Abstract
Background and objectives
Nephrolithiasis is a prevalent condition that affects 10%–15% of adults in their lifetime. It is associated with high morbidity due to colicky pain, the necessity for surgical intervention, and sometimes progression to CKD. In recent years, multiple monogenic causes of nephrolithiasis and nephrocalcinosis have been identified. However, the prevalence of each monogenic gene in a pediatric renal stone cohort has not yet been extensively studied.
Design, setting, participants, & measurements
To determine the percentage of cases that can be explained molecularly by mutations in one of 30 known nephrolithiasis/nephrocalcinosis genes, we conducted a high-throughput exon sequencing analysis in an international cohort of 143 individuals <18 years of age, with nephrolithiasis (n=123) or isolated nephrocalcinosis (n=20). Over 7 months, all eligible individuals at three renal stone clinics in the United States and Europe were approached for study participation.
Results
We detected likely causative mutations in 14 of 30 analyzed genes, leading to a molecular diagnosis in 16.8% (24 of 143) of affected individuals; 12 of the 27 detected mutations were not previously described as disease causing (44.4%). We observed that in our cohort all individuals with infantile manifestation of nephrolithiasis or nephrocalcinosis had causative mutations in recessive rather than dominant monogenic genes. In individuals who manifested later in life, causative mutations in dominant genes were more frequent.
Conclusions
We present the first exclusively pediatric cohort examined for monogenic causes of nephrolithiasis/nephrocalcinosis, and suggest that important therapeutic and preventative measures may result from mutational analysis in individuals with early manifestation of nephrolithiasis or nephrocalcinosis.
Keywords: genetic renal disease; kidney stones; hypercalciuria; nephrocalcinosis; mutation; child; Europe; exons; genes, dominant; humans
Introduction
The incidence of pediatric nephrolithiasis and nephrocalcinosis (NL/NC) has significantly increased over the last several decades (1). Furthermore, this condition is associated with high morbidity due to episodes of colicky pain, the necessity for surgical intervention, and sometimes progression to CKD. Nephrolithiasis (NL) and nephrocalcinosis (NC) share a recognized degree of heritability. In twin studies, heritability accounted for nearly half of all NL/NC prevalence (2,3). According to the Online Mendelian Inheritance in Man (OMIM) database, mutations in at least 30 genes can cause monogenic forms of NL/NC by autosomal recessive, autosomal dominant, or X-linked transmission. Recently, we studied 166 adults and 106 children with NC or NL and detected causative mutations in 11.4% of adult and 20.8% of early-onset cases (4). This result confirmed a significant occurrence of heritable NL/NC, while also indicating that there are additional unidentified monogenic causes of NL/NC.
However, the contribution of monogenic causes of NL/NC has yet to be extensively studied. Because pediatric patients are more likely to have a monogenic cause of disease, mutational analysis is particularly relevant in this cohort. Furthermore, early detection of disease-causing mutations is of great importance in early-onset NL/NC because genetic diagnoses allow finely tailored treatment plans that may prevent recurrent disease or progression to ESRD.
Until recently, mutation analysis for individuals with NL/NC has not been widely accessible. However, through the availability of high-throughput multiplex PCR and next-generation sequencing, rapid mutation analysis of multiple genes in large cohorts has become an efficient and cost-effective screening method (5–7).
To identify the prevalence of monogenic causes in early-onset NL/NC (onset before 18 years of age), we analyzed all coding exons and adjacent splice sites of 30 known NL/NC-causing genes with a defined OMIM phenotype in a cohort of 143 children recruited at three renal stone clinics with at least one episode of NL or the presence of NC upon renal ultrasound before 18 years of age. This patient cohort had no patient overlap with the previously published cohort (4).
Materials and Methods
Study Cohort
This study was approved by the Institutional Review Board of Boston Children’s Hospital. Study inclusion criteria were defined as: first clinical manifestation of NL and/or presence of NC on renal ultrasound before 18 years of age. Individuals with conditions or medication that might have caused secondary renal stone disease were excluded. To avoid selection bias, all patients seen at three renal stone clinics (Boston Children’s Hospital, University Clinic Skopje, and University Clinic Zagreb) over a finite period (February–August 2014) were approached for study participation. The exact percentage of individuals who declined study participation was not recorded. After obtaining informed consent, clinical data, pedigree information, and DNA samples were collected from 143 individuals. All individuals were recruited after the conclusion of the study of Halbritter et al. (4), thereby excluding enrollment overlap between the two studies. The cohort consisted of 72 male and 71 female participants. Of these individuals, 123 had NL, 20 had NC based on renal ultrasound, and three with NC also had reported NL.
Mutation Analysis
Mutation analysis was performed using a barcoded multiplex PCR-based approach, as previously described (5,6). We designed 518 target-specific primers for 381 coding exons and the adjacent splice sites of 30 genes that are known monogenic causes of NL/NC (defined by OMIM; www.ncbi.nlm.nih.gov/omim). The genes screened were: ADCY10, AGXT, PRT, ATP6V0A4, ATP6V1B1, CA2, CASR, CLCN5, CLCNKB, CLDN16, CLDN19, CYP24A1, FAM20A, GRHPR, HNF4A, HOGA1, HPRT1, KCNJ1, OCRL, SLC12A1, SLC22A12, SLC2A9, SLC34A1, SLC34A3, SLC3A1, SLC4A1, SLC7A9, SLC9A3R1, VDR, and XDH (Supplemental Tables 1 and 2). Amplicon sizes were chosen to range from 250 to 300 bp (primer sequences are available from the authors). The use of barcoded multiplex PCR (Fluidigm 48.48-Access Arrays system) allowed parallel amplification of all 518 amplicons in 48 individuals at a time (5,6). Subsequently, the pooled libraries were sequenced on an Illumina MiSeq instrument using the v2 chemistry. Sequence reads were aligned to the human reference sequence using CLC Genomics Workbench (CLC-bio, Aarhus, Denmark) (5). Prior to further evaluation, we excluded synonymous variants and variants that occur with minor allele frequency >1% in the dbSNP (version 138) database. Remaining variants were validated as previously described (7). Briefly, all variants that had previously been described in individuals with NL/NC were considered as likely to be disease causing. Novel variants were ranked based on their likelihood to be deleterious for the function of the encoded protein considering protein truncation and obligatory splice site mutations as likely to be disease causing. For missense alleles, evolutionary conservation among orthologs across phylogeny, and bioinformatics prediction programs (PolyPhen-2 [8], SIFT [9], and MutationTaster [10]) were taken into consideration. All variants that were present in the homozygous state in healthy control cohorts (Exome Aggregation Consortium and Exome Variant Server) were excluded. The remaining variants were confirmed in original patient DNA by Sanger sequencing. Whenever parental DNA was available, segregation analysis was performed. Final calling of variant pathogenicity was performed by geneticists together with physician scientists, who had knowledge of the clinical phenotypes and pedigree structure.
Coverage Statistics
We achieved a median sequencing coverage of 194× per individual, and 197× per amplicon. As previously published, median coverage values >20× are sufficient to exclude false-negative results in high-throughput exon sequencing (5,6). In this study, only eight of 143 individuals (5.7%), and 37 of 518 amplicons (7.1%) had a median coverage <20×. This value is average for next-generation sequencing; hence, this experiment fulfilled the necessary quality criteria.
Web Resources
Online resources used were as follows:
UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway;
1000 Genomes Browser, http://browser.1000genomes.org;
Ensembl Genome Browser, http://www.ensembl.org;
Exome Variant Server, http://evs.gs.washington.edu/EVS;
Exome Aggregation Consortium, exac.broadinstitute.org;
OMIM, http://www.omim.org;
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2 (8);
SIFT, http://sift.jcvi.org (9); and
MutationTaster http://www.mutationtaster.org (10).
Results
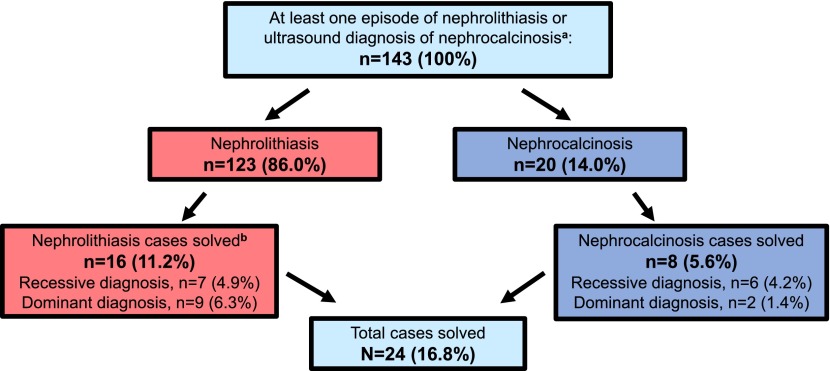
We examined an international cohort of 143 individuals with early-onset NL or NC for the presence of mutations in 30 genes that cause NL/NC if mutated. We detected mutations in 14 of these 30 genes and established a molecular diagnosis likely to explain the disease phenotype in 24 of 143 unrelated individuals with NL/NC (16.8%) (Tables 1 and 2). Pathogenic mutations were detected in nine recessive genes in 13 individuals: ATP6V1B1 (one individual), ATP6V0A4 (one individual), CLDN16 (one individual), CLDN19 (one individual), SLC3A1 (three individuals), CYP24A1 (two individuals), SLC12A1 (one individual), AGXT (one individual), and OCRL (two individuals) (Figure 1, Table 1). We also detected pathogenic mutations in five dominant genes in 11 individuals: ADCY10 (two individuals), SLC4A1 (one individual), SLC9A3R1 (one individual), SLC34A1 (five individuals), and VDR (two individuals) (Figure 1, Table 2).
Table 1.
Molecular genetic diagnoses established in 13 of 143 (9.1%) individuals from 143 families with NL/NC in one of nine recessive or X-linked genes
| Gene [Protein] (Individual with Mutation) | Nucleotide Change | Amino Acid Change | Zygosity State | PPh2, Evolutionary Conservation | Reference | Sex | Age of Onset, yr | NL/NC | Stone Analysis | Clinical Diagnosis (Before Mutational Analysis) | Genetic Diagnosis (After Mutational Analysis) | Practical Implication (Following Genetic Diagnosis)a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATP6V1B1 [ATPase, H+ transporting, lysosomal 56/58 kDa, V1 subunit B1] (B482)b | c.242T>C | p.Leu81Pro | hom | 1.00, X.t. | 14 | M | 7 | NL | CaOx | Incomplete dRTA | dRTA, deafness | Hearing screen, monitor for ↑ K+, no diagnostics for secondary causes, genetic counseling |
| ATP6V0A4 [ATPase, H+ transporting, lysosomal V0 subunit a4] (B329)c | c.292–1G>A | obligatory splice | comp het | N/A | Novel | F | <1 | NL+NC | N/A | HC, HK, dRTA, metabolic acidosis | dRTA | Monitor for ↑ K+, no diagnostics for secondary causes, genetic counseling |
| c.1346G>T | p.Arg449Leu | 1.00, X.t. | 34 | |||||||||
| CLDN16 [Claudin 16] (B604)b | c.453G>T | p.Leu151Phe | hom | 0.99, X.t. | 17 | M | 13 | NC | N/A | FHHNC | FHHNC | Monitor for tetany and seizures, no diagnostics for secondary causes, genetic counseling |
| CLDN19 [Claudin 19] (A4592)c | c.59G>A | p.Gly20Asp | hom | 0.99, D.r. | 18 | F | 15 | NL+NC | N/A | Medullary NC, HC, HM | HOMG5 | Evaluate for eye disease, monitor for NC, genetic counseling |
| SLC3A1 [Cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic, and neutral amino acid transport] | c.647C>T | p.Thr216Met | hom | 0.98, D.r. | 35 | F | 2 | NL | Cystine | Cystinuria, HO | Cystinuria | Very high fluid intake, treatment with tiopronin and potassium citrate, alkalinization of urine, limit animal protein intake, genetic counseling |
| c.647C>T | p.Thr216Met | hom | 0.98, D.r. | 35 | M | 1 | NL | Cystine | Cystinuria | Cystinuria | ||
| c.1094G>A | p.Arg365Gln | comp | 0.94, D.r. | 36 | F | 17 | NL | Cystine | Cystinuria | Cystinuria | ||
| c.1400T>C | p.Met467Thr | het | 0.29, D.r. | 24 | ||||||||
| (B425)b | ||||||||||||
| (B458)b | ||||||||||||
| (B499)c | ||||||||||||
| CYP24A1 [Cytochrome P450, family 24, subfamily A, polypeptide 1] | c.1186C>T | p.Arg396Trp | hom | 1.00, D.r. | 19 | M | 5 | NL | N/A | NL | HC/NL | Avoidance of exogenous Vitamin D and extreme sunlight, monitor for hypercalcemia with peptic ulcers and pancreatitis, genetic counseling |
| c.1147G>C | p.Glu383Gln | comp | 1.00, D.r. | Novel | F | 0.8 | NC | CaOx | Idiopathic HC | HC/NL | ||
| c.428_430del | p.Glu143del | het | N/A, D.r. | 19 | ||||||||
| (B540)b | ||||||||||||
| (B607)b | ||||||||||||
| SLC12A1 [Sodium/potassium/chloride transporter] (B446)b | c.2755G>C | p.Asp919His | hom | 1.00, G.g. | Novel | F | 0.3 | NC | N/A | BS | BS | Treatment with indomethacin and electrolyte substitution, genetic counseling |
| AGXT [Alanine-gloxylate aminotransferase] (B424)b | c.508G>A | p.Gly170Arg | hom | 1.00, D.r. | 12 | F | 0.6 | NC | CaOx | Primary hyperoxaluria | PH1 | Trial treatment with pyridoxine, ophthalmology screen, cardiac screen, monitor for oxalates and tissue calcification, genetic counseling |
| OCRL [Oculocerebrorenal syndrome of Lowe] | c.1484C>T | p.Pro495Leu | hem | 1.00, D.r. | 37 | M | 10 | NL | N/A | LS | LS/DD2 | Consult with ophthalmology, monitor for seizures, genetic counseling |
| c.2510G>A | p.Arg837His | hem | 0.23, D.r. | Novel | M | 7 | NL | CaOx | Idiopathic HC | LS/DD2 | ||
| (B422)b | ||||||||||||
| (B199)b |
PPh2, Polyphen2-HumVar (http://genetics.bwh.harvard.edu/pph2/); NL, nephrolithiasis; NC, nephrocalcinosis; H+, proton; hom, homozygous; X.t., Xenopus tropicalis; M, male; CaOx, calcium oxalate; dRTA, distal renal tubular acidosis; ↑, increased; K+, potassium; comp het, compound heterozygous; F, female; n/a, not available; HC, hypercalciuria; HK, hypokalemia; FHHNC, familial hypomagnesemia with hypercalciuria and nephrocalcinosis; D.r., Dario rerio; HM, hypomagnesemia; HOMG5, hypomagnesemia 5, renal, with ocular involvement; HO, hyperoxaluria; N/A, not available; G.g., Gallus gallus; BS, Bartter syndrome; PH1, primary hyperoxaluria, type 1; hem, hemizygous; LS, Lowe syndrome; DD2, Dent disease 2.
Practical implications are based off the defined Online Mendelian Inheritance of Man database phenotype (http://www.omim.org).
Patients derive from the Balkan region.
Patients are American.
Table 2.
Molecular genetic diagnoses established in 11 of 143 (7.7%) individuals from 143 families with NL/NC in one of five dominant genes
| Gene [Protein] (Individual with mutation) | Nucleotide Change | Amino Acid Change | Zygosity State | PPh2, Evolutionary Conservation | Reference | Sex | Age of Onset, yr | NL/NC | Stone Analysis | Clinical Diagnosis (Before Mutational Analysis) | Genetic Diagnosis (After Mutational Analysis) | Practical Implication (Following Genetic Diagnosis)a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADYC10 [Adenylate cyclase 10 (soluble)] | c.1052C>A | p.Pro351His | het | 1.00, G.g | Novel | M | 15 | NL | CaOx | HC | Absorptive HC | Genetic counseling |
| c.758G>A | p.Cys253Tyr | het | 0.69, M.m. | Novel | M | 1.5 | NL+NC | N/A | HC | Absorptive HC | ||
| (B580)b | ||||||||||||
| (B599)b | ||||||||||||
| SLC4A1 [Anion exchanger (Diego blood group)] (B280)b | c.1766G>A | p.Arg589His | het | 0.95, X.t. | 25 | F | 11 | NC | N/A | dRTA + cysts | Primary dRTA | Monitor for hereditary spherocytosis, osteomalacia, hypokalemia, and periodic paralysis, genetic counseling |
| SLC9A3R1 [NHE3, cation proton antiporter 3] (B529)c | c.328C>G | p.Leu110Val | het | 0.10, M.m. | 27 | F | 11 | NL | N/A | HPh | NPHLOP2 | Monitor bone density, genetic counseling |
| SLC34A1 [Type 2 sodium/phosphate cotransporter] | c.458G>T | p.Gly153Val | het | 1.00, D.r. | Novel | M | 4 | NL | N/A | HPh, idiopathic HC | NPHLOP1/FS | Screen for deafness, genetic counseling |
| c.398C>T | p.Ala133Val | het | 0.99, D.r. | 30 | M | 3 | NL | N/A | HPh, HC | NPHLOP1/FS | ||
| c.437C>T | p.Pro146Leu | het | 0.95, D.r. | Novel | M | 3 | NL | N/A | HPh | NPHLOP1/FS | ||
| c.1367T>A | p.Ile456Asn | het | 0.99, D.r. | Novel | F | 9 | NL | CaOx, CaPh | HSP, HC | NPHLOP1/FS | ||
| c.1348G>A | p.Gly450Ser | het | 0.99, D.r. | Novel | M | 7 | NL | CaOx | HPh, idiopathic HC | NPHLOP1/FS | ||
| (B491)c | ||||||||||||
| (B523)c | ||||||||||||
| (B484)c | ||||||||||||
| (B610)b,d | ||||||||||||
| (B417)c | ||||||||||||
| VDR [Vitamin D (1, 25-dihydroxyvitamin D3) receptor] | c.260A>G | p.Asn87Ser | het | 0.99, D.r. | Novel | M | 12 | NL | N/A | HPh | VDDR2A | Calcium supplements, genetic counseling |
| c.1207G>A | p.Glu403Lys | het | 0.90, D.r. | Novel | F | 4 | NL | N/A | HPh | VDDR2A | ||
| (B481)c | ||||||||||||
| (B447)c |
PPh2, Polyphen2-HumVar (http://genetics.bwh.harvard.edu/pph2/); NL, nephrolithiasis; NC, nephrocalcinosis; het, heterozygous; G.g., Gallus gallus; M, male; CaOx, calcium oxalate; HC, hypercalciuria; M.m., Mus musculus; N/A, not available; X.t., Xenopus tropicalis; F, female; dRTA, distal renal tubular acidosis; HPh, hypophosphatemia; NPHLOP2, hypophosphatemic nephrolithiasis/osteoporosis, 2; D.r., Danio rerio; NPHLOP1, hypophosphatemic nephrolithiasis/osteoporosis, 1; FS, Fanconi syndrome; CaPh, calcium phosphate; HSP, Henoch-Schonlein purpura; VDDR2A, Vitamin D-dependent rickets, type 2A.
Practical implications are based off the defined Online Mendelian Inheritance of Man database phenotype (http://www.omim.org).
Patients are American.
Patients derive from the Balkan region.
B610 is an adopted individual and biologic family information is unavailable.
Figure 1.
Established molecular diagnoses in 24 of 143 (16.8%) individuals with nephrolithiasis/nephrocalcinosis (NL/NC). A flow chart showing the distribution for molecular diagnoses of nephrolithiasis (NL) or nephrocalcinosis (NC), and for recessive and dominant inheritance. aBy renal ultrasound. b“Solved” denotes that two recessive or one dominant mutation(s) were/was detected that explain the disease phenotype of NL or NC.
No pathogenic mutations were detected in genes APRT, CA2, CASR, CLCN5, CLCNKB, FAM20A, GRHPR, HNF4A, HOGA1, HPRT1, KCNJ1, SLC2A9, SLC22A12, SLC34A3, SLC7A9, and XDH. Of the 27 detected mutations, 12 (44.4%) were novel pathogenic variants that have not been previously reported in databases of human disease-causing mutations. In this study, no additional functional studies were performed to validate the pathogenicity of these alleles.
To determine a possible correlation between sex and monogenic causes of disease, we analyzed the sex of the molecularly solved individuals normalized to that of the cohort. The cohort consisted of 72 males and 71 females. Among individuals with pathogenic mutations, 13 were male and 11 were female (Tables 1 and 2), resulting in no significant difference in the detection of pathogenic mutations between sexes.
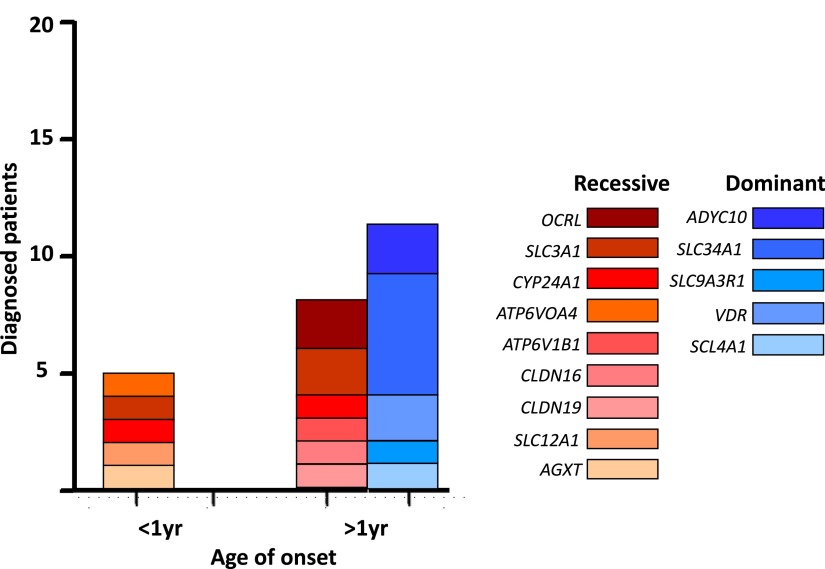
We observed that in the infantile subgroup of five individuals <1 year of age when the disease first manifested, all individuals (five of five) had pathogenic mutations in a recessive gene (Figure 2), including ATP6V0A4, SLC3A1, CYP24A1, SLC12A1, and AGXT. No individual in this age group had a pathogenic mutation in a dominant gene (Figure 2). Among the subgroup of 19 individuals >1 year, eight of 19 (42.1%) individuals had a pathogenic mutation in a recessive gene (Figure 2), including ATP6V1B1, CLDN16, CLDN19, SLC3A1, CYP24A1, or OCRL. The remaining 11 of 19 (57.9%) individuals had a pathogenic mutation in a dominant gene, including ADYC10, SLC4A1, SLC9A3R1, SLC34A1, or VDR (Figure 2). These results show that in our cohort recessive causes of disease were more frequent in infantile-onset NL/NC, whereas dominant causes were more frequent thereafter.
Figure 2.
Distribution of established molecular genetic causes of nephrolithiasis/nephrocalcinosis (NL/NC). Number of individuals with molecular diagnoses grouped by age of onset, and genetic heritability (recessive versus dominant). Note that, in the infantile subgroup, recessive diagnoses are more frequent, whereas in patients >1 year at onset of disease dominant diagnoses are more frequent.
Discussion
Pediatric-onset NL and NC are often overlooked clinical conditions that frustrate both clinicians and families. In recent years, multiple monogenic causes of NL/NC have been identified (11–28), but the prevalence of mutations in each monogenic gene in a pediatric NL/NC cohort has not yet been extensively studied.
Here, we performed mutational analysis in a cohort of 143 individuals with NL or NC onset before 18 years of age. We sequenced the coding regions of 30 genes known to cause monogenic NL/NC, and identified a causative mutation in 24 of 143 individuals (16.8%). This percentage confirms the findings of Halbritter et al. who identified a monogenic cause of NL/NC in 20.8% of children in a separate/nonoverlapping mixed adult and pediatric cohort (4). However, participants for both studies were recruited in specialized renal stone clinics at tertiary care centers where most children with NL/NC are seen. Therefore, the cohort may have a certain bias toward severe cases and future studies might show that the percentage of monogenic causes in isolated NL/NC is lower than observed in this study.
The monogenic causes of NL/NC reported in the literature are very heterogeneous (11–28). This heterogeneity is evident in our cohort, in which causative mutations were distributed among 14 of the 30 genes screened. This finding demonstrates that broad genetic screening, as performed in this study, is necessary to establish a molecular diagnosis in monogenic cases of early-onset NL/NC.
We analyzed the age distribution of individuals in whom we identified a causative mutation. Recessive monogenic diseases typically manifest earlier in life than dominant monogenic diseases (4) and our results reflect this finding for monogenic causes of NL/NC. As shown in Figure 2, all individuals with infantile manifestation of NL/NC in our cohort harbored causative mutations in recessive rather than dominant monogenic genes. In contrast, causative mutations in dominant monogenic genes were more frequently observed in individuals in whom the disease manifested later in life.
In recessive cases, we observed a surprisingly high number of homozygous (ten of 13) as compared with compound heterozygous (three of 13) mutations. We therefore revisited all respective cases to specifically exclude parental consanguinity. However, none of these children were knowingly born of consanguineous unions. Consequently, at this time we cannot provide a satisfying explanation for this phenomenon.
A major challenge of high-throughput mutational analysis is to differentiate between pathogenic mutations and benign variants. We distinguished between alleles that had previously been described in individuals with NL/NC, and thus were likely to be causative, and novel variants. Novel variants were only considered as likely to be disease causing if they were protein-truncating, affected highly conserved amino acid residues, were predicted to be damaging in bioinformatics prediction programs, and were not present in the homozygous state in healthy control individuals.
The gene SLC34A1 was originally reported as an autosomal-dominant disease gene (23). However, multiple groups later questioned the pathogenicity of single heterozygous alleles in SLC34A1 (29–31). At this point, the pathogenicity of single heterozygous alleles in SLC34A1, as identified in five individuals with NL/NC in this study, cannot be clarified definitely.
Heritability has been suggested to account for nearly 50% of NL/NC cases (2,3). The study of Halbritter et al. has shown that known NL/NC genes account for 11.4% of adult-onset NL/NC and 20.8% of early-onset NL/NC (4). These percentages indicate that many more NL/NC-associated genes remain to be identified. Monogenic conditions, particularly recessive conditions, are commonly not appreciated as being of genetic origin because many appear as sporadic cases, since the parents will be healthy heterozygous carriers of one mutated allele. However, with approximately one in five NL/NC patients harboring monogenic mutations, there are many indicators of inherited disease that clinicians should be aware of. Examples of such indicators are: early onset, familial prevalence, familial consanguinity, multiple or recurrent stones, and NC. An in-depth discussion of when to suspect a genetic condition in NL/NC is outside the scope of this publication, however, other groups have reviewed this in great detail (32).
A molecular genetic diagnosis has vast implications for both affected individuals and unaffected family members. As addressed in Tables 1 and 2, genetic screening of asymptomatic relatives may identify individuals carrying the same disease-causing mutation. Because molecular genetic screening in healthy relatives of individuals with monogenic disease is generally discouraged, we initially refrained from performing mutation analysis in healthy relatives. However, following identification of certain causative mutations in affected individuals, the resulting knowledge might have important prophylactic implications for currently unaffected family members. In these situations we recommend that affected individuals initiate discussion on genetic counseling and/or mutation analysis for healthy relatives at risk (Tables 1 and 2). Ultimately, information resulting from mutation analysis will guide clinicians to monitor individuals for development of disease and to institute preventative treatment when possible.
Furthermore, consensus guidelines recommend standard treatment for NL/NC, such as increased fluid intake, limited sodium intake, treatment with thiazide diuretics, and potassium citrate therapy (33), that may not directly address the pathophysiology of a particular molecular diagnosis. For example, although standard measures may address NC associated with CLDN16 mutations, with a definite molecular diagnosis clinicians will also know to monitor for tetany and seizures, which have been reported for certain CLDN16 mutations (Table 1). Therefore, we suggest “Practical Implications” (Tables 1 and 2) for each gene in which we detected a likely disease-causing mutation.
In conclusion, we have shown that mutations in a heterogeneous array of genes can be identified in 16.8% of individuals with early-onset NL/NC, and we suggest that specific genetic diagnoses in such cases hold vast potential for personalization of treatment plans. Therefore, genetic screening should be implemented in the clinical practice of pediatric patients with NL or NC, because knowledge of the molecular diagnosis may change the approach to prophylaxis and treatment.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank the physicians and the participating families for their contribution. F.H. is an Investigator of the Howard Hughes Medical Institute, a Doris Duke Distinguished Clinical Scientist, and the Warren E. Grupe Professor of Pediatrics.
This research was supported by grants from the National Institutes of Health (DK1069274, DK1068306, and DK064614 to F.H.), and by the March of Dimes Foundation (6-FY11-241 to F.H.). H.Y.G. was supported by the Basic Science Research Program through the National Research Foundation of Korea by the Ministry of Education (2015R1D1A1A01056685).
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
This article contains supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.07540715/-/DCSupplemental.
References
- 1.Dwyer ME, Krambeck AE, Bergstralh EJ, Milliner DS, Lieske JC, Rule AD: Temporal trends in incidence of kidney stones among children: a 25-year population based study. J Urol 188: 247–252, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter DJ, Lange M, Snieder H, MacGregor AJ, Swaminathan R, Thakker RV, Spector TD: Genetic contribution to renal function and electrolyte balance: a twin study. Clin Sci (Lond) 103: 259–265, 2002 [DOI] [PubMed] [Google Scholar]
- 3.Goldfarb DS, Fischer ME, Keich Y, Goldberg J: A twin study of genetic and dietary influences on nephrolithiasis: a report from the Vietnam Era Twin (VET) Registry. Kidney Int 67: 1053–1061, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Halbritter J, Baum M, Hynes AM, Rice SJ, Thwaites DT, Gucev ZS, Fisher B, Spaneas L, Porath JD, Braun DA, Wassner AJ, Nelson CP, Tasic V, Sayer JA, Hildebrandt F: Fourteen monogenic genes account for 15% of nephrolithiasis/nephrocalcinosis. J Am Soc Nephrol 26: 543–551, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halbritter J, Diaz K, Chaki M, Porath JD, Tarrier B, Fu C, Innis JL, Allen SJ, Lyons RH, Stefanidis CJ, Omran H, Soliman NA, Otto EA: High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J Med Genet 49: 756–767, 2012 [DOI] [PubMed] [Google Scholar]
- 6.Halbritter J, Porath JD, Diaz KA, Braun DA, Kohl S, Chaki M, Allen SJ, Soliman NA, Hildebrandt F, Otto EA, GPN Study Group : Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum Genet 132: 865–884, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadowski, CE, Lovric, S, Ashraf, S, Pabst, WL, Gee, HY, Kohl, S, Engelmann, S, Vega-Warner, V, Fang, H, Halbritter, J, Somers, MJ, Tan, W, Shril, S, Fessi, I, Lifton, RP, Bockenhauer, D, El-Desoky, S, Kari, JA, Zenker, M, Kemper, MJ, Mueller, D, Fathy, HM, Soliman, NA, Hildebrandt, F: A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 26: 1279–1289, 2015. [DOI] [PMC free article] [PubMed]
- 8.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR: A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar P, Henikoff S, Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081, 2009 [DOI] [PubMed] [Google Scholar]
- 10.Schwarz JM, Cooper DN, Schuelke M, Seelow D: MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11: 361–362, 2014 [DOI] [PubMed] [Google Scholar]
- 11.Reed BY, Gitomer WL, Heller HJ, Hsu MC, Lemke M, Padalino P, Pak CY: Identification and characterization of a gene with base substitutions associated with the absorptive hypercalciuria phenotype and low spinal bone density. J Clin Endocrinol Metab 87: 1476–1485, 2002 [DOI] [PubMed] [Google Scholar]
- 12.Purdue PE, Allsop J, Isaya G, Rosenberg LE, Danpure CJ: Mistargeting of peroxisomal L-alanine:glyoxylate aminotransferase to mitochondria in primary hyperoxaluria patients depends upon activation of a cryptic mitochondrial targeting sequence by a point mutation. Proc Natl Acad Sci U S A 88: 10900–10904, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith AN, Skaug J, Choate KA, Nayir A, Bakkaloglu A, Ozen S, Hulton SA, Sanjad SA, Al-Sabban EA, Lifton RP, Scherer SW, Karet FE: Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat Genet 26: 71–75, 2000 [DOI] [PubMed] [Google Scholar]
- 14.Karet FE, Finberg KE, Nelson RD, Nayir A, Mocan H, Sanjad SA, Rodriguez-Soriano J, Santos F, Cremers CW, Di Pietro A, Hoffbrand BI, Winiarski J, Bakkaloglu A, Ozen S, Dusunsel R, Goodyer P, Hulton SA, Wu DK, Skvorak AB, Morton CC, Cunningham MJ, Jha V, Lifton RP: Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet 21: 84–90, 1999 [DOI] [PubMed] [Google Scholar]
- 15.Pearce SH, Williamson C, Kifor O, Bai M, Coulthard MG, Davies M, Lewis-Barned N, McCredie D, Powell H, Kendall-Taylor P, Brown EM, Thakker RV: A familial syndrome of hypocalcemia with hypercalciuria due to mutations in the calcium-sensing receptor. N Engl J Med 335: 1115–1122, 1996 [DOI] [PubMed] [Google Scholar]
- 16.Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, McCredie D, Milford D, Sanjad S, Lifton RP: Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 285: 103–106, 1999 [DOI] [PubMed] [Google Scholar]
- 17.Weber S, Hoffmann K, Jeck N, Saar K, Boeswald M, Kuwertz-Broeking E, Meij II, Knoers NV, Cochat P, Suláková T, Bonzel KE, Soergel M, Manz F, Schaerer K, Seyberth HW, Reis A, Konrad M: Familial hypomagnesaemia with hypercalciuria and nephrocalcinosis maps to chromosome 3q27 and is associated with mutations in the PCLN-1 gene. Eur J Hum Genet 8: 414–422, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Konrad M, Schaller A, Seelow D, Pandey AV, Waldegger S, Lesslauer A, Vitzthum H, Suzuki Y, Luk JM, Becker C, Schlingmann KP, Schmid M, Rodriguez-Soriano J, Ariceta G, Cano F, Enriquez R, Juppner H, Bakkaloglu SA, Hediger MA, Gallati S, Neuhauss SC, Nurnberg P, Weber S: Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet 79: 949–957, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlingmann KP, Kaufmann M, Weber S, Irwin A, Goos C, John U, Misselwitz J, Klaus G, Kuwertz-Bröking E, Fehrenbach H, Wingen AM, Güran T, Hoenderop JG, Bindels RJ, Prosser DE, Jones G, Konrad M: Mutations in CYP24A1 and idiopathic infantile hypercalcemia. N Engl J Med 365: 410–421, 2011 [DOI] [PubMed] [Google Scholar]
- 20.Reilly DS, Lewis RA, Ledbetter DH, Nussbaum RL: Tightly linked flanking markers for the Lowe oculocerebrorenal syndrome, with application to carrier assessment. Am J Hum Genet 42: 748–755, 1988 [PMC free article] [PubMed] [Google Scholar]
- 21.Simon DB, Karet FE, Hamdan JM, DiPietro A, Sanjad SA, Lifton RP: Bartter’s syndrome, hypokalaemic alkalosis with hypercalciuria, is caused by mutations in the Na-K-2Cl cotransporter NKCC2. Nat Genet 13: 183–188, 1996 [DOI] [PubMed] [Google Scholar]
- 22.Enomoto A, Kimura H, Chairoungdua A, Shigeta Y, Jutabha P, Cha SH, Hosoyamada M, Takeda M, Sekine T, Igarashi T, Matsuo H, Kikuchi Y, Oda T, Ichida K, Hosoya T, Shimokata K, Niwa T, Kanai Y, Endou H: Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature 417: 447–452, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Prié D, Huart V, Bakouh N, Planelles G, Dellis O, Gérard B, Hulin P, Benqué-Blanchet F, Silve C, Grandchamp B, Friedlander G: Nephrolithiasis and osteoporosis associated with hypophosphatemia caused by mutations in the type 2a sodium-phosphate cotransporter. N Engl J Med 347: 983–991, 2002 [DOI] [PubMed] [Google Scholar]
- 24.Calonge MJ, Gasparini P, Chillarón J, Chillón M, Gallucci M, Rousaud F, Zelante L, Testar X, Dallapiccola B, Di Silverio F, Barceló P, Estivill X, Zorzano A, Nunes V, Palacín M: Cystinuria caused by mutations in rBAT, a gene involved in the transport of cystine. Nat Genet 6: 420–425, 1994 [DOI] [PubMed] [Google Scholar]
- 25.Bruce LJ, Cope DL, Jones GK, Schofield AE, Burley M, Povey S, Unwin RJ, Wrong O, Tanner MJ: Familial distal renal tubular acidosis is associated with mutations in the red cell anion exchanger (Band 3, AE1) gene. J Clin Invest 100: 1693–1707, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feliubadaló L, Font M, Purroy J, Rousaud F, Estivill X, Nunes V, Golomb E, Centola M, Aksentijevich I, Kreiss Y, Goldman B, Pras M, Kastner DL, Pras E, Gasparini P, Bisceglia L, Beccia E, Gallucci M, de Sanctis L, Ponzone A, Rizzoni GF, Zelante L, Bassi MT, George AL, Jr, Manzoni M, De Grandi A, Riboni M, Endsley JK, Ballabio A, Borsani G, Reig N, Fernández E, Estévez R, Pineda M, Torrents D, Camps M, Lloberas J, Zorzano A, Palacín M, International Cystinuria Consortium : Non-type I cystinuria caused by mutations in SLC7A9, encoding a subunit (bo,+AT) of rBAT. Nat Genet 23: 52–57, 1999 [DOI] [PubMed] [Google Scholar]
- 27.Karim Z, Gérard B, Bakouh N, Alili R, Leroy C, Beck L, Silve C, Planelles G, Urena-Torres P, Grandchamp B, Friedlander G, Prié D: NHERF1 mutations and responsiveness of renal parathyroid hormone. N Engl J Med 359: 1128–1135, 2008 [DOI] [PubMed] [Google Scholar]
- 28.Scott P, Ouimet D, Valiquette L, Guay G, Proulx Y, Trouvé ML, Gagnon B, Bonnardeaux A: Suggestive evidence for a susceptibility gene near the vitamin D receptor locus in idiopathic calcium stone formation. J Am Soc Nephrol 10: 1007–1013, 1999 [DOI] [PubMed] [Google Scholar]
- 29.Wagner CA, Rubio-Aliaga I, Biber J, Hernando N: Genetic diseases of renal phosphate handling. Nephrol Dial Transplant 29[Suppl 4]: iv45–iv54, 2014 [DOI] [PubMed] [Google Scholar]
- 30.Lapointe JY, Tessier J, Paquette Y, Wallendorff B, Coady MJ, Pichette V, Bonnardeaux A: NPT2a gene variation in calcium nephrolithiasis with renal phosphate leak. Kidney Int 69: 2261–2267, 2006 [DOI] [PubMed] [Google Scholar]
- 31.Magen D, Berger L, Coady MJ, Ilivitzki A, Militianu D, Tieder M, Selig S, Lapointe JY, Zelikovic I, Skorecki K: A loss-of-function mutation in NaPi-IIa and renal Fanconi’s syndrome. N Engl J Med 362: 1102–1109, 2010 [DOI] [PubMed] [Google Scholar]
- 32.Ferraro PM, D’Addessi A, Gambaro G: When to suspect a genetic disorder in a patient with renal stones, and why. Nephrol Dial Transplant 28: 811–820, 2013 [DOI] [PubMed] [Google Scholar]
- 33.Pearle MS, Goldfarb DS, Assimos DG, Curhan G, Denu-Ciocca CJ, Matlaga BR, Monga M, Penniston KL, Preminger GM, Turk TM, White JR, American Urological Association : Medical management of kidney stones: AUA guideline. J Urol 192: 316–324, 2014 [DOI] [PubMed] [Google Scholar]
- 34.Stover EH, Borthwick KJ, Bavalia C, Eady N, Fritz DM, Rungroj N, Giersch AB, Morton CC, Axon PR, Akil I, Al-Sabban EA, Baguley DM, Bianca S, Bakkaloglu A, Bircan Z, Chauveau D, Clermont MJ, Guala A, Hulton SA, Kroes H, Li Volti G, Mir S, Mocan H, Nayir A, Ozen S, Rodriguez Soriano J, Sanjad SA, Tasic V, Taylor CM, Topaloglu R, Smith AN, Karet FE: Novel ATP6V1B1 and ATP6V0A4 mutations in autosomal recessive distal renal tubular acidosis with new evidence for hearing loss. J Med Genet 39: 796–803, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bisceglia L, Calonge MJ, Dello Strologo L, Rizzoni G, de Sanctis L, Gallucci M, Beccia E, Testar X, Zorzano A, Estivill X, Zelante L, Palacin M, Gasparini P, Nunes V: Molecular analysis of the cystinuria disease gene: identification of four new mutations, one large deletion, and one polymorphism. Hum Genet 98: 447–451, 1996 [DOI] [PubMed] [Google Scholar]
- 36.Schmidt C, Vester U, Hesse A, Lahme S, Lang F, Zerres K, Eggermann T, Arbeitsgemeinschaft Pädiatrische Nephrologie : The population-specific distribution and frequencies of genomic variants in the SLC3A1 and SLC7A9 genes and their application in molecular genetic testing of cystinuria. Urol Res 32: 75–78, 2004 [DOI] [PubMed] [Google Scholar]
- 37.Tasic V, Lozanovski VJ, Korneti P, Ristoska-Bojkovska N, Sabolic-Avramovska V, Gucev Z, Ludwig M: Clinical and laboratory features of Macedonian children with OCRL mutations. Pediatr Nephrol 26: 557–562, 2011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.