Abstract
We used molecular dynamics simulations to explore the effects of asymmetric transbilayer distribution of anionic phosphatidylserine (PS) lipids on the structure of a protein on the membrane surface and subsequent protein–lipid interactions. Our simulation systems consisted of an amyloidogenic, beta-sheet rich dimeric protein (D42) absorbed to the phosphatidylcholine (PC) leaflet, or protein-contact PC leaflet, of two membrane systems: a single-component PC bilayer and double PC/PS bilayers. The latter comprised of a stable but asymmetric transbilayer distribution of PS in the presence of counterions, with a 1-component PC leaflet coupled to a 1-component PS leaflet in each bilayer. The maximally asymmetric PC/PS bilayer had a non-zero transmembrane potential (TMP) difference and higher lipid order packing, whereas the symmetric PC bilayer had a zero TMP difference and lower lipid order packing under physiologically relevant conditions. Analysis of the adsorbed protein structures revealed weaker protein binding, more folding in the N-terminal domain, more aggregation of the N- and C-terminal domains and larger tilt angle of D42 on the PC leaflet surface of the PC/PS bilayer versus the PC bilayer. Also, analysis of protein-induced membrane structural disruption revealed more localized bilayer thinning in the PC/PS versus PC bilayer. Although the electric field profile in the non-protein-contact PS leaflet of the PC/PS bilayer differed significantly from that in the non-protein-contact PC leaflet of the PC bilayer, no significant difference in the electric field profile in the protein-contact PC leaflet of either bilayer was evident. We speculate that lipid packing has a larger effect on the surface adsorbed protein structure than the electric field for a maximally asymmetric PC/PS bilayer. Our results support the mechanism that the higher lipid packing in a lipid leaflet promotes stronger protein–protein but weaker protein–lipid interactions for a dimeric protein on membrane surfaces.
Keywords: Molecular dynamics simulations, Asymmetric lipid membrane, Protein structures on surfaces, Protein–lipid interactions, Protein aggregation, Beta-amyloid
1. Introduction
Protein structural transitions on cell membrane surfaces are important molecular events in regulating normal and pathogenic cellular processes (Gething and Sambrook, 1992; Bucciantini and Cecchi, 2010). Upon binding to a surface, a protein undergoes surface-induced and localized secondary structural changes, such as folding or unfolding of ordered helices or beta-sheets in different domains. On the membrane surface, these folded or unfolded domains may further interact and undergo complex higher order tertiary and quaternary structural changes, hydrodynamic shape alterations, and intra- or inter-peptide domain aggregations (Vymetal and Vondrasek, 2011; Stefani, 2012). Membrane-orientational transitions of protein, such as tilted or non-tilted conformation of certain motifs, may also occur (Basyn et al., 2001) on the surface. All of these altered protein structures may provide the necessary functional motifs for receptor-based binding, such as cell signaling and immunological responses (Martin and Hartl, 1997; Kurre et al., 2001; Kanekiyo et al., 2014). For an intrinsically disordered and aggregation-prone, or amyloidogenic, protein, the altered protein structure on the surface may trigger irreversible self-aggregation in several membrane-associated cytotoxic aggregation cascade pathways, e.g., beta-amyloid cascade in Alzheimer's pathogenesis (Ross and Poirier, 2004; Ellis, 2006). At present, the role of transbilayer lipid distribution in regulating the above protein transitional events of the amyloidogenic protein is unclear.
It is commonly thought that lipids are not randomly distributed but form compositionally distinct domains along the normal of the planar lipid membrane. These lipid domains are commonly known as asymmetric transbilayer lipid domains (van Meer et al., 2008). Various asymmetric transbilayer lipid distributions, resulting from two different kinds of lipids in a bilayer, can elicit very different properties than those of symmetric one-component lipid distribution. Examples of asymmetric lipid distributions include zwitterionic phosphatidylcholine (PC) lipids in one leaflet and charged, anionic or cationic, lipids in the other leaflet. Such lipid arrangements represent the maximally asymmetric transbilayer distribution possible in a biological membrane. This highly asymmetric lipid arrangement may have different lipid order packing and transmembrane potential distribution (Gurtovenko and Vattulainen, 2008) when compared with the symmetric bilayer. Among the various key lipids types, anionic phosphatidylserine (PS) is of interest because of its distinct roles in cell surface signaling. PS can trigger apoptosis, protein trafficking, and immunological responses, in various cells (Williamson and Schlegel, 1994; Fadok et al., 2001; Yeung et al., 2008; Fadeel and Xue, 2009; Lingwood and Simons, 2010). Here, two bilayer systems: an asymmetric double bilayer PC/PS bilayer system consisting of a PC monolayer coupled to a PS monolayer in each bilayer and a symmetric, one-component PC bilayer control, were investigated.
For the maximally asymmetric PC/PS double bilayer system, the asymmetric charge arrangement of the PS lipids in the presence of counterions has been shown to create a non-zero transmembrane potential (TMP) difference of approximately −240 mV measured from the PS side towards the PC side of the bilayer (Gurtovenko and Vattulainen, 2008). This is the same sign as the local TMP difference found in cells. In contrast, no TMP difference occurs in the control PC bilayer system. Interestingly, the large difference in the melting temperatures of PC and PS lipids in one-component bilayers implies a higher order packing of the PC monolayer in the highly coupled PC/PS bilayer than in the PC bilayer. How the TMP and order packing lipid properties affect the structures of an amyloidogenic protein dimer, a simplest model multimer, is unclear.
A beta-sheet rich beta-amyloid dimer (Luhrs et al., 2005) was chosen as our model amyloidogenic protein multimer in this study. Recent atomic force microscopy studies (Jang et al., 2010; Jang et al., 2013) have indicated that amyloidogenic beta-amyloid multimers exhibit beta-sheet rich fibril morphology. This fibril structure has a U-shaped beta-strand-loop-beta-strand motif with an exposed hydrophobic surface in the C-terminal domain that interacts with the cell membranes. Recent molecular dynamics (MD) simulations (Jang et al., 2010; Poojari and Strodel, 2013; Nasica-Labouze et al., 2015) further established the capability of these fibrillar beta-amyloid multimers to form pores in a simple one-component lipid bilayer. At present, the knowledge of early events of protein structural transitions and protein–lipid interactions of any beta-amyloid multimers on asymmetric lipid surfaces that may lead to membrane pore formation is unclear.
We used molecular dynamics (MD) simulations to investigate the effects of asymmetric transbilayer lipid distribution on the structure of D42 on the membrane surface of PC/PS and PC bilayers. In addition, the effects of the absorbed protein on the bilayer structure of the PC/PS and PC systems were compared. Our goal was to elucidate the lipid–protein interaction mechanisms of how the transbilayer lipid asymmetry regulates the complex protein structural transitions and the membrane structural disruption behaviors of an amyloidogenic protein on membrane surfaces that may lead to membrane pore formation.
2. Materials and methods
2.1. Starting structures
2.1.1. Protein
A beta-sheet rich, fibril-like beta-amyloid dimer (D42) was used. This protein has two chains (A and B), both with 42 residues and identical sequence (Hardy and Higgins, 1992) as given below.
H–ASP–ALA–GLU–PHE–(ARG-5)–HIS–SER–GLY–TYR–GLU–VAL–HIS–HIS–GLN–(LYS-16)–LEU–VAL–PHE–PHE–ALA–GLU–ASP–VAL–GLY–SER–ASN–(LYS-28)–GLY–ALA–ILE–ILE–GLY–LEU-–MET–VAL–GLY–GLY–VAL–VAL–ILE–ALA–OH
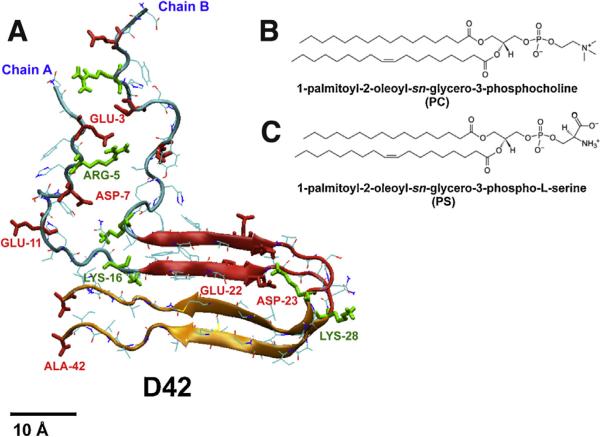
At neutral pH, the sequence has 7 negative charges (ASP-1, GLU-3, ASP-7, GLU-11, GLU-22, ASP-23 and C-terminal end carboxylate) and 4 positive charges (N-terminal end amine, ARG-5, LYS-16 and LYS-28). Hence, D42 (chain A and chain B) has a net charge of −6. Each chain has a distinct N-terminal domain and a C-terminal domain. Therefore, D42 has a total of four structural domains: N-terminal domain of chain A, N-terminal domain of chain B, C-terminal domain of chain A, and C-terminal domain of chain B. As shown in Fig. 1A, each N-terminal domain has an unstructured peptide fragment (residues 1–17) exhibiting random coil structure. In contrast, each C-terminal domain has a more structured U-shaped motif (residues 18–42) with a beta-strand-loop-beta-strand-coil structure, and its sequence is highlighted above in bold.
Fig. 1. Structures of protein and lipids.
(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) The initial protein structure of D42 (A) consists of two 42-residue long peptide chains, chain A and chain B, with identical sequence (see Section 2). Each chain has a random-coil N-terminal domain (residues 1–17) and an U-shaped C-terminal domain (residues 18–42). The latter contains a compact beta-strand-to-loop-to-beta-strand-random coil (residues 18–28 in red and residues 29–42 in orange) motif. Four positively charged residues (N-terminal end amine, ARG-5, LYS-16 and LYS-28) and seven negatively charged residues (ASP–1, GLU–3, ASP–7, GLU–11, GLU–22, ASP–23 and C-terminal end carboxylate) in each chain are rendered in thick green and red licorice representations, respectively, and the non-charged residues are in thin color lines. A scale bar of 10 Å is shown. The chemical structures of the lipids, PC (B) and PS (C), are given. Here PC is zwitterionic and PS anionic, according to the charge distribution in the polar headgroup of each lipid.
To construct the D42 dimer, the atomic coordinates of the C-terminal domains were extracted from the experimental NMR fibrillar structure (PDB file: 2BEG) of a beta-amyloid pentamer (Luhrs et al., 2005). The atomic coordinates of the unstructured N-terminal domain were obtained from the same N-terminal domain region (residues 1–16) of the experimental NMR structure (PDB file: 1BA4) of a beta-amyloid monomer in a water/micelle environment (Coles et al., 1998). The four parallel beta-strands in the C-terminal domain are stabilized by the parallel inter-chain hydrogen bonds along the peptide backbones. A region of the C-terminal domain (LYS-28 to ALA-42), marked in orange in Fig.1A, is beta-sheet rich and has a hydrophobic sub-sequence (residues 29–41). This region is defined as the lipid insertion domain. However, in our recent studies on monomeric beta-amyloid peptide in various lipid bilayers, the lipid insertion domain was alpha-helix rich (Qiu et al., 2014; Cheng et al., 2015c).
2.1.2. Lipid
Two phospholipids: 1-palmitoyl-2-oleoyl-sn-glycero-phospho-choline (PC) and 1-palmitoyl-2-oleoyl-sn-glycero-phospho-l-serine (PS) were used. PC (Fig. 1B) has a zwitterionic polar headgroup, a glycerol backbone, and two acyl chains. PS (Fig.1C) has an anionic polar headgroup, a glycerol backbone, and two acyl chains. The chemical compositions of the acyl chains (C16:0 in sn-1 and C18:1 in sn-2) of PC and PS are identical. Here, we focus on the spatial distribution of the phosphate group of PC (PO4-PC), phosphate group of PS (PO4-PS), and the orientational order of the carbon atoms of the acyl chains of either PC or PS with respect to the normal of the lipid bilayer.
2.1.3. Protein/lipid bilayer systems
We have created two initial simulation structures of protein/lipid bilayer systems using the previously published, pre-equilibrated symmetric PC and asymmetric PC/PS lipid bilayer systems (Gurtovenko and Vattulainen, 2008; Qiu et al., 2011). The initial structures consisted of the D42 near the PC surface of the PC bilayer (Fig. 2A) and of the D42 near the PC surface of one of the PC/PS bilayers (Fig. 2C) in the presence of water and counterions. The steps used to construct these initial simulation structures of protein/lipid bilayer systems are given below.
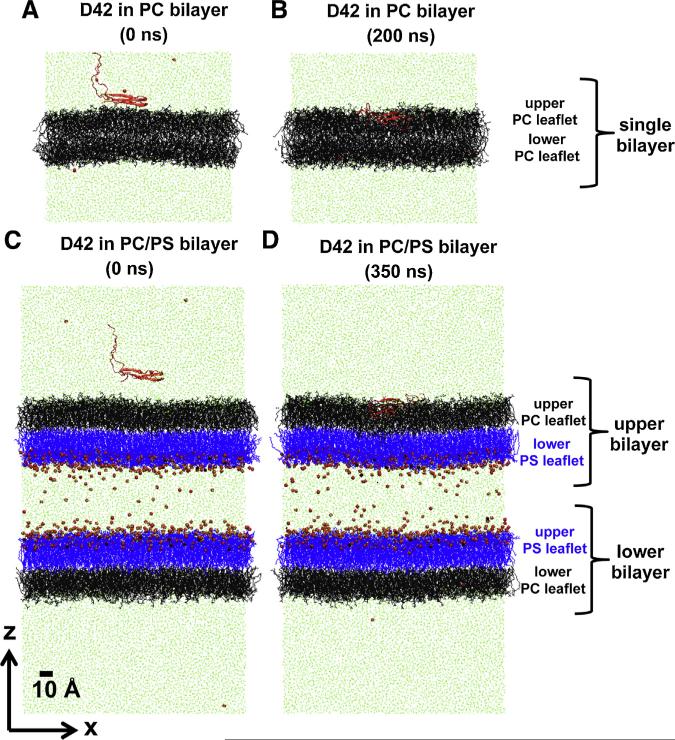
Fig. 2. Protein/lipid bilayer systems before and after simulation.
(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Representative initial (A, C) and final (B, D) simulated structures of protein/lipid bilayer systems containing PC (A, B) and PC/PS (C, D) lipid bilayers in the transverse-view (x–z plane view with z along the normal of the lipid bilayer) are shown at indicated simulation times. For clarity, a thin x–z slice with a thickness of Δy ≈ 5 nm of water/ion solution is presented to illustrate the total system size of each system. PC and PS lipids are in black and blue lines, respectively. Sodium ion and water molecules captured in the thin x–z slice are in small red spheres and light green lines, respectively. The protein D42 is shown in red ribbons. The definition of upper or lower lipid leaflet, and single, upper or lower bilayer is given. The upper PC leaflet of either the single PC bilayer (B) or the upper PC/PS bilayer (D) is defined as the protein-contact layer of this study. The scale bar indicates 10 Å.
First, two lipid bilayer systems, PC and PC/PS, were constructed in the absence of protein. A pre-equilibrated PC lipid bilayer system (see Fig. 2A) was obtained from a 200 ns-MD-simulated structure published previously (Qiu et al., 2011). For the PC/PS lipid bilayer system, an asymmetric PC/PS double-bilayer structure consisting of two asymmetrically orientated bilayers, i.e., two PC/PS bilayers separated by a slab of counterions and water, was used (see Fig. 2C). Each PC/PS bilayer comprised of a single-component PC leaflet coupled to a single-component PS leaflet. The two PS leaflets were facing the counterions and water slab. This PC/PS lipid bilayer system was constructed from a smaller 100 ns-MD-simulated asymmetric PC/PS double-bilayer structure (Gurtovenko and Vattulainen, 2008) using a self-replicating method. Essentially, the smaller system was replicated along the x- and y-directions, or along the plane of the bilayer, 3 times. The resulting 9 times larger asymmetric system was then simulated for 500 ns using the published simulation procedure (Gurtovenko and Vattulainen, 2008) to relax the replicated structure to produce the final pre-equilibrated PC/PS lipid bilayer system.
Second, the protein D42 was added to the pre-equilibrated lipid bilayer system. The initial membrane orientation of the protein was that the long axis of the U-shaped C-terminal domain (Fig. 1) was parallel to the plane of the membrane surface with the hydrophobic region (residues 29–41) directly facing the membrane surface. The methods of incorporating D42 to the bilayer systems are shown below.
For the symmetric PC bilayer system, D42 was placed near the surface of the lipid bilayer, the water molecules that overlapped the protein were then removed, and counterions were added to the aqueous phase. For the asymmetric PC/PS bilayer system, multiple steps were performed. First, D42 was solvated in a separated water box with counterions of the same lateral dimensions as the PC/PS bilayer system to form a pre-built D42/water/ion box. Second, water above the upper bilayer of the pre-equilibrated solvated PC/PS lipid bilayer system was removed so that only a thin layer of interfacial water of a few water molecules thick remained on the top PC lipid leaflet of the upper bilayer. The interfacial water layer covered all the headgroups of the PC lipids. Third, the pre-built D42/water/ion box from the first step was placed near the trimmed PC/PS lipid bilayer system from the second step. Finally, an extra box of randomized water was added to the combined system from the previous step. The separated protein hydration step was necessary to maintain the stability of the complex and fragile PC/PS lipid bilayer system. For both systems, the center of mass of the C-terminal domains of D42 was initially placed within 1 nm from the surface of the PC leaflet, also defined as the protein-contact PC-leaflet.
Both bilayer systems with D42 near the protein-contact PC-leaflet were energy minimized. Thereafter, each bilayer system was subjected to a few nanosecond of protein-restrained pre-production MD simulation run. During this pre-production run, all the atoms of D42 were position-restrained in the presence of 6 counterions in the water phase (Qiu et al., 2011). The water and lipid bilayer were free to move around the position-restrained protein during this pre-production run. The protein-restrained MD pre-production MD run was very critical to allow the solvent to relax properly around the protein after the construction and to improve the stability of the unrestrained MD production run of the system. During the protein-restrained pre-production MD simulation run, the protein-contact PC surface drifted ~2–3 nm away from the protein in the PC/PS bilayer system but not in the PC bilayer as illustrated in Fig. 2A and C, respectively. After the pre-production run, an unconstrained full-production MD run was performed on each bilayer system.
The global compositions of the lipid and water molecules in our systems are: 576 PC, and 65,854 water in the PC bilayer system, and 954 PC, 1152 PS, and 235,900 water in the PC/PS bilayer system. The number of Na+ counterions in the PC bilayer system is 6 to balance the −6e charge of D42. The number of Na+ counts ions in the PC/PS bilayer is 1158, which balances the charge of each −1e charged PS lipid and the −6e charged D42. The system or simulation box, sizes along the x-, y- and z-directions were ~13 × 14 × 14 nm3 for the PC bilayer system and 17 × 17 × 33 nm3 for the PC/PS bilayer system (see Fig. 2). The z-direction was aligned along the normal of the bilayer. Note that the z-dimension of the PC/PS bilayer system was more than twice the z-dimension of the PC bilayer system due to the presence of two lipid bilayers and the extra water/ion slab between them. Therefore, the symmetric PC bilayer system has two PC leaflets and the asymmetric PC/PS bilayer system has two PC leaflets and two PS leaflets. Our design rationale was that D42 was placed on the top, or protein-contact PC, leaflet of either the PC bilayer or the PC/PS bilayer system (Fig. 2). In the above initial configuration design, the protein D42 was facing the 1-component PC leaflet in both bilayer systems.
The design of the above protein/bilayer systems allowed us to examine the effects of transbilayer lipid head group distribution asymmetry on protein–lipid interactions in well-characterized lipid and aqueous environments. In both bilayer systems, the net −6e charged D42 experienced the same chemical composition of the lipid monolayer, i.e., a single-component PC leaflet, and the same aqueous environment with +6 counterions in both PC/PS and PC bilayer systems. The middle counterion and water slab facing the two PS leaflets served the purpose to balance the large negative charge of PS headgroups, but did not come into contact with the protein. Therefore, the protein was affected solely by the biophysical properties of the contacting single-component PC leaet in both bilayers. The differences in the biophysical properties of the PC/PS and PC bilayers were due to the transbilayer lipid head group distribution asymmetry in the design, since the composition of the contacting lipid leaflet and number of counterions surrounding the protein were identical in each bilayer system.
The structure and binding behavior of beta-amyloid protein on membrane surface are sensitive to the ionic strength of the medium (Nasica-Labouze et al., 2015). In this work, we have not included additional salt in our simulation. This is because we would like to focus on key interactions involving lipid and protein. This approach of not including salt in protein/lipid interaction simulation is well accepted in various studies using either united atom or all-atom force fields (Herce and Garcia, 2007; Dunkin et al., 2011; Hu et al., 2013; Huang and Garcia, 2013; Pourmousa and Karttunen, 2013; Park et al., 2015).
2.2. Molecular dynamics simulations
Molecular dynamics (MD) simulations on the protein/lipid bilayer systems were performed at a constant temperature (310 K) and a constant 1 atm pressure using procedures published previously (Gurtovenko and Vattulainen, 2008; Qiu et al., 2011). To improve phase space sampling of our simulations, 4 independent simulation replicates, each with a different initial velocity distribution but with identical initial coordinates, were generated for each bilayer system. The procedures of the MD simulations have been published elsewhere (Gurtovenko and Vattulainen, 2008; Qiu et al., 2011). Briey, molecular dynamics simulations were performed under NPT conditions using Gromacs 4.5.3 (Bekker et al., 1993; Berendsen et al., 1995; Lindahl et al., 2001; Van Der Spoel et al., 2005) with Berger et al. (1997) and Holtje et al. (2001) lipid parameters and a modified GROMOS87 force field (van Buuren et al., 1993; van der Spoel et al., 1996) for both lipid and protein, called GROMOS87/Berger lipid force field. Periodic boundary conditions were applied on each x,y and z direction. A simple point charge (SPC) model (Berendsen et al., 1981) was used for water. The non-bonded van der Waals interactions were estimated using a twin-range cutoff Lennard-Jones potential (Berendsen et al., 1995) with interactions within 1.0 nm evaluated every step and interactions between 1.0 and 1.5 nm evaluated every 10 steps. Electrostatic interactions were estimated by Particle-Mesh-Ewald method (Darden et al., 1993; Essmann et al., 1995). Bond lengths were constrained by a linear constraint solver (LINCS) algorithm (Hess et al., 1997). A leapfrog integrator with a 2 fs time step was used to integrate the motion of the systems. Temperature baths of 310 K were coupled to water, lipids and protein separately, using a v-rescale thermostat (Bussi et al., 2007) with a coupling time of 0.05 ps. A Berendsen barostat (Berendsen et al., 1984) with a coupling time of 1 ps kept the systems at an isotropic pressure of 1 atm.
For the PC bilayer, a 200 ns-simulation per replicate was performed (Fig. 2B). For the larger PC/PS double-bilayer system, a 350 ns-simulation per replicate was performed (Fig. 2D). With four replicates for each bilayer system, a total of 2.2 μs of MD simulations were performed in this study.
2.3. Data analysis
2.3.1. Protein secondary, tertiary and quaternary structure
The protein secondary structure of D42 was determined by a DSSP algorithm (Kabsch and Sander, 1983) frame-by-frame for each simulation replicate. Here, 6 hydrogen-bonded structures: 3-turn helix (G), 4-turn helix (H) and 5-turn helix (I), beta-sheet (E), isolated beta-bridge (B), and turn (T), and two non-hydrogen bonded structures: coil (C) and bend (S), at each time frame, were assigned to all the residues of D42. In this work, we used the reduced secondary structures: helix (GHI), beta-sheet and-bridge (BE), turn (T), and loop (SC), to analyze the hydrogen-bonded and non-hydrogen bonded behavior of the residues in D42. The extraction of the reduced secondary structures from the complete DSSP structures was performed frame-by-frame using an in-house script in Python. The Visual Molecular Dynamics (VMD) program (Humphrey et al., 1996) was also used to create snapshots of proteins with detailed secondary structures as well as images of lipids, ions and solvent surrounding the protein.
Quantitative residue-contact map and radii of gyration measurements were employed to investigate the tertiary and quaternary structure of D42. Tertiary structure usually refers to the three-dimensional arrangements of the secondary structures within a peptide chain, while quaternary structures are the arrangement of the tertiary structures in a protein multimer, the dimer in our study.
For the residue-contact map measurements, a frame-by-frame residue-contact matrix of protein on each membrane surface was first generated using the tool, g_mdmat from GROMACS (Hess et al., 2008) and an in-house Python script. This residue-contact matrix describes the minimum distance among the residues of the protein in the form of a two-dimensional matrix with the row or column positions matching the sequence positions of the residues of the protein at a given time frame. Specifically, each matrix element gives the minimum distance between a pair of residues (residue i, residue j) in the ith row and the jth column at each simulation frame. This residue-contact matrix is then plotted in a three-dimensional residue-contact map with the minimum distance color coded in the z-dimension, and pixel locations representing the sequence positions of the residues along the x- (column) and y-(row) dimensions. In this study, the time-averaged residue-contact matrix over the last 50 ns was first generated in each replicate. Finally, average residue-contact matrix across all replicates in each complex was calculated. The calculations were performed using an in-house script written in Python. Since each chain of D42 has two domains, N-terminal domain and C-terminal domain, the residue-contact matrix can be divided into zones. Each zone represents contact regions among the four domains, i.e., N-terminal and C-terminal domains in chain A and chain B. An average of the matrix elements in each zone provides a quantitative measure of the interactions among residues in the interacting domains that further quantify the ternary and quaternary structures of D42.
A frame-by-frame radii of gyration calculation of D42 was generated using the tool, g_gyrate from GROMAS (Hess et al., 2008). This tool generates the radii of gyration, RgX, RgY, and RgZ of the protein along three principal axes, as well as the average radius of gyration Rg. The theory of the radii of gyration calculations has been described elsewhere (Theodorou and Suter, 1985; Vymetal and Vondrasek, 2011). A useful geometrical shape parameter called “asphericity” or b is useful to characterize the hydrodynamic shape of a protein, and its definition is given by
| (1) |
Note that b is zero for a spherical shape or isotropic object when all three radii of gyration are identical (Theodorou and Suter, 1985; Vymetal and Vondrasek, 2011). Again, the radii of gyration and the b factor were averaged over the last 50 ns of each simulation replicate and across all replicates using an in-house R-script.
2.3.2. protein–lipid minimum distance
Protein/lipid interactions were analyzed using the analysis tool, g_mindist from GROMACS (Hess et al., 2008). This tool calculates the minimum distance between any atom of a protein to any atom of a lipid (dmin) as a function of simulation time and as a function of residue location over a given simulation time range. The plot of dmin versus time provides the protein binding kinetics of D42 to the membrane surface, and dmin versus residue location provides a residue-specific protein/lipid binding profile. In this work, the average protein/lipid binding profile over the last 50 ns and across all replicates was determined.
2.3.3. Annular and non-annular regions
To analyze the composition and structure of the lipid and solvent molecules near the protein, the annular (AL) region containing the lipids and solvent (w) surrounding the protein was identified. A two-dimensional molecule sorting tool, Surround (Qiu et al., 2011), was used to identify the molecules that were within a threshold distance of 0.5 nm from any atom of the protein as projected on the x–y plane. In this study, AL–PC, AL–PS, and AL–w groups were identified at each time frame. Molecules not selected for the AL region were assigned to the non-annular region (nAL), and were labeled as nAL–PC, nAL–PS, nAL–w, accordingly. The Surround computational package, written in C and Python. A demonstration of the sorting of AL and nAL for the current bilayer systems is given in Fig. 12 of Data in Brief (Cheng et al., 2015b).
2.3.4. Transbilayer density and mass profile
The transbilayer number density ρN(z) profiles of different lipid structural groups: PO4 of PC, PO4 of PS, solvent (w), and protein were calculated using the tool, g_density from GROMACS (Hess et al., 2008) at each time frame and in separated AL and nAL regions surrounding the protein. The number density ρN(z) profile for each group is given by
| (2) |
Here, ni(z) is the number of a selected group within a thin z-slice at a given z-coordinate, A is the cross-sectional or x–y area of the simulation system, N is the total number of time frames for the calculations. In this study, Δz = Zs/100 for the PC bilayer and = Zs/300 for the larger PC/PS double bilayers, where Zs is the Z-dimension of the simulation box which depended on the amount of explicit solvent on the top and bottom of the lipid bilayer.
A similar transbilayer mass density ρm(z) profiles of different molecules, protein, lipid, water and ion, were also calculated using a similar definition as in Eq. (2) by replacing ni(z) with mi(z), which is defined as the mass of each molecule within the z-slice.
The time-averages ρN(z) or ρm(z) profile was over the last 50 ns of each simulation replicate in each complex. Finally, the average density profile across all four replicates of each system was calculated.
2.3.5. Order parameter of lipid and protein chain carbon backbone
The orientational order parameter S(n) for a given lipid carbon number position (n) or the C-alpha atom of D42 backbone of the nth residue was calculated using the tool, g_order from GROMACS (Hess et al., 2008). Lipids in each leaflet were grouped into AL and nAL (see above). Here, the time-averaged S(n) measures the segmental orientation order of the PC or PS acyl chains or the C-alpha atom of the peptide chain of D42 with respect to the normal (z-axis) of the bilayer over a given time period
| (3) |
where θn is the angle between the z-axis and a vector Cn (= rn+1 – rn−1) joining the nearest-neighbor carbons on each side of the carbon atom n, where rn+1 and rn−1 are the position vectors of the n + 1 and n − 1 carbon positions of the lipid chain or the peptide backbone, respectively. N is the number of time frames for the averaging. Angle brackets denote averaging over all participating lipids in the AL or nAL region. The time-average was over the last 50 ns of each simulation replicate in this work. Average across all simulation replicates for each bilayer system was also performed.
2.3.6. Bilayer thickness map
Time-averaged bilayer thickness maps of a lipid bilayer were calculated over a given length of time using a grid-based membrane analysis tool, GridMAT-MD (Allen et al., 2009), for all our simulations. This tool reads the coordinates of all the phosphorus atoms in the lipid bilayer, determines the shortest z-distances between those from the top layer and those from the bottom layer, and generates a 2-dimensional thickness matrix for each time frame. Upon averaging the matrix over a given time range, the time-averaged thickness map was created for a given trajectory using an in-house R script. In addition, we identified the location of the protein by assigning a large “thickness” value e.g., 6nm, for each identified protein atom on the x–y plane and overlaid them on the thickness map frame-by-frame. The time-averaged thickness and protein location maps were created over the last 50 ns of each trajectory. The average thickness maps were also generated from the average pixel-by-pixel calculations of all repeated replicates of each system. To identify the thickness near the periodic boundary, a larger, repeated or tiled image along the x- and y- directions, is presented for each thickness map.
2.3.7. Charge density, transmembrane potential and electric field profiles
The electrostatic transmembrane potential profile across the lipid bilayer V(z) depends on the system charge density profile ρq(z) along the z-axis (bilayer normal),
| (4) |
where εo is the permittivity in vacuum. Therefore, V(z) can be calculated by integrating Eq. (4) using the ρq(z) of protein, lipids, water and ions (Gurtovenko and Vattulainen, 2008). The z-component of force acting on a unit positive charge is given by the electric field E(z), which is just the negative gradient of the potential
| (5) |
Potential and electric field calculations were performed at each time frame using the tool, g_potential, from (Hess et al., 2008). To be consistent, the same slice thickness for the density profile calculation was used, Δz = Zs/100 for the PC bilayer and = Zs/300, where Zs is the Z-dimension of the simulation box. Finally, the average V(z) and E(z) profiles were determined over the last 50 ns of each replicate and across the four replicates of each system.
3. Results
3.1. Protein binding
The binding behavior of D42 on the PC surfaces of symmetric PC and asymmetric PC/PS bilayers was first examined. Although D42 attached firmly to the PC surface in all 8 independent simulation replicates, the behavior of membrane attachment of D42 was different between bilayer systems. Due to the larger separation between the protein and the PC leaflet of the PC/PS lipid bilayer during the pre-production protein-restrained MD simulation (see Section 2), the protein underwent translation and rotation before establishing stable contact with the membrane surface. In contrast, no significant protein rotation was detected for the PC bilayer. Here, the protein–lipid minimum distance plot (see Section 2) was used to quantify the binding kinetics of D42 of each simulation replicate, The protein made its first contact with the PC leaflet at the initial attachment time. This initial attachment time was determined from the intercept of two extrapolated lines of the sharp decline and the stabilized value of ~0.14 Å in the protein–lipid minimum distance versus time plot (see Fig. 1 of Data in Brief (Cheng et al., 2015b). As summarized in Table 1, the initial attachment time of D42 was 3–4 ns for the PC bilayer but varied over a range from 5 to 28 ns for the PC/PS bilayer for different simulation replicates.
Table 1. Surface area of lipid and protein attachment times in protein/lipid bilayer systems.
The surface area of PC or PS in each lipid leaflet of the symmetric PC and asymmetric PC/PS bilayer systems is shown. The surface area of lipid was determined from the area of the x-y plane of the system box simulated under the constant number, pressure and temperature (NPT) conditions divided by the number of lipid molecules in each lipid leaflet. Also shown are the initial and final attachment times of the protein on the lipid surface determined from the protein-lipid minimum distance plot and VMD visualization (see Section 2). Results from four independent simulation replicates (rep1 to rep4) of each bilayer system are given. The area result represents average over the last 50 ns of each replicate. Also, the average area result across all four repeating replicates of each bilayer system with mean and standard error is given for each bilayer system.
| Bilayer system | Simulation replicate | Area per PC (Å2) | Area per PS (Å2) | Initial attachment timea (ns) | Final attachment timeb (ns) |
|---|---|---|---|---|---|
| PC (0-200ns) | rep1 | 66.62 | 4.0 | 5.0 | |
| rep2 | 66.62 | 3.5 | 4.5 | ||
| rep3 | 66.64 | 2.5 | 3.5 | ||
| rep4 | 66.62 | 4.0 | 5.0 | ||
| average | 66.62 ± 0.01 | 3.5 ± 0.4 | 4.5 ± 0.4 | ||
| PC/PS (0-350ns) | rep1 | 62.41 | 51.68 | 28.0 | 38.5 |
| rep2 | 62.52 | 51.77 | 5.0 | 17.0 | |
| rep3 | 62.51 | 51.77 | 5.0 | 20.0 | |
| rep4 | 62.37 | 51.65 | 14.5 | 25.0 | |
| average | 62.45 ± 0.04 | 51.72 ± 0.03 | 13.3 ± 5.6 | 25.1 ± 4.8 |
The initial attachment time is defined as the time at which the plot of protein-lipid minimum distance versus time has reached a steady state value of ~0.14Å from the sharp decline as demonstrated in Fig. 1 of Data in Brief (Cheng et al., 2015b)).
The final attachment time is defined as the time at which the membrane-orientation vector of the hydrophobic region (LYS-28 to ALA-42) of chain A reached a stable value with respect to the normal of the contact PC layer after the initial attachment as demonstrated in Fig. 2 of Data in Brief (Cheng et al., 2015b)).
The hydrophobic region between LYS-28 to ALA-42 or residues 29–41 of the chain A of D42 established the initial contact with the PC leaflet (protein-contact leaflet) during the protein adsorption process in both PC and PC/PS lipid bilayer system. A membrane-orientation vector, starting from ALA-42 (C-terminal end) and ending at LYS-28 (tip of the loop), was used to represent the membrane-orientation of D42 during the protein-binding event. The angle between this vector and the z-axis (along the normal of the bilayer), or membrane-orientation angle, was examined as a function of time. This membrane-orientation angle changed only slightly for the PC bilayer system. For example, the angle varied from ~92° immediately after the initial attachment at 4 ns to ~94° at 5 ns and remained stable for the simulation replicate rep1 (see Fig. 2 of Data in Brief (Cheng et al., 2015b)). However, a broad range of initial angles, from 76° at 5 ns for rep1 to 119° at 28 ns for rep2, were evident for the PC/PS lipid bilayer system (see Fig. 2 of Data in Brief (Cheng et al., 2015b)). In addition, it took 10–15 ns for the angle to reach a stable value, of 90–95°. Therefore, the time at which the membrane-orientation angle reached a stable value is defined as the final attachment time. As summarized in Table 1, this final attachment time varied from 17 to 39 ns.
3.2. Surface area of lipid
Average surface areas of PC and PS were calculated over the last 50 ns of each replicate and over all replicates of each bilayer system from the size of the simulation box (see Section 2). As shown in Table 1, the average surface area of PC in the upper and lower PC leaflets of the PC bilayer was ~67 Å2 as compared with a smaller value of ~62 Å2 in the upper PC leaflet of the PC/PS bilayer. The average surface area of PS in the PS leaflet of the PC/PS bilayer was ~52 Å2, much smaller than the average surface area of PC in either PC or PC/PS bilayer.
3.3. Transmembrane electrostatic properties
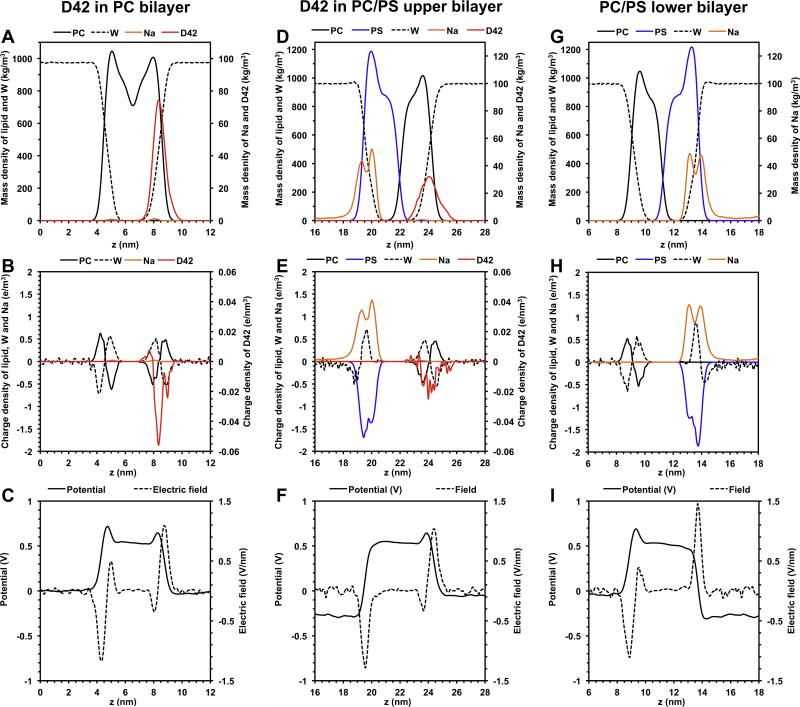
The average transmembrane electrostatic properties of the PC and PC/PS bilayer systems during the last 50 ns and over all replicates were calculated (see Fig. 3). The average charge density distributions ρq(z) sorted by molecule types (protein, lipid, water and ion) across the z-axis (normal of the bilayer) of the PC and the upper PC/PS bilayer are shown in Fig. 3B and E, respectively. In comparison, the charge distributions of the lipid, water and ion of the lower PC/PS bilayer in the absence of the absorbed D42 are given in Fig. 3H. Also shown in Fig. 3A, D and G are the mass density distribution plots, also sorted by molecule types. These mass density plots allow a direct comparison of the molecule locations and their electrostatic properties across the bilayer.
Fig. 3. Average transbilayer mass, charge, potential and electric field distribution of protein/lipid bilayer systems.
The average mass (A, D), charge (B, E), potential (C, F) and electric field (C, F) distribution as a function of z distance of the protein/lipid bilayer systems containing PC bilayer (A–C) and PC/PS upper bilayer (D–F). The z-axis is along the normal of the lipid bilayer. The distributions of lipid bilayer/water system but without D42 for the PC/PS lower bilayer (G–I) are also shown. The calculatons were averaged over the last 50 ns and across all repeating replicates of each system.
The average transmembrane potential V(z) versus z-axis plots for the PC (Fig. 3C), upper PC/PS (Fig. 3F) and lower PC/PS (Fig. 3I) bilayers were calculated using Eq. (3) of Section 2. The reference potential was set to zero at z = 0 in all the potential plots. As expected, for the PC bilayer, the V(z) profile in the aqueous regions (below the lower PC leaflet and above the upper PC leaflet) was zero as shown in Fig. 3C. In contrast, in the PC/PS bilayer, the V(z) was ~−240 mV in the central counterion/water slab region (z ≈ 14–19 nm) and was zero in the aqueous regions (above the upper bilayer and below the lower bilayer as shown in Fig. 3F and I).
The average electric field E(z) versus z-axis plots for the PC (Fig. 3C), upper PC/PS (Fig. 3F) and lower PC/PS (Fig. 3I) bilayers were also calculated using Eq. (4) of Section 2. The average electric field E(z) was localized around the polar regions of each bilayer. Referring to Fig. 3C and 3F, the E(z) profile of the upper PC leaflet in the PC (z ≈ 7–10 nm) and PC/PS (z ≈ 23–26 nm) exhibited an identical pattern, i.e., a small negative peak followed by a large positive peak. In contrast, the E(z) profile of the lower PC leaflet (z ≈ 3–6 nm) in the PC bilayer (Fig. 3C) exhibited a large negative peak followed by a small positive peak that was very different from a single negative peak in the lower PS leaflet (z ≈ 19–22 nm) of the upper PC/PS bilayer (Fig. 3F). Cross comparisons of Fig. 3B, C, E and F revealed that the localized electric field profile was similar across the protein for both PC and PC/PS bilayers. Since the protein was absorbed on the upper PC surface of both bilayers (Fig. 2), the electric field in the lower PC leaflet or the lower PS leaflet has negligible effects on the protein.
3.4. Higher order protein structure
3.4.1. Secondary structure
Upon binding, the secondary structures of D42 on the PC surface of the symmetric PC and asymmetric PC/PS bilayers were investigated. Fig. 3 of Data in Brief (Cheng et al., 2015b) illustrates the time evolutions of the secondary structure of D42 from rep 4 of PC and rep 2 of PC/PS bilayer systems according to the DSSP algorithm. Using the reduced secondary structure representations (see Section 2), the time evolutions of the number of residues involved in hydrogen-bonds (BE, GHI, and T) in both N- and C-terminal domains of D42 on the surface of PC and PC/PS bilayers were extracted. Figs. 4–6 of Data in Brief (Cheng et al., 2015b) demonstrate the time evolution data from rep 1 of the PC bilayer system and rep 1 and rep 2 of PC/PS bilayer system. A control plot of the protein in the absence of lipid membrane (Fig. 7 of Data in Brief (Cheng et al., 2015b)) is shown for comparison. The plots demonstrate significant folding of the N-terminal domain after protein binding, and that the C-terminal domain unfolded more on the surface of the PC bilayer than on the PC/PS bilayer.
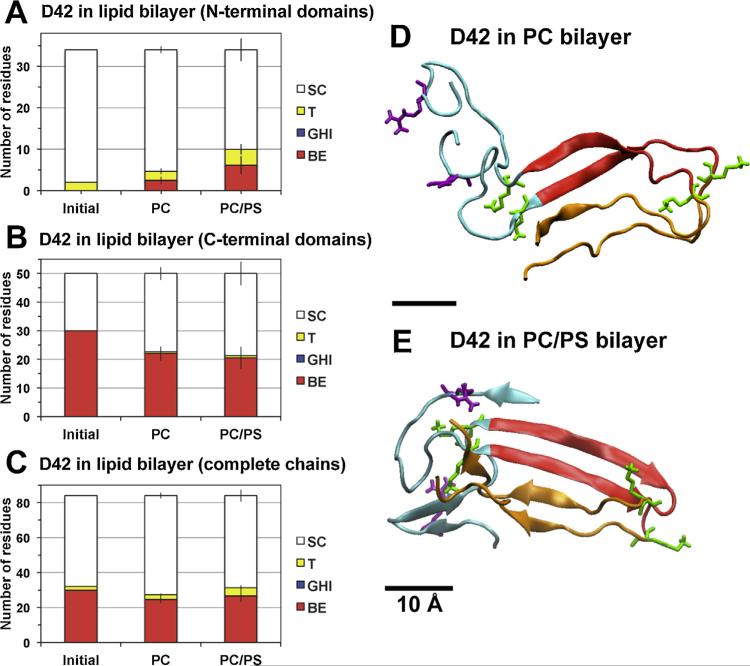
Fig. 4. Average secondary structure of protein in protein/lipid bilayer systems.
(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The average numbers of residues involved in reduced secondary structures (SC, T, GHI and BE) calculated from DSSP (see Section 2) for D42 in PC and PC/PS lipid bilayers in the N-terminal domains (A), C-terminal domains (B) and complete chains (C) are shown. The structural calculations were averaged over the last 50 ns of the simulation of each simulation replicate. The average was then across all four independent replicates of each lipid bilayer system with means and standard errors (bars) as given. For comparison, the initial (0 ns) reduced secondary structures of D42 are also presented. Protein conformations of representative replicates of D42 in PC (D) and PC/PS (E) bilayers at 200 and 350 ns simulation times, respectively, are illustrated. The N-terminal domain (residues 1–17 in blue ribbon) and C-terminal domain (residues 18–28 in red and residues 29–42 in orange) of each chain are shown. The six positively charged residues (lysine in green and arginine in purple) are in thick licorice lines. A scale bar of 10 Å is shown.
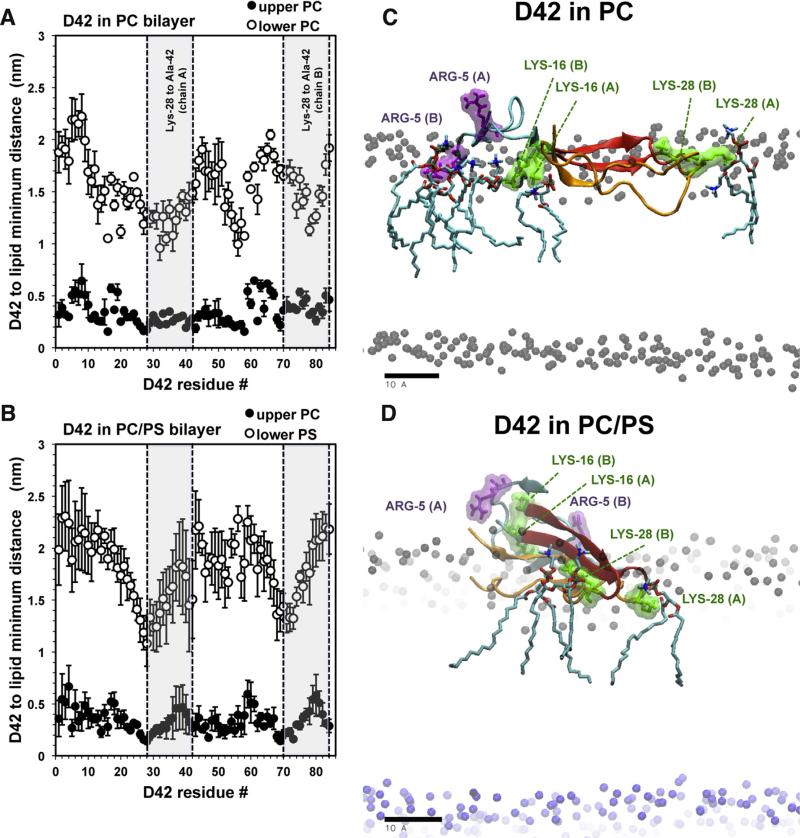
Fig. 6. Average residue-specific protein–lipid interactions in protein/lipid bilayer systems.
(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Average minimum distance between the atoms of D42 and the lipids in the upper (filled circles) and lower (open circles) layers of the PC (A) and PC/PS (B) bilayers versus residue number plots are shown. The average was over the last 50 ns of each simulation replicate and across all four repeating replicates with means and standard errors of the means (error bars) given. Note that residues numbers 1–42 and 43–84 represent chain A and chain B, respectively. Shaded areas highlighted the hydrophobic regions bound by Lys-28 and Ala-42 of chain A and chain B. Representative orientations of D42 on PC (C) and PC/PS (D) surfaces on the transverse (x–z) plane, with z-axis along the normal of the lipid bilayer, are shown. Also shown are the nearest PC lipids surrounding the positive residues (ARG-5 in purple and LYS-16 or LYS-28 in green) rendered in licorice lines and colored surfaces. To visualize the thickness of the bilayer, the polar phosphate groups of PC (black sphere) and PS (blue sphere) lipids highlight the upper and lower polar regions of the lipid bilayers, respectively. A scale bar indicates 10 Å.
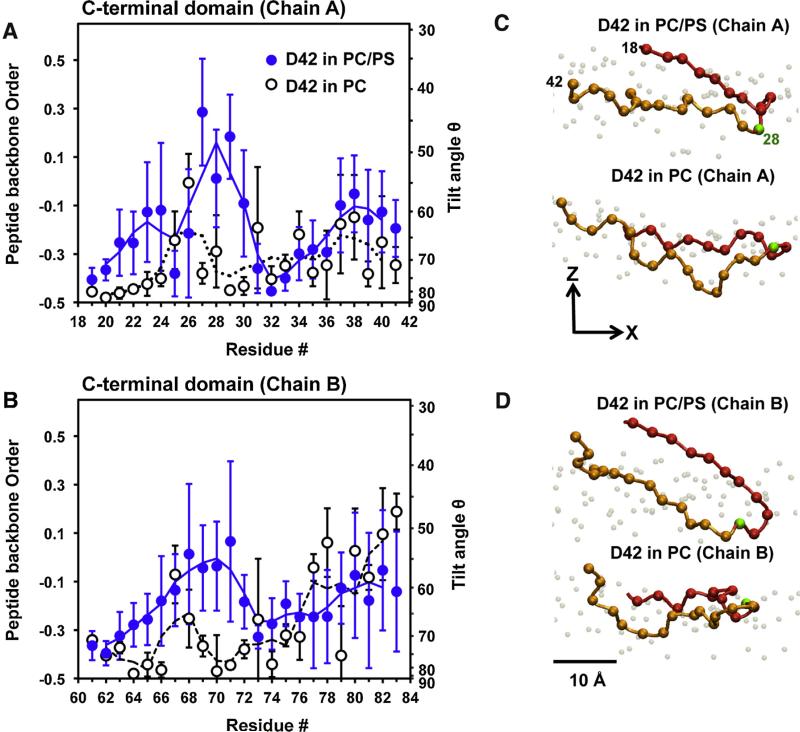
Fig. 7. Average residue-specific peptide backbone orientational order in protein/lipid bilayer systems.
(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Average orientational order and tilt angle of the C-alpha backbone in the C-terminal domain of chain A (A) and chain B (B) of D42 versus residue number in PC (black circles) and PC/PS (blue circles) bilayers are shown. See Section 2 for the details of the calculations of chain orientational order and tilt angle of the protein. The average was over the last 50 ns of each simulation replicate and across all four repeating replicates with the means and standard errors of the means (error bars) given. In panel B, residues numbers 60 to 84 of chain B correspond to the residues numbers 18 to 42 of chain A. For examples, residue number 70 and 84 in chain B correspond to residues 28 and 42 in chain B. The representative C-alpha carbon backbone orientations in the C-terminal domain of chain A (C) and chain B (D) of D42 in the PC and PC/PS bilayer on the transverse (x–z) plane, where z is the normal of the bilayer, are illustrated. For clarity, the C-alpha atoms from residues 17–27 (red balls), residue 28 (green ball) and residues 29–42 (orange balls) are color-coded. Also, the lipid phosphate atoms (gray balls) are shown to identify the polar regions of the PC leaflet in both PC and PC/PS bilayer. A scale bar indicates 10 Å
To further quantify the above folding/unfolding patterns, the average number of residues involved in hydrogen-bonds (BE, GHI, and T) and non-hydrogen bonds (SC) in the N-terminal and C-terminal domains were calculated over the last 50 ns of all replicates for each bilayer system. The number of secondary structures before the protein binding is also presented for comparison.
For the N-terminal domains (Fig. 4A), there were ~5 and 10 (mostly BE and T) hydrogen bonds for the PC and PC/PS bilayer systems, respectively. These numbers were significantly larger than ~2, the number of hydrogen bonds in the T structure before the protein binding. For the C-terminal domains (Fig. 4B), there was a reduction of hydrogen-bonds from ~30, before protein binding, to ~20 after protein binding for both bilayer systems. The structures before and after the protein binding were mostly BE. As shown in Fig. 4C, the total number of residues involved in hydrogen-bonds of the entire protein in both the N- and C-terminal domains dropped from ~32 to ~27 in the PC bilayer system but the number of hydrogen bonds in the PC/PS bilayer system changed little after protein binding.
Representative secondary structures of D42 on the PC and PC/PS bilayer surfaces are illustrated in Fig. 4D and E, respectively. Compared to the initial structures before the membrane binding (Fig. 1A), the beta-sheet structure of the U-shaped C-terminal domains was partially disrupted on both PC and PC/PS surfaces. Isolated beta-sheets were evident in the original random coil region of the N-terminal domains and the extent of beta-sheet structure was less on the PC surface than on PC/PS surfaces. The above observations agree with the average secondary structure profiles tabulated in Fig. 4A–C.
The surface aggregation behavior of the N-terminal and C-terminal domains exhibited an interesting dependence with the lipid composition of the bilayer. The N-terminal and the C-terminal regions of D42 were mostly segregated on the PC (Fig. 4D) bilayer but were aggregated on the PC/PS (Fig. 4E) bilayer. The structural domain aggregation behavior is investigated quantitatively below.
3.4.2. Tertiary and quaternary structure
Tertiary and quaternary structures of D42 were quantified using average residue-contact map and average radii of gyration analysis. The average was over the last 50 ns of each simulation and across all replicates in each bilayer system. The detailed procedures of the analysis are given in Section 2. The 3D residue-contact maps of D42 in both bilayer systems are presented in Fig. 10 of Data in Brief (Cheng et al., 2015b).
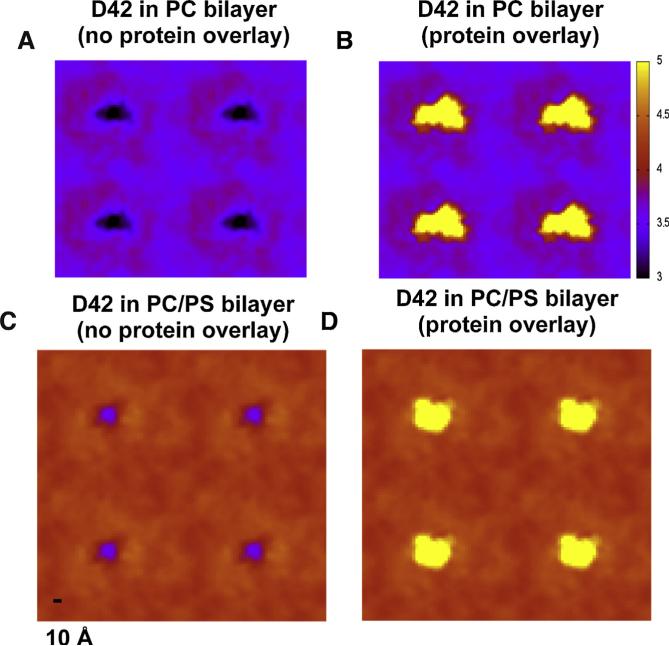
Fig. 10. Average lipid bilayer thickness maps in protein/lipid bilayer systems.
(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Average three-dimensional thickness (color coded in z-dimension) maps of the PC bilayer (A, B) and the PC/PS bilayer (C, D), with (B, D) and without overlays (A, C) of protein locations (fixed at 6 nm thickness or yellow color-coded) are shown. The map calculations were averaged over the last 50 ns of each replicate and across all four repeating replicates in each system. A color scale bar from 3 to 5 nm (black to yellow) and a spatial scale bar of 1 nm on the lateral or x–y plane are given. See Section 2 for details of the thickness map calculations.gr1
Based on the residue-contact maps, average separation of residues from the same C- or N-terminal domains (Zones 1, 2, 4, and 5) was examined first (Fig. 5A). No significant differences among the average separation in either Zone 2 or 5, corresponding to the residues within the C-terminal domain, were detected for either bilayer. However, the residues of Zone 1 or 4, those within the N-terminal domain, were significantly closer for D42 in the PC/PS bilayer.
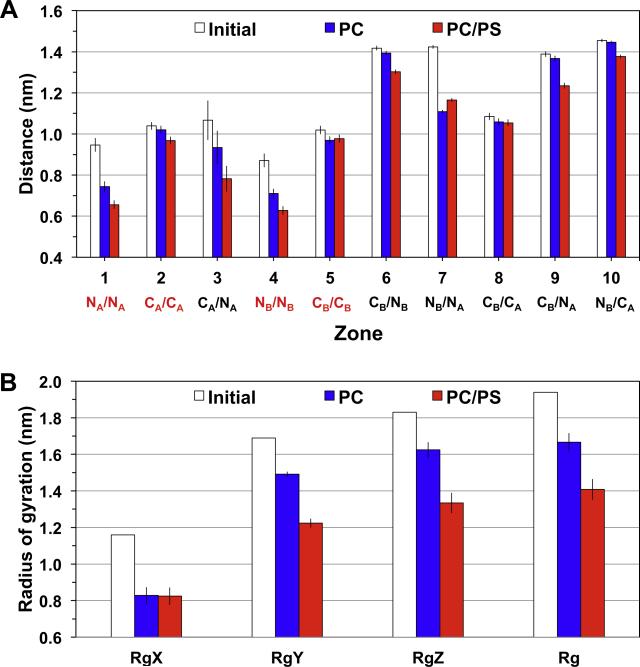
Fig. 5. Average residue-contact distance and radii of gyration of D42 in protein/lipid bilayer systems.
(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Average residue-contact distances in 10 different residue-contact zones (A) and radii of gyration (B), along three principal components, RgX, RgY and RgZ, and their average, Rg, for the PC (blue) and PC/PS (red) bilayers are shown. The average was over the last 50 ns of each replicate and across all four repeating replicates with means and standard errors of the means (error bars) given. The values before the simulations, i.e., initial or 0 ns (white), are presented for comparison. The four contact zones (NA/NA, CA/CA, NB/NB and CB/CB) among residues within the same chain, i.e., chain A or chain B, are marked in red to facilitate the comparison among different zones in panel A. The definitions of the contact zones and radii of gyrations are given in Section 2.
Average distances among residues from different domains (Zones 3, 5, 6, 7, 8, 9, and 10) were then examined. There were small differences in the average distances in Zones corresponding to the residues within the N-terminal domain or C-terminal domain from different chains, for all bilayer surfaces. However, residues from Zone 3, 6, 9, and 10, corresponding to the N- and C-terminal domain from different chains were significantly closer in the PC/PS than the PC bilayer. Similar observations of the average residue-contact distance results were found in D40, a beta-amyloid dimer similar to D42 but without the last two residues in each chain. See Figs. 8–11 of Data in Brief (Cheng et al., 2015b) for the comparison of the structures and the residue contact maps and results for D40 and D42.
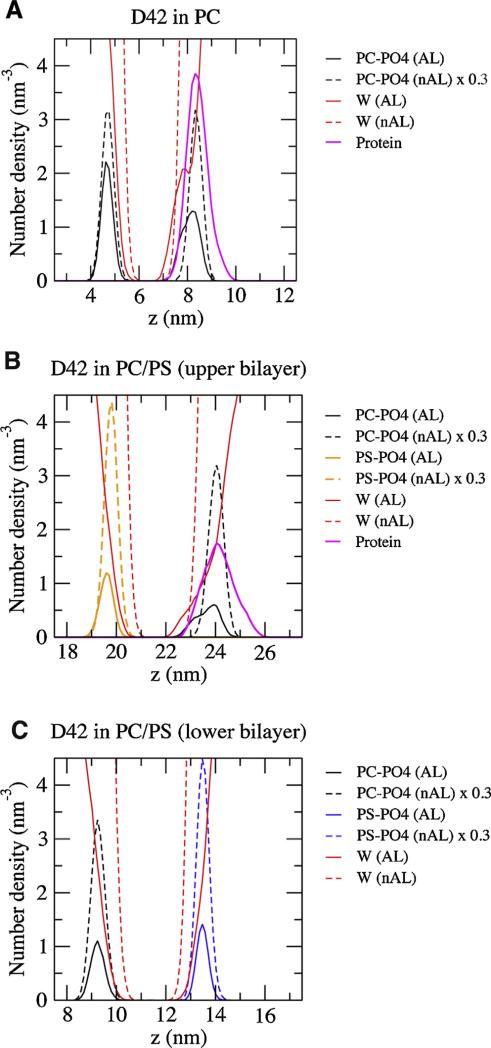
Fig. 8. Average transbilayer density profile in protein/lipid bilayer systems.
(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Average number density of groups of molecules versus z-axis (normal of the bilayer) of the complexes containing the single PC bilayer (A), PC/PS upper bilayer (B), and PC/PS lower bilayer are shown. The average was over the last 50 ns of each simulation replicate and across all four repeating replicates of each replicate. The results for the upper PC/PS bilayer in which D42 was attached are shown. The non-annular (nAL) and annular (AL) lipid and water groups are in dashed and solid lines, respectively. The groups represent the phosphate of PC (PC–PO4) in black, the phosphate of PS (PS–PO4) in orange, the water (W) in red, and the protein in thick pink. The number densities of the lipids in the nAL region are scaled by a factor of 0.3 to facilitate the comparison of the density peak locations. The method of separation into AL and nAL regions has been described in Section 2.
The average radii of gyration about the principal axes, RgX, RgY and RgZ, as well as the average radius of gyration, Rg, of D42 on different membrane surfaces were calculated, and the results are shown in Fig. 5B. The smallest radius of gyration, RgX, was associated with the principal axis X along the long axis of the U-shaped C-terminal domain. The larger RgY or RgZ was with the principal axis Y or Z perpendicular to X. Note that RgY was only slightly smaller than RgZ. The radii of gyration of D42 were always smaller when the protein was on membrane surfaces than before protein binding. The RgX of D42 was ~0.8 nm for all membrane surfaces. In contrast, the RgY or RgZ of D42 on the PC/PS surface was always smaller than that on PC control surfaces. The asphericity or b parameter (see Eq. (1) of Section 2), which measures the deviation of the spherical shape of the protein, was calculated for D42 on each membrane surface. The b parameters were 1.18 ± 0.16 and 0.69 ± 0.19 for PC and PC/PS bilayers, respectively, as compared with 1.24 before the simulations.
3.5. Protein membrane orientation
Protein orientation on PC and PC/PS bilayer surfaces was systematically investigated by use of the average minimum distance between individual residue of protein and lipid (Fig. 6) and peptide backbone chain orientational order (Fig. 7). Similar to the notations used in the DSSP plots (see Fig. 3 of Data in Brief (Cheng et al., 2015b)), the residue number runs from 1 to 42 and 43 to 84 for depicting chain A and chain B, respectively, in the plots. The average was from the last 50 ns of each replicate and across all four replicates from each bilayer system.
Fig. 6 shows the average minimum distance between the atoms of the protein residue and lipid as a function of the residue number of D42 on PC (Fig. 6A) and PC/PS (Fig. 6B) bilayers. As demonstrated in Fig. 2, D42 contacted only the upper PC leaflet in either system. The PC bilayer (Fig. 6A) maintained a minimum distance between the protein and PC atoms in the upper leaflet around 0.25 nm and showed little dependence on the residue location except some small dips of depths between 0.1 to 0.3 nm. Interestingly, well-separated minima were observed in the lower leaflet for residue numbers ~16 and 32 of chain A and ~58 and 80 of chain B, corresponding LYS-16 and ILE-32 of the chains, respectively. For the PC/PS bilayer (Fig. 6B), more clearly defined small dips for the upper PC leaflet and significant dips for the lower PS leaflet at residue numbers ~28 and 72, corresponding to the vicinities of LYS-28 of both chains, were evident.
Fig. 6C and D demonstrates the nearest lipids within 0.25 nm of the positively charged residues, ARG-5, LYS-16, and LYS-28 of D42 for representative replicates. For the PC bilayer (Fig. 6C), ARG-5 of Chain A, LYS-16 in both Chain A and Chain B, and LYS-28 of Chain A were surrounded by PC molecules. For the PC/PS bilayer (Fig. 6D), only the LYS-28 residues from both Chain-A and Chain-B were surrounded by PC lipids, while LYS-16 and ARG-5 residues faced away from the bilayer surface.
Fig. 7 shows the average residue-resolved orientational order of the C-alpha atoms along the peptide backbone of D42 on PC and PC/PS surfaces. It is clear that the values of orientational order of D42 around the residue number 28, or LYS-28 of chain A (Fig. 7A), and residue number 70, or LYS-28 of chain B (Fig. 7B), on PC/PS bilayers were consistently higher than those on PC bilayers. The average tilt angle of LYS-28 backbone with respect to the bilayer normal was around ~50° for the PC/PS bilayer when compared with ~80° for the control PC bilayer. Fig. 7C and D illustrate the orientation of the C-alpha atoms of the C-terminal domains of chain A and chain B of D42 on the leaflet surfaces.
3.6. Protein-induced membrane structural disruptions
Binding-induced membrane structural disruption was examined by use of transbilayer density profile plots, lipid acyl chain order parameter plots and bilayer thickness maps.
The two-dimensional Surround algorithm (see Section 2) was used to sort the lipid, water and protein molecules into annular (AL) and non-annular (nAL) regions. Fig. 12 in Data in Brief (Cheng et al., 2015b) demonstrates this separation. The average transbilayer density profile of molecules (Fig. 8) and lipid acyl chain order parameter (Fig. 9) were subsequently calculated in both AL and nAL regions. No sorting is required for the bilayer thickness maps. Averaging over the last 50 ns and across all replicates was performed in all the plots and maps (see Section 2).
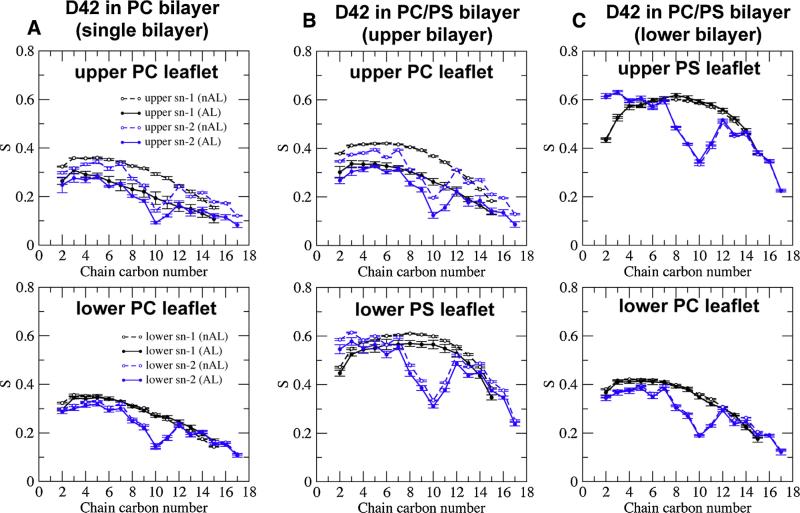
Fig. 9. Average orientational order parameter of lipid acyl chains in protein/lipid bilayer systems.
(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Average orientational order parameter (S) of the sn-1 (black) and sn-2 (blue) acyl chains of phospholipid versus chain carbon number in the upper (top row) or lower (bottom row) leaflet of the PC single bilayer (A), PC/PS upper bilayer (B) and PC/PS lower bilayer (C). The average was over the last 50 ns of each simulation replicate and across all four repeating replicates with means and standard errors of the means (error bars) shown. Order parameters of lipids in the annular (AL) and non-annular (nAL) regions labeled in filled and open circles, respectively, are given. The method of sorting the lipids into AL and nAL regions is described in Section 2. The definitions of upper and lower lipid leaflets are given in Fig. 2.
The average transbilayer lipid density profile of AL and nAL phosphate groups: PC (PO4-PC) and PS (PO4-PS) are presented. A broadening of PO4-PC peaks in the AL region versus nAL region of the upper leaflet was observed for both bilayers. However, the peak locations of the lipid density profiles in the AL region were similar to those in the nAL region. Note that the peak-to-peak z-distance between two matched lipid phosphate peaks defines the thickness of the lipid bilayer. Here the thickness of the PC bilayer (Fig. 8A) was ~3.5 nm while that of PC/PS was ~4.3 nm (Fig. 8B) in both the AL or nAL region. As expected, no broadening of the PO4-PC peak was detected in the lower PC/PS bilayer, where there was no protein (Fig. 8C).
For the transbilayer protein density profile, the protein peak overlapped the PO4-PC peak for the PC and PC/PS bilayers. These results agree with the deep penetration of the protein in the upper PC leaflet of the PC and PC/PS bilayers as shown in Fig. 2C and D. The transbilayer water density profile was higher near the center of the bilayer for the AL region of the PC bilayer but not forthe PC/PS bilayer.
In the absence of protein (nAL region), the average order parameter of the lipid acyl chain in the upper PC leaflet of the PC bilayer was significantly lower than that of the PC/PS bilayer (see Fig. 9A and B). In the presence of protein, the average order parameter of the lipid acyl chain shows significantly less lipid chain order for both chains in the upper PC bilayer in the AL region versus the nAL region (Fig. 9A and B) for both bilayer systems. The PC/PS bilayer showed significantly less lipid chain order in the upper PC leaflet than the lower PC leaflet; the PC/PS bilayer also showed slight drop in the lipid chain order in the upper PS leaflet as compared with the lower PS leaflet (Fig. 9B and C). This small but significant disorder in the lower PS leaflet of the PC/PS bilayers suggests that the binding of protein to the upper PC leaflet influenced the structural properties of the lower PS leaflet. This indicates coupling between the two leaflets in the asymmetric PC/PS bilayer. As expected, no significant difference in the average order parameter of the AL versus the nAL region for both PC and PS leaflets was detected in the lower PC/PS bilayer where the protein was not present (Fig. 9C).
The average color-coded bilayer thickness maps of the PC (Fig. 10A and B) and PC/PS (Fig. 10C and D) bilayers were determined from the last 50 ns of each simulation and across all replicates for all bilayers. Periodic images along the x and y directions are shown to highlight the periodic boundaries of the thickness maps. As shown in Fig. 10A, the thickness of the PC bilayer was deformed to ~3 nm in an extended ellipsoid-shaped region of 4 nm × 2 nm and surrounded by a region of thickness ~3.5 nm within the boundary of the absorbed protein, as marked by the yellow pixels in Fig. 10B. Outside the protein boundary, the thickness increased abruptly to ~4 nm in an annular domain of around 2 to 3 nm, and nally dropped to ~3.5 nm for regions farther away from the center of the protein. On the other hand, as shown in Fig. 10C, the thickness of the PC/PS bilayer was deformed to ~3.5 nm in a more localized and less extended circle-like region ~2 nm in diameter and surrounded by a region of thickness ~4.3 within the boundary of the absorbed protein as shown in Fig. 10D. Outside the protein boundary, the thickness remained at ~4.3 nm with no obvious annular domain of increased thickness as found in the PC bilayer system. The thicknesses of the bilayers away from the protein calculated from these thickness maps agree well with those from the peak-to-peak lipid phosphate density profile measurements (Fig. 8). Overall, the thickness disruption region was significantly more spread out laterally (along x–y plane), or more extended (including the thicker or overshoot annular domain) on the bilayer PC than on the PC/PS bilayer.
4. Discussion
In this work binding of D42, protein conformational transitions, and subsequent protein–lipid interactions on the upper PC leaflet of the asymmetric PC/PS and symmetric PC bilayers have been systematically investigated.
We observed weaker binding, more folding in the N-terminal domain, more aggregation of the N- and C-terminal domains, and a larger tilt angle of the protein on the surface of the PC/PS bilayer versus the PC bilayer. The differences in the surface structures of the protein resulted in more localized bilayer thinning of the PC/PS bilayer versus the PC bilayer. Our results were based on several simulation replicates totaling up to 2.2 μs of total simulation time. Below we examine how the biophysical properties of the lipids: membrane potential and lipid packing order, regulate the protein structures and protein–lipid interactions in our model membrane systems.
Membrane potential was a key parameter investigated in our simulations. We observed that the protein experienced a similar local electric field profile in both PC and PC/PS bilayers. The large difference in the local electric field in the lower leaflet simply did not affect the adsorbed protein to the bilayer. This is because the protein interacted with the upper PC leaflet exclusively in both bilayers. Interestingly, a different simulation study (results not shown) carried out in our lab on a monomeric alpha-helix rich beta-amyloid, which was partially inserted into the asymmetric PC/PS bilayer with the negatively charged C-terminus (Ala-42) near the middle of the bilayer, showed a strong repulsion of the protein from the PS leaflet that eventually drove the peptide to the surface. Those results indicate the presence of the strong negative electric field, effectively pushed the negatively charged C-terminus and inhibited the insertion of monomeric peptide across the bilayer.
Lipid order packing of the upper PC leaflet was higher in the PC/PS bilayer than the PC bilayer, as shown by smaller area per PC of 63 Å2 in the PC/PS bilayer versus 67 Å2 in the PC bilayer (Table 1). These values were based on unconstrained NPT simulations. These surface areas of PC and PS agree reasonably well with published values from both simulations and experiment (MacDonald et al., 1976; Kucerka et al., 2005; Gurtovenko and Vattulainen, 2008; Cordomi et al., 2009). Further, we found significantly higher acyl chain orientational order in the PC/PS bilayer than in the PC bilayer (Fig. 9). Therefore, the protein experienced a more ordered lipid environment in the protein-contact PC leaflet in the asymmetric PC/PS bilayer than in the symmetric PC bilayer. Both PC and PC/PS bilayer systems were planar and stable throughout the entire simulation for all replicates. Similar conclusions of an enhancement in the order packing of a PC leaflet by the opposing leaflet containing lipids of higher packing, e.g., PS and GM1, a glycolipid with a ceramide moiety, have been reported in two other asymmetric bilayer systems (Lopez Cascales et al., 2006; Patel and Balaji, 2008). The effect of lipid packing on protein structural changes and protein–lipid interactions in the two bilayer systems is examined below.
We observed weaker protein binding to the PC/PS bilayer versus the PC bilayer. Electrostatic interactions between the charged residues of the protein and lipid polar headgroups have been proposed to drive the initial membrane surface binding of beta-sheet rich beta-amyloid oligomers in various bilayer systems (Davis and Berkowitz, 2010; Jang et al., 2013; Yu et al., 2013; Tofoleanu et al., 2015). Those studies investigated single- and multiple-component lipid bilayers consisting of PC, PS, phosphatidylethanolamine (PE), and phosphatidylglycerol (PG) of different acyl chain compositions. Here, the dimeric protein interacted exclusively with the PC leaflet of both asymmetric PC/PS and PC bilayers. However, an initial repulsion of D42 from the surface (see Fig. 2) of the PC/PS bilayer and a longer protein binding lag time (see Table 1) in the PC/PS than in PC bilayer were detected. These observations suggest that the tighter PC polar headgroup packing in the PC/PS bilayer provides a less conformationally favorable binding surface to the charged residues of the protein when compared to the relatively looser PC polar headgroup packing in the PC bilayer. Weak binding of a beta-amyloid p3 pentamer, which contains identical C-terminal domains as in this work but no N-terminal domains on the same mono-unsaturated PC as used in this work, has been reported (Yu et al., 2013). However, another binding study using the same beta-amyloid p3 pentamer, but on membrane surfaces of unsaturated PC, i.e.,18:1 in both sn-1 and sn-2, reported strong protein surface binding. Note that the mono-unsaturated PC should be more tightly packed than the all-unsaturated PC (Silvius, 1982). Therefore, the above observations further support the notion that tightly packed PC surfaces are less favourable for promoting protein surface binding. The same mechanisms may operate for binding of other amyloidogenic proteins of different initial structures (Gorbenko and Kinnunen, 2006; Gorbenko and Trusova, 2011).
We observed significantly larger folding from random coil to beta-sheet in the N-terminal domain of D42 on the PC surface of the PC/PS bilayer than the PC bilayer. We speculate that the more ordered PC surface in the PC/PS bilayer also promotes protein folding more effectively than the less ordered PC surface in the symmetric PC bilayer. However, unfolding of the C-terminal domain of similar extent was evident in both bilayers, suggesting that protein unfolding (beta-sheet to random coil) is probably not sensitive to the lipid packing. Previous simulation studies on beta-sheet-rich beta-amyloid p3 dimer (Jang et al., 2013) on mono-unsaturated PC surfaces and beta-amyloid pentamer (Tofoleanu et al., 2015) on all-unsaturated PC surfaces have also reported spontaneous unfolding of beta-sheets, and therefore agree with our results. Previous studies (McLaurin and Chakrabartty, 1997; Wong et al., 2009; Nag et al., 2013) have revealed that the presence of anionic lipid supports and promotes beta-sheet structures on membrane surfaces. However, in our study, the anionic PS lipids in the PC/PS asymmetric bilayer were not in contact with the protein. Their effect was to promote a more ordered PC leaflet for the protein to interact. Therefore, the PS effect on surface protein folding is indirect in this study.
We observed significantly more aggregation of C- and N-terminal domains in the PC/PS than in the PC bilayer. Although more folding of the N-terminal domains helped promote and stabilize the aggregated structures of the protein in the PC/PS bilayer compared with the PC bilayer, only a small fraction of the N-terminal domain was folded regardless of bilayer (Fig. 5A). We speculate that the larger domain aggregation in the PC/PS bilayer may be explained by stronger protein–protein and the concomitant weaker lipid–protein interactions in the PC/PS bilayer versus the PC bilayer. From the protein–lipid minimum distance plot (Fig. 6), we discovered that several hydrophobic residues in the lipid insertion domain (residues 29–41) and two charged LYS-16 residues inserted deeper towards the lower PC leaflet than other residues for the PC bilayer. In other words, these residues helped stabilize, or anchored the protein on the surface. The partially unfolded C-terminal domain around the lipid insertion domain may further help the anchoring (Tofoleanu and Buchete, 2012). Loosely packed PC molecules likely provided a better hydrophobic and electrostatic binding environment for the adsorbed protein to interact in the PC bilayer. In contrast, only two LYS-28 residues anchored the protein to the tighter packed PC leaflet of the PC/PS bilayer. Tightly packed PC molecules in the PC/PS bilayer presented an environment that discouraged the hydrophobic interactions and provided fewer anchoring sites towards the lower leaflet. Fewer and relatively weaker hydrophobic anchoring sites in the PC/PS bilayer therefore gave rise to stronger protein–protein interaction among the residues of the adsorbed protein.
Stronger protein–protein and weaker protein–lipid interactions may also explain the larger protein tilt on the PC/PS versus PC bilayer surfaces (Figs. 6 and 7). Additionally, the observed longer time lag of the protein attachment (~10 ns for the PC/PS bilayer surface versus ~1 ns for the PC bilayer surface) implies weaker protein–lipid interaction during the attachment phase of protein binding on membrane surface as shown in Table 1 and Fig. 2 of Data in Brief (Cheng et al., 2015b). However, after the protein was firmly attached to the bilayer, we observed more localized protein-induced bilayer thinning involving only a few residues of the protein (Fig. 6) in the PC/PS bilayer versus PC bilayer. This further supports the weaker protein–lipid interaction mechanism of the protein on the PC/PS surface.
The asymmetric PC/PS and the control symmetric PC membranes remained in the bilayer form throughout the entire simulation time. Neither lipid flip-flop (lipid exchange between the two lipid leaflets) nor pore formation within the bilayer was detected. In this study, the PC (protein-contact) and PS (non-protein-contact) leaflets in the asymmetric PC/PS membrane were extracted from stable simulated single-component PC and single-component PS bilayers (Gurtovenko and Vattulainen, 2008). These separate PC and PS bilayers were simulated under the same NPT simulation protocols and force fields as in this study. Due to the difference in the melting temperatures of the PS (Tm ~ 14 °C) and PC (Tm ~ −2 °C) bilayers (Silvius, 1982), PS has an intrinsically smaller surface area per lipid than PC. An independent simulation study (Esteban-Martin et al., 2009) using similar simulation protocols has investigated asymmetric one-component PC bilayers containing different numbers of lipids between two lipid leaflets, or lipid-number asymmetric PC membranes. For these lipid-number asymmetric PC membranes, an increase in the lipid-number mismatch percentage gives rise to a concomitant increase in the surface area per lipid mismatch percentage between the two lipid leaflets (see Fig. 13 of Data in Brief (Cheng et al., 2015b)). Here, the lipid-number mismatch percentage is defined as the difference in the lipid-numbers between two leaflets divided by the larger of the lipid-numbers in one of the leaflets and multiplied by 100. Similarly, the surface area per lipid mismatch percentage is defined as the difference in the surface area per lipid between two leaflets divided by the larger of the surface area per lipid in one of the leaflets and multiplied by 100. Stable and unperturbed lipid bilayers with surface area mismatch as large as 60% in lipid-number asymmetric PC membranes (Esteban-Martin et al., 2009) were observed. Our lipid-headgroup asymmetric PC/PS bilayer system had a lipid-number mismatch of ~17% giving rise to a surface area mismatch of ~17% (see Fig. 13 of Data in Brief (Cheng et al., 2015b)), well below the 60% threshold.
Due to the mismatch in the surface area per lipid betweenthe two leaflets, the lipid-number asymmetric PC membranes (Esteban-Martin et al., 2009; Ollila et al., 2009; Kasson et al., 2013) were not at equilibrium but in metastable, or kinetically trapped, bilayer states with imbalanced lateral pressure and surface tension among the lipids. Interestingly, several lipid-number asymmetric PC membranes with surface area mismatch greater than 30% became porated after a temperature jump (Esteban-Martin et al., 2009), suggesting the existence of an energy barrier separating the metastable bilayer state and the porated membrane state. In our PC/PS system, we have both lipid-number asymmetry and lipid-headgroup asymmetry. Unlike previous studies (Esteban-Martin et al., 2009; Ollila et al., 2009; Kasson et al., 2013; Park et al., 2015), we did not manually remove the lipids in one leaflet of a simulated bilayer to create the lipid-number asymmetric membrane. The lipid-numberasymmetry in our PC/PS system was necessary to the construction of the lipid bilayer because PC and PS have intrinsically different surface areas in separate single-component bilayers (see above). Interestingly, a recent simulation work, using the all-atom CHARMM C36 force fields, confirmed that the lipid-number asymmetric PC membranes were also in a metastable bilayer state with surface area mismatch as large as 25% (Park et al., 2015), the calculated 17% mismatch of our asymmetric PC/PS membranes is below this threshold. Our lipid-number mismatch of 17% and surface area mismatch of 17%, in the PC/PS membranes follow the same linear relationship in the surface area mismatch versus lipid-number mismatch plots from two independent simulation studies (Esteban-Martin et al., 2009; Park et al., 2015) on asymmetric PC membranes (see Fig. 13 of Data in Brief (Cheng et al., 2015b)). Therefore, we speculate that our maximally lipid-headgroup asymmetric PC/PS membranes were also in metastable bilayer states, as in the previously reported lipid-number asymmetric PC membranes.
Since the protein adsorbed to the same one-component PC leaflet in both symmetric PC and maximally asymmetric PC/PS bilayers, the difference in the lipid packing between the two systems is likely an important membrane property that influenced the protein structural transitions on the membrane surfaces. However, the observed effects may not exclusively be due to the lipid packing difference. Other membrane properties associated with the asymmetric lipid distribution, such as transmembrane potential, coupling of the lower leaflet to the upper leaflet and the local pressure may be involved. Examination of the local electric field suggests that the transmembrane potential has no significant effects on the adsorbed protein. The coupling of the lower PS leaflet with smaller surface area to the upper PC leaflet may affect the adsorbed protein structures. As matter of fact, we see strong coupling of the PC and PS leaflet from the acyl chain order parameter plots when compared the AL lipids with the nAL lipids in the lower leaflet (see Fig. 9). Recent studies suggested the importance of the local pressure in affecting the protein surface structures (Esteban-Martin et al., 2009; Kasson et al., 2013). Those studies also used asymmetric bilayer created with different numbers of lipids in the leaflets of the bilayer, or different surface areas. The increase in the lateral pressure is consistent with the decrease in the surface area per lipid in the leaflet. Therefore, the change in lateral pressure is a consequence of the change in surface area, and may affect the protein transitions on surface. More work is still needed to understand the effects of various membrane properties on absorbed protein structures in asymmetric bilayers.
As with any unconstrained NPT MD simulations of a protein/lipid bilayer, the results can depend strongly on the initial configurations of the protein and membrane assemblies. This is a result of poor phase space sampling, limited computing resources, and high free energy barriers separating various protein secondary and higher order structures (Buchete and Hummer, 2007; Stefani, 2008; Derreumaux, 2013; Poojari and Strodel, 2013). The use of efficient simulation strategies for effective phase space sampling like multiscale simulations (Derreumaux, 2013; Cheng et al., 2015a; Nasica-Labouze et al., 2015) are needed to further explore protein structures on surfaces, and to uncover deeper insertion states of membrane-active beta-sheet rich proteins.
Finally, the results of any MD simulations depend on the force fields (Poger and Mark, 2012; Kukol, 2015; Sun et al., 2015). Our primary incentive of choosing GROMOS87/Berger lipid force field was to compare our results with the published studies using the identical forces fields. These studies include lipid-headgroup asymmetric charged membranes (Gurtovenko and Vattulainen, 2008), the lipid-number asymmetric membranes (Esteban-Martin et al., 2009), monomeric beta-amyloid protein interactions with symmetric membranes (Qiu et al., 2011; Manna and Mukhopadhyay, 2013; Qiu et al., 2014; Cheng et al., 2015a), and other non-beta-amyloid protein interactions with symmetric membranes proteins (Herce and Garcia, 2007; Dunkin et al., 2011; Huang and Garcia, 2013; Pourmousa and Karttunen, 2013). Furthermore, since we were interested in the effect of an electric field induced on the asymmetric distribution of charged lipid headgroups, it was important to use a united atom force field. This is because all atom force field overestimates the dipole or intramembrane potential from the partial charges on the aliphatic lipid atoms (Pastor and Mackerell, 2011). In addition, the inaccuracy estimations of interactions of ions with lipid headgroup and the surface tension of bilayer versus monolayer have been identified (Pastor and Mackerell, 2011). No force field is perfect. Continuous adjustments of the force field parameters to deal with the limitations and validations of the results with experiments are required (Venable et al., 2013; Kukol, 2015; Torres-Sanchez et al., 2015; Vanommeslaeghe and MacKerell, 2015). In addition, we do not add additional salt in our simulations. This is because our focus is on the interactions between lipid and proteins without the complications of salt–lipid and salt–protein interactions. As indicated in a recent review on force fields (Sun et al., 2015), there is ongoing debates and refinement of force field parameter for ion or salt interactions with lipids, particularly charged lipids (Pandit and Berkowitz, 2002; Bockmann et al., 2003; Pandit et al., 2003; Mukhopadhyay et al., 2004; Cordomi et al., 2009; Pastor and Mackerell, 2011; Hu et al., 2013; Venable et al., 2013).
Since there is no perfect force field, it is important to gauge our results in the context of the limitations or weaknesses of the GROMOS87/Berger lipid force field. This force field may have two primary defects or concerns that may affect our results and interpretations. First, for model octanol and lipid bilayer systems, the positively charged amino acid residues, ARG and LYS, may have higher interaction potential, and the potential associated with LYS is larger than ARG (Tieleman et al., 2006; MacCallum et al., 2008; Sun et al., 2015). Second, the lipid-peptide interactions may be overestimated (Tieleman et al., 2006; Kukol, 2009, 2015).
To address the first concern of charged residues, we notice that the LYS residues (LYS-16 and LYS-28) are exclusively located in the C-terminal domains and the ARG residues (ARG-5) in the N-terminal domains of D42 dimer. A large fraction of D42 interacted with the PC-leaflet, and we did not see unusual attachment or appreciably different behavior for these residues, as evident from the residue-lipid minimum distances of ~0.2 or less as given in Fig. 6.
The second concern of the stronger lipid-peptide interactions could affect our results in three ways. First, the possible increase free energy for binding means that there is a higher probability of dimer attaching to the membrane surface. We do not consider this a deficiency; the stronger lipid–protein interactions make it easier to sample surface states. However, this issue is probably why all our replicate dimers attached to the bilayer surface. Otherwise, we might expect some replicates to show the dimer leaving the bilayer and diffuse away. Second, the increased interaction strength would affect the time scale of events, although it is not clear in what way. If the interfacial interaction is strong, the interaction may slow penetration of dimer towards the center of the bilayer. Alternatively, since a large fraction of the protein interacts with the bilayer surface, the speed of dimer penetration may be enhanced. Third, the dimer could be less stable in the membrane, which would increase the probability of the unfolded state and decrease the probability of the formation of an ordered state, like beta-sheet structure. Overall, it is very important to note that differential effect between the two PC and PC/PS bilayer systems will not be significantly affected by the choice of force field; if there is a difference between the two bilayer systems, that will still exist when investigated by other force fields, and the relative magnitude of the difference should be independent of the force fields.
Comparison of different force fields is a continuous effort in the field of simulations and modeling of various membrane systems. A united atom GROMOS87/Berger force field (see Section 2) used in this work represents a reasonable force field to study complex systems, particularly those involving ions and anionic lipids. All atom force fields, like CHARMM and AMBER, united atom force fields, like CHARMM—united atom, GROMOS and mixed all atom and united atom force fields are constantly developed and parameterized with experiments. At present, all atom force fields have limitations in simulating membrane potential and ion-lipid interactions involving protein, and are therefore not ready to simulate large, multiple component membrane systems containing anionic lipids, ions and protein (Cordomi et al., 2009; Pastor and Mackerell, 2011; Hu et al., 2013; Venable et al., 2013).
The ultimate test of force fields is how well they agree with experiments. GROMOS87/Berger force field indicates good agreement of simulation results with a broad range of available experimental results, e.g., surface area per lipid, membrane potential and ion-lipid interactions (Mukhopadhyay et al., 2004; Gurtovenko and Vattulainen, 2008). GROMOS87/Berger force field has successfully been used for investigating membranes and protein–lipid interactions in various systems, particularly mixed lipid bilayers containing cholesterol and charged lipids (Polyansky et al., 2005; Herce and Garcia, 2007; Gurtovenko and Vattulainen, 2008; Esteban-Martin et al., 2009; Dunkin et al., 2011; Manna and Mukhopadhyay, 2011a; Manna and Mukhopadhyay, 2011b; Qiu et al., 2011; Huang and Garcia, 2013; Pourmousa and Karttunen, 2013; Gurtovenko and Lyulina, 2014; Qiu et al., 2014; Cheng et al., 2015a). Various studies using the GROMOS87/Berger lipid force field (Herce and Garcia, 2007; Dunkin et al., 2011; Huang and Garcia, 2013; Pourmousa and Karttunen, 2013) have demonstrated the pore-forming behaviors of various cationic proteins interacting with symmetric membranes. These pore-forming behaviors agree well with experiments. Beta-amyloid proteins are highly polymorphic, both in solution and in model lipid membranes. Aggregates of beta-amyloid proteins are known to bind to membranes (Stefani, 2012; Nasica-Labouze et al., 2015). However, no high-resolution protein structures of D42 dimer, or any membrane-active proteins, on any symmetric or asymmetric membrane surfaces are experimentally available. Our predictions of binding kinetics and protein structures of beta-amyloid dimer on membrane surface await validations from future high-resolution, single-molecule experiments.
In any MD simulation study, it is critical to carefully consider the system size and the associated effects of periodic boundary condition imposed on the simulations. If the simulation system size is too small, interactions between atoms at the edges of the simulation box can lead to artifacts, which could bias the results of the simulation study (Hu et al., 2013). In our work, we constructed a large asymmetric double bilayer PC/PS system (~17 × 17 × 33 nm3) by tiling 9 times of a small stable asymmetric bilayer system of identical structures from a previously published work (Gurtovenko and Vattulainen, 2008). In addition, the symmetric PC bilayer of comparable size ~13 × 14 × 14 nm3 was built as a control. The surface area per lipid and membrane potential of our membrane systems agree favorably with previous simulation studies (Gurtovenko and Vattulainen, 2008; Esteban-Martin et al., 2009), and the surface area per lipid agrees with experimental studies (MacDonald et al., 1976; Kucerka et al., 2005). This agreement indicates that system size effects and periodic boundary condition issues are unlikely to generate artifacts in our study. Additionally, previous work has been done to examine the effect of system size on packing order of lipids. For example, a four times (Esteban-Martin et al., 2009) or two times larger (Park et al., 2015) lipid-number asymmetric PC membrane system has been simulated to compare with the smaller system. In both cases, no appreciable difference in the packing order or surface area per lipid mismatch was detected. In our work, similar packing order mismatch was found in both the smaller and the larger asymmetric PC/PS system.
There are many ways to design lipid headgroup asymmetric bilayer membranes. Matching the surface area per lipid in a leaflet of an asymmetric bilayer to that of the single component symmetric bilayer by the adjusting the number of lipids in one leaflet is a critical step in designing the initial structure of any stable lipid headgroup asymmetric bilayer (Lopez Cascales et al., 2006; Gurtovenko and Vattulainen, 2008; Patel and Balaji, 2008). There is always uncertainty in the number of lipids that should be assigned to each bilayer leaflet, and how this uncertainty is resolved is part of the design consideration of the system. This is true in the maximally asymmetric PC/PS (Gurtovenko and Vattulainen, 2008) and other partially asymmetric bilayer systems (Lopez Cascales et al., 2006; Patel and Balaji, 2008). If there is a surplus or deficit of lipids in one leaflet the packing density of both leaflets may be affected, because the leaflet with a deficit of lipids will tend to exert forces that compress the leaflet that contains a relative surplus. The partially asymmetric bilayers were constructed by substituting a single-component bilayer with another lipid type, e.g., PS (Lopez Cascales et al., 2006) or GM1 (Patel and Balaji, 2008). As shown in Section 2, we used equilibrated leaflets from single component PS and PC membranes to construct the maximally asymmetric PC/PS bilayer without substituting any lipids in both leaflets.
How do our results provide insights into understanding the beta-amyloid oligomers interactions with cell membranes? These interactions are associated with the early events of pathogenic amyloid cascade pathways, particularly membrane pore formation (Nasica-Labouze et al., 2015). A neurotoxic beta-amyloid dimer, the smallest unit of beta-amyloid oligomers, has recently been isolated from the brain of Alzheimer's patients (Jin et al., 2011). Due to their highly polymorphic and aggregation-prone behavior in solution, the exact structure of this dimer or any other pathogenic multimers of beta-amyloid, on membrane surfaces are unknown (Nasica-Labouze et al., 2015). The U-shaped fibrillar dimer in this study represents a possible structure, or subunit of a larger structure, that may lead to membrane pore formation (Jang et al., 2007). In this work, the dimer was able to generate significant bilayer thinning in both PC and PC/PS bilayers, a possible first step in membrane pore formation (Fig. 10). A more localized bilayer thinning in the PC/PS bilayer was detected. Although, no pore formation was detected in any of our simulated replicates, the dimer was more aggregated and tilted in the PC/PS versus the PC bilayer system. This more tightly packed and tilted structure may have higher propensity to form membrane pores in the PC/PS bilayer than in the control PC bilayer.
It is important to note that a beta-amyloid pore model consists of multiple U-shaped beta-amyloid chains with the loop and N-terminal domain regions on opposite sides of the bilayer (Jang et al., 2013). Therefore, the dimer with a tightly packed and tilted orientation in PC/PS bilayer is more likely to migrate from the protein-contact leaflet to the non-protein-contact leaflet, when compared to the dimer with loosely packed and non-tilted orientation in the PC bilayer. Of course, a large free energy barrier exists in the protein translocation process. Hence, the negative electric field in the non-protein-contact PS leaflet (Fig. 3) can significantly lower this energy barrier in the PC/PS bilayer, due to electrostatic interactions with the positively charged Lys-28 residue in the loop region of the dimer (Fig. 2). In other words, once the loop region of the tilted dimer reaches the PS leaflet, the negative electric field will promote the anchoring of the loop region of the dimer towards the headgroup region, which may eventually form a membrane pore in the bilayer. However, more work is needed to uncover this possible membrane pore formation mechanism, which may involve single or multiple dimers in the asymmetric PC/PS bilayer.
Previous studies have also suggested the importance of anionic lipids on the surface in promoting beta-amyloid binding, folding, and aggregation (Wong et al., 2009; Davis and Berkowitz, 2010; Yu et al., 2013). Our preliminary study of D42 binding to lipid surface containing PS revealed a similar, but slightly stronger, domain aggregation effect than the ordered PC surface (results not shown). Future work is still needed to study the regulatory effects of lipid order and anionic lipids.
Conclusions drawn from this work may also apply to other isoforms of beta-amyloid multimers. A parallel study using D40, a beta-amyloid with an almost identical structure to D42, but containing only 40 residues in each chain, revealed similar aggregation behavior as found in D42 (see Figs. 9–11 of Data in Brief (Cheng et al., 2015b)). Additionally, membrane-active proteins, such as the large family of aggregation-prone amyloid forming proteins of different primary sequences (Gorbenko and Kinnunen, 2006) and anti-microbial proteins (Jang et al., 2010), share similar structural motifs rich in beta-sheets. Therefore, the lipid–protein interaction mechanisms of beta-sheet containing aggregation-prone D42 on the surface of asymmetric and symmetric bilayers may also be operative on those proteins as well.
5. Conclusion
We have used MD simulations to study the effects of maximally asymmetric transbilayer distribution of anionic lipids on the structures and protein–lipid interactions on a membrane surface. We speculate that the lipid packing rather than the electric field has a larger effect in promoting stronger protein–protein interactions on the surface of the asymmetric PC/PS bilayer versus the symmetric PC bilayer. Additionally, the more aggregated and tilted D42 has higher propensity in promoting membrane pore formation in the asymmetric PC/PS bilayer.
Acknowledgements
This work was supported by the Robert A. Welch Research Foundation grant (D-1158), NIH grant (GM090897-02), National Science Foundation (ACI 1531594), Williams Endowment fund of Trinity University, Texas Advanced Computing Center (TACC) for the use of Lonestar Cluster under the project (G-803132) “Protein Unfolding in Lipid Membranes”, and Texas Tech University High Performance Computing Center (TTU-HPCC) for supporting in computing. The authors would like to acknowledge the valuable help of Dr. Andrey A. Gurtovenko of the Institute of Macromolecular Compounds, Russian Academy of Sciences for allowing us to use the published asymmetric PC/PS bilayer system and valuable advice on the simulation details of the PC/PS bilayer system.
References
- Allen WJ, Lemkul JA, Bevan DR. GridMAT-MD: a grid-based membrane analysis tool for use with molecular dynamics. J. Comput. Chem. 2009;30:1952–1958. doi: 10.1002/jcc.21172. [DOI] [PubMed] [Google Scholar]
- Basyn F, Charloteaux B, Thomas A, Brasseur R. Prediction of membrane protein orientation in lipid bilayers: a theoretical approach. J. Mol. Graph Model. 2001;20:235–244. doi: 10.1016/s1093-3263(01)00114-0. [DOI] [PubMed] [Google Scholar]
- Bekker H, Berendsen HJC, Dijkstra EJ, Achterop S, van Drunen R, van der Spoel D, Sijbers A, Keegstra H, Reitsma B, Renardus MKR. Physics Computing 92. World Scientific; Singapore: 1993. Gromacs: A Parallel Computer for Molecular Dynamics Simulations. [Google Scholar]
- Berendsen HD, van der Spoel D, van Drunen R. GROMACS: a messagepassing parallel molecular dynamics implementation. Comp. Phys. Comm. 1995;91:43–56. [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction Models for Water in Relation to Protein Hydration. In: Pullman B, editor. Intermolecular Forces. Reidel; Dordrecht, The Netherlands: 1981. pp. 331–342. [Google Scholar]
- Berger O, Edholm O, Jahnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmann RA, Hac A, Heimburg T, Grubmuller H. Effect of sodium chloride on a lipid bilayer. Biophys. J. 2003;85:1647–1655. doi: 10.1016/S0006-3495(03)74594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucciantini M, Cecchi C. Biological membranes as protein aggregation matrices and targets of amyloid toxicity. Methods Mol. Biol. 2010;648:231–243. doi: 10.1007/978-1-60761-756-3_15. [DOI] [PubMed] [Google Scholar]
- Buchete NV, Hummer G. Structure and dynamics of parallel beta-sheets, hydrophobic core, and loops in Alzheimer's A beta fibrils. Biophys. J. 2007;92:3032–3039. doi: 10.1529/biophysj.106.100404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity- rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Cheng KH, Qiu L, Cheng SY, Vaughn MW. Lipid insertion domain unfolding regulates protein orientational transition behavior in a lipid bilayer. Biophys. Chem. 2015a;206:22–39. doi: 10.1016/j.bpc.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SY, Chou G, Buie C, Vaughn MW, Compton C, Cheng KH. Data supporting beta-amyloid dimer structural transitions and protein-lipid interactions on asymmetric lipid bilayer surfaces using MD simulations on experimentally derived NMR protein structures. Data in Brief. 2015b doi: 10.1016/j.dib.2016.03.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SY, Duong HV, Compton C, Vaughn MW, Nguyen H, Cheng KH. Characterization of 3D Voronoi tessellation nearest neighbor lipid shells provides atomistic lipid disruption profile of protein containing lipid membranes. Biophys. Chem. 2015c;198:22–35. doi: 10.1016/j.bpc.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Solution structure of amyloid beta-peptide(1–40) in a water-micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry. 1998;37:11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- Cordomi A, Edholm O, Perez JJ. Effect of force field parameters on sodium and potassium ion binding to dipalmitoylphosphatidylcholine bilayers. J. Chem. Theory Comput. 2009;5:2125–2134. doi: 10.1021/ct9000763. [DOI] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L. Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- Davis CH, Berkowitz ML. A molecular dynamics study of the early stages of amyloid-beta(1–42) oligomerization: the role of lipid membranes. Proteins. 2010;78:2533–2545. doi: 10.1002/prot.22763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derreumaux P. Coarse-grained models for protein folding and aggregation. Methods Mol. Biol. 2013;924:585–600. doi: 10.1007/978-1-62703-017-5_22. [DOI] [PubMed] [Google Scholar]
- Dunkin CM, Pokorny A, Almeida PF, Lee HS. Molecular dynamics studies of transportan 10 (tp10) interacting with a POPC lipid bilayer. J. Phys. Chem. B. 2011;115:1188–1198. doi: 10.1021/jp107763b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ. Molecular chaperones: assisting assembly in addition to folding. Trends Biochem. Sci. 2006;31:395–401. doi: 10.1016/j.tibs.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen L. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- Esteban-Martin S, Risselada HJ, Salgado J, Marrink SJ. Stability of asymmetric lipid bilayers assessed by molecular dynamics simulations. J. Am. Chem. Soc. 2009;131:15194–15202. doi: 10.1021/ja904450t. [DOI] [PubMed] [Google Scholar]
- Fadeel B, Xue D. The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Crit. Rev. Biochem. Mol. Biol. 2009;44:264–277. doi: 10.1080/10409230903193307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, de Cathelineau A, Daleke DL, Henson PM, Bratton DL. Loss of phospholipid asymmetry and surface exposure of phosphatidylserine is required for phagocytosis of apoptotic cells by macrophages and fibroblasts. J. Biol. Chem. 2001;276:1071–1077. doi: 10.1074/jbc.M003649200. [DOI] [PubMed] [Google Scholar]
- Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Gorbenko G, Trusova V. Protein aggregation in a membrane environment. Adv. Protein Chem. Struct. Biol. 2011;84:113–142. doi: 10.1016/B978-0-12-386483-3.00002-1. [DOI] [PubMed] [Google Scholar]
- Gorbenko GP, Kinnunen PK. The role of lipid–protein interactions in amyloid-type protein fibril formation. Chem. Phys. Lipids. 2006;141:72–82. doi: 10.1016/j.chemphyslip.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Gurtovenko AA, Lyulina AS. Electroporation of asymmetric phospholipid membranes. J. Phys. Chem. B. 2014;118:9909–9918. doi: 10.1021/jp5028355. [DOI] [PubMed] [Google Scholar]
- Gurtovenko AA, Vattulainen I. Membrane potential and electrostatics of phospholipid bilayers with asymmetric transmembrane distribution of anionic lipids. J. Phys. Chem. B. 2008;112:4629–4634. doi: 10.1021/jp8001993. [DOI] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- Herce HD, Garcia AE. Molecular dynamics simulations suggest a mechanism for translocation of the HIV–1 TAT peptide across lipid membranes. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20805–20810. doi: 10.1073/pnas.0706574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Bekke H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J. Comp. Chem. 1997;13:952–962. [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. Gromacs 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Holtje M, Forster T, Brandt B, Engels T, von Rybinski W, Holtje HD. Molecular dynamics simulations of stratum corneum lipid models: fatty acids and cholesterol. Biochim. Biophys. Acta. 2001;1511:156–167. doi: 10.1016/s0005-2736(01)00270-x. [DOI] [PubMed] [Google Scholar]
- Hu Y, Ou S, Patel S. Free energetics of arginine permeation into model DMPC lipid bilayers: coupling of effective counterion concentration and lateral bilayer dimensions. J. Phys. Chem. B. 2013;117:11641–11653. doi: 10.1021/jp404829y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Garcia AE. Free energy of translocating an arginine-rich cellpenetrating peptide across a lipid bilayer suggests pore formation. Biophys. J. 2013;104:412–420. doi: 10.1016/j.bpj.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(33–38):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jang H, Connelly L, Arce FT, Ramachandran S, Kagan BL, Lal R, Nussinov R. Mechanisms for the insertion of toxic, fibril-like beta-amyloid oligomers into the membrane. J. Chem. Theory Comput. 2013;9:822–833. doi: 10.1021/ct300916f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Teran Arce F, Ramachandran S, Capone R, Lal R, Nussinov R. Structural convergence among diverse, toxic beta-sheet ion channels. J. Phys. Chem. B. 2010;114:9445–9451. doi: 10.1021/jp104073k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Zheng J, Nussinov R. Models of beta-amyloid ion channels in the membrane suggest that channel formation in the bilayer is a dynamic process. Biophys. J. 2007;93:1938–1949. doi: 10.1529/biophysj.107.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Shepardson N, Yang T, Chen G, Walsh D, Selkoe DJ. Soluble amyloid beta-protein dimers isolated from Alzheimer cortex directly induce Tau hyperphosphorylation and neuritic degeneration. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5819–5824. doi: 10.1073/pnas.1017033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kanekiyo T, Xu H, Bu G. ApoE and Abeta in Alzheimer's disease: accidental encounters or partners? Neuron. 2014;81:740–754. doi: 10.1016/j.neuron.2014.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasson PM, Hess B, Lindahl E. Probing microscopic material properties inside simulated membranes through spatially resolved three-dimensional local pressure fields and surface tensions. Chem. Phys. Lipids. 2013;169:106–112. doi: 10.1016/j.chemphyslip.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucerka N, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J. Membr. Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- Kukol A. Lipid models for united-atom molecular dynamics simulations of proteins. J. Chem. Theory Comput. 2009;5:615–626. doi: 10.1021/ct8003468. [DOI] [PubMed] [Google Scholar]
- Kukol A. Lipid membranes for membrane proteins. Methods Mol. Biol. 2015;1215:73–90. doi: 10.1007/978-1-4939-1465-4_4. [DOI] [PubMed] [Google Scholar]
- Kurre P, Morris J, Miller AD, Kiem HP. Envelope fusion protein binding studies in an inducible model of retrovirus receptor expression and in CD34(+) cells emphasize limited transduction at low receptor levels. Gene Ther. 2001;8:593–599. doi: 10.1038/sj.gt.3301438. [DOI] [PubMed] [Google Scholar]
- Lindahl E, Hess B, van der Spoel D. GROMACS 3. 0: a package for molecular simulation and trajectory analysis. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Lopez Cascales JJ, Otero TF, Smith BD, Gonzalez C, Marquez M. Model of an asymmetric DPPC/DPPS membrane: effect of asymmetry on the lipid properties. A molecular dynamics simulation study. J. Phys. Chem. B. 2006;110:2358–2363. doi: 10.1021/jp0562680. [DOI] [PubMed] [Google Scholar]
- Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Dobeli H, Schubert D, Riek R. 3D structure of Alzheimer's amyloid-beta(1–42) fibrils. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum JL, Bennett WF, Tieleman DP. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys. J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald RC, Simon SA, Baer E. Ionic influences on the phase transition of dipalmitoylphosphatidylserine. Biochemistry. 1976;15:885–891. doi: 10.1021/bi00649a025. [DOI] [PubMed] [Google Scholar]
- Manna M, Mukhopadhyay C. Cholesterol driven alteration of the conformation and dynamics of phospholamban in model membranes. Phys. Chem. Chem. Phys. 2011a;13:20188–20198. doi: 10.1039/c1cp21793c. [DOI] [PubMed] [Google Scholar]
- Manna M, Mukhopadhyay C. Molecular dynamics simulations of the interactions of kinin peptides with an anionic POPG bilayer. Langmuir. 2011b;27:3713–3722. doi: 10.1021/la104046z. [DOI] [PubMed] [Google Scholar]
- Manna M, Mukhopadhyay C. Binding, conformational transition and dimerization of amyloid-beta peptide on GM1-containing ternary membrane: insights from molecular dynamics simulation. PLoS One. 2013;8:e71308. doi: 10.1371/journal.pone.0071308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Hartl FU. The effect of macromolecular crowding on chaperoninmediated protein folding. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1107–1112. doi: 10.1073/pnas.94.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaurin J, Chakrabartty A. Characterization of the interactions of Alzheimer beta-amyloid peptides with phospholipid membranes. Eur. J. Biochem. 1997;245:355–363. doi: 10.1111/j.1432-1033.1997.t01-2-00355.x. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P, Monticelli L, Tieleman DP. Molecular dynamics simulation of a palmitoyl-oleoyl phosphatidylserine bilayer with Na+ counterions and NaCl. Biophys. J. 2004;86:1601–1609. doi: 10.1016/S0006-3495(04)74227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag S, Sarkar B, Chandrakesan M, Abhyanakar R, Bhowmik D, Kombrabail M, Dandekar S, Lerner E, Haas E, Maiti S. A folding transition underlies the emergence of membrane affinity in amyloid-beta. Phys. Chem. Chem. Phys. 2013;15:19129–19133. doi: 10.1039/c3cp52732h. [DOI] [PubMed] [Google Scholar]
- Nasica-Labouze J, Nguyen PH, Sterpone F, Berthoumieu O, Buchete NV, Cote S, De Simone A, Doig AJ, Faller P, Garcia A, Laio A, Li MS, Melchionna S, Mousseau N, Mu Y, Paravastu A, Pasquali S, Rosenman DJ, Strodel B, Tarus B, Viles JH, Zhang T, Wang C, Derreumaux P. Amyloid beta protein and Alzheimer's disease: when computer simulations complement experimental studies. Chem. Rev. 2015;115:3518–3563. doi: 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollila OH, Risselada HJ, Louhivuori M, Lindahl E, Vattulainen I, Marrink SJ. 3D pressure field in lipid membranes and membrane-protein complexes. Phys. Rev. Lett. 2009;102:078101. doi: 10.1103/PhysRevLett.102.078101. [DOI] [PubMed] [Google Scholar]
- Pandit SA, Berkowitz ML. Molecular dynamics simulation of dipalmitoylphosphatidylserine bilayer with Na+ counterions. Biophys. J. 2002;82:1818–1827. doi: 10.1016/S0006-3495(02)75532-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit SA, Bostick D, Berkowitz ML. Molecular dynamics simulation of a dipalmitoylphosphatidylcholine bilayer with NaCl. Biophys. J. 2003;84:3743–3750. doi: 10.1016/S0006-3495(03)75102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Beaven AH, Klauda JB, Im W. How tolerant are membrane simulations with mismatch in area per lipid between leaflets? J. Chem. Theory Comput. 2015;11:3466–3477. doi: 10.1021/acs.jctc.5b00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor RW, Mackerell AD., Jr. Development of the CHARMM force field for lipids. J. Phys. Chem. Lett. 2011;2:1526–1532. doi: 10.1021/jz200167q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RY, Balaji PV. Characterization of symmetric and asymmetric lipid bilayers composed of varying concentrations of ganglioside GM1 and DPPC. J. Phys. Chem. B. 2008;112:3346–3356. doi: 10.1021/jp075975l. [DOI] [PubMed] [Google Scholar]
- Poger D, Mark AE. Lipid bilayers: the effect of force field on ordering and dynamics. J. Chem. Theory Comput. 2012;8:4807–4817. doi: 10.1021/ct300675z. [DOI] [PubMed] [Google Scholar]
- Polyansky AA, Volynsky PE, Nolde DE, Arseniev AS, Efremov RG. Role of lipid charge in organization of water/lipid bilayer interface: insights via computer simulations. J. Phys. Chem. B. 2005;109:15052–15059. doi: 10.1021/jp0510185. [DOI] [PubMed] [Google Scholar]
- Poojari C, Strodel B. Stability of transmembrane amyloid beta-peptide and membrane integrity tested by molecular modeling of site-specific Abeta42 mutations. PLoS One. 2013;8:e78399. doi: 10.1371/journal.pone.0078399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourmousa M, Karttunen M. Early stages of interactions of cell-penetrating peptide penetratin with a DPPC bilayer. Chem. Phys. Lipids. 2013;169:85–94. doi: 10.1016/j.chemphyslip.2013.02.011. [DOI] [PubMed] [Google Scholar]
- Qiu L, Buie C, Cheng KH, Vaughn MW. Scaling and alpha-helix regulation of protein relaxation in a lipid bilayer. J. Chem. Phys. 2014;141:225101. doi: 10.1063/1.4902229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Buie C, Reay A, Vaughn MW, Cheng KH. Molecular dynamics simulations reveal the protective role of cholesterol in beta-amyloid protein- induced membrane disruptions in neuronal membrane mimics. J. Phys. Chem. B. 2011;115:9795–9812. doi: 10.1021/jp2012842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Silvius JR. In: Thermotropic Phase Transitions of Pure Lipids in Model Membranes and Their Modifications by Membrane Proteins. lipid–protein Interactions. Jost PC, Griffith OH, editors. John Wiley and Sons; New York: 1982. [Google Scholar]
- Stefani M. Protein folding and misfolding on surfaces. Int. J. Mol. Sci. 2008;9:2515–2542. doi: 10.3390/ijms9122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani M. Structural features and cytotoxicity of amyloid oligomers: implications in Alzheimer's disease and other diseases with amyloid deposits. Prog. Neurobio. 2012;99:226–245. doi: 10.1016/j.pneurobio.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Sun D, Forsman J, Woodward CE. Evaluating force fields for the computational prediction of ionized arginine and lysine side-chains partitioning into lipid bilayers and octanol. J. Chem. Theory Comput. 2015;11:1775–1791. doi: 10.1021/ct501063a. [DOI] [PubMed] [Google Scholar]
- Theodorou DN, Suter UW. Geometrical considerations in model systems with periodic boundaries. J. Chem. Phys. 1985;82:955–966. [Google Scholar]
- Tieleman DP, Maccallum JL, Ash WL, Kandt C, Xu Z, Monticelli L. Membrane protein simulations with a united-atom lipid and all-atom protein model: lipid–protein interactions, side chain transfer free energies and model proteins. J. Phys. Condens. Matter. 2006;18:S1221–S1234. doi: 10.1088/0953-8984/18/28/S07. [DOI] [PubMed] [Google Scholar]
- Tofoleanu F, Brooks BR, Buchete NV. Modulation of Alzheimer's Abeta protofilament-membrane interactions by lipid headgroups. ACS Chem. Neurosci. 2015;6:446–455. doi: 10.1021/cn500277f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofoleanu F, Buchete NV. Molecular interactions of Alzheimer's Abeta protofilaments with lipid membranes. J. Mol. Biol. 2012;421:572–586. doi: 10.1016/j.jmb.2011.12.063. [DOI] [PubMed] [Google Scholar]
- Torres-Sanchez A, Vanegas JM, Arroyo M. Examining the mechanical equilibrium of microscopic stresses in molecular simulations. Phys. Rev. Lett. 2015;114:258102. doi: 10.1103/PhysRevLett.114.258102. [DOI] [PubMed] [Google Scholar]
- van Buuren AR, Marrink SJ, Berendsen HJC. A molecular dynamics study of the decane/water interface. J. Phys. Chem. 1993;97:9206–9212. [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- van der Spoel D, van Buuren AR, Tieleman DP, Berendsen HJC. Molecular dynamics simulations of peptides from BPTI: a closer look at amide-aromatic interactions. J. Biomol. NMR. 1996;8:229–238. doi: 10.1007/BF00410322. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K, MacKerell AD., Jr. CHARMM additive and polarizable force fields for biophysics and computer-aided drug design. Biochim. Biophys. Acta. 2015:861–871. doi: 10.1016/j.bbagen.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venable RM, Luo Y, Gawrisch K, Roux B, Pastor RW. Simulations of anionic lipid membranes: development of interaction-specific ion parameters and validation using NMR data. J. Phys. Chem. B. 2013;117:10183–10192. doi: 10.1021/jp401512z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vymetal J, Vondrasek J. Gyration- and inertia-tensor-based collective coordinates for metadynamics. Application on the conformational behavior of polyalanine peptides and Trp-cage folding. J. Phys. Chem. A. 2011;115:11455–11465. doi: 10.1021/jp2065612. [DOI] [PubMed] [Google Scholar]
- Williamson P, Schlegel RA. Back and forth: the regulation and function of transbilayer phospholipid movement in eukaryotic cells. Mol. Membr. Biol. 1994;11:199–216. doi: 10.3109/09687689409160430. [DOI] [PubMed] [Google Scholar]
- Wong PT, Schauerte JA, Wisser KC, Ding H, Lee EL, Steel DG, Gafni A. Amyloid-beta membrane binding and permeabilization are distinct processes influenced separately by membrane charge and fluidity. J. Mol. Biol. 2009;386:81–96. doi: 10.1016/j.jmb.2008.11.060. [DOI] [PubMed] [Google Scholar]
- Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang Q, Pan Q, Zhou F, Zheng J. Molecular interactions of Alzheimer amyloid-beta oligomers with neutral and negatively charged lipid bilayers. Phys. Chem. Chem. Phys. 2013;15:8878–8889. doi: 10.1039/c3cp44448a. [DOI] [PMC free article] [PubMed] [Google Scholar]