Crystal structures of a leucine-rich repeat domain of IpaH9.8 from S. flexneri were determined in two crystal forms: P212121 and C2221.
Keywords: Shigella flexneri, effector, leucine-rich repeats, IpaH9.8, E3 ligase
Abstract
Infectious diseases caused by bacteria have significant impacts on global public health. During infection, pathogenic bacteria deliver a variety of virulence factors, called effectors, into host cells. The Shigella effector IpaH9.8 functions as an ubiquitin ligase, ubiquitinating the NF-κB essential modulator (NEMO)/IKK-γ to inhibit host inflammatory responses. IpaH9.8 contains leucine-rich repeats (LRRs) involved in substrate recognition and an E3 ligase domain. To elucidate the structural basis of the function of IpaH9.8, the crystal structure of the LRR domain of Shigella IpaH9.8 was determined and this structure was compared with the known structures of other IpaH family members. This model provides insights into the structural features involved in substrate specificity.
1. Introduction
Pathogenic bacteria, including Shigella, deliver a number of effectors into the host cell through their type III secretion systems (Büttner, 2012 ▸; Parsot, 2009 ▸; Kim et al., 2014 ▸). S. flexneri secretes ten IpaH family proteins, all of which have leucine-rich repeats (LRRs) involved in substrate recognition and an E3 ligase domain located in the C-terminal region (CTD), which is distinct from either the RING or HECT domains and has been termed a ‘novel E3 ligase’ (NEL) domain (Hicks & Galán, 2010 ▸; Singer et al., 2008 ▸; Zhu et al., 2008 ▸; Ashida et al., 2014 ▸). NEL enzymes comprise a large family of bacterial effector proteins, including those from Yersinia, Salmonella, Edwardsiella, Bradyrhizobium, Rhizobium and some Pseudomonas species (Chou et al., 2012 ▸; Zouhir et al., 2014 ▸). The NEL domain forms a ubiquitin thioester intermediate via a catalytic cysteine in a manner analogous to that of the structurally unrelated eukaryotic HECT domain (Quezada et al., 2009 ▸). The crystal structures of the IpaH family proteins Shigella IpaH3 (Zhu et al., 2008 ▸) and Salmonella SspH2 (Quezada et al., 2009 ▸) reveal an architecture consisting of two structural elements: an N-terminal LRR domain linked by a short stretch of residues to a novel C-terminal helical domain with NEL activity. Each structure showed that autoinhibition of IpaH can occur by two distinct mechanisms that disrupt the catalytic domain.
The Shigella effector IpaH9.8 has been demonstrated to ubiquitinate the NF-κB essential modulator (NEMO)/IKK-γ, an essential component of the IκB kinase complex. As a result, NEMO is degraded by the host proteasome, and NF-κB activation and the subsequent inflammatory response to Shigella infection are attenuated (Ashida et al., 2010 ▸). IpaH9.8 interacts not only with NEMO but also with ABIN-1 (A20-binding inhibitor of NF-κB), which acts as an adaptor for the IpaH9.8-mediated ubiquitination of NEMO. The IpaH9.8 LRRs are required for the interaction with NEMO, and the CTD is involved in the interaction with ABIN-1. The structure of the NEL domain of IpaH9.8 has been reported and shows a domain-swapped dimeric structure under nonreducing conditions (Seyedarabi et al., 2010 ▸). Under nonreducing conditions, IpaH9.8 undergoes a domain swap driven by the formation of a disulfide bond involving the catalytic cysteine, with this dimer unable to catalyze ubiquitination. However, structures of the NEL domains of IpaH3 (Zhu et al., 2008 ▸) and IpaH1.4 (Singer et al., 2008 ▸) reveal a monomeric structure and other studies demonstrate that IpaH9.8 functions in the same way as the other types of dimers (Edwards et al., 2014 ▸).
Although the structure of the LRRs from IpaH3 has been determined, each IpaH family protein targets distinct host proteins and has a different role in Shigella pathogenesis. For example, IpaH7.8 degrades glomulin and induces macrophage cell death (Suzuki et al., 2014 ▸). IpaH4.5 modulates host inflammation via interaction with the p65 subunit of NF-κB (Wang et al., 2013 ▸). NEMO directly interacts with the LRR domain of IpaH9.8 and is ubiquitinated. The interaction between the IpaH9.8 LRR domain and NEMO involves the region between the leucine-zipper (LZ) and the zinc-finger (ZF) domains of NEMO (residues 347–396). IpaH9.8 targets both the Lys309 and Lys321 residues of NEMO for ubiquitination (Ashida et al., 2010 ▸). However, the structural basis for the targeting of specific substrates by the LRRs of IpaH9.8 is currently unknown. To understand the mechanistic details underlying IpaH9.8-mediated NEMO ubiquitination, we determined crystal structures of the IpaH9.8 LRRs.
2. Materials and methods
2.1. Macromolecule production
DNA encoding the LRR domain of IpaH9.8 (IpaH9.8 LRR; residues 22–244) was cloned into the pCold I vector to create a construct encoding N-terminally hexahistidine–small ubiquitin-related modifier 1 (SUMO1)-tagged IpaH9.8 LRR. The protein construct was transformed and expressed in Escherichia coli BL21 (DE3) cells. The culture was grown at 37°C until the OD600 reached 0.6–0.8, at which point expression was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and the temperature was reduced to 16°C for 15 h. Tagged proteins were affinity-purified using nickel resin. The tag moiety was proteolytically removed by the addition of ubiquitin-like-specific protease 1 (Ulp1) overnight at 4°C. Further purifications were performed using anion-exchange (HiTrap Q FF, GE Healthcare) and gel-filtration (HiLoad Superdex 75, GE Healthcare) chromatography.
2.2. Crystallization
Purified IpaH9.8 LRR was concentrated to 22.1 mg ml−1 in a buffer consisting of 25 mM Tris–HCl pH 7.5, 1.0 mM DTT (Table 1 ▸). Crystallization conditions were screened by the sitting-drop vapour-diffusion method at 293 K with 2 µl drops (1 µl IpaH9.8 LRR at 22.1 mg ml−1 and 1 µl reservoir solution) using screening kits from Hampton Research (Index, Crystal Screen and Crystal Screen 2). Plate-shaped crystals were grown from solutions consisting of 0.1 M bis-tris pH 5.5, 0.2 M lithium sulfate monohydrate, 25%(w/v) PEG 3350 (form 1) and 0.1 M bis-tris pH 5.5, 0.2 M ammonium sulfate, 25%(w/v) PEG 3350 (form 2).
Table 1. Crystallization.
| Form 1 | Form 2 | |
|---|---|---|
| Method | Sitting-drop vapour diffusion | Sitting-drop vapour diffusion |
| Temperature (K) | 293 | 293 |
| Protein concentration (mg ml−1) | 22.1 | 22.1 |
| Buffer composition of protein solution | 25 mM Tris–HCl pH 7.5, 1 mM DTT | 25 mM Tris–HCl pH 7.5, 1 mM DTT |
| Composition of reservoir solution | 0.1 M bis-tris pH 5.5, 0.2 M lithium sulfate monohydrate, 25%(w/v) PEG 3350 | 0.1 M bis-tris pH 5.5, 0.2 M ammonium sulfate, 25%(w/v) PEG 3350 |
| Volume and ratio of drop | 1 µl:1 µl | 1 µl:1 µl |
| Volume of reservoir (µl) | 100 | 100 |
2.3. Data collection and processing
The crystals were cooled without cryoprotective additives. X-ray diffraction data sets were collected at 100 K on beamline BL44XU at SPring-8, Hyogo, Japan. The data sets were processed using HKL-2000 (Otwinowski & Minor, 1997 ▸). Although the data [R p.i.m. and 〈I/σ(I)〉] allowed us to include higher resolution data, we did not collect high-resolution data because we judged the resolution limit by eye and determined the camera distance by referring to the high-resolution limit. Data-collection and processing statistics for the crystals are given in Table 2 ▸.
Table 2. Data collection and processing.
Values in parentheses are for the outer shell.
| Form 1 | Form 2 | |
|---|---|---|
| Diffraction source | BL44XU, SPring-8 | BL44XU, SPring-8 |
| Wavelength (Å) | 0.9 | 0.9 |
| Temperature (K) | 100 | 100 |
| Detector | MX300-HE CCD | MX300-HE CCD |
| Crystal-to-detector distance (mm) | 270 | 300 |
| Rotation range per image (°) | 1.0 | 1.0 |
| Total rotation range (°) | 180 | 180 |
| Exposure time per image (s) | 1.0 | 1.0 |
| Space group | P212121 | C2221 |
| a, b, c (Å) | 60.8, 66.2, 105.2 | 68.3, 105.0, 61.6 |
| Resolution range (Å) | 50.0–1.80 (1.83–1.80) | 50.0–2.00 (2.03–2.00) |
| Total No. of reflections | 288954 | 102926 |
| No. of unique reflections | 39927 | 15296 |
| Completeness (%) | 99.7 (100.0) | 99.8 (100.0) |
| Multiplicity | 7.3 (7.4) | 6.7 (6.9) |
| 〈I/σ(I)〉 | 49.4 (6.3) | 44.3 (6.4) |
| R merge | 0.066 (0.472) | 0.074 (0.483) |
| R p.i.m. | 0.026 (0.185) | 0.031 (0.199) |
| Overall B factor from Wilson plot (Å2) | 22.8 | 24.8 |
2.4. Structure solution and refinement
The structure of IpaH9.8 LRR was determined using molecular replacement in MOLREP (Vagin & Teplyakov, 2010 ▸) from the CCP4 suite with the LRR-domain structure of IpaH3 (Zhu et al., 2008 ▸) as the search model {PDB entry 3cvr; 51% sequence identity to the IpaH9.8 LRR domain [IpaH9.8 (22–244) versus IpaH3 (24–270)] using BLAST2 (Tatusova & Madden, 1999 ▸)}. The IpaH9.8 LRR model from crystal form 1 was built in Coot (Emsley et al., 2010 ▸) and refined in REFMAC5 (Murshudov et al., 2011 ▸). The structure of IpaH9.8 LRR from crystal form 2 was determined by molecular replacement using MOLREP with the refined model of form 1. Structure refinement of IpaH9.8 LRR form 2 was guided by referencing the structure from form 1. Structural homologues of IpaH9.8 were identified using the DaliLite server (Holm & Park, 2000 ▸). Structural validations were performed using PROCHECK (Laskowski et al., 1993 ▸). Structure-solution and refinement statistics for the crystals are given in Table 3 ▸. Structural figures were generated using PyMOL (DeLano, 2002 ▸) and CCP4mg (McNicholas et al., 2011 ▸).
Table 3. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Form 1 | Form 2 | |
|---|---|---|
| Resolution range (Å) | 34.1–1.80 (1.85–1.80) | 34.1–2.00 (2.05–2.00) |
| No. of reflections, working set | 37748 (2523) | 14484 (943) |
| No. of reflections, test set | 1996 (133) | 763 (54) |
| Final R cryst | 0.210 (0.262) | 0.211 (0.249) |
| Final R free | 0.259 (0.330) | 0.265 (0.296) |
| No. of non-H atoms | ||
| Protein | 3578 | 1779 |
| Ligand | 0 | 0 |
| Solvent | 143 | 52 |
| Total | 3721 | 1831 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.011 | 0.014 |
| Angles (°) | 1.574 | 1.725 |
| Average B factor (Å2) | ||
| Protein | 30.4 | 35.4 |
| Ramachandran plot | ||
| Most favoured (%) | 81.1 | 80.5 |
| Allowed (%) | 18.9 | 19.5 |
3. Results and discussion
3.1. Overall structure of IpaH9.8 LRR
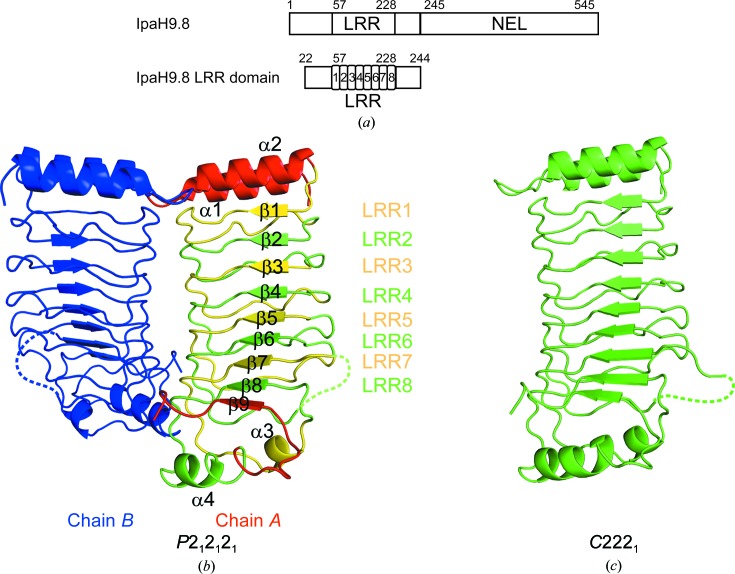
The structure of IpaH9.8 LRR was determined in two different crystal forms. The first structure was determined from a P212121 crystal form (form 1), whereas the other structure was determined from a C2221 crystal form (form 2) (Fig. 1 ▸). The form 1 and form 2 structures have two and one molecules in the asymmetric unit, respectively. The final refined models consist of residues 21–175 and 182–244 for chain A and 21–175 and 183–244 for chain B in the asymmetric unit (molecules A and B) of the form 1 structure and residues 21–175 and 180–244 of the form 2 structure, respectively. The structures of residues 176–181 in chain A of form 1, 176–182 in chain B of form 1 and 176–179 of form 2 could not be constructed because of weak electron density. The overall structures of these molecules have similar root-mean-square deviations (r.m.s.d.s) of 0.298 Å for 212 Cα atoms for form 1 chain A and form 2, 0.452 Å for 213 Cα atoms for form 1 chain B and form 2 and 0.312 Å for 213 Cα atoms for form 1 chain A and form 1 chain B. The quality of the model (resolution and peptide length) of form 1 chain A is better than the others; hence, we will use it in the discussion below. IpaH9.8 LRR is composed of eight tandemly repeated LRR motifs (LRR1–LRR8) and folds into a solenoid-like arrangement. Each repeat unit of LRR1–LRR6 consists of β-strand–turn–short segmental loop structures, whereas the β-strand–turns in LRR7 and LRR8 are followed by helix α3 and helix α4, respectively. The N-terminus of the LRR domain is capped by α1 and α2, whilst the C-terminus is flanked by α3, α4 and β9.
Figure 1.
Structural overview of IpaH9.8 LRR. (a) Schematic diagram of IpaH9.8. (b) The IpaH9.8 LRR structure in the P212121 form (form 1). Two chains are present in the asymmetric unit. Each LRR motif of chain A is coloured yellow and green alternately, whereas the N- and C-terminal regions of chain A are coloured red. Chain B is coloured blue. Loops without a determined structure are indicated by dashed lines. (c) The IpaH9.8 LRR structure in the C2221 form (form 2).
3.2. Comparison of IpaH9.8 LRR with other IpaH family proteins
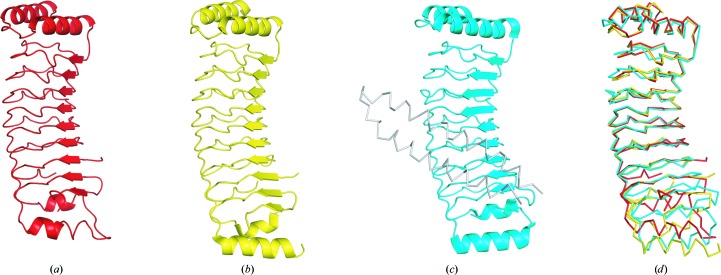
The structures of IpaH family proteins from Shigella and Salmonella have been determined, and a DALI search (Holm & Rosenström, 2010 ▸) with IpaH9.8 LRR revealed this family of proteins to be the top-scoring structural homologues. The LRR domains of IpaH3 (Zhu et al., 2008 ▸), Yersinia YopM (Evdokimov et al., 2001 ▸) and the Salmonella IpaH family member SspH1 (Keszei et al., 2014 ▸) are composed of tandemly repeated LRR motifs and α-helices at the N-terminus (Fig. 2 ▸). Although the fundamental structure of IpaH9.8 LRR shares structural similarity with that of the LRR domain of IpaH3 (r.m.s.d. of 2.3 Å for 205 Cα atoms from DaliLite), a number of differences were observed. IpaH9.8 LRR consists of eight repeat units (LRR1–LRR8), whereas the LRR domain of IpaH3 consists of nine (LRR1–LRR9). The N-terminus–LRR6 and LRR8–C-terminus portions of IpaH9.8 are remarkably similar to the N-terminus–LRR6 and LRR9–C-terminus portions of IpaH3, with r.m.s.d. values of 0.7 and 0.9 Å, respectively.
Figure 2.
Comparison of IpaH9.8 LRR with homologous structures. (a) The IpaH9.8 LRR structure as determined in this research. (b) The IpaH3 LRR domain structure from full-length IpaH3 (PDB entry 3cvr). (c) The SspH1 structure with its substrate PKN1 as a ribbon diagram (PDB entry 4nkg). (d) Structural superposition of the LRR domains from IpaH9.8, IpaH3 and SspH1.
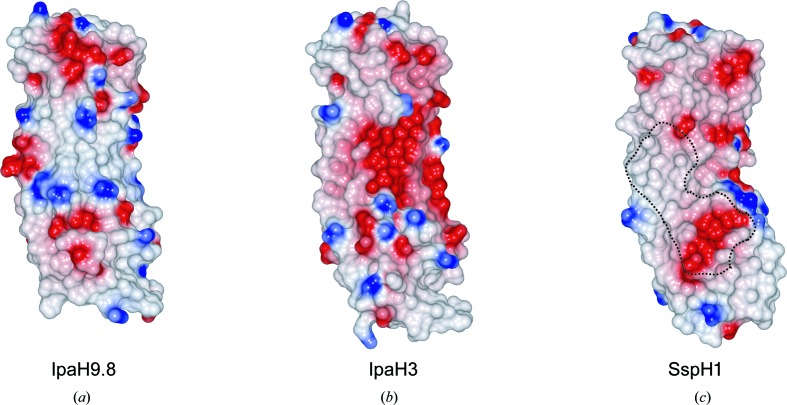
YopM contains 15 LRR repeats and the structure of the N-terminal portion of YopM (PDB entry 4ow2; A. Rumm, M. Perbandt & M. Aepfelbacher, unpublished work) resembles that of IpaH9.8, with an r.m.s.d. of 1.7 Å. The LRR domain of SspH1 (PDB entry 4nkh; Keszei et al., 2014 ▸) is composed of eight repeat units (LRR1–LRR8) and N- and C-terminal helices, and its overall structure resembles that of IpaH9.8, with an r.m.s.d. of 2.0 Å for 197 Cα atoms. In IpaH3, the regions corresponding to the disordered loop in IpaH9.8 (residues 176–181) were not assigned in the model, whereas the corresponding loop of SspH1 (residues 315–318) was determined. The IpaH family has been shown to interact with specific substrates through its LRR domain. The concave face of the LRR β-sheet is involved in substrate interaction in SspH1. We compared the charge distribution on the substrate-recognition interface of SspH1 and the corresponding surfaces of IpaH9.8 and IpaH3 (Fig. 3 ▸). IpaH9.8 LRR has a large positively charged patch at the centre of the concave face. However, the surface-charge environments of the concave faces differ significantly between IpaH9.8 and IpaH3 or SspH1, despite their structural similarities. This analysis suggested that each IpaH family protein recognizes substrates in a different way.
Figure 3.
Surface-potential representation of IpaH family proteins. Surface-potential representations of IpaH9.8 (a), IpaH3 (b) and SspH1 (c) are shown. The dashed line shown in (c) surrounds the surface within 4 Å of PKN1 in the complex structure. Red, blue and white represent acidic, basic and neutral residues, respectively. The surface potentials were calculated and mapped using CCP4mg (McNicholas et al., 2011 ▸).
4. Discussion
In this study, we determined the crystal structures of two crystal forms of IpaH9.8 LRR at 1.8 and 2.0 Å resolution, respectively. These structures present the novel LRR domain of the IpaH family, allowing potential functional differences in substrate binding to be identified. IpaH9.8 was shown to specifically interact with NEMO. The residues on IpaH9.8 that contact NEMO may be unique across the IpaH family. In the structure of the SspH1 LRR–PKN1 complex (Keszei et al., 2014 ▸), the residues on SspH1 that contact PKN1 were unique across the IpaH family. On the basis of the above results, mapping of the conserved residues on the surface of IpaH LRR domains was performed using a ClustalW (Larkin et al., 2007 ▸) multi-sequence alignment of IpaH LRR domains from nine different Shigella IpaH proteins and the ConSurf program (Celniker et al., 2013 ▸) (Fig. 4 ▸). The unique patches of residues are mainly located on the concave surface of the LRR domain and the edge of the LRR domain around the LRR6–LRR7 region. The unique surface of the molecule is relatively abundant in positively charged residues, i.e. Arg62, Lys101, Arg163 (concave surface), Arg166 (LRR6) and Arg190 (LRR7). IpaH9.8 LRR interacts with the region between the LZ and ZF domains of NEMO (residues 347–396). The sequence of NEMO (residues 347–396) showed two clusters of acidic residues (347-CQESARIEDMRKRHVEVSQAPLPPAPAYLSSPLALPSQRRSPPEEPPDFC-396). Hence, we favour the idea that the IpaH9.8–NEMO interaction occurs through complementary charge interaction, with the basic surface of IpaH9.8 accommodating negatively charged residues of NEMO, although we were unable to detect any significant changes in the interaction by mutations of the positive surface of IpaH9.8 (data not shown).
Figure 4.
Molecular-surface conservation of IpaH9.8 LRR. (a) Amino-acid conservation in IpaH family proteins mapped onto the surface of the IpaH9.8 LRR structure. Cyan indicates variable residues in the IpaH family and purple indicates residues with high conservation. (b) Sequence alignment of the IpaH LRR domain.
Supplementary Material
PDB reference: LRR domain of Shigella IpaH9.8, P212121 form, 5b0n
PDB reference: C2221 form, 5b0t
Acknowledgments
This work was performed using synchrotron beamline BL44XU at SPring-8 under the Cooperative Research Program of the Institute for Protein Research, Osaka University. Diffraction data were collected on the Osaka University beamline BL44XU at SPring-8, Harima, Japan under proposal Nos. 2014A6952 and 2014B6952. This work was supported in part by the following grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan: a Grant-in-Aid for Specially Promoted Research (23000012 to CS), Grants-in-Aid for Scientific Research on Innovative Areas (24112009 to TM, 25121711 and 15H01174 to MK) and a Grant-in-Aid for Scientific Research (B) (15H04341 to TM). Part of this work was supported by grants from the Naito Foundation and the Terumo Foundation for Life Sciences and Arts (MK).
References
- Ashida, H., Kim, M. & Sasakawa, C. (2014). Nature Rev. Microbiol. 12, 399–413. [DOI] [PubMed]
- Ashida, H., Kim, M., Schmidt-Supprian, M., Ma, A., Ogawa, M. & Sasakawa, C. (2010). Nature Cell Biol. 12, 66–73. [DOI] [PMC free article] [PubMed]
- Büttner, D. (2012). Microbiol. Mol. Biol. Rev. 76, 262–310. [DOI] [PMC free article] [PubMed]
- Celniker, G., Nimrod, G., Ashkenazy, H., Glaser, F., Martz, E., Mayrose, I., Pupko, T. & Ben-Tal, N. (2013). Isr. J. Chem. 53, 199–206.
- Chou, Y.-C., Keszei, A. F. A., Rohde, J. R., Tyers, M. & Sicheri, F. (2012). J. Biol. Chem. 287, 268–275. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- Edwards, D. J., Streich, F. C. Jr, Ronchi, V. P., Todaro, D. R. & Haas, A. L. (2014). J. Biol. Chem. 289, 34114–34128. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evdokimov, A. G., Anderson, D. E., Routzahn, K. M. & Waugh, D. S. (2001). J. Mol. Biol. 312, 807–821. [DOI] [PubMed]
- Hicks, S. W. & Galán, J. E. (2010). Curr. Opin. Microbiol. 13, 41–46. [DOI] [PMC free article] [PubMed]
- Holm, L. & Park, J. (2000). Bioinformatics, 16, 566–567. [DOI] [PubMed]
- Holm, L. & Rosenström, P. (2010). Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed]
- Keszei, A. F., Tang, X., McCormick, C., Zeqiraj, E., Rohde, J. R., Tyers, M. & Sicheri, F. (2014). Mol. Cell. Biol. 34, 362–373. [DOI] [PMC free article] [PubMed]
- Kim, M., Otsubo, R., Morikawa, H., Nishide, A., Takagi, K., Sasakawa, C. & Mizushima, T. (2014). Cells, 3, 848–864. [DOI] [PMC free article] [PubMed]
- Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J. & Higgins, D. G. (2007). Bioinformatics, 23, 2947–2948. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- McNicholas, S., Potterton, E., Wilson, K. S. & Noble, M. E. M. (2011). Acta Cryst. D67, 386–394. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Parsot, C. (2009). Curr. Opin. Microbiol. 12, 110–116. [DOI] [PubMed]
- Quezada, C. M., Hicks, S. W., Galán, J. E. & Stebbins, C. E. (2009). Proc. Natl Acad. Sci. USA, 106, 4864–4869. [DOI] [PMC free article] [PubMed]
- Seyedarabi, A., Sullivan, J. A., Sasakawa, C. & Pickersgill, R. W. (2010). FEBS Lett. 584, 4163–4168. [DOI] [PubMed]
- Singer, A. U., Rohde, J. R., Lam, R., Skarina, T., Kagan, O., Dileo, R., Chirgadze, N. Y., Cuff, M. E., Joachimiak, A., Tyers, M., Sansonetti, P. J., Parsot, C. & Savchenko, A. (2008). Nature Struct. Mol. Biol. 15, 1293–1301. [DOI] [PMC free article] [PubMed]
- Suzuki, S., Mimuro, H., Kim, M., Ogawa, M., Ashida, H., Toyotome, T., Franchi, L., Suzuki, M., Sanada, T., Suzuki, T., Tsutsui, H., Núñez, G. & Sasakawa, C. (2014). Proc. Natl Acad. Sci. USA, 111, E4254–E4263. [DOI] [PMC free article] [PubMed]
- Tatusova, T. A. & Madden, T. L. (1999). FEMS Microbiol. Lett. 174, 247–250. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wang, F. et al. (2013). Cell. Microbiol. 15, 474–485. [DOI] [PubMed]
- Zhu, Y., Li, H., Hu, L., Wang, J., Zhou, Y., Pang, Z., Liu, L. & Shao, F. (2008). Nature Struct. Mol. Biol. 15, 1302–1308. [DOI] [PubMed]
- Zouhir, S., Bernal-Bayard, J., Cordero-Alba, M., Cardenal-Muñoz, E., Guimaraes, B., Lazar, N., Ramos-Morales, F. & Nessler, S. (2014). Biochem. J. 464, 135–144. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: LRR domain of Shigella IpaH9.8, P212121 form, 5b0n
PDB reference: C2221 form, 5b0t