. The purity of the fractions was assessed by 10% SDS/PAGE. Pooled fractions were run through an Amicon 30K Ultracentrifugation filter and the flow thru was dialyzed at 4 °C against 50 mM TrisHCL, pH 8.0, and 100 mM NaCl. Ubiquitin concentrations were determined by A280, assuming an extinction coefficient of ε280 = 1,280 M−1⋅cm−1. Protein stock was diluted with 50 mM Tris⋅HCl, pH 8.0, 100 mM NaCl, and 50% glycerol (vol/vol) to a final concentration of 10% glycerol (vol/vol), divided into aliquots, frozen in liquid N2, and stored at −80 °C.
The ORF for UBA1 was cloned into Pet28a plasmid between the BamHI and NHEI restriction sites using forward (5′-ATTTTAGCTAGCATGTCCAGCTCGCCGCTG-3′) and reverse (5′-ATTTTAGGATCCTTAGCGGATGGTGTATCG-3′) primers. Human Uba1 was purified by modifications to a published protocol (25). The plasmid for expression of human Uba1 was transformed into the E. coli strain BL21(DE3) (Novagen) by electroporation, and transformants were selected on LB plates containing 30 μg/mL kanamycin. After a 24-h growth at 37 °C, a single colony was suspended in 40 mL LB media supplemented with 30 μg/mL kanamycin and grown at 37 °C overnight. This culture was diluted with 4 L terrific broth media supplemented with 30 μg/mL kanamycin and grown at 37 °C to A600 = 0.3, at which point the incubator was decreased to 15 °C and growth continued until A600 = 0.5. IPTG was added to a final concentration of 0.25 mM and growth continued overnight at 15 °C. Cells were collected by centrifugation at 6,340 × g for 15 min and frozen at −80 °C. Cells were resuspended in buffer A (20 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 0.5 mM TCEP [tris(2-carboxyethyl)phosphine], and 1 mM PMSF) supplemented with 20 mM imidazole and two complete, EDTA-free protease inhibitor mixture tablets (Roche) and lysed by five cycles of 1-min sonication and cooling to <10 °C. The cell suspension was centrifuged at 47,807.6 × g for 30 min. Supernatant was applied to 10 mL high-density cobalt beads (GoldBio.com) that had been pre-equilibrated with buffer A supplemented with 20 mM imidazole. The suspension was rocked at 4 °C for 1 h. Beads were collected by centrifugation (660 × g for 10 min), and the supernatant was discarded. The beads were resuspended in 100 mL buffer A supplemented with 20 mM imidazole and packed into a column, collecting the flow through. The column was washed in succession with 50 mL each of buffer A supplemented with 50, 100, 150, and 200 mM imidazole. Bound protein was eluted with a 100 mL linear gradient of imidazole (200–500 mM) in buffer A, collecting 2.5-mL fractions. Column was washed with 50 mL buffer A supplemented with 500 mM imidazole, collecting 2.5-mL fractions. Fractions were analyzed by SDS/PAGE, pooled, concentrated via Amicon 50 K Ultracentrifugation filter, and applied to a 5-mL QFF column (GE Healthcare) that had been pre-equilibrated with buffer B (20 mM Tris⋅HCl, pH 8.0, 0.5 mM TCEP, and 1 mM PMSF) supplemented with 150 mM NaCl, collecting flow through. Column was washed with 12.5 mL buffer B supplemented with 150 mM NaCl, collecting flow through. Bound protein was eluted with a 100-mL linear gradient of NaCl (150 mM to 1 M) in buffer, collecting 1.25-mL fractions. Column was washed with 20 mL buffer B supplemented with 1 M NaCl, collecting 1.25-mL fractions. Fractions were analyzed by SDS/PAGE, and desired protein was eluted between 192.5 and 585.625 mM NaCl. Pooled fractions were diluted with buffer B (without PMSF) to 150 mM NaCl, concentrated to <1 mL via Amicon 50 K Ultracentrifugation filter, and applied to Hi-load Superdex 16/60 200 size exclusion column that had been pre-equilibrated with buffer B supplemented with 150 mM NaCl (without PMSF). Protein was eluted with buffer B supplemented with 150 mM NaCl (without PMSF), collecting 1.0-mL fractions. Fractions were analyzed by SDS/PAGE. Pooled fractions were diluted with an equal volume of buffer B (without PMSF) supplemented with 150 mM NaCl and 20% glycerol (vol/vol), concentrated via Amicon 50 K Ultracentrifugation filter, divided into aliquots, frozen in liquid N2, and stored at −80 °C. The concentration of Uba1 was determined by Bradford Assay using BSA as a standard.
Human UbcH5c (S22R) was purified by modifications to a published protocol (49). The plasmid for expression of human UbcH5c (S22R) was introduced into BL21/DE3 E. coli cells by electroporation, and plasmid-producing colonies were selected for on LB agar plages supplemented with 50 μg/mL kanamycin. After overnight growth at 37 °C, a single colony was suspended in 100 mL LB media supplemented with 50 μg/mL kanamycin and grown at 37 °C overnight. This culture was diluted with 8 L LB media supplemented with 50 μg/mL kanamycin and grown at 37 °C to A600 = 0.8, at which point IPTG was added to 0.8 mM and incubation continued overnight (15.5 h) at 15 °C. Cell pellet was collected by centrifugation, resuspended in 3.6 g/mL buffer C (30 mM Mes, 1 mM EDTA, pH 6.0), and lysed by sonification. The supernatant was clarified by centrifugation and loaded onto a 20-mL SP Sepharose column pre-equilibrated with buffer C. After washing with 5 CV of buffer C, bound protein was eluted with a 9-CV linear NaCl gradient (0–500 mM) in buffer C, collecting 5-mL fractions. Fractions were analyzed by SDS/PAGE, and peak fractions (∼200–300 mM NaCl) were pooled, dialyzed overnight in 4 L buffer D (25 mM sodium phosphate, pH 7.0, and 150 mM NaCl), concentrated via Amicon 10K ultracentrifugation, and applied to Hi-load Superdex 16/60 200 size exclusion column that had been pre-equilibrated in buffer D. Protein was eluted with buffer D, collecting 1.0-mL fractions. Fractions were analyzed by SDS/PAGE. Pooled fractions were concentrated via an Amicon 10 K ultracentrifugation filter, divided into aliquots, frozen in liquid N2, and stored at −80 °C. The concentration was determined by the calculated extinction coefficient (ε280 = 24,750 M−1⋅cm−1).
For monoubiquitination of PCNA, 32 μM ubiquitin, 16 μM PCNA, 520 nM Uba1, 32 μM UbcH5c (S22R), and 3 mM ATP were incubated in buffer containing 50 mM MMT, pH 9.0, 25 mM NaCl, 3 mM MgCl2, and 1 mM TCEP for 24 h at 37 °C. The reaction was diluted >10-fold with buffer E (10 mM Hepes, pH 7.5, and 0.5 mM TCEP) supplemented with 100 mM NaCl and applied to a 5-mL QFF (Q Sepharose Fast Flow, GE Healthcare) that had been pre-equilibrated with buffer E supplemented with 100 mM NaCl. After washing the column with 20 CV of buffer E supplemented with 100 mM NaCl, bound protein was eluted with a 20-CV linear NaCl gradient (100–500 mM) in buffer E, collecting 1.25-mL fractions. Fractions were analyzed via SDS/PAGE, and peak fractions (∼280–440 mM NaCl) were pooled, diluted with buffer E to 150 mM NaCl, concentrated via Amicon 10K ultracentrifugation, and applied to a Hi-load Superdex 16/60 200 size exclusion column that had been pre-equilibrated in buffer E supplemented with 150 mM NaCl. Protein was eluted with buffer E supplemented with 150 mM NaCl, collecting 1.0-mL fractions. Fractions were analyzed by SDS/PAGE. Pooled fractions were concentrated via an Amicon 10 K ultracentrifugation filter, divided into aliquots, frozen in liquid N2, and stored at −80 °C (25).
The concentrations of the four-subunit DNA polymerase δ complex was expressed as the concentration of the p125 subunit. This was determined by slight modifications to a published protocol (27, 56). Purified four-subunit DNA polymerase δ complexes were separated on 10% denaturing SDS/PAGE together with known amounts of BSA to generate standard curves after Coomassie blue staining and densitometry of the gels. Digital images of the stained gels were analyzed with Image Lab 4.0 software (Bio-Rad).
Exonuclease Activity Assay.
Exonuclease reactions were carried out at room temperature and contained 1× replication buffer supplemented with 0.1 mg/mL BSA, 1 mM DTT, 1 mM ATP, and 200 nM of 5′-end–labeled 29-mer primer (either alone or annealed in the P/T substrate with 800 nM neutravidin). The reaction was initiated by addition of 200 nM of polymerase (either WT or exonuclease deficient), and aliquots were removed at variable times, quenched with 250 mM EDTA, pH 8.0, and diluted 20-fold with 70% formamide/25 mM EDTA/0.01% (wt/vol) tracking dyes. Exonuclease products were analyzed on 8% sequencing gels. Before loading onto gel, samples were heated at 95 °C for 5 min and immediately chilled in ice water for 5 min. Gel images were obtained on a Typhoon Model 9410 imager and quantitated with ImageQuant software (GE Healthcare). Although the polymerase and labeled primer were present in equimolar amounts, less than 20% of the substrate was degraded over the time course for most conditions. In these circumstances, the data were fit to a linear regression, and exonuclease activity was expressed as nanomolar primer degraded per minute. At the initial time point (1.5 min) for WT polymerase with single-stranded primer, all of the substrate was degraded, setting a lower limit for exonuclease activity of WT human pol δ at 200 nM/1.5 min = 133.33 nM/min.
Steady-State ATPase Activity.
ATPase activity of replication factor C (hRFCp140ΔN555) was assayed spectrophotometrically via an NADH oxidation enzyme-coupled assay as previously described (57, 58). The ATPase activity was determined at 25 °C in an assay solution containing 1× replication buffer [25 mM TrisOAc, pH 7.5, 10 mM Mg(OAc)2, 125 mM KOAc], 0.1 mg/mL BSA, 1 mM DTT, 1 mM ATP, 125 nM RFC, 125 nM forked DNA (500 nM Neutravidin), and 125 nM of either WT PCNA (wtPCNA) or monoubiquitinated PCNA [(Ub)3-PCNA]. The initial rates of ATP hydrolysis are reported.
Stopped-Flow Fluorescence Spectroscopy.
Stopped-flow studies were performed on an Applied Photophysics SX20 stopped-flow machine equipped with a fluorescence detector. PCNA unloading experiments were performed as described previously (5). In short, RFC•Cy5-PCNA•ATP•Cy3-P/T DNA in one syringe was mixed with unlabeled mutant PCNA in the other syringe. PCNA reloading experiments were performed exactly as described previously (5). In short, RFC•PCNA•ATP•Cy3-P/T DNA in one syringe was mixed with Cy5-PCNA in the other syringe. Loading and unloading was monitored by exciting the donor (Cy3) at 514 nm and following the resulting FRET signal using a 645-nm cutoff filter (Andover Corporation). All traces were fit to double exponential equations using Applied Photophysics ProData software. Conditions for each experiment are detailed in the figure captions.
Acknowledgments
We thank Dr. Senthil K. Perumal, a former member of the S.J.B. group, who expressed and purified the human replication protein A (RPA) used in all studies described in this report. This work was supported by National Institutes of Health Grant GM13306 (to S.J.B.). M.H. is supported by the National Cancer Institute of the National Institutes of Health under Award F32CA165471.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523653113/-/DCSupplemental.
References
- 1.Hedglin M, Kumar R, Benkovic SJ. Replication clamps and clamp loaders. Cold Spring Harb Perspect Biol. 2013;5(4):a010165. doi: 10.1101/cshperspect.a010165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hedglin M, Benkovic SJ. Regulation of Rad6/Rad18 activity during DNA damage tolerance. Annu Rev Biophys. 2015;44:207–228. doi: 10.1146/annurev-biophys-060414-033841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruning JB, Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure. 2004;12(12):2209–2219. doi: 10.1016/j.str.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Balakrishnan L, Bambara RA. Okazaki fragment metabolism. Cold Spring Harb Perspect Biol. 2013;5(2):a010173. doi: 10.1101/cshperspect.a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedglin M, Perumal SK, Hu Z, Benkovic S. Stepwise assembly of the human replicative polymerase holoenzyme. eLife. 2013;2:e00278. doi: 10.7554/eLife.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langston LD, O’Donnell M. DNA polymerase delta is highly processive with proliferating cell nuclear antigen and undergoes collision release upon completing DNA. J Biol Chem. 2008;283(43):29522–29531. doi: 10.1074/jbc.M804488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhuang Z, et al. Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc Natl Acad Sci USA. 2008;105(14):5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narita T, et al. Human replicative DNA polymerase δ can bypass T-T (6-4) ultraviolet photoproducts on template strands. Genes Cells. 2010;15(12):1228–1239. doi: 10.1111/j.1365-2443.2010.01457.x. [DOI] [PubMed] [Google Scholar]
- 9.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13(3):141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11(2):96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Washington MT, Carlson KD, Freudenthal BD, Pryor JM. Variations on a theme: Eukaryotic Y-family DNA polymerases. Biochim Biophys Acta. 2010;1804(5):1113–1123. doi: 10.1016/j.bbapap.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 13.Zhou JQ, Tan CK, So AG, Downey KM. Purification and characterization of the catalytic subunit of human DNA polymerase delta expressed in baculovirus-infected insect cells. J Biol Chem. 1996;271(47):29740–29745. doi: 10.1074/jbc.271.47.29740. [DOI] [PubMed] [Google Scholar]
- 14.Ducoux M, et al. Mediation of proliferating cell nuclear antigen (PCNA)-dependent DNA replication through a conserved p21(Cip1)-like PCNA-binding motif present in the third subunit of human DNA polymerase delta. J Biol Chem. 2001;276(52):49258–49266. doi: 10.1074/jbc.M106990200. [DOI] [PubMed] [Google Scholar]
- 15.Lee MY, Jiang YQ, Zhang SJ, Toomey NL. Characterization of human DNA polymerase delta and its immunochemical relationships with DNA polymerase alpha and epsilon. J Biol Chem. 1991;266(4):2423–2429. [PubMed] [Google Scholar]
- 16.Zhang P, et al. Expression of the catalytic subunit of human DNA polymerase delta in mammalian cells using a vaccinia virus vector system. J Biol Chem. 1995;270(14):7993–7998. doi: 10.1074/jbc.270.14.7993. [DOI] [PubMed] [Google Scholar]
- 17.Zhang SJ, et al. A conserved region in the amino terminus of DNA polymerase delta is involved in proliferating cell nuclear antigen binding. J Biol Chem. 1995;270(14):7988–7992. doi: 10.1074/jbc.270.14.7988. [DOI] [PubMed] [Google Scholar]
- 18.Shikata K, et al. The human homologue of fission Yeast cdc27, p66, is a component of active human DNA polymerase delta. J Biochem. 2001;129(5):699–708. doi: 10.1093/oxfordjournals.jbchem.a002909. [DOI] [PubMed] [Google Scholar]
- 19.Zhang P, et al. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase delta. J Biol Chem. 1999;274(38):26647–26653. doi: 10.1074/jbc.274.38.26647. [DOI] [PubMed] [Google Scholar]
- 20.Xu H, Zhang P, Liu L, Lee MY. A novel PCNA-binding motif identified by the panning of a random peptide display library. Biochemistry. 2001;40(14):4512–4520. doi: 10.1021/bi010103+. [DOI] [PubMed] [Google Scholar]
- 21.Li H, et al. Functional roles of p12, the fourth subunit of human DNA polymerase delta. J Biol Chem. 2006;281(21):14748–14755. doi: 10.1074/jbc.M600322200. [DOI] [PubMed] [Google Scholar]
- 22.Acharya N, Klassen R, Johnson RE, Prakash L, Prakash S. PCNA binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. Proc Natl Acad Sci USA. 2011;108(44):17927–17932. doi: 10.1073/pnas.1109981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Meng X, Zhang S, Lee EY, Lee MY. Characterization of human DNA polymerase delta and its subassemblies reconstituted by expression in the MultiBac system. PLoS One. 2012;7(6):e39156. doi: 10.1371/journal.pone.0039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendel A, et al. PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet. 2011;7(9):e1002262. doi: 10.1371/journal.pgen.1002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hibbert RG, Sixma TK. Intrinsic flexibility of ubiquitin on proliferating cell nuclear antigen (PCNA) in translesion synthesis. J Biol Chem. 2012;287(46):39216–39223. doi: 10.1074/jbc.M112.389890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc Natl Acad Sci USA. 2005;102(51):18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chilkova O, et al. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007;35(19):6588–6597. doi: 10.1093/nar/gkm741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng X, Zhou Y, Lee EY, Lee MY, Frick DN. The p12 subunit of human polymerase delta modulates the rate and fidelity of DNA synthesis. Biochemistry. 2010;49(17):3545–3554. doi: 10.1021/bi100042b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgers PM. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J Biol Chem. 1991;266(33):22698–22706. [PubMed] [Google Scholar]
- 30.Georgescu RE, et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21(8):664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, et al. A novel DNA damage response: Rapid degradation of the p12 subunit of dna polymerase delta. J Biol Chem. 2007;282(21):15330–15340. doi: 10.1074/jbc.M610356200. [DOI] [PubMed] [Google Scholar]
- 32.Rahmeh AA, et al. Phosphorylation of the p68 subunit of Pol δ acts as a molecular switch to regulate its interaction with PCNA. Biochemistry. 2012;51(1):416–424. doi: 10.1021/bi201638e. [DOI] [PubMed] [Google Scholar]
- 33.Podust VN, Chang LS, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase delta using recombinant baculoviruses: The p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J Biol Chem. 2002;277(6):3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- 34.Masuda Y, et al. Dynamics of human replication factors in the elongation phase of DNA replication. Nucleic Acids Res. 2007;35(20):6904–6916. doi: 10.1093/nar/gkm822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Z, Perumal SK, Yue H, Benkovic SJ. The human lagging strand DNA polymerase δ holoenzyme is distributive. J Biol Chem. 2012;287(46):38442–38448. doi: 10.1074/jbc.M112.404319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kochaniak AB, et al. Proliferating cell nuclear antigen uses two distinct modes to move along DNA. J Biol Chem. 2009;284(26):17700–17710. doi: 10.1074/jbc.M109.008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beck M, et al. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140(6):1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conti C, et al. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol Biol Cell. 2007;18(8):3059–3067. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462(7270):231–234. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kubota T, Katou Y, Nakato R, Shirahige K, Donaldson AD. Replication-Coupled PCNA Unloading by the Elg1 Complex Occurs Genome-wide and Requires Okazaki Fragment Ligation. Cell Reports. 2015;12(5):774–787. doi: 10.1016/j.celrep.2015.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mezzina M, Menck CF, Courtin P, Sarasin A. Replication of simian virus 40 DNA after UV irradiation: Evidence of growing fork blockage and single-stranded gaps in daughter strands. J Virol. 1988;62(11):4249–4258. doi: 10.1128/jvi.62.11.4249-4258.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berger CA, Edenberg HJ. Pyrimidine dimers block simian virus 40 replication forks. Mol Cell Biol. 1986;6(10):3443–3450. doi: 10.1128/mcb.6.10.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi JY, et al. Translesion synthesis across 1,N2-ethenoguanine by human DNA polymerases. Chem Res Toxicol. 2006;19(6):879–886. doi: 10.1021/tx060051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91(9):1163–1172. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11(2):96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hishiki A, et al. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J Biol Chem. 2009;284(16):10552–10560. doi: 10.1074/jbc.M809745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bomar MG, Pai MT, Tzeng SR, Li SS, Zhou P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase eta. EMBO Rep. 2007;8(3):247–251. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acharya N, Yoon JH, Hurwitz J, Prakash L, Prakash S. DNA polymerase eta lacking the ubiquitin-binding domain promotes replicative lesion bypass in humans cells. Proc Natl Acad Sci USA. 2010;107(23):10401–10405. doi: 10.1073/pnas.1005492107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svoboda DL, Vos JM. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: Fork uncoupling or gap formation. Proc Natl Acad Sci USA. 1995;92(26):11975–11979. doi: 10.1073/pnas.92.26.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Durando M, Tateishi S, Vaziri C. A non-catalytic role of DNA polymerase η in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res. 2013;41(5):3079–3093. doi: 10.1093/nar/gkt016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergoglio V, et al. DNA synthesis by Pol η promotes fragile site stability by preventing under-replicated DNA in mitosis. J Cell Biol. 2013;201(3):395–408. doi: 10.1083/jcb.201207066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson RE, Haracska L, Prakash S, Prakash L. Role of DNA polymerase eta in the bypass of a (6-4) TT photoproduct. Mol Cell Biol. 2001;21(10):3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon JH, Prakash L, Prakash S. Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci USA. 2009;106(43):18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kusumoto R, Masutani C, Shimmyo S, Iwai S, Hanaoka F. DNA binding properties of human DNA polymerase eta: Implications for fidelity and polymerase switching of translesion synthesis. Genes Cells. 2004;9(12):1139–1150. doi: 10.1111/j.1365-2443.2004.00797.x. [DOI] [PubMed] [Google Scholar]
- 56.Haracska L, et al. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol Cell Biol. 2001;21(21):7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kunkel TA, Pavlov YI, Bebenek K. Functions of human DNA polymerases eta, kappa and iota suggested by their properties, including fidelity with undamaged DNA templates. DNA Repair (Amst) 2003;2(2):135–149. doi: 10.1016/s1568-7864(02)00224-0. [DOI] [PubMed] [Google Scholar]
- 58.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell. 2008;30(4):519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 59.Temviriyanukul P, et al. Temporally distinct translesion synthesis pathways for ultraviolet light-induced photoproducts in the mammalian genome. DNA Repair (Amst) 2012;11(6):550–558. doi: 10.1016/j.dnarep.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 60.Schmutz V, et al. Role of the ubiquitin-binding domain of Polη in Rad18-independent translesion DNA synthesis in human cell extracts. Nucleic Acids Res. 2010;38(19):6456–6465. doi: 10.1093/nar/gkq403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor JS, O’Day CL. cis-syn thymine dimers are not absolute blocks to replication by DNA polymerase I of Escherichia coli in vitro. Biochemistry. 1990;29(6):1624–1632. doi: 10.1021/bi00458a038. [DOI] [PubMed] [Google Scholar]
- 62.Meneghini R. Gaps in DNA synthesized by ultraviolet light-irradiated WI38 human cells. Biochim Biophys Acta. 1976;425(4):419–427. doi: 10.1016/0005-2787(76)90006-x. [DOI] [PubMed] [Google Scholar]
- 63.Meneghini R, Cordeiro-Stone M, Schumacher RI. Size and frequency of gaps in newly synthesized DNA of xeroderma pigmentosum human cells irradiated with ultraviolet light. Biophys J. 1981;33(1):81–92. doi: 10.1016/S0006-3495(81)84873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Binz SK, Dickson AM, Haring SJ, Wold MS. Functional assays for replication protein A (RPA) Methods Enzymol. 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
- 65.Jónsson ZO, Podust VN, Podust LM, Hübscher U. Tyrosine 114 is essential for the trimeric structure and the functional activities of human proliferating cell nuclear antigen. EMBO J. 1995;14(22):5745–5751. doi: 10.1002/j.1460-2075.1995.tb00261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21(6):873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 67.Clausen AR, Zhang S, Burgers PM, Lee MY, Kunkel TA. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Repair (Amst) 2013;12(2):121–127. doi: 10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140(1):1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 69.Rey L, et al. Human DNA polymerase eta is required for common fragile site stability during unperturbed DNA replication. Mol Cell Biol. 2009;29(12):3344–3354. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Baranovskiy AG, Tahirov TH, Pavlov YI. The C-terminal domain of the DNA polymerase catalytic subunit regulates the primase and polymerase activities of the human DNA polymerase α-primase complex. J Biol Chem. 2014;289(32):22021–22034. doi: 10.1074/jbc.M114.570333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen B, et al. Diffusion of human replication protein A along single-stranded DNA. J Mol Biol. 2014;426(19):3246–3261. doi: 10.1016/j.jmb.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]