Significance
Here we report on the development of a modular terpene production platform as a screening tool for the functional characterization of next-generation sequencing information and on the application of this system in the characterization of the biosynthetic pathway leading to the potent antioxidant carnosic acid in Lamiaceae. In addition to revealing an important plant natural product biosynthetic pathway, this work highlights the usefulness of synthetic biology approaches in the elucidation of plant natural product biosynthetic pathways and develops yeast as a tool to exploit the wealth of next-generation sequencing data that are constantly being produced through transcriptomic, genomic, or metagenomic studies. The approach described here will be useful to numerous researchers studying terpene biosynthesis in plants and microorganisms.
Keywords: cytochrome P450, metabolic engineering, yeast, terpene
Abstract
Synthetic biology approaches achieving the reconstruction of specific plant natural product biosynthetic pathways in dedicated microbial “chassis” have provided access to important industrial compounds (e.g., artemisinin, resveratrol, vanillin). However, the potential of such production systems to facilitate elucidation of plant biosynthetic pathways has been underexplored. Here we report on the application of a modular terpene production platform in the characterization of the biosynthetic pathway leading to the potent antioxidant carnosic acid and related diterpenes in Salvia pomifera and Rosmarinus officinalis. Four cytochrome P450 enzymes are identified (CYP76AH24, CYP71BE52, CYP76AK6, and CYP76AK8), the combined activities of which account for all of the oxidation events leading to the biosynthesis of the major diterpenes produced in these plants. This approach develops yeast as an efficient tool to harness the biotechnological potential of the numerous sequencing datasets that are increasingly becoming available through transcriptomic or genomic studies.
Chemical synthesis of industrially important plant secondary metabolites is often hampered by their complex stereochemistry and environmental concerns regarding the use of toxic catalysts and solvents. Production of the compounds of interest in heterologous microbial systems can provide a sustainable and environmentally friendly alternative. Several groundbreaking efforts have been reported, including the production of the antimalarial drug artemisinin (1–3), the fragrances sclareol and santalol (4–7), the flavoring agent vanillin (8, 9), and the antioxidant resveratrol (10). Recently, the complete pathway of opioid biosynthesis comprising more than 20 enzymatic activities was reconstructed in yeast, demonstrating the potential of synthetic biology to contribute in the sustainable production of highly complex chemicals (11). In addition to the development of dedicated platform strains (12), such approaches also require detailed knowledge of the biosynthetic pathways and the genes involved. Elucidation of plant secondary biosynthetic pathways can be a demanding process, involving the acquisition of transcriptomic or genomic sequence information and the analysis of a large number of candidate proteins for the identification of the activity responsible for a specific biosynthetic step. Frequently, this approach also requires the time-consuming chemical synthesis of precursors and the structure elucidation of pathway intermediates. Synthetic biology can expedite this process by enabling functional characterization of gene products and synthesis of substrates or intermediates on the same microbial platform. Here, we report on the application of a modular yeast terpene production platform in the elucidation of the biosynthesis of carnosic acid and related diterpenes.
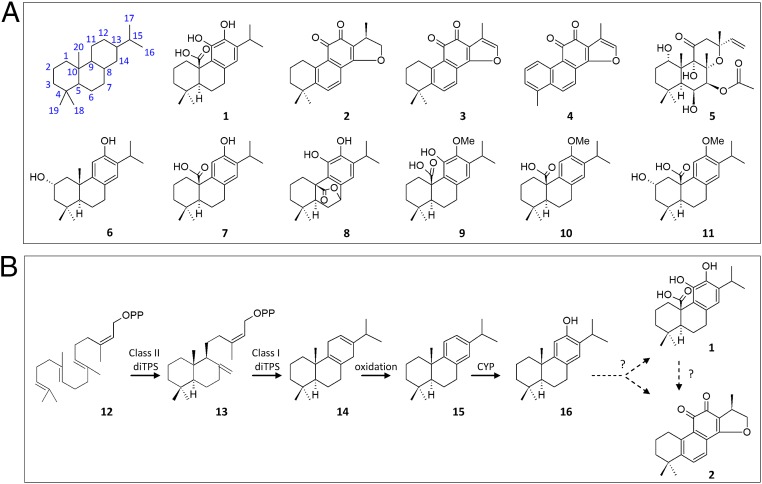
Carnosic acid (1) and carnosol (8) are potent antioxidant (13), antiadipogenic (14), and anticancer agents (15, 16), whereas several carnosic-acid–related diterpenes, such as 12-methoxy-carnosic acid (9) and pisiferic acid (7; Fig. 1A) have strong antiparasitic, antifungal, and antibacterial activities (17–19). These compounds belong to the group of labdane-type diterpenes (20), which also includes several other members with bioactive properties, such as tanshinones (2–4), which exhibit strong antiinflammatory activity (21) and are effective against various cardiovascular and cerebrovascular disorders (22, 23), and forskolin (5), which activates adenylyl cyclase and increases the intracellular levels of cAMP (24–26) (Fig. 1A). Despite their industrial importance, these compounds still remain largely inaccessible by chemical synthesis, calling for the development of biotechnological methods for their production.
Fig. 1.
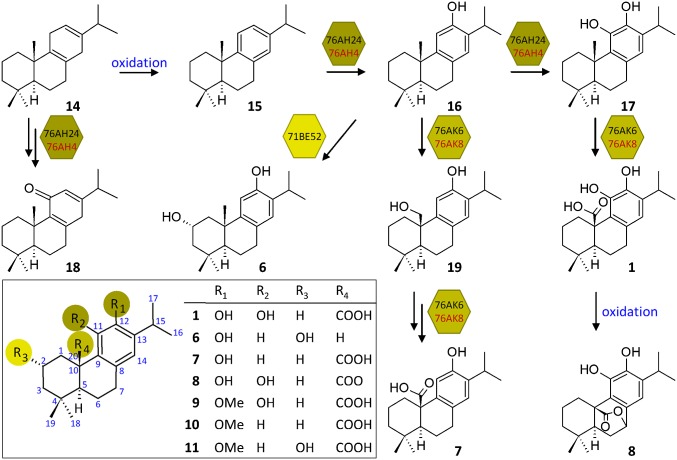
Chemical structures and biosynthesis of labdane-type diterpenes. (A) Chemical structures of the bioactive labdane-related diterpenes carnosic acid (1), cryptotanshinone (2), tanshinone IIA (3), tanshinone I (4), and forskolin (5) (Top). The main diterpenes isolated from S. pomifera leaves include salviol (6), pisiferic acid (7), carnosol (8), 12-methoxy-carnosic acid (9), O-methyl-pisiferic acid (10), and 2α-hydroxy-O-methyl-pisiferic acid (11) (Bottom). (B) The biosynthesis of tanshinone and carnosic acid begins with the cyclization of GGPP (12) by a class II diTPS to produce (+) copalyl diphosphate (CPP) (13), which is in turn converted to miltiradiene (14) by a class I diTPS. Spontaneous oxidation of 14 gives rise to abietatriene (15), which is oxidized to ferruginol (16) by a CYP enzyme. However, uncharacterized subsequent events lead to carnosic acid (1) and tanshinones (2–4).
The 20 carbon atom skeletons of labdane diterpenes, which are characterized by a bicyclic decalin core, are synthesized from geranylgeranyl diphosphate (GGPP; 12) in a modular fashion. Initially, protonation-initiated cyclization by class II diterpene synthases (diTPSs) gives rise to a diphosphate precursor, which is then converted to the basic diterpene skeleton through a divalent-cation–dependent ionization reaction catalyzed by a class I diTPS (reviewed in ref. 20). Subsequent decoration of the various skeletons by modifying enzymes, such as cytochrome P450s (CYPs) (27), gives rise to a plethora of secondary metabolites (28, 29). In Rosmarinus officinalis and Salvia fruticosa, carnosic acid and carnosol are synthesized from (+)-copalyl diphosphate (CPP; 13) through the intermediates miltiradiene (14), abietatriene (15), and ferruginol (16) (Fig. 1B) (30–32). In Salvia miltiorrhiza, the same precursors give rise to the different members of the tanshinone group (33, 34). However, the steps that follow synthesis of 16 and the point where the tanshinone and carnosic acid pathways disentangle are not yet understood, hampering efforts for heterologous pathway reconstruction.
Aiming to elucidate carnosic acid biosynthesis, we set out to develop yeast as a tool for the rapid functional characterization and screening of terpene-targeting CYP genes revealed by next generation transcriptomic analyses. The yeast Saccharomyces cerevisiae offers a convenient host for the functional expression of membrane-bound CYPs due to the resemblance of its intracellular compartments with those of higher eukaryotes. Heterologous expression and isolation of yeast microsomal fractions has provided a valuable tool for in vitro characterization of numerous CYPs (35). In addition, the presence of only three endogenous CYPs (involved in sterol metabolism) prevents unwanted side products in heterologous CYP-expressing engineered yeast. Recently, we developed a terpene production system to recreate the diversity of labdane-related structures in yeast (36). Here, we use this modular platform to set up a rapid and efficient screening system by which different CYPs can be tested for activity against a predefined pathway product and applied it to the characterization of a next-generation transcriptome sequencing dataset obtained from analysis of glandular trichomes of the Eastern Mediterranean species of sage Salvia pomifera.
Results
Whole-leaf extracts of S. pomifera are rich in carnosic-acid–related compounds, including 9, 8, 7, salviol (6), O-methyl-pisiferic acid (10), and 2α-hydroxy-O-methyl-pisiferic acid (11) (Fig. 1A and ref. 37). To elucidate the pathway leading to carnosic-acid–related diterpenes, the transcriptome of S. pomifera glandular trichomes was analyzed by next generation sequencing, revealing the class II and class I terpene synthases responsible for the synthesis of 14 [CPP-synthase (CDS) and miltiradiene synthase (SpMilS), respectively] and 81 contigs with similarity to CYP genes (37). To analyze the biosynthetic steps following formation of 14, 15 of these contigs were shortlisted for further analysis based on two criteria: transcript abundance in the trichome cDNA library (SI Appendix, Table S1), and similarity with other plant CYPs belonging to the CYP71 and CYP76 families, members of which have previously been implicated in terpene biosynthesis (28).
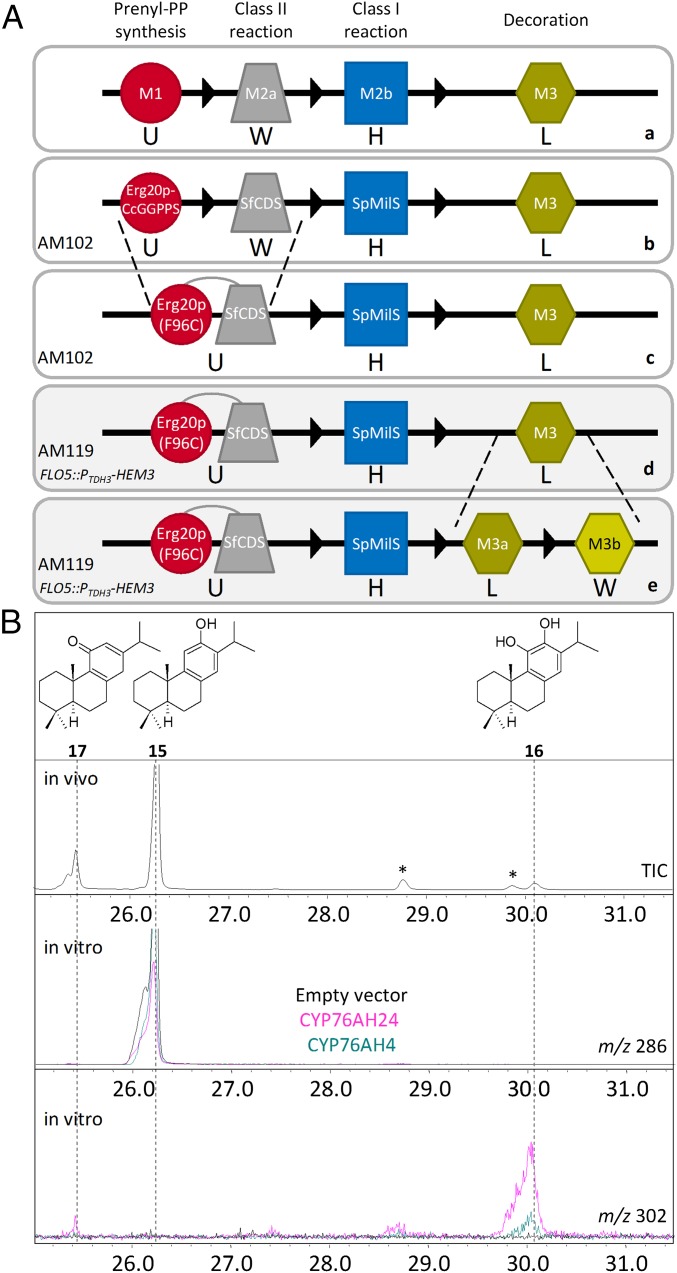
A platform aiming to reconstruct the chemical diversity of diterpene biosynthesis in yeast using module-specific parts has recently been reported (36). In this platform, terpene biosynthesis is split into three conceptual modules: prenyl diphosphate substrate synthesis (M1), terpene skeleton synthesis (M2), and terpene skeleton decoration (M3). In the case of labdane diterpenes, M2 is composed of two submodules, one responsible for the synthesis of the labdane diphosphate precursor by a class II diTPS (M2a) and one for the conversion of this diphosphate to the labdane skeleton by a class I diTPS (M2b) (Fig. 2A, a). Using standardized vectors, module-specific parts can be exchanged to give rise to a diverse array of structures. To functionally characterize the selected CYPs, this platform was configured to produce 14 by using fixed M1, M2a, and M2b parts (SI Appendix, Fig. S1). The fusion between the yeast farnesyl diphosphate synthase (Erg20p) and the GGPP synthase from Cistus creticus (Erg20p-CcGGPPS) (5) was used as the M1-specific part (under uracil selection), the S. fruticosa CPP synthase (SfCDS) as the M2a part (tryptophan selection), and S. pomifera miltiradiene synthase (SpMilS) as the M2b part (histidine selection). Strain AM102 (Mat a/α, PGAL1-HMG2(K6R)::HOX2, ura3, trp1, his3, PTDH3-HMG2(K6R)X2-::leu2 ERG9/erg9, UBC7/ubc7, SSM4/ssm4) (38) was used as the “chassis” (SI Appendix, Table S2).
Fig. 2.
Characterization of S. pomifera CYP76AH24 ortholog as a bifunctional monooxygenase. (A) Diagrammatic representation of the modular design and the platform configuration used. (a) General design of the modular terpene production platform. (b–e) Diagrammatic illustratrations of the steps taken to engineer the platform to facilitate the rapid functional characterization of CYP enzymes. (B) Expression of the CYP76AH24 ortholog in optimized miltiradiene-producing yeast cells (A, b–d) resulted in the production of 16, as main compound, and the formation of 17 and 18 minor products. The two peaks indicated by asterisks correspond to degradation products of 17. In vitro enzymatic assay using a microsomal preparation of yeast cells expressing CPR2 and the S. pomifera CYP76AH24 ortholog (pink) or R. officinalis ferruginol synthase, CYP76AH4 (teal) confirmed the hydroxylation of 16 at C-11. Microsomal preparations of cells expressing only CPR2 are used as negative control (black).
To test for oxidation of 14, the selected CYPs were introduced into the yeast dual-expression vector pESC-LEU, under the control of the galactose-inducible PGAL10 promoter, together with the poplar CYP reductase CPR2 (39) under PGAL1, and exchanged at M3 (Fig. 2A, b). GC-MS analysis of miltiradiene-producing AM102 cells expressing the different CYPs revealed only one active enzyme, CYP76AH24, which produced 16, as confirmed by comparison with authentic standard and characterization of the purified yeast-produced compound (Fig. 2B and SI Appendix, Figs. S4–S6). CYP76AH24 was the most abundant CYP transcript in the glandular trichome cDNA library used in the transcriptomic study and exhibits almost 80% identity at the amino acid level with CYP76AH1, the ferruginol synthase from S. miltiorrhiza (34) and 91% identity with CYP76AH4, the ferruginol synthase from R. officinalis (32).
Examination of the structures of the main S. pomifera compounds (Fig. 1A) suggested that 16 is likely their common precursor and that additional CYPs are possibly responsible for the further modification of 16 to the different compounds. To enable analysis of the steps downstream of synthesis of 16, the yeast modular platform was modified to improve production of 16 and to allow for the simultaneous expression of multiple CYPs. To improve overall diterpene titers and to make better use of the available auxotrophic selection markers, the Erg20p-CcGGPPS fusion (used as the M1 part) and SfCDS (used as the M2a-specific part) were replaced by a fusion between a mutant form of the yeast FPP synthase [Erg20p(F96C)] and SfCDS. Erg20p(F96C) has been shown to be more efficient in supporting diterpene production than CcGGPPS (5), whereas fusion of a class II synthase N terminally to Erg20p(F96C) was found to facilitate substrate channeling and improve coupling of the subsequent biosynthetic step (5). Coexpression of SfCDS-Erg20p(F96C) (M2a-M1) with SpMilS (M2b) and CPR/CYP76AH24 (M3) resulted in a threefold increase in the ferruginol titer, from 2.7 to 8.4 mg/L (Fig. 2A, c and SI Appendix, Fig. S2 A and B).
High-level expression of CYPs in yeast may impose stress by heme depletion, and engineering the endogenous heme biosynthetic pathway has been found to improve CYP activity (40). To optimize the platform for CYP expression, one copy of the yeast HEM3 gene, encoding for the rate-limiting enzyme in the pathway, was introduced in the FLO5 locus of AM102 yeast cells, under the control of the strong PTDH3 promoter, to generate strain AM119 (Mat a/α, PGAL1-HMG2(K6R)::HOx2, ura3, trp1, his3, PTDH3-HMG2(K6R)X2-::leu2 ERG9/erg9, UBC7/ubc7, SSM4/ssm4, PTDH3-HEM3::FLO5) (SI Appendix, Table S2). Using this strain, an additional 2.5-fold increase in the titer of 16 was obtained, reaching 21.2 mg/L (Fig. 2A, d, Fig. 2B, and SI Appendix, Fig. S2 A and B). The substrate of ferruginol synthase is 15, a spontaneous oxidation product of 14 (32). Quantification of 15 and 16 revealed that, whereas in AM102 cells, 32% of 15 was converted to 16, in AM119 cells, the efficiency of this conversion increased to 62%, suggesting an improvement in the activity of CYP76AH24 in the modified strain (SI Appendix, Fig. S2B). This improvement allowed the identification of additional compounds synthesized by CYP76AH24 (Fig. 2B), which were not detectable using the initial platform configuration (SI Appendix, Fig. S2A). Isolation and structure elucidation revealed two compounds: 11-hydroxy ferruginol (8,11,13-podocarpatriene-11,12-diol) (17) (SI Appendix, Figs. S7–S9) and 11-keto miltiradiene (18) (SI Appendix, Figs. S10–S16 and Table S3). To the best of our knowledge, 18 is reported for the first time in this work. The ability of CYP76AH24 to catalyze additional oxidation events on 16 was confirmed by in vitro studies in which microsomal membranes isolated from yeast cells expressing CYP76AH24 and CPR2 were incubated with 16 in the presence of NADPH (Fig. 2B). Production of 17 was confirmed by GC-MS analysis, using comparison with the isolated structurally characterized compound. In vitro experiments with 14 as substrate also confirmed the formation of 18 by CYP76AH24, albeit with lower efficiency than synthesis of 17 (kcat = 1.97 min−1 and KM = 72.63 μM for the reaction with 14 compared with kcat = 6.11 min−1 and KM = 45.84 μM for the reaction with 16; kinetic data provided in SI Appendix, Table S4 and Fig. S25). The significant levels of 18 produced by yeast cells expressing CYP76AH24 are thus possibly due to the high amounts of 14 produced by the platform. In agreement with previous observations (41), 17 was found to be unstable under chromatographic conditions, giving rise to several oxidation products during GC-MS analysis (indicated by asterisks in Fig. 2B and SI Appendix, Fig. S2A). These findings strongly suggest that CYP76AH24 is a bifunctional enzyme that catalyzes successive oxidation events on C-12 and C-11 of the labdane skeleton. All other ferruginol synthase enzymes isolated to date (from S. miltiorrhiza, S. fruticosa, or R. officinalis) have been reported to have single functionality. To investigate whether CYP76AH24 is the only enzyme able to catalyze additional oxidation events on 16 or this activity has previously been unnoticed with the other enzymes, the ferruginol synthase from rosemary, CYP76AH4, was analyzed in vitro. Microsomal preparations of CYP76AH4 were assayed in parallel with CYP76AH24 and both enzymes catalyzed production of 17 (Fig. 2B). Kinetic analysis revealed that the two enzymes exhibit similar affinity for 16 (KM = 45.8 ± 9.4 μM in the case of CYP76AH24 and 49.5 ± 11.1 μM in the case of CYP76AH4), but also for 14 and 15 as substrates (SI Appendix, Table S4 and Fig. S25).
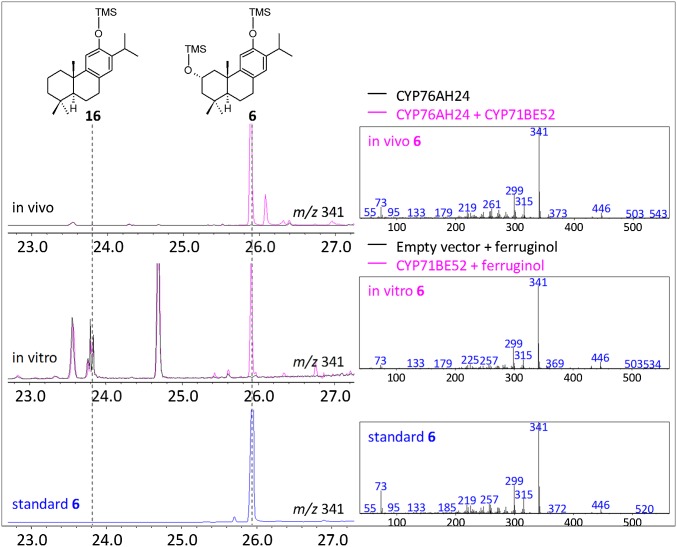
These results suggest that synthesis of 17 is likely the event that follows formation of 16 en route to 1 in S. pomifera and R. officinalis. To elucidate the subsequent steps in this pathway, the ability of additional CYPs to act on the products of CYP76AH24 was investigated. To this end, the remaining 14 CYPs were cloned in the yeast vector pWTDH3myc and introduced under tryptophan selection as a second M3 module, M3b, alongside CYP76AH24 (M3a) (Fig. 2A, e). Using this reconfigured platform, two additional CYPs acting on CYP76AH24 products were identified. The first one, CYP71BE52, was found to catalyze oxidation of 16 at position 2α, giving rise to 6, one of the major diterpenes of S. pomifera. In yeast culture extracts, 6 was identified by comparison of retention time and mass spectrum with authentic standard (Fig. 3, Top and Bottom and SI Appendix, Figs. S17 and S18), whereas microsomal preparations of CYP71BE52 and CPR2-producing cells were used in enzymatic assays to confirm conversion of 16 to 6 in vitro (Fig. 3, Middle; kinetic parameters for the in vitro reaction of CYP71BE56 with 16 are provided in SI Appendix, Table S4 and Fig. S25). CYP71BE52 was the third most abundant transcript among the identified CYP family genes, suggesting that it likely encodes for the main enzyme responsible for the production of 6 in S. pomifera glandular trichomes.
Fig. 3.
CYP71BE52 is a salviol synthase. Expression of S. pomifera CYP71BE52 (pink) as an M3b-specific part in the ferruginol-producing yeast platform resulted in the production of 6, as identified by GC-MS analysis of the TMS-derivatized solvent extract (Top). A microsomal preparation of CYP71BE52 converted 16 to 6 in the presence of NADPH as cofactor (Middle). Formation of 6 was confirmed by comparison with authentic standard (Bottom).
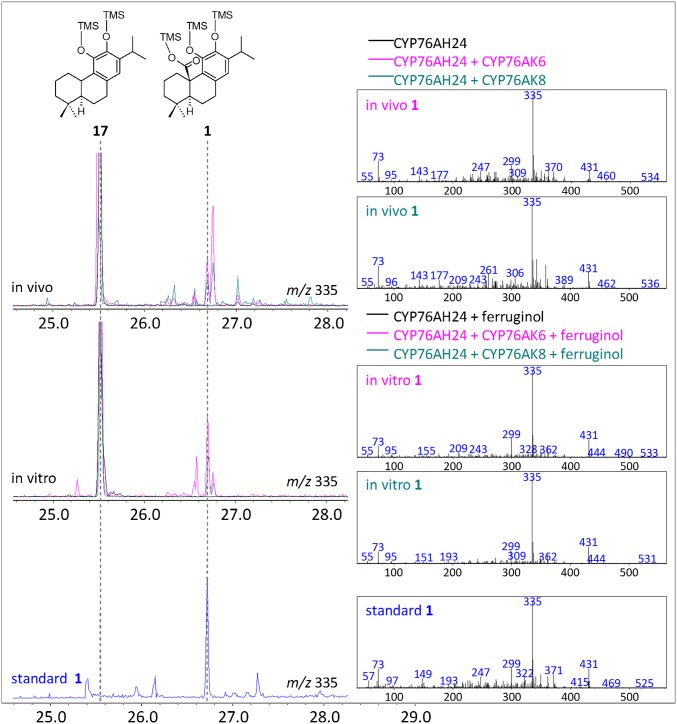
In addition to CYP71BE52, the screen also revealed CYP76AK6, the second most abundant transcript after CYP76AH24, as an enzyme able to oxidize several compounds produced by CYP76AH24 in S. cerevisiae. Three compound groups were detected by GC-MS analysis of trimethylsilyl (TMS) derivatized ethyl acetate extracts of culture media (SI Appendix, Supplementary Methods 1.4). The first group of products displayed characteristic fragment ions with 335 and 431 m/z (Fig. 4, Top). Comparison of retention time and mass spectrum with authentic standard confirmed one of these products as 1 (Fig. 4, Bottom), likely synthesized by successive oxidation events at C-20 of 17. Structure elucidation of the additional compounds in this group was not possible due to the low amounts produced in yeast. However, based on the similarity of their mass spectra with that of 1, and the fact that oxidized C-20 of the labdane skeleton is labile and frequently undergoes elimination during GC-MS analysis (42), it is likely that these are the corresponding C-20 aldehyde and alcohol intermediates. The second group of compounds included two peaks exhibiting strong 348 and 430 m/z fragment ions, which are characteristic of 8 (SI Appendix, Fig. S3A, a), a potent antioxidant resulting from the oxidation of 1. One of these compounds was confirmed to be 8 by comparison with authentic standard (SI Appendix, Figs. S3A, b, S19, and S20). The remaining compound in this group could result from oxidative degradation and rearrangement of carnosic acid to a molecule bearing a catechol, quinone, or semiquinone feature at C-12–C-11 (13, 43, 44), but insufficient amounts precluded characterization at this stage. The last group of compounds displayed 343 m/z as the most abundant fragment ion (SI Appendix, Fig. S3B, a). One of these compounds matched the retention time and mass spectrum of TMS-derivatized 7 standard (SI Appendix, Figs. S3B, a and d, S21, and S22). Purification of the yeast products followed by structure elucidation also revealed production of pisiferol (19) (SI Appendix, Figs. S3B, a and c, S23, and S24), the product of the first oxidation event on C-20 of ferruginol en route to 7. Identification of the remaining compound in this group (indicated by an asterisk, SI Appendix, Fig. S3B, a) was not possible at this stage, but this peak likely corresponds to pisiferal, the intermediate between 19 and 7. Taken together, these results suggest that CYP76AK6 is a multifunctional enzyme that catalyzes successive oxidation events at C-20 of the labdane skeleton. An enzymatic assay using a combination of microsomal preparations of CYP76AH24 and CYP76AK6 in the presence of 16 as substrate confirmed the ability of CYP76AK6 to produce in vitro a similar blend of molecules to those obtained from yeast cells (Fig. 4 and SI Appendix, Fig. S3B, b). However, production of 8 was not detected in the in vitro reaction, suggesting that enzymatic or nonenzymatic oxidation events occurring in the process of yeast cultivation are likely responsible for the conversion of 1 to 8 and other oxidative degradation products.
Fig. 4.
CYP76AK6 and its homolog, CYP76AK8, catalyze successive oxidation events at C-20. Formation of 1 by CYP76AK6 (pink) and CYP76AK8 (teal) in yeast cells coexpressing CYP76AH24 and SpMilS. TMS-derivatized extracts of yeast cultures analyzed by GC-MS revealed the formation of 1 (Top), identified by comparison with authentic standard (blue) (Bottom). Enzymatic assays containing microsomal preparations of CYP76AH24 ortholog and CYP76AK6 (pink) or CYP76AK8 (teal), 16 and NADPH revealed the formation of 1 in vitro (Middle).
To explore whether the same steps are also responsible for carnosic acid biosynthesis in R. officinalis, a transcriptome database available online (medicinalplantgenomics.msu.edu) was used to identify the closest rosemary homolog of CYP76AK6 through blastx analysis. CYP76AK8, which is encoded by the ORF of R. officinalis locus 4766 isotig 5 and exhibits 75% similarity at the amino acid sequence level with CYP76AK6, was amplified from R. officinalis cDNA and tested for the production of 1 in the 11-hydroxy-ferruginol-producing yeast cells. The product profile of S. cerevisiae cells expressing CYP76AK8 was similar to that of CYP76AK6, including 1, 8, 7, and 19 (Fig. 4 and SI Appendix, Fig. S3 A and B, a). Moreover, in vitro reaction combining individual microsomal preparations of CYP76AH24 and CYP76AK8 with 16 as substrate and NADPH cofactor resulted in the production of the same compounds, except for 8 (Fig. 4 and SI Appendix, Fig. S3 A and B, b), whereas kinetic parameters for the in vitro reactions of CYP76AK6 and CYP76AK8 with 16 and 17 were found to be comparable (SI Appendix, Table S4 and Fig. S25), suggesting that CYP76AK8 is the rosemary ortholog of CYP76AK6. Investigation of the order of biosynthetic events using in vitro assays revealed that although CYP76AK6 and CYP76AK8 can oxidize C-20 of both 16 and 17, CYP76AH24 or CYP76AH4 are unable to modify 7. The titer of 1 obtained in shake-flask cultivation using the described configuration was in the range of 1 mg/L of culture. Considering the number of biosynthetic steps introduced downstream of the mevalonate pathway in this platform, further optimization through state-of-the-art metabolic engineering approaches is expected to achieve significant improvement in product yields.
Discussion
The combined activities of the CYPs identified here, targeting four positions of the abietane skeleton, C-2, C-11, C-12, and C-20, are sufficient to explain all of the oxidation events involved in the biosynthesis of the major diterpenes identified in S. pomifera (Fig. 5), and to clarify the pathway leading to 1 and 8 in both S. pomifera and R. officinalis. In the first step of this mechanism, 15 is converted to 16 by CYP76AH24 or CYP76AH4, which is then taken up by CYP76AK6 or CYP76AK8 to produce 7. Further oxidation of 16 by CYP76AH24 or CYP76AH4 produces 17, which is converted to 1 by CYP76AK6 or CYP76AK8. In yeast cultures, 1 is converted to 8, either by nonenzymatic oxidation or through a yet unidentified yeast enzymatic system. In S. pomifera, CYP71BE52 oxidizes 16 at position 2α to produce salviol. O-methylation at position 12 of 1, 7, or 2α-hydroxy-pisiferic acid produces the corresponding methoxy derivatives. Further studies will extend this approach to the identification of the methyltransferase(s) responsible for this step, using the already available transcriptomic information.
Fig. 5.
Proposed mechanism for the biosynthesis of carnosic-acid–related diterpenes in S. pomifera and R. officinalis. The biosynthesis of labdane-type diterpenes in S. pomifera (black) and R. officinalis (red) is initiated by the action of bifunctional enzymes, CYP76AH24 or CYP76AH4, respectively, which are responsible for the hydroxylation of 15 initially at position C-12 to produce 16 and subsequently at position C-11 to yield 17. The same bifunctional enzymes can also catalyze two successive oxidation events on 14 to yield 18. A S. pomifera enzyme, CYP71BE52, catalyzes oxidation of 16 at position 2α to synthesize 6. CYP76AK6 or CYP76AK8 catalyze successive oxygenations at position C-20 of 16 and 17 to yield 7 via 19, and 1, respectively. In the yeast platform, 1 is further oxidized to 8 by a yet undefined mechanism. The combined activities of CYP76AH24, CYP71BE52, and CYP76AK6 on the abietatriene skeleton (Inset) are sufficient to explain the biosynthesis of the main diterpenes (1, 6-11) isolated from S. pomifera and R. officinalis.
Although the example presented in this report only goes as far as the characterization of CYP activities responsible for the oxidation of miltiradiene-derived structures, the developed approach has broader application. The modular structure of the platform allows its facile reconfiguration for the production of other terpene skeletons, so that, for example, when a library of M3 parts is available, this can be tested in parallel on different mono-, sesqui-, di-, or triterpenoid substrates. This is particularly useful in the analysis of libraries derived from transcriptomic studies, which encode biosynthetic activities responsible for the production of a diverse group of terpene compounds. The design is expandable, allowing the incorporation of additional module-specific parts to study events beyond oxidation (e.g., acetylation, methylation, etc).
Transcription of genes involved in secondary metabolite biosynthesis is frequently influenced by environmental factors or developmental phase. Such variations could influence the efficiency of pathway elucidation approaches based on transcriptional analysis. The effectiveness of the overall effort can be greatly improved if transcriptomic analysis is carried out in parallel to metabolomic studies, and, if possible, on the same sample or under very similar conditions. In the case study presented here, metabolomic and transcriptomic analysis was combined in the same plants (37). Following next-generation sequencing, genes were prioritized for analysis on the basis of their transcription levels and by increasing the representation of the CYP71 and CYP76 subfamilies, members of which have previously been found to be involved in terpene oxidation (29). Rational short listing of candidate genes can help minimize time and reduce cost. In the current effort, the three CYPs involved in carnosic-acid–related diterpenes in S. pomifera corresponded to the top three expressed CYP transcripts in the glandular trichome library. Because these CYPs also belong to the targeted CYP subfamilies, it is unclear at this stage whether a selection of candidates based on transcript abundance would, in general, be advantageous over selection, based on sequence relatedness. For major metabolites, prioritization based on gene expression levels is likely to be more successful, whereas sequence-based selection may work better for minor compounds or for transcriptional datasets obtained under different conditions than those of the metabolic profile.
Taken together, these results highlight the effectiveness of using engineered yeast as a tool for the elucidation of biosynthetic pathways. The yeast system can expedite structure elucidation of products or pathway intermediates by readily providing sufficient amounts of compounds through the high titers achieved using the engineered yeast strains. This also makes adequate quantities of substrates and standards available for subsequent confirmation and characterization of the pathway steps through in vitro studies or for the evaluation of the biological activity of isolated compounds. Yeast is a commonly used microorganism that does not require dedicated culture systems or specialized equipment, whereas scaling up can be achieved using established infrastructure. Due to its relatively short growth cycle, yeast allows for fast turnaround times, enabling analysis of a large number of samples in a short time. Thus, in combination with DNA synthesis and rapid cloning techniques, this system can serve as a tool to exploit the wealth of next-generation sequencing data that is constantly being produced through transcriptomic, genomic, or metagenomic studies. Additionally, because many of these compounds have industrial importance, coupling pathway identification with production in the same organism minimizes transfer to an industrial setting. As yeast is a favorable host for industrial fermentation, once a specific pathway is analyzed, the system is already set up for further optimization of heterologous production of specific compounds using metabolic engineering approaches, such as flux analysis, pathway balancing, or metabolic channeling.
Materials and Methods
Yeast Strains Construction.
Yeast strain AM119 was generated using AM102 (38) as follows. The HEM3 gene was cloned into EcoRI-XhoI restriction sites of the plasmid construct COD7 (PTDH3-HEM3-CYC1t, LoxP-HIS5-LoxP) (38) using primers 5-HEM3-EcoRI and 3-HEM3-XhoI (SI Appendix, Table S5). The COD7/HEM3 construct was PCR amplified with primers 5-FLO5-COD7 and 3-FLO5-COD7 (SI Appendix, Table S5), which incorporate flanking sequences with complementarity to the 3′UTR of FLO5 gene. Selection marker excision gave rise to strain AM119.
In Vitro Enzymatic Assay and Kinetic Analysis.
The kinetic parameters of the different CYPs were determined as previously described (36) using varying concentrations (1–75 μM) of each substrate (SI Appendix, Table S4). The enzymatic reactions were incubated with mild shaking at 30 °C for 3 h and terminated by extraction with 100 μL of decane or ethyl acetate containing 10 μg/mL sclareol as internal standard. A total of 2 μL of extracts was analyzed by GC-MS using the conditions described in SI Appendix. To evaluate the combined enzymatic activity of CYPs in vitro, a 0.5-mL reaction containing 90 mM Tris⋅HCl (pH 7.5), 1 mM NADPH, 80 pmol of CYP76AH24 or CYP76AH4 microsomal protein, and 100 μM ferruginol was incubated with shaking at 30 °C for 18 h. Subsequently, 80 pmol of CYP76AK6 or CYP76AK8 was added and the reaction was incubated for an additional 18 h and terminated by extraction with 200 μL pentane or ethyl acetate. The extract was gently evaporated and derivatized with Sylon HTP. A total of 2 μL of derivatized extract was analyzed by GC-MS. All assays were carried out in duplicates.
Supplementary Material
Acknowledgments
We thank Prof. Reuben Peters for the plasmid bearing the CYP76AH4 from rosemary and Prof. David R. Nelson for annotation of the CYP sequences. This work was funded by the European Commission FP7 Program TransPOT (FP7-2011-REGPOT, Contract 285948). The page charges for this article have been kindly covered by the Special Account for Research (ELKE) of the University of Crete.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523787113/-/DCSupplemental.
References
- 1.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 2.Westfall PJ, et al. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci USA. 2012;109(3):E111–E118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paddon CJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496(7446):528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 4.Schalk M, et al. Toward a biosynthetic route to sclareol and amber odorants. J Am Chem Soc. 2012;134(46):18900–18903. doi: 10.1021/ja307404u. [DOI] [PubMed] [Google Scholar]
- 5.Ignea C, et al. Efficient diterpene production in yeast by engineering Erg20p into a geranylgeranyl diphosphate synthase. Metab Eng. 2015;27:65–75. doi: 10.1016/j.ymben.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Scalcinati G, et al. Combined metabolic engineering of precursor and co-factor supply to increase α-santalene production by Saccharomyces cerevisiae. Microb Cell Fact. 2012;11:117. doi: 10.1186/1475-2859-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Daviet L, Schalk M, Siewers V, Nielsen J. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab Eng. 2013;15:48–54. doi: 10.1016/j.ymben.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Brochado AR, et al. Improved vanillin production in baker’s yeast through in silico design. Microb Cell Fact. 2010;9:84. doi: 10.1186/1475-2859-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Evolva (2016) Vanilin. A sustainable production route. Available at www.evolva.com/vanillin/. Accessed February 23, 2016.
- 10. Evolva (2016) Everesveratrol. Better, naturally. Available at res.evolva.com/. Accessed February 23, 2016.
- 11.Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD. Complete biosynthesis of opioids in yeast. Science. 2015;349(6252):1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen J. BIOENGINEERING. Yeast cell factories on the horizon. Science. 2015;349(6252):1050–1051. doi: 10.1126/science.aad2081. [DOI] [PubMed] [Google Scholar]
- 13.Birtić S, Dussort P, Pierre FX, Bily AC, Roller M. Carnosic acid. Phytochemistry. 2015;115:9–19. doi: 10.1016/j.phytochem.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 14.Gaya M, et al. Antiadipogenic effect of carnosic acid, a natural compound present in Rosmarinus officinalis, is exerted through the C/EBPs and PPARγ pathways at the onset of the differentiation program. Biochim Biophys Acta. 2013;1830(6):3796–3806. doi: 10.1016/j.bbagen.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 15.de la Roche M, et al. An intrinsically labile α-helix abutting the BCL9-binding site of β-catenin is required for its inhibition by carnosic acid. Nat Commun. 2012;3:680. doi: 10.1038/ncomms1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danilenko M, et al. Carnosic acid potentiates the antioxidant and prodifferentiation effects of 1alpha,25-dihydroxyvitamin D3 in leukemia cells but does not promote elevation of basal levels of intracellular calcium. Cancer Res. 2003;63(6):1325–1332. [PubMed] [Google Scholar]
- 17.Mokoka TA, et al. Antileishmanial activity of 12-methoxycarnosic acid from Salvia repens Burch. ex Benth. (Lamiaceae) S Afr J Bot. 2014;90:93–95. [Google Scholar]
- 18.Kobayashi K, Nishino C, Tomita H, Fukushima M. Antifungal activity of pisiferic acid-derivatives against the rice blast fungus. Phytochemistry. 1987;26(12):3175–3179. [Google Scholar]
- 19.Fukui HKK, Egawa H. A new diterpene with antimicrobial activity from Chamaecyparis pisifera Endle. Agric Biol Chem. 1978;42(7):1419–1423. [Google Scholar]
- 20.Peters RJ. Two rings in them all: The labdane-related diterpenoids. Nat Prod Rep. 2010;27(11):1521–1530. doi: 10.1039/c0np00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson AL, et al. A zebrafish compound screen reveals modulation of neutrophil reverse migration as an anti-inflammatory mechanism. Sci Transl Med. 2014;6(225):225ra29. doi: 10.1126/scitranslmed.3007672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng TO. Cardiovascular effects of Danshen. Int J Cardiol. 2007;121(1):9–22. doi: 10.1016/j.ijcard.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Xu S, et al. Tanshinone II-A attenuates and stabilizes atherosclerotic plaques in apolipoprotein-E knockout mice fed a high cholesterol diet. Arch Biochem Biophys. 2011;515(1-2):72–79. doi: 10.1016/j.abb.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Seamon KB, Padgett W, Daly JW. Forskolin: Unique diterpene activator of adenylate cyclase in membranes and in intact cells. Proc Natl Acad Sci USA. 1981;78(6):3363–3367. doi: 10.1073/pnas.78.6.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daly JW. Forskolin, adenylate cyclase, and cell physiology: An overview. Adv Cyclic Nucleotide Protein Phosphorylation Res. 1984;17:81–89. [PubMed] [Google Scholar]
- 26.Shoback DM, Brown EM. Forskolin increases cellular cyclic adenosine monophosphate content and parathyroid hormone release in dispersed bovine parathyroid cells. Metabolism. 1984;33(6):509–514. doi: 10.1016/0026-0495(84)90004-0. [DOI] [PubMed] [Google Scholar]
- 27.Ortiz de Montellano PR. Cytochrome P-450 Structure, Mechanism, and Biochemistry. Plenum; New York: 1986. [Google Scholar]
- 28.Bak S, et al. Cytochromes p450. Arabidopsis Book. 2011;9:e0144. doi: 10.1199/tab.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pateraki I, Heskes AM, Hamberger B. Cytochromes P450 for terpene functionalisation and metabolic engineering. Adv Biochem Eng Biotechnol. 2015;148:107–139. doi: 10.1007/10_2014_301. [DOI] [PubMed] [Google Scholar]
- 30.Božić D, et al. Towards elucidating carnosic acid biosynthesis in Lamiaceae: Functional characterization of the three first steps of the pathway in Salvia fruticosa and Rosmarinus officinalis. PLoS One. 2015;10(5):e0124106. doi: 10.1371/journal.pone.0124106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brückner K, et al. Characterization of two genes for the biosynthesis of abietane-type diterpenes in rosemary (Rosmarinus officinalis) glandular trichomes. Phytochemistry. 2014;101:52–64. doi: 10.1016/j.phytochem.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 32.Zi J, Peters RJ. Characterization of CYP76AH4 clarifies phenolic diterpenoid biosynthesis in the Lamiaceae. Org Biomol Chem. 2013;11(44):7650–7652. doi: 10.1039/c3ob41885e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao W, et al. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett. 2009;11(22):5170–5173. doi: 10.1021/ol902051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo J, et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci USA. 2013;110(29):12108–12113. doi: 10.1073/pnas.1218061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- 36.Ignea C, et al. Reconstructing the chemical diversity of labdane-type diterpene biosynthesis in yeast. Metab Eng. 2015;28:91–103. doi: 10.1016/j.ymben.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Trikka FA, et al. Combined metabolome and transcriptome profiling provides new insights into diterpene biosynthesis in S. pomifera glandular trichomes. BMC Genomics. 2015;16(1):935. doi: 10.1186/s12864-015-2147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ignea C, et al. Positive genetic interactors of HMG2 identify a new set of genetic perturbations for improving sesquiterpene production in Saccharomyces cerevisiae. Microb Cell Fact. 2012;11(1):162. doi: 10.1186/1475-2859-11-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ro DK, Ehlting J, Douglas CJ. Cloning, functional expression, and subcellular localization of multiple NADPH-cytochrome P450 reductases from hybrid poplar. Plant Physiol. 2002;130(4):1837–1851. doi: 10.1104/pp.008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michener JK, Nielsen J, Smolke CD. Identification and treatment of heme depletion attributed to overexpression of a lineage of evolved P450 monooxygenases. Proc Natl Acad Sci USA. 2012;109(47):19504–19509. doi: 10.1073/pnas.1212287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tada M, Ishimaru K. Efficient ortho-oxidation of phenol and synthesis of anti-MRSA and anti-VRE compound abietaquinone methide from dehydroabietic acid. Chem Pharm Bull (Tokyo) 2006;54(10):1412–1417. doi: 10.1248/cpb.54.1412. [DOI] [PubMed] [Google Scholar]
- 42.Hossain MB, Rai DK, Brunton NP, Martin-Diana AB, Barry-Ryan C. Characterization of phenolic composition in Lamiaceae spices by LC-ESI-MS/MS. J Agric Food Chem. 2010;58(19):10576–10581. doi: 10.1021/jf102042g. [DOI] [PubMed] [Google Scholar]
- 43.Masuda T, et al. Recovery mechanism of the antioxidant activity from carnosic acid quinone, an oxidized sage and rosemary antioxidant. J Agric Food Chem. 2002;50(21):5863–5869. doi: 10.1021/jf025605o. [DOI] [PubMed] [Google Scholar]
- 44.Masuda T, Inaba Y, Takeda Y. Antioxidant mechanism of carnosic acid: Structural identification of two oxidation products. J Agric Food Chem. 2001;49(11):5560–5565. doi: 10.1021/jf010693i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.