Abstract
Cyclotides are fascinating naturally occurring micro-proteins (≈30 residues long) present in several plant families, and display various biological properties such as protease inhibitory, anti-microbial, insecticidal, cytotoxic, anti-HIV and hormone-like activities. Cyclotides share a unique head-to-tail circular knotted topology of three disulfide bridges, with one disulfide penetrating through a macrocycle formed by the two other disulfides and interconnecting peptide backbones, forming what is called a cystine knot topology. This cyclic cystine knot (CCK) framework gives the cyclotides exceptional rigidity, resistance to thermal and chemical denaturation, and enzymatic stability against degradation. Interestingly, cyclotides have been shown to be orally bioavailable, and other cyclotides have been shown to cross the cell membranes. Moreover, recent reports have also shown that engineered cyclotides can be efficiently used to target extracellular and intracellular protein-protein interactions, therefore making cyclotides ideal tools for drug development to selectively target protein-protein interactions. In this work we will review all the available methods for production of these interesting proteins using chemical or biological methods.
Keywords: intein, protein splicing, native chemical ligation, expressed protein ligation, sortase, butelase
1. Introduction
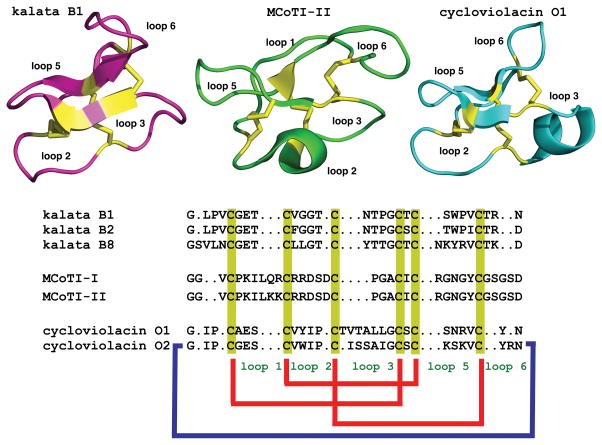
Cyclotides are fascinating micro-proteins (≈30 residues long) present in several families of plants (Poth, Colgrave, Lyons, Daly, & Craik, 2011). They share a unique head-to-tail circular knotted topology of three disulfide bridges, with one disulfide penetrating through a macrocycle formed by the two other disulfides and inter-connecting peptide backbones, forming what is called a cyclic cystine knot (CCK) topology (Fig. 1). This cyclic cystine knot (CCK) framework gives cyclotides a compact, highly rigid structure (Daly et al., 2013; Puttamadappa, Jagadish, Shekhtman, & Camarero, 2010), which confers exceptional resistance to thermal/chemical denaturation, and enzymatic degradation (Colgrave & Craik, 2004; Garcia & Camarero, 2010). In fact, the use of cyclotide-containing plants in indigenous medicine first highlighted the fact that these peptides are resistant to boiling and are apparently orally bioavailable (Saether et al., 1995).
Figure 1.
Primary and tertiary structures of cyclotides from the Möbius (kalata B1, pdb code: 1NB1), bracelet (cycloviolacin O1, pdb code: 1NBJ) and trypsin inhibitor (MCoTI-II, pdb code: 1IB9) subfamilies. The sequence of kalata B8, a novel hybrid cyclotide isolated from the plant O. affinis is also shown. Conserved cysteine residues are marked in yellow and disulfide connectivities in red. The circular backbone topology is shown with a blue line.
Despite the sequence diversity all cyclotides share the same CCK motif (Fig. 1). Hence, cyclotides can be considered as natural combinatorial peptide libraries structurally constrained by the cystine-knot scaffold and head-to-tail cyclization but in which hypermutation of essentially all residues is permitted with the exception of the strictly conserved cysteines that comprise the knot (Austin, Kimura, Woo, & Camarero, 2010; Huang, Colgrave, Clark, Kotze, & Craik, 2010; Simonsen et al., 2008).
More than 280 cyclotides have been isolated from different plant families, including the Rubiaceae, Vioalceae, Cucurbitaceae, Fabaceae and Solanaceae families (Chiche et al., 2004; Gruber et al., 2008; Nguyen et al.; Poth et al., 2011; Poth et al.; Poth et al., 2012). It has been estimated, however, that around 50,000 cyclotides might exist (Gruber et al., 2008; Zhang et al., 2014). All of the cyclotides reported so far from the Violaceae and Rubiaceae families are biosynthesized via processing from dedicated genes that in some cases encode multiple copies of the same cyclotide, and in others, mixtures of different cyclotide sequences (Dutton et al., 2004). Cyclotides from the Fabaceae family, however, are biosynthesized from evolved albumin-1 genes (Nguyen et al.; Poth et al., 2011). These precursor genes undergo post-translational processing to generate a circular peptide by a mechanism that likely involves a transpeptidation reaction mediated by asparaginyl endoproteinase (AEP)-like proteases (Gillon et al., 2008; Mylne et al., 2012; Mylne et al., 2011; Saska et al., 2007). For example, an AEP-related asparagine/aspartate (Asx) peptide ligase isolated from Clitoria ternatea was recently shown to be able to cyclize polypeptides, including cyclotides, in vitro (Nguyen et al., 2014). Cyclotides can be also produced chemically using solid-phase peptide synthesis, chemoenzymatically, or expressed in microrganisms using DNA recombinant techniques. The present review will describe the advantages and disadvantages of these approaches as well as their most recent applications (Tables 1 and 2).
Table 1.
Summary of the chemical approaches used to produce cyclotides.
| Cyclization | Chemistry | Linker | Pros and Cons | Application | Reference |
|---|---|---|---|---|---|
| NCL | Boc | Mercato- propionamide |
|
|
(Daly et al., 1999; Tam et al., 1999) |
| NCL | Fmoc | Sulfonamide |
|
|
(Ingenito et al., 1999),(Shin et al., 1999) |
| NCL | Fmoc | Ddz |
|
|
(Blanco- Canosa & Dawson, 2008; Gunasekera et al., 2013; Koehbach et al., 2013) |
| NCL | Fmoc | Trityl |
|
|
(T.L. Aboye et al., 2008) |
| Head-to- tail chemical ligation | Fmoc | 2-Chlorotrityl |
|
|
(Cheneval et al., 2014) |
| SrtA | Fmoc | 2-Chlorotrityl |
|
|
(Jia et al., 2014) |
| Trypsin | Fmoc | Wang |
|
|
(Thongyoo et al., 2008) |
| Butelase 1 | Fmoc | Wang |
|
|
(Nguyen et al., 2014)] |
Table 2.
Examples of cyclotides produced using ribosomal expression methods.
| Cyclotide | Method | In vitro/in vivo | Pros and Cons | Application | Reference |
|---|---|---|---|---|---|
| Kalata B1 | Thiol- induced N->S acyl shift | In vitro |
|
|
(Cowper et al., 2013) |
| Kalata B1 | EPL | Both |
|
|
(R. H. Kimura et al., 2006) |
| MCoTI-I | EPL | Both |
|
|
(Austin et al., 2009; Jagadish et al., 2013) |
| MCoTI-II | EPL | Both |
|
|
(J. A. Camarero et al., 2007) |
| MCo-PMI | EPL | in vitro |
|
|
(Ji et al., 2013) |
| MCoTI-I | PTS | in vivo |
|
|
(Jagadish et al., 2013) |
| rMCoTI-II | SrtA | In vitro |
|
|
(Stanger et al., 2014) |
2. Chemical synthesis of cyclotides
Cyclotides are small peptides, approximately 30 amino acids long, and therefore can be readily synthesized by chemical methods using solid-phase peptide synthesis (SPPS) (Marglin & Merrifield, 1970). Chemical synthesis using a solid-phase approach has been utilized to generate native cyclotide structures as well as grafted analogues (T. L. Aboye et al., 2012; T.L. Aboye, Clark, Craik, & Göransson, 2008; Daly, Love, Alewood, & Craik, 1999; Ji et al., 2013; Tam & Lu, 1998; Tam & Lu, 1997; Thongyoo, Tate, & Leatherbarrow, 2006). Table 1 summarizes the different synthetic approaches used thus far for the chemical synthesis of cyclotides.
2.1. Cyclization by intramolecular Native Chemical Ligation (NCL)
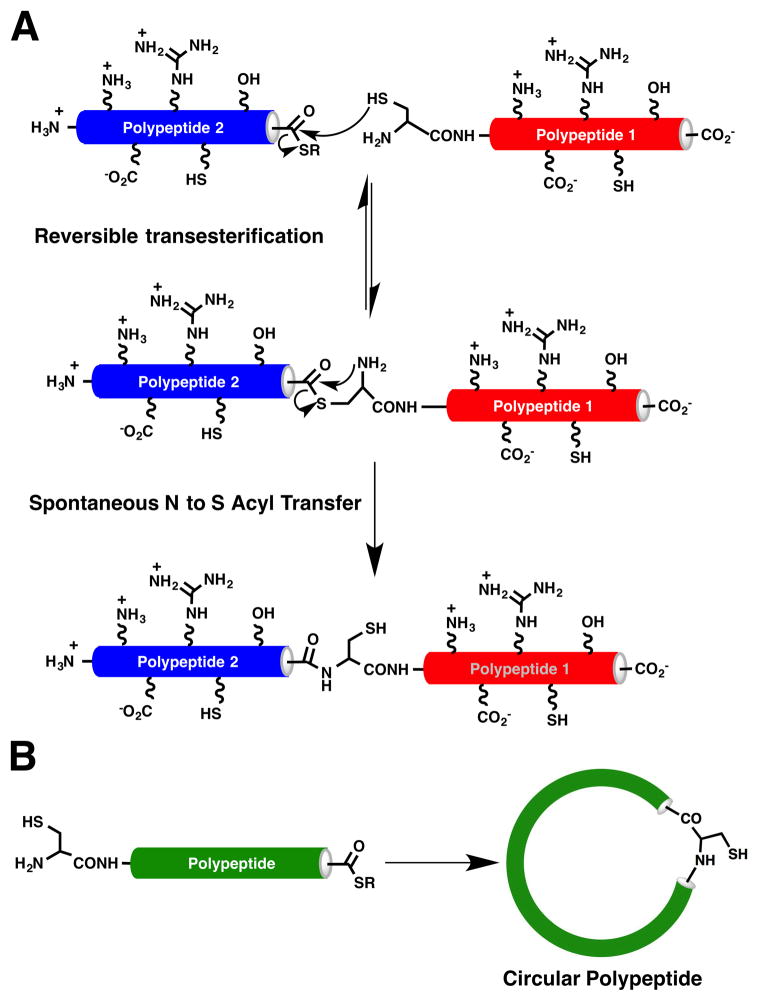
The most commonly used method for the backbone cyclization of the linear cyclotide precursor employs an intramolecular NCL (Dawson, Muir, Clark-Lewis, & Kent, 1994), in which the peptide sequence contains an N-terminal cysteine and an α-thioester group at the C-terminus (Fig. 2) (J. A. Camarero & Mitchell, 2005; J. A. Camarero & Muir, 1999; J.A. Camarero & Muir, 1997). This ligation does not compromise the chirality of the C-terminal residue at the ligation site and more importantly can be accomplished in aqueous buffers at physiological pH. Although in principle any of the six Cys residues present in the cyclotide can be used, some studies on the naturally occurring cyclotide kalata B1 and MCoTI-I/II have indicated that the Cys residues located in loop 3 and 6 usually provide better yields (Ji et al., 2013; R. H. Kimura, Tran, & Camarero, 2006). Both tert-butyloxycarbonyl (Boc)- and 9-fluorenylmethoxycarbonyl (Fmoc)-based chemistries have been used to incorporate C-terminal thioesters during peptide chain assembly (Beligere & Dawson, 1999; J. A. Camarero, Adeva, & Muir, 2000; J. A. Camarero, Cotton, Adeva, & Muir, 1998) or using safety-catch based linkers (Fmoc) (Blanco-Canosa & Dawson, 2008; J. A. Camarero, Hackel, de Yoreo, & Mitchell, 2004; J. A. Camarero & Mitchell, 2005; Ingenito, Bianchi, Fattori, & Pessi, 1999; Shin et al., 1999). Once the peptide is cleaved from the resin, the linear cyclotide precursors can be cyclized and folded sequentially. More recently, it has been shown that the cyclization and folding can be carried out in a single pot reaction by using glutathione (GSH) as a thiol additive (Contreras, Elnagar, Hamm-Alvarez, & Camarero, 2011).
Figure 2.
Backbone cyclization of polypeptides using native chemical Ligation. A. Principle of Native Chemical Ligation (NCL). B. Intramolecular NCL leads to the formation of a backbone cyclized polypeptide.
2.1.1 Synthesis of α-thioester peptides by Boc-based chemistry
The most commonly used approach for the production of α-thioester linear cyclotide precursors makes use of the 3-mercaptopropionamide linker (J.A. Camarero & Muir, 1997). This linker is stable to the conditions employed during Boc-based peptide synthesis and provides the desired α-thioester peptide in good yields. This scheme was first used by Craik and Tam for the chemical synthesis of cyclotides kalata B1, circulins A/B and cyclopsychotride to study the folding pathways and antimicrobial properties of cyclotides, respectively (Daly et al., 1999; Tam, Lu, Yang, & Chiu, 1999). Synthetic cyclotides were used to study their antimicrobial activities (Tam et al., 1999) and to analyze the folding pathways of kalata B1 (Daly et al., 1999). Despite the limitations of Boc-based peptide synthesis requiring repeated use of corrosive trifluoroacetic acid (TFA) and highly toxic anhydrous hydrogen fluoride (HF) in the final cleavage step, which make it difficult to parallelize for automated high-throughput synthesis, this approach is still widely used for the synthesis of cyclotides. For example, this approach was recently used to engineer an orally active cyclotide based on the sequence cyclotide kalata B1 for treating inflammatory pain in animal models (Tam et al., 1999). Other bioactive cyclotides engineered to target extracellular receptors have also been recently synthesized using this approach (Chan et al., 2011; Eliasen et al., 2012; Gunasekera et al., 2008).
2.1.2 Synthesis of α-thioester peptides by Fmoc-based chemistry
The limitations associated with the use of Boc-based chemistry for the production of α-thioester linear cyclotide precursors spurred the development of different approaches for the synthesis of α-thioester peptides using the milder Fmoc-based chemistry (J. A. Camarero & Mitchell, 2005). The use of thioester-based linkers, however, is seriously limited due to the poor stability of the thioester functionality to strong nucleophiles such as piperidine, which are used repeatedly for the deprotection the Nα-Fmoc group during the peptide assembly.
Trityl resin
The first attempt to synthesize peptide α-thioesters using Fmoc-based SPPS was reported by Futaki (Futaki, Sogawa, Maruyama, Asahara, & Niwa, 1997). In this approach, the corresponding peptide α-thioesters were prepared in solution using a partially protected precursor. The protected peptide precursor was assembled on a 4-chlorotrityl (4-Cl-Trt) resin using standard Fmoc-based chemistry. After cleavage from the acid-labile resin, the C-terminal carboxylic group of the partially protected peptide was thioesterified with the appropriate thiol and coupling reagent. This approach was recently used for the synthesis of the bracelet cyclotide cycloviolacin O2 (T.L. Aboye et al., 2008). In this work the α-thioester peptide obtained by Fmoc-SPPS was cyclized by NCL and oxidatively folded using redox buffer containing GSH/cysteamine and 35% dimethylsulfoxide at pH 8.5. The folding pathway was characterized and was found to include predominately fully oxidized intermediates that slowly converted to the native peptide structure. It is important to note, however, that a major drawback to this method, besides the poor solubility of protected peptides, is epimerization of C-terminal residues other than Gly during the thioesterification reaction. A recent study (von Eggelkraut-Gottanka, Klose, Beck-Sickinger, & Beyermann, 2003) has shown that epimerization strongly depends on the conditions as well as the coupling reagent used. The best results were obtained using phosphonium salts, which only gave 2% epimerization at the C-terminal amino acid.
Sulfonamide linker
Ingenito (Ingenito et al., 1999) and Shin (Shin et al., 1999) independently reported in 1999 the Fmoc-based SPPS of C-terminal thioesters using Kenner’s acyl-sulfonamide “safety catch” linker (Kenner, McDermott, & Sheppard, 1971) as modified by Backes (Backes & Ellman, 1999; Backes, Virgilio, & Ellman, 1996). This acyl-sulfonamide linker is completely stable to basic or strongly nucleophilic conditions and can be activated by treatment with either trimethylsilyl-diazomethane (TMS-CHN2) or iodoacetonitrile to provide a Nα-alkyl acylsulfonamide, which is susceptible to nucleophilic attack. Activation with TMS-CHN2 gives usually better thiolysis yields (Ingenito et al., 1999). This approach has been used for the generation of α-thioester linear cyclotide precursors. For example, Leatherbarrow used this approach to synthesize cyclotide MCoTI-II. The cyclization site in this work was chosen at the end of loop 3 (Fig. 1). Cyclotides MCoTI-I/II are powerful trypsin inhibitors which have recently been isolated from the dormant seeds of Momordica cochinchinensis, a plant member of the Cucurbitaceae family (Hernandez et al., 2000). Although MCoTI-cyclotides do not share significant sequence homology with other cyclotides beyond the presence of the three cystine bridges, structural analysis by NMR has shown that they adopt a similar backbone-cyclic cystine-knot topology (Felizmenio-Quimio, Daly, & Craik, 2001; Heitz et al., 2001). MCoTI cyclotides, however, show high sequence homology with related cystine-knot squash trypsin inhibitors (STIs) (Hernandez et al., 2000) and therefore represent interesting molecular scaffolds for drug design (Reiss et al., 2006; Werle, Kafedjiiski, Kolmar, & Bernkop-Schnurch, 2007). The same group also used this approach to generate MCoTI-based cyclotides with potent β-tryptase and human leukocyte elastase (Gray et al., 2014; Thongyoo, Bonomelli, Leatherbarrow, & Tate, 2009).
Our group has also recently used a similar approach for the chemical synthesis of several engineered MCoTI-cyclotides able to target both extracellular (CXCR4 antagonist) (T. L. Aboye et al., 2012) and intracellular (p53 agonist) (Ji et al., 2013) protein-protein interactions with low nM activities. In both cases the N-terminal region of loop 6 was used for the cyclization. The linear thioester precursors were cyclized and folded directly in a “one-pot” reaction using reduced GSH as redox agent and thiol cofactor in phosphate buffer at pH 7.2 resulting in very good yields (T. L. Aboye et al., 2012; Ji et al., 2013). We have also recently used this synthetic scheme for the rapid parallel synthesis of cyclotides using a “tea-bag” approach (T. Aboye, Kuang, Neamati, & Camarero, 2015). This work also includes an efficient purification procedure to rapidly remove non-folded or partially folded cyclotides from the cyclization-folding crude. This procedure can be easily used in parallel for the purification of individual compounds, but also and more importantly for the purification of cyclotide mixtures. This makes this approach potentially compatible with the synthesis of positional scanning libraries in order to perform efficient screening of large libraries. These protocols were successfully used for the production of a small library of bioactive cyclotides based on the cyclotide MCo-CVX-5c (WT), which is a potent CXCR4 antagonist recently developed in our group (T. L. Aboye et al., 2012).
3,4-Diaminobenzoic (Dbz) linker
This “safety catch” linker was recently developed by Dawson for the production of α-thioester peptides by Fmoc-based SPPS (Blanco-Canosa & Dawson, 2008). This linker is stable to the conditions used in Fmoc-based SPPS and can be activated at the end of the synthesis to yield the corresponding α-thioester after thiolytic cleavage followed by TFA deprotection of the side-chain protecting groups. This approach has been recently used for the chemical synthesis of cyclotides kalata B1 and B7 (Gunasekera, Aboye, Madian, El-Seedi, & Goransson, 2013; Koehbach et al., 2013).
2.2. Cyclization by intramolecular head-to-tail chemical ligation
Craik has recently described the chemical synthesis of cyclotides by Fmoc-based SPPS by direct chemical coupling of the Nα-amino and C-terminal carboxylic groups using uronium salts as coupling reagent. Due to the lack of chemoselectivity, this reaction requires to be performed with all the side-chain protecting groups. In this approach, the linear precursor is assembled on the acid-labile 2-chlorotrityl chloride (2-CTC) resin. Treatment of the peptide-resin with mild acid conditions (1% TFA) allows cleaving the peptide with all the side-chain protecting groups intact. This partially protected peptide is then cyclized in solution using uronium salts as coupling reagent and dimethylformamide (DMF) as solvent. Once the backbone cyclization has been performed the side-chain protecting groups are removed and the unprotected reduced cyclotides are folded oxidatively in aqueous buffers. This approach has been used for the synthesis of different cyclotides, including kalata B1 and its enantiomer D-kalata B1, MCoTI-II and paridigin-br-1.
One of the main advantages of this approach is its simplicity as it employs standard SPPS reagents. It also allows choosing different cyclization sites without the need to have an extra N-terminal Cys residue, required for NCL-mediated cyclization. On the other hand, protected peptides are difficult to handle and characterize and they have tendency to aggregate and precipitate. The cyclization using uronium salts can also provide unwanted reactions between the Nα-amino group and the uroniun salt leading to the capping of the amino functionality by forming a tetramethylguanidine (TMG) adduct. The use of phosphonium-based coupling reagents (e.g. PyBOP) should minimize this unwanted side-reaction. In addition, special precaution should be taken when choosing the cyclization site as racemization (epimerization) of the C-terminal residue during the cyclization can happen. This should be easily prevented by using cyclization sites such as Gly-Xaa or Pro-Xaa (where Xaa denotes any amino acid).
2.3 Cyclization by chemoenzymatic-mediated ligation
Proteases and trans-peptidases have been recently used for the production of backbone cyclized polypeptides, including cyclotides among other circular polypeptides, using linear synthetic peptides as substrates (T. L. Aboye & Camarero, 2012).
2.3.1. Trypsin-mediated backbone cyclization
The serine-protease trypsin has been shown to be able to cyclize the naturally occurring cyclic peptide sun-flower trypsin inhibitor 1 (SFTI-1) (Marx et al., 2003). SFTI-1 is a potent trypsin inhibitor with a Ki value in the nanomolar range. The enzyme trypsin was able to catalyze the formation of a peptide bond between the α-amino and α-carboxylic groups of the P1′ and P1 residues, respectively. A similar approach has also been reported for the enzymatic cyclization of MCoTI-cyclotides (Thongyoo, Roque-Rosell, Leatherbarrow, & Tate, 2008). In this work a folded linear cyclotide precursor bearing the P1 and P1′ residues at the C- and N-termini, respectively, was used as a viable substrate for trypsin-mediated cyclization, thus enabling synthesis of the cyclic backbone without the need for a C-terminal α-thioester. The linear peptide was produced by standard Fmoc-based SPPS and was folded in 1 mM GSH, 100 mM NH4HCO3 buffer at pH 7.8 containing 50% MeCN. The cyclization step was performed using trypsin immobilized on sepharose beads in 100 mM sodium phosphate buffer at pH 7.4 for 15 min at 37° C. After washing the trypsin-beads with the same buffer, a simple treatment of the beads with 0.1% aqueous TFA (pH ≈3) yielded cyclotide MCoTI-II in 92% yield (Thongyoo et al., 2008). More recently, Craik has also used this approach for the synthesis of different MCoTI-based cyclotide mutants that were used to study their mechanism of cellular uptake (Cascales et al., 2011).
The use of trypsin-mediated cyclization provides a very efficient route for the production of cyclotides with trypsin inhibitory properties. It is worth noting, however, that although the cyclization yield for naturally occurring cyclotide MCoTI-II was extremely efficient (≈92%), the introduction of mutations that affect the binding to the proteolytic enzyme may lower the cyclization yield. For example, it has been shown that introduction of mutations into the trypsin binding loop of SFTI-1 can affect the equilibrium between the cyclized and linear form of SFTI-1 when incubated with trypsin (Austin et al., 2010), which should affect the cyclization yield of this peptide.
2.3.2. Butelase-mediated backbone cyclization
For the last few years mounting evidence has suggested the biosynthesis of cyclotides involves an aspariginyl endopeptidase-mediated transpeptidation step during the cyclization process of these circular peptides in plants (Mylne et al., 2011; Saska et al., 2007). Tam has recently provided in an important step towards fully understanding how cyclotides are biosynthesized in plants. His group reported the isolation and characterization of a new Asx-specific peptide ligase from the cyclotide-producing plant Clitoria ternatea of the Fabaceae family (Nguyen et al., 2014). This new ligase, called butelase 1, was able to efficiently cyclize different polypeptides, including cyclotide kalata B1, with yields greater than 95%. Butelase 1 shares 71% sequence identity and the same catalytic triad with aspariginyl endopeptidase legumain but was unable to hydrolyze the protease substrate of legumain. Instead, butelase 1 was an efficient peptide ligase with Kcat values of up to 17 s−1 and catalytic efficiencies as high as 542,000 M−1s−1, making it one of fastest peptide ligase known so far. Butelase 1 also displayed a broad specificity for the N-terminal amino acids of the peptide substrate requiring only the C-terminal NHV or DHV tripeptide motif for efficient backbone cyclization. The ligase, however, was more efficient with substrates bearing the NHV tripeptide motif (Nguyen et al., 2014). Butelase 1-mediated cyclization of kalata B1 was carried out using the natural Asn residue in loop 6, which has been suggested as the natural cyclization site in plants. It is worth noting that in contrast to trypsin-mediated cyclizations (see above), butelase 1 does not require conformational assistance by disulfide bonds for efficient cyclization. For example, an S-alkylated version of linear kalata B1 peptide bearing the tripeptide NHV was also efficiently cyclized by the ligase. Intriguingly, kinetic analysis revealed that the S-alkylated version of linear kalata B1 was cyclized around 50-times faster than the corresponding natively folded linear polypeptide. The use of butelase 1 to cyclize cyclotides has only been tested for the cyclization of kalata B1, and surprisingly not with any of the naturally occurring cyclotides found in C. ternatea. However, the high sequence homology of kalata B1 with the different cyclotides isolated from C. ternatea (Poth et al., 2011; Poth et al., 2010), especially at the ligation site in loop 6, suggests that cyclization should also be efficient with C. ternatea cyclotides. In fact, butelase 1 has shown to efficiently cyclize different peptides including SFTI-1, conotoxin MrlA, the insect antimicrobial peptide thanatin and the human salivary antimicrobial peptide histatin (Nguyen et al., 2014).
Butelase 1-mediated cyclizations are very efficient and have minimal sequence requirements at the ligation site, and together with all the features mentioned above make this ligase a very promising tool for the production of cyclic polypeptides. All the studies performed with butelase 1, however, have been carried out using enzyme isolated from natural sources. Attempts to produce biologically active butelase using recombinant heterologous expression have not been successful so far. This may limit its future potential in biotechnological applications.
2.3.3. Sortase-mediated backbone cyclization
It is well known that prokaryotes use protease-catalyzed protein splicing to attach proteins to peptidoglycan in their cell wall envelope (Marraffini, DeDent, & Schneewind, 2006). For example, sortases are transpeptidase enzymes found in most Gram-positive bacteria that are specialized in this task. Among the different sortases characterized so far, the Staphylococcus aureus sortase A (SrtA) is one of the most widely used for biotechnological applications (Katoh, Goto, Reza, & Suga, 2011; Popp & Ploegh, 2011). SrtA specifically recognizes the hexa-peptide motif LPTXTG (where X can be any residue) motif, cleaving the peptide bond between the Thr and Gly residues. SrtA active site contains a Cys residue that generates a transient thio-acyl enzyme intermediate during the cleavage reaction. This branched thioester intermediate then reacts with the α-amino group of poly-Gly (a tri-Gly sequence in the case of SrtA) substrates. In the absence of any poly-Gly substrate the thioester group is simply hydrolyzed.
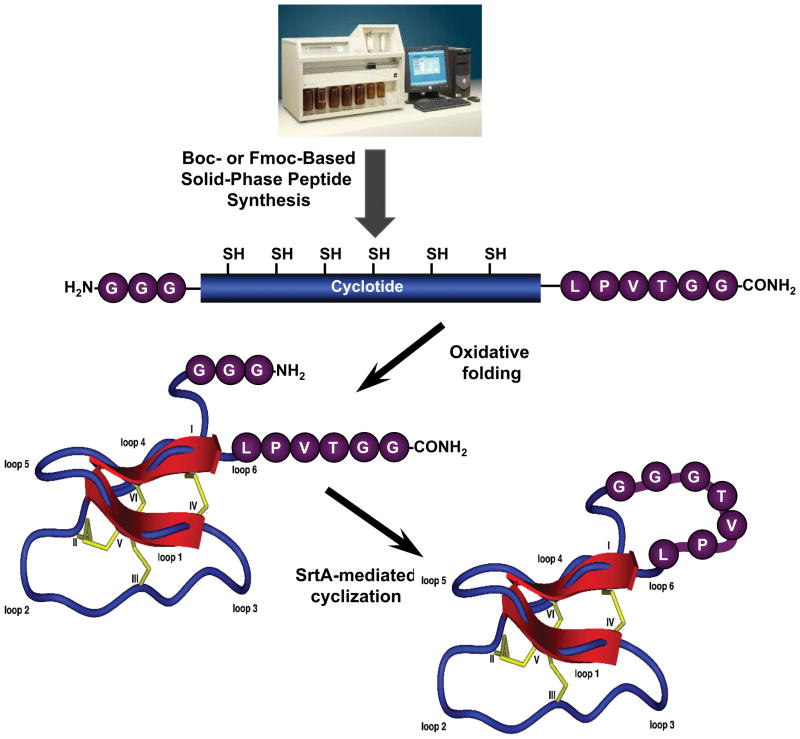
SrtA has been recently used for the generation of circular GFP using an intramolecular version of sortase-mediated ligation (SML) (Fig. 3) (Antos et al., 2009; Parthasarathy, Subramanian, & Boder, 2007). Other proteins/polypeptides cyclized using SML include dihydrofolate reductase (DHFR) and pleckstrin homology (PH) domains (Tsukiji & Nagamune, 2009), as well as circular versions of cytokines and histatin 1 (Bolscher et al., 2011). Craik has also recently used this approach for the backbone cyclization of different classes of disulfide-rich peptides including an analog of the cyclotide kalata B1 (Jia et al., 2014). It is worth noting, however, that due to the sequence requirements for SrtA, the heptapeptide motif (LPVTGGG) is left at the ligation site (Fig. 3). This should be taken into consideration when using this approach for the cyclization of small cyclic polypeptides. In this work the corresponding kalata B1 linear precursor was chemically synthesized using standard Fmoc-based SPPS using loop 6 for the cyclization site. This loop has been shown to be relatively more flexible than the rest of the cyclotide loops (Daly et al., 2013; Puttamadappa et al., 2010; Puttamadappa, Jagadish, Shekhtman, & Camarero, 2011) and it has been shown to tolerate loop extension without negatively affecting the native cyclotide fold (Gunasekera et al., 2008; Wong et al., 2012). To facilitate the SrtA-mediated cyclization, the tripeptides GGG and TGG were added to the N- and C-temini of the linear precursor, respectively. The linear precursor was oxidatively folded and cyclized using SrtA to provide a natively folded kalata B1 analog.
Figure 3.
Sortase-mediated backbone cyclization of a kalata B1 based cyclotide. The linear precursor peptide obtained by SPPS contains a SrtA recognition at the C- (LPVTG) and a N-termini (GGG). The linear precursor is oxidatively folded and backbone-cyclized using SrtA. Note that after the ligation the sequence LPVTTGGG remains on the cyclotide.
3. Biological synthesis of cyclotides
Recent advances in the fields of protein engineering and molecular biology make possible the biological synthesis of backbone-cyclized polypeptides using standard heterologous expression systems (T. L. Aboye & Camarero, 2012; Sancheti & Camarero, 2009). The different approaches described thus far for the biological synthesis of folded cyclotides include the use of thioester-mediated cyclization generated by modified inteins (also called expressed protein ligation) (J. A. Camarero, Kimura, Woo, Shekhtman, & Cantor, 2007; R. Kimura & Camarero, 2005) or thiol-induced selective N->S acyl-transfer (Cowper, Craik, & Macmillan, 2013) and intein-mediated protein trans-splicing (Borra, Dong, Elnagar, Woldemariam, & Camarero, 2012) (see reference (Perler, 2002) for an excellent searchable database of intein literature including circular peptides and protein) (Table 2).
3.1. Intein-mediated backbone cyclization
Developments in the field of protein engineering and the discovery of intein-mediated protein splicing have made possible the generation of recombinant proteins containing an N-terminal Cys residue and/or a C-terminal α-thioester functionality. These important developments have made possible to perform NCL reactions between recombinant and/or synthetic polypeptides, hence allowing access to a multitude of chemically engineered proteins including backbone-cyclized polypeptides (see reference (Berrade & Camarero, 2009) for a recent review on the use of inteins for protein engineering).
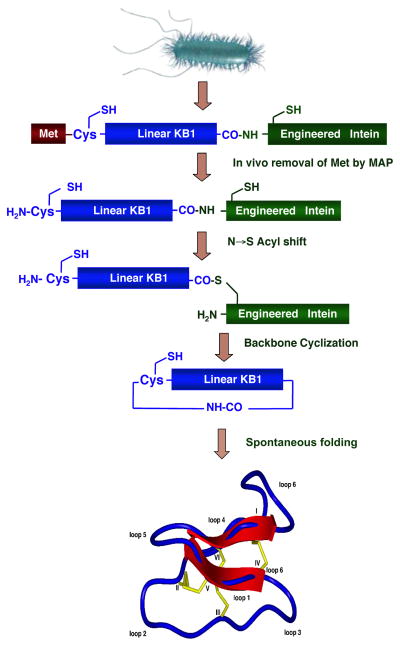
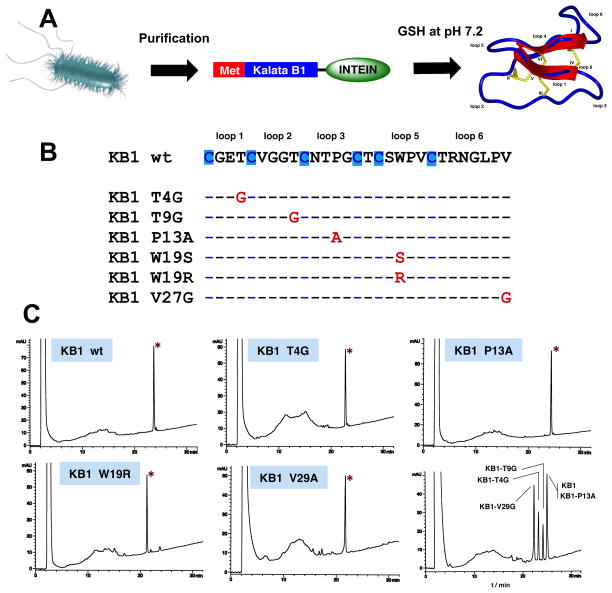
An intramolecular version of intein-mediated ligation (also called Expressed Protein Ligation (EPL)) has been used for the generation of many different types of backbone-cyclized polypeptides and proteins (see reference (T. L. Aboye & Camarero, 2012) for a recent review on the use of EPL for protein cyclization). Our group has recently used this approach for the recombinant expression of the cyclotide kalata B1 in E. coli (Fig. 4) (R. H. Kimura et al., 2006). In this work, several cyclotide linear precursors were fused to a modified Saccharomyces cerevisiae (Sce) vacuolar membrane ATPase (VMA) intein at the C-terminus and to a methionine residue at the N-terminus. The N-terminal Met residue is removed in situ by the endogenous bacterial methionine aminopeptidase (MAP) providing the required N-terminal Cys residue for the cyclization step. The intein fusion construct was then expressed in E. coli and purified. The recombinant precursor was cyclized and oxidatively folded in a “one-pot” reaction in aqueous buffer at pH 7.2 containing reduced GSH (Fig. 5). This approach was also used for the production of different kalata B1 mutants (Fig. 5). Some of the mutants were designed to test the effect of replacement of different β-branched residues adjacent to Cys residues in loops 1, 2, and 6 by the highly flexible Gly residue. The replacement of the hydrophobic residue Trp in loop 5 by the more hydrophilic Arg and Ser residues and the less hydrophobic Ala residue was also explored. Finally, the Pro residue in loop 3, which has been found to be important for defining the β turn in this loop (Rosengren, Daly, Plan, Waine, & Craik, 2003), was also mutated to Ala. As shown in Fig. 5, the “one-pot” cyclization/folding reaction was very efficient in all the cases with yields ranging from 20–60% hence highlighting the robustness of this methodology. Our group has also pioneered the use of intein-mediated backbone cyclization for the biosynthesis of fully folded cyclotides inside bacterial cells using standard heterologous expression systems (Austin, Wang, Puttamadappa, Shekhtman, & Camarero, 2009; J. A. Camarero et al., 2007). We have recently used this approach for the generation of a small library based on the cyclotide MCoTI-I where all of the residues except Cys and those located in loop 6 were replaced by different residues. In this work, we used the more efficient Mycobacterium xenopi (Mxe) GyrA intein instead. The resulting library was screened for biological activity and the ability to adopt a cyclotide fold indicating that most of the residues in loops 1, 2, 3 and 5 could be replaced without significantly affecting the ability to adopt a native cyclotide topology (Austin et al., 2009). Again, these results highlight the robustness of the cyclotide molecular framework, demonstrating the feasibility of in-cell generation of large genetically encoded libraries of cyclotides that could be screened inside the cell using high throughput selection techniques.
Figure 4.
Intein-mediated backbone cyclization of a cyclotide linear precursor. The intein-cyclotide construct is recombinantly expressed in E. coli and can be cyclized and folded either in vitro (R. H. Kimura et al., 2006) or in vivo (Austin et al., 2009; J. A. Camarero et al., 2007).
Figure 5.
In vitro production of different kalata B1 cylotides using an intein-mediated backbone cyclization (R. H. Kimura et al., 2006). A. The intein-kalata constructs were expressed in E. coli, purified and cyclized/folded in aqueous buffer at pH 7.2 containing reduced GSH for 24 h. B. Sequences of all the katala B mutants cyclized using this approach. C. Analytical HPLC traces of the cyclization/folding crude after 18 h. The peak corresponding to the folded cyclotide is marked with an asterisk.
Intein-mediated backbone cyclization of polypeptides has also also been used for the biosynthesis of other naturally occurring disulfide-rich backbone-cyclized polypeptides such as the Bowman-Birk inhibitor SFTI-1 (Austin et al., 2010), and the θ-defensins RTD-1 (Gould et al., 2012) and HTD-2 (Conibear et al., 2014), hence showing the general applicability of this method for the production of naturally occurring or engineered circular polypeptides.
3.2. Thiol-induced N->S acyl backbone cyclization
Recent work by Macmillan has shown that a polypeptide having a single C-terminal Cys residue is capable under special conditions of initiating an N->S acyl shift to provide an α-thioester intermediate that can be used for NCL (Kang, Richardson, & Macmillan, 2009). This reaction has been shown to be selective depending on the nature of the preceding Cys residue, being favored in cases the C-terminal Cys is preceded by a Gly, Cys or His residue. According to the same authors, the rate of the molecular rearrangement is also significantly slower when the Cys residue is not positioned at the C-terminus, which could allow regioselective control for the formation of α-thioesters in polypeptides containing multiple Cys residues (Richardson, Chan, Blanc, Saadi, & Macmillan, 2010). To test it, Macmillan and Craik used this approach to generate the cyclotide kalata B1 from a linear precursor produced by heterologous expression in E. coli (Cowper et al., 2013). In this work the cyclization site was selected at the C-terminal of loop 3 given the presence of required GC dipeptide motif in this loop (Fig. 1). The linear cyclotide precursor was fused to the C-terminus of the of a thioredoxin (Trx) tag to improve its expression yield and solubility. To allow the generation of the required N-terminal Cys reside for the cyclization step, the Trx tag was linked through a tobacco etch virus (TEV) protease recognition sequence. After purification of the precursor Trx-kalata B1 construct, the Trx tag was remove with TEV protease. Subsequent in vitro cyclization of the purified linear kalata B1 precursor was accomplished by incubating at 45° C in the presence of 10% (w/v) sodium 2-mercaptoethane sulfonate (MESNA) for 48 h. The reduced cyclotide was then oxidatively folded. The final product was fully characterized by NMR to show that was adopting a native cyclotide fold. Although this approach seems promising, it still remains to be seen how applicable it is for the synthesis of other cyclotides that do not contain a suitable Xaa-Cys motif, where Xaa is a Gly, Cys or His residue.
3.3. Sortase-induced backbone cyclization
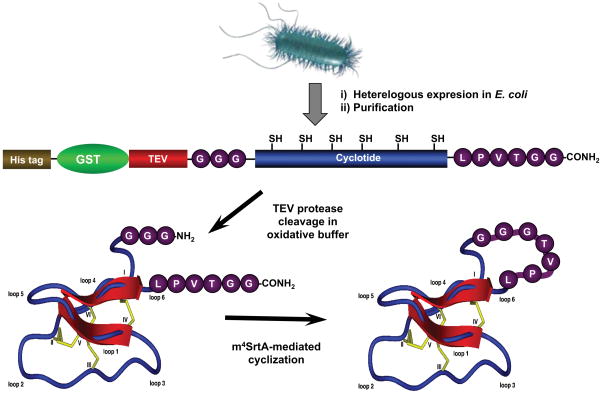
As mentioned earlier, SrtA has been used for the backbone cyclization of synthetic linear cyclotide precursors (see section 2.3.3). In a similar fashion Hannoush recently expressed a glutathione S-transferase (GST)-fusion protein containing a linearized version of the cyclotide MCoTI-II (Stanger et al., 2014). To allow the SrtA-mediated backbone cyclization, the SrtA recognition motifs GGG and LPETGG were added the N- and C-termini of the linear cyclotide sequence, respectively. A TEV protease cleavage site was also inserted between GST tag and the GGG motif in order to remove the His-GST tag after Ni2+-nitriloacetic acid (NTA) purification and to enable subsequent cyclization by Srt A (Fig. 6). After expression of the GST construct in E. coli, the GST tag was removed by treatment with TEV protease under oxidative conditions. This step provided the required N-terminal GGG motif for SrtA-mediated cyclization and folded the linear precursor into a native Cys-knot fold. It is worth noting, that in this work the cyclization was carried out with an optimized SrtA enzyme with improved catalytic activity (m4SrtA) (Chen, Dorr, & Liu, 2011). This evolved enzyme has a 140-fold increase in kcat/Km compared to the wild-type form. Given that the cyclization using SrtA leaves the heptapeptide motif LPETGGG at the cyclization, the cyclization was performed at loop 6. As in other cyclotides, the loop 6 in MCoTI-cyclotides has been shown to be more flexible than the rest of cyclotide framework (Puttamadappa et al., 2010, 2011) and therefore able to accept long peptide grafts without negatively affecting the cyclotide fold (T. L. Aboye et al., 2012; Chan et al., 2011; Ji et al., 2013).
Figure 6.
Strategy used for the biosynthesis of a cyclotide using SrtA-induced backbone cyclization (Stanger et al., 2014).
3.4. Backbone cyclization using intein-mediated protein trans-splicing (PTS)
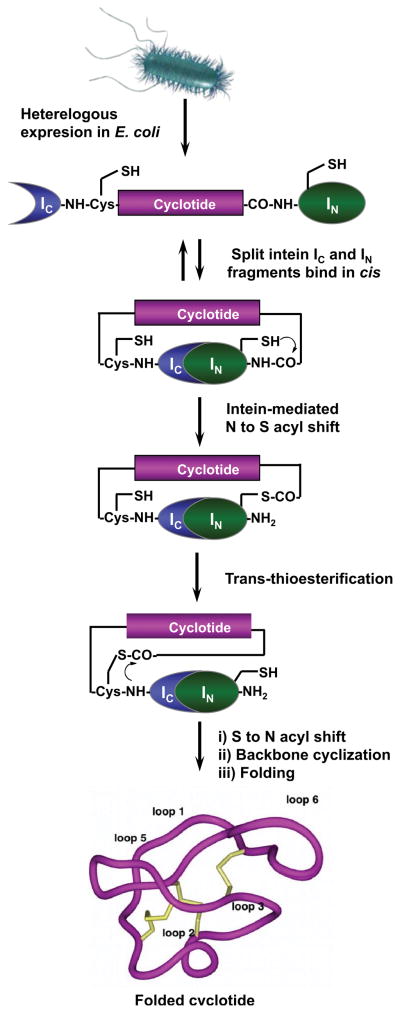
Our group has recently used PTS for the production of naturally occurring and engineered cyclotides (Jagadish et al., 2013). Protein trans-splicing is a post-translational modification similar to protein splicing with the difference that the intein self-processing domain is split into two fragments (N-intein and C-intein). The split-intein fragments are not active individually, however, they can bind to each other with high specificity under appropriate conditions to form an active protein splicing or intein domain in trans. By rearranging the order of the intein fragments, i.e. by fusing the N- and C-intein fragments to the C- and N-terminus of the polypeptide to cyclize, the trans-splicing reaction results in the formation of a backbone-cyclized polypeptide (Fig. 7).
Figure 7.
In-cell expression of native folded cyclotides using intein-mediated protein trans-splicing (PTS) (Jagadish et al., 2013).
This approach was first reported by the Benkovic group using the naturally occurring Synechocystis sp. (Ssp) PCC6803 DnaE split intein, and it has been used for the preparation of many small backbone-cyclized polypeptides as well as large genetically-encoded libraries of small cyclic peptides in bacterial cells (Abel-Santos, Scott, & Benkovic, 2003; Scott, Abel-Santos, Jones, & Benkovic, 2001; Scott, Abel-Santos, Wall, Wahnon, & Benkovic, 1999). PTS has also been used for the production of recombinant backbone-cyclized proteins. In a recent study, the artificially split Ssp PCC6803 DnaB mini-intein was used for the cyclization of the TEM-1 β-lactamase in the bacterial periplasm (Deschuyteneer et al., 2010).
It should be noted that use of the Ssp DnaE or DnaB split inteins requires the presence of specific amino acids at both extein-intein junctions for efficient protein splicing to happen (Tavassoli & Benkovic, 2007). Our group has overcome this limitation by using the Nostoc punctiforme PCC73102 (Npu) DnaE split intein (Iwai, Zuger, Jin, & Tam, 2006). The Npu DnaE split intein is one of the most efficient split inteins reported thus far. This intein has one of the highest reported rates of protein trans-splicing (τ1/2 ~ 60 s), a high splicing efficiency (≈80%), and high sequence tolerance at the extein-intein junctions (Borra et al., 2012; Jagadish et al., 2013; Zettler, Schutz, & Mootz, 2009). We have used this split intein for the in-cell production of several MCoTI-based cyclotides (Jagadish et al., 2013). In cell expression was highly efficient providing intracellular concentrations in the range of 20–40 μM. The high efficiency of Npu DnaE PTS-mediated cyclization in combination with the use of nonsense suppressing orthogonal tRNA/synthetase technology has also made possible the production of cyclotides containing unnatural amino acids (Uaa’s) inside live bacterial cells (Jagadish et al., 2013). We used this is approach to incorporate p-azido-phenylalanine into loop 2 of cyclotide MCoTI-I, which reacted in-cell through copper-free click chemistry with dibenzocyclooctyne-containing fluorescent probes to provide in-cell fluorescently-labeled cyclotides (Jagadish et al., 2013). This exciting result opens the possibility for in-cell screening of genetically-encoded libraries of cyclotides for the rapid selection of novel cyclotide sequences able to bind a specific bait protein using high throughput cell-based optical screening approaches.
More recently, we have shown that this approach can be also used for the expression of folded cyclotides in eukaryotic microorganisms such as yeast S. cerevisiae (manuscript submitted). In this work, in-cell PTS was used to generate a novel bioactive cyclotide that was able to inhibit α-synuclein-induced cytotoxicity in live yeast cells, therefore allowing us for the first time to perform a phenotypic screen to select cyclotide sequences with biological activity from inactive cyclotides in a yeast synucleopathy model. These results make possible to perform in-cell phenotypic screens against other cellular pathologies using cyclotide-based libraries. Using eukaryotic microorganisms for screening purposes should not only provide a more biologically relevant cellular background for phenotypic screening but also facilitate the production of cyclotide-based libraries containing post-translational modifications such as phosphorylation and/or glycosylation, which are not available in bacterial expression systems.
4. Summary and concluding remarks
In this book chapter we have reviewed different chemical and biological methods for the production of cyclotides. Most of the methods reviewed make use of an intramolecular version of NCL. NCL requires only the presence of an N-terminal Cys residue and a α-thioester function. Cyclotides present six Cys residues that in theory could be used as cyclization sites. According to the published data, the Cys residues at the C- and N-terminal regions of loop 3 and loop 6 have been successfully used in the production of several cyclotides. The NCL reaction is also fully compatible with physiological conditions, i.e. aqueous buffers at pH ≈ 7, and does not compromise the chirality of the C-terminal residue during the cyclization step. In addition, if GSH is used as thiol co-factor, the cyclization and folding can be accomplished in a single “one-pot” reaction (Austin et al., 2010; R. H. Kimura et al., 2006). The linear precursors required for NCL-mediated cyclization can be also obtained by either pure synthetic (using both Boc- and Fmoc-based SPPS) or recombinant (using modified inteins or a thiol-induced N->S acyl shift reaction) methods. Synthetic methods allow the synthesis of cyclotides containing non-natural amino acids (T. L. Aboye et al., 2012) or even the production of D-cyclotides (Sando et al., 2011). On the other hand, the use of pure recombinant expression methods allows scalability for cyclotide production and more importantly in-cell production of cyclotides (Austin et al., 2010; J. A. Camarero et al., 2007; Jagadish et al., 2013). In cell production makes possible the generation of large genetically-encoded libraries of cyclic peptides inside live cells that can be rapidly screened for the selection of novel sequences able to modulate or inhibit the biological activities of particular biomolecular targets.
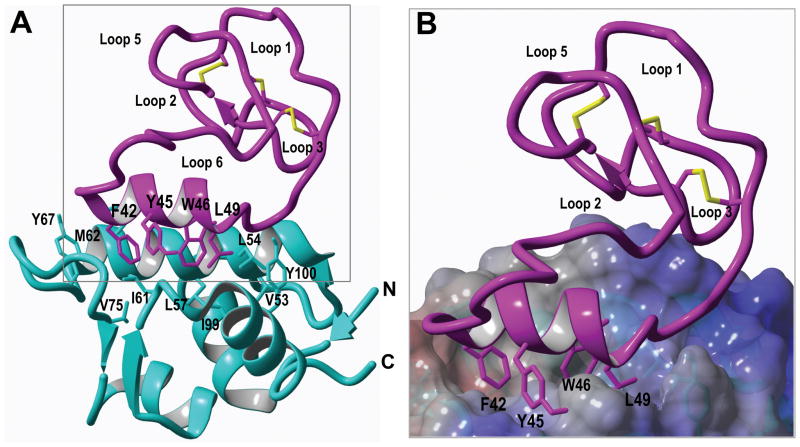
Having access to the recombinant production of cyclotides using bacterial heterologous expression also facilitates the introduction of NMR active isotopes such as 15N and/or 13C in a very inexpensive fashion (Puttamadappa et al., 2010, 2011). This should facilitate the use of heteronuclear NMR spectroscopy to study structure-activity relationships of any biologically active circular polypeptides and their molecular targets. This was recently demonstrated in the elucidation of the solution structure of the first cyclotide engineered to bind the p53 binding domain of the oncogene Hdm2 (Fig. 8) (Ji et al., 2013).
Figure 8.
Solution structure of the complex formed by engineered cyclotide MCo-PMI and the p53 binding domain of Hdm2 (pdb code: 2M86) (Ji et al., 2013). A. Ribbon representation of MCo-PMI (purple) and Hdm2 (cyan blue) complex. The side-chains of Phe42, Trp46 and Leu49 in MCo-PMI and the Hdm2 residues shaping the hydrophobic binding pocket are shown as sticks in purple and cyan blue respectively. B. Close-up view of the binding interface within the Hdm2-MCo-PMI complex. The electrostatic potential at the molecular surface of Hdm2 is shown as positive in blue, negative in red and non-charged in white.
The use of PTS-mediated backbone cyclization in combination with nonsense suppressing orthogonal tRNA/synthetase technology has also made possible the introduction of non natural amino acids into in-cell expressed backbone-cyclized polypeptides (Jagadish et al., 2013). Expansion of the genetic code allows for the incorporation of novel chemical functionalities that can be used in molecular evolution and cell based assays for the selection of novel cyclic peptides with improved biological or novel biophysical activities (Jagadish et al., 2013; Young et al., 2011). More recently PTS has also been used for in-cell expression of folded bioactive cyclotides in eukaryotic microorganisms such as yeast S. cerevisiae (manuscript submitted). S. cerevisiae is a versatile and robust model system for eukaryotic cellular biology and several human pathologies derived from protein misfolding have been successfully modeled in this microorganism. Using a yeast synucleopathy model (Kritzer et al., 2009) in combination with PTS we were recently able to express a cyclotide inhibitor of α-synuclein cytotoxicity. This made possible for the first time to perform a phenotypic screen to select bioactive cyclotides in a eukaryotic system. The use of eukaryotic microorganisms, such as yeast, for screening purposes should provide a more biologically relevant cellular background for phenotypic screening but also facilitate the production of cyclotide-based libraries containing post-translational modifications such as phosphorylation and/or glycosylation, which are not available in bacterial expression systems.
Cyclotides are small globular microproteins with a unique head-to-tail cyclized backbone, which is stabilized by three disulfide bonds. The number and positions of cysteine residues are conserved throughout the family, forming a CCK motif (Craik, Daly, Bond, & Waine, 1999) that acts as a highly stable and versatile framework on which hyper-variable loops are arranged (Fig. 1). This CCK scaffold affords exceptional resistance to thermal, chemical denaturation as well as enzymatic degradation. This is particularly important for oral bioavailability. In fact, the use of cyclotide-containing plants in indigenous medicine first highlighted the fact that the peptides are resistant to boiling and are orally bioavailable (Saether et al., 1995). An orally bioavailable bradykinin B1 cyclotide receptor antagonist has also been recently reported (Wong et al., 2012). Cyclotides have also shown excellent tolerance to sequence variations. This combined with the possibility to produce them using heterologous expressions systems makes them ideal substrates for molecular evolution strategies to enable generation and selection of compounds with optimal binding and inhibitory characteristics. Finally, cyclotides can be also efficiently produced using standard SPPS techniques thereby allowing the introduction of non-natural amino acids and chemical modifications to improve their pharmacological properties.
In summary, all these features open new opportunities for the development of the cyclotide molecular scaffold for novel and exciting biotechnological and biomedical applications.
Acknowledgments
This work was partially supported by National Institutes of Health Research Grant R01-GM090323 (JAC) and Bristol-Myers Squibb.
References
- Abel-Santos E, Scott CP, Benkovic SJ. Use of inteins for the in vivo production of stable cyclic peptide libraries in E. coli. Methods Mol Biol. 2003;205:281–294. doi: 10.1385/1-59259-301-1:281. [DOI] [PubMed] [Google Scholar]
- Aboye T, Kuang Y, Neamati N, Camarero JA. Rapid Parallel Synthesis of Bioactive Folded Cyclotides by Using a Tea-Bag Approach. Chembiochem. 2015 doi: 10.1002/cbic.201402691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboye TL, Camarero JA. Biological synthesis of circular polypeptides. J Biol Chem. 2012;287(32):27026–27032. doi: 10.1074/jbc.R111.305508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboye TL, Ha H, Majumder S, Christ F, Debyser Z, Shekhtman A, … Camarero JA. Design of a novel cyclotide-based CXCR4 antagonist with anti-human immunodeficiency virus (HIV)-1 activity. J Med Chem. 2012;55(23):10729–10734. doi: 10.1021/jm301468k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboye TL, Clark RJ, Craik DJ, Göransson U. Ultra-stable peptide scaffolds for protein engineering-synthesis and folding of the circular cystine knotted cyclotide cycloviolacin O2. Chembiochem. 2008;9(1):103–113. doi: 10.1002/cbic.200700357. [DOI] [PubMed] [Google Scholar]
- Antos JM, Popp MW, Ernst R, Chew GL, Spooner E, Ploegh HL. A straight path to circular proteins. J Biol Chem. 2009;284(23):16028–16036. doi: 10.1074/jbc.M901752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J, Kimura RH, Woo YH, Camarero JA. In vivo biosynthesis of an Ala-scan library based on the cyclic peptide SFTI-1. Amino Acids. 2010;38(5):1313–1322. doi: 10.1007/s00726-009-0338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J, Wang W, Puttamadappa S, Shekhtman A, Camarero JA. Biosynthesis and biological screening of a genetically encoded library based on the cyclotide MCoTI-I. Chembiochem. 2009;10(16):2663–2670. doi: 10.1002/cbic.200900534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes BJ, Ellman JA. An alkanesulfonamide “safety-catch” linker for solid-phase synthesis. J Org Chem. 1999;64(64):2322–2330. [Google Scholar]
- Backes BJ, Virgilio AV, Ellman JA. Activation Method to Prepare a Highly Reactive Acylsulfonamide “Safety-Catch’ Linker for Solid-Phase Synthesis. J Am Chem Soc. 1996;118:3055–3056. [Google Scholar]
- Beligere GS, Dawson PE. Conformationally assisted ligation using C-terminal thioester peptides. J Am Chem Soc. 1999;121:6332–6333. [Google Scholar]
- Berrade L, Camarero JA. Expressed protein ligation: a resourceful tool to study protein structure and function. Cell Mol Life Sci. 2009;66(24):3909–3922. doi: 10.1007/s00018-009-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Canosa JB, Dawson PE. An efficient Fmoc-SPPS approach for the generation of thioester peptide precursors for use in native chemical ligation. Angew Chem Int Ed Engl. 2008;47(36):6851–6855. doi: 10.1002/anie.200705471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolscher JG, Oudhoff MJ, Nazmi K, Antos JM, Guimaraes CP, Spooner E, … Veerman EC. Sortase A as a tool for high-yield histatin cyclization. Faseb J. 2011;25(8):2650–2658. doi: 10.1096/fj.11-182212. [DOI] [PubMed] [Google Scholar]
- Borra R, Dong D, Elnagar AY, Woldemariam GA, Camarero JA. In-cell fluorescence activation and labeling of proteins mediated by FRET-quenched split inteins. J Am Chem Soc. 2012;134(14):6344–6353. doi: 10.1021/ja300209u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarero JA, Adeva A, Muir TW. 3-Thiopropionic acid as a highly versatile multidetachable thioester resin linker. Lett Pept Sci. 2000;7(1):17–21. [Google Scholar]
- Camarero JA, Cotton GJ, Adeva A, Muir TW. Chemical ligation of unprotected peptides directly from a solid support. J Pept Res. 1998;51(4):303–316. doi: 10.1111/j.1399-3011.1998.tb00428.x. [DOI] [PubMed] [Google Scholar]
- Camarero JA, Hackel BJ, de Yoreo JJ, Mitchell AR. Fmoc-based synthesis of peptide alpha-thioesters using an aryl hydrazine support. J Org Chem. 2004;69(12):4145–4151. doi: 10.1021/jo040140h. [DOI] [PubMed] [Google Scholar]
- Camarero JA, Kimura RH, Woo YH, Shekhtman A, Cantor J. Biosynthesis of a fully functional cyclotide inside living bacterial cells. Chembiochem. 2007;8(12):1363–1366. doi: 10.1002/cbic.200700183. [DOI] [PubMed] [Google Scholar]
- Camarero JA, Mitchell AR. Synthesis of proteins by native chemical ligation using Fmoc-based chemistry. Protein Pept Lett. 2005;12(8):723–728. doi: 10.2174/0929866054864166. [DOI] [PubMed] [Google Scholar]
- Camarero JA, Muir TW. Biosynthesis of a Head-to-Tail Cyclized Protein with Improved Biological Activity. J Am Chem Soc. 1999;121:5597–5598. [Google Scholar]
- Camarero JA, Muir TW. Chemoselective Backbone Cyclization of Unprotected Peptides. Chem Comm. 1997;1997:202–219. [Google Scholar]
- Cascales L, Henriques ST, Kerr MC, Huang YH, Sweet MJ, Daly NL, Craik DJ. Identification and characterization of a new family of cell-penetrating peptides: cyclic cell-penetrating peptides. J Biol Chem. 2011;286(42):36932–36943. doi: 10.1074/jbc.M111.264424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LY, Gunasekera S, Henriques ST, Worth NF, Le SJ, Clark RJ, … Daly NL. Engineering pro-angiogenic peptides using stable, disulfide-rich cyclic scaffolds. Blood. 2011;118(25):6709–6717. doi: 10.1182/blood-2011-06-359141. [DOI] [PubMed] [Google Scholar]
- Chen I, Dorr BM, Liu DR. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc Natl Acad Sci U S A. 2011;108(28):11399–11404. doi: 10.1073/pnas.1101046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheneval O, Schroeder CI, Durek T, Walsh P, Huang YH, Liras S, … Craik DJ. Fmoc-based synthesis of disulfide-rich cyclic peptides. J Org Chem. 2014;79(12):5538–5544. doi: 10.1021/jo500699m. [DOI] [PubMed] [Google Scholar]
- Chiche L, Heitz A, Gelly JC, Gracy J, Chau PT, Ha PT, … Le-Nguyen D. Squash inhibitors: from structural motifs to macrocyclic knottins. Curr Protein Pept Sci. 2004;5(5):341–349. doi: 10.2174/1389203043379477. [DOI] [PubMed] [Google Scholar]
- Colgrave ML, Craik DJ. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry. 2004;43(20):5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- Conibear AC, Wang CK, Bi T, Rosengren KJ, Camarero JA, Craik DJ. Insights into the Molecular Flexibility of theta-Defensins by NMR Relaxation Analysis. J Phys Chem B. 2014 doi: 10.1021/jp507754c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras J, Elnagar AY, Hamm-Alvarez SF, Camarero JA. Cellular uptake of cyclotide MCoTI-I follows multiple endocytic pathways. J Control Release. 2011;155(2):134–143. doi: 10.1016/j.jconrel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowper B, Craik DJ, Macmillan D. Making ends meet: chemically mediated circularization of recombinant proteins. Chembiochem. 2013;14(7):809–812. doi: 10.1002/cbic.201300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik DJ, Daly NL, Bond T, Waine C. Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol. 1999;294(5):1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- Daly NL, Love S, Alewood PF, Craik DJ. Chemical synthesis and folding pathways of large cyclic polypeptides: studies of the cystine knot polypeptide kalata B1. Biochemistry. 1999;38(32):10606–10614. doi: 10.1021/bi990605b. [DOI] [PubMed] [Google Scholar]
- Daly NL, Thorstholm L, Greenwood KP, King GJ, Rosengren KJ, Heras B, … Craik DJ. Structural insights into the role of the cyclic backbone in a squash trypsin inhibitor. J Biol Chem. 2013;288(50):36141–36148. doi: 10.1074/jbc.M113.528240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson PE, Muir TW, Clark-Lewis I, Kent SBH. Synthesis of Proteins by Native Chemical Ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- Deschuyteneer G, Garcia S, Michiels B, Baudoux B, Degand H, Morsomme P, Soumillion P. Intein-mediated cyclization of randomized peptides in the periplasm of Escherichia coli and their extracellular secretion. ACS Chem Biol. 2010;5(7):691–700. doi: 10.1021/cb100072u. [DOI] [PubMed] [Google Scholar]
- Dutton JL, Renda RF, Waine C, Clark RJ, Daly NL, Jennings CV, … Craik DJ. Conserved Structural and Sequence Elements Implicated in the Processing of Gene-encoded Circular Proteins. J Biol Chem. 2004;279(45):46858–46867. doi: 10.1074/jbc.M407421200. [DOI] [PubMed] [Google Scholar]
- Eliasen R, Daly NL, Wulff BS, Andresen TL, Conde-Frieboes KW, Craik DJ. Design, synthesis, structural and functional characterization of novel melanocortin agonists based on the cyclotide kalata B1. J Biol Chem. 2012;287(48):40493–40501. doi: 10.1074/jbc.M112.395442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felizmenio-Quimio ME, Daly NL, Craik DJ. Circular proteins in plants: solution structure of a novel macrocyclic trypsin inhibitor from Momordica cochinchinensis. J Biol Chem. 2001;276(25):22875–22882. doi: 10.1074/jbc.M101666200. [DOI] [PubMed] [Google Scholar]
- Futaki S, Sogawa K, Maruyama J, Asahara T, Niwa M. Preparation of Peptide Thioesters using Fmoc-Solid-Phase Peptide Synthesis and its Application to the Construction of a Template-Assembled Synthetic Protein (TASP) Tetrahedron Lett. 1997;38(35):6237–6240. [Google Scholar]
- Garcia AE, Camarero JA. Biological activities of natural and engineered cyclotides, a novel molecular scaffold for peptide-based therapeutics. curr mol pharmacol. 2010;3(3):153–163. doi: 10.2174/1874467211003030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillon AD, Saska I, Jennings CV, Guarino RF, Craik DJ, Anderson MA. Biosynthesis of circular proteins in plants. Plant J. 2008;53(3):505–515. doi: 10.1111/j.1365-313X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- Gould A, Li Y, Majumder S, Garcia AE, Carlsson P, Shekhtman A, Camarero JA. Recombinant production of rhesus theta-defensin-1 (RTD-1) using a bacterial expression system. Mol Biosyst. 2012;8(4):1359–1365. doi: 10.1039/c2mb05451e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray K, Elghadban S, Thongyoo P, Owen KA, Szabo R, Bugge TH, … Ellis V. Potent and specific inhibition of the biological activity of the type-II transmembrane serine protease matriptase by the cyclic microprotein MCoTI-II. Thromb Haemost. 2014;112(2):402–411. doi: 10.1160/TH13-11-0895. [DOI] [PubMed] [Google Scholar]
- Gruber CW, Elliott AG, Ireland DC, Delprete PG, Dessein S, Goransson U, … Craik DJ. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell. 2008;20(9):2471–2483. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera S, Aboye TL, Madian WA, El-Seedi HR, Goransson U. Making Ends Meet: Microwave-Accelerated Synthesis of Cyclic and Disulfide Rich Proteins Via In Situ Thioesterification and Native Chemical Ligation. Int J Pept Res Ther. 2013;19(1):43–54. doi: 10.1007/s10989-012-9331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera S, Foley FM, Clark RJ, Sando L, Fabri LJ, Craik DJ, Daly NL. Engineering stabilized vascular endothelial growth factor-A antagonists: synthesis, structural characterization, and bioactivity of grafted analogues of cyclotides. J Med Chem. 2008;51(24):7697–7704. doi: 10.1021/jm800704e. [DOI] [PubMed] [Google Scholar]
- Heitz A, Hernandez JF, Gagnon J, Hong TT, Pham TT, Nguyen TM, … Chiche L. Solution structure of the squash trypsin inhibitor MCoTI-II. A new family for cyclic knottins. Biochemistry. 2001;40(27):7973–7983. doi: 10.1021/bi0106639. [DOI] [PubMed] [Google Scholar]
- Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, … Le Nguyen D. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry. 2000;39(19):5722–5730. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- Huang YH, Colgrave ML, Clark RJ, Kotze AC, Craik DJ. Lysine-scanning mutagenesis reveals an amendable face of the cyclotide kalata B1 for the optimization of nematocidal activity. J Biol Chem. 2010;285(14):10797–10805. doi: 10.1074/jbc.M109.089854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingenito R, Bianchi E, Fattori D, Pessi A. Solid phase synthesis of peptide C-terminal thioesters by Fmoc/t-Bu chemistry Source. J Am Chem Soc. 1999;121(49):11369–11374. [Google Scholar]
- Iwai H, Zuger S, Jin J, Tam PH. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 2006;580(7):1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- Jagadish K, Borra R, Lacey V, Majumder S, Shekhtman A, Wang L, Camarero JA. Expression of fluorescent cyclotides using protein trans-splicing for easy monitoring of cyclotide-protein interactions. Angew Chem Int Ed Engl. 2013;52(11):3126–3131. doi: 10.1002/anie.201209219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Majumder S, Millard M, Borra R, Bi T, Elnagar AY, … Camarero JA. In Vivo Activation of the p53 Tumor Suppressor Pathway by an Engineered Cyclotide. J Am Chem Soc. 2013;135(31):11623–11633. doi: 10.1021/ja405108p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Kwon S, Wang CI, Huang YH, Chan LY, Tan CC, … Craik DJ. Semienzymatic cyclization of disulfide-rich peptides using Sortase A. J Biol Chem. 2014;289(10):6627–6638. doi: 10.1074/jbc.M113.539262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Richardson JP, Macmillan D. 3-Mercaptopropionic acid-mediated synthesis of peptide and protein thioesters. Chem Commun (Camb) 2009;(4):407–409. doi: 10.1039/b815888f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Goto Y, Reza MS, Suga H. Ribosomal synthesis of backbone macrocyclic peptides. Chem Commun (Camb) 2011;47(36):9946–9958. doi: 10.1039/c1cc12647d. [DOI] [PubMed] [Google Scholar]
- Kenner GW, McDermott JR, Sheppard RC. The Safety Catch Principle in Solid Phase Peptide Synthesis. Chem Comm. 1971:636–637. [Google Scholar]
- Kimura R, Camarero JA. Expressed protein ligation: a new tool for the biosynthesis of cyclic polypeptides. Protein Pept Lett. 2005;12(8):789–794. doi: 10.2174/0929866054864274. [DOI] [PubMed] [Google Scholar]
- Kimura RH, Tran AT, Camarero JA. Biosynthesis of the cyclotide kalata B1 by using protein splicing. Angew Chem Int Ed. 2006;45(6):973–976. doi: 10.1002/anie.200503882. [DOI] [PubMed] [Google Scholar]
- Koehbach J, O’Brien M, Muttenthaler M, Miazzo M, Akcan M, Elliott AG, … Gruber CW. Oxytocic plant cyclotides as templates for peptide G protein-coupled receptor ligand design. Proc Natl Acad Sci U S A. 2013;110(52):21183–21188. doi: 10.1073/pnas.1311183110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritzer JA, Hamamichi S, McCaffery JM, Santagata S, Naumann TA, Caldwell KA, … Lindquist S. Rapid selection of cyclic peptides that reduce alpha-synuclein toxicity in yeast and animal models. Nat Chem Biol. 2009;5(9):655–663. doi: 10.1038/nchembio.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marglin A, Merrifield RB. Chemical synthesis of peptides and proteins. Annu Rev Biochem. 1970;39:841–866. doi: 10.1146/annurev.bi.39.070170.004205. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, DeDent AC, Schneewind O. Sortases and the Art of Anchoring Proteins to the Envelopes of Gram-Positive Bacteria. Microbiol Mol Biol Rev. 2006;70(1):192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx UC, Korsinczky ML, Schirra HJ, Jones A, Condie B, Otvos L, Jr, Craik DJ. Enzymatic cyclization of a potent bowman-birk protease inhibitor, sunflower trypsin inhibitor-1, and solution structure of an acyclic precursor peptide. J Biol Chem. 2003;278(24):21782–21789. doi: 10.1074/jbc.M212996200. [DOI] [PubMed] [Google Scholar]
- Mylne JS, Chan LY, Chanson AH, Daly NL, Schaefer H, Bailey TL, … Craik DJ. Cyclic peptides arising by evolutionary parallelism via asparaginyl-endopeptidase-mediated biosynthesis. Plant Cell. 2012;24(7):2765–2778. doi: 10.1105/tpc.112.099085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne JS, Colgrave ML, Daly NL, Chanson AH, Elliott AG, McCallum EJ, … Craik DJ. Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat Chem Biol. 2011;7(5):257–259. doi: 10.1038/nchembio.542. [DOI] [PubMed] [Google Scholar]
- Nguyen GK, Wang S, Qiu Y, Hemu X, Lian Y, Tam JP. Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat Chem Biol. 2014;10(9):732–738. doi: 10.1038/nchembio.1586. [DOI] [PubMed] [Google Scholar]
- Nguyen GK, Zhang S, Nguyen NT, Nguyen PQ, Chiu MS, Hardjojo A, Tam JP. Discovery and characterization of novel cyclotides originated from chimeric precursors consisting of albumin-1 chain a and cyclotide domains in the fabaceae family. J Biol Chem. 2011 doi: 10.1074/jbc.M111.229922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy R, Subramanian S, Boder ET. Sortase A as a novel molecular “stapler” for sequence-specific protein conjugation. Bioconjug Chem. 2007;18(2):469–476. doi: 10.1021/bc060339w. [DOI] [PubMed] [Google Scholar]
- Perler FB. InBase: the Intein Database. Nucleic Acids Res. 2002;30(1):383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Ploegh HL. Making and breaking peptide bonds: protein engineering using sortase. Angew Chem Int Ed Engl. 2011;50(22):5024–5032. doi: 10.1002/anie.201008267. [DOI] [PubMed] [Google Scholar]
- Poth AG, Colgrave ML, Lyons RE, Daly NL, Craik DJ. Discovery of an unusual biosynthetic origin for circular proteins in legumes. Proc Natl Acad Sci U S A. 2011;108(25):1027–1032. doi: 10.1073/pnas.1103660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poth AG, Colgrave ML, Philip R, Kerenga B, Daly NL, Anderson MA, Craik DJ. Discovery of cyclotides in the fabaceae plant family provides new insights into the cyclization, evolution, and distribution of circular proteins. ACS Chem Biol. 2010;6(4):345–355. doi: 10.1021/cb100388j. [DOI] [PubMed] [Google Scholar]
- Poth AG, Mylne JS, Grassl J, Lyons RE, Millar AH, Colgrave ML, Craik DJ. Cyclotides associate with leaf vasculature and are the products of a novel precursor in petunia (Solanaceae) J Biol Chem. 2012;287(32):27033–27046. doi: 10.1074/jbc.M112.370841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttamadappa SS, Jagadish K, Shekhtman A, Camarero JA. Backbone Dynamics of Cyclotide MCoTI-I Free and Complexed with Trypsin. Angew Chem Int Ed Engl. 2010;49(39):7030–7034. doi: 10.1002/anie.201002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttamadappa SS, Jagadish K, Shekhtman A, Camarero JA. Erratum in: Backbone Dynamics of Cyclotide MCoTI-I Free and Complexed with Trypsin. Angew Chem Int Ed Engl. 2011;50(31):6948–6949. doi: 10.1002/anie.201002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S, Sieber M, Oberle V, Wentzel A, Spangenberg P, Claus R, … Losche W. Inhibition of platelet aggregation by grafting RGD and KGD sequences on the structural scaffold of small disulfide-rich proteins. Platelets. 2006;17(3):153–157. doi: 10.1080/09537100500436663. [DOI] [PubMed] [Google Scholar]
- Richardson JP, Chan CH, Blanc J, Saadi M, Macmillan D. Exploring neoglycoprotein assembly through native chemical ligation using neoglycopeptide thioesters prepared via N-->S acyl transfer. Org Biomol Chem. 2010;8(6):1351–1360. doi: 10.1039/b920535g. [DOI] [PubMed] [Google Scholar]
- Rosengren KJ, Daly NL, Plan MR, Waine C, Craik DJ. Twists, knots, and rings in proteins. Structural definition of the cyclotide framework. J Biol Chem. 2003;278(10):8606–8616. doi: 10.1074/jbc.M211147200. [DOI] [PubMed] [Google Scholar]
- Saether O, Craik DJ, Campbell ID, Sletten K, Juul J, Norman DG. Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry. 1995;34(13):4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- Sancheti H, Camarero JA. “Splicing up” drug discovery. Cell-based expression and screening of genetically-encoded libraries of backbone-cyclized polypeptides. Adv Drug Deliv Rev. 2009;61(11):908–917. doi: 10.1016/j.addr.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando L, Henriques ST, Foley F, Simonsen SM, Daly NL, Hall KN, … Craik DJ. A Synthetic mirror image of kalata B1 reveals that cyclotide activity is independent of a protein receptor. Chembiochem. 2011;12(16):2456–2462. doi: 10.1002/cbic.201100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saska I, Gillon AD, Hatsugai N, Dietzgen RG, Hara-Nishimura I, Anderson MA, Craik DJ. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J Biol Chem. 2007;282(40):29721–29728. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- Scott CP, Abel-Santos E, Jones AD, Benkovic SJ. Structural requirements for the biosynthesis of backbone cyclic peptide libraries. Chem Biol. 2001;8(8):801–815. doi: 10.1016/s1074-5521(01)00052-7. [DOI] [PubMed] [Google Scholar]
- Scott CP, Abel-Santos E, Wall M, Wahnon D, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci USA. 1999;96(24):13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Winans KA, Backes BJ, Kent SBH, Ellman JA, Bertozzi CR. Fmoc-Based Synthesis of Peptide-aThioesters: Application to the Total Chemical Synthesis of a Glycoprotein by Native Chemical Ligation. J Am Chem Soc. 1999;121:11684–11689. [Google Scholar]
- Simonsen SM, Sando L, Rosengren KJ, Wang CK, Colgrave ML, Daly NL, Craik DJ. Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J Biol Chem. 2008;283(15):9805–9813. doi: 10.1074/jbc.M709303200. [DOI] [PubMed] [Google Scholar]
- Stanger K, Maurer T, Kaluarachchi H, Coons M, Franke Y, Hannoush RN. Backbone cyclization of a recombinant cystine-knot peptide by engineered Sortase A. FEBS Lett. 2014;588(23):4487–4496. doi: 10.1016/j.febslet.2014.10.020. [DOI] [PubMed] [Google Scholar]
- Tam JP, Lu YA. A biomimetic strategy in the synthesis and fragmentation of a cyclic protein. Prot Sci. 1998;7(7):1583–1592. doi: 10.1002/pro.5560070712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam JP, Lu YA, Yang JL, Chiu KW. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc Natl Acad Sci U S A. 1999;96(16):8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam JP, Lu YA. Synthesis of Large Cyclic Cystine-Knot Peptide by Orthogonal Coupling Strategy Using Unprotected Peptide Precursor. Tetrathedron Lett. 1997;38(32):5599–5602. [Google Scholar]
- Tavassoli A, Benkovic SJ. Split-intein mediated circular ligation used in the synthesis of cyclic peptide libraries in E. coli. Nat Protoc. 2007;2(5):1126–1133. doi: 10.1038/nprot.2007.152. [DOI] [PubMed] [Google Scholar]
- Thongyoo P, Bonomelli C, Leatherbarrow RJ, Tate EW. Potent inhibitors of beta-tryptase and human leukocyte elastase based on the MCoTI-II scaffold. J Med Chem. 2009;52(20):6197–6200. doi: 10.1021/jm901233u. [DOI] [PubMed] [Google Scholar]
- Thongyoo P, Roque-Rosell N, Leatherbarrow RJ, Tate EW. Chemical and biomimetic total syntheses of natural and engineered MCoTI cyclotides. Org Biomol Chem. 2008;6(8):1462–1470. doi: 10.1039/b801667d. [DOI] [PubMed] [Google Scholar]
- Thongyoo P, Tate EW, Leatherbarrow RJ. Total synthesis of the macrocyclic cysteine knot microprotein MCoTI-II. Chem Commun (Camb) 2006;(27):2848–2850. doi: 10.1039/b607324g. [DOI] [PubMed] [Google Scholar]
- Tsukiji S, Nagamune T. Sortase-Mediated Ligation: A Gift from Gram-Positive Bacteria to Protein Engineering. Chembiochem. 2009;10(5):787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- von Eggelkraut-Gottanka R, Klose A, Beck-Sickinger AG, Beyermann M. Peptide (alpha)thioester formation using standard Fmoc-chemistry. Tetrahedron Lett. 2003;44(17):3551–3554. [Google Scholar]
- Werle M, Kafedjiiski K, Kolmar H, Bernkop-Schnurch A. Evaluation and improvement of the properties of the novel cystine-knot microprotein McoEeTI for oral administration. Int J Pharm. 2007;332(1–2):72–79. doi: 10.1016/j.ijpharm.2006.09.028. [DOI] [PubMed] [Google Scholar]
- Wong CT, Rowlands DK, Wong CH, Lo TW, Nguyen GK, Li HY, Tam JP. Orally active peptidic bradykinin B1 receptor antagonists engineered from a cyclotide scaffold for inflammatory pain treatment. Angew Chem Int Ed Engl. 2012;51(23):5620–5624. doi: 10.1002/anie.201200984. [DOI] [PubMed] [Google Scholar]
- Young TS, Young DD, Ahmad I, Louis JM, Benkovic SJ, Schultz PG. Evolution of cyclic peptide protease inhibitors. Proc Natl Acad Sci USA. 2011;108(27):11052–11056. doi: 10.1073/pnas.1108045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettler J, Schutz V, Mootz HD. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009;583(5):909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hua Z, Huang Z, Chen Q, Long Q, Craik DJ, Liao B. Two Blast-independent tools, CyPerl and CyExcel, for harvesting hundreds of novel cyclotides and analogues from plant genomes and protein databases. Planta. 2014 doi: 10.1007/s00425-014-2229-5. [DOI] [PubMed] [Google Scholar]