Abstract
Overexpression or activation of cyclic AMP-response element-binding protein (CREB) has been known to be involved in several human malignancies, including lung cancer. Genes regulated by CREB have been reported to suppress apoptosis, induce cell proliferation, inflammation, and tumor metastasis. However, the critical target genes of CREB in lung cancer have not been well understood. Here, we identified GSK-3α as one of the CREB target genes which is critical for the viability of lung cancer cells. The CREB knockdown significantly reduced the expression of GSK-3α and the direct binding of CREB on the promoter of GSK3A was identified. Kaplan-Meier analysis with a public database showed a prognostic significance of aberrant GSK-3α expression in lung cancer. Inhibition of GSK-3α suppressed cell viability, colony formation, and tumor growth. For the first time, we demonstrated that GSK-3α is regulated by CREB in lung cancer and is required for the cell viability. These findings implicate CREB-GSK-3α axis as a novel therapeutic target for lung cancer treatment.
Introduction
The transcription factor cyclic AMP-response element-binding protein (CREB) regulates diverse cellular processes which include cell differentiation, proliferation, survival, glucose metabolism, immune regulation, and synaptic plasticity associated with memory [1–7]. Previously, we showed that CREB is critical for the regulation of mucous differentiation of normal human tracheobronchial epithelial (NHTBE) cells [8]. In addition, the decreased survival duration was significantly associated with overexpression of CREB or activated CREB (p-CREB) in never smokers with non-small cell lung cancer (NSCLC)[9] and the knockdown of CREB suppresses the viability of lung cancer cells [10]. Several serine-threonine kinases can activate CREB and p-CREB induces the expression of multiple cAMP response element-containing genes, those of which play important roles in the function of CREB. Several approaches for identifying CREB target genes have been reported [11–14], but distinct target genes of CREB in lung cancer remain largely unknown.
GSK-3, which has two isoforms of GSK-3α and GSK-3β, is a serine/threonine protein kinase that is involved in cell-cycle progression, differentiation, and apoptosis. GSK-3 is constitutively active in resting cells and it phosphorylates and inhibits oncogenic signaling such as β-catenin/WNT pathway [15–21]. Although GSK-3 has been studied as a tumor suppressor [22–24], there is increasing evidence that GSK-3 plays an oncogenic role in various human cancers. Most studies have focused on the role of total GSK-3 or GSK-3β [25–27], but recent studies implicated the oncogenic role of GSK-3α in acute myeloid leukemia (AML) [28], prostate cancer [29], and pancreatic cancer [30]. Interestingly, CREB overexpression or its increased activity has been linked to the progression of those human cancers [31–36]. In particular, CREB functions as a proto-oncogene in AML [31, 37] and GSK-3α is also a critical target for AML therapy [28]. Recently, GSK-3α and GSK-3β have been reported to be new kinase targets of tivantinib, which is a potent selective inhibitor of the receptor tyrosine kinase c-MET, in lung cancer cells. Tivantinib showed higher potency for GSK-3α more than for GSK-3β and the inhibition of GSK-3α or GSK-3β expression caused apoptosis in lung cancer cells [38].
Here, we first identified that GSK-3α, not GSK-3β, is regulated by CREB in lung cancer cells. Furthermore, we examined that there is a positive correlation between high GSK-3α expression and shorter survival of lung cancer patients. Knockdown of GSK-3α attenuates cell viability, colony formation, and tumor growth. Together, these findings implicates that GSK-3α is a critical target gene of CREB and CREB-GSK-3α signaling is a potential therapeutic target for lung cancer.
Materials and Methods
Cell culture
Human lung cancer cell lines (H1993, H1437, H1734, and A549) were obtained from the American Type Culture Collection. Lung cancer cells were cultured in RPMI-1640 medium (Invitrogen), supplemented with 10% (volume/volume) heat-inactivated fetal bovine/serum (FBS; Sigma Aldrich), 2 mM L-glutamine, 100 U/ml of penicillin G sodium and 100 μg/ml of streptomycin sulfate (Invitrogen). Normal human tracheobronchial epithelial cells (NHTBE) were obtained from the Lonza Walkersville, Inc. and cultured in BEGM™ with several supplements. All cells have been passaged directly from original low-passage stocks and were used before passage 30. The cells were also tested within the last three months for correct morphology by microscope and to detect mycoplasma contamination using a MycoAlert mycoplasma detection kit (Lonza Walkersville, Inc.). All cells were cultured at 37°C in humidified atmosphere of 95% air and 5% CO2.
Antibodies/Chemicals
Monoclonal anti-β-actin antibody (A2228) was purchased from Sigma Aldrich. Rabbit polyclonal antibody against GSK-3α (ab28833) was purchased from Abcam. Forskolin (3828), anti-CREB (9197), anti-p-CREB (9198), anti-cyclin A2 (4656), anti-cyclin B1 (4138), anti-cyclin E2 (4132), and GSK-3β (12456) were obtained from Cell Signaling Technology. Rabbit monoclonal cyclin D1 antibody (2261–1) was purchased from Epitomics.
Knockdown/ Overexpression of Genes
Silencer CREB siRNA and GSK-3α siRNAs were purchased from Thermo scientific or Invitrogen; CREB siRNA (109994, Invitrogen), GSK-3α siRNA-1 (L-003009-00-0005, ON-TARGETplus SMARTpool, ThermoScientific), and GSK-3α siRNA-2 (145366, Invitrogen). BLOCK-it Fluorescent Oligo (Invitrogen) was used as a control. Each siRNA was transfected using Lipofectamine RNAiMAX (Invitrogen). Also, cells were infected with lentiviral shScrambled or shRNAs targeting CREB or GSK-3α with 8 μg/ml polybrene and the infected cells were selected with puromycin. The sequences of shRNAs are listed in the S1 Table (available online). For CREB overexpression, H1437 and A549 cells were transfected with plasmid DNA of pCMV-empty or pCMV-CREB (Clontech Laboratories, Inc.) using Lipofectamine 2000 (Invitrogen).
Cell Viability
Cell viability was evaluated by MTT assay. After cells were transfected with siRNAs for 72 h, the cells were incubated with MTT (final concentration 0.5 mg/ml) for 4 h at 37°C incubator. Following MTT incubation, 150 μl of 100% DMSO was added to dissolve the crystals. Viable cells were counted by reading the absorbance at 570 nm using a microplate reader SpectraMax (Molecular Devices).
Colony Formation Assay
At 24 h after transfection by the indicated siRNAs, 2 x 103 cells were transferred in the 6-well plates and allowed to grow for 7–14 days. The medium was removed, fixed with 10% formalin for 15 min, and followed by staining with crystal violet to visualize the colonies.
Quantitative Real-time PCR
Total RNA was purified from cells using an RNeasy Mini Kit (Qiagen). Reverse transcription of total RNA was performed using the M-MLV reverse transcriptase (Promega). Quantitative PCR (qPCR) was performed using SYBR Green PCR Core Reagents (Applied Biosystems) and iCycler thermal cycler (Bio-Rad Laboratories). Primer sequences are listed in the S1 Table (available online).
Western Blot Analysis
Standard SDS-PAGE and western blotting procedures were used to analyze the expression of various proteins. Whole cell lysates from each of the lung cancer cell lines tested were prepared using SDS lysis buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, and 0.02% bromophenol blue) containing protease inhibitors and phosphatase. All proteins were visualized using a horseradish peroxidase-conjugated secondary antibody and Amersham ECL™ Western Blotting Detection Reagents (GE Healthcare Life Sciences). Intensity of individual bands was quantified using ImageJ densitometry software and expressed relative to actin signal, as a measure of protein relative abundance in the different samples.
Chromatin Immunoprecipitation (ChIP)
The SimpleChIP Enzymatic kit (Cell Signaling) was used as described by the manufacturer. PCR was performed with primers specific for the indicated promoter regions and the reactions were performed in triplicate and 1% of the total input sample was used as a control. Primer sequences are listed in the S1 Table (available online).
Immunostaining
For immunofluorescence, detection of primary antibodies was done using fluorescent conjugates of Alexa Fluor® 488 antibody (Invitrogen) along with ProLong® Gold Antifade Reagent with DAPI (Invitrogen). Before staining of fixed paraffin-embedded tissues, we followed the standard protocol which included steps such as deparaffinization, antigen retrieval, and permeabilization.
Flow Cytometry
For cell cycle flow cytometry, the cells were fixed in 70% ethanol and stained with propidium iodine staining (BD Pharmingen) for DNA content. Apoptosis was measured using the FITC Annexin V Apoptosis Detection Kit (BD Pharmingen) following the manufacturer’s instruction.
In Vivo Studies
All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Yale University and conformed to the legal mandates and federal guidelines for the care and maintenance of laboratory animals (protocol #: 2012–11464). Female J:NU nude mice were obtained from Jackson Laboratory and used when 6–7 weeks old. H1993 cells were pre-treated with 20 nM of control siRNA or GSK-3α siRNA for 24 h, followed by transplantation (2 x 106 cells/flank, xenograft n = 7/ group) into the flank of mice. Also, H1993-shScrambled, H1993-shGSK3A#2, or H1993-shGSK3A#4 cells were inoculated at dorsal flanks (6 x 105 cells/flank; shScrambled n = 9, shGSK3A#2 n = 4, and shGSK3A#4 n = 5). All xenografts were transplanted in both the right and left dorsal flanks of mice. Tumor volume was measured with digital calipers and calculated by the formula 0.52 x length x width2. Mice were sacrificed at the end of the study by being placed in a carbon dioxide chamber.
Statistical Methods
The quantitation results are presented as means ± standard deviation (SD). The statistical significance of differences among the groups was determined by Student’s t-test, with a P value below 0.01 considered to be statistically significant. The Kaplan-Meier method was used to analyze univariate survival, and comparisons of the survival distributions among groups were performed using the log-rank test.
Results
GSK-3α is regulated by CREB in lung cancer cells
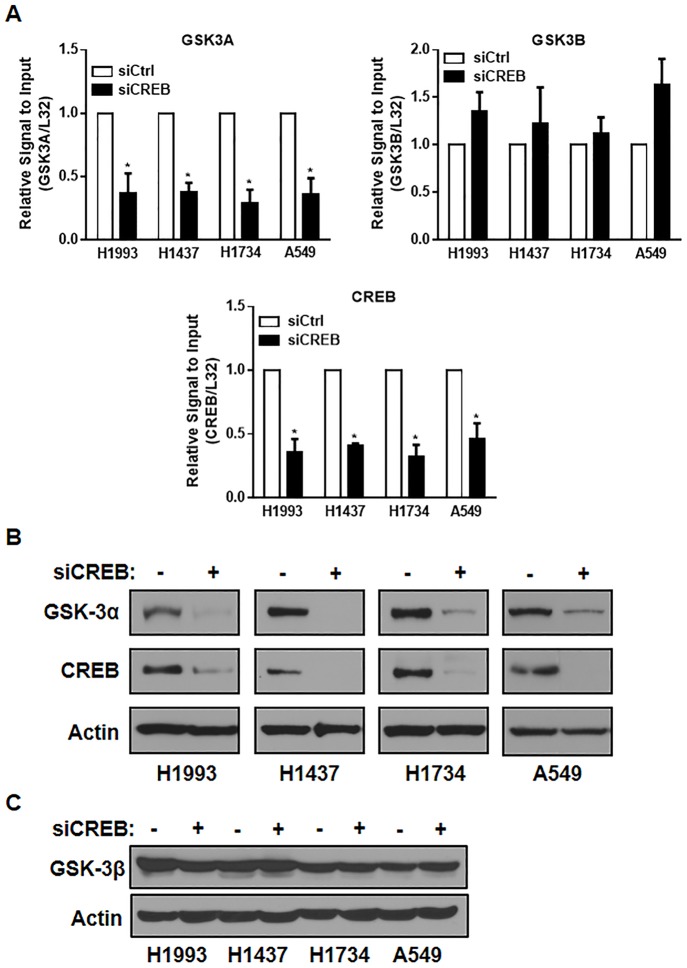
To investigate the critical CREB target genes in lung cancer, we performed qPCR analysis using specific primers against a subset of genes related to cell survival, proliferation, and viability in CREB knockdown cells. We found that the mRNA level of GSK-3α, not the level of GSK-3β, was significantly downregulated by CREB siRNA in all lung cancer cell lines we tested (H1993, H1437, H1734, and A549) (Fig 1A). In addition, the protein expression of GSK-3α was dramatically suppressed by CREB knockdown in all four lung cancer cell lines we tested (Fig 1B) but the protein level of GSK-3β was not changed by CREB knockdown (Fig 1C).
Fig 1. CREB inhibition suppresses the expression of GSK-3α.
(A) Effect of CREB knockdown on the mRNA level of GSK3A, GSK3B, and CREB. Each of the indicated cells were transfected with control siRNA or CREB siRNA (40 nM, each) for 48 h, followed by qPCR analysis. All values in the graphs represent mean ± SD of three independent experiments. Two-sided t-test. *, P < 0.01. (B-C) Effect of CREB knockdown on the protein levels of GSK-3α (B) and GSK-3β (C). Each of the indicated cells were transfected with control siRNA or CREB siRNA (40 nM, each) for 72 h, followed by western blot analysis.
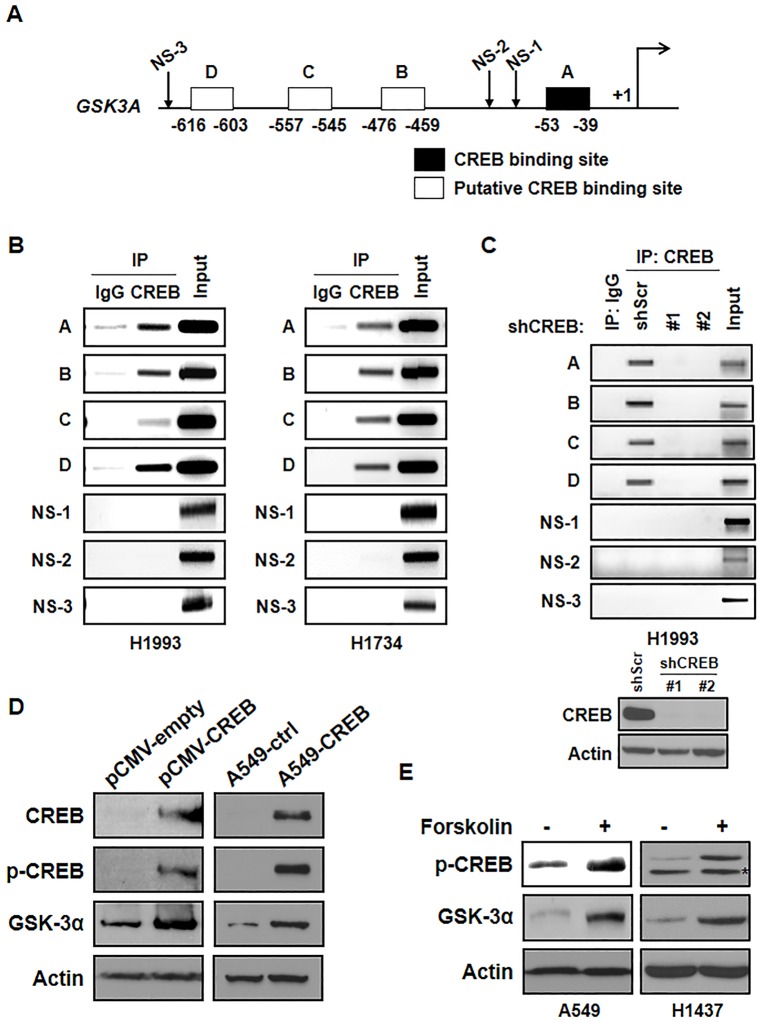
We noticed that the GSK3A promoter contained several putative CREB binding sites as determined by TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) (Fig 2A). To examine whether CREB binds to the human GSK3A promoter, we performed ChIP assay by using immunoprecipitation of CREB antibody and IgG as a negative control. Not only primer set A which covers the CREB consensus binding site (-39 to -53), but also three other primer sets (B: -459 to -476, C: -545 to -557, and D: -603 to -616) showed the binding of CREB on the GSK3A promoter. However, non-specific regions (NS-1, NS-2, and NS-3) of the promoter did not show any binding of CREB (Fig 2B). In addition, the knockdown of CREB markedly suppressed the association of CREB with the GSK3A promoter (Fig 2C). Consistent with previous data which showed CREB knockdown suppressed the expression of GSK-3α, overexpression of CREB strongly induced GSK-3α expression at the protein level after transient or stable overexpression of CREB (Fig 2D). In fact, the expressions of p-CREB and GSK-3α were increased by forskolin, which activates the enzyme adenylyl cyclase and increases intracellular levels of cyclic AMP (Fig 2E). Taken together, we suggest that CREB is a potential upstream regulator of GSK-3α in lung cancer cells.
Fig 2. CREB binds to the promoter of GSK-3α and regulates the expression of GSK-3α.
(A) Schematic diagram showing the positions of CREB binding elements located in the gene promoter of human GSK3A (TFSEARCH). A-D: the specific regions for primers which cover CREB binding elements, A: -39 to -53, B: -459 to -476, C: -545 to -557, D: -603 to -616, NS-1,-2, and -3: the regions for primers which include non-specific binding elements NS-1: -126 to -230, NS-2: -183 to -378, and NS-3: -1214 to -1356. Primer sequences are in the S1 Table (available online). (B) Direct binding of CREB on the GSK3A promoter. A ChIP assay was done with chromatins prepared from H1993 and H1734 cells. The binding of CREB to the GSK3A promoter was detected by visualization of the PCR product. The single bands detected in input samples indicate the specificity of the PCR primers. (C) Direct binding of CREB on the GSK3A promoter in CREB-knockdown cells. The protein level of CREB in each stable cell line was confirmed by western blot analysis. (D) Effect of CREB overexpression on the expression of GSK-3α. H1437 cells were transfected with the same amount of pCMV-empty or pCMV-CREB expression vector and the expression of CREB, p-CREB, and GSK-3α was examined by western blot analysis (left). A549-control (A549-ctrl) or A549-CREB cells were transfected with pCMV vectors and selected by puromycin (1 μg/ml). The effect of CREB on the expression of GSK-3α was also confirmed (right). (E) Effect of forskolin on the induction of the level of p-CREB and GSK-3α. A549 and H1437 cells were treated with forskolin (10 μM, 30 min) and the expression of each protein was examined by western blot analysis.
GSK-3α is a poor prognosis factor in lung cancer
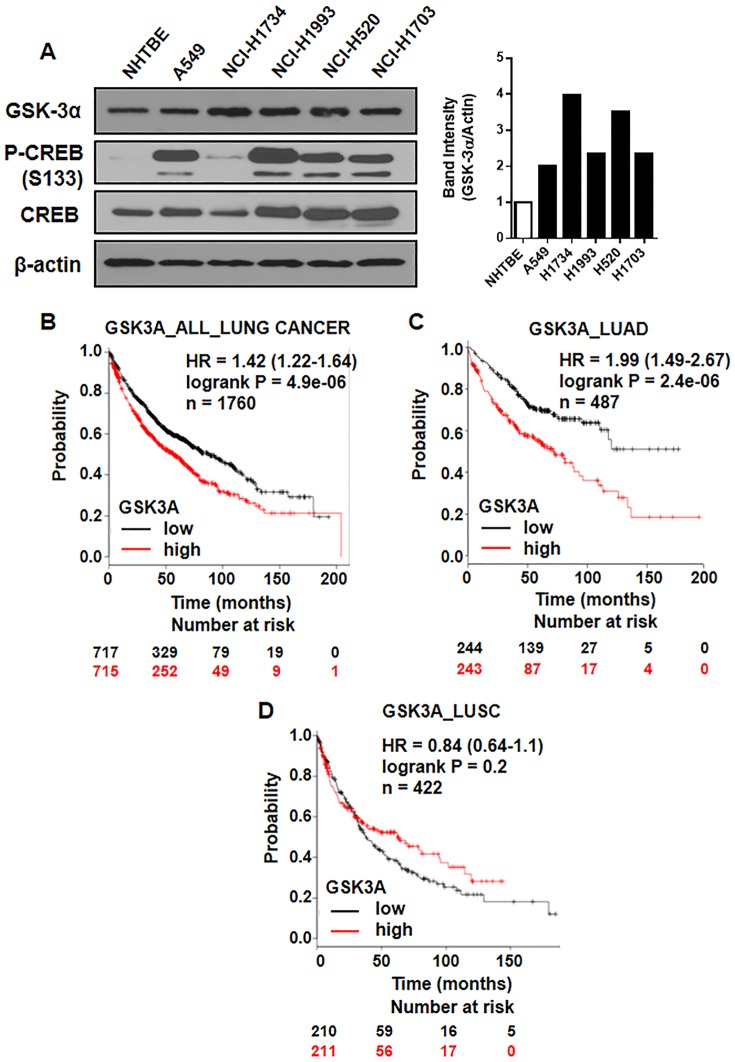
There is evidence that GSK-3β is overexpressed in lung cancer. The overexpression of GSK-3β serves as an independent marker of poor prognosis for NSCLC and its inhibition suppresses cell proliferation in NSCLC cells [39]. However, the role of GSK-3α in lung cancer still need more investigation. Here, we found that GSK-3α is overexpressed in multiple lung cancer cell lines compared with NHTBE cells (Fig 3A).
Fig 3. GSK-3α overexpression indicates poor prognosis of lung cancer.
(A) Protein levels of GSK-3α, p-CREB, and CREB in multiple lung cancer cell lines compared with NHTBE cells. (B-D) Kaplan-Meier analysis of overall survival by low or high GSK3A (GSK3A probe set 202210_x_at) expression in (B) 1760 lung cancer patients, (C) 487 lung adenocarcinoma patients, and (D) 422 lung squamous cell carcinoma with adjuvant treatment. Overall survival analysis of the patients was performed by using Cox proportional hazard models and follow-up data for the indicated period.
To further assess whether our finding of GSK3A overexpression is relevant to human lung cancer, we used publically available Kaplan-Meier plotter (http://kmplot.com/analysis), which is consist of 1760 lung cancer patients who receive chemo/radiotherapy based on the databases (CARRAY: n = 504; GSE14814: n = 90; GSE19188: n = 156; GSE29013: n = 55; GSE31210: n = 246; GSE3141: n = 111; GSE37745: n = 196; GSE4573: n = 131; GSE8894: n = 138; and TCGA: n = 133). To determine whether GSK3A mRNA (202210_x_at) abundance in tumors was associated with overall survival, we performed on overall survival analysis of the patients using Cox proportional hazard models and follow-up data for 200 months after surgery. Overexpression of GSK3A mRNA levels was associated with poor overall survival of lung cancer patients (harzard ratio (HR) = 1.42, logrank P = 4.9e-06) (Fig 3B). Interestingly, GSK3A mRNA levels were more strongly associated with poor overall survival of lung cancer patients with adenocarcinoma (HR = 1.99, logrank P = 2.4e-06) (Fig 3C). However, positive GSK3A expression was not significantly correlated with shorter survival time of the patients with squamous cell carcinoma histology type (Fig 3D). We also found that overexpression of CREB mRNA level was associated with poor overall survival of lung cancer patients with very similar pattern for GSK3A mRNA (S2 Fig). Our results on clinical tumor samples demonstrate that aberrant activation of GSK-3α is associated with human mortality of lung cancer patients, especially with lung adenocarcinoma.
Inhibition of GSK-3α suppresses the viability of lung cancer cells
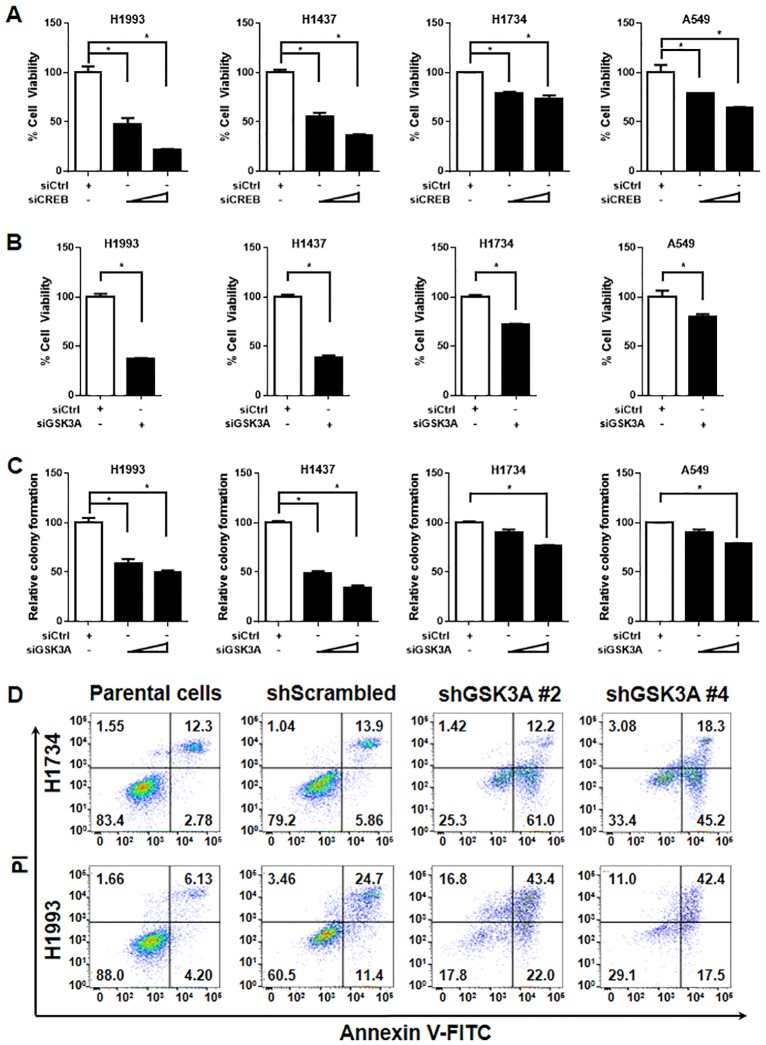
To determine the effect of GSK-3α, which is a CREB target gene, we confirmed the effect of CREB knockdown on the cell viability in multiple lung cancer cell lines. Consistent with our previous reports [10], CREB inhibition suppressed the viability of lung cancer cells (Fig 4A). Next, knockdown of GSK-3α with its specific siRNA resulted in a decrease in the viability of all lung cancer cell lines we tested (Fig 4B). In addition, knockdown of GSK-3α led to reduced colony formation of the cells (Fig 4C). Consequently, GSK-3α knockdown significantly suppressed the cell viability of KRAS-WT lung cancer cell lines (H1993 and H1437), compared to KRAS-mutant lung cancer cell lines (H1734 and A549). We further examined the effect of GSK-3α knockdown on the cell death using FACS analysis. As presented in Fig 4D, the cells which were expressing shGSK3A showed the increased cell death compared with each control cell line. The validation of siRNAs or shRNAs on the expression of GSK-3α was performed by qPCR or western blot analysis in multiple lung cancer cell lines (S1 Fig). These results suggest that GSK-3α positively regulates the viability of lung cancer cells.
Fig 4. GSK-3α knockdown suppresses lung cancer cell viability.
(A) Effect of CREB knockdown on the cell viability. Lung cancer cell lines (H1993, H1437, H1734, and A549) were transiently transfected with control siRNA (50 nM), or CREB siRNA (10, 50 nM) for 72 h, followed by MTT assay. All values in the graphs represent mean ± SD of three independent experiments. Two-sided t-test. *, P < 0.01. (B) Effect of GSK-3α knockdown on the cell viability. The cells were transiently transfected with control siRNA, or GSK-3α siRNA (40 nM, each) for 72 h and the quantitative data was followed by MTT assay. All values in the graphs represent mean ± SD of three independent experiments. Two-sided t-test. *, P < 0.01. (C) Effect of GSK-3α knockdown on colony formation. One day after transfection of control siRNA (40 nM), or GSK-3α siRNA (10, 40 nM), the cells were seeded again in 6-wells with low density (2 x 103/ well) and incubated for 7–14 days. Mean ± SD in three independent experiments. Two-sided t-test. *, P < 0.01. (D) Effect of GSK-3α knockdown on the cell death. H1734 and H1993 cells were infected with lentiviral expressing shRNA targeting GSK-3α (two sequences; shGSK3A#2 and shGSK3A#4) with 8 μg/ml polybrene. After 48 h infection, cells were selected with 0.5 μg/ml puromycin for 3 days. The selected cells were performed by staining of annexin V and PI to analysis apoptotic cell death by LSRII.
CREB-GSK-3α signaling is critical for the regulation of cyclins
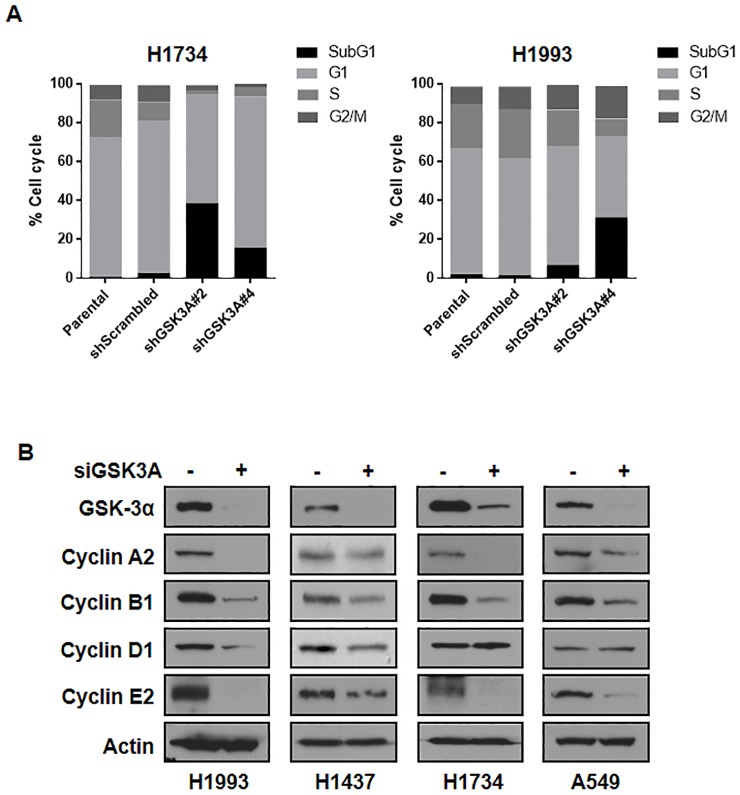
To gain insight into the role of GSK-3α in lung cancer cell viability, we first examined whether GSK-3α regulates cell cycle using FACS analysis. Interestingly, GSK-3α knockdown suppressed S phase of the cell cycle, which might contribute the reduced cell viability of lung cancer cells (Fig 5A). As shown in Fig 5B, the protein expression of several cyclins including cyclin A2, cyclin B1, cyclin D1, and cyclin E2 was markedly suppressed by GSK-3α knockdown in lung cancer cell lines. In addition, these results were consistent with previous reports that CREB can regulate the expression of cyclins [12, 31, 40, 41]. Taken together, these results suggested that GSK-3α might function positively in the viability of lung cancer cells by regulating the expression of cyclins.
Fig 5. Effects of GSK-3α knockdown on the expression of cyclins.
(A) Effect of GSK-3α knockdown on the cell cycle. Indicated cells were starved in serum-free RPMI medium for 24 h and replenished with RPMI supplemented with 10% FBS for another 24 h. Harvested cells were stained with PI to analysis cell cycle by LSRII. (B) Effect of GSK-3α knockdown on the gene expressions related to cell viability or cell cycle including cyclin A2, cyclin B1, cyclin D1, and cyclin E2. The lung cancer cells were transiently transfected with control siRNA, or GSK-3α siRNA (40 nM, each) for 48 h and were followed by western blot analysis.
GSK-3α is critical for tumor growth
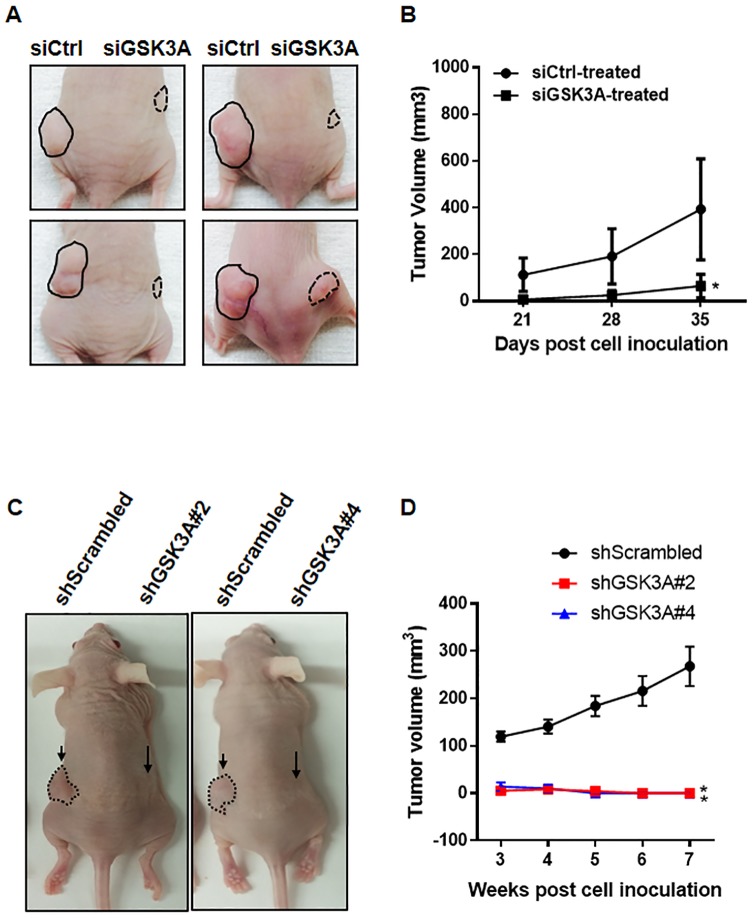
We next addressed the role GSK-3α in tumor growth in vivo using subcutaneous injections of control or GSK-3α-depleted cells. The growth of tumors was monitored over five weeks after the cells were explanted in nude mice. Representative photographs of mice at the end of five weeks showed that the development of tumors derived from the GSK-3α-depleted cells was markedly suppressed compared with tumors derived from control cells (Fig 6A). Consistent with our hypothesis and data, GSK-3α knockdown resulted in a significant suppression in tumor growth and tumor volume (Fig 6B). We confirmed the effect of GSK-3α depletion on the expression of GSK-3α or cyclin B1 in the tissues derived from xenografts (S5 Fig). Repeatedly, we noticed that the complete deletion of GSK3A in the cells could not develop the tumors in nude mice (Fig 6C and 6D). Overall, these data clearly demonstrate that the decreased levels of GSK-3α impair lung cancer tumor growth in vivo.
Fig 6. Effects of GSK-3α-deficient cells on the capability of tumor growth in vivo.
(A) Representative images of xenografts derived from control siRNA or GSK-3α siRNA-treated H1993 cells. After 24 h siRNA transfection (20 nM, each), the cells were inoculated subcutaneously into the right and left side dorsal flanks of female nude mice (xenograft n = 7/group). (B) Tumor volume of xenografts derived from control or GSK-3α-deficient H1993 cells were evaluated at each time point as indicated. Tumor volume was measured with digital calipers and calculated by the formula 0.52 x length x width2. Two-sided t-test. *, P < 0.01. (C) Representative images of xenografts derived from H1993-shScrambled, shGSK3A#2, or shGSK3A#4 cells. The cells were inoculated subcutaneously into dorsal flanks of nude mice (left: shScrambled cells, right: shGSK3A cells) and tumor volume was measured over the indicated time points (shScrambled n = 9, shGSK3A#2 n = 4, and shGSK3A#4 n = 5). Two-sided t-test. *, P < 0.05.
Discussion
In this study, we first identified GSK-3α as a novel target of CREB in lung cancer cells. Our study reveals oncogenic roles of GSK-3α as a CREB target gene and as a novel prognostic biomarker in lung cancer. GSK-3α has been shown to be a therapeutic target in multiple human cancers including AML, pancreatic cancer, and prostate cancer. However, the role of GSK-3α in lung cancer still largely remains unknown. Here, our results show that decreased levels of GSK-3α impair the viability of lung cancer cells in vitro and in vivo. GSK-3α is overexpressed in multiple lung cancer cell lines and lung tumor tissues. Also, the aberrant overexpression of GSK-3α was associated with a shorter survival time especially in patients with lung adenocarcinoma. More importantly, CREB regulates the expression of GSK-3α but not GSK-3β, suggesting that there is a specific arm of CREB-GSK-3α signaling in lung cancer.
The activity of CREB is regulated by multiple phosphorylation mechanisms and the phosphorylation of CREB at serine-133 is required for recruitment of the co-activator CREB-binding protein (CBP)/p300 and its transcriptional activity. In fact, GSK-3 has been known as a repressor of CREB activity. CREB DNA binding activity is inhibited by GSK-3β overexpression and increased by lithium or sodium valproate which are GSK-3 inhibitors in human neuroblastoma cells [42]. Moreover, GSK-3β has been reported to repress multiple CREB-target genes [43, 44]. Conversely, recent study has been reported that GSK-3 promotes the association of CREB and its co-activators with MEIS1, a homeobox (HOX) DNA-binding cofactor, to induce HOX-mediated transcription and transformation in MLL leukemias [45]. Although the functional consequence of CREB activity by GSK-3 is not still clear, our study strongly suggests that CREB positively regulates GSK-3α, not GSK-3β, in lung cancer cells, thus providing a new concept of CREB-GSK-3α signaling.
Although the overexpression of GSK-3β and its function as a tumor promoter in lung cancer has been demonstrated [39], the role of GSK-3α in lung cancer remains elusive. In our current study, we found that GSK-3α is overexpressed in lung cancer cell lines compared to normal bronchial epithelial cells. Knockdown of GSK-3α in lung cancer cells causes suppression of cell proliferation and also an induction of substantial apoptosis. These results indicate that GSK-3α also plays a critical role in the growth of lung cancer cells.
The mechanism of GSK-3α as a tumor promoter in lung cancer is virtually unknown. It has been shown that GSK-3α promotes oncogenic KRAS function via IKK-NF-κB activity in pancreatic cancer. The authors suggested GSK-3α as a key downstream effector of mutant KRAS to regulate NF-κB signaling pathways [30]. Interestingly, our study showed that the GSK-3α knockdown highly suppressed the cell viability of KRAS-WT lung cancer cell lines, H1993 and H1437 cells, compared to KRAS-mutant lung cancer cell lines, H1734 (G13C) and A549 (G12S) cells. Although it remains unclear whether KRAS status is directly related to the role of GSK-3α in lung cancer cells, we suggest that the function of GSK-3α in KRAS signaling might be regulated in a cell-type specific manner.
To better understand the role of GSK-3α and examine the critical genes affected by GSK-3α in lung cancer, we initially screened a subset of NF-κB target genes such as MYC, WT1, BIRC2, IL-9, HMOX1, and TERT, which were regulated by a pan-GSK-3 inhibitor AR-014418 in pancreatic cancer [30]. Our data was not fully consistent with the previous study, but we observed a significant decrease in the mRNA level of TERT by GSK-3α knockdown and little difference in the other NF-κB targets the analyzed (EDN1, CYP19A1, HMOX1, WT1, and BIRC2) (S3 Fig). Additionally, we found that the expression of several cyclins which are known as CREB target gens was markedly downregulated by GSK-3α knockdown in multiple lung cancer cell lines. Though our study did not directly identify how cyclins are regulated by GSK-3α, these results strongly support the novel role of GSK-3α as an active regulator in the viability of lung cancer cells.
Theoretically, the inhibition of GSK-3 can lead to β-catenin stabilization and hyperactivation of Wnt/β-catenin signaling [46–49]. However, Doble et al. showed that both isoforms of GSK-3 need to be inhibited for β-catenin stabilization. GSK-3α/β double-knockout mouse embryonic stem cell (ESC) displayed hyperactivated Wnt/β-catenin signaling [50]. Consistent with this report, there is increasing evidence that the single loss of either GSK-3α or GSK-3β isoform does not lead to β-catenin stabilization [28, 39]. Our data also demonstrated that inhibition of GSK-3α does not induce the level of β-catenin and AXIN2, an Axin-related protein which plays an important role in Wnt/β-catenin signaling pathway (S4 Fig). These results support that GSK-3α might function independent of the β-catenin stabilization in lung cancer but further analysis needs to be completed to fully understand the roles of GSK-3α and GSK-3β in β -catenin stabilization in lung cancer.
The observations that GSK-3α expression plays a causal role in survival of lung cancer patients, suggest GSK-3α could be useful as a prognostic biomarker in lung cancer, especially lung adenocarcinoma. Further studies examining the mechanisms for targeting CREB-GSK-3α pathway in lung cancer will be essential for determining and developing the GSK-3α specific inhibitors. In conclusion, we propose that GSK-3α is a promising therapeutic target as a novel target gene of CREB to diagnose tumor and develop the therapy, with relevance for lung cancer.
Supporting Information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Cancer Institute grant R01-CA126801 (to Ja Seok Koo) and Cancer Center Support Grant CA-16359 (to Yale Cancer Center). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wen AY, Sakamoto KM, Miller LS. The role of the transcription factor CREB in immune function. Journal of immunology. 2010;185(11):6413–9. 10.4049/jimmunol.1001829 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlezon WA Jr., Duman RS, Nestler EJ. The many faces of CREB. Trends in neurosciences. 2005;28(8):436–45. 10.1016/j.tins.2005.06.005 . [DOI] [PubMed] [Google Scholar]
- 3.Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annual review of neuroscience. 1998;21:127–48. 10.1146/annurev.neuro.21.1.127 . [DOI] [PubMed] [Google Scholar]
- 4.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nature reviews Molecular cell biology. 2001;2(8):599–609. 10.1038/35085068 . [DOI] [PubMed] [Google Scholar]
- 5.Arnould T, Vankoningsloo S, Renard P, Houbion A, Ninane N, Demazy C, et al. CREB activation induced by mitochondrial dysfunction is a new signaling pathway that impairs cell proliferation. The EMBO journal. 2002;21(1–2):53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaudry D, Stork PJ, Lazarovici P, Eiden LE. Signaling pathways for PC12 cell differentiation: making the right connections. Science. 2002;296(5573):1648–9. 10.1126/science.1071552 . [DOI] [PubMed] [Google Scholar]
- 7.Bender RA, Lauterborn JC, Gall CM, Cariaga W, Baram TZ. Enhanced CREB phosphorylation in immature dentate gyrus granule cells precedes neurotrophin expression and indicates a specific role of CREB in granule cell differentiation. The European journal of neuroscience. 2001;13(4):679–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SW, Hong JS, Ryu SH, Chung WC, Yoon JH, Koo JS. Regulation of mucin gene expression by CREB via a nonclassical retinoic acid signaling pathway. Molecular and cellular biology. 2007;27(19):6933–47. 10.1128/MCB.02385-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo HS, Liu DD, Bekele BN, Kim MK, Pisters K, Lippman SM, et al. Cyclic AMP response element-binding protein overexpression: a feature associated with negative prognosis in never smokers with non-small cell lung cancer. Cancer research. 2008;68(15):6065–73. 10.1158/0008-5472.CAN-07-5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aggarwal S, Kim SW, Ryu SH, Chung WC, Koo JS. Growth suppression of lung cancer cells by targeting cyclic AMP response element-binding protein. Cancer research. 2008;68(4):981–8. 10.1158/0008-5472.CAN-06-0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conkright MD, Guzman E, Flechner L, Su AI, Hogenesch JB, Montminy M. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Molecular cell. 2003;11(4):1101–8. . [DOI] [PubMed] [Google Scholar]
- 12.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119(7):1041–54. 10.1016/j.cell.2004.10.032 . [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(12):4459–64. 10.1073/pnas.0501076102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pellegrini M, Cheng JC, Voutila J, Judelson D, Taylor J, Nelson SF, et al. Expression profile of CREB knockdown in myeloid leukemia cells. BMC cancer. 2008;8:264 10.1186/1471-2407-8-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen P, Frame S. The renaissance of GSK3. Nature reviews Molecular cell biology. 2001;2(10):769–76. 10.1038/35096075 . [DOI] [PubMed] [Google Scholar]
- 16.Woodgett JR. Molecular cloning and expression of glycogen synthase kinase-3/factor A. The EMBO journal. 1990;9(8):2431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Remedi MS, Pappan KL, Kwon G, Rohatgi N, Marshall CA, et al. Glycogen synthase kinase-3 and mammalian target of rapamycin pathways contribute to DNA synthesis, cell cycle progression, and proliferation in human islets. Diabetes. 2009;58(3):663–72. 10.2337/db07-1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alao JP, Stavropoulou AV, Lam EW, Coombes RC. Role of glycogen synthase kinase 3 beta (GSK3beta) in mediating the cytotoxic effects of the histone deacetylase inhibitor trichostatin A (TSA) in MCF-7 breast cancer cells. Molecular cancer. 2006;5:40 10.1186/1476-4598-5-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esfandiari F, Fathi A, Gourabi H, Kiani S, Nemati S, Baharvand H. Glycogen synthase kinase-3 inhibition promotes proliferation and neuronal differentiation of human-induced pluripotent stem cell-derived neural progenitors. Stem cells and development. 2012;21(17):3233–43. 10.1089/scd.2011.0678 . [DOI] [PubMed] [Google Scholar]
- 20.Jacobs KM, Bhave SR, Ferraro DJ, Jaboin JJ, Hallahan DE, Thotala D. GSK-3beta: A Bifunctional Role in Cell Death Pathways. International journal of cell biology. 2012;2012:930710 10.1155/2012/930710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beurel E, Jope RS. The paradoxical pro- and anti-apoptotic actions of GSK3 in the intrinsic and extrinsic apoptosis signaling pathways. Progress in neurobiology. 2006;79(4):173–89. 10.1016/j.pneurobio.2006.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Lin HK, Hu YC, Xie S, Yang L, Chang C. Suppression of androgen receptor-mediated transactivation and cell growth by the glycogen synthase kinase 3 beta in prostate cells. The Journal of biological chemistry. 2004;279(31):32444–52. 10.1074/jbc.M313963200 . [DOI] [PubMed] [Google Scholar]
- 23.Farago M, Dominguez I, Landesman-Bollag E, Xu X, Rosner A, Cardiff RD, et al. Kinase-inactive glycogen synthase kinase 3beta promotes Wnt signaling and mammary tumorigenesis. Cancer research. 2005;65(13):5792–801. 10.1158/0008-5472.CAN-05-1021 . [DOI] [PubMed] [Google Scholar]
- 24.Ma C, Wang J, Gao Y, Gao TW, Chen G, Bower KA, et al. The role of glycogen synthase kinase 3beta in the transformation of epidermal cells. Cancer research. 2007;67(16):7756–64. 10.1158/0008-5472.CAN-06-4665 . [DOI] [PubMed] [Google Scholar]
- 25.Mishra R. Glycogen synthase kinase 3 beta: can it be a target for oral cancer. Molecular cancer. 2010;9:144 10.1186/1476-4598-9-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shakoori A, Mai W, Miyashita K, Yasumoto K, Takahashi Y, Ooi A, et al. Inhibition of GSK-3 beta activity attenuates proliferation of human colon cancer cells in rodents. Cancer science. 2007;98(9):1388–93. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li D. GSK-3beta as a driving force in ovarian cancer. Cell research. 2006;16(7):609 10.1038/sj.cr.7310082 . [DOI] [PubMed] [Google Scholar]
- 28.Banerji V, Frumm SM, Ross KN, Li LS, Schinzel AC, Hahn CK, et al. The intersection of genetic and chemical genomic screens identifies GSK-3alpha as a target in human acute myeloid leukemia. The Journal of clinical investigation. 2012;122(3):935–47. 10.1172/JCI46465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darrington RS, Campa VM, Walker MM, Bengoa-Vergniory N, Gorrono-Etxebarria I, Uysal-Onganer P, et al. Distinct expression and activity of GSK-3alpha and GSK-3beta in prostate cancer. International journal of cancer Journal international du cancer. 2012;131(6):E872–83. 10.1002/ijc.27620 . [DOI] [PubMed] [Google Scholar]
- 30.Bang D, Wilson W, Ryan M, Yeh JJ, Baldwin AS. GSK-3alpha promotes oncogenic KRAS function in pancreatic cancer via TAK1-TAB stabilization and regulation of noncanonical NF-kappaB. Cancer discovery. 2013;3(6):690–703. 10.1158/2159-8290.CD-12-0541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shankar DB, Cheng JC, Kinjo K, Federman N, Moore TB, Gill A, et al. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer cell. 2005;7(4):351–62. 10.1016/j.ccr.2005.02.018 . [DOI] [PubMed] [Google Scholar]
- 32.Crans-Vargas HN, Landaw EM, Bhatia S, Sandusky G, Moore TB, Sakamoto KM. Expression of cyclic adenosine monophosphate response-element binding protein in acute leukemia. Blood. 2002;99(7):2617–9. . [DOI] [PubMed] [Google Scholar]
- 33.Cheng JC, Kinjo K, Judelson DR, Chang J, Wu WS, Schmid I, et al. CREB is a critical regulator of normal hematopoiesis and leukemogenesis. Blood. 2008;111(3):1182–92. 10.1182/blood-2007-04-083600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park MH, Lee HS, Lee CS, You ST, Kim DJ, Park BH, et al. p21-Activated kinase 4 promotes prostate cancer progression through CREB. Oncogene. 2013;32(19):2475–82. 10.1038/onc.2012.255 . [DOI] [PubMed] [Google Scholar]
- 35.Deng X, Liu H, Huang J, Cheng L, Keller ET, Parsons SJ, et al. Ionizing radiation induces prostate cancer neuroendocrine differentiation through interplay of CREB and ATF2: implications for disease progression. Cancer research. 2008;68(23):9663–70. 10.1158/0008-5472.CAN-08-2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Yang J, Cui X, Chen Y, Zhu VF, Hagan JP, et al. A novel epigenetic CREB-miR-373 axis mediates ZIP4-induced pancreatic cancer growth. EMBO molecular medicine. 2013;5(9):1322–34. 10.1002/emmm.201302507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakamoto KM, Frank DA. CREB in the pathophysiology of cancer: implications for targeting transcription factors for cancer therapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15(8):2583–7. 10.1158/1078-0432.CCR-08-1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remsing Rix LL, Kuenzi BM, Luo Y, Remily-Wood E, Kinose F, Wright G, et al. GSK3 alpha and beta are new functionally relevant targets of tivantinib in lung cancer cells. ACS chemical biology. 2014;9(2):353–8. 10.1021/cb400660a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng J, Liu D, Qiu Z, Huang Y, Chen B, Wang L, et al. GSK3beta overexpression indicates poor prognosis and its inhibition reduces cell proliferation and survival of non-small cell lung cancer cells. PloS one. 2014;9(3):e91231 10.1371/journal.pone.0091231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beier F, Lee RJ, Taylor AC, Pestell RG, LuValle P. Identification of the cyclin D1 gene as a target of activating transcription factor 2 in chondrocytes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(4):1433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pigazzi M, Manara E, Baron E, Basso G. miR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer research. 2009;69(6):2471–8. 10.1158/0008-5472.CAN-08-3404 . [DOI] [PubMed] [Google Scholar]
- 42.Grimes CA, Jope RS. CREB DNA binding activity is inhibited by glycogen synthase kinase-3 beta and facilitated by lithium. Journal of neurochemistry. 2001;78(6):1219–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tullai JW, Chen J, Schaffer ME, Kamenetsky E, Kasif S, Cooper GM. Glycogen synthase kinase-3 represses cyclic AMP response element-binding protein (CREB)-targeted immediate early genes in quiescent cells. The Journal of biological chemistry. 2007;282(13):9482–91. 10.1074/jbc.M700067200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen T, Rehfeld JF, Nielsen FC. GSK-3beta reduces cAMP-induced cholecystokinin gene expression by inhibiting CREB binding. Neuroreport. 2004;15(5):841–5. . [DOI] [PubMed] [Google Scholar]
- 45.Wang Z, Iwasaki M, Ficara F, Lin C, Matheny C, Wong SH, et al. GSK-3 promotes conditional association of CREB and its coactivators with MEIS1 to facilitate HOX-mediated transcription and oncogenesis. Cancer cell. 2010;17(6):597–608. 10.1016/j.ccr.2010.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. The EMBO journal. 1998;17(5):1371–84. 10.1093/emboj/17.5.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108(6):837–47. . [DOI] [PubMed] [Google Scholar]
- 48.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. Journal of cell science. 2003;116(Pt 7):1175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nature medicine. 2004;10(1):55–63. 10.1038/nm979 . [DOI] [PubMed] [Google Scholar]
- 50.Doble BW, Patel S, Wood GA, Kockeritz LK, Woodgett JR. Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Developmental cell. 2007;12(6):957–71. 10.1016/j.devcel.2007.04.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.