Significance
The regulation of ubiquitylation is critical to maintain proteomic and cellular homeostasis. The cullin-RING E3 ubiquitin ligases (CRLs) mediate one-fifth of all ubiquitylation, but their regulation is largely unknown. Here, we describe how the generation of a small metabolite, inositol hexakisphosphate (IP6), locally switches two CRLs from their active to their inactive state by means of stabilizing their interaction with an inhibitor: the constitutive photomorphogenesis 9 signalosome. Furthermore, we demonstrate the physiologic consequences of IP6 depletion on CRL dysregulation, CRL substrate levels, and global cellular phenotypes. Targeting IP6 synthesis synergizes with the cytotoxic effect of CRL inhibition, which may have therapeutic relevance.
Keywords: inositol hexakisphosphate, cullin RING E3 ligases, COP9 signalosome, IP5K, CRL
Abstract
The family of cullin-RING E3 Ligases (CRLs) and the constitutive photomorphogenesis 9 (COP9) signalosome (CSN) form dynamic complexes that mediate ubiquitylation of 20% of the proteome, yet regulation of their assembly/disassembly remains poorly understood. Inositol polyphosphates are highly conserved signaling molecules implicated in diverse cellular processes. We now report that inositol hexakisphosphate (IP6) is a major physiologic determinant of the CRL–CSN interface, which includes a hitherto unidentified electrostatic interaction between the N-terminal acidic tail of CSN subunit 2 (CSN2) and a conserved basic canyon on cullins. IP6, with an EC50 of 20 nM, acts as an intermolecular “glue,” increasing cullin–CSN2 binding affinity by 30-fold, thereby promoting assembly of the inactive CRL–CSN complexes. The IP6 synthase, Ins(1,3,4,5,6)P5 2-kinase (IPPK/IP5K) binds to cullins. Depleting IP5K increases the percentage of neddylated, active Cul1 and Cul4A, and decreases levels of the Cul1/4A substrates p27 and p21. Besides dysregulating CRL-mediated cell proliferation and UV-induced apoptosis, IP5K depletion potentiates by 28-fold the cytotoxic effect of the neddylation inhibitor MLN4924. Thus, IP5K and IP6 are evolutionarily conserved components of the CRL–CSN system and are potential targets for cancer therapy in conjunction with MLN4924.
Cullin-RING ligases (CRLs), comprising cullins (Cul 1–3, 4A/B, 5, 7, 9), RING finger Roc1/2, and cullin-specific adaptors, form a prominent family of multiprotein E3 ubiquitin ligases that together mediate 20% of proteasomal degradation (1, 2), and are emerging therapeutic targets (3). CRLs require neddylation, the attachment of an ubiquitin-like NEDD8 molecule, for optimal function (4, 5) and are tightly regulated by the constitutive photomorphogenesis 9 (COP9) signalosome (CSN), an eight-subunit deneddylase complex conserved from plants to humans (6, 7). CSN biochemically inhibits but genetically activates CRLs (8–14). Elucidating the mechanism of binding and dynamic disassembly of the CRL–CSN complexes is therefore key to understand physiological functions of CRLs (6). Despite recent progress in solving the crystal structures of CRLs (1, 15), CSN (16), and the electron microscopy structure of the CSN–CRL1 complex (10), structural elements mediating CRL–CSN interactions remain elusive. Moreover, the molecular switches underlying CRL–CSN complex dynamics are unclear.
The inositol polyphosphate pathway interacts with the CRL–CSN complexes via yet unspecified molecular mechanisms. Inositol polyphosphates (IP4, IP5, IP6) are highly conserved signaling molecules generated from the second messenger inositol 1,4,5-trisphosphate (IP3) by a family of inositol phosphate kinases (IPKs), including inositol 1,4,5-triphosphate 3-kinases, inositol 1,3,4-trisphosphate 5/6-kinase (ITPK1), inositol polyphosphate multikinase (IPMK), and inositol 1,3,4,5,6-pentakisphosphate 2-kinase (IP5K) (17). Majerus and colleagues reported that CSN copurifies with ITPK1 (18), which catalyzes the first committed step in forming higher inositol polyphosphates. However, the physiological significance of the ITPK1–CSN interaction remains unclear (19). IP6 can be further phosphorylated by IP6 kinases (IP6Ks) and diphosphoinositol pentakisphosphate kinases (PPIP5Ks) to generate highly energetic inositol pyrophosphate species (IP7 and IP8) (20). Recently, we reported that IP6 kinase-1 (IP6K1) binds both CSN and CRL4A to promote CRL4A–CSN complex formation in a catalytic activity-independent manner (21). Both kinase-dead IP6K1 and the IP6K inhibitor TNP [N(2)-(m-(trifluoromethy)lbenzyl) N(6)-(p-nitrobenzyl)purine] further stabilize the CRL4A–CSN complex, suggesting that the catalytic activity of IP6K1 (i.e., the turnover of IP6), negatively regulates CRL4A–CSN complex stability. However, the mechanism of action of IP6 and its related metabolites remains unknown.
In the present study, we delineate CRL–CSN interaction mechanisms, demonstrating an essential electrostatic interaction between the N-terminal acidic tail of CSN subunit 2 (CSN2) and a conserved basic canyon on cullins. Furthermore, we report that IP5K and IP6 physiologically mediate interactions between CSN2 and Cul1/4A to facilitate the assembly of CRL–CSN complexes and regulate their downstream functions. Together with earlier studies on IP6K1 (21), these findings establish that IP6, in conjunction with its metabolizing enzymes, determines the stability, activity, and dynamics of CRL–CSN complexes.
Results
The N-Terminal Acidic Tail of CSN2 Binds to the C-Terminal Basic Canyon of Cullins.
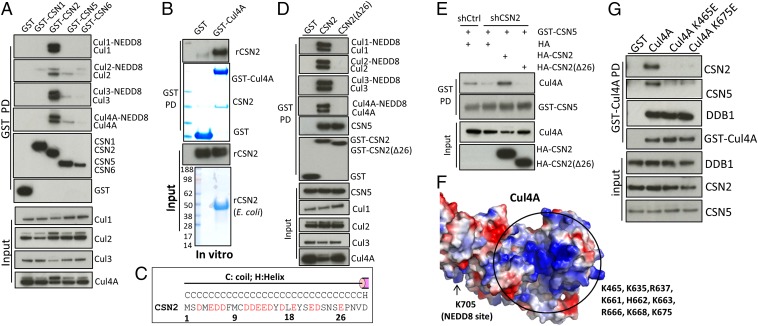
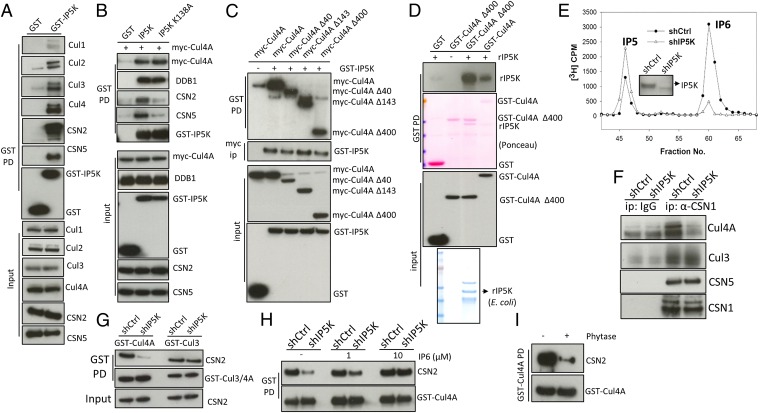
To investigate how higher inositol polyphosphates influence stability of the inactive CRL–CSN complexes, we first characterized the biochemical elements underlying CRL–CSN interactions. We previously showed that the N-terminal 189-aa region of CSN2 interacts with Cul1 in cells (22), a finding supported by recent electron microscopy data showing that CSN2 is located in proximity to Cul1 in the CRL1–CSN complex (10), although direct binding between cullins and CSN2 remains to be confirmed. GST pulldown of CSN2, but not CSN subunits 1/5/6, leads to robust coprecipitation of Cul1, -2, -3, and -4A (Fig. 1A), consistent with CSN2 being the major signalosome subunit interacting with cullins. Overexpressing CSN2, but not CSN1/5/6, enhances cullin neddylation (Fig. 1A), suggesting that overexpressed CSN2 acts as a dominant-negative to shield cullins from binding to and being deneddylated by endogenous CSN, consistent with CSN2 mediating contact between CSN and CRL. To validate direct CSN2–cullin interactions in vitro (Fig. 1B), we established a binding assay using bacterially purified CSN2 and HEK293-purified, salt-stripped (to remove endogenously bound CSN) GST-Cul4A. This direct binding does not require cullin neddylation, as the unneddylatable Cul4A K705A and K705R mutants binds CSN2 with similar affinity (Fig. S1A). Cullins form cognate heterodimers with RING finger proteins Roc1/2 (1, 2). Coexpression of Roc1 enhances cullin binding to CSN2 (Fig. S1B). Therefore, in subsequent experiments preparing purified cullins, we cotransfected Roc1.
Fig. 1.
The N-terminal acidic tail of CSN2 binds to the C-terminal basic canyons of cullins. (A) Pulldown of GST-CSN1/2/5/6 reveals coprecipitation of Cul1, -2, -3, and -4A with CSN2. (B) Direct interaction between E. coli-purified CSN2 and HEK293-purified GST-Cul4A. (C) Sequence the CSN2 N-terminal tail, the corresponding secondary structure was predicted by Psipred. Acidic residues in HsCSN2 are highlighted in red. (D) GST pulldown of CSN2 wild-type and its N-terminal deletion mutant Δ26. (E) The N-terminal tail of CSN2 is required for its ability to assemble CRL4A–CSN complexes. The shRNA targets the 3′-UTR of CSN2 and therefore does not influence transfected CSN2 (21). (F) Structure of Cul4A/Roc1 (PDB ID code 2HYE) shown in surface mode. The basic canyon is circled. (G) Mutations K465E and K675E abolish Cul4A binding to CSN2/5 but not to DDB1.
Fig. S1.
The N-terminal acidic tail of CSN2 binds to cullins. (A) Direct in vitro binding between CSN2 and GST-Cul4A wild-type, K705A, and K705R. (B) GST-Cul4A pulldown of recombinant CSN2 in the presence/absence of HA-Roc1. (C) High salt treatment markedly diminishes Cul4A binding to CSN subunits, with minimal effects on Cul4A–DDB1 interactions. (D) Sequence alignment of the CSN2 N-terminal tail, the corresponding secondary structure was predicted by Psipred. Acidic residues in HsCSN2 are highlighted in red. At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus. (E) Deletion of the N-terminal 17, but not the N-terminal 8, residues abolishes binding of CSN2 to cullins. (F) Pulldown of CSN2 wild-type and various mutants. (G) Amino acids 1–40 of CSN2 are sufficient to bind to Cul1, -2, -3, -4A. After transfection of the myc-cullin constructs, recombinant GST-rCSN21–40 on beads was incubated with cell lysates. (H) Recombinant CSN2 Δ26 is not pulled down by purified GST-Cul4A in vitro.
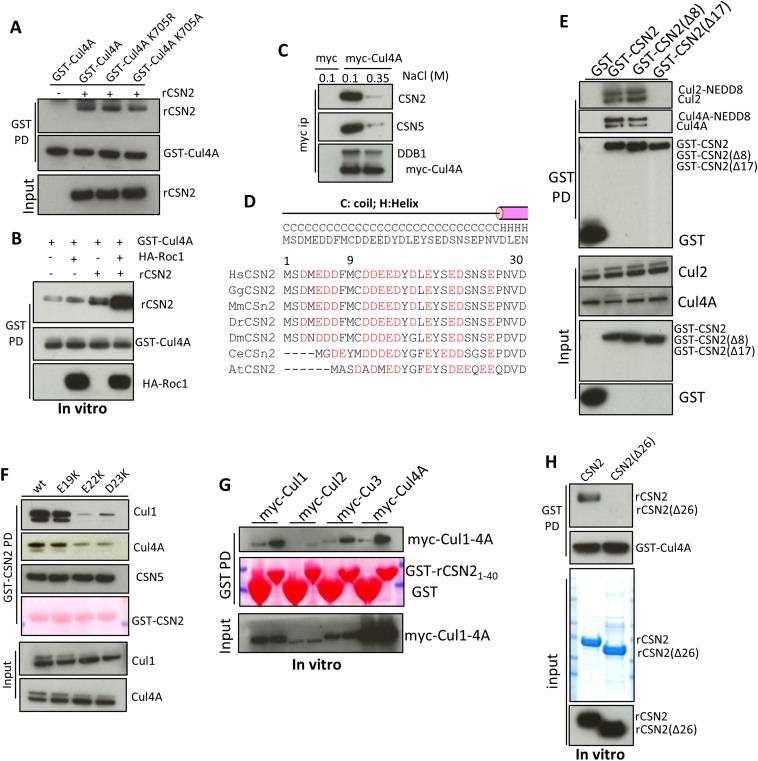
The CSN2–Cul4A interaction is markedly disrupted by increasing concentrations of NaCl (Fig. S1C), implying a primarily electrostatic linkage. Cullins contain a basic canyon (Fig. 1F and Fig. S2 A–D) that is adjacent to CSN in the CRL1–CSN electron microscopy structure (10). CSN2 displays a conserved, highly acidic N terminus (Fig. 1C and Fig. S1D), which is absent in the crystal structure of holo-signalosome (16), consistent with its predicted disposition as an unstructured tail (Fig. 1C). We wondered whether CSN2’s N-terminal tail binds cullins, and so examined its deletion. Deleting the N-terminal 26 aa of CSN2 does not influence its integration into the signalosome, reflected by equal binding to CSN5 (Fig. 1D). However, deleting the N-terminal 26 aa (Fig. 1D) or 17 aa (Fig. S1E) abolishes binding to Cul1, -2, -3, and -4A, whereas deleting the N-terminal 8 aa does not (Fig. S1E). Mutating residues between amino acids 17–26, including E19, E22, and D23 (Fig. S1F), also diminishes CSN2 binding to cullins. The purified, GST-tagged CSN2 N-terminal tail (amino acids 1–40) is sufficient to pull down cullins 1, -2, -3, and -4A from cell lysates (Fig. S1G), whereas recombinant CSN2 lacking the N-terminal 26 aa fails to bind purified Cul4A in vitro (Fig. S1H), consistent with the notion that the N terminus of CSN2 directly binds cullins. We therefore conclude that amino acids 9–26 of CSN2, conserved from plants to humans (Fig. S1D), are essential for direct cullin binding.
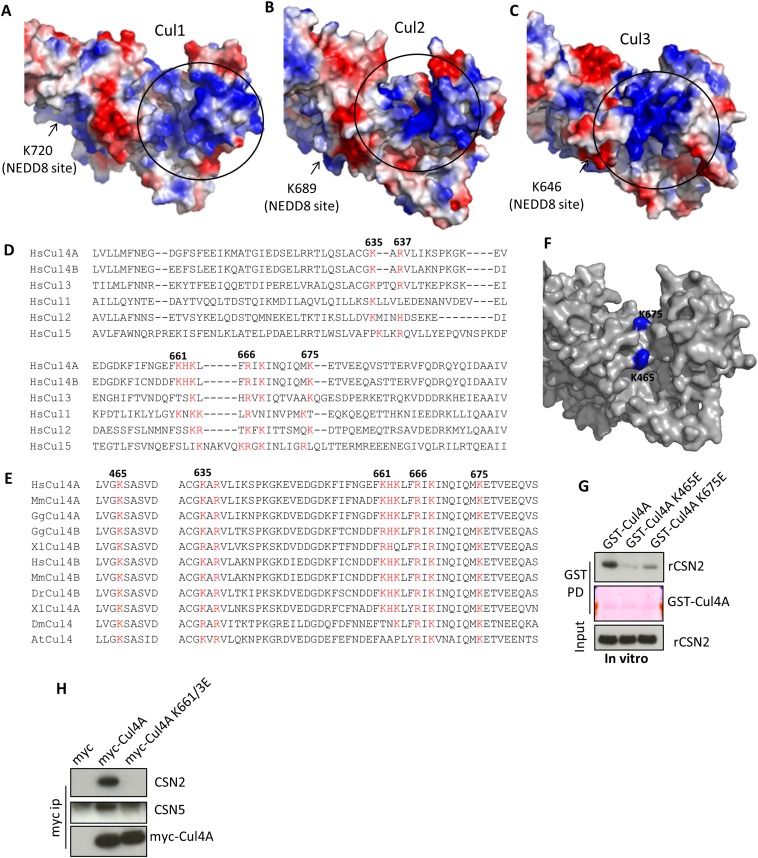
Fig. S2.
The basic canyon of cullins binds to CSN2. (A–C) Structure of Cul1 (A) (PDB ID code 1LDK) (15) and homology models of Cul2 (B) and Cul3 (C) shown in surface mode. The circled blue region is the basic canyon. (D) Sequence alignment of human Cul1–5. Residues highlighted in red are part of the basic canyon. (E) Sequence alignment Cul4A/B or the single Cul4 from different species. At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus; Xl, Xenopus laevis. (F) Lysines 465 and 675 (blue) are located in the groove the canyon. (G) In vitro binding of wildtype K465E or K675E mutants of Cul4A to recombinant CSN2. (H) Myc immunoprecipitation of Cul4A wild-type, the K661/663E mutant.
To examine whether CSN2’s N-terminal tail is critical for physiologic cullin–CSN assembly, we conducted rescue experiments using cells with CSN2 stably depleted as previously reported (21). In these cells, binding of cullins to other CSN subunits, such as CSN5, is markedly reduced (Fig. 1E). This defect is rescued by shRNA-resistant full-length but not N-terminal deleted CSN2 (Fig. 1E), consistent with a critical role for CSN2’s N-terminal tail in forming CRL–CSN complexes.
The requirement for the acidic N-terminal tail of CSN2 to bind all examined cullins suggests a common mechanism of interaction. We hypothesized that the basic canyon (23), conserved in all cullins (Fig. 1F and Fig. S2 A–D) and in cullin orthologs from plants to humans (Fig. S2E), interacts with the acidic tail (amino acids 9–26) of CSN2. To examine this possibility, we focused on Cul4A for in vitro studies because of its higher yield (among Cul1, -2, -3, and -4A) when purified from HEK293 cells as a GST-tagged protein. We hypothesize that Cul4A lysines 465 and 675, located in the bottom of the canyon (Fig. S2F), could mediate electrostatic contact with CSN2. Cul4A K465E and K675E mutants no longer bind CSN2 or CSN5, whereas Cul4A N-terminal binding to adaptor DDB1 (24) is not affected (Fig. 1G). Purified Cul4A K465E and K675E proteins manifest diminished ability to directly pull down recombinant CSN2 in vitro (Fig. S2G). Mutating other residues (K661/3E) in the basic canyon also abolishes interactions with CSN (Fig. S2H). Taken together, these data establish the importance of the basic canyon in Cul4A for binding to CSN2.
IP6 Augments Binding of Cullins to CSN2.
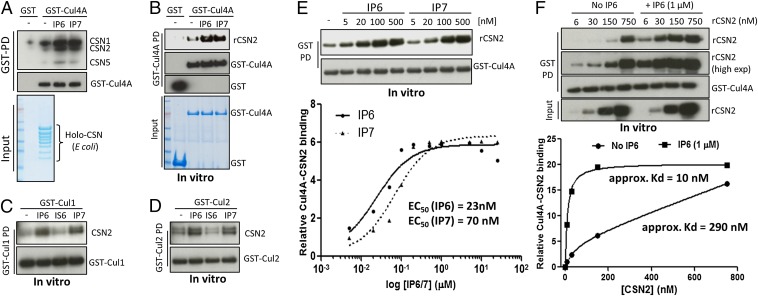
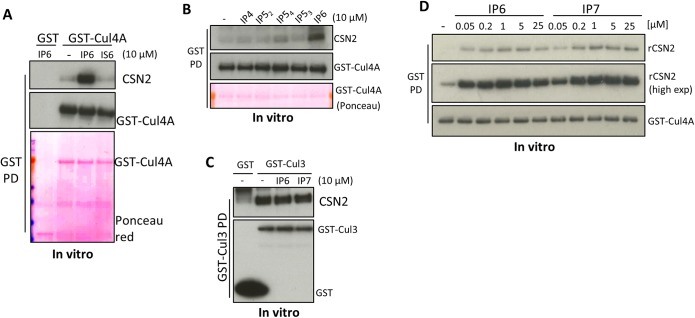
We hypothesized that highly charged inositol polyphosphates disrupt the electrostatic CRL–CSN2 interactions, thereby providing a regulatory means to control CRL–CSN complex disassembly. Unexpectedly, IP6 and IP7 markedly enhance Cul4A binding to the purified CSN holo-complex (Fig. 2A). IP6/7 acts directly on the cullin–CSN2 interface, because Cul4A–CSN2 binding is also augmented by IP6/7 (Fig. 2B), but not by inositol hexakis-sulfate (IS6) (Fig. S3A), or IP4 and IP5 isomers, except for some modest effects of Ins(1,2,3,5,6)P5 (IP54) (Fig. S3B). IP6/7 also augments in vitro binding of CSN2 to Cul1 and to some extent Cul2 (Fig. 2 C and D), but not Cul3 (Fig. S3C).
Fig. 2.
IP6 stimulates CSN2–Cul4A binding in vitro. (A) IP6 and IP7 stimulate in vitro binding between HEK293-purified Cul4A and recombinant holo-signalosome. GST-Cul4A pulldown was blotted for CSN1, -2, and -5 simultaneously. (B) IP6 and IP7 stimulate in vitro binding between purified Cul4A and recombinant CSN2. (C and D) IP6 stimulates binding between recombinant CSN2 and HEK293-purified Cul1 (C) and Cul2 (D). (E) Concentration-dependent analysis of IP6 in stimulating CSN2–Cul4A binding. (F) CSN2 concentration-dependent analysis of CSN2–Cul4A binding with/without IP6.
Fig. S3.
IP6 stimulates CSN2–Cul4A binding in vitro. (A) IP6, but not IS6, stimulates in vitro binding between purified Cul4A and CSN2. (B) IP6 but not IP4, or the various IP5 isomers, stimulate in vitro binding between purified Cul4A and CSN2. IP52: Ins(1,3,4,5,6)P5, IP53: Ins(1,2,4,5,6)P5, IP54: Ins(1,2,3,5,6)P5. (C) IP6 does not stimulate CSN2-Cul3 binding in vitro. (D) IP6/7 dose-dependent analysis for the potency of IP6/7 to stimulate CSN2–Cul4A binding.
IP6 dose–response analysis reveals significant effects at 5 nM IP6 and maximal actions at 200 nM (Fig. 2E and Fig. S3D), concentrations substantially lower than physiologic, intracellular IP6 levels, which range from 10 to 80 μM (25). Other IP6-binding modules, where IP6 often acts as intermolecular bridge, have similarly high affinity (26–30). IP7 is somewhat less potent than IP6 (Fig. 2E), with EC50 values for IP7 (70 nM) about threefold higher than for IP6 (23 nM). Because intracellular IP7 concentration is more than 10-fold lower than IP6 (20), the weaker potency of IP7 suggests that IP6 is the physiologic mediator. To estimate the efficacy of IP6 at promoting CSN2–Cul4A binding, we fixed IP6 concentration at 1 µM (saturating) and performed rCSN2 concentration-response analysis, which reveals a marked left-shift in the presence of IP6, with a 29-fold increase in affinity (Fig. 2F).
IP5K Binds to CRLs and Generates Physiologic IP6 to Mediate CRL–CSN Complex Stability.
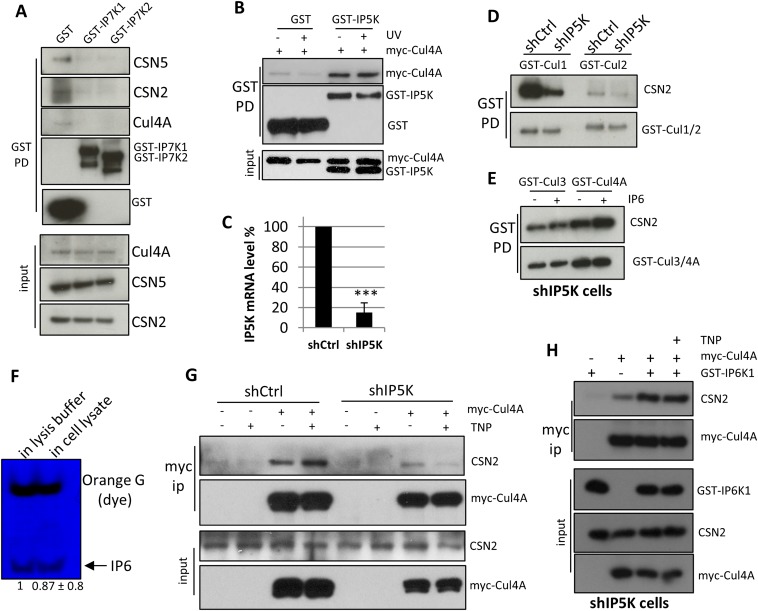
Thus far we have shown that IP6 potently and efficiently increases the affinity of certain cullin–CSN2 complexes in purified systems. To assess whether IP6 is physiologically required for CSN2–cullin binding, we examined Ins(1,3,4,5,6)P5 2-Kinase (IP5K/IPPK), the only enzyme that generates IP6 in human cells (17). IP5K (Fig. 3A), but not the inositol pyrophosphate-generating PPIP5Ks (Fig. S4A), coprecipitates with CSN and cullins. Kinase-dead IP5K, IP5K-K138A (31) pulls down Cul4A and DDB1 equally well but only weakly pulls down CSN2/5 (Fig. 3B), suggesting that IP5K binds directly to CRLs but not CSN. Further fragment mapping experiments suggest that the N-terminal domain of Cul4A, which mediates binding to the adaptor protein DDB1 (24), is not required for binding IP5K (Fig. 3C). Indeed, using purified recombinant proteins, we observe direct interactions between the C-terminal domain of Cul4A (amino acids 401–end) and IP5K (Fig. 3D). Taken together, these data indicate that IP5K binds to CRL in a kinase-independent fashion, locally generating IP6, which augments CRL–CSN binding in a kinase-dependent fashion.
Fig. 3.
IP5K binds CRL, generating IP6 to assemble CRL–CSN complexes in vivo. (A) IP5K pulls down CSN subunits and the cullins. (B) Coexpression of myc-Cul4A and GST-IP5K or its kinase-dead K138A mutant, followed by GST pulldown. (C) Coexpression of GST-IP5K and myc-Cul4A or its various N-terminal truncation mutants, followed by GST pulldown and myc immunoprecipitation. (D) Direct in vitro binding between recombinant IP5K and HEK293-purified Cul4A wild-type or the C-terminal domain (Δ400). (E) Inositol profiling of cells with shRNA-mediated knockdown of IP5K. (Inset) Western blotting of IP5K. (F) CSN1 immunoprecipitation in control and IP5K knockdown cells. (G) GST pulldown of Cul4A and Cul3 in control and IP5K knockdown cells. (H) IP6 addition to cell lysates increases binding between Cul4A and CSN in IP5K knockdown but not control cells. (I) Bacterial phytase treatment (10 µg/mL, 12 h, 4 °C) disrupts the CRL4A–CSN complex.
Fig. S4.
IP5K binds CRL, generating IP6 to assemble CRL–CSN complexes in vivo. (A) GST-PPIP5K1/2 pulldown does not coprecipitate with CSN subunits or cullins. (B) GST-IP5K pulldown of Cul4A and CSN2 with/without UV radiation (100 J/m2). (C) Real-time PCR analysis of IP5K knockdown. PCR was performed as previously described (47). (D) shRNA-mediated knockdown of IP5K diminishes binding between CSN2 and Cul1/2. (E) IP6 (1 µM) increases Cul4A–CSN, but not Cul3–CSN, complex formation in IP5K knockdown cells. (F) PAGE imaging of IP6 after incubation with cell lysis buffer or cell lysate. (G) TNP (10 µM) increases Cul4A–CSN2 binding in wild-type but not IP5K knockdown cells. (H) Overexpressing IP6K increases Cul4A–CSN2 binding in IP5K knockdown cells, whereas further addition of TNP has no effect on Cul4A–CSN2 interactions in IP5K knockdown cells.
We previously reported that the IP6 metabolizing enzyme IP6K1 binds to CRL4A to promote the formation of the inactive CRL4A–CSN complex (21). Furthermore, this interaction is disrupted by UV radiation, leading to CRL activation (21). In contrast to IP6K1, IP5K binding to Cul4A is not altered upon UV stimulation (Fig. S4B), suggesting that IP5K constitutively associates with Cul4A, and that IP6 is basally required for CRL4A–CSN interaction.
To further validate the role of IP5K, we examined its depletion. shRNA targeting IP5K depletes IP5K mRNA (Fig. S4C), protein, and IP6 levels (Fig. 3E). This depletion of IP5K also markedly diminishes binding of CSN to both endogenous (Fig. 3F) and overexpressed (Fig. 3G) Cul4A, but not Cul3, without affecting interactions among CSN subunits (Fig. 3F). Binding of CSN2 to Cul1 and Cul2 is also decreased with IP5K depletion (Fig. S4D). Taken together, these data suggest that IP5K differentially influences specific cullin–CSN complexes in a manner consistent with the observed effects of IP6 on these interactions in vitro (Fig. 2 B–D and Fig. S3C).
To confirm that IP5K knockdown acts by depleting IP6, we supplemented cell lysates with exogenous IP6. Adding IP6 to cell lysates with IP5K knockdown, but not control cell lysates, dose-dependently rescues Cul4A–CSN binding (Fig. 3H), with minimal effect on Cul3–CSN interactions (Fig. S4E). The effect of IP6 is unlikely to be caused by its downstream metabolites because it is stable under the assay conditions (Fig. S4F). These data establish that IP6 physiologically mediates specific CSN–CRL interactions. Finally, treating affinity-precipitated GST-Cul4A with bacterial phytase, an IP6 phosphatase, dissociates CSN from CRL4A (Fig. 3I), further supporting IP6 but not its metabolites as a physiologic cofactor in the CRL–CSN complex.
Because both IP5K and IP6K1 interact with Cul4A and metabolize inositol phosphates, we next examined how IP6K1 influences CRL4A–CSN dynamics in the absence of IP5K (21, 32). Consistent with previous data, the IP6K inhibitor TNP increases CRL4A–CSN2 binding in IP5K-proficient cells (Fig. S4G). However, TNP fail to increase CRL4A-CSN2 binding in IP5K knockdown cells (Fig. S4G). Overexpressing IP6K1 in IP5K knockdown cells still rescues the diminished CRL4A–CSN2 interaction, but TNP no longer has any effect (Fig. S4H). Taken together, these data further support our previous conclusion that IP6K1’s stabilization of CRL4A–CSN interaction is kinase activity-independent, whereas its role in dissociating CRL4A–CSN complexes is kinase activity-dependent (21), and thus dependent on IP6 generated by IP5K.
IP5K Regulates CRL1/4A Neddylation and Physiological Function.
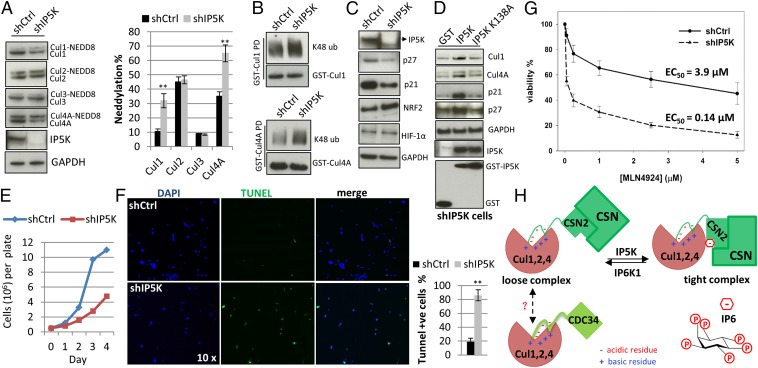
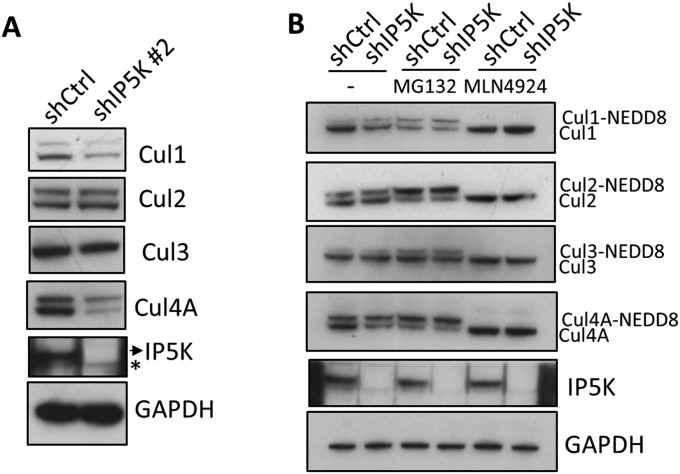
To understand the role of IP5K/IP6 in cullin physiology, we examined the functional consequences of IP5K knockdown and the resulting loss of CSN binding to cullins. Consistent with CSN being a cullin deneddylase, IP5K knockdown leads to an increased ratio of neddylated to unneddylated Cul1 and Cul4A, without notable effects on Cul2 and Cul3 (Fig. 4A). Similar results are obtained using a different shRNA (Fig. S5A). Total levels of Cul1 and Cul4A are lower in the absence of IP5K (Fig. 4A and Fig. S5A), consistent with the protective role of CSN-associated deubiquitinases in preventing self-ubiquitylation and -destruction of neddylated, active CRLs (33, 34). Indeed, ubiquitylation of Cul1 and Cul4A is augmented in IP5K knockdown cells (Fig. 4B). Moreover, the proteasome inhibitor MG132 and the neddylation inhibitor MLN4924 (35) both nullify the difference between control and IP5K knockdown cells for both neddylation/unneddylation ratio and cullin protein abundance for Cul1/Cul4A (Fig. S5B). These findings imply that the defects in IP5K knockdown cells reflect altered neddylation and self-ubiquitylation.
Fig. 4.
IP5K regulates Cul1/4A neddylation and function. (A) Levels of neddylated cullins in control and shIP5K cells. Lysis buffer contains 1 mM N-ethylmaleimide to minimize deneddylation during processing. (B) Increased levels of K48-linkage–specific polyubiquitination of Cul1 and Cul4A in IP5K knockdown cells. (C) Levels of cullin substrates. (D) Rescue of shIP5K cells with wildtype or kinase-dead IP5K that are resistant to the shRNA because of synonymous mutations. (E) Growth curve of control and shIP5K cells. (F) IP5K depletion exacerbates UV-induced apoptosis, as measured by TUNEL assay. Cells were treated with UV (100 J/m2). After 20 h, cells were stained using the terminal deoxynucleotidyl transferase (TdT)-mediated TUNEL assay system. Fragmented apoptotic cell nuclei were visualized by TUNEL (TdT, green), and the nucleus was stained with DAPI (blue). (G) Viability of control and shIP5K cells after treatment with MLN4924 (0.05, 0.25, 1, 2.5, 5 µM) for 48 h. (H) Scheme depicting the interconversion of cullins between a CSN-bound, inactive state and a CDC34-bound, active state. In this model, CSN2 and E2 ligase CDC34 both bind to cullins’ basic canyon via electrostatic interactions (23). IP6 generated by IP5K mediates high-affinity, inactive cullin–CSN complexes, whereas IP6K1 metabolizes IP6 leading to decreased complex affinity (21), which may be followed by cullin activation and CDC34 binding.
Fig. S5.
IP5K regulates Cul1/4A neddylation and function. (A) Levels of neddylated cullins in control and shIP5K (#2) cells. (B) Levels of neddylated cullins with/without MG132 (10 µM) and MLN4924 (0.2 µM) treatments (4 h).
The increased ratio of neddylated to unneddylated Cul1/4A with IP5K loss suggests that CRL1 and CRL4A are more active in IP5K knockdown cells. Accordingly, we examined the abundance of cullin substrates. Protein levels of p27 and p21, well-established CRL1 and CRL4A substrates, respectively, are significantly diminished upon IP5K depletion (Fig. 4C), whereas HIF1-α and NRF2, substrates of CRL2 and CRL3, respectively, are unaffected. The altered Cul4A neddylation ratio and diminished p21 and p27 levels in IP5K knockdown cells are rescued by the expression of shRNA-resistant IP5K, but not its K138A mutant (Fig. 4D). Taken together, these studies indicate that depleting IP6 levels by IP5K knockdown leads to CSN dissociation from CRL1 and CRL4A, resulting in aberrant CRL1 and CRL4A activation.
CRLs target numerous substrates and contribute to a major portion of proteome homeostasis (2). CRL1 and CRL4A, in particular, regulate cell growth and death, with a well-known role for CRL4A in UV-induced apoptosis (2). Accordingly, we examined these traits in cells following IP5K knockdown. Under basal conditions, IP5K knockdown results in pronounced growth defects (Fig. 4E). Moreover, these cells are highly susceptible to UV-induced apoptosis (Fig. 4F). MLN4924-elicited cytotoxicity is also synergized by IP5K depletion (Fig. 4G), consistent with a central role for IP5K/IP6 in regulating CRL1 and CRL4A neddylation and function.
Discussion
How the assembly of CRL–CSN complexes is dynamically regulated has been enigmatic, which in part explains why few stimuli triggering CRL E3 ligase activity have been identified. Given the mutually exclusive binding of substrate and CSN to substrate receptor (10–12), CSN dissociation and CRL activation may depend on substrate encounter (12). However, the rapid degradation of certain substrates (e.g., CDT1) in response to certain stimuli (e.g., UV light) suggests a complementary, more proactive mechanism. Here, we provide evidence that IP6, with nanomolar potency, participates in CRL–CSN dynamics, particularly for CRL1/4A. The CRLs affect fundamental cellular processes such as cell growth/death and are deregulated in diverse diseases, especially in cancer (3, 36). Targeting IP5K/IP6 and its interface with CRL–CSN in combination therapy may alleviate the dose-limiting toxicity of MLN4924 observed in clinical trials (37).
Prior data have suggested that CSN2 is the subunit of CSN that binds cullins (10, 22). We now demonstrate their direct binding in vitro. Moreover, we define the precise structural elements mediating the formation of CRL–CSN complexes. Binding of the acidic tail (amino acids 9–26) of CSN2 to the basic canyon of cullins is reminiscent of interactions between cullins and the E2 ligase CDC34, which also binds to the basic canyon via its C-terminal acidic tail (23). Because the CDC34-bound and CSN-bound forms of cullins represent active and inactive state, respectively, competition between CSN2 and CDC34 could be a novel mechanism whereby CSN inhibits CRL (Fig. 4H).
IP6 is commonly found at protein interaction interfaces, where it usually acts as a “glue” molecule enhancing binding affinity. However, previous studies of IP6’s complex-stabilizing role rarely extended to its synthase, IP5K (26, 27, 30). We found that genetic manipulation of IP5K alters CRL1/4A–CSN complex stability, CRL1/4A neddylation status, and downstream CRL1/4A function in a cellular context. By directly binding to the C-terminal domain of cullins, IP5K provides a local pool of IP6 to coordinate CRL–CSN complexes. Prior work has also shown that ITPK1, another enzyme involved in IP6 synthesis, associates with CSN via direct binding to CSN1 (38). Taken together, these data indicate that a local cluster of IPKs may position inositol polyphosphate intermediates for efficient IP6 generation, analogous to the coupling of classic metabolic enzymes (39).
The role of IP7 in the CRL–CSN complex remains enigmatic. Recently, we reported that, although IP6K1 protein itself stabilizes CRL4A–CSN complexes, the catalytic activity of IP6K1 is required to dissociate CSN from CRL4A in vivo (21). In vitro, IP7 also promotes CRL4A–CSN2 binding, but with threefold lower potency than IP6 (Fig. 2E). Together with the known instability and low cellular concentration of IP7 relative to IP6, the data strongly suggest that IP7 is not a physiologic “glue,” but rather is behaving differently. IP7 uniquely pyrophosphorylates proteins that have undergone casein kinase 2 (CK2)-mediated phosphorylation (40). CK2 is associated with purified CSN and phosphorylates CNS2 and CSN5 (41). Whether physiologic pyrophosphorylation events underlie IP7’s putative involvement in inhibiting CRL–CSN complex formation remains to be examined, pending the development of tools necessary to probe pyrophosphorylation in a cellular context. Moreover, as IP6K1 can also dephosphorylate IP6 to IP5 (42), which is ineffective in assembling CRL–CSN complexes (Fig. S3B), whether the cellular IP6K1 data reflects IP6’s turnover to IP7 or IP5 needs to be examined. Nonetheless, our finding that IP6 promotes CRL–CSN complex assembly, whereas its metabolism by IP6K1 disassembles the complex, offers a new perspective on IP6/IP6K biology: namely, IP6Ks catalyze local conversion of IP6 to IP7/IP5 to oppose IP6-mediated effects (Fig. 4H). Consistently, UV-elicited cell death is enhanced by IP5K depletion, but decreased by IP6K1 deletion, possibly because of a differential effect on cullin neddylation; loss of IP5K, but not IP6K1 increases the percentage of neddylated cullins (Fig. 4A) (21). This antagonism between upstream and downstream kinases of IP6 metabolism is also found in regulating the tumor suppressor LKB1. Catalytic activity of IPMK activates LKB1 (43), whereas that of IP6K2 suppresses it (44). IP6 plays structural roles in a number of important cellular machineries (26, 27, 30, 45), which might also be regulated by one of the three known IP6Ks.
Materials and Methods
Materials.
Cell culture, cell counting, and cell death assays (TUNEL and MTT), [3H] inositol profiling, and lentiviral knockdown, Western blotting, real-time PCR, and IP6 imaging by the PAGE method were as described previously (46, 47). The primary antibodies used were: Cul1, Cul2, HIF-1α, p21, p27 (Santa Cruz); Cul3 Cul4A, CSN5, K48-specific polyubiquitin, and DDB1 (Cell Signaling); GAPDH (Roche); IP5K and CSN2 (ProteinTech); NRF2 (R&D Systems); and GST (Sigma). Polyclonal CSN1 antibody was as described previously (22).
Expression and Purification of Recombinant Protein from Escherichia coli.
The CSN holo-complex and CSN2 was purified and reconstituted as previously described (32, 48).
In Vitro Binding.
GST-cullins were expressed and purified from HEK293 cells using glutathione beads. Every single binding experiment used cullins purified from one 10-cm plate. Endogenous CSN was stripped away by washing in high salt buffer (50 mM Tris⋅HCl, pH 7.5, 350–400 mM NaCl, l% Triton X-100). After washing and equilibration with binding buffer [50 mM Tris (pH 7.4), 100 mM NaCl, 0.8% Triton X-100, and 3% (vol/vol) glycerol], the glutathione beads with bound cullins was used for in vitro binding to purified recombinant CSN2 purified. The EC50 for IP6/7 was determined by fitting the data with the dose–response (stimulation) Equation: Y = Bottom + (Top − Bottom)/[1+10(LogEC50-x)] (GraphPad). The approximate Kd for CSN2 with/without IP6 was estimated using a one-site binding (with saturation) equation (GraphPad).
Computational Modeling.
Protein secondary structure was predicted using the Psipred serve r (49). Three-dimensional homology models of full-length Cul2 and Cul3 were generated by I-Tasser (50), using Cul4A structure as a template (PDB ID code 2HYE) (24). Images were generated by using Pymol.
Statistical Analysis.
All results are presented as the mean and SE of at least three independent experiments. Statistical significance was calculated by Student’s t test (*P < 0.05, **P < 0.01).
GST pulldown, coimmunoprecipitation, phytase treatment, computational modeling, and a complete list of antibodies and constructs are described in SI Materials and Methods.
SI Materials and Methods
Materials.
HEK293 cells were cultured in DMEM/10% FBS/1% glutamine/1% Penstrep at 5% CO2 (vol/vol). Cell counting and cell death assays (TUNEL and MTT) were as described previously (46). The primary antibodies used were: Cul1, Cul2, HIF-1α, p21, p27 (Santa Cruz); Cul3 Cul4A, CSN5, K48-specific polyubiquitin, and DDB1 (Cell Signaling); GAPDH (Roche); IP5K and CSN2 (ProteinTech); NRF2 (R&D Systems); and GST (Sigma). Polyclonal CSN1 antibody was as described previously (22). shRNA for CSN2 (id: 1773s21c1), IP5K (#1: 1399s1c1; #2: 667s21c1), control scrambled shRNA and TNP were purchased from Sigma. The CSN2 knockdown cell line was generated previously (32). IgG beads were purchased from GE Healthcare. UV cross-linker was from UVP, LLC. All other reagents were purchased from Sigma unless otherwise specified.
Plasmids.
pcDNA3-myc3-CuL1, -2, -3, -4A were from Addgene and were then subcloned into pCMV-GST. CSN1, CSN2, CSN5, CSN6 were cloned into pGEX6p2, pCMV-HA, and pCMV-GST vectors at SalI/NotI restriction sites. IP5K gene was from Harvard Plasmid ID Database and was cloned into pCMV-GST vector. All point mutants were made using a site-directed mutagenesis kit (Stratagene). The lentiviral knockdown system used was as described previously (46).
GST Pulldown and Coimmunoprecipitation.
Cells were lysed in lysis buffer (50 mM Tris⋅HCl, pH 7.5, 125 mM NaCl, l% Triton X-100, 1× phosphatase inhibitor and 1× protease inhibitor). For GST pulldown experiments, cell lysate was incubated with glutathione beads at 4 °C for 2 h. For immunoprecipitation experiments, cell lysate was first incubated with desired antibody at 4 °C overnight, followed by incubation with protein A/G plus beads (Santa Cruz) at 4 °C for another 1 h. Beads were then washed three times with lysis buffer and boiled in SDS sample buffer. Western blotting was performed as previously described (46).
Expression and Purification of Recombinant Protein from Escherichia coli.
The CSN2 holo-complex was purified and reconstituted as previously described (48). For GST-tagged CSN2, the plasmids were transformed into BL21 (DE3). Initial purification used Glutathione Sepharose 4B (GE Healthcare). The beads were then washed and either incubated with 15 mM reduced glutathione (Sigma) for elution, or with Precission Protease (GE) to remove the GST tag. The supernatants were collected and analyzed by SDS/PAGE, followed by staining using SimplyBlue SafeStain (Invitrogen).
In Vitro Binding.
GST-cullins were expressed and purified from HEK293 cells using glutathione beads. Every single binding experiment used cullins purified from one 10-cm plate. Endogenous CSN was stripped away by washing in high salt buffer (50 mM Tris⋅HCl, pH 7.5, 350–400 mM NaCl, l% Triton X-100). After washing and equilibration with binding buffer [50 mM Tris (pH 7.4), 100 mM NaCl, 0.8% Triton X-100, and 3% glycerol], the glutathione beads with bound cullins were used for in vitro binding to recombinant CSN2 purified as stated above. To determine the stimulatory effects of IP6/7, the EC50 was determined by fitting the data with the dose–response (stimulation) Equation: Y = Bottom + (Top − Bottom)/[1+10(LogEC50−x)] (GraphPad). The approximate Kd for CSN2 with/without IP6 was estimated using a one-site binding (with saturation) equation (GraphPad).
Computational Modeling.
Protein secondary structure was predicted using the Psipred serve r (49). Three-dimensional homology models of full-length Cul2 and Cul3 were generated by I-Tasser (50), using Cul4A structure as a template (PDB ID code 2HYE) (24). Images were generated by using Pymol.
[3H] Inositol Profiling.
Radiolabeling with [3H] Inositol and HPLC detection inositol phosphates were performed as described previously (46).
IP6 Stability Measurement.
To determine the metabolic fate of exogenous IP6 after incubation with cell lysate, samples were analyzed for IP6 levels using the PAGE method (51), with IP6 in lysis buffer as control.
Phytase Treatment.
Cell lysates pulled down on glutathione beads were treated with bacterial phytase (10 µg/mL) for 12 h at 4 °C with rotation.
Statistical Analysis.
All results are presented as the mean and SE of at least three independent experiments. Statistical significance was calculated by Student’s t test (*P < 0.05, **P < 0.01).
Acknowledgments
We thank F. Chang for technical assistance. This work was supported by US Public Health Service Grant DA-000266 (to S.H.S.), and by grants from the Chinese Ministry of Science and Technology and the Beijing municipal government (to F.R.).
Footnotes
Conflict of interest statement: Dr. York and the authors have independently supplied reagents to the same laboratory and have therefore appeared as coauthors in a paper published as Pulloor NK, et al. (2014) Human genome-wide RNAi screen identifies an essential role for inositol pyrophosphates in type-I interferon response. PLoS Pathog 10(2):e1003981, although we did so independently and without knowledge of the other’s participation in the work.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525580113/-/DCSupplemental.
References
- 1.Lydeard JR, Schulman BA, Harper JW. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013;14(12):1050–1061. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 3.Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov. 2014;13(12):889–903. doi: 10.1038/nrd4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: Conformational control of conjugation. Cell. 2008;134(6):995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32(1):21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolf DA, Zhou C, Wee S. The COP9 signalosome: An assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol. 2003;5(12):1029–1033. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- 7.Wei N, Chamovitz DA, Deng XW. Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell. 1994;78(1):117–124. doi: 10.1016/0092-8674(94)90578-9. [DOI] [PubMed] [Google Scholar]
- 8.Lyapina S, et al. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292(5520):1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 9.Cope GA, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298(5593):608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 10.Enchev RI, et al. Structural basis for a reciprocal regulation between SCF and CSN. Cell Reports. 2012;2(3):616–627. doi: 10.1016/j.celrep.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emberley ED, Mosadeghi R, Deshaies RJ. Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. J Biol Chem. 2012;287(35):29679–29689. doi: 10.1074/jbc.M112.352484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer ES, et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147(5):1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 13.Schwechheimer C, et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292(5520):1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- 14.Doronkin S, Djagaeva I, Beckendorf SK. The COP9 signalosome promotes degradation of Cyclin E during early Drosophila oogenesis. Dev Cell. 2003;4(5):699–710. doi: 10.1016/s1534-5807(03)00121-7. [DOI] [PubMed] [Google Scholar]
- 15.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416(6882):703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 16.Lingaraju GM, et al. Crystal structure of the human COP9 signalosome. Nature. 2014;512(7513):161–165. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- 17.Hatch AJ, York JD. SnapShot: Inositol phosphates. Cell. 2010;143(6):1030–1030.e1. doi: 10.1016/j.cell.2010.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson MP, Sun Y, Cao L, Majerus PW. Inositol 1,3,4-trisphosphate 5/6-kinase is a protein kinase that phosphorylates the transcription factors c-Jun and ATF-2. J Biol Chem. 2001;276(44):40998–41004. doi: 10.1074/jbc.M106605200. [DOI] [PubMed] [Google Scholar]
- 19.Qian X, et al. The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase. Biochem J. 2005;389(Pt 2):389–395. doi: 10.1042/BJ20050297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shears SB. Inositol pyrophosphates: Why so many phosphates? Adv Biol Regul. 2015;57:203–216. doi: 10.1016/j.jbior.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao F, et al. Inositol hexakisphosphate kinase-1 mediates assembly/disassembly of the CRL4-signalosome complex to regulate DNA repair and cell death. Proc Natl Acad Sci USA. 2014;111(45):16005–16010. doi: 10.1073/pnas.1417900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, et al. The COP9 signalosome inhibits p27(kip1) degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr Biol. 2002;12(8):667–672. doi: 10.1016/s0960-9822(02)00791-1. [DOI] [PubMed] [Google Scholar]
- 23.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139(5):957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angers S, et al. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443(7111):590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 25.Irvine RF. Inositide evolution—Towards turtle domination? J Physiol. 2005;566(Pt 2):295–300. doi: 10.1113/jphysiol.2005.087387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macbeth MR, et al. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309(5740):1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcázar-Román AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8(7):711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 28.Wang Q, et al. Autoinhibition of Bruton’s tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate. eLife. 2015;4:4. doi: 10.7554/eLife.06074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee WK, et al. Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140. Proc Natl Acad Sci USA. 2013;110(48):19360–19365. doi: 10.1073/pnas.1304670110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446(7136):640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 31.González B, Baños-Sanz JI, Villate M, Brearley CA, Sanz-Aparicio J. Inositol 1,3,4,5,6-pentakisphosphate 2-kinase is a distant IPK member with a singular inositide binding site for axial 2-OH recognition. Proc Natl Acad Sci USA. 2010;107(21):9608–9613. doi: 10.1073/pnas.0912979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao F, et al. Inositol hexakisphosphate kinase-1 mediates assembly/disassembly of the CRL4-signalosome complex to regulate DNA repair and cell death. Proc Natl Acad Sci USA. 2014;111(45):16005–16010. doi: 10.1073/pnas.1417900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu JT, Chan YR, Chien CT. Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol. 2006;16(7):362–369. doi: 10.1016/j.tcb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Wee S, Geyer RK, Toda T, Wolf DA. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol. 2005;7(4):387–391. doi: 10.1038/ncb1241. [DOI] [PubMed] [Google Scholar]
- 35.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Sun Y. Cullin-RING Ligases as attractive anti-cancer targets. Curr Pharm Des. 2013;19(18):3215–3225. doi: 10.2174/13816128113199990300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swords RT, et al. Pevonedistat (MLN4924), a first-in-class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: A phase 1 study. Br J Haematol. 2015;169(4):534–543. doi: 10.1111/bjh.13323. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y, Wilson MP, Majerus PW. Inositol 1,3,4-trisphosphate 5/6-kinase associates with the COP9 signalosome by binding to CSN1. J Biol Chem. 2002;277(48):45759–45764. doi: 10.1074/jbc.M208709200. [DOI] [PubMed] [Google Scholar]
- 39.Castellana M, et al. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol. 2014;32(10):1011–1018. doi: 10.1038/nbt.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhandari R, et al. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc Natl Acad Sci USA. 2007;104(39):15305–15310. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhle S, et al. Protein kinase CK2 and protein kinase D are associated with the COP9 signalosome. EMBO J. 2003;22(6):1302–1312. doi: 10.1093/emboj/cdg127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wundenberg T, Grabinski N, Lin H, Mayr GW. Discovery of InsP6-kinases as InsP6-dephosphorylating enzymes provides a new mechanism of cytosolic InsP6 degradation driven by the cellular ATP/ADP ratio. Biochem J. 2014;462(1):173–184. doi: 10.1042/BJ20130992. [DOI] [PubMed] [Google Scholar]
- 43.Bang S, Chen Y, Ahima RS, Kim SF. Convergence of IPMK and LKB1-AMPK signaling pathways on metformin action. Mol Endocrinol. 2014;28(7):1186–1193. doi: 10.1210/me.2014-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao F, et al. Inositol pyrophosphates promote tumor growth and metastasis by antagonizing liver kinase B1. Proc Natl Acad Sci USA. 2015;112(6):1773–1778. doi: 10.1073/pnas.1424642112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montpetit B, et al. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472(7342):238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao F, et al. Inositol pyrophosphates mediate the DNA-PK/ATM-p53 cell death pathway by regulating CK2 phosphorylation of Tti1/Tel2. Mol Cell. 2014;54(1):119–132. doi: 10.1016/j.molcel.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu R, et al. Inositol polyphosphate multikinase is a coactivator of p53-mediated transcription and cell death. Sci Signal. 2013;6(269):ra22. doi: 10.1126/scisignal.2003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharon M, et al. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009;17(1):31–40. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 49.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41(Web Server issue):W349-57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Losito O, Szijgyarto Z, Resnick AC, Saiardi A. Inositol pyrophosphates and their unique metabolic complexity: Analysis by gel electrophoresis. PLoS One. 2009;4(5):e5580. doi: 10.1371/journal.pone.0005580. [DOI] [PMC free article] [PubMed] [Google Scholar]