Abstract
The immutans (im) variegation mutation of Arabidopsis has green- and white- sectored leaves due to action of a nuclear recessive gene. IM codes for PTOX, a plastoquinol oxidase in plastid membranes. Previous studies have revealed that the green and white sectors develop into sources (green tissues) and sinks (white tissues) early in leaf development. In this report we focus on white sectors, and show that their transformation into effective sinks involves a sharp reduction in plastid number and size. Despite these reductions, cells in the white sectors have near-normal amounts of plastid RNA and protein, and surprisingly, a marked amplification of chloroplast DNA. The maintenance of protein synthesis capacity in the white sectors might poise plastids for their development into other plastid types. The green and white im sectors have different cell wall compositions: whereas cell walls in the green sectors resemble those in wild type, cell walls in the white sectors have reduced lignin and cellulose microfibrils, as well as alterations in galactomannans and the decoration of xyloglucan. These changes promote susceptibility to the pathogen Pseudomonas syringae. Enhanced susceptibility can also be explained by repressed expression of some, but not all, defense genes. We suggest that differences in morphology, physiology and biochemistry between the green and white sectors is caused by a reprogramming of leaf development that is coordinated, in part, by mechanisms of retrograde (plastid-to-nucleus) signaling, perhaps mediated by ROS. We conclude that variegation mutants offer a novel system to study leaf developmental programming, cell wall metabolism and host-pathogen interactions.
Introduction
Variegated plants provide an excellent system to gain insight into mechanisms of chloroplast biogenesis and nuclear-organelle communication [1, 2, 3]. These plants have green- and white- (or yellow-) sectored leaves due to mutations in nuclear, chloroplast or mitochondrial genes. Whereas cells in the green sectors resemble wild type with respect to morphology and pigment composition (carotenoids and chlorophylls), plastids in cells of the white sectors lack pigments and organized lamellae. Over the years our research has focused on nuclear gene-induced variegations in which the cells have a uniform (mutant) genotype. This is in contrast to variegations in which the green sectors have a wild type genotype and the white sectors a mutant genotype [2].
One of the best-studied nuclear gene-induced variegations is the Arabidopsis immutans (im) mutant [4–6]. This mutation defines the gene for PTOX (plastid terminal oxidase), a plastid membrane enzyme that transfers electrons from plastoquinol (PQH2) to molecular oxygen, generating water and plastoquinone (PQ) [7–8]. PTOX activity is important for proper thylakoid development (chloroplast biogenesis), as well as for optimal functioning of mature photosynthetic membranes [5]. During early chloroplast biogenesis, when the PETC is not yet fully-assembled and functional, PTOX activity is the primary (and perhaps only) way to oxidize the PQ pool, e.g., in response to overreduction that might occur as a byproduct of redox reactions that utilize PQ as a substrate [9]. A striking example of this is the requirement for PTOX as a cofactor of the two, successive desaturation reactions of carotenoid biosynthesis, mediated by phytoene desaturase (PDS) and zeta-carotene desaturase (ZDS). A hallmark of chloroplast biogenesis is the progressive accumulation of carotenoids, and in the absence of PTOX, phytoene (the colorless substrate of PDS) accumulates because electron transfer into the PQ pool is inhibited, presumably because transfer into an overreduced pool is energetically unfavorable and/or that PQ becomes a limiting substrate for PDS as the PQH2/PQ ratio of the membrane increases [10–11]. Regardless of the precise cause, one consequence of phytoene accumulation is that carotenogenesis is blocked at the PDS step, thus preventing the accumulation of downstream, colored (photoprotective) carotenoids. This results in photooxidation of the contents of the developing organelle—the principle cause of white sector formation in im [11].
Under conditions of steady-state photosynthesis, PTOX is not able to compete with P700+ as an electron acceptor when mature leaves of wild type Arabidopsis are compared to im or to transgenic plants that overexpress PTOX [12]. Other studies, however, have established that PTOX does indeed play a role in mature chloroplasts: a) it serves as the terminal oxidase of chlororespiration (oxidation of the PQ pool in the dark) [13–14]; it helps balance the redox state of the PQ pool during cyclic electron flow (CEF) around photosystem one (PSI) [15–16]; and it acts as a photosynthetic safety valve during conditions of stress. As a safety valve, PTOX dissipates excess electron flow and thereby prevents the formation of high excitation pressures (1-qP, a relative measure of the reduction state of QA) that would otherwise promote photooxidative damage and photoinhibition [5, 17].
Whereas photooxidation is likely the primary cause of white sector formation in im, the question arises why the green sectors have normal-appearing chloroplasts, despite lacking PTOX; all im alleles isolated to date are null [5]. Our current working hypothesis is that im variegation is governed by membrane redox and excitation pressures (EPs) during the early stages of chloroplast biogenesis, when thylakoid membranes are elaborated from precursor plastids, i.e., from etioplasts in dark-grown seedlings or from proplastids in meristems [11]. According to the threshold hypothesis, developing im plastids with above-threshold redox states (overreduced, high EPs) become photooxidized, while plastids with below-threshold redox states (low EPs) develop into normal chloroplasts [5, 10]. Support for this hypothesis comes the observation that plastids do not have intermediate phenotypes, and that cells in the white sectors are heteroplastidic, i.e., they contain rare, normal-appearing chloroplasts in addition to the white plastids, which have a uniform size (decreased) and morphology. This indicates that im plastids are capable of developing independently from one another and display plastid autonomous behavior [10]. We surmise that this independence is a reflection of unique plastid biochemistries (and redox states) caused by factors such as the non-uniform distribution of photosynthetic substrates (light and CO2) across the leaf [18].
One assumption of our model is that the chaotic pattern of im variegation in the mature leaf is largely a reflection of the pattern of dicot leaf development, during which nearly all chloroplast differentiation and division processes occur in the leaf primordium (conversion of proplastids to chloroplasts), with subsequent development involving primarily cell and leaf expansion [19–20]. If this is the case, we hypothesize that the phenotypes of im plastids (green or white) are generally determined early in the leaf differentiation process, and that plastids do not change from white to green (or vice versa) in the expanding leaf. One purpose of the present research was to test this hypothesis.
Another purpose of the current investigations arose from early observations that the green leaf sectors of im have higher than normal photosynthetic rates and anatomies reminiscent of leaves adapted to growth in high-light conditions [21–22]. The white leaf sectors, on the other hand, have a normal thickness, although their palisade cells fail to expand. Accompanying these changes, the white sectors have downregulated expression of nuclear genes for plastid proteins involved in photosynthesis, and both tissue types have upregulated expression of genes for oxidative stress [23]. There are also tissue-specific alterations in the expression of genes that mediate source-sink interactions. These changes are consistent with experiments showing that the green sectors feed the white ones via apoplastic movement of sucrose from the green cells to the white ones, where it is hydrolyzed by invertase to yield products that can be utilized for energy [22].
In addition to genes for photosynthesis, oxidative stress and sink-source interactions, the global transcriptomic analyses revealed that the green and white sectors have dramatic tissue-specific alterations in the expression of genes for cell wall biosynthesis [22–23]. This suggests that cell wall remodeling plays a central role in the biochemical, physiological and morphological adaptions of each tissue-type to an absence of PTOX. Interestingly, these changes were also accompanied by modified patterns of expression of genes for pathogen response. Therefore, the second aim of our research was to gain insight into the poorly-understood cell wall determinants that promote pathogen susceptibility and response by determining the composition of cell walls in im green and white sectors, and second, by examining the response of these tissue-types to infection by the pathogen P. syringae.
Materials and Methods
Plant materials and growth condition
Arabidopsis plants with the spotty allele of im [15] were germinated and grown at 22°C under continuous illumination, first three weeks at 15 μmol m−2s−1 for 5 days, then at 50–60 μmol m−2s−1 for the rest of their growth. Col-0 served as wild type controls.
Determination of protein and RNA
Leaves were harvested from 7-week-old Col-0 and im plants (green and white sectors) and weighed. These tissues were used for both protein and RNA determinations as described previously [22–23]. For the protein analyses, leaf samples were frozen in liquid nitrogen, homogenized and resuspended in extraction buffer [10 mM NaCl; 10 mM MgCl2; 5 mM EDTA; 10 mM β-mercaptoethanol; 25 mM Tris-HCl (pH 7.5); and 1 mM PMSF]. After centrifugation (5,000g for 15 minutes), equal fresh weights of the protein extracts were subjected to 10% SDS-PAGE analysis, followed by Coomassie Blue staining (Fisher Scientific).
Total RNA was extracted using the Trizol Isolation kit (Invitrogen). The RNA samples were then loaded onto denaturing gels on a fresh weight basis; the gels contained 1% agarose and 3% formaldehyde in 10 mM phosphate buffer (pH 7.0). Following electrophoresis, the gels were stained with ethidium bromide.
cDNA preparation and quantitative real-time PCR
Total RNA was assessed on a 0.7% agarose gel, quantified and normalized by NanoDrop ND-1000 spectrophotometer. For qRT-PCR analysis, first-strand cDNAs were synthesized from 3 μg of total RNA using Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's recommendations.
Real-time qPCR was performed on an iCycler Real-Time system (Bio-Rad). Each reaction was done in a final volume of 25 μl containing 12.5 μl of SYBR Green Master Mix reagent (Fermentas), 300 ng of cDNA (for gene expression analysis) or 200 ng of total DNA (for chloroplast DNA quantification) sample, and 200 nM gene-specific primers (S1 Table), designed to generate products of 150–200 bp using Vector NTI 9.0 software (Informax). The qPCR conditions were as follows: 50°C for 2 min, 95°C for 10 min, 45 cycles of 95°C for 15 s, 60°C for 30 sec, and 72°C for 45 sec. At the end of the 45 cycles, a melting curve was generated to analyze the specificity of the reactions. Each sample was tested in three technical replicates. Expression level of the Actin 2 gene was used as the endogenous control. The relative expression level or chloroplast DNA concentration was calculated as 2–ΔΔCT [ΔCT = CT, gene of interest − CT, Actin2. ΔΔCT = ΔCT, treatment − ΔCT, Col-0]. Absolute numbers were calculated as described previously (Lu et al., 2012).
Histochemical procedures
Cell viability
Cell viability was monitored using the Evans Blue [24]. For this assay, excised leaflets from im and Col-0 (7-week-old) were vacuum infiltrated with 0.1% (w/v) Evans blue (Sigma-Aldrich) for 10 min, then destained by washing three times with distilled water. The leaves were observed with a Zeiss Axioskop 2 plus (Carl Zeiss) microscope. In this assay, necrotic cells are stained, but living cells exclude the dye.
Chloroplast DNA
Chloroplast DNA was visualized by confocal microscopy of DAPI-stained tissues [25]. DAPI (4',6-diamidino-2-phenylindole) is a fluorescent dye that has high affinity for AT-rich regions of DNA. For this procedure, green and white segments were dissected from 7-week-old im and Col-0 leaves, fixed in 2.5% (v/v) glutaraldehyde, then stained for 30 min with 1 ug/mL DAPI. The destained samples were examined with a Carl Zeiss LSM 510 META confocal microscope, using 440 nm excitation and 685 nm emission wavelengths for chlorophyll and 410 nm excitation and 460 nm emission wavelengths for DAPI.
Determinations of plastid number
To measure plastid numbers, light microscopy was performed on toluidine blue or Schiff’s reagent stained sections from fixed 4 and 8 week-old im and Col-0 Arabidopsis using previously described procedures [26]. Toluidine blue is a basic dye with high affinity for acidic components (DNA, polysaccharides) [27] and Schiff’s reagent preferentially binds to the starch [28]. In brief, leaf segments (3 to 5 mm2) were incubated for 3 h in 1.6% (w/v) paraformaldehyde and 0.2% (w/v) glutaraldehyde in 25 mM sodium phosphate buffer (pH 7.1), washed twice with water (15 min each), and dehydrated in a graded ethanol series (20 min at each step). The dehydrated samples were moved to 4°C and then gradually infiltrated with cold LR White embedding resin (Fisher Scientific), first in 33% (v/v) then 66% (v/v) resin in 100% ethanol (24 h for each infiltration), and finally in 100% resin (three times, 24 h each time). The infiltrated samples were transferred to gelatin capsules containing 100% resin for embedding, and the resin was polymerized by incubation at 65°C for 20 h. Sections of 300 nm were prepared using a Leica VT1000S vibratome, stained either with 0.05% toluidine blue or Schiff's reagent, and visualized with a Zeiss Axioskop 2 plus (Carl Zeiss) microscope. Images were captured with a Nikon DXM 1200 digital camera, and plastid numbers were counted using ImageJ software available from the NCBI website.
Callose deposition
To examine callose deposition, expanded leaves from six independent 7-week-old plants were cleared with 96% ethanol and fixed in an acetic acid:ethanol (1:3) solution for 2 hours. The samples were incubated sequentially for 15 min each in 75% ethanol, 50% ethanol, and 150 mM phosphate buffer (pH 8.0), then stained for 2 h at 25°C in 150 mM phosphate buffer (pH 8.0) containing 0.01% (w/v) aniline blue. After staining, the leaves were examined by UV fluorescence microscopy using a Leica DMIRE2 microscope.
Reactive oxygen species
For hydrogen peroxide visualization, leaves were cut from 7-week-old plants and dipped for 16 hours in a solution containing 1 mg/mL DAB (3,3’-diaminobenzidine), pH 5.0. Chlorophyll was extracted with hot 96% ethanol, and the leaves were photographed with a Canon EOS300D camera.
Transmission electron microscopy (TEM) and chlorophyll fluorescence
Leaf tissues were fixed in a solution of 2% glutaraldehyde and 2% paraformaldehyde in a 0.1 mM cacodylate buffer (pH 7.2). Subsequent steps were performed as described previously [29]. Images were captured on a JEOL 2100 STEM. Chlorophyll fluorescence was observed with a Carl Zeiss LSM 510 META confocal microscope using 440 nm excitation and 685 nm emission wavelength for chlorophyll.
Cell wall composition analyses
Cell walls were isolated from plant tissues as described by Zabotina et al. [30]. Cell wall extracts were prepared separately from white im, green im, and Col-0 wild type leaves. Monosaccharide compositions were determined as described by Pogorelko et al. [31].
To assess lignin contents, cell wall extracts (3 mg) were treated with 800 uL of fresh acetyl bromide in acetic acid (1:16) for 2 hours at 50°C. An aliquot (15 uL) of each sample was then mixed with 85 uL of acetic acid containing 0.05M sodium hydroxide and 0.03M hydroxylamine hydrochloride. UV absorbance was measured at 280 nm to estimate lignin contents.
To determine cellulose contents, cell wall extracts (10 mg) were suspended in 3 mL of a nitric-acetic acid mixture (545 uL water, 2.2 mL glacial acetic acid, 255 uL of 16 M nitric acid) and heated at 100°C for 30 minutes. After incubation, the solid phase was sedimented by centrifugation and washed 3 times with 3 mL of deionizer water, once with 3 mL of 100% acetone, and then dried at 50°C for 48 hours. Dried residues were weighed to measure cellulose content.
Infection assay, in planta growth and disease development
Cultures of Pseudomonas syringae DC3000 were grown overnight in LB medium at 25°C up to OD600 = 1. Cells were collected by centrifugation at 4000g for 10 minutes and re-suspended in infiltration buffer (10 mM MES, 10 mM MgCl2, pH = 5.8) at a concentration of 104 cfu/mL; the resuspended cells were pressure-infiltrated into the abaxial side of leaves. To assay bacterial growth, a circle of 7 mm in diameter was cut from the leaf, ground in 500 uL of a 10 mM MgCl2 solution, and a series of dilutions (from 10−1 up to 10−8) were plated on LB media containing 50 mg/L of rifampicin. Calculations were performed using the dilution that gave approximately 50–70 colonies.
Results
The threshold hypothesis: plastid composition of sectors is fixed early in leaf development
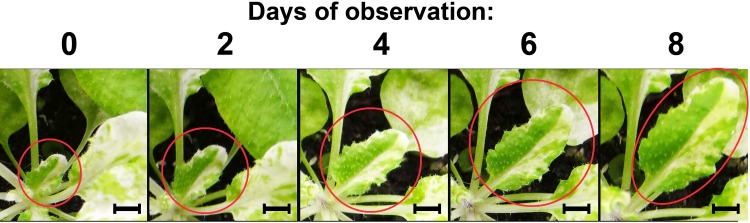
One element of the threshold hypothesis that warrants examination is the assumption that the pattern of variegation in mature im leaves is primarily a reflection of dicot leaf development. According to this scenario, each proplastid in an im meristem cell differentiates into either a chloroplast or a white plastid depending on its redox state, then divides to generate a clone of plastids and cells (representing a sector) in the leaf primordium. Consistent with this model, green and white sectors have been observed in im leaf primordia by confocal microscopy [6]. Fig 1A further reveals that sector identity does not change during leaf expansion phase; sectors merely increase in size.
Fig 1. Sector identity is maintained during im leaf expansion.
Images of the same im leaf were captured by light microscopy every two days from early expansion (day 0) to the attainment of full expansion (day 8). Bar = 5 mm.
This suggests that plastid identity and sector boundaries become fixed during the primordium stage, and that white plastids do not differentiate into chloroplasts (or vice versa) during or after the expansion phase. One reason for the relatively limited number of sectors in the mature leaf is that leaf development begins with a small number of proplastids and meristem cells (fewer than 20 proplastids per meristem cell,) and gives rise to a large number of cells and chloroplasts (100 or more chloroplasts in a typical mesophyll cell) [19–20]. Thus, early differentiation events that flip a plastid into one state or another can be magnified by division and expansion into a large sector in the mature leaf.
In contrast to the above generalization, we note that the white sectors of mature im leaves occasionally have very small green spots that correspond (via UV and light spectroscopy) to heteroplastidic cells or to a small clone of chloroplast-containing cells. We propose that these arise from the limited number of plastid and cell divisions that occur in expanding and mature leaves [19], perhaps as a consequence of the ability of dividing plastids to alter their developmental fate according ambient EP thresholds, as early suggested by Wetzel et al. [10] in the developmental race model of plastid differentiation.
The threshold hypothesis: im white sectors have sharply decreased plastid numbers
As described in the Introduction, the green and white leaf sectors of im display alterations in biochemistry, physiology and morphogenesis caused, in part, by differential reprogramming of gene expression in the two tissue types [21–23, 32]. This reprogramming results in the acquisition of tissue-specific strategies to cope with oxidative stress, as well as alterations that promote sink-source interactions between the green (source) and white cells (sinks). As part of an ongoing effort to gain insight into these processes, the present studies sought to examine plastid numbers in the white sectors. These experiments were prompted by the chance observation of relatively few plastids in some cells of these sectors.
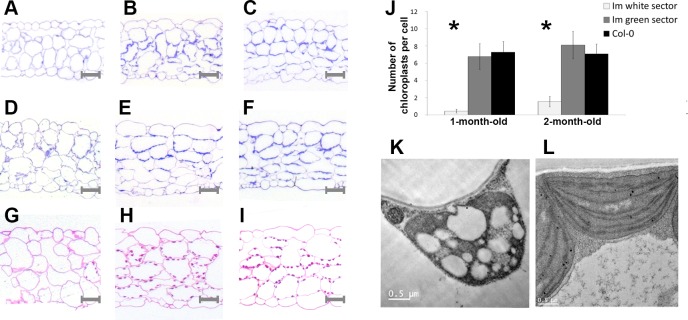
For these analyses, we counted plastid numbers in toluidine blue-stained sections of fully-expanded leaves from 4- and 8-week-old plants growing under continuous light; the samples included wild type leaves, and tissues dissected from im green and white sectors (Fig 2).
Fig 2. Tissue anatomy and plastid numbers.
Leaves from 4- and 8-week-old Col-0 and im (white and green sectors) were fixed, stained and examined by light microscopy. (A, D, G) im white; (B, E, H) im green; (C, F, I) Col-0; (A, B, C) 1 month-old; (D–I) 2 month-old. (A-F) Sections were stained with toluidine blue and plastids were counted using Image J software (NCBI website); (J) the sections were 500 nm thick and approximately 150 cells were analyzed for each tissue-type. Asterisks indicate significant difference (t-test, p < 0.01). (G-I) Sections from 2-month-old leaf tissues were stained with Schiff’s reagent for starch. TEM images of representative plastids from fully-expanded im white (K) and wild type leaves (L).
In terms of gross anatomy, the data are consistent with earlier observations [21] showing that the green sectors (Fig 2B, 2E and 2H) have enlarged air spaces and larger than normal mesophyll and epidermal cells, whereas the white sectors (Fig 2A, 2D and 2G) have smaller than normal cells and a lack of palisade cell expansion. Ultrastructural analyses reveal that chloroplasts in the im green and wild type tissues have normal morphologies (Fig 2L), whereas plastids in the white sectors are highly vacuolated, lack lamellae and are much smaller than normal (Fig 2K); the TEM data confirm previous findings [10].
Fig 2J reveals that the toluidine blue-stained im green sectors have wild type chloroplast numbers per cell at both developmental time points, while plastid numbers in the white sectors are dramatically reduced 5- to 10-fold, with many sections lacking a plastid. The three tissue-types were also stained with Schiff’s reagent (specific for starch) as an alternate way to estimate plastid numbers (Fig 2G, 2H and 2I). The light micrographs show that cells in both green tissues have an abundance of starch-containing bodies (i.e., chloroplasts) in their peripheries, consistent with the quantitative analyses (Fig 2J). As expected, the white tissues are not stained because they are not photosynthetic and do not accumulate starch (Fig 2G). Considered together, the data in Fig 2 indicate that im white sectors have drastic reductions in plastid number and size, compared to wild type or im green sectors. These reductions could be caused by decreased rates of plastid division and/or by decreased plastid viability.
The threshold hypothesis: chloroplast DNAs are amplified in im white sectors
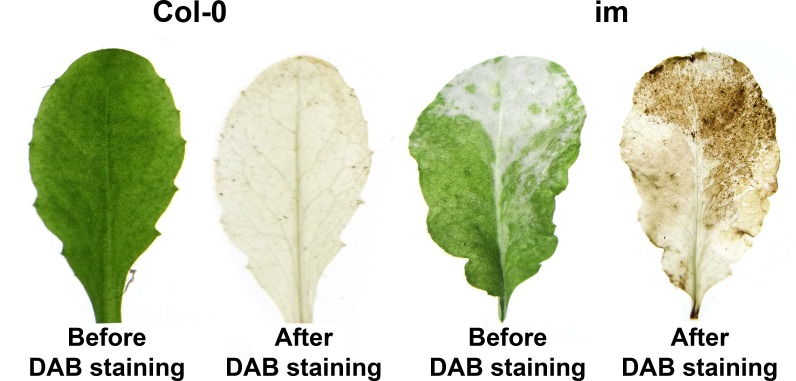
To address this question, we first monitored ROS accumulation in im and wild type leaves by staining the tissues with DAB (3′, 3′- diaminobenzidine), a reagent that detects H2O2. Fig 3 shows that im white sectors stain strongly with DAB, compared to wild type leaves, which are not stained.
Fig 3. ROS accumulation.
Leaves were detached from 7 week-old Col-0 and im and stained with DAB, specific for H2O2, by methods described in the Materials and Methods.
This observation confirms previous findings that white, but not green, im sectors generate ROS [33]. This raises the possibility that ROS accumulation, either directly or indirectly, affects plastid viability, division and/or size.
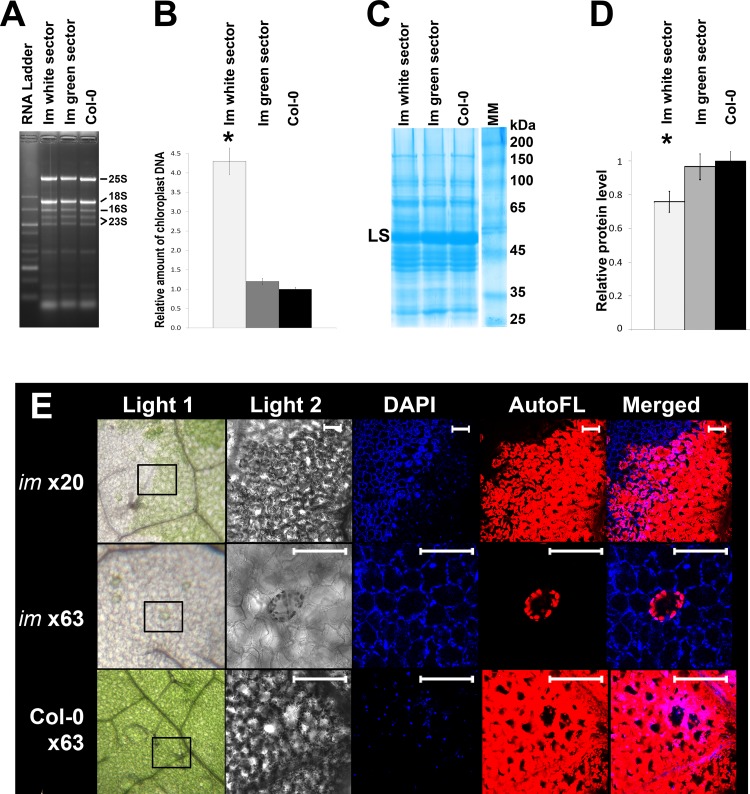
We next examined profiles of total cell RNA and total cell protein accumulation in expanded leaves from im (dissected green and white sectors) and wild type. Fig 4A shows that all three tissue-types have similar RNA profiles with respect to their rRNA binding patterns.
Fig 4. RNA, protein and chloroplast DNA.
(A) Ethidium bromide-stained denaturing gel of total cell RNAs extracted from 7 week-old wild type Col-0 and im leaves (white and green sectors). The RNAs were loaded on the gel on a fresh weight basis. The rRNA bands are labeled. (B) Chloroplast DNA-specific primers. Real-time qPCR revealed 4 times the amount of chloroplast DNA in white tissues compared to green and wild-type tissues. Same samples as in (A). (C) Coomassie blue-stained 12.5% SDS-polyacrylamide gel of total cell proteins using the same samples as in (A). The proteins were loaded on a fresh weight basis. The large subunit (LS) of Rubisco is indicated; it comprises nearly all of the band migrating at 55 kDa. (D) Protein amounts in the gel lanes were quantified using ImageJ software. Asterisks indicate a significant difference (t-test, p < 0.05). (E) Confocal fluorescence microscopy. Leaves from 2 month-old im and Col-O were fixed and stained with DAPI (1 ug/ml), then certain leaf area (indicated on Light 1) examined by confocal microscopy (Light 2, DAPI, AutoFL, and Merged, as described in the Materials and Methods. Auto FL is chlorophyll. Bar = 20 μm.
These include the cytosolic 80S rRNAs (25S and 18S) and the plastid 70S rRNA species (16S rRNA and the two 23S rRNA breakdown products). All of these rRNAs are present in roughly equal amounts in the three samples. The three samples also have similar total cell protein profiles, as monitored by SDS PAGE analysis (Fig 4C), with the exception that protein abundance is ~20% reduced in the white sectors (Fig 4D). This reduction was evident when the gels were loaded on an equal fresh weight basis (as in Fig 4C and 4D) or on an equal protein basis. At least part of this decrease can be attributed to Rubisco, as indicated by the decreased intensity of the LS band; Rubisco can constitute up to ~50% of the total cell protein in Arabidopsis leaves [34].
Taken together, the data in Figs 2 and 4 indicate that sharp reductions in plastid number and size in the white sectors are not matched by similar reductions in plastid ribosome concentrations and protein biosynthetic capacities. To examine why this might be the case, we considered the possibility that chloroplast DNA is amplified in the white sectors. To test this hypothesis, we first performed confocal fluorescence microscopy on im and wild type leaves stained with DAPI (4',6-diamidino-2-phenylindole), a DNA-specific fluorescent dye (Fig 4E). Fig 4E reveals that DAPI-stained white sectors fluoresce more intensely than im green or wild type tissues, suggesting that they have more chloroplast DNA. To confirm this, we examined the three tissue types by Real-Time qPCR, using primers specific for chloroplast DNA. Fig 4B shows that the amount of chloroplast DNA relative to the amount of genomic DNA is similar in the wild type and im green samples, but amplified about 5-fold in the white sectors.
In summary, the data in Figs 2 and 4 are consistent with the idea that chloroplast RNA and protein accumulation are significantly different from normal on a per plastid basis, perhaps because mechanisms of plastid DNA amplification are deployed in cells of white sectors.
Cell wall remodeling in im
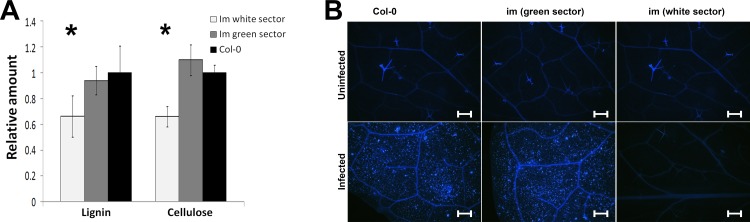
As described in the Introduction, the green and white im sectors display striking, tissue-specific alterations in the expression of genes whose products play a role in cell wall metabolism. This reprogramming likely reflects a whole plant growth strategy that involves, primarily, the optimization of tissue-specific responses to oxidative stress, as well as interactions between the green and white cells that promote their ability to serve as sinks (white cells) or sources (green cells) [22, 23]. One of the purposes of the present research was to test this hypothesis and investigate cell wall compositions of the im green and white sectors. In Fig 5A we assayed cell wall extracts from Col-0 and im (white and green sectors) for their content of lignin and cellulose -the two predominant polymers in cell walls.
Fig 5. Composition of cell wall polymers.
(A) Lignin and cellulose contents in the cell walls of Col-0 and im (white and green sectors). The contents were determined on a per mg basis of cell wall extracts. (B) Callose accumulation in Col-0 and im (white and green sectors) before and after infection with P. syringae. Expanded leaves from 7 week-old plants were cleared with ethanol and stained with aniline blue. The images were captured via UV fluoresecence microscopy. Bars = 200 μm.
The data show that the relative amount of each polymer is similar in both green tissues, but that each is much reduced in the im white sectors. The monosaccharide composition of other cell wall polysaccharides is also similar in each of the two green tissues, but altered in cell walls from the white sectors (Table 1).
Table 1. Monosaccharide composition (mol%) of cell wall from immutans and Col-0 rosette leaves.
| Fuc | Rha | Ara | Gal | Glc | Xyl | Man | GalA | GlcA | |
|---|---|---|---|---|---|---|---|---|---|
| Col-0 | 2.37±0.02 | 6.44±0.37 | 9.61±0.97 | 23.08±0.26 | 33.10±1.71 | 18.12±0.51 | 5.93±0.03 | 0.40±0.05 | 0.81±0.07 |
| im (white sector) | 2.10±0.04* | 5.69±0.19 | 7.08±0.48 | 15.01±1.59* | 50.61±4.50* | 15.24±2.04 | 3.13±0.41* | 0.39±0.03 | 0.74±0.05 |
| im (green sector) | 2.36±0.07 | 6.74±0.15 | 9.59±0.46 | 21.02±0.20 | 34.05±1.99 | 19.39±0.75 | 5.49±0.60 | 0.56±0.03 | 0.80±0.05 |
*Significantly different from wild type (t-test, p<0.01, n = 3)
Thus, while non-cellulosic glucose content is increased by 50%, the amounts of fucose (12%), galactose (35%), and mannose (47%) are reduced in the white sectors compared to the green sectors and Col-O. Taken together, the data in Fig 5A and Table 1 indicate that the cell walls of im white differ significantly from wild type, and that while cell walls of im green resemble those of wild type more closely, they are not the same.
Cell wall remodeling in im: responses to Pseudomonas syringae infection
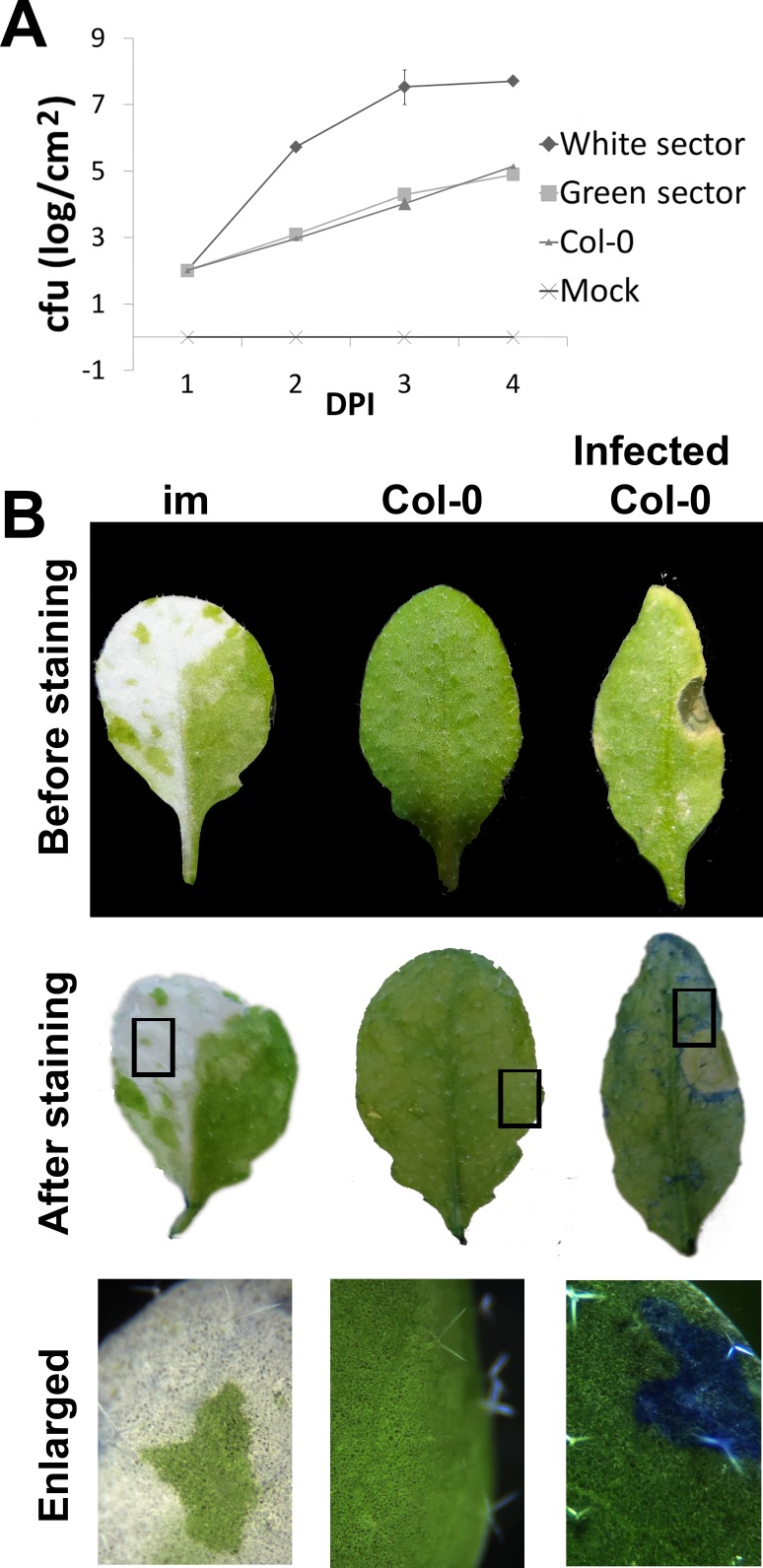
The next question we asked was whether the cell wall modifications in im affect pathogen response. To address this question, we analyzed susceptibility of im to P. syringae DC3000. This pathogen causes necrosis and cell death, as illustrated by Evans blue staining of infected tissues (Fig 6).
Fig 6. Evans blue exclusion test.
(A) Leaves of im and Col-0 were infiltrated with a P. syringae cell culture at a density of 104 colony forming units (cfu), and bacterial growth was monitored daily for 4 days after infection (DPI). (B) Leaves were detached from 7 week-old Col-0 and im and stained with Evans blue. In some experiments, detached wild type leaves were infiltrated with a P. syringae cell culture (density of 104 cfu) prior to staining. In the Evans blue exclusion test, living cells exclude the dye, while dead cells take it up.
This dye is taken up by dead cells, but not living ones, as shown by control non-infected wild type leaves, which do not retain the stain. Non-infected im do not retain the stain either. This indiactes that the white im sectors are composed of living rather than dead cells, in contrast to necrotic cells and tissues that are found in the white sectors of some variegations [35].
To obtain a quantitative estimate of the susceptibility of im to P. syringae, bacterial cultures were infiltrated into the leaves of im and Col-0 plants, and bacterial cell growth was monitored over the course of four days post-infection. Fig 6A shows there is a marked increase in susceptibility of the white tissues to the pathogen, while susceptibility of the green sectors resembles that of wild type. To investigate the source of these differences in susceptibility, we analyzed the expression of several diagnostic defense-related genes by Real-Time qPCR of RNAs from non-infected and infected leaves of im (green and white sectors) and wild type. The genes included MAPK/ERK kinase kinase member A1 (MEKK1), PHYTOALEXIN DEFICIENT 4 (PAD4), ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1), PATHOGEN RELATED 1 and 5 (PR1, PR5), and β-GLUCAN SYNTHASE 2 (bGS2): MEKK1 is involved in responses to ROS; PAD4 and EDS1 mediate responses to multiple stresses; PR1 and PR5 are well-known pathogenesis-related genes; and bGS2 is involved in callose deposition.
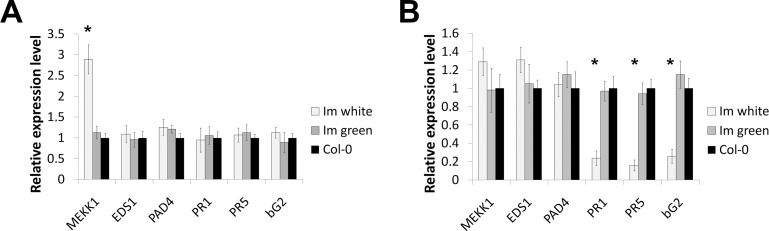
Fig 7A and 7B show the results of the Real-Time qPCR experiments.
Fig 7. Expression of pathogen response genes.
Real-time qPCR evaluation of pathogen-related genes involved in immune response in (A) non-infected and (B) infected plants at 4 DPI. The samples were as in Fig 6B, and the genes are described in the text.
In these experiments we calculated the relative RNA amounts for each treatment (infected versus non-infected) across the six genes. Although these analyses preclude comparisons between treatments, it has been established that all six genes are sharply upregulated in infected versus non-infected wild type Arabidopsis leaves (see references above). Given these considerations, our experiments revealed that the six genes have similar levels of expression in non-infected wild type and im (white and green sectors), with the exception of a striking upregulation of MEKK1 in im white sectors (Fig 7A). In the infected samples, it can be inferred that all six genes are significantly upregulated to a similar extent in both green tissues. PR1, PR5 and bG2 expression, on the other hand, are less responsive to the pathogen in the im white sectors. This correlates with the enhanced susceptibility of these sectors to P. syringae infection.
To examine this correlation, we monitored callose deposition in the three tissue types of infected and non-infected leaf tissues by aniline blue staining. Callose is a (1,3)-β-glucan polymer, and its induction in response to pathogen infection causes cell wall thickening and (perhaps) resistance to further infection [36]. Fig 5B shows that callose is not induced in any of the non-infected samples, but that upon infection, high levels of callose are induced in both im green and wild type. By contrast, there is no callose deposition in the white sectors. These observations are consistent with the idea that im white tissues are more susceptible to P. syringae than the green tissues because of defects in defense gene expression.
Discussion
White sectors and sink demand
One purpose of the present studies was to gain insight into the threshold mechanism of im variegation. Previous studies have demonstrated that the green and white im sectors differ significantly with respect to their morphology, biochemistry and physiology. Transcript profiling has indicated these differences are due, in part, to a reprogramming of development in the green and white sectors that optimizes whole plant growth [4, 21, 22, 23]. This reprogramming is strongly influenced by at least two factors, both of which occur very early in leaf development. The first involves tissue-specific adaptations to oxidative stress that are likely elicited in response to overreduced plastid membranes (the primary lesion in im). The second factor occurs in response to sector formation (in the leaf primordium) and involves the fashioning of the sectors as effective sources (green sectors) or sinks (white sectors).
Previous studies have revealed that the green sectors of im have a gross anatomy reminiscent of wild type leaves that have been adapted to growth in high light, despite growth of the mutants under low light (permissive) conditions [21]. We presume this morphology is a response to oxidative stress in the developing leaf. Cells in the green sectors also have elevated photosynthetic rates, perhaps as a consequence of sink demand; photosynthetic rates in source tissues are often feedback controlled by sink strength [22]. Although the reason for the increased rates is not understood, the present data show they are not associated with an increase in chloroplast number or size, both of which are similar to those in wild type leaf cells.
In contrast to the green sectors, plastid numbers (and sizes) are drastically reduced in cells of the white sectors. These reduction are not accompanied by significant cell and/or tissue necrosis, as revealed by Evans blue staining. Surprisingly, RNA amounts and protein synthetic capacities are only marginally affected in the white sectors, but chloroplast DNAs are dramatically amplified. These alterations are accompanied by reductions in cell size, confirming previous FACS analyses of protoplasts from im (white and green sectors) and wild type leaves [32]. Taken together, these observations suggest that white im tissues are fashioned into effective sinks during leaf development, and that this process involves 1) a decrease in plastid division and/or viability; 2) the maintenance of near-normal levels of protein synthetic capacity; and 3) an elevated potential for chloroplast gene expression (enhanced chloroplast DNA amounts).
Although mechanisms underlying sink demand are poorly understood, we suggest that the characteristics we have discovered play an important role in the growth strategy of the plant. We assume that this strategy attempts to maximize the potential of the green and white sectors to serve as sources (manipulation of source strength, e.g. via regulating photosynthetic capacities) or sinks (manipulation of sink demand, e.g., via regulating rates of plastid division) by calibrating their relative proportion throughout leaf development. It might be noted that the characteristics of im white sectors we have described are not universally applicable to white leaf tissues; e.g., the white sectors of some variegations have significant cell and tissue necrosis [35]. This indicates there are probably many different strategies to optimize growth and that these depend on responses to the primary lesion in the mutant, directly and indirectly.
In addition to serving as determinants of sink demand, the unique characteristics of im white sectors might be advantageous for another reason. It is interesting that plastids in the white sectors resemble etioplasts in dark grown seedling inasmuch as they contain nearly the full complement of proteins required for photosynthesis [37]. One reason for this is to prime the organelle for subsequent light development, thus avoiding a developmental delay that might be toxic for the developing chloroplast when mechanisms of light capture and use are still not fully operational and integrated. In an analogous manner, we propose that elevated chloroplast DNA levels and near-normal protein synthetic capacities are a strategy to poise the white plastid for further development. This could occur in cases where a white plastid divides and the progeny find themselves with redox thresholds compatible with normal chloroplast biogenesis; differentiation into chloroplasts could commence immediately. Alternatively, white and green im plastids are precursors of plastids that are transmitted to progeny via selfing or outcrossing, and during these developmental transformations (e.g., gametogenesis, seed and embryo development), an individual plastid undergoes a series of dedifferentiation and redifferentiation events. In fact, branches with nearly all white plastids can give rise to all-green plants in the next generation under permissive (low light) conditions [10, 38]. This suggests that maintenance of high protein synthetic capacities might help poise a white plastid for its conversion into another plastid type.
Retrograde signaling and ROS
It is well-established that retrograde (plastid-to-nucleus) signaling pathways play an important role in coordinating the early events of chloroplast biogenesis [39–42]. We have also proposed that plastid signals relay the metabolic and developmental state of developing im plastids to the nucleus, where they elicit changes in gene expression that, in turn, modify plastid and cell phenotypes, as well as patterns of leaf development (termed plastid developmental signals) [3, 43]. While a number of retrograde pathways have been identified in chloroplasts, nearly all poorly-defined, one of the best-studied is elicited in response to alterations in chloroplast redox [44–48]. Our finding that developing im membranes have overreduced PQ pools suggests that one or more pathways responsive to redox are operational in developing im leaves [6, 11].
In addition to signals emanating from the redox state of the PQ pool, it might be noted that PTOX itself might be a generator of plastid signals. This possibility comes from studies showing that ROS are produced as a side product of the PTOX reaction in liposomes containing purified PTOX; in E. coli membranes that contain PTOX; in tobacco PTOX overexpression plants; and in PSII-enriched membrane fragments that contain PTOX [49–50]. A precedent for ROS signaling comes from the well-known singlet oxygen retrograde pathway [51–52].
Although we recognize that high concentration of ROS are toxic (and might explain the phenotype of plastids in the white cells), our observation that white cells produce ROS is consistent with the idea that ROS signaling might play a role in coordinating developmental, physiological and biochemical processes in im leaves. It is also well-established that ROS mediate signaling pathways initiated in response to pathogen invasion [53–55]. In this context it is interesting that MEKK1 is the only diagnostic defense gene that is significantly upregulated in the white tissues. This might be because it is the only one directly responsive to ROS; we do not know whether MEKKI upregulation has any physiological significance in the white sectors. On the other hand, a coordinated upregulation of MEKK1, EDS1 and PAD4 in infected im white tissues is indicative of their ability to respond to pathogen attack: EDS1 and PAD4 are induced by MEKK1 [56], and they form a complex in both the nucleus and cytoplasm [57] that provides a basal immune response. In this context it is noteworthy that pathogenesis-related defense genes (PR1, PR5 and bG2) do not respond to pathogen infection at wild type levels in white sector tissues. This finding is striking, and might perhaps be related to the significant cell wall remodeling that occurs during the development of the green and white sectors.
Cell wall remodeling
Glucose is the main structural constituent of cellulose and hemicellulosic xyloglucan, while mannose is a constituent of the backbone of hemicellulosic polysaccahride galactomannan, a subsidiary component of cell walls. Galactose is present in the side chains of galactomannan and its reduction correlates to a decrease in mannose content (Table 1). This reduction might be caused by alterations in pectic rhamnogalacturonan side chains, since galactomannans are not widely present in common plant cell walls, such as rhamnogalacturonans. In addition, pectins are known to be targeted by plant pathogens’ hydrolases to accelerate infection processes [58]. Reductions in fucose might also reflect a reduction of galactose in xyloglucan where the disaccharide Gal-Fuc forms a side chain attached to a xylosilated backbone. Considered together, these data suggest that the biosynthesis of galactomannans and decoration of xyloglucan are affected in the im white sectors.
Lignin and cellulose form the primary mechanical barriers preventing pathogen invasion [59–60]. Lignin is composed of polyphenol residues and not polysaccharides; however, the lignin biosynthesis phenylpropanoid pathway involves the sequential conversion of the amino acid phenylalanine into sinapoylglucose [61]. Since mutations affecting sinapoylglucose accumulation in Arabidopsis cells also exhibit reduced lignin deposition, it correlates with our finding of decreased lignin content in white sectors of im [62]. A reduction of lignin and cellulose microfibrils (Fig 5A) together with modification of some hemicellulosic polysaccharides in the cell wall causes, perhaps, rearrangement of the whole cell wall structure, thereby increasing the lytic efficiency of bacterial hydrolases directed to the plant cell wall as well as the penetration and infection efficiency of P.syringae cells.
Conclusions
In summary, we propose that tissue-specific alterations in cell wall composition in im are a reflection of alterations in gene expression that remodel leaf development in response to oxidative stress and that also promote sink-source interactions between the green and white sector, both of these efforts are initiated very early in leaf development. According to this scenario, enhanced pathogen susceptibility in the white sectors is a secondary consequence of this development reprogramming. If so, it is not clear why some defense pathways are operational in the white sectors and others are not (pathogen related genes), although considerable cross talk between pathways can be envisioned in fashioning each tissue-type. Regardless, we conclude that both a lack of activation of pathogen related genes and modification of cell wall composition are a consequence of impaired chloroplast biogenesis in im, and that these result in compromised resistance to pathogen invasion. This conclusion is consistent with previous observations showing that alterations in cell wall composition lead to changes in plant signaling mechanisms and response to environmental conditions including phytopathogen invasion [37–40]. We conclude that im offers a novel model system to explore leaf developmental programming, cell wall metabolism and pathogen response.
Supporting Information
(DOCX)
Acknowledgments
This work was supported by the U.S. Department of Energy (Energy Biosciences; Grant DF-FG02-94ER20147 to S.R.R.). We thank Marena Bartz for her technical assistance in the manuscript preparation.
Data Availability
Data are available at https://figshare.com/s/409adc4a80c647819343.
Funding Statement
This work was supported by Energy Biosciences, Grant DF-FG02-94ER20147.
References
- 1.Aluru MR, Yu F, Fu A, Rodermel S. Arabidopsis variegation mutants: New insights into chloroplast biogenesis. J Exp Bot 2006; 57: 1871–1881. [DOI] [PubMed] [Google Scholar]
- 2.Yu F, Fu A, Aluru M, Park S, Xu Y, Liu H, Liu X, Foudree A, Nambogga M, Rodermel S. Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ 2007; 30(3):350–65. [DOI] [PubMed] [Google Scholar]
- 3.Rodermel S. Pathways of plastid-to-nucleus signaling. Trends Plant Sci 2001; 6(10):471–8. [DOI] [PubMed] [Google Scholar]
- 4.McDonald AE, Ivanov AG, Bode R, Maxwell DP, Rodermel SR, Hüner NP. Flexibility in photosynthetic electron transport: the physiological role of plastoquinol terminal oxidase (PTOX). Biochim Biophys Acta 2011; 1807(8):954–67. 10.1016/j.bbabio.2010.10.024 [DOI] [PubMed] [Google Scholar]
- 5.Putarjunan A, Liu X, Nolan T, Yu F, Rodermel S. Understanding chloroplast biogenesis using second-site suppressors of immutans and var2. Photosynth Res 2013; 116(2–3):437–53. 10.1007/s11120-013-9855-9 [DOI] [PubMed] [Google Scholar]
- 6.Foudree A, Putarjunan A, Kambakam S, Nolan T, Fussell J, Pogorelko G, Rodermel S. The Mechanism of Variegation in immutans Provides Insight into Chloroplast Biogenesis. Front Plant Sci 2012;3:260 10.3389/fpls.2012.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu D, Wright DA, Wetzel C, Voytas DF, Rodermel S. The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 1999; 11(1):43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carol P, Stevenson D, Bisanz C, Breitenbach J, Sandmann G, Mache R, Coupland G, Kuntz M. Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 1999;11(1):57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nawrocki WJ, Tourasse NJ, Taly A, Rappaport F, Wollman FA. The plastid terminal oxidase: its elusive function points to multiple contributions to plastid physiology. Annu Rev Plant Biol 2015; 66:49–74. 10.1146/annurev-arplant-043014-114744 [DOI] [PubMed] [Google Scholar]
- 10.Wetzel CM, Jiang CZ, Meehan LJ, Voytas DF, Rodermel SR. Nuclear-organelle interactions: the immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J 1994; 6(2):161–75. [DOI] [PubMed] [Google Scholar]
- 11.Rosso D, Bode R, Li W, Krol M, Saccon D, Wang S, Schillaci LA, Rodermel SR, Maxwell DP, Hüner NP. Photosynthetic redox imbalance governs leaf sectoring in the Arabidopsis thaliana variegation mutants immutans, spotty, var1, and var2. Plant Cell 2009; 21(11):3473–92. 10.1105/tpc.108.062752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosso D, Ivanov AG, Fu A, Geisler-Lee J, Hendrickson L, Geisler M, Stewart G, Krol M, Hurry V, Rodermel SR, Maxwell DP, Hüner NP. IMMUTANS does not act as a stress-induced safety valve in the protection of the photosynthetic apparatus of Arabidopsis during steady-state photosynthesis. Plant Physiol 2006; 142(2):574–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rumeau D, Peltier G, Cournac L. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ 2007; 30(9):1041–51. [DOI] [PubMed] [Google Scholar]
- 14.Peltier G, Cournac L. Chlororespiration. Annu Rev Plant Biol 2002; 53:523–50. [DOI] [PubMed] [Google Scholar]
- 15.Peng L, Fukao Y, Fujiwara M, Shikanai T. Multistep assembly of chloroplast NADH dehydrogenase-like subcomplex A requires several nucleus-encoded proteins, including CRR41 and CRR42, in Arabidopsis. Plant Cell 2012; 24(1):202–14. 10.1105/tpc.111.090597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okegawa Y, Kobayashi Y, Shikanai T. Physiological links among alternative electron transport pathways that reduce and oxidize plastoquinone in Arabidopsis. Plant J 2010; 63(3):458–68. 10.1111/j.1365-313X.2010.04252.x [DOI] [PubMed] [Google Scholar]
- 17.Ivanov AG, Rosso D, Savitch LV, Stachula P, Rosembert M, Oquist G, Hurry V, Hüner NP. Implications of alternative electron sinks in increased resistance of PSII and PSI photochemistry to high light stress in cold-acclimated Arabidopsis thaliana. Photosynth Res 2012;113(1–3):191–206. 10.1007/s11120-012-9769-y [DOI] [PubMed] [Google Scholar]
- 18.Smith LG, Hake S. The Initiation and Determination of Leaves. Plant Cell 1992;4(9):1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullet JE. Chloroplast development and gene expression. Annu. Rev. Plant Physiol. Plant Mol Biol 1988;39:475–50210. [Google Scholar]
- 20.Pyke K, López-Juez E. Cellular differentiation and leaf morphogenesis in Arabidopsis. CRC Crit Rev Plant Sci 1999;18:527–54610. [Google Scholar]
- 21.Aluru MR, Bae H, Wu D, Rodermel SR. The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol 2001;127(1):67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aluru MR, Stessman DJ, Spalding MH, Rodermel SR. Alterations in photosynthesis in Arabidopsis lacking IMMUTANS, a chloroplast terminal oxidase. Photosynth Res 2007; 91(1):11–23. [DOI] [PubMed] [Google Scholar]
- 23.Aluru MR, Zola J, Foudree A, Rodermel SR. Chloroplast photooxidation-induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiol 2009;150(2):904–23. 10.1104/pp.109.135780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Windholm JM. The use fluorescein diacetate and phenosafranine for determining viability of cultured plant cells. Stain Technology 1972;47(4):89–94. [DOI] [PubMed] [Google Scholar]
- 25.Kapuscinski J. "DAPI: a DNA-specific fluorescent probe". Biotech Histochem 1995;70(5):220–33. [DOI] [PubMed] [Google Scholar]
- 26.Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol 2010; 153:514–525. 10.1104/pp.109.151985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sridharan G, Shankar A. Toluidine blue: a review of its chemistry and clinical utility. JOMFP 2012;16:251–255. 10.4103/0973-029X.99081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiff H. Eine neue Reihe organischen Diaminen. Justus Liebigs Ann Chemie 1866; 140: 92–137. [Google Scholar]
- 29.Horner HT, Wagner BL. The association of druse crystals with the developing stomium of Capsicum annum (Solanaceae) anthers. Am J Bot 1980; 67:1347–1360. [Google Scholar]
- 30.Zabotina OA, van de Ven WT, Freshour G, Drakakaki G, Cavalier D, Mouille G, et al. Arabidopsis XXT5 gene encodes a putative alpha-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J 2008; 56(1):101–15. 10.1111/j.1365-313X.2008.03580.x [DOI] [PubMed] [Google Scholar]
- 31.Pogorelko G, Fursova O, Lin M, Pyle E, Jass J, Zabotina OA. Post-synthetic modification of plant cell walls by expression of microbial hydrolases in the apoplast. Plant Mol Biol 2011; 77(4–5):433–45. 10.1007/s11103-011-9822-9 [DOI] [PubMed] [Google Scholar]
- 32.Meehan L, Harkins K, Chory J, Rodermel S. Lhcb transcription is coordinated with cell size and chlorophyll accumulation. Studies on fluorescence-activated, cell-sorter-purified single cells from wild-type and immutans Arabidopsis thaliana. Plant Physiol 1996; 112:953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Putarjunan A, Rodermel S. gigantea suppresses immutans variegation by interactions with cytokinin and gibberellin signaling pathways. Plant Physiol 2014; 166(4):2115–32. 10.1104/pp.114.250647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stessman D, Miller A, Spalding M, Rodermel S. Regulation of photosynthesis during Arabidopsis leaf development in continuous light. Photosynth Res 2002; 72(1):27–37. [DOI] [PubMed] [Google Scholar]
- 35.Kato Y, Miura E, Matsushima R, Sakamoto W. White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiol 2007; 144(2):952–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellinger D, Naumann M, Falter C, Zwikowics C, Jamrow T, Manisseri C, Somerville SC, Voigt CA. Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol 2013; 161(3):1433–44. 10.1104/pp.112.211011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plöscher M, Reisinger V, Eichacker LA. Proteomic comparison of etioplast and chloroplast protein complexes. J Proteomics 2011; 74(8):1256–65. 10.1016/j.jprot.2011.03.020 [DOI] [PubMed] [Google Scholar]
- 38.Rédei GP. Somatic Instability Caused by a Cysteine-Sensitive Gene in Arabidopsis. Science 1963; 139(3556):767–9. [DOI] [PubMed] [Google Scholar]
- 39.Ruckle ME, DeMarco SM, Larkin RM. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 2007; 19(12):3944–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.León P, Gregorio J, Cordoba E. ABI4 and its role in chloroplast retrograde communication. Front Plant Sci. 2013;3:304 10.3389/fpls.2012.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terry MJ, Smith AG. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci 2013;4:14 10.3389/fpls.2013.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jarvis P, López-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 2013; 14(12):787–802. 10.1038/nrm3702 [DOI] [PubMed] [Google Scholar]
- 43.López-Juez E. Steering the solar panel: plastids influence development. New Phytol 2009; 182(2):287–90. 10.1111/j.1469-8137.2009.02808.x [DOI] [PubMed] [Google Scholar]
- 44.Pfannschmidt T, Schütze K, Fey V, Sherameti I, Oelmüller R. Chloroplast redox control of nuclear gene expression—a new class of plastid signals in interorganellar communication. Antioxid Redox Signal 2003; 5(1):95–101. [DOI] [PubMed] [Google Scholar]
- 45.Bräutigam A, Weber AP. Proteomic analysis of the proplastid envelope membrane provides novel insights into small molecule and protein transport across proplastid membranes. Mol Plant 2009; 2(6):1247–61. 10.1093/mp/ssp070 [DOI] [PubMed] [Google Scholar]
- 46.Jung HS, Crisp PA, Estavillo GM, Cole B, Hong F, Mockler TC, Pogson BJ, Chory J. Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc Natl Acad Sci U S A 2013; 110(35):14474–9. 10.1073/pnas.1311632110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrillo E, Godoy Herz MA, Barta A, Kalyna M, Kornblihtt AR. Let there be light: regulation of gene expression in plants. RNA Biol 2014; 11(10):1215–20. 10.4161/15476286.2014.972852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dietzel L, Gläßer C, Liebers M, Hiekel S, Courtois F, Czarnecki O, Schlicke H, Zubo Y, Börner T, Mayer K, Grimm B, Pfannschmidt T. Identification of Early Nuclear Target Genes of Plastidial Redox Signals that Trigger the Long-Term Response of Arabidopsis to Light Quality Shifts. Mol Plant 2015; 8(8):1237–52. 10.1016/j.molp.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 49.Heyno E, Gross CM, Laureau C, Culcasi M, Pietri S, Krieger-Liszkay A. Plastid alternative oxidase (PTOX) promotes oxidative stress when overexpressed in tobacco. J Biol Chem 2009; 284(45):31174–80. 10.1074/jbc.M109.021667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Q, Feilke K, Krieger-Liszkay A, Beyer P. Functional and molecular characterization of plastid terminal oxidase from rice (Oryza sativa). Biochim Biophys Acta 2014; 1837(8):1284–92. 10.1016/j.bbabio.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 51.Kim C, Apel K. Singlet oxygen-mediated signaling in plants: moving from flu to wild type reveals an increasing complexity. Photosynth Res 2013; 116(2–3):455–64. 10.1007/s11120-013-9876-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim C, Apel K. 1O2-mediated and EXECUTER-dependent retrograde plastid-to-nucleus signaling in norflurazon-treated seedlings of Arabidopsis thaliana. Mol Plant 2013; 6(5):1580–91. 10.1093/mp/sst020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonak C, Okresz L, Bogre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signaling. Curr Opin Plant Biol 2002; 5:415–24. [DOI] [PubMed] [Google Scholar]
- 54.Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci 2004. October;9(10):490–8. [DOI] [PubMed] [Google Scholar]
- 55.Bailey-Serres J, Mittler R. The roles of reactive oxygen species in plant cells. Plant Physiol 2006; 141(2):311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pitzschke A, Djamei A, Bitton F, Hirt H. A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol Plant 2009; 2(1):120–37. 10.1093/mp/ssn079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu S, Jeong RD, Venugopal SC, Lapchyk L, Navarre D, Kachroo A, Kachroo P. SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathog 2011; 7(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lionetti V, Cervone F, Bellincampi D. Methyl esterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases. J Plant Physiol 2012; 169(16):1623–30. 10.1016/j.jplph.2012.05.006 [DOI] [PubMed] [Google Scholar]
- 59.Miedes E, Vanholme R, Boerjan W, Molina A. The role of the secondary cell wall in plant resistance to pathogens. Front Plant Sci 2014; 5:358 10.3389/fpls.2014.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pang SL, Ong SS, Lee HH, Zamri Z, Kandasamy KI, Choong CY, Wickneswari R. Isolation and characterization of CCoAOMT in interspecific hybrid of Acacia auriculiformis x Acacia mangium—a key gene in lignin biosynthesis. Genet Mol Res 2014; 13(3):7217–38. 10.4238/2014.September.5.7 [DOI] [PubMed] [Google Scholar]
- 61.Fraser CM, Chapple C. The phenylpropanoid pathway in Arabidopsis Arabidopsis Book; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 2009; 60(5):771–82. 10.1111/j.1365-313X.2009.03996.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
Data are available at https://figshare.com/s/409adc4a80c647819343.