Significance
This work investigates the metabolic basis of the interactions between the cosmopolitan bloom-forming alga Emiliania huxleyi and its specific large DNA virus. We demonstrate some of the basic metabolic principles used by the EhV virus to “engineer” its host sphingolipid metabolism, to produce a unique suite of virus-specific lipids. These sphingolipids were essential for the virus assembly and infectivity. These results present novel insight into the chemical “arm race” at sea that coevolved around a unique metabolic pathway. Furthermore, it may provide important evolutionary and biochemical insights into sphingolipid biosynthesis and its functional role in other host–pathogen interactions (e.g. HIV, hepatitis C virus, dengue virus).
Keywords: marine viruses, algal blooms, Emiliania huxelyi, sphingolipids, serine palmitoyltransferase
Abstract
Marine viruses are the most abundant biological entities in the oceans shaping community structure and nutrient cycling. The interaction between the bloom-forming alga Emiliania huxleyi and its specific large dsDNA virus (EhV) is a major factor determining the fate of carbon in the ocean, thus serving as a key host-pathogen model system. The EhV genome encodes for a set of genes involved in the de novo sphingolipid biosynthesis, not reported in any viral genome to date. We combined detailed lipidomic and biochemical analyses to characterize the functional role of this virus-encoded pathway during lytic viral infection. We identified a major metabolic shift, mediated by differential substrate specificity of virus-encoded serine palmitoyltransferase, a key enzyme of sphingolipid biosynthesis. Consequently, unique viral glycosphingolipids, composed of unusual hydroxylated C17 sphingoid bases (t17:0) were highly enriched in the infected cells, and their synthesis was found to be essential for viral assembly. These findings uncover the biochemical bases of the virus-induced metabolic rewiring of the host sphingolipid biosynthesis during the chemical “arms race” in the ocean.
Marine photosynthetic microorganisms are the basis of oceanic food webs. Despite the fact that their biomass represents only approximately 0.2% of the photosynthetic biomass on earth, they fix nearly 50% of the biosphere's net carbon and greatly influence the global biogeochemical cycles of major nutrients (1). The coccolithophore Emiliania huxleyi (Prymnesiophyceae, Haptophyta) is a cosmopolitan eukaryotic unicellular alga that dominates the modern oceans, forming large oceanic blooms that cover thousands of square kilometers (2). E. huxleyi accounts for approximately one-third of the total marine CaCO3 production, derived from its intricate calcite skeleton (3). In addition, E. huxleyi is a major emitter of the climate-bioactive gas dimethyl sulfide, which can affect cloud formation (4, 5).
E. huxleyi’s massive annual blooms in the North Atlantic are routinely infected and terminated by E. huxleyi virus (EhV), a specific nucleocytoplasmic large double-stranded DNA virus. EhV belongs to the coccolithoviruses (6), a subset of the Phycodnaviridae that infect a wide range of microalgae (7). These viruses, with genomes encoding up to 600 proteins and a high burst size, require substantial amounts of building blocks to facilitate their replication and assembly. To fulfill these metabolic requirements, a coordinated modulation of host metabolism is induced during viral infection to produce the required building blocks for viral production (8).
A unique emerging feature, from recent marine metagenomics studies, revealed high prevalence of viral encoded auxiliary metabolic genes (AMGs). These viral genes may expand their host metabolic capabilities during infection (9, 10). AMGs are thought to represent critical, rate-limiting steps of host metabolism, and expression of these genes during infection is believed to be critical for the ecological success of the virus (11). AMGs have been found to be involved in photosynthesis (12, 13), the pentose phosphate pathway (14), phosphate regulation (15), sulfur metabolism (16), sphingolipid (SL) metabolism (17, 18), and DNA/RNA processing (19, 20). Although a wealth of genomic resources of marine microbial origin is now available, a fundamental understanding of the biochemical characteristics of these virally encoded metabolic genes is lacking, and their functional role in rewiring host metabolism and in viral replication cycles remains underexplored.
SLs are essential structural components of eukaryotic membranes and critical signaling lipids in diverse cellular pathways (21). In addition, SLs are potential key regulators in the life cycle of obligatory intracellular pathogens, such as viruses (22–25). The de novo synthesis of SLs is initiated by the rate-limiting enzyme pyridoxal 5′-phosphate (PLP)-dependent serine palmitoyl transferase (SPT; EC 2.3.1.50). SPT catalyzes the condensation of palmitoyl-CoA and l-serine to yield 3-ketosphinganine (3KDS; 3-ketodihydrosphingosine), a committed intermediate of the SL biosynthesis pathway (26). 3KDS is subsequently reduced to sphinganine (SA; dihydrosphingosine), followed by N-acylation by a (dihydro)-ceramide synthase (CerS). Ceramide is formed by the desaturation of dihydroceramide (27) and can be further modified to form complex SLs, including glycosphingolipids (GSLs). The predominance of 18 carbon sphingoid bases in most plants and mammalian SLs is consistent with SPT’s preference for saturated fatty acyl-CoAs with 16 carbon atoms, combined with the abundance of palmitoyl-CoA (28–31).
SLs form an integral structural component of membranes of numerous viruses, including EhV (24, 32–36). The EhV virion comprises at least two internal lipid bilayers (37, 38), which are composed largely of virus-specific GSLs (vGSLs) (8, 34, 35). Genome analysis of EhV has revealed a set of putative SL biosynthetic genes that have not been described in any other viral genome (17, 18, 39). These virus-encoded genes are close homologs of the host genes responsible for the de novo synthesis of ceramide. Transcriptome analysis during lytic viral infection of E. huxleyi indicated rapid induction of these viral genes at very early stages of infection and accumulation of the lipid product vGSLs (8). These findings suggest the involvement of virus-encoded enzymes in the synthesis of vGSLs. These lipids are produced exclusively during viral infection and are known to act as a signaling lipid to induce host programmed cell death (PCD) (35).
The co-occurrence of host and virus-encoded SL biosynthetic pathways raises fundamental questions regarding their biochemical regulation during infection and their functional role in the viral replication cycle. In the present work, we identified some of the basic metabolic strategies used by EhV to manipulate its host SL metabolism that facilitate viral assemble and egress.
Results
Viral SPT Is Expressed at Early Stages of Infection.
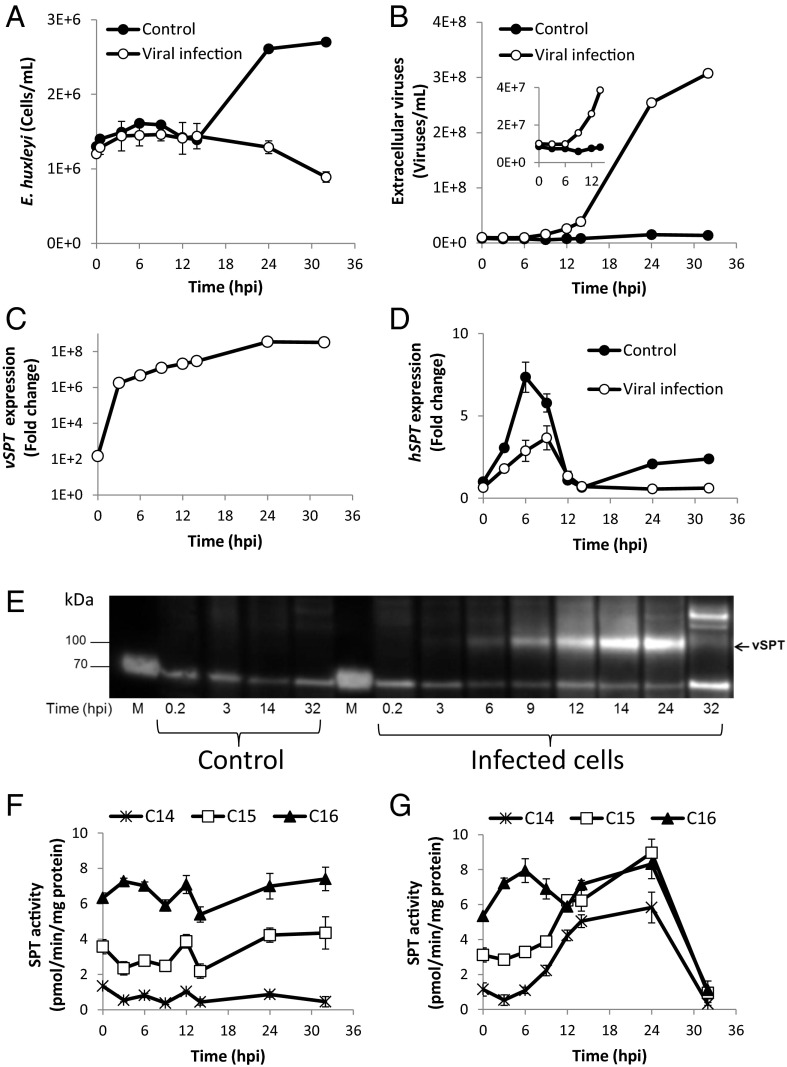
To gain a better understanding of the regulation and coordination of de novo SL synthesis during viral infection, we assessed the coexpression of viral SPT (vSPT) and host SPT (hSPT). Exponential cultures of E. huxleyi (strain CCMP374) were infected with E. huxleyi-specific virus EhV201 to induce a lytic infection. Cultures infected with EhV201 showed growth arrest at 24 h post infection (hpi) and subsequent lysis, whereas the control cells continued to grow exponentially (Fig. 1A). The release of viruses to the culture medium, as assessed by flow cytometry, was apparent already at 9 hpi and peaked at 32 hpi (Fig. 1B). Quantitative RT-PCR (qRT-PCR) analysis revealed a rapid induction of vSPT transcription at early stages of infection (Fig. 1C). A 100-fold increase in vSPT gene expression was detected in the cells a few minutes after infection (T = 10 min). This rapid induction was followed by a profound accumulation of the transcript within the first 3 h of infection, then a continuous rise to 108-fold at 24 hpi. In contrast, a significant decrease in the hSPT mRNA level (P < 0.05, t test) was observed already at 3 hpi (Fig. 1D).
Fig. 1.
Viral SPT is highly expressed at early stages of infection. (A and B) E. huxleyi cultures were infected with EhV201 lytic virus and compared with noninfected control cells. Host cell abundance (A) and extracellular viral production (B) are shown. (C and D) Temporal expression patterns analyzed by qRT-PCR during EhV infection of the viral gene vSPT (C) or the host E. huxleyi-encoded hSPT (D). (E) Western blot analysis using vSPT antibody against protein extracts from control or throughout the course of viral infection. M, protein ladder. (F and G) SPT enzymatic activity assay performed in vitro using three different fatty acyl-CoAs (C14-CoA, C15-CoA, and C16-CoA) and radiolabeled Ser as substrates of protein extracts from control (F) or viral-infected cultures (G). Results are the average ± SD of two biological replicates of one experiment. A similar trend was observed in two additional experiments with similar settings.
To examine the expression level of the vSPT protein, we raised a polyclonal antibody against a synthetic peptide of the vSPT (Materials and Methods). Western blot analysis using protein extracts from microsomal fractions revealed a ∼100-kDa band corresponding to the predicted size of vSPT (96 kDa), which was present exclusively in cell lysates of infected cells (Fig. 1E). To validate the vSPT antibody, we subjected the excised band to MS analysis (Materials and Methods and SI Appendix, Fig. S1). The specific vSPT band was visible in protein samples from the early stages of infection starting at 3 hpi, and reached a maximum level at 14–24 hpi (SI Appendix, Fig. S2). At later stages of infection (32 hpi), rapid turnover of vSPT was observed, possibly implying that vSPT expression is highly controlled during infection (Fig. 1E).
Taken together, the foregoing findings indicate that a key step in modulation of the de novo SL pathway of the infected cell is mediated by the induction of vSPT at both the RNA and protein levels.
Viral Infection Induces a Change in the Substrate Specificity of SPT.
To determine the possible metabolic role of vSPT in rewiring the SL biosynthetic pathway, we examined SPT enzymatic activity during the course of viral infection. Activity and substrate utilization of hSPT and vSPT enzymes were evaluated using radiolabeled serine and nonlabeled fatty acyl-CoAs with different carbon chain lengths as substrates. Constitutive SPT activity was detected in microsomal fractions of control cell extracts using palmitoyl-CoA (C16) as a substrate (Fig. 1F). Interestingly, hSPT also used pentadecanoyl-CoA (C15) and heptadecanoyl CoA (C17) as substrates, albeit to a lesser extent (<50%) compared with palmitoyl-CoA (C16) (SI Appendix, Fig. S3A). In contrast, viral infection induced a shift in substrate preference by SPT, with a significant increase in activity with C14-CoA and C15-CoA as substrates (P < 0.01). This elevated activity gradually increased and correlated with vSPT expression level, reaching a maximum at 24 hpi (Fig. 1E).
At the time of maximal vSPT expression (14–24 hpi), the activity with C15-CoA was twofold to threefold higher in infected cells compared with controls. The activity with C14-CoA was also markedly induced during the peak of vSPT expression, albeit at significantly lower activity levels than with C15-CoA (P < 0.01, t test). No activity was detected after the addition of myriocin, a potent inhibitor of SPT (40), verifying the specific SPT enzymatic activity (SI Appendix, Fig. S3 A and B). No major change in activity was detected when C16-CoA and C17-CoA were used as substrates during the first 24 hpi (Fig. 1G and SI Appendix, Fig. S3B). At later stages of viral infection, SPT activity was markedly reduced in all substrates, concomitant with a sharp decrease in vSPT protein abundance.
To identify the metabolic products of SPT enzymatic activity, we incubated the microsomal fractions of cell extracts from either control cells or infected cells (12 hpi) with different acyl-CoAs and D3-15N-serine. The resulting sphingoid bases from this in vitro enzymatic activity assay were extracted and analyzed for the production of labeled 3KDS by LC-MS (SI Appendix, Fig. S3 C and D). The production of labeled C16-3KDS was evident only in samples from infected cells. Both control and viral-infected samples produced C17-3KDS and C18-3KDS, although the C17-3KDS–to–C18-3KDS ratio was higher in infected cells, indicating greater SPT activity with C15-CoA as a substrate in these samples. The incorporation of the D3-15N-serine into 3KDS was completely abolished in the presence of myriocin, further validating the involvement of SPT enzymatic activity in the synthesis of these compounds. These results indicate the involvement of vSPT in the remodeling of host SL synthesis during viral infection through the use of shorter acyl-CoAs as substrates for long-chain base (LCB) synthesis. This may suggest that viral infection leads to the preferential formation of C16 and C17 sphingoid bases by SPT in infected cells.
Viral Infection Is Dependent on SPT Activity.
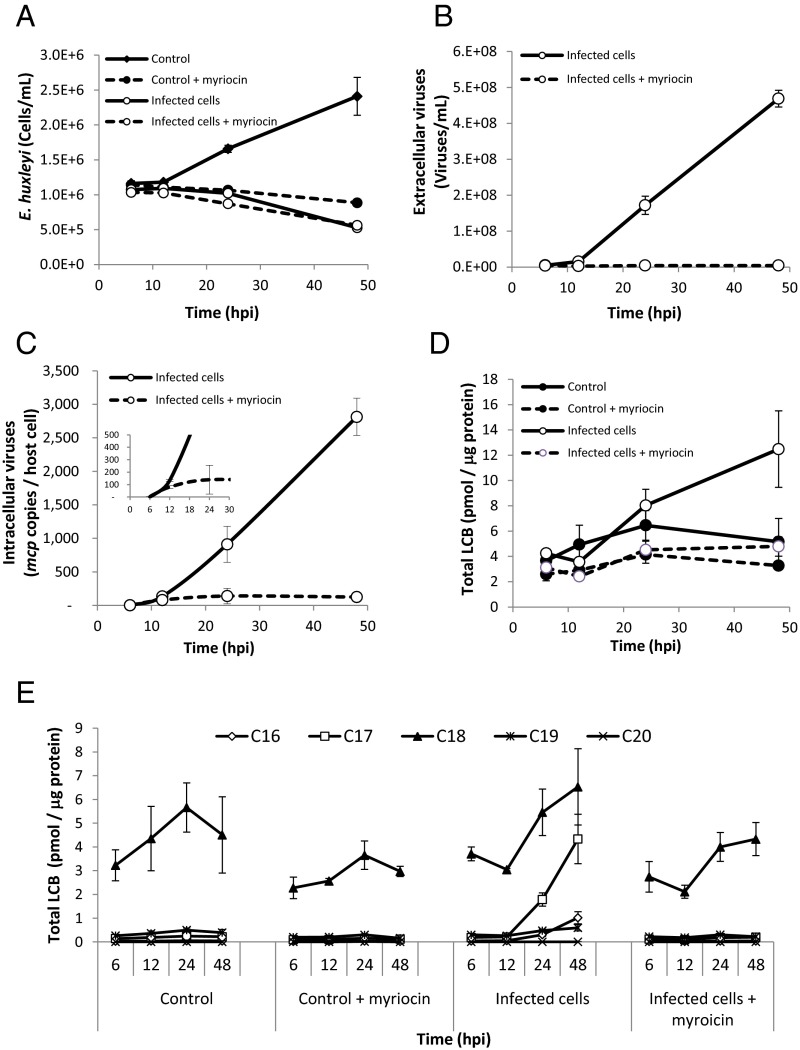
To determine whether SPT has a functional role in viral replication, we inhibited de novo SL biosynthesis at different time points of viral infection by myriocin. In the presence of myriocin, the growth of E. huxleyi control cells was arrested during the first 48 h, but resumed after 72 h (Fig. 2A). Preincubation with myriocin led to complete inhibition of extracellular viral accumulation and synthesis of viral DNA, as assessed by the presence of the mcp gene (Fig. 2 B and C). Interestingly, during the first 12 h of infection, myriocin did not suppress viral replication (Fig. 2 C, Inset) or the growth pattern of the infected host (Fig. 2A). To further clarify the specific role of the vSPT during viral infection, we added myriocin to the infected culture at 20 hpi, when vSPT is highly expressed and active. Extracellular accumulation of virions was strongly suppressed in a dose-dependent manner (SI Appendix, Fig. S4). These results indicate that de novo SL biosynthesis is required for EhV virion assembly or egress.
Fig. 2.
Viral infection induced a shift in the substrate specificity of in vivo SPT activity, which is required for viral assembly. (A and B) E. huxleyi cultures were infected with EhV201 lytic virus and compared with noninfected control cells in the presence or absence of 1 μM myriocin. Host cell abundance (A) and extracellular viral production (B) are presented. (C) Quantification of viral infection by intracellular viral DNA, as estimated by qRT-PCR using primers for the mcp gene with (dotted line) and without (solid line) 1 μM myriocin. (D) Quantification of total LCB during the time course of infection. (E) LCBs grouped according to carbon chain length during infection with (dotted line) and without (solid line) 1 μM myriocin, as estimated by LC-MS. The results presented are the average ± SD of three biological replicates of one representative experiment. A similar trend was observed in an additional experiment.
SPT is the key rate-limiting enzyme in SL metabolism, responsible for the de novo synthesis of LCBs (sphingoid bases), which form the backbone of all diverse SLs species. The length of the sphingoid base carbon chain is determined by the fatty acyl-CoA substrate used by the SPT enzyme. To assess SPT activity in vivo during viral infection, we quantified the chain lengths of the LCBs in infected cells compared with controls. The amount of LCBs (5.05 ± 1.15 pmol/μg protein) did not change significantly during the exponential growth of control host cells (Fig. 2D). In contrast, viral infection led to a 2.5-fold increase in LCBs (12.5 ± 3 pmol/μg protein) at 48 hpi. Interestingly, the increase in LCBs during infection was highly specific for C16 and C17 LCBs, accounting for 8% and 35% of the total LCBs, respectively, produced after 48 h of viral infection (Fig. 2E). These data corroborate the results obtained in the in vitro SPT activity assay (Fig. 1G). In contrast, 95% of the LCBs from uninfected E. huxleyi were composed of C18 (Fig. 2E), in agreement with the high activity detected using palmitoyl-CoA (C16) as a substrate (Fig. 1F). An additional 4% was attributed to C17 LCBs, consistent with the moderate SPT activity observed with pentadecanoyl-CoA (C15). Application of myriocin reduced C18 LCB levels by 35%, and completely abolished virus-induced C16 and C17 LCBs (Fig. 2E).
Taken together, these data indicate that viral infection is dependent on SPT enzymatic activity and leads to a shift in the substrate specificity of SPT to form unique sphingoid base products composed of C16 and C17 chains.
Viral Infection Induces Metabolic Remodeling Toward the Production of Hydroxylated Sphingoid Bases.
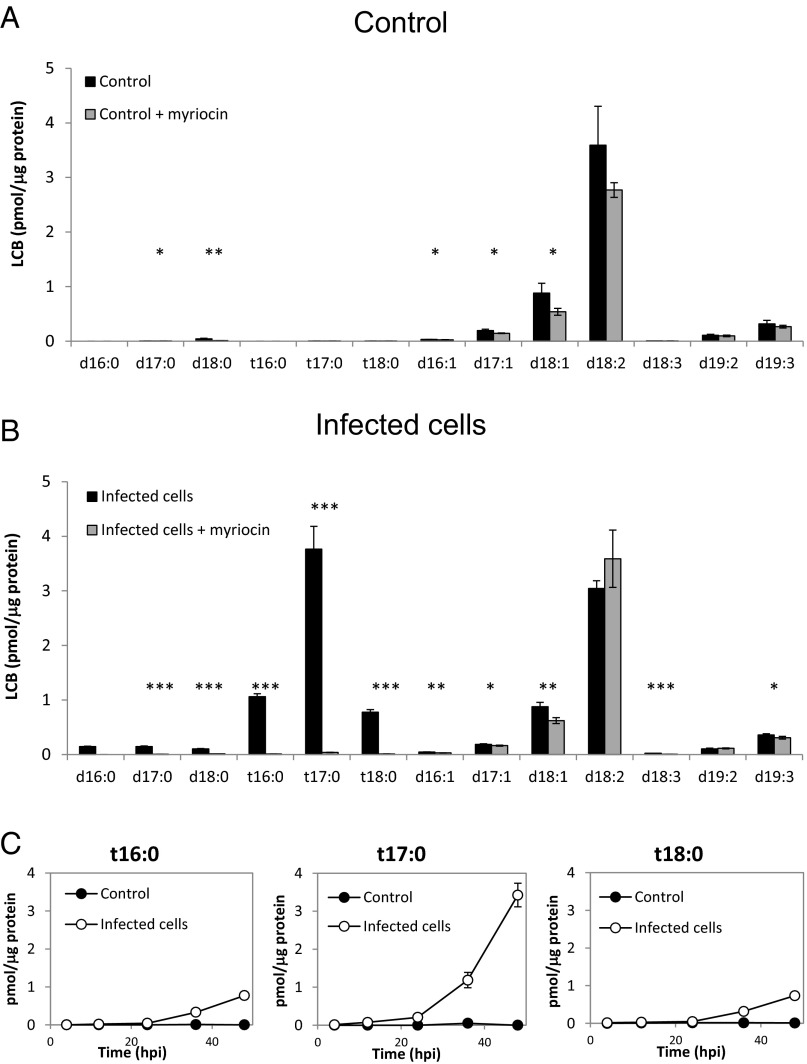
To further characterize the chemical nature of virus-induced C17 sphingoid bases, we analyzed the various LCB species during viral infection. LC-MS analysis indicated a clear induction of trihydroxylated C17 LCB (t17:0) in infected cells, which was completely absent in the control (Fig. 3 A and B and SI Appendix, Table S1). At 24 hpi, t17:0 accounted for 10% of the total LCB, and continued to accumulate to the level of 27.5% at 48 hpi, reaching levels of 4 pmol/μg of protein (Fig. 3C). The induction of t16:0 and t18:0 was also apparent, although at lower levels (6% of total LCB at 48 hpi). Myriocin treatment completely abolished the production of t16:0, t17:0, and t18:0 LCBs in infected cells (Fig. 3B and SI Appendix, Table S1), indicating that these LCBs are the result of de novo LCB synthesis rather than derived from the recycling of SLs by the salvage pathway. In contrast, the abundance of the host LCBs was only mildly affected by myriocin (Fig. 3A and SI Appendix, Fig. S5), suggesting a low turnover rate for these lipids. The most abundant sphingoid base in E. huxleyi control cells was d18:2, which accounted for 66% of the identified LCBs (Fig. 3A and SI Appendix, Table S1). This is in line with a previous study describing sialic GSL, which is composed of a d18:2 LCB conjugated to a C22:0 fatty acid and a monosaccharide sialic acid head group in E. huxleyi (34). In addition, we detected significant amounts (6%) of d19:3, which was previously identified as the abundant LCB backbone of E. huxleyi host GSL (41). The levels of these sphingoid bases did not change significantly during the exponential growth of E. huxleyi cells or during viral infection (SI Appendix, Fig. S5).
Fig. 3.
PhytoSOs are uniquely produced during lytic viral infection. (A and B) LC-MS quantification of different LCB species detected in E. huxleyi control (A) and EhV-infected cultures (B) at 39 hpi. Myriocin (1 μM) was applied at 15 hpi (gray bars). (C) LC-MS quantification of phytoSO with different carbon chain lengths, detected in E. huxleyi control (closed circles) and EhV-infected (open circles) cultures during the course of infection. Results are the average ± SD of three biological replicates. Statistical significance was determined using the unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001.
The virus-induced t17:0 was putatively identified as C17-phytosphingosine (C17-phytoSO). To elucidate its putative structure, we compared the LC-MS properties of this LCB with a commercial standard of C17-phytoSO (Avanti Polar Lipids). Both the sample and the standard were derivatized with o-phathaldelyde (OPA) and analyzed by high-resolution LC-MS (SI Appendix, Fig. S6). The two compounds had a similar mass ([M + H] 480.3131 vs. 480.3129), similar retention time (15.38 vs. 15.36), and similar MS fragments (SI Appendix, Fig. S6).
We further determined that the C17 LCBs are a linear chain rather than an iso-branched chain (42). In our LC-MS analysis, iso-branched C17-SA had a significantly different retention time than straight-chain C17 SA (SI Appendix, Fig. S7A). When known standards were added to the sample, the virus-induced C17:0 SA showed the same retention time as the straight chain C17 SA (and different than the iso-branched C17-SA). In addition, the virus-induced t17:0 LCB had an identical retention time to the C17-phytoSO (SI Appendix, Fig. S7B). These data strongly support C17-phytoSO as the suggested structure.
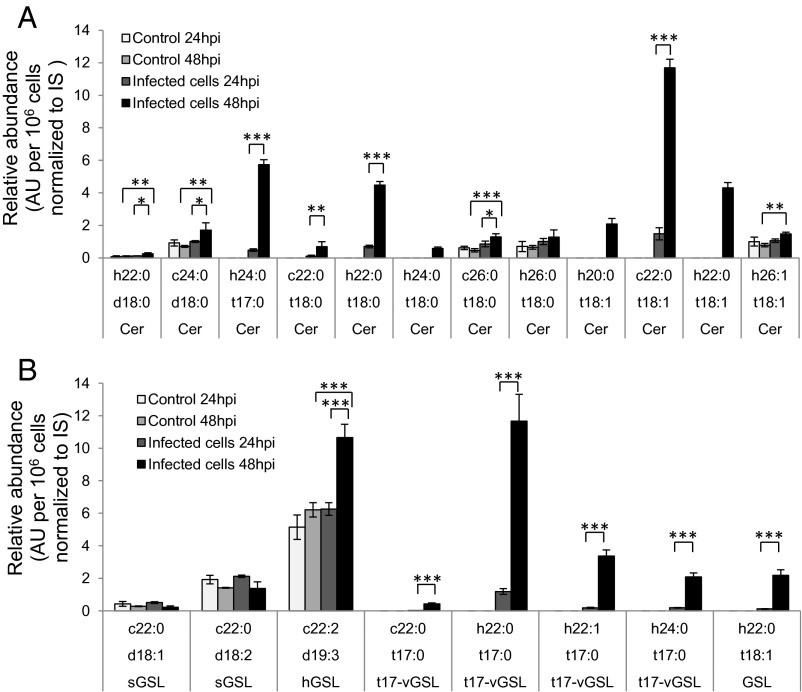
We further analyzed the composition of ceramides and GSL by LC-MS to examine incorporation of the virus-induced t17:0 into complex SLs. We observed an induction of phytoceramides (Fig. 4A) and glycosyl phytoceramides (Fig. 4B) during viral infection. The analysis revealed a marked induction of viral GSLs (t17-vGSLs) composed of a t17:0 LCB and N-acylated with FAs composed of h22:0, h22:1, or h24:0. Interestingly, although t17:0/h22:0 was the most abundant t17-vGSL (SI Appendix, Figs. S8–S10), its respective ceramide could not be detected, suggesting rapid glycosylation of this viral ceramide intermediate.
Fig. 4.
Glycosyl phytoceramides are specifically produced during lytic viral infection. LC-MS quantification of different ceramides (A) and GSLs (B) detected in control and EhV-infected E. huxleyi cultures at 24 and 48 hpi. The results presented are the average ± SD of three biological replicates. Internal standards (IS) of ceramide (d18:1/C12:0) for A, and glucosyl ceramide (d18:1/C12:0) for B were used to normalize the data. Statistical significance was calculated using ANOVA followed by Bonferroni correction. *P < 0.05; **P < 0.01; ***P < 0.001.
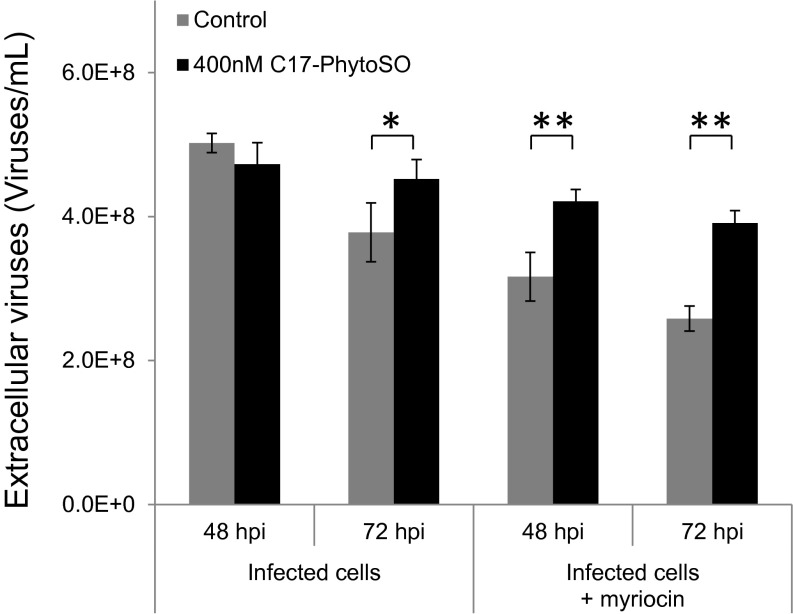
To assess the functional role of C17-phytoSO in the viral replication cycle, we inhibited the de novo SL synthesis pathway and supplemented the infected cells with exogenous C17-phytoSO. Cells with myriocin (250 μM) were supplemented with exogenous C17-phytoSO (400 nM) and then exposed to viral infection. Although myriocin treatment resulted in reduced viral release, application of C17-phytoSO led to a recovery of viral production (Fig. 5). The finding that C17-phytoSO could rescue viral assembly while SPT was inhibited, indicates the unique ability of the virus to use C17-phytoSO to complete its replication cycle.
Fig. 5.
Exogenous application of C17-phytoSO can alleviate viral production in myriocin-treated infected cultures. Myriocin-treated control and infected cultures were supplemented with either DMSO (gray bars) or 400 nM C17-phytoSO (black bars) and assessed for extracellular viral production at 48 and 72 hpi. Results are the average ± SD of three biological replicates. Statistical significance was determined using the Student t test. *P < 0.05; **P < 0.01.
Taken together, these results demonstrate that viral infection is mediated by a rewiring of SL biosynthesis to produce unique C17-phytoSO as the main backbone for vGSLs.
Discussion
The recent wealth of genomic information derived from the marine environment has revealed the high prevalence of virus-encoded AMG, previously thought to be restricted to the genomes of their hosts (9, 10). EhV is an exceptional case, because it encodes for an almost complete biosynthetic pathway for the de novo synthesis of ceramides that is homologous to its host. This remarkable phenomenon, not seen in any other known viral genome (17, 18), prompted us to explore the nature of the metabolic rewiring imposed by the virus expressing these genes. Specifically, we aimed to explore the mode of action and coordination by which the viral-encoded enzymes act orthogonally to the host pathway, to gain insight into the function of SLs during host–virus dynamics. Since these host–virus interactions control the fate of vast E. huxleyi blooms in the ocean, the outcome of this biochemical “arms race” has a profound ecological and biogeochemical imprint.
We characterized the metabolic rewiring of SL biosynthesis during viral infection and uncovered the biochemical basis for the production of novel virus-specific lipids (Fig. 6). This metabolic shift induced by viral infection was based on down-regulation of host gene expression while the viral genes are highly up-regulated, along with alterations in the substrate specificity of SPT activity. The outcome of this metabolic remodeling is the production of virus-specific SLs composed of t17:0 sphingoid bases. These virus-specific SLs are essential for viral assembly and infectivity.
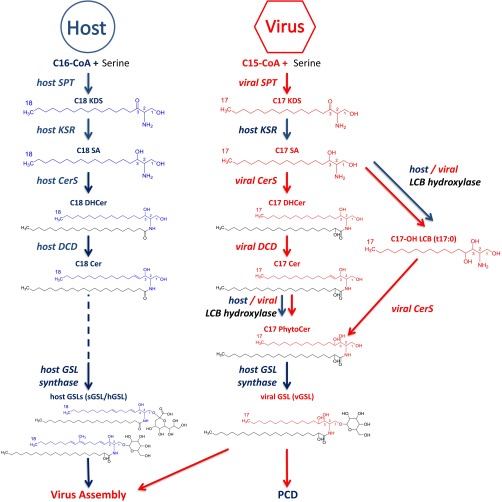
Fig. 6.
A proposed model describing the metabolic remodeling in SL biosynthetic pathways during viral infection. The metabolic basis for the interactions between the bloom-forming alga E. huxleyi (blue) and its specific virus EhV (red) is mediated by the expression of several virus-encoded enzymes (red, italics) involved in the SL biosynthesis pathway acting orthogonal to the host encoded enzymes (blue, italics). The viral-encoded SPT induces a shift in substrate specificity which allows the efficient utilization of C15-CoA to produce C17 sphingoid bases (compared with C18 LCB produced by non-infected host cells). These virally induced odd carbon chain sphingoid bases are subsequently hydroxylated, possibly by a virus-encoded sphingoid base hydroxylase (LCB hydroxylase) and further metabolized to synthesize unique vGSLs. These vGSLs, combined with the host GSLs, are required for virus assembly and are enriched in the virion, suggesting a structural role for these molecules. vGSLs are also involved in inducing the host PCD, likely affecting a timed cell lysis to allow proper viral egress. KSR, 3-ketodihydrosphingosine reductase; DHCer, dihydroceramide; DCD, dihydroceramide desaturase; PhytoCer, phytoceramide.
Virus-Encoded SPT Is Responsible for Biosynthesis of Unique SLs.
Our data show that viral infection induces a shift in LCB profile of the host cell. We suggest that the virus-encoded SPT is responsible for this redirection of the host SL metabolism to produce C17 LCB during lytic viral infection. We provide several lines of evidence to support these conclusions. First, the vSPT gene is up-regulated by orders of magnitude higher than hSPT at early phase of infection, whereas its host counterpart is down-regulated. Second, SPT of uninfected cells has no activity toward C14-CoA as a substrate and only marginal activity with C15-CoA, suggesting that hSPT cannot use these substrates efficiently. It was previously shown that the heterologous expression of vSPT from EhV in yeast results in greater in vitro activity with C14-CoA than with C16-CoA (43). Third, there is a clear correlation between the expression of vSPT protein, the in vitro SPT activity by utilization of C14- and C15-CoA, the accumulation of newly produced short-chain (C16-C17) sphingoid bases, and the dynamics of viral infection. The specific involvement of vSPT in the formation of viral SLs is further corroborated by its sensitivity to myriocin (SI Appendix, Fig. S3), which also suppressed viral assembly and release in a dose-dependent manner (SI Appendix, Fig. S4).
The rapid induction of vSPT expression at early stages of infection (Fig. 1C) is in line with previously reported transcriptomics data (8, 44, 45). Interestingly, during later stages of lytic infection (>32 hpi), transcription levels of vSPT remained high, whereas vSPT protein levels and activity were both markedly decreased (Fig. 1 E and G). This decoupling may imply a possible host defense mechanism to suppress virus-induced SL synthesis, but requires further work to resolve the role of vSPT as a potential target.
The “Missing Link”: Viral-Encoded Sphingolipid Pathway Lacks 3KDS Reductase.
In the present study, we have demonstrated the direct involvement of the vSPT enzyme in rewiring host ceramide synthesis. However, the viral genome also encodes for CerS and dihydroceramide desaturase, which awaits further characterization. The missing link in this pathway is the enzyme 3KDS reductase, which is absent from the viral genome (18). Interestingly, our transcriptomics data indicated that the host 3KDS reductase is not down-regulated as is the rest of the pathways, but rather up-regulated during viral infection. Thus, we suggest that the host 3KDS reductase can reduce short-chain KDS, albeit with somewhat lower efficiency, as inferred from myriocin-sensitive accumulation of 3KDS with C16 and C17 carbon chains only during viral infection (SI Appendix, Fig. S11). It is noteworthy that whereas 3KDS is usually rapidly metabolized to SA by 3KDS reductase, it accumulates when this enzyme is partially inactivated in Arabidopsis, leading to the accumulation of free phytoSO (t18:0) and 4-hydroxy-3KDS (46).
Virus-Induced SLs Are Composed of Unique Hydroxylated Odd Chain Sphingoid Bases.
We have putatively identified t17:0, the most pronounced metabolic product during viral infection, as C17-phytoSO. Previous reports (34, 35) have suggested a different ceramide backbone for vGSLs from the proposed t17:0/h22:0, which is composed of C16 LCBs; however, recent developments in the sensitivity of targeted lipidomic methodologies that enable accurate determination of vGSL mass, fragments, and retention time, along with our biochemical analysis of SPT activity, have led us to suggest this t17:0-based structure. Nevertheless, our data clearly show the use of C14-CoA as an alternative potential substrate for SPT during viral infection. Thus, even though we cannot exclude the production of vGSLs composed of C16 LCBs, we present two main lines of evidence indicating that the t17-vGSLs (composed of t17:0 LCBs) is indeed a highly abundant form of this class of SLs. These include (i) a detailed LC-MS analysis of both vGSLs and LCBs after hydrolysis of the complex SLs, showing an accurate mass of expected fragments (<5 ppm error) when compared against known standards, and (ii) SPT activity data showing high utilization of C15-CoA as a preferential substrate during the course of viral infection.
Furthermore, a previous report indicated a fourfold increase in C15 saturated fatty acids during EhV infection (47), indicating a redirection of host metabolism to the synthesis of C15-FA. This metabolic shift enables the availability of the required substrate (C15-CoA) for vSPTs to synthesize C17 LCBs. Interestingly, we also observed an elevation of C18-phytoSO during viral infection. C18-phytoSO was recently shown to be catabolized in a unique pathway into C15 fatty acids in yeast and mammalian cells (48). We identified the E. huxleyi homologs of the two enzymes, DPL1 and MPO1, involved in the recycling of C18-phytoSO into C15 acyl-CoA, and our transcriptome data (4, 8) indicated that both genes are expressed during E. huxleyi growth and infection. Thus, we propose this recycling pathway as a potential metabolic route, enabling the production of C17 LCB during viral infection.
Proposed Functional Role for the Virus-Specific SLs.
The production of unique short-chain sphingoid bases also has been reported for the human SPT subunit SPTLC3, which confers altered substrate specificity toward C14-CoA, thus producing C16-sphingoid bases (49, 50). Sphingoid bases with shorter carbon chains are less hydrophobic and thus are likely to exhibit different biophysical properties, which might influence their subcellular localization and distribution in membranes.
Another important feature of the unique viral SLs compared with the host-derived SLs is their hydroxylated LCBs. Interestingly, E. huxleyi and EhV encode for several putative LCB hydroxylases that are highly expressed at early stages of infection (8, 39). Hydroxylated LCBs as phytoSOs are highly abundant in plants and fungal membranes (31), and have a significant impact on the biophysical characteristics of the membrane in which they are embedded. Multiple hydroxylations of the ceramide backbones of SLs have been associated with increased stability and decreased permeability of membranes, as well as better membrane resilience to environmental stress conditions (51–53). The EhV-specific vGSL has a ceramide core consisting of a trihydroxy LCB (t17:0) and C22-C24 hydroxylated fatty acids. Thus, the enrichment of GSLs with a multihydroxylated ceramide backbone in the EhV membranes may confer a structural requirement to maintain the integrity of the viral particles and may affect their decay rate under UV stress in the upper ocean and atmosphere (54). Furthermore, these bioactive lipids can regulate other cellular processes, such as induction of PCD, which has been shown to be structurally and stereochemically specific (55–58). In agreement, vGSLs have been found to act as signaling lipids to induce algal host PCD (35). Moreover, EhV have been shown to interact with E. huxleyi lipid rafts, which are membrane microdomains enriched with GSLs, sterols, and proteins (59). These findings provide an interesting link between the unique structure of SLs induced by EhV and their involvement in the timely induction of cell death processes during viral infection.
SLs may represent a general requirement for the life cycle of other viruses. This is in line with the up-regulation of host SL metabolism by the hepatitis C, dengue, and West Nile viruses, which is known to be essential in their replication cycle (32, 60, 61). Because EhV is the only virus known to encode SPT, its dependence on virus-specific SLs provides a unique case of coevolution of a biosynthetic pathway and diversification of its metabolic products. The unique bioactive lipids derived from EhV-infected cells provide sensitive biomarkers for assessing this host–pathogen interactions during algal blooms in the oceans and its possible impact on marine food webs (34, 35, 41, 62, 63). Given that E. huxleyi–EhV interactions have an important ecological role in controlling the fate and cycling of carbon and sulfur in the ocean (4, 64), our results may provide new insights into the chemical arms race during this interaction, and its potential impact on large-scale biogeochemical processes.
Materials and Methods
Chemicals and Internal Standards.
All chemicals were purchased from Sigma-Aldrich unless specified otherwise. Liquid chromatography-grade solvents were purchased from Merck and Bio-Lab. Lipids and internal standards were purchased from Avanti Polar Lipids. The iso-C17 SA was a kind gift from Howard Riezman (University of Geneva) and Andreas Zumbuehl (University of Fribourg). The lipid internal standards were added to the extraction solution used in the initial extraction step.
Culture Growth and Viral Infection Dynamics.
Cells of the noncalcifying E. huxleyi strain CCMP374 were cultured in F/2 medium (65) and incubated at 18 °C under a 16:8-h light/dark illumination cycle. A light intensity of 100 μM photons·m−2·s−1 was provided by cool-white LED lights. All experiments were performed with exponential-phase cultures (5·105 to 106 cells·mL−1). The virus used for this study was E. huxleyi lytic virus EhV201 (6). In all infection experiments, E. huxleyi was infected with a 1:100 volumetric ratio of viral lysate to culture (multiplicity of infection of 1:1 viral particle per cell). For time course experiments, sampling was performed every few hours (as specified), starting at 10 min after infection (i.e., 0.2 hpi).
Inhibition of SL Biosynthesis.
For the SPT inhibition experiments, myriocin (Enzo Life Sciences) in DMSO was added to cultures directly at different time points (1 h before infection, at infection, or at different times after infection up to 20 hpi) along with infection with EhV201 to reach a final concentration of 1 μM unless specified otherwise. An equal volume of DMSO was added to all control cultures. Culture conditions were identical to those described above.
In Vitro SPT Activity Assay.
The SPT activity assay was done as described previously (66) with minor modifications. Pellets of a 300-mL culture of 106 cells/mL was sonicated with 400 μL of lysis buffer without sucrose monolaurate (SML) but with PLP, then centrifuged at 30,000 × g for 30 min at 4 °C. The resulting pellet was resuspended in lysis buffer with SML. The protein content of the microsomal fraction was determined by the BCA assay (Pierce). For each reaction, 250–300 μg of protein was used, and the assay was performed at 18 °C with gentle shaking for 1 h, followed by lipid extraction as described previously (66). Samples were analyzed for 14C incorporation into the SL fraction by a scintillation counter. The data shown are the average of at least two technical repeats of three biological replicates.
To determine the carbon chain length of the LCB produced by the vSPT, the in vitro activity assay was performed with 5 mM D3-15N serine (DNLM-6863; Cambridge Isotope Laboratories) and 50 μM acyl-CoA (C14-, C15-, or C16-CoA), with or without myriocin (1 μM) in the reaction. The in vitro reaction was performed with protein extracts from noninfected cells (control) and from EhV201-infected cells (at 12 hpi). The resulting LCBs were extracted as described previously (66) and directly subjected to MS analysis (LC-MS analysis without derivatization).
LCB Analysis.
Cellular sphingoid bases are usually N-acylated and conjugated to different head groups. Because we were interested mainly in the cellular sphingoid base profile as a surrogate marker for SPT activity, the analysis of the LCB profile was simplified by performing an acid hydrolysis before the LC/MS analysis (66). This step releases the LCBs by hydrolyzing the N-acyl chains and head groups. Therefore, the reported concentrations of sphingoid base reflect not the free LCBs, but rather the total sphingoid base content for all individual SL subclasses, including ceramides and GSLs. Total LCBs (Fig. 2) was defined as the sum of all of the measured individual LCBs.
To determine the LCB in cell cultures, cultures were sampled for 48 h, in addition to cultures in which SPT enzymatic activity was inhibited by 1 μM myriocin. The inhibitor was added to the culture at time 0 (i.e., with the virus), and the cultures were sampled at 6, 12, 24, and 48 hpi. The lipids were analyzed as described previously (67) with some modifications (SI Appendix, Materials and Methods).
SL Analysis.
Lipids were extracted from E. huxleyi cells infected with EhV201 and from noninfected cells harvested at 12, 24, 32, and 48 hpi in three biological replicates of 50 mL. Lipid analysis was performed as described previously (68) with some modifications (SI Appendix, Materials and Methods).
Putative Identification of the t17:0 Structure.
Identification of the t17:0 LCB was based on a comparison with a commercially available standard in terms of accurate mass (<2 ppm difference between them), same retention time, and similar fragmentation pattern of the OPA derivatives (SI Appendix, Fig. S6), as well as the free LCB (SI Appendix, Fig. S7). The retention time of the virus-induced t17:0 matched the log retention time correlation with the carbon chain lengths of C16-, C17-, and C18-phytoSO STD (SI Appendix, Fig. S6D).
C17 LCB was previously identified as the main LCB in Caenorhabditis elegans, and the structure of these LCBs was resolved by NMR and shown to be a C17 iso-branched chain (42). Whereas the mass of C17 iso-branched SA is identical to that of linear C17 SA, the different structure affected the retention time of the molecules in the LC analysis (SI Appendix, Fig. S7A). These results also may indicate a different retention time for straight and iso-branched C17-phytoSO. The fact that OPA derivative of the t17:0 from the viral-infected sample was similar to the OPA derivative of the C17-phytoSO strongly points to the suggested structure. To further support this conclusion, we performed a spiking experiment in which known standards were added to the sample and analyzed together at the same matrix of compounds (SI Appendix, Fig. S7B). As shown in the chromatograms, C17:0 SA produced during viral infection had the same retention time as the commercial straight-chain C17 SA, but different from that of the iso-branched C17 SA. Furthermore, the virus-induced t17:0 (nonderivatized) showed an identical retention time to the commercial C17-phytoSO.
The putative identification of the most abundant t17-vGSL, monohexosylcer(t17:0/h22:0) (SI Appendix, Figs. S8–S10) was verified according to the fragmentation pattern of the molecule in both negative and positive ionization modes of the LC-MS, as well as by MS/MS in the positive ion mode showing the expected 304 fragment of the LCB t17:0, and clearly demonstrated the loss of three hydroxyl groups from the LCB (SI Appendix, Fig. S9), further validating hydroxylation of the sphingoid base. The hydroxylation of the fatty acid in the ceramide backbone was clearly evident in the fragments identified by the negative ion mode, where the loss of hydroxyl group from the fatty acid was observed (SI Appendix, Fig. S10).
Supplementary Material
Acknowledgments
We thank Howard Riezman, Joerg Thomas Hannich, and Andreas Zumbuehl for kindly providing the iso-branched C17-SA standard. This research was supported by the European Research Council Starting Grant (INFOTROPHIC Grant 280991, to A.V.), the Swiss National Science Foundation (Grant 31003A_153390/1, to T.H.), the 7th Framework Program of the European Commission (RESOLVE Project 3057, to T.H.), and the Zurich Center of Integrated Human Physiology, University of Zurich (T.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523168113/-/DCSupplemental.
References
- 1.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281(5374):237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 2.Holligan PM, et al. A biogeochemical study of the coccolithophore, Emiliania huxleyi, in the North Atlantic. Global Biogeochem Cycles. 1993;7(4):879–900. [Google Scholar]
- 3.Rost B, Riebesell U. Coccolithophores and the biological pump: Responses to environmental changes. In: Thierstein HR, Young JR, editors. Coccolithophores: From Molecular Processes to Global Impact. Springer; Berlin: 2004. pp. 99–125. [Google Scholar]
- 4.Alcolombri U, et al. Identification of the algal dimethyl sulfide-releasing enzyme: A missing link in the marine sulfur cycle. Science. 2015;348(6242):1466–1469. doi: 10.1126/science.aab1586. [DOI] [PubMed] [Google Scholar]
- 5.Simó R. Production of atmospheric sulfur by oceanic plankton: Biogeochemical, ecological and evolutionary links. Trends Ecol Evol. 2001;16(6):287–294. doi: 10.1016/s0169-5347(01)02152-8. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder DC, Oke J, Malin G, Wilson WH. Coccolithovirus (Phycodnaviridae): Characterisation of a new large dsDNA algal virus that infects Emiliana huxleyi. Arch Virol. 2002;147(9):1685–1698. doi: 10.1007/s00705-002-0841-3. [DOI] [PubMed] [Google Scholar]
- 7.Van Etten JL, Graves MV, Müller DG, Boland W, Delaroque N. Phycodnaviridae: Large DNA algal viruses. Arch Virol. 2002;147(8):1479–1516. doi: 10.1007/s00705-002-0822-6. [DOI] [PubMed] [Google Scholar]
- 8.Rosenwasser S, et al. Rewiring host lipid metabolism by large viruses determines the fate of Emiliania huxleyi, a bloom-forming alga in the ocean. Plant Cell. 2014;26(6):2689–2707. doi: 10.1105/tpc.114.125641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurwitz BL, Hallam SJ, Sullivan MB. Metabolic reprogramming by viruses in the sunlit and dark ocean. Genome Biol. 2013;14(11):R123. doi: 10.1186/gb-2013-14-11-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enav H, Mandel-Gutfreund Y, Béjà O. Comparative metagenomic analyses reveal viral-induced shifts of host metabolism towards nucleotide biosynthesis. Microbiome. 2014;2(1):9. doi: 10.1186/2049-2618-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitbart M. Marine viruses: Truth or dare. Annu Rev Mar Sci. 2012;4:425–448. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- 12.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438(7064):86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- 13.Sharon I, et al. Photosystem I gene cassettes are present in marine virus genomes. Nature. 2009;461(7261):258–262. doi: 10.1038/nature08284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson LR, et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA. 2011;108(39):E757–E764. doi: 10.1073/pnas.1102164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng Q, Chisholm SW. Marine viruses exploit their host’s two-component regulatory system in response to resource limitation. Curr Biol. 2012;22(2):124–128. doi: 10.1016/j.cub.2011.11.055. [DOI] [PubMed] [Google Scholar]
- 16.Anantharaman K, et al. Sulfur oxidation genes in diverse deep-sea viruses. Science. 2014;344(6185):757–760. doi: 10.1126/science.1252229. [DOI] [PubMed] [Google Scholar]
- 17.Wilson WH, et al. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309(5737):1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- 18.Monier A, et al. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 2009;19(8):1441–1449. doi: 10.1101/gr.091686.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci USA. 2011;108(42):17486–17491. doi: 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yutin N, Koonin EV. Evolution of DNA ligases of nucleo-cytoplasmic large DNA viruses of eukaryotes: A case of hidden complexity. Biol Direct. 2009;4:51. doi: 10.1186/1745-6150-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilera-Romero A, Gehin C, Riezman H. Sphingolipid homeostasis in the web of metabolic routes. Biochim Biophys Acta. 2014;1841(5):647–656. doi: 10.1016/j.bbalip.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 22.Hanada K. Sphingolipids in infectious diseases. Jpn J Infect Dis. 2005;58(3):131–148. [PubMed] [Google Scholar]
- 23.Sakamoto H, et al. Host sphingolipid biosynthesis as a target for hepatitis C virus therapy. Nat Chem Biol. 2005;1(6):333–337. doi: 10.1038/nchembio742. [DOI] [PubMed] [Google Scholar]
- 24.Brügger B, et al. The HIV lipidome: A raft with an unusual composition. Proc Natl Acad Sci USA. 2006;103(8):2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider-Schaulies J, Schneider-Schaulies S. Sphingolipids in viral infection. Biol Chem. 2015;396(6-7):585–595. doi: 10.1515/hsz-2014-0273. [DOI] [PubMed] [Google Scholar]
- 26.Hanada K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim Biophys Acta. 2003;1632(1-3):16–30. doi: 10.1016/s1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 27.Futerman AH, Riezman H. The ins and outs of sphingolipid synthesis. Trends Cell Biol. 2005;15(6):312–318. doi: 10.1016/j.tcb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Merrill AH, Jr, Williams RD. Utilization of different fatty acyl-CoA thioesters by serine palmitoyltransferase from rat brain. J Lipid Res. 1984;25(2):185–188. [PubMed] [Google Scholar]
- 29.Haynes CA, et al. Quantitation of fatty acyl-coenzyme As in mammalian cells by liquid chromatography-electrospray ionization tandem mass spectrometry. J Lipid Res. 2008;49(5):1113–1125. doi: 10.1194/jlr.D800001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimberlin AN, et al. Arabidopsis 56-amino acid serine palmitoyltransferase-interacting proteins stimulate sphingolipid synthesis, are essential, and affect mycotoxin sensitivity. Plant Cell. 2013;25(11):4627–4639. doi: 10.1105/tpc.113.116145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markham JE, Lynch DV, Napier JA, Dunn TM, Cahoon EB. Plant sphingolipids: Function follows form. Curr Opin Plant Biol. 2013;16(3):350–357. doi: 10.1016/j.pbi.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Martín-Acebes MA, et al. The composition of West Nile virus lipid envelope unveils a role of sphingolipid metabolism in flavivirus biogenesis. J Virol. 2014;88(20):12041–12054. doi: 10.1128/JVI.02061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merz A, et al. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem. 2011;286(4):3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fulton JM, et al. Novel molecular determinants of viral susceptibility and resistance in the lipidome of Emiliania huxleyi. Environ Microbiol. 2014;16(4):1137–1149. doi: 10.1111/1462-2920.12358. [DOI] [PubMed] [Google Scholar]
- 35.Vardi A, et al. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science. 2009;326(5954):861–865. doi: 10.1126/science.1177322. [DOI] [PubMed] [Google Scholar]
- 36.Scheiffele P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Biol Chem. 1999;274(4):2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 37.Schatz D, et al. Hijacking of an autophagy-like process is critical for the life cycle of a DNA virus infecting oceanic algal blooms. New Phytol. 2014;204(4):854–863. doi: 10.1111/nph.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackinder LC, et al. A unicellular algal virus, Emiliania huxleyi virus 86, exploits an animal-like infection strategy. J Gen Virol. 2009;90(Pt 9):2306–2316. doi: 10.1099/vir.0.011635-0. [DOI] [PubMed] [Google Scholar]
- 39.Michaelson LV, Dunn TM, Napier JA. Viral trans-dominant manipulation of algal sphingolipids. Trends Plant Sci. 2010;15(12):651–655. doi: 10.1016/j.tplants.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 40.Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun. 1995;211(2):396–403. doi: 10.1006/bbrc.1995.1827. [DOI] [PubMed] [Google Scholar]
- 41.Vardi A, et al. Host–virus dynamics and subcellular controls of cell fate in a natural coccolithophore population. Proc Natl Acad Sci USA. 2012;109(47):19327–19332. doi: 10.1073/pnas.1208895109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chitwood DJ, Lusby WR, Thompson MJ, Kochansky JP, Howarth OW. The glycosylceramides of the nematode Caenorhabditis elegans contain an unusual, branched-chain sphingoid base. Lipids. 1995;30(6):567–573. doi: 10.1007/BF02537032. [DOI] [PubMed] [Google Scholar]
- 43.Han G, et al. Expression of a novel marine viral single-chain serine palmitoyltransferase and construction of yeast and mammalian single-chain chimera. J Biol Chem. 2006;281(52):39935–39942. doi: 10.1074/jbc.M609365200. [DOI] [PubMed] [Google Scholar]
- 44.Allen MJ, et al. Locus-specific gene expression pattern suggests a unique propagation strategy for a giant algal virus. J Virol. 2006;80(15):7699–7705. doi: 10.1128/JVI.00491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pagarete A, et al. Unveiling the transcriptional features associated with coccolithovirus infection of natural Emiliania huxleyi blooms. FEMS Microbiol Ecol. 2011;78(3):555–564. doi: 10.1111/j.1574-6941.2011.01191.x. [DOI] [PubMed] [Google Scholar]
- 46.Chao DY, et al. Sphingolipids in the root play an important role in regulating the leaf ionome in Arabidopsis thaliana. Plant Cell. 2011;23(3):1061–1081. doi: 10.1105/tpc.110.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans C, Pond DW, Wilson WH. Changes in Emiliania huxleyi fatty acid profiles during infection with E. huxleyi virus 86: Physiological and ecological implications. Aquat Microb Ecol. 2009;55(3):219–228. [Google Scholar]
- 48.Kondo N, et al. Identification of the phytosphingosine metabolic pathway leading to odd-numbered fatty acids. Nat Commun. 2014;5:5338. doi: 10.1038/ncomms6338. [DOI] [PubMed] [Google Scholar]
- 49.Hornemann T, et al. The SPTLC3 subunit of serine palmitoyltransferase generates short-chain sphingoid bases. J Biol Chem. 2009;284(39):26322–26330. doi: 10.1074/jbc.M109.023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russo SB, Tidhar R, Futerman AH, Cowart LA. Myristate-derived d16:0 sphingolipids constitute a cardiac sphingolipid pool with distinct synthetic routes and functional properties. J Biol Chem. 2013;288(19):13397–13409. doi: 10.1074/jbc.M112.428185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pascher I. Molecular arrangements in sphingolipids: Conformation and hydrogen bonding of ceramide and their implication on membrane stability and permeability. Biochim Biophys Acta. 1976;455(2):433–451. doi: 10.1016/0005-2736(76)90316-3. [DOI] [PubMed] [Google Scholar]
- 52.Karlsson KA. On the character and functions of sphingolipids. Acta Biochim Pol. 1998;45(2):429–438. [PubMed] [Google Scholar]
- 53.Lynch DV, Dunn TM. An introduction to plant sphingolipids and a review of recent advances in understanding their metabolism and function. New Phytol. 2004;161(3):677–702. doi: 10.1111/j.1469-8137.2004.00992.x. [DOI] [PubMed] [Google Scholar]
- 54.Sharoni S, et al. Infection of phytoplankton by aerosolized marine viruses. Proc Natl Acad Sci USA. 2015;112(21):6643–6647. doi: 10.1073/pnas.1423667112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pata MO, Hannun YA, Ng CK. Plant sphingolipids: Decoding the enigma of the Sphinx. New Phytol. 2010;185(3):611–630. doi: 10.1111/j.1469-8137.2009.03123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi L, et al. Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res. 2007;17(12):1030–1040. doi: 10.1038/cr.2007.100. [DOI] [PubMed] [Google Scholar]
- 57.Cheng J, Park T-S, Chio L-C, Fischl AS, Ye XS. Induction of apoptosis by sphingoid long-chain bases in Aspergillus nidulans. Mol Cell Biol. 2003;23(1):163–177. doi: 10.1128/MCB.23.1.163-177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thevissen K, et al. Fungicidal activity of truncated analogues of dihydrosphingosine. Bioorg Med Chem Lett. 2008;18(13):3728–3730. doi: 10.1016/j.bmcl.2008.05.067. [DOI] [PubMed] [Google Scholar]
- 59.Rose SL, et al. Isolation and characterization of lipid rafts in Emiliania huxleyi: A role for membrane microdomains in host–virus interactions. Environ Microbiol. 2014;16(4):1150–1166. doi: 10.1111/1462-2920.12357. [DOI] [PubMed] [Google Scholar]
- 60.Hirata Y, et al. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog. 2012;8(8):e1002860. doi: 10.1371/journal.ppat.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perera R, et al. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012;8(3):e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pond DW, Bell MV, Harris RP, Sargent JR. Microplanktonic polyunsaturated fatty acid markers: A mesocosm trial. Estuar Coast Shelf Sci. 1998;46(2):61–67. [Google Scholar]
- 63.Bidle KD, Vardi A. A chemical arms race at sea mediates algal host–virus interactions. Curr Opin Microbiol. 2011;14(4):449–457. doi: 10.1016/j.mib.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 64.Lehahn Y, et al. Decoupling physical from biological processes to assess the impact of viruses on a mesoscale algal bloom. Curr Biol. 2014;24(17):2041–2046. doi: 10.1016/j.cub.2014.07.046. [DOI] [PubMed] [Google Scholar]
- 65.Keller MD, Selvin RC, Claus W, Guillard RRL. Media for the culture of oceanic ultraphytoplankton. J Phycol. 1987;23(4):633–638. [Google Scholar]
- 66.Rütti MF, Richard S, Penno A, von Eckardstein A, Hornemann T. An improved method to determine serine palmitoyltransferase activity. J Lipid Res. 2009;50(6):1237–1244. doi: 10.1194/jlr.D900001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Penno A, et al. Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J Biol Chem. 2010;285(15):11178–11187. doi: 10.1074/jbc.M109.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hummel J, et al. Ultra-performance liquid chromatography and high- resolution mass spectrometry for the analysis of plant lipids. Front Plant Sci. 2011;2:54. doi: 10.3389/fpls.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.