Abstract
One bioactive compound, identified as alternariol 9-methyl ether, was isolated from the crude extract of the endophytic fungus Alternaria sp. Samif01 residing in the roots of Salvia miltiorrhiza Bunge. Alternariol 9-methyl ether was active against bacteria with minimum inhibitory concentration values ranging from 25 to 75 μg/mL and median inhibitory concentration (IC50) values ranging from 16.00 to 38.27 μg/mL. The IC50 value of alternariol 9-methyl ether against spore germination of Magnaporthe oryzae was 87.18 μg/mL. Alternariol 9-methyl ether also showed antinematodal activity against Bursaphelenchus xylophilus and Caenorhabditis elegans with IC50 values of 98.17 μg/mL and 74.62 μg/mL, respectively. This work is the first report on alternariol 9-methyl ether and its biological activities from the endophytic fungus Alternaria sp. Samif01 derived from S. miltiorrhiza Bunge. The results indicate the potential of Alternaria sp. Samif01 as a source of alternariol 9-methyl ether and also support that alternariol 9-methyl ether is a natural compound with high potential bioactivity against microorganisms.
Keywords: Salvia miltiorrhiza Bunge; Endophytic fungus; Alternaria sp., Alternariol 9-methyl ether; Bioactivity
Introduction
Plant endophytic fungi are defined as fungi that live asymptomatically within plant tissues.1 They are rich potential resources for producing bioactive metabolites such as antimicrobial, insecticidal, anti-viral, anti-tumor and antioxidant compounds.2, 3 Some endophytic fungi have the ability to produce the same or similar bioactive compounds as the ones that originate from their host plants.4 Isolation of the endophytic fungi that produce bioactive substances has also become an efficient method to screen broad-spectrum, stable agents with low phytotoxicity.5, 6
Salvia miltiorrhiza Bunge (Lamiaceae) has been widely used in traditional oriental medicine for improving body functions, treatment of cardiovascular diseases, and liver diseases.7, 8 Some endophytic fungi isolated from S. miltiorrhiza were determined to contain tanshinones by TLC, HPLC, and LC-MS analyses,9, 10 though these tanshinone-producing endophytic fungi should be further verified.
In this work, we report the endophytic AME-producing fungus Alternaria sp. Samif01 derived from S. miltiorrhiza. Both the fungus and the AME structure were identified. The antimicrobial and antinematodal activities of AME were also evaluated to provide data supporting the development and utilization of Alternaria sp. Samif01.
Materials and methods
General
The melting point (m.p.) of the compound was determined on an XT4-100B microscopic melting-point apparatus (Tianjin Tianguang Optical Instruments Company, China) and uncorrected. NMR spectra were recorded on a Bruker Avance DRX-400 (1H at 400 MHz and 13C at 100 MHz) NMR spectrometer using tetramethylsilane (TMS) as an internal standard, and chemical shifts were recorded as δ values. A high-resolution electrospray mass spectrometry (HR-ESI-MS) spectrum was measured using a Bruker Apex IV FTMS mass spectrometer. A microplate spectrophotometer (PowerWave HT, BioTek Instruments, USA) was applied to measure light absorption values of the samples. The silica gel (200–300 mesh) for column chromatography (CC) and silica gel GF254 for thin layer chromatography (TLC) were purchased from Qingdao Marine Chemical Company, China. Sephadex LH-20 and silica gel RP-18 were obtained from Pharmacia Biotech, Sweden. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) was purchased from Amresco (USA). Streptomycin sulfate and carbendazim were purchased from Sigma–Aldrich (USA). Avemectin with a purity of 97.2% was kindly provided by Dr. Shankui Yuan at the Institute for the Control of Agrochemicals, Chinese Ministry of Agriculture. All other chemicals used in the study were of analytical grade.
Plant materials and isolation of endophytic fungi
The three-year old healthy roots of S. miltiorrhiza Bunge were collected from the Institute of Medicinal Plant Development (116°16′27″ E, 40°1′59″ N), Chinese Academy of Medical Sciences, Beijing, China, in July 2011. The plant was identified according to its morphological features by Prof. Yuhai Guo, a botanist from the College of Agronomy and Biotechnology at China Agricultural University. The voucher specimen (BSMPMI-201107001) of this plant was deposited in the Herbarium of the Institute of Chinese Medicinal Materials, China Agricultural University. The plant samples were stored in sealed plastic bags at 4 °C for processing within 24 h of collection. The isolation of endophytic fungi was performed according to previous reports11, 12 with some modifications.
The root samples were rinsed thoroughly with tap water to remove soil residues and dust, sterilized with 75% ethanol for 2 min and immersed in 0.2% mercuric chloride for 20 min, then rinsed 4 times with sterile distilled water. After surface sterilization, both root epidermis and remnant tissues were cut into small pieces of 0.5 cm × 0.5 cm and placed under aseptic conditions on potato dextrose agar (PDA) plates containing 500 μg/mL of streptomycin sulfate and incubated at 25 °C until the mycelia were apparent on PDA plates. Pure cultures were finally isolated by hyphal tip isolation on PDA plates without antibiotics and stored at 4 °C.
Morphological characterization
The morphological characteristics of the fungal isolate Samif01 were observed and described according to the methods of Ainsworth et al.,13 Photita et al.14 and Li et al.,11 including colony morphology and microscopic observation of mycelia and asexual/sexual spores.
DNA extraction, ITS-rDNA amplification and sequence analysis
Total genomic DNA of the fungal isolate Samif01 was prepared according to the protocols described by Wang et al.15 and Jasalavich et al.16 The ITS region was amplified by polymerase chain reaction (PCR) with the primer pair ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) as described previously.11, 12, 17 For identification, the PCR product was purified using the QIA quick Gel Purification Kit (Qiagen, Hilden, Germany) as described by the manufacturer's protocol and sequenced using the primer pair ITS1 and ITS4 on the ABI PRISM 3730 sequencer (Applied Biosystem, USA). Then, the sequence was run by the BLASTN program against the database (National Center for Biotechnology Information website: http://www.ncbi.nlm.nih.gov) and submitted to GenBank, where the accession number was obtained.
Mycelia suspension culture and crude extract preparation
After growing on a PDA plate at 25 °C for 5 days, two or three mycelia plugs from the actively growing colony edge of the endophytic fungus Samif01 were inoculated into 1000 mL Erlenmeyer flasks containing 300 mL potato dextrose broth (PDB). All flasks were incubated in a rotary shaker at 150 rpm and 25 °C for 15 days. Afterwards, a total of 30 L fermentation broth was harvested. The fermented broth was filtered under vacuum to separate the mycelia from the filtrate. The mycelia were dried and powdered (40.2 g), then extracted in methanol with ultrasound. The methanol solution was concentrated in vacuum at 50 °C to obtain a crude methanol extract (12.6 g), which was further thoroughly mixed with water and fractionated with ethyl acetate to obtain a concentrated ethyl acetate fraction (4.6 g). The filtrate was fractionated with ethyl acetate to afford a concentrated ethyl acetate fraction (3.0 g). As the TLC profiles of the mycelia and filtrate fractions were similar, they were combined to obtain a crude total ethyl acetate extract (7.6 g).
Separation and identification of alternariol 9-methyl ether
The crude ethyl acetate extract (7.0 g) was first subjected to chromatography over a silica gel column (200–300 mesh) eluted with petroleum ether-ethyl acetate (3:2, v/v) to obtain eight fractions (fractions A–H) using TLC detection. According to the TLC-bioautography assay of these eight fractions, fraction H presented a strong antimicrobial activity. It was selected for further separation and purification of the antimicrobial metabolites. Thus, fraction H (2.0 g) was further fractionated on a silica gel column eluted with petroleum ether-ethyl acetate (from 100:0–0:100, v/v) to give five subfractions (fractions I–V). Fraction III (0.16 g) obtained by elution with petroleum ether-ethyl acetate (100:3, v/v) was further purified by Sephadex LH-20 (CHCl3–MeOH = 1:1, v/v) and recrystallized to obtain one compound (30 mg), which was identified using physicochemical and spectrometric methods (i.e., crystal shape, m.p., HR-ESI-MS, and NMR) as well as the data in the literature.
Biological activity assay
A modified broth dilution-colorimetric assay using the chromogenic reagent 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT, purchased from Amresco, USA) was used to detect the antibacterial activity of the samples according to our previous reports.18, 19 The antimicrobial activity of the samples was evaluated by both antibacterial and antifungal activity assays. Six bacterial strains including two Gram-positive (Bacillus subtilis ATCC 11562 and Staphylococcus haemolyticus ATCC 29970) and four Gram-negative (Agrobacterium tumefaciens ATCC 11158, Pseudomonas lachrymans ATCC 11921, Ralstonia solanacearum ATCC 11696, and Xanthomonas vesicatoria ATCC 11633) bacteria were selected to detect the antibacterial activity of the samples. The minimum inhibitory concentration (MIC) value for the bacteria was defined as the lowest sample concentration that inhibits visible growth, as indicated by MTT staining. In addition, the determination of AME antifungal activity was performed by the spore germination assay.18 Spores were prepared from 7-day-old cultures of Magnaporthe oryzae according to our previous report.19 The antinematodal activity of the samples was determined according to the method of Wang et al.20 Both Bursaphelenchus xylophilus and Caenorhabditis elegans were used as test nematodes. The median inhibitory concentration (IC50) value of each sample was calculated using the linear relation between the inhibitory probability and the concentration logarithm according to the method of Sakuma.21 All of these bacterial, fungal, and nematode strains were provided by the Department of Plant Pathology of China Agricultural University.
Results and discussion
Identification of the endophytic fungus Samif01
A total of 57 endophytic fungal isolates were separated from the root epidermis and remnant tissues of S. miltiorrhiza. According to their morphological characters (e.g., the shape of conidia, type of conidiophores, growth rate, colony color and texture), 14 representative fungal isolates were selected for further bioactivity screening.22 Among these fungi, the extract isolated from the Samif01 strain by TLC-bioautography assay showed the greatest antimicrobial activity.23
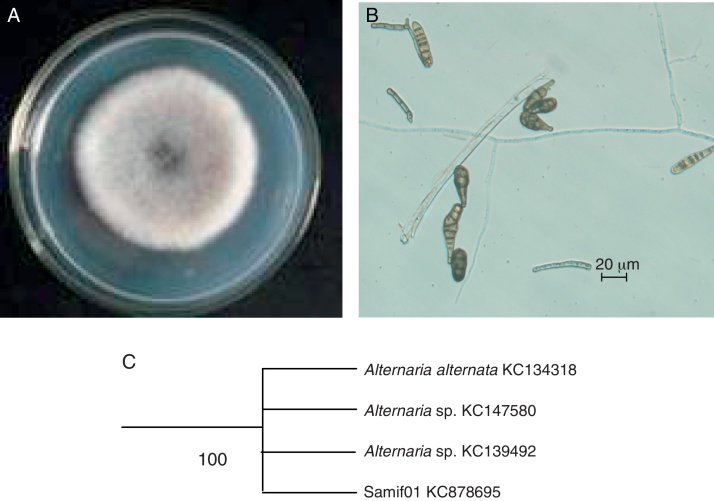
The colony of isolate Samif01 grew rapidly on a PDA plate at 25 °C, reaching 60–70 mm in diameter after 5 days of incubation. The color around the colony varied from initially white to dark green (Fig. 1A), whereas the back side of the colony displayed a yellowish color. The conidia were obclavate, obpyriform with three to seven transverse septa (commonly four), zero to three longitudinal septa, smooth, brown to dark brown (Fig. 1B). These morphological characteristics enabled the identification of isolate Samif01 as a species in the genus Alternaria. The ITS1-5.8S-ITS4 (ITS) partial sequence of isolate Samif01 was submitted to NCBI GenBank to obtain its accession number, KC878695. The sequence of the isolate Samif01 showed 100% similarity to the Alternaria sp. (KC139492), Alternaria sp. (KC147580) and Alternaria alternata (KC134318) fungi in GenBank. The phylogenetic tree of the endophytic fungus Samif01 was shown in Fig. 1C. On the basis of the ITS sequence and morphological characteristics, the fungus Samif01 was considered a member of the genus Alternaria and identified as Alternaria sp. Samif01.11, 13, 14
Fig. 1.
The colony front view (A), conidiospores (B) and phylogenetic tree (C) of the endophytic fungus Alternaria sp. Samif01 from S. miltiorrhiza.
Structural identification of alternariol 9-methyl ether
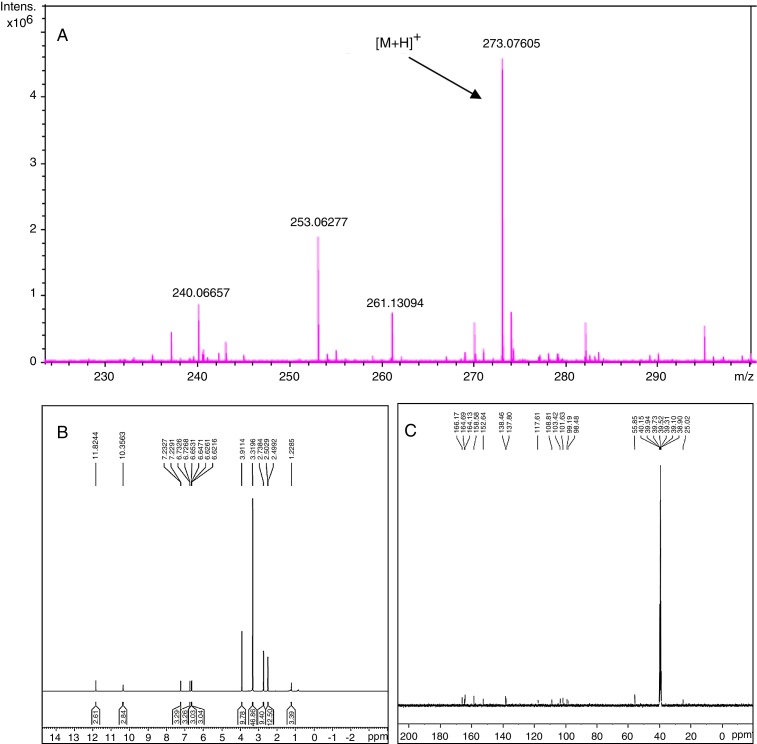
A very effective antimicrobial compound was successfully obtained from the ethyl acetate fraction of the crude methanol extract of the endophytic fungus Samif01 based on bioassay-guided fractionation. Its characterization data were given as follows: yellowish needle-like crystals (acetone); m.p. 267–270 °C. The molecular formula was assigned as C15H12O5 by HR-ESI-MS (m/z 273.0761 [M+H]+; calculated for C15H13O5 as 273.0758) shown in Fig. 2A, which just matched the results for alternariol 9-methyl ether previously reported in the literature.24 1H NMR (400 MHz, DMSO-d6, Fig. 2B) δ (ppm), 2.74 (3H, s, CH3-1), 6.73 (1H, d, J = 2.3 Hz, H-2), 10.36 (1H, s, OH-3), 6.65 (1H, d, J = 2.4 Hz, H-4), 11.82(1H, s, OH-7), 6.62(1H, d, J = 1.8 Hz, H-8), 3.91 (3H, s, CH3O-9), 7.23(1H, d, J = 1.6 Hz, H-10); 13C NMR (100 MHz, DMSO-d6, Fig. 2C) δ (ppm), 138.46 (C-1), 25.02 (CH3-1), 117.61 (C-2), 158.58 (C-3), 101.63 (C-4), 152.64 (C-4a), 164.69 (C-6), 98.48 (C-6a), 164.13 (C-7), 99.19 (C-8), 166.17 (C-9), 55.85 (CH3O-9), 103.42 (C-10), 137.80 (C-10a), 108.81 (C-10b). After comparing the data with reports in the literature,19, 24 the compound was identified as alternariol 9-methyl ether (also called AME, djalonensone, and 3,7-dihydroxy-9-methoxy-1-methyl-6H-dibenzo [b,d] pyran-6-one), and the structure is shown in Fig. 3.
Fig. 2.
HR-ESI-MS spectrum (A), 1H NMR spectrum (B) at 400 MHz and 13C NMR spectrum (C) at 100 MHz of alternariol 9-methyl ether in DMSO-d6. The chemical shifts are shown in ppm with TMS as an internal standard.
Fig. 3.
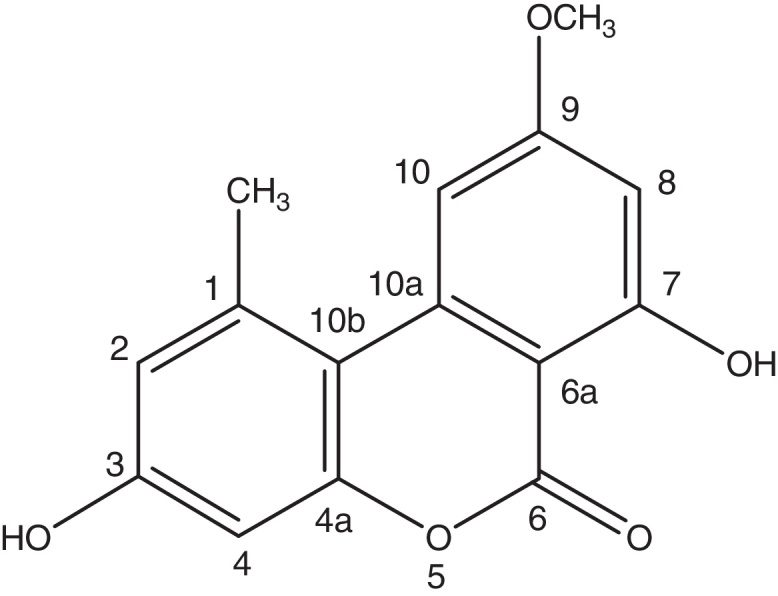
Chemical structure of alternariol 9-methyl ether.
Alternariol 9-methyl ether (AME) is a dibenzo-α-pyrone analog. It was reported previously as a mycotoxin produced by the phytopathogenic fungi in the genus Alternaria.25 Recently, it has been isolated from a series of endophytic fungi, mainly including Alternaria fungi, such as Alternaria sp. from Polygonum senegalense,24 Alternaria brassicicola ML-P08 from Malus halliana,26 Alternaria sp. from Datura stramonium,27 and Alternaria sp. N.SBA10 from Scutellaria baicalensis.28 Other AME-producing endophytic fungi include Cephalosporium acremonium IFB-E007 from Trachelospermum jasminoides,29 Hyalodendriella sp. Ponipodef12 from the hybrid ‘Neva” of Populus deltoides Marsh x P. nigra L.,19 Nigrospora sphaerica from the lichen Parmelinella wallichiana,30 and Phialophora sp. from the lichen Cetrelia braunsiana.30
Bioactivity of alternariol 9-methyl ether
The antimicrobial activity of alternariol 9-methyl ether (AME) was evaluated by broth-dilution-colorimetric and spore germination assays, with the results shown in Table 1. AME was found to display antibacterial activity toward six different bacteria strains with IC50 values varying from 16.00 to 38.27 μg/mL. AME showed stronger antibacterial activity against R. solanacearum than the conventional antibiotic streptomycin sulfate. This result indicates that the R. solanacearum strain might be a streptomycin-resistant strain. The IC50 value of AME against spore germination of M. oryzae was 87.18 μg/mL. AME also showed antinematodal activity against C. elegans and B. xylophilus with IC50 values as 74.62 μg/mL and 98.17 μg/mL, respectively.
Table 1.
Biological activities of alternariol 9-methyl ether.
| Test organism | Alternariol 9-methyl ether (μg/mL) |
CK+ (μg/mL) |
||
|---|---|---|---|---|
| MIC | IC50 | MIC | IC50 | |
| A. tumefaciens | 75.0 | 38.07 ± 2.78 | 25.0 | 11.96 ± 1.25 |
| B. subtilis | 50.0 | 22.83 ± 1.06 | 37.5 | 20.58 ± 0.88 |
| P. lachrymans | 25.0 | 16.00 ± 0.30 | 25.0 | 4.69 ± 0.79 |
| R. solanacearum | 75.0 | 38.27 ± 1.80 | 100.0 | 41.26 ± 2.41 |
| S. haemolyticus | 37.5 | 19.90 ± 1.62 | 12.5 | 4.24 ± 0.94 |
| X. vesicatoria | 75.0 | 31.99 ± 1.34 | 50.0 | 22.83 ± 1.37 |
| M. oryzae | nd | 87.18 ± 1.39 | nd | 6.25 ± 0.19 |
| B. xylophilus | nd | 74.62 ± 0.62 | nd | 12.90 ± 0.27 |
| C. elegans | nd | 98.17 ± 0.92 | nd | 14.00 ± 0.19 |
Note: The positive controls (CK+) for bacteria, fungus and nematodes were streptomycin sulfate, carbendazim and avermectin, respectively. The ‘nd’ means not detected.
Previous reports showed that AME exhibited strong antifungal activity against Microbotryum violacerum,31 induced mitochondrial apoptosis in human colon carcinoma cells,32 and exhibited DNA strand breaks, micronuclei, and gene mutations in various cultured mammalian cells.33 The results in this work further supported the antimicrobial activity of AME. However, the mechanisms of action of AME need to be further studied. These findings have attracted the attention of many researchers all over the world regarding the synthesis and applications of AME.25
Conclusion
This work is the first report of the AME-producing endophytic fungus Alternaria sp. Samif01 from the medicinal plant S. miltiorrhiza. In this study, AME was found to be active against bacteria, fungus and nematodes. The results indicate the potential of Alternaria sp. Samif01 as a source of AME and also support that AME is a natural compound with strong antimicrobial activity.
The remarkable antimicrobial and antinematodal activity of AME suggests that endophytic fungus such as Alternaira sp. Samif01 could protect S. miltiorrhiza plants by producing bioactive metabolites that are toxic or even lethal to phytopathogens. From TLC-bioautography examination, additional antimicrobial compounds have not yet been isolated from Alternaria sp. Samif01. The separation and purification of these compounds are still in progress. In addition, the phylogenetic identification of the Samif01 strain requires complementary investigation.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by grants from the National Basic Research Program of China (2013CB127805) and the Hi-Tech R&D Program of China (2011AA10A202).
Associate Editor: Eleni Gomes
References
- 1.Saikkonen K., Faeth S.H., Helander M., Sullivan T.J. Fungal endophytes: a continuum of interactions with host plants. Annu Rev Ecol Syst. 1998;29:319–343. [Google Scholar]
- 2.Strobel G.A. Endophytes as sources of bioactive products. Microbes Infect. 2003;5:535–544. doi: 10.1016/s1286-4579(03)00073-x. [DOI] [PubMed] [Google Scholar]
- 3.Kharwar R.N., Mishra A., Gond S.K., Stierle A., Stierle D. Anticancer compounds derived from fungal endophytes: their importance and future challenges. Nat Prod Rep. 2011;28:1208–1228. doi: 10.1039/c1np00008j. [DOI] [PubMed] [Google Scholar]
- 4.Zhao J., Shan T., Mou Y., Zhou L. Plant-derived bioactive compounds produced by endophytic fungi. Mini Rev Med Chem. 2011;11:159–168. doi: 10.2174/138955711794519492. [DOI] [PubMed] [Google Scholar]
- 5.Gimenez C., Cabrera R., Reina M., Gonzalez-Coloma A. Fungal endophytes and their role in plant protection. Curr Org Chem. 2007;11:707–720. [Google Scholar]
- 6.Vaz A.B.M., Brandao L.R., Vieira M.L.A. Diversity and antimicrobial activity of fungal endophyte communities associated with plants of Brazilian savanna ecosystems. Afr J Microbiol Res. 2012;6:3173–3185. [Google Scholar]
- 7.Wang B.Q. Salvia miltiorrhiza: chemical and pharmacological review of a medicinal plant. J Med Plants Res. 2010;4:2813–2820. [Google Scholar]
- 8.Wu Y.B., Ni Z.Y., Shi Q.W. Constituents from Salvia species and their biological activities. Chem Rev. 2012;112:5967–6026. doi: 10.1021/cr200058f. [DOI] [PubMed] [Google Scholar]
- 9.Wei X.Y., Jing M.B., Wang J.C., Yang X.J. Preliminary study on Salvia miltiorrhiza Bunge endophytic fungi. Acad J Xian Jiaotong Univ. 2010;22:241–246. [Google Scholar]
- 10.Ming Q., Han T., Li W. Tanshinone IIA and tanshinone I production by Trichoderma atroviride D16, an endophytic fungus in Salvia miltiorrhiza. Phytomedicine. 2012;19:330–333. doi: 10.1016/j.phymed.2011.09.076. [DOI] [PubMed] [Google Scholar]
- 11.Li J., Zhao J., Xu L., Zhou L., Li X., Wang J. Endophytic fungi from rhizomes of Paris polyphylla var. yunnanensis. World J Microbiol Biotechnol. 2008;24:733–737. [Google Scholar]
- 12.Xu L., Zhou L., Zhao J., Li J., Li X., Wang J. Fungal endophytes from Dioscorea zingiberensis rhizomes and their antibacterial activity. Lett Appl Microbiol. 2008;46:68–72. doi: 10.1111/j.1472-765X.2007.02264.x. [DOI] [PubMed] [Google Scholar]
- 13.Ainsworth G.C., Sparrow F.K., Sussman A.S. Academic Press; New York: 1973. The Fungi: An Advanced Treatise. Edition Vol IV(A), A Taxonomic Review with Keys Ascomycetes and Fungi Imperfecti. [Google Scholar]
- 14.Photita W., Taylor P.W.J., Ford R., Hyde K.D., Lumyong S. Morphological and molecular characterization of Colletotrichum species from herbaceous plants in Thailand. Fungal Divers. 2005;18:117–133. [Google Scholar]
- 15.Wang H., Qi M., Cutler A.J. A simple method of preparing plant samples for PCR. Nucleic Acids Res. 1993;21:4153–4154. doi: 10.1093/nar/21.17.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jasalavich C.A., Ostrofsky A., Jellison J. Detection and identification of decay fungi in spruce wood by restriction fragment length polymorphism analysis of amplified genes encoding rRNA. Appl Environ Microbiol. 2000;66:4725–4734. doi: 10.1128/aem.66.11.4725-4734.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong L., Zhou Y., Gao S. Endophytic fungi from the hybrid ‘Neva’ of Populus deltoides Marsh x Populus nigra L. and their antimicrobial activity. Afr J Microbiol Res. 2011;5:3924–3929. [Google Scholar]
- 18.Liu H., Mou Y., Zhao J. Flavonoids from Halostachys caspica and their antimicrobial and antioxidant activities. Molecules. 2010;15:7933–7945. doi: 10.3390/molecules15117933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng X., Mao Z., Lou J. Benzopyranones from the endophytic fungus Hyalodendriella sp. Ponipodef12 and their bioactivities. Molecules. 2012;17:11303–11314. doi: 10.3390/molecules171011303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Luo C., Liu H., Xu J., Sun W., Zhou L. Nematicidal activity of the alkaloids from Macleaya cordata against certain nematodes. Afr J Agric Res. 2012;7:5925–5929. [Google Scholar]
- 21.Sakuma M. Probit analysis of preference data. Appl Entomol Zool (Jpn) 1998;33:339–347. [Google Scholar]
- 22.Lou J., Fu L., Luo R., Wang X., Luo H., Zhou L. Endophytic fungi from medicinal herb Salvia miltiorrhiza Bunge and their antimicrobial activity. Afr J Microbiol Res. 2013;7:5343–5349. [Google Scholar]
- 23.Zhao J., Xu L., Huang Y., Zhou L. Detection of antimicrobial components from extracts of the endophytic fungi associated with Paris polyphylla var. yunnanensis using TLC-bioautography-MTT assay. Nat Prod Res Dev. 2008;20:28–32. [Google Scholar]
- 24.Aly A.H., Edrada-Ebel R., Indriani I.D. Cytotoxic metabolites from the fungal endophyte Alternaria sp. and their subsequent detection in its host plant Polygonum senegalense. J Nat Prod. 2008;71:972–980. doi: 10.1021/np070447m. [DOI] [PubMed] [Google Scholar]
- 25.Lou J., Fu L., Peng Y., Zhou L. Metabolites from Alternaria fungi and their bioactivities. Molecules. 2013;18:5891–5935. doi: 10.3390/molecules18055891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu W. Bioactive metabolites from Alternaria brassicicola ML-P08, an endophytic fungus residing in Malus halliana. World J Microbiol Biotechnol. 2009;25:1677–1683. [Google Scholar]
- 27.Sun J., Awakawa T., Noguchi H., Abe I. Induced production of mycotoxins in an endophytic fungus from the medicinal plant Datura stramonium L. Bioorg Med Chem Lett. 2012;22:6397–6400. doi: 10.1016/j.bmcl.2012.08.063. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Z.H., Liu Y.Y., Fan L. Antioxidant activity and structure identification of metabolites of an endophytic fungus Alternaria sp. N.SBA10 isolated from Scutellaria baicalensis. Mycosystema. 2012;31:917–923. [Google Scholar]
- 29.Zhang H.W., Huang W.Y., Song Y.C., Chen J.R., Tan R.X. Four 6H-dibenzo [b,d] pyran-6-one derivatives produced by the endophyte Cephalosporium aceremonium IFB-E007. Helv Chim Acta. 2005;88:2861–2864. [Google Scholar]
- 30.He J.W., Chen G.D., Gao H. Heptaketides with antiviral activity from three endolichenic fungal strains Nigrospora sp., Alternaria sp. and Phialophora sp. Fitoterapia. 2012;83:1087–1091. doi: 10.1016/j.fitote.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Hussain H., Krohn K., Ullah Z., Draeger S., Schulz B. Bioactive chemical constituents of two endophytic fungi. Biochem Syst Ecol. 2007;35:898–900. [Google Scholar]
- 32.Bensassi F., Gallerne C., el Dein O.S., Hajlaoui M.R., Bacha H., Lemaire C. Mechanism of alternariol monomethyl ether-induced mitochondrial apoptosis in human colon carcinoma cells. Toxicology. 2011;290:230–240. doi: 10.1016/j.tox.2011.09.087. [DOI] [PubMed] [Google Scholar]
- 33.Pfeiffer E., Eschbach S., Metzler M. Alternaria toxins: DNA strand-breaking activity in mammalian cellsin vitro. Mycotoxin Res. 2007;23:152–157. doi: 10.1007/BF02951512. [DOI] [PubMed] [Google Scholar]