Abstract
The human BCL6 gene, which is involved in the pathogenesis of certain human lymphomas, encodes a transcriptional repressor that is needed for germinal center B cell development and T follicular helper cell differentiation. Our goal was to identify BCL6 target genes using a cell system in which BCL6 repressive effects are inhibited followed by subtractive hybridization, and we detected the RUVBL1 (Pontin, TIP49) gene as a potential target of BCL6 repression. Here we show that the BCL6 protein significantly represses RUVBL1 transcription (6.8-fold). Knockdown of endogenous BCL6 in a human B cell lymphoma line leads to significant upregulation of RUVBL1, and there is an inverse expression pattern between the BCL6 and RUVBL1 proteins in certain human lymphomas. RUVBL1 is part of the AAA+ superfamily and participates in multiple processes, including gene transcription regulation, chromatin remodeling, and DNA repair, which, if dysregulated, may promote lymphoma development. A further understanding of the relationship between RUVBL1 and BCL6 should improve our understanding of the pathogenesis of human lymphomas.
Keywords: RUVBL1, Pontin, TIP49, NMP238, BCL6 target, Lymphomagenesis
Highlights
-
•
BCL6, a transcriptional repressor, is deregulated in human lymphomas.
-
•
The RUVBL1 (Pontin, TIP49) gene is a target of BCL6 repression.
-
•
Regulation of RUVBL1 by BCL6 may be important in lymphomagenesis.
1. Introduction
The BCL6 gene on chromosome 3, band q27 accompanies the development of certain human lymphomas and encodes a nuclear BTB/POZ zinc finger protein – a transcriptional repressor that is important in the formation of germinal center B cells and T follicular helper cells [1], [2], [3], [4]. BCL6 regulates gene expression via interactions with corepressors that recruit histone deacetylases and induce epigenetic remodeling and heterochromatin formation [5], [6]. To identify BCL6 target genes, we developed a dominant-negative cell system in which BCL6 repressive effects are inhibited, permitting the detection of genes that are ordinarily repressed. We used subtractive hybridization to detect up-regulated messages [7] and describe here the identification of the RUVBL1 gene (also called Pontin 52, Pontin, TIP49, NMP238) as a potential novel target of BCL6 repression.
The RUVBL1 protein, expressed virtually ubiquitously, is evolutionarily conserved and localizes to the cell nucleus and the cytoplasm [8]. It is an ATPase that is part of the AAA+(ATPases associated with diverse cellular activities) superfamily, which encompasses a large group of ring-shaped complexes involved in diverse cellular processes [9], including gene transcription regulation (although RUVBL1 is not a transcription factor), chromatin remodeling, sensing of DNA damage and repair, and the assembly of protein and ribonucleoprotein complexes [10], any of which, if perturbed, could potentially lead to lymphoma development. RUVBL1 is overexpressed in many cancers [11] and its role in carcinogenesis is under investigation. For example, in a hepatocyte-conditional RUVBL1+/− mouse model and Diethylnitrosamine cancer induction protocol, tumor size was significantly larger in RUVBL1+/− (as compared with RUVBL1+/+) mice after 9 and 12 months of cancer progression, indicating the potential risks of prolonged RUVBL1 inhibition [10].
2. Materials and methods
2.1. Identification of BCL6 repression targets by subtractive hybridization and validation by Northern blotting
As described [7], we converted the BCL6 zinc fingers (BCL6ZF), which lack repressive effects but bind DNA, into a transcriptional activator and used this construct to compete with wild-type BCL6 in BJAB cells (an Epstein-Barr virus-negative Burkitt lymphoma cell line expressing high levels of BCL6) [12]. To convert the BCL6ZF into a transcriptional activator, we fused nucleotides 1418–2181 of the BCL6 cDNA, which comprise the ZF domain but not the upstream repressor domains, to the VP16 activating domain of herpes simplex virus that had been hemagglutinin-tagged and subcloned in pcDNA3 (gift of Dr. J. Leiden, then at Abbott Laboratories, North Chicago, IL). The cells were transiently co-transfected with the construct or the vector in which it was cloned by electroporation along with p-Hook-1 (Invitrogen), a vector that directs synthesis of a single-chain antibody, sFv, which is expressed on the surface of transfected cells. Transfected cells were separated from the total cell population with magnetic beads (Invitrogen) coated with the hapten (phOx) toward which the sFv is specifically directed. Subtractive hybridization was performed; inserts of cDNA clones obtained following PCR were sequenced.
Potential BCL6 targets were 32P-labeled and hybridized to eight Northern blots which were prepared from the total RNA of BJAB cells that had been transiently cotransfected with the study construct (BCL6ZF) or the control construct (vector) and pHook-1 and selected on magnetic beads as described above. The blots were washed under stringent conditions and autoradiographed, then stripped and rehybridized with a 32P-labeled human β-actin control probe (Clontech Laboratories, Inc., Mountain View, CA), washed under stringent conditions, and reautoradiographed. Scanning densitometry was used to normalize relative band intensity to β-actin. Differential expression was compared by an unpaired t test.
2.2. Construction of RUVBL1 promoter reporter plasmids
We identified the promoter sequences of the RUVBL1 gene (Accession NM_003707), and then we compared the sequences within this region with the preferential binding site for BCL6 ([A/T]TC[C/T][A/T][A/C]GA) [13]. We found an exact match to 7 of these 8 bases in the RUVBL1 promoter region. To clone the sequences surrounding this site, 0.705 kb of human genomic DNA from the RUVBL1 promoter region was amplified by PCR (forward primer, 5′-CGGTGTGAGACTGTGGAACTAC-3′; reverse primer, 5′-CCATCCTCCACCGAGAGATAA-3′). The PCR product was a single band of the correct size on a 0.9% agarose gel; 1 μl of the PCR product was blunted and ligated to 50 ng of the pJET 1.2/Blunt Cloning Vector (Thermo Scientific, Waltham, MA) per the manufacturer's instructions. The appropriate insert was isolated from this vector with BglII restriction enzyme digests and ligated to the BglII site of the pGL3 basic luciferase reporter vector (Promega, Madison, WI) (pGL3RUVBL1). Multiple restriction enzyme digests confirmed the correct size and proper orientation of the insert in the vector.
2.3. Transient transfections/functional analysis
We used 293 T (human embryonic kidney) cells which are readily transfectable and have minimal expression of endogenous BCL6 [14] as well as RUVBL1 [www.abnova.com, Western blotting with RUVBL1 monoclonal antibody, H00008607- M01]. They were grown under standard conditions [15], plated in a six-well dish, transfected at 50–60% confluence by ViaFect™ Transfection Reagent (Promega) per the manufacturer's instructions, and harvested at 50 h. The constructs transfected included the BCL6 consensus binding site in the promoter region of RUVBL1 (pGL3RUVBL1 described above, 0.6 µg) and full-length BCL6 cDNA subcloned in the pCGN expression vector (1.25 µg) or an equivalent amount of a truncated BCL6 expression construct (control) subcloned in pCGN. We used this construct in previous experiments as a control [16] because it lacks the BCL6 zinc finger DNA-binding region and therefore cannot bind DNA. A CMV-driven β-galactosidase construct was cotransfected. Luciferase levels were normalized for transfection efficiency by using the values of β-galactosidase assays as described [16]. Relative luciferase activity was defined as the luciferase levels obtained with the BCL6 construct divided by the luciferase levels obtained with the control. An unpaired t test was used to determine whether the mean relative luciferase activity of the RUVBL1 consensus binding site from three independent experiments (triplicate wells in each) was significantly different from 1 in cells transfected with BCL6 vs. those transfected with the control.
2.4. BCL6 siRNA transfections
We used electroporation to transfect BJAB cells with human BCL6 siGENOME SMARTpool reagent or CONTROL nontargeting siRNA 1 (Dharmacon, LaFayette, CO) as previously reported [16]. Whole cell extracts prepared from the transfected cells were boiled and sheared in a reducing buffer (10% glycerol, 2 g% SDS, 1% 1 m Tris–HCl [pH 6.8], 5% 2-mercaptoethanol, and 0.005 gm% bromophenol blue) and used to prepare nine Western blots as described [16]. The antibodies employed were rabbit polyclonal antibodies to BCL6 (sc-858 or sc-368, Santa Cruz Biotechnology, Santa Cruz, CA), a validated affinity-isolated Prestige antibody to RUVBL1 produced in rabbit (Sigma-Aldrich Co. LLC, Saint Louis, MO, #HPA019947), and affinity-isolated actin antibody produced in rabbit (#A2066, Sigma-Aldrich). Membranes were washed, incubated with anti-rabbit IgG (Fc), alkaline phosphatase conjugate (Promega, Madison, WI), and washed again. Western Blue Stabilized Substrate for Alkaline Phosphatase (Promega) was used to detect protein bands. Beta-actin was used to ascertain the amount of protein loaded, and relative band intensity was normalized to the intensity of β-actin expression by scanning densitometry. BCL6 and RUVBL1 protein levels in the BCL6 siRNA-transfected cells were compared with the control nontargeting siRNA-transfected cells by an unpaired t test.
2.5. ChIP assays
Two ChIP assays were performed with an EZ-ChIP™ Chromatin Immunoprecipitation Kit (#17-371, EMD Millipore Corp, Billerica, MA) as described previously [17]. Briefly, BJAB cells, 4.7×106 per sample, were cross-linked with 1% formaldehyde for 5 min at room temperature. Cross-linked chromatin was sonicated to ∼200–1000 bp with a Microson™ XL 2000 sonicator (Qsonica, LLC, Newtown, CT), then immunoprecipitated with antibodies to BCL6 (sc-858 X, Santa Cruz Biotechnology, Dallas, TX) and anti-rabbit IgG as a negative control. ChIP DNA was amplified by PCR. One primer set (see Section 2.2) amplified the BCL6 consensus site in the promoter of the RUVBL1 gene, and another primer set amplified a coding region of RUVBL1 which did not contain putative BCL6 binding sites. These primers amplified 162 bp within a 199 bp exon that is present in all coding transcripts of RUVBL1 (forward primer, 5′-ACAAACTTCGAGGGGAGATT-3′; reverse primer, 5′-CGATGGGAGCGATAGAAGAC-3′). Bands were detected on 0.8% and 1% agarose gels, respectively.
2.6. Immunohistochemistry
Under an Institutional Review Board-approved protocol we obtained human paraffin-embedded tissues from the Surgical Pathology archives. Sections from the same block (not necessarily consecutive sections) were stained with various antibodies. BCL6 staining was performed as previously described [16], [17], [18] with mouse monoclonal anti-human BCL6 (#M7211, DAKO or clone LN22, Novocastra). After antigen retrieval for 40 min in a 98° C waterbath with pH 6 antigen retrieval buffer (DAKO), staining for RUVBL1 was carried out overnight with an affinity purified Prestige antibody produced in rabbit (Sigma-Aldrich, St. Louis, MO, #HPA019947) that was diluted 1:265 to 1:350. Antigen-antibody binding was detected with DAB chromogen. Tissues were counterstained with hematoxylin. Appropriate positive and negative controls were used, including a negative isotype-matched control.
3. Results
3.1. Subtractive hybridization of BCL6 constructs identifies RUVBL1 as a target of BCL6 repression in BJAB cells
After subtractive hybridization was performed, PCR products of the experimental subtracted sample showed differentially expressed bands as compared with the unsubtracted control on a 2% agarose gel. The finding that sequences from two clones were identical to overlapping regions of BCL6ZF (indicating identification of BCL6ZF mRNA that had been overexpressed in the study cells) implied that subtractive hybridization was successful. The sequences from another clone matched the RUVBL1 gene (GenBank ID NM_003707).
3.2. Differential expression of RUVBL1 confirmed by Northern blotting
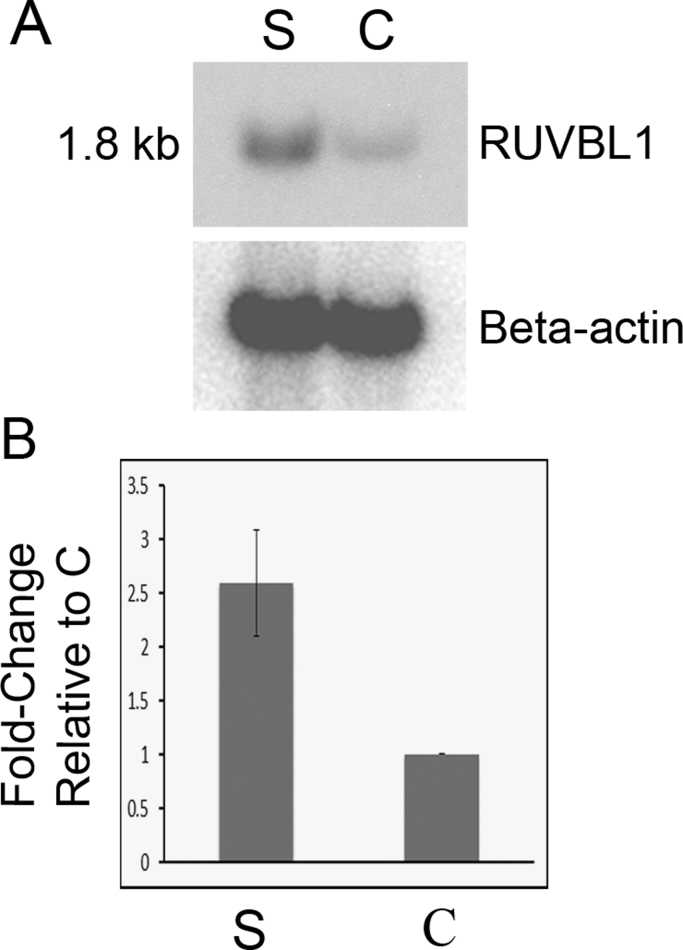
A representative Northern blot hybridized with the cDNA fragment of RUVBL1 obtained from cDNA subtraction and amplification is depicted in Fig. 1A. The expected 1.8-kb transcript [8] was identified. Relative band intensity normalized to β-actin by scanning densitometry showed that differential expression of RUVBL1 (S lane) as compared with the vector control (C) was, by unpaired t test, mean±SEM, 2.59±0.50 (range, 1.18–5.72)-fold, p<0.006 (Fig. 1B).
Fig. 1.
Northern blotting confirms differential expression of RUVBL1. (A) Representative autoradiograph of a Northern blot made from total RNA of BJAB cells (human B cell lymphoma line) transiently transfected with an expression construct (S, study) containing the BCL6 fingers or the vector (control, C) in which the S construct was cloned. Top: Hybridization with a cDNA fragment of RUVBL1 obtained from subtractive hybridization shows its 1.8 kb transcript. Bottom: The blot was stripped and hybridized with a human β-actin probe. (B) Densitometry data showing differential expression of RUVBL1 normalized to β-actin. For eight Northern blots, differential expression of RUVBL1 (S lane) as compared with the control (C) is significant by unpaired t test; mean±SEM, 2.593±0.497 (range 1.18–5.72)-fold (p<0.006). The data were normalized with results for the control designated as 1.
3.3. The BCL6 protein represses transcription from the BCL6 consensus site in the RUVBL1 promoter
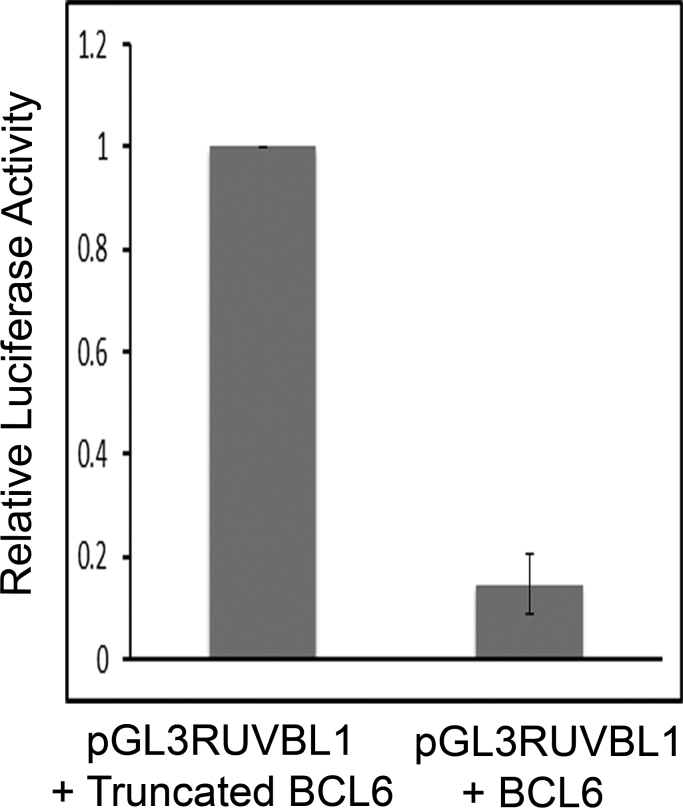
Transient transfections in 293 T cells showed that relative luciferase activity was significantly repressed (6.8-fold, p<0.0001) from the BCL6 consensus binding site in the promoter of RUVBL1 by the full-length BCL6 protein (mean±SEM=0.15±0.06) as compared with a truncated construct (control) that cannot bind DNA (Fig. 2; for comparison, the absolute value of the control was designated 1). The repression is presumed to be from the BCL6 consensus sequence in the RUVBL1 promoter, but we cannot exclude the possibility that the transfected BCL6 protein might be driving an intermediary transcription factor acting on another site in the RUVBL1 promoter.
Fig. 2.
The BCL6 protein represses transcription from the RUVBL1 promoter (transfection assays). Relative luciferase levels in 293 T cells show that RUVBL1 promoter activity is significantly repressed (6.8-fold, p<0.0001) by the full-length BCL6 protein (mean±SEM=0.146±0.059) as compared with a truncated control that lacks the BCL6 zinc fingers and thus cannot bind DNA. The data were normalized with results for the control set at 1.
3.4. Knockdown of the BCL6 protein increases endogenous RUVBL1 expression
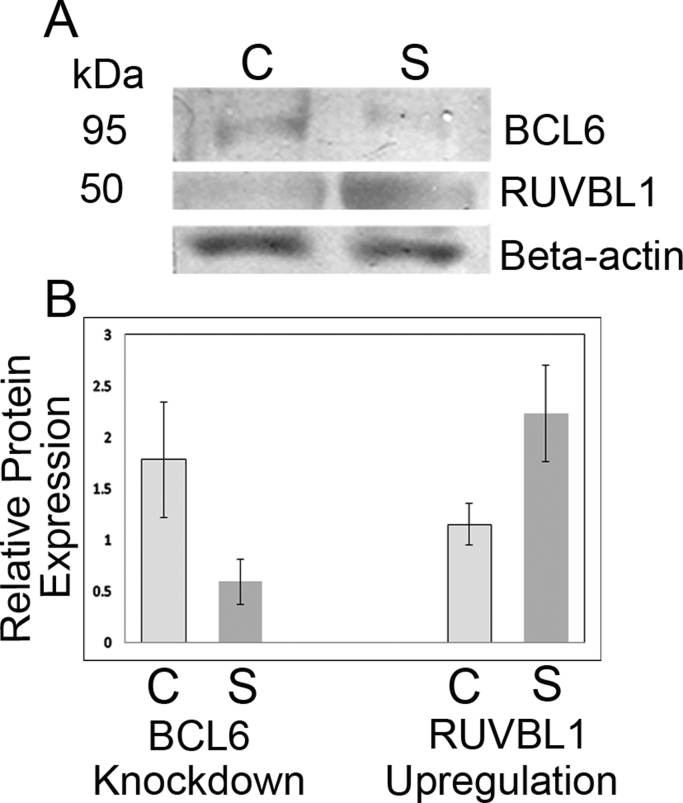
Analysis of the Western blots showed that knockdown of BCL6 protein levels and upregulation of RUVBL1 (∼50 kDa) occurred as early as 18 h after transfection of siRNA duplexes targeting endogenous BCL6; these effects lasted until at least 47 h (Fig. 3A). Knockdown of the BCL6 protein was 3-fold in the BCL6 siRNA-transfected cells as compared with controls (p<0.06); upregulation of RUVBL1 in the study-transfected cells as compared with controls was 2-fold (p<0.05), Fig. 3B. These data indicate that upregulation of endogenous RUVBL1 expression is an effect of BCL6 knockdown.
Fig. 3.
Knockdown of BCL6 protein levels by siRNA increases RUVBL1 protein expression. (A) Representative Western blot depicting BCL6 (95 kDa), RUVBL1 (50 kDa), and β-actin protein expression in BJAB cells 45 h after transfection with siRNA duplexes targeting BCL6 [C, control siRNA; S, study (BCL6) siRNA]. All bands are from the same blot. (B) The graph indicates the relative protein expression of BCL6 and RUVBL1 after normalization for β-actin expression. BCL6 protein expression (left): mean±SEM for control siRNA 1.783±0.556, and for BCL6 siRNA 0.595±0.216, p=0.064. RUVBL1 protein expression (right): mean±SEM for control siRNA 1.146±0.205, and for BCL6 siRNA 2.229±0.47, p<0.05.
3.5. BCL6 binding site in the promoter region of the RUVBL1 gene
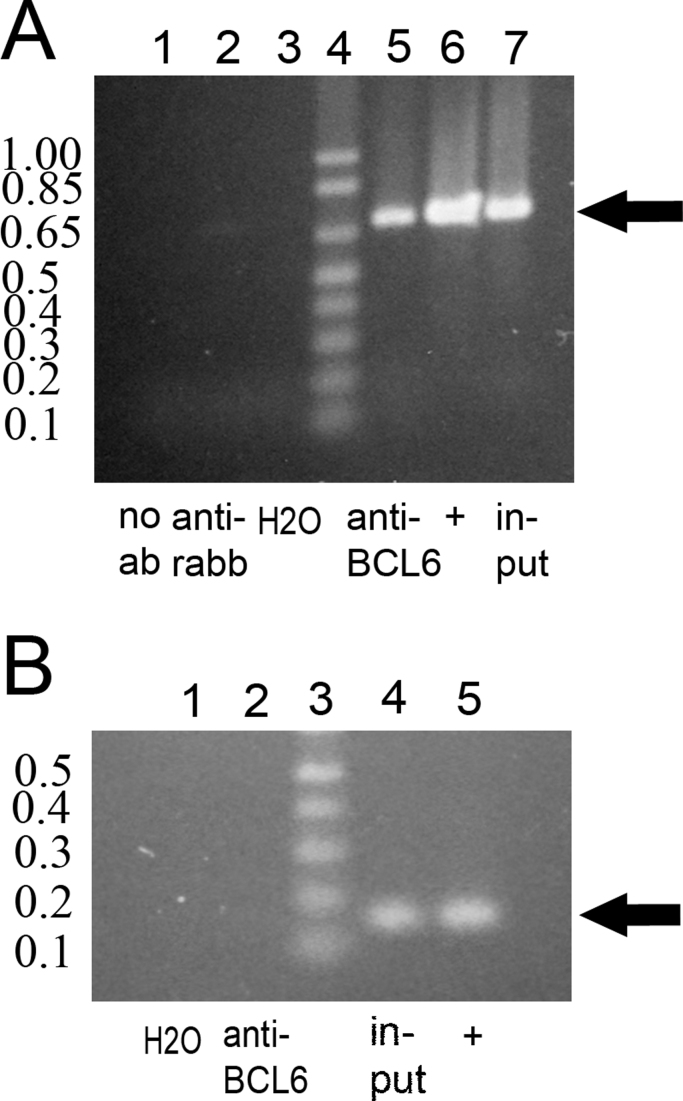
Tiling ChIP-on-chip studies showed that BCL6 preferentially localizes to the 8-base consensus binding site [A/T]TC[C/T][A/T][A/C]GA [13]. We found an exact match to 7 of these bases (TTCTAAG) in the promoter region of RUVBL1. Two ChIP assays showed that in BJAB cells endogenous BCL6 was bound, perhaps as part of a complex [19], [20], [21], to the BCL6 consensus site that we identified in the promoter region of RUVBL1 because DNA from this region was enriched in chromatin immunoprecipitated with anti-BCL6 (Fig. 4A, lane 5). Two controls (absence of antibody, Fig. 4A, lane 1) and an unrelated antibody (anti-rabbit IgG, Fig. 4A, lane 2) did not enrich DNA from this region. Bands of the correct size were noted with input (lane 7) and the positive control (genomic DNA, lane 6). Binding of BCL6 was not detected in a coding region of RUVBL1 which did not contain putative BCL6 binding sites (Fig. 4B, lane 2), whereas bands in the correct location (Fig. 4B, lanes 4 and 5) were noted with input and the positive control (genomic DNA).
Fig. 4.
ChIP assay. (A) RUVBL1 promoter region containing the high-affinity BCL6 binding site (0.705 kb). In BJAB cells BCL6 is bound (perhaps as part of a complex) to the region of the RUVBL1 promoter containing the putative BCL6 binding site (arrow) because DNA from this region is enriched in chromatin immunoprecipitated with anti-BCL6 (lane 5). Controls – no antibody (lane 1) and unrelated antibody, anti-rabbit IgG (lane 2) – do not enrich this genomic region. The positive (+) control is genomic DNA amplified with the same primers (lane 6). Lane 4 is 1 Kb Plus DNA Ladder (Invitrogen™, Life Technologies, Grand Island, NY); lane 7 is input. (B) RUVBL1 coding region (0.162 kb, arrow). Binding of BCL6 to a part of the coding region of RUVBL1 is not observed (lane 2) because there is no putative binding site. Input (lane 4) and the positive (+) control, genomic DNA amplified with the same primers (lane 5), show the anticipated 162 bp product (arrow). Lane 3 is 1 Kb Plus DNA Ladder (Invitrogen™).
3.6. Relationship between BCL6 and RUVBL1 expression in Human lymphomas and benign lymphoid tissue
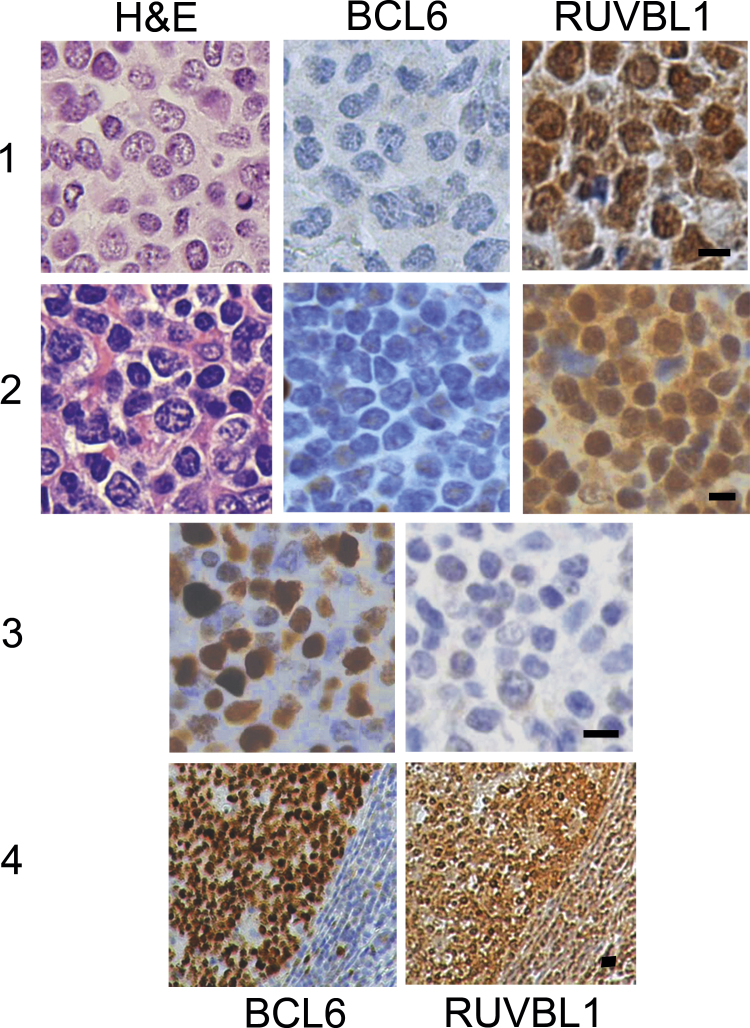
We studied 22 randomly selected human lymphomas by immunohistochemistry (9 T cell and 13 B cell malignancies, termed “T” or “B”, respectively). If 30% or fewer of the tumor cells stained with anti-BCL6, lymphomas were considered BCL6 negative [22]. Similarly, if 30% or fewer of the tumor cells stained with anti-RUVBL1, lymphomas were considered RUVBL1 negative. The data are summarized in Table 1. Briefly, 13 lymphomas were BCL6 positive (3 T, 10 B); nine were BCL6 negative (6 T, 3 B). All of the tumors had some areas that were positive for RUVBL1, and in 20 of 22 lymphomas studied, RUVBL1 expression was greater than that of BCL6. In two B cell tumors (#13 and #14, Table 1) the reverse was true. Interpretation could be difficult because of variable expression of BCL6 and RUVBL1 that sometimes occurred within the same lymphoma (e.g., tumor #18 in Table 1 and Fig. 5, panels 2 and 3, in which BCL6 staining was negative in part of the lymphoma, where RUVBL1 staining was strong – Fig. 5, panel 2, but BCL6 positive in another part of the lymphoma, where RUVBL1 staining was negative – panel 3). We called this tumor BCL6 negative overall, as most of the tissue studied was BCL6 negative.
Table 1.
BCL6 and RUVBL1 staining in lymphomas.
| Tumor | Lymphoma type/stage | RUVBL1% staining/average intensity | BCL6% staining/average intensity | % staining×average intensity |
|
|---|---|---|---|---|---|
| RUVBL1 | BCL6 | ||||
| T cell | Localized (stage 1) | ||||
| 1 | Anaplastic large cell | 80/3+ | 50/2+ | 240 | 100 |
| 2 | Anaplastic large cell | 85/3+ | 50/2+ | 255 | 100 |
| Advanced (stages 3 and 4) | |||||
| 3 | Anaplastic large cell | 80/2+ | 0 | 160 | 0 |
| 4 | Peripheral | 80/2+ | 0 | 160 | 0 |
| 5 | Large cell (immunoblastic), peripheral | 85/3+ | 10/2.5+ | 255 | 25 |
| 6 | Large cell | 80/2+ | 25/2+ | 160 | 50 |
| 7 | Delta/gamma peripheral | 90/3+ | 25/2+ | 270 | 50 |
| 8 | Peripheral | 67/3+ | 30/2+ | 201 | 60 |
| 9 | Anaplastic large cell | 90/3+ | 40/2+ | 270 | 80 |
| Avr 219 | 51.67 | ||||
| B cell | Localized (stage 1) | ||||
| 10 | High grade, EBV-related | 70/2+/mostly big cells | 40/1.5+/mostly small cells | 140 | 60 |
| 11 | DLBL, follicular origin | 50/2+ | 50/1.5+ | 100 | 75 |
| 12 | Large cell, follicular origin | 85/2.5+ | 50/3+ | 212.5 | 150 |
| 13 | DLBL | 60/2+ | 70/2+ | 120 | 140 |
| 14 | DLBL, follicular center type | 80/1.5+ | 80/3+ | 120 | 240 |
| 15 | DLBL, non-germinal center-like | 90/3+ | 85/2.5+ | 270 | 212.5 |
| Advanced (stages 3 and 4) | |||||
| 16 | Mantle cell | 95/3+ | 5/2.5+ | 285 | 12.5 |
| 17 | DLBL | 70/1.5+ | 25/1.5+ | 105 | 37.5 |
| 18 | Large cell, follicular origin | 60/2.5+ | 25/2.5+ | 150 | 62.5 |
| 19 | DLBL, CD5+ | 80/2.5+ | 40/2+ | 200 | 80 |
| 20 | DLBL with anaplastic features | 70/3+ | 50/2+ | 210 | 100 |
| 21 | Large cell, activated B cell type | 90/3+ | 50/2.5+ | 270 | 125 |
| 22 | DLBL, germinal center-like | 90/3+ | 60/2.5+ | 270 | 150 |
| Avr 188.65 | 111.15 | ||||
DLBL: Diffuse large B cell lymphoma; EBV: Epstein-Barr Virus.
Fig. 5.
Immunohistochemistry of representative randomly chosen human lymphomas stained for BCL6 and RUVBL1. The H&E stain shows effacement of normal tissue architecture by malignant cells. Panel 1: T cell tumor, BCL6 negative, RUVBL1 positive. Panel 2: B cell lymphoma, BCL6 negative area, where RUVBL1 is positive. Panel 3: Same B cell lymphoma as in panel 2, BCL6 positive area, where RUVBL1 staining is negative. Panel 4: Germinal center of human tonsil; BCL6 and RUVBL1 are both expressed within the germinal center, but many more lymphocytes outside the germinal center express RUVBL1 as compared with BCL6. The bars in the lower right photos indicate 20 µm for each image in the corresponding panel.
In Table 1 we have indicated the percent staining times the average intensity for both RUVBL1 and BCL6. For T cell tumors, a paired t test comparing this product for RUVBL1 staining with BCL6 staining was statistically significant (mean±SEM=219±16.25 for RUVBL1 and 51.67±12.69 for BCL6, p<0.0001). This also was true for B cell tumors (mean±SEM=188.65±19.35 for RUVBL1 and 111.15±18.51 for BCL6, p=0.0097). An unpaired t test showed that BCL6 staining was significantly less in the T cell lymphomas than in the B cell tumors (mean±SEM=51.67±12.69 in the T cell tumors and 111.15±18.51 in the B cell tumors, p<0.03), but there was no statistically significant difference in RUVBL1 staining between T and B cell tumors. We did not find a statistically significant increase in RUVBL1 expression in the advanced (as compared with early stage) tumors.
Expression of RUVBL1 usually was nuclear, but cytoplasmic expression also was noted. Intensity of staining is shown in Table 1 and is depicted as an average; it was generally 2+(moderate) to 3+(strong) with both the BCL6 and RUVBL1 antibodies, with the average for RUVBL1 being 2.5+ and for BCL6 2+. The intensity of RUVBL1 staining was similar in T and B cell tumors (average 2.7+ and 2.4+, respectively), but the intensity of BCL6 expression was generally less in the T cell tumors than in the B cell tumors (average 1.6+ vs. 2.2+, respectively).
Two lymphomas (one B cell, one T cell) occurred within lymph nodes that contained some residual germinal centers, where, as expected, BCL6 expression was strong (3+). RUVBL1 staining within the germinal centers was nuclear and/or cytoplasmic and weaker (1+ to 2+) than in the surrounding tumor tissue.
Sections of six tonsils from patients ranging in age from 3 to 30 years were studied with BCL6 and RUVBL1 antibodies. These tissues were from non-lymphoma patients and showed lymphoid follicular hyperplasia. As we previously reported [7], BCL6 stains the nuclei of lymphocytes in germinal centers most heavily in larger cells (centroblasts). RUVBL1 expression within germinal centers was strongest where BCL6 expression also was strong and could be nuclear or cytoplasmic. However, RUVBL1 expression also was noted in many lymphocytes in the follicular mantle (though expression in this area was variable), where BCL6 stained only scattered cells (Fig. 5, panel 4). A negative isotype-matched control showed no staining.
4. Discussion
The RUVBL1 protein, first called TIP49, was isolated from rat liver nuclear extracts by Kanemaki et al. [23]. They identified it as a 49-kDa protein that was complexed with TATA-binding protein and had high homology with the RuvB bacterial recombination factors. The following year Qiu et al. [24] identified a 50 kDa human protein in a yeast two-hybrid system (the same protein called Pontin 52 and TIP49), which they named RUVBL1. Fluorescence in situ hybridization localized the RUVBL1 gene to 3q21, a region often rearranged in leukemia and deleted in solid tumors [25]. RUVBL1 was detected in the RNA polymerase II holoenzyme complex and was essential for yeast viability [24].
Much has been written about RUVBL1. It is a transcriptional cofactor with ATPase and helicase activities [26], a binding partner of β-catenin [27], an essential nuclear cofactor for c-MYC oncogenicity [28], and an essential regulator in pre-TCR signaling during T cell development [29]. Further, it regulates β-catenin-mediated malignant transformation and T-cell factor target gene induction via functions on chromatin remodeling [30], and, as a constituent of chromatin modification complexes, may facilitate the access of repair machinery to DNA-damaged sites [31]. It has been proposed that interaction of RUVBL1/Reptin with transcription-related factors may be important for the initial localization of these factors to acetylated promoter regions [32]. Working with a leukemia model, Breig et al. [33] found that RUVBL1 is needed for cell proliferation rather than cell survival. Others have reported increased expression of RUVBL1 in advanced (as compared with early stage) diffuse large B cell lymphomas [34], but BCL6 expression was not studied. We did not observe this phenomenon in our study, but the number of tumors that we studied is small.
We previously described other targets of the BCL6 repressive effects that can induce apoptosis, e.g., the programmed cell death-2 gene and the integral membrane 2B gene [7], [15], [17]. Similarly, Jha et al. suggested that RUVBL1 is required for p53-mediated pathways of apoptosis following DNA damage [35]. However, Bereshchenko et al. [36] showed that, in the mouse, embryonic stem cells and hematopoietic tissues contain high expression of RUVBL1, which is essential for early murine embryogenesis and adult hematopoiesis. Conditional ablation of RUVBL1 in hematopoietic tissues led to bone marrow failure, and studies indicated that this included the loss of hematopoietic stem cells via apoptosis. They postulate that inhibition of RUVBL1 expression may inhibit tumor growth and induce apoptosis in cancer cells. Finally, Taniue et al. [37] found that RUVBL1 binds to the p53 promoter, repressing its transcription, and blocks p53-mediated apoptosis in a human colon cancer cell line. As with BCL6, whether apoptosis is induced or inhibited by RUVBL1 may depend on its cellular context and expression level [7].
We have presented the following findings in a B lymphoma cell line which support the notion that RUVBL1 is a target of BCL6 repression: subtractive hybridization, using a dominant negative system in which the BCL6 repressive effects were inhibited, followed by Northern blotting, indicated differential expression of RUVBL1; in transient transfection assays the full-length BCL6 protein led to significant repression (6.8-fold) from the BCL6 consensus binding site in the promoter of RUVBL1 as compared with a truncated control that cannot bind DNA (although, as mentioned, we cannot exclude the possibility that the transfected BCL6 protein may be driving an intermediary transcription factor); knockdown of the BCL6 protein by siRNAs increases expression of the endogenous RUVBL1 protein; and ChIP assays showed that the BCL6 protein is likely part of a complex that is bound to the promoter region of RUVBL1 in vivo. It has been reported previously that the BCL6 protein interacts with several corepressors, e.g., N-CoR, SMRT, mSIN3A, BCoR, and it is thought that the repressive effects of BCL6 are mediated through multiprotein repression complexes [19], [20], [21].
We did not observe an inverse relationship between BCL6 and RUVBL1 within the germinal centers of human tonsils, but expression of RUVBL1 was more prominent in follicular mantle cells than BCL6, though the degree of RUVBL1 staining in this area was variable. In human lymphomas that did not express BCL6, RUVBL1 expression was high, and in some BCL6-negative lymphomas that showed focal BCL6 expression, we could identify RUVBL1-negative areas where BCL6 expression was noted. All of the lymphomas studied expressed RUVBL1, and in both T and B cell tumors RUVBL1 expression was greater than that of BCL6 in the majority of the tumors studied (Table 1), although in two B cell tumors the reverse was true. Studies performed in the BJAB cell line and transient transfection assays indicate a repressive effect of BCL6 on its RUVBL1 target. However, in lymphoma tissues, multiple factors may modulate their respective expression. Others [38] have reported that recruitment of RUVBL1/Reptin complexes is a common phenomenon in cancer and, at least one mechanism by which this occurs is via accumulation of the transcription factor E2f1. Thus, it is possible that the high expression of RUVBL1 we observed in most of the lymphomas that we studied is a secondary phenomenon, following its recruitment by this protein or by other proteins or mechanisms that have not yet been identified.
The importance of ATPase/helicase motifs has been shown for several cofactors that function in transcription [28]. Bauer et al. [27] postulate that the role of RUVBL1 may be to unwind nuclear duplexes, a step known to be essential for transcription initiation. As BCL6 is a transcriptional repressor, its relationship with RUVBL1 is likely to be relevant at the transcriptional level, with effects on additional downstream events that are yet to be ascertained. The potential risks of prolonged RUVBL1 inhibition (e.g., secondary to the repressive effects of BCL6), which led to enhanced cancer progression in a murine model of hepatocellular carcinoma [10] (see Introduction) may be relevant also to lymphomagenesis. As in the case of Notch [39], regulation of gene expression by BCL6 is likely to encompass complex interactions involving co-activators, cooperating transcription factors, chromatin regulators, and other factors. How these are coordinated to control the effects of transcription, including the interactions between activator and repressor complexes, is not known. An understanding of the relationship between BCL6 and RUVBL1 during lymphomagenesis requires further investigation, which may provide insights for the development of new molecular tools for the targeted treatment of lymphomas.
Acknowledgments
We thank Dr J. Turner for the use of his cell culture facilities, luminometer, and sonicator, Dr L. Shen (Turner laboratory) for helpful discussions, and D. Kane (Hematopathology Laboratory) for BCL6 immunohistochemistry. This work was supported by the Department of Pathology and The University of Chicago Cancer Center Support Grant P30 CA014599 (to B.W.B.).
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.02.006.
Contributor Information
Beverly W. Baron, Email: Beverly.Baron@uchospitals.edu.
Rebecca M. Baron, Email: RBARON@PARTNERS.ORG.
Joseph M. Baron, Email: jbaron@medicine.bsd.uchicago.edu.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Baron B.W., Nucifora G., McCabe N. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc. Natl. Acad. Sci. USA. 1993;90:5262–5266. doi: 10.1073/pnas.90.11.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang C.C., Ye B.H., Chaganti R.S. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc. Natl. Acad. Sci. USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dent A.L., Shaffer A.L., Yu X. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 4.Liu X., Yan X., Zhong B. Bcl6 expression specifies the T follicular helper cell program in vivo. J. Exp. Med. 2012;209:1841–1852. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dent A.L., Vasanwala F.H., Toney L.M. Regulation of gene expression by the proto-oncogene BCL-6. Crit. Rev. Oncol. Hematol. 2002;41:1–9. doi: 10.1016/s1040-8428(01)00164-0. [DOI] [PubMed] [Google Scholar]
- 6.Green M.R., Vicente-Duenas C., Romero-Camarero I. Transient expression of BCL6 is sufficient for oncogenic function and induction of mature B-cell lymphoma. Nat. Commun. 2014;5:3904–3916. doi: 10.1038/ncomms4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baron B.W., Anastasi J., Thirman M.J. The Human programmed cell death-2 (PDCD2) gene is a target of BCL6 repression: implications for a role of BCL6 in the down- regulation of apoptosis. Proc. Natl. Acad. Sci. USA. 2002;99:2860–2865. doi: 10.1073/pnas.042702599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holzmann K., Gerner C., Korosec T. Identification and characterization of the ubiquitously occurring nuclear matrix proteIn NMP238. Biochem. Biophys. Res. Commun. 1998;252:39–45. doi: 10.1006/bbrc.1998.9604. [DOI] [PubMed] [Google Scholar]
- 9.Ammelburg M., Prickey T., Lupas A.N. Classification of AAA+ proteins. J. Struct. Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Matias P.M., Baek S.H., Bandeiras T.M. The AAA+ proteins Pontin and Reptin enter adult age: from understanding their basic biology to the identification of selective inhibitors. Front. Mol. Biosci. 2015;2:17. doi: 10.3389/fmolb.2015.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grigoletto A., Lestienne P., Rosenbaum J. The multifaceted proteins Reptin and Pontin as major players in cancer. Biochim. Biophys. Acta. 1815;2011:147–157. doi: 10.1016/j.bbcan.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Allman D., Jain A., Dent A. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. [PubMed] [Google Scholar]
- 13.Ci W., Polo J.M., Cherchietti L. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113:5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohtani M., Miyadai T. Functional analysis of fish BCL-6 and Blimp-1 in vitro: transcriptional repressors for B-cell terminal differentiation in fugu (Takifugu rubripes) Mol. Immunol. 2011;48:818–825. doi: 10.1016/j.molimm.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Baron B.W., Hyjek E., Gladstone B. PDCD2, a protein whose expression is repressed by BCL6, induces apoptosis in Human cells by activation of the caspase cascade. Blood Cells Mol. Dis. 2010;45:169–175. doi: 10.1016/j.bcmd.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Baron B.W., Zeleznik-Le N., Baron M.J. Repression of the PDCD2 gene by BCL6 and the implications for the pathogenesis of Human B and T cell lymphomas. Proc. Natl. Acad. Sci. USA. 2007;104:7449–7454. doi: 10.1073/pnas.0701770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baron B.W., Baron R.M., Baron J.M. The ITM2B (BRI2) gene is a target of BCL6 repression: implications for lymphomas and neurodegenerative diseases. Biochim. Biophys. Acta. 1852;2015:742–748. doi: 10.1016/j.bbadis.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baron B.W., Anastasi J., Hyjek E.M. PIM1 gene cooperates with Human BCL6 gene to promote the development of lymphomas. Proc. Natl. Acad. Sci. USA. 2012;109:5735–5739. doi: 10.1073/pnas.1201168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhordain P., Albagli O., Lin R.J. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhordain P., Lin R.J., Quief S. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26:4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huynh K.D., Fischle W., Verdin E., Bardwell V.J. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 22.Gualco G., Weiss L.M., Harrington W.J., Jr., Bacchi C.E. Nodal diffuse large B-cell lymphomas in children and adolescents: immunohistochemical expression patterns and C- MYC translocation in relation to clinical outcome. Am. J. Surg. Pathol. 2009;33:1815–1822. doi: 10.1097/PAS.0b013e3181bb9a18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanemaki M., Makino Y., Yoshida T. Molecular cloning of a rat 49-kDa TBP- interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem. Biophys. Res. Commun. 1997;235:64–68. doi: 10.1006/bbrc.1997.6729. [DOI] [PubMed] [Google Scholar]
- 24.Qiu X.B., Lin Y.L., Thome K.C. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J. Biol. Chem. 1998;273:27786–27793. doi: 10.1074/jbc.273.43.27786. [DOI] [PubMed] [Google Scholar]
- 25.Kashuba V.I., Gizatullin R.Z., Protopopov A.I. NotI linking/jumping clones of Human chromosome 3: mapping of the TFRC, RAB7 and HAUSP genes to regions rearranged in leukemia and deleted in solid tumors. FEBS Lett. 1997;419:181–185. doi: 10.1016/s0014-5793(97)01449-x. [DOI] [PubMed] [Google Scholar]
- 26.Makino Y., Kanemaki M., Kurokawa Y. A rat RuvB-like protein, TIP49a, is a germ-cell enriched novel DNA helicase. J. Biol. Chem. 1999;274:15329–15335. doi: 10.1074/jbc.274.22.15329. [DOI] [PubMed] [Google Scholar]
- 27.Bauer A., Huber O., Kemler R. Pontin 52, An interaction partner of β-catenin, binds to the TATA box binding protein. Proc. Natl. Acad. Sci. USA. 1998;95:14787–14792. doi: 10.1073/pnas.95.25.14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood M.A., McMahon S.B., Cole M.D. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- 29.Boo K., Baek S.H., Lee H. Pontin is required for pre-TCR signaling at the β-selection checkpoint in T cell development. Biochem. Biophys. Res. Commun. 2014;447:44–50. doi: 10.1016/j.bbrc.2014.03.092. [DOI] [PubMed] [Google Scholar]
- 30.Feng Y., Lee N., Fearon E.R. TIP49 regulates β-catenin-mediated neoplastic transformation and T-cell factor target gene induction via effects on chromatin remodeling. Cancer Res. 2003;63:8726–8734. [PubMed] [Google Scholar]
- 31.Gospodinov A., Tsaneva I., Anachkova B. RAD51 foci formation in response to DNA damage is modulated by TIP49. Int. J. Biochem. Cell Biol. 2009;41:925–933. doi: 10.1016/j.biocel.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Choi J., Heo K., An W. Cooperative action of TIP48 and TIP49 in H2A.Z exchange catalyzed by acetylation of nucleosomal H2A. Nucleic Acids Res. 2009;37:5993–6007. doi: 10.1093/nar/gkp660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breig O., Bras S., Martinez Soria S. Pontin is a critical regulator for AML1-ETO- induced leukemia. Leukemia. 2014;28:1271–1279. doi: 10.1038/leu.2013.376. [DOI] [PubMed] [Google Scholar]
- 34.Nishiu M., Yanagawa R., Nakatsuka S. Microarray analysis of gene-expression profiles in diffuse large B-cell lymphoma: identification of genes related to disease progression. Jpn. J. Cancer Res. 2002;93:894–901. doi: 10.1111/j.1349-7006.2002.tb01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jha. S., Shibata E., Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol. Cell. Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bereshchenko O., Mancini E., Luciani L. Pontin is essential for murine Hematopoietic stem cell survival. Haematologica. 2012;97:1291–1294. doi: 10.3324/haematol.2011.060251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniue K., Oda T., Hayashi T. A member of the ETS family, EHF, and the ATPase RUVBL1 inhibit p53-mediated apoptosis. EMBO Rep. 2011;12:682–689. doi: 10.1038/embor.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarangelo A., Lo N., Teng R. Recruitment of Pontin/Reptin by E2f1 amplifies E2f transcriptional response during cancer progression. Nat. Commun. 2015;(6) doi: 10.1038/ncomms10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H., Zang C., Liu X.S., Aster J.C. The role of notch receptors in transcriptional regulation. J. Cell Physiol. 2015;230:982–988. doi: 10.1002/jcp.24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material