The crystal structure of CotA complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) in a hole motif has been solved; this novel binding site could be a potential structure-based target for protein engineering of CotA laccase.
Keywords: 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate); laccase; mutagenesis; substrate binding; protein engineering
Abstract
The CotA laccase from Bacillus subtilis is an abundant component of the spore outer coat and has been characterized as a typical laccase. The crystal structure of CotA complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) (ABTS) in a hole motif has been solved. The novel binding site was about 26 Å away from the T1 binding pocket. Comparison with known structures of other laccases revealed that the hole is a specific feature of CotA. The key residues Arg476 and Ser360 were directly bound to ABTS. Site-directed mutagenesis studies revealed that the residues Arg146, Arg429 and Arg476, which are located at the bottom of the novel binding site, are essential for the oxidation of ABTS and syringaldazine. Specially, a Thr480Phe variant was identified to be almost 3.5 times more specific for ABTS than for syringaldazine compared with the wild type. These results suggest this novel binding site for ABTS could be a potential target for protein engineering of CotA laccases.
1. Introduction
Spore coat protein A (CotA) is a protein isolated from the bacterium Bacillus subtilis. It is able to oxidize specific laccase substrates and is characterized as a typical laccase with one mononuclear copper centre (T1 copper) and one trinuclear centre (TNC) comprised of one T2 copper and two T3 coppers (Bertrand et al., 2002 ▸; Martins et al., 2002 ▸). This enzyme can catalyze the oxidation of a wide variety of substrates by using dioxygen as an electron acceptor. The substrate-binding pocket of CotA is near the T1 copper. Substrate oxidation occurs at the T1 binding pocket and the electrons are then shuttled to the TNC, where the reduction of dioxygen occurs. The water channel is near the T2 copper and points to the protein surface; the water molecules produced by the reduction of dioxygen are released through this channel (Enguita et al., 2003 ▸). As a component of the spore coat, CotA laccase has the dual advantages of both thermostability and thermoactivity. It is believed that a higher proline content and compact packing may be the factors the promote the increased thermostability of CotA (Enguita et al., 2003 ▸). To date, CotA has been used as a model protein in studying the relationship between protein structure and function and in the protein engineering of laccases (Gupta & Farinas, 2010 ▸; Koschorreck et al., 2009 ▸; Mollania et al., 2011 ▸; Jia et al., 2014 ▸; Enguita et al., 2003 ▸, 2004 ▸; Bento et al., 2005 ▸, 2010 ▸; Chen et al., 2010 ▸; Fernandes et al., 2011 ▸; Silva, Damas et al., 2012 ▸; Durão et al., 2008 ▸).
The overall structure of CotA comprises three cupredoxin-type domains, characterized by a Greek-key β-barrel topology, which is similar to other multi-copper oxidases (Messerschmidt & Huber, 1990 ▸; Messerschmidt et al., 1992 ▸; Zaitseva et al., 1996 ▸). However, structural comparisons reveal that the CotA protein contains some unique structural features. One unique feature is that CotA has a region of positive charge on its surface at the interface between domain 1 and domains 2 and 3 (Enguita et al., 2003 ▸). More than a dozen positively charged residues (ten lysines and five arginines) are located in this area, which leads to a highly positive patch on the surface of CotA. No similar positively charged area has been found in fungal and other bacterial laccases with known structures (Roberts et al., 2003 ▸; Silva, Durão et al., 2012 ▸; Piontek et al., 2002 ▸; Garavaglia et al., 2004 ▸). The biological significance of this positively charged region is not known, but it has been speculated that it may be involved in the assembly of CotA into the spore outer coat layer (Enguita et al., 2003 ▸; McKenney et al., 2013 ▸; Bauer et al., 1999 ▸).
Another interesting feature of CotA is located close to the positively charged region of the protein. An extruding loop comprising residues 359–365 and adjacent residues form a hole near to the water channel. The positive residues Arg146, Arg429 and Arg476 are located on one side of the hole. The significance of this hole is currently unknown, but it has been reported that a mutation of Thr480 to alanine at the entrance to the hole results in increased activity of CotA on the spore surface (Jia et al., 2014 ▸). Therefore, it is of interest to investigate the properties of this hole in CotA.
Here, we report the crystal structure of CotA complexed with a nonphenolic substrate, 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) (ABTS). The ABTS molecule unexpectedly binds to the positively charged hole in the protein. The novel substrate-binding site is described and the biological significance of this motif is discussed.
2. Materials and methods
2.1. Protein crystallization and data collection
Purification of recombinant CotA and the mutant proteins was performed as described previously (Xie et al., 2015 ▸). Crystals of the CotA proteins were obtained at 18°C by the vapour-diffusion method from a reservoir solution consisting of 30–42%(v/v) ethylene glycol, 100 mM sodium citrate pH 5.6 (Table 1 ▸). Crystals were grown over a period of 5–7 d and were then soaked in a solution consisting of 5 mM ABTS for approximately 20 s. It was observed that the colour of the crystal changed from light blue to dark green. The crystals were then transferred into a cryoprotectant solution consisting of 25%(v/v) glycerol in the crystal mother liquor before being flash-cooled in liquid nitrogen. Diffraction data were collected on the 3W1A beamline at the Beijing Synchrotron Radiation Facility (BSRF). Data sets were processed and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▸) (Table 2 ▸). All crystals were flash-cooled and maintained at 100 K in a flow of cold nitrogen gas during data collection.
Table 1. Crystallization.
| Method | Vapour diffusion |
| Plate type | Hanging drop, 24-well |
| Temperature (K) | 291 |
| Protein concentration (mg ml−1) | 6 |
| Buffer composition of protein solution | 100 mM sodium citrate pH 5.6 |
| Composition of reservoir solution | 30–42%(v/v) ethylene glycol |
| Volume and ratio of drop | 2 µl, 1:1 |
| Volume of reservoir (µl) | 200 |
Table 2. Data collection and processing.
Values in parentheses are for the outer shell.
| ABTS complex | Native | |
|---|---|---|
| Diffraction source | 3W1A, BSRF | 3W1A, BSRF |
| Wavelength (Å) | 1.0 | 1.0 |
| Temperature (K) | 100 | 100 |
| Detector | MAR CCD 165 mm | MAR CCD 165 mm |
| Crystal-to-detector distance (mm) | 150 | 150 |
| Rotation range per image (°) | 1 | 1 |
| Total rotation range (°) | 140 | 120 |
| Exposure time per image (s) | 15 | 25 |
| Space group | P3121 | P3121 |
| a, b, c (Å) | 101.99, 101.99, 135.79 | 101.73, 101.73, 135.57 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Mosaicity (°) | 0.296 | 0.375 |
| Resolution range (Å) | 29.4–2.30 (2.34–2.30) | 29.4–2.30 (2.34–2.30) |
| Total No. of reflections | 160013 (7883) | 135883 (6623) |
| No. of unique reflections | 36864 (1811) | 36605 (1784) |
| Completeness (%) | 99.9 (100) | 100 (100) |
| Multiplicity | 4.3 (4.4) | 3.7 (3.7) |
| 〈I/σ(I)〉 | 9.6 (6.5) | 12.2 (4.6) |
| R merge (%) | 14.7 (25.5) | 9.7 (28.1) |
| Overall B factor from Wilson plot (Å2) | 34.9 | 35.0 |
2.2. Structure determination and refinement
The structures of native CotA and of the CotA–ABTS complex were elucidated by molecular replacement using Phaser (McCoy et al., 2007 ▸) from the CCP4 program suite (Winn et al., 2011 ▸). The starting model was the native structure of CotA (PDB entry 1gsk; Enguita et al., 2003 ▸), from which the solvent molecules, as well as the copper ions, had been removed. Only one solution was evident. Refinement of the structural model was completed using the maximum-likelihood functions implemented in REFMAC5 (Murshudov et al., 2011 ▸) from the CCP4 program suite. Model building and improvement was performed with Coot (Emsley et al., 2010 ▸) in combination with σA-weighted 2F o − F c and F o − F c maps. Subsequent electron-density calculation enabled the location of three of the four copper ions in the native and complex structures. No obvious positive peaks for the T2 copper were observed in the F o − F c maps at a σ level of 1.0 for the native protein or its ABTS complex; therefore, the T2 copper was not included in the final models. The ABTS molecule was included in the complex after a few refinement cycles because of clear density in both 2F o − F c and F o − F c maps. After these rounds of refinement, solvent molecules were added to the models based on standard geometrical and chemical restraints. The occupancies of the copper ions were refined by REFMAC5 individually. Residues 91–95 in the native structure and residues 91–94 in the complex structure were not built in the final models because of the poor quality of the electron density. The initial methionine and the final two residues in both structures were disordered and were not included. PROCHECK (Laskowski et al., 1993 ▸) was used for validation. Details of the overall refinement and final quality of the models are shown in Table 3 ▸. PyMOL (http://www.pymol.org) was used to prepare structural figures.
Table 3. Structure solution and refinement.
Values in parentheses are for the outer shell.
| ABTS complex | Native | |
|---|---|---|
| Resolution range (Å) | 29.46–2.30 (2.360–2.300) | 29.38–2.30 (2.360–2.300) |
| No. of reflections, working set | 34956 (2584) | 34656 (2473) |
| No. of reflections, test set | 1837 (121) | 1824 (108) |
| Final R cryst | 0.158 (0.186) | 0.159 (0.185) |
| Final R free | 0.194 (0.217) | 0.187 (0.205) |
| No. of non-H atoms | ||
| Protein | 4070 | 4080 |
| Ion | 3 | 3 |
| Ligand | 58 | 22 |
| Solvent | 394 | 405 |
| Total | 4525 | 4517 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.015 | 0.014 |
| Angles (°) | 1.647 | 1.594 |
| Average B factors (Å2) | ||
| Protein | 21.7 | 21.0 |
| Ion | 15.2 | 14.8 |
| Ligand | 33.4 | 29.7 |
| Water | 28.8 | 30.3 |
| Ramachandran plot | ||
| Most favoured (%) | 96.2 | 96.6 |
| Allowed (%) | 3.8 | 3.4 |
2.3. Enzyme assay
The laccase activity towards ABTS and syringaldazine (SGZ) was measured at 40°C. The ABTS assay mixture contained 1 mM ABTS and 100 mM citrate–phosphate buffer pH 4.0. Oxidation of ABTS was followed by the increase in absorbance at 420 nm (∊420 = 36 000 M −1 cm−1). The SGZ assay mixture contained 0.1 mM SGZ (dissolved in ethanol) and 100 mM citrate–phosphate buffer pH 7.0. Oxidation of SGZ was followed by the increase in absorbance at 525 nm (∊525 = 65 000 M −1 cm−1). The protein concentration was determined by the Bradford method (Bradford, 1976 ▸) using bovine serum albumin as the standard. All assays were performed in triplicate to calculate the standard error.
2.4. Solvent-accessible surface calculation
Calculation of the solvent-accessible surface was performed using the CASTp server (Dundas et al., 2006 ▸).
2.5. Copper-content determination
To determine the copper content of CotA proteins, 500 µl of a solution containing 5 mg ml−1 protein was mixed with 500 µl nitric acid and hydrolyzed at 100°C for 2 h. The sample was diluted to a suitable volume and measured using an atomic absorption spectrometer (PinAAcle 900T, PerkinElmer).
3. Results and discussion
3.1. The hole structure in CotA laccase
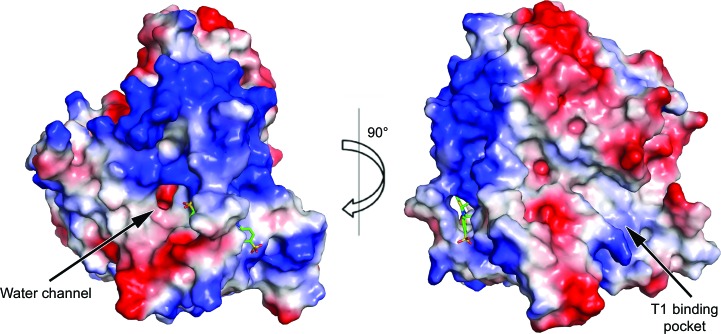
Crystal structures of native CotA and the CotA–ABTS complex have been determined at 2.3 Å resolution. The asymmetric units of the crystals contained one molecule in space group P3121. At the interface between domain 1 and domains 2 and 3 there was a positively charged surface region. Moreover, the linking loop (residues 359–365) between domains 2 and 3 and four β-strands of domain 3 formed a hole with dimensions of 8 × 5 × 4 Å and a calculated surface area of 159 Å2. In the structure of the CotA–ABTS complex, ABTS was observed sitting within the hole. The novel binding site was approximately 26 Å away from the T1 binding pocket and was near the exit of the water channel (Fig. 1 ▸).
Figure 1.
Surface electrostatic potential of CotA. A region with a large positive potential was found in the connection area between domains. The water channel and the T1 binding pocket are labelled. The ABTS molecule is represented as a green stick model.
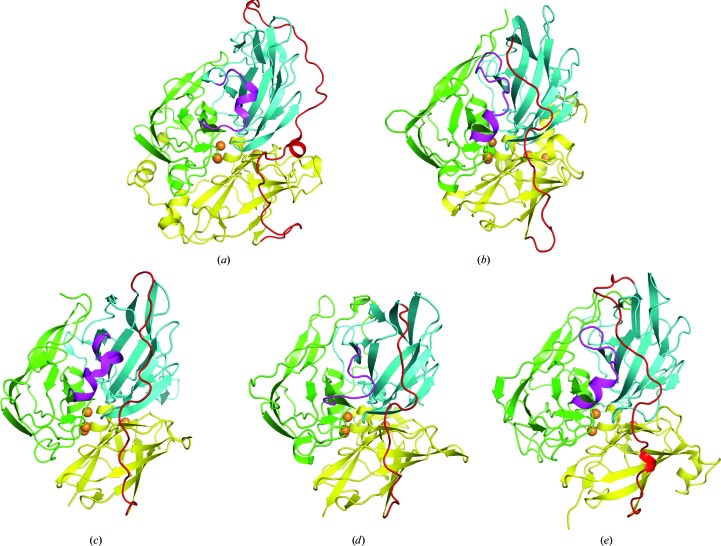
Structural comparisons were performed to determine whether fungal and bacterial laccases contained a similar hole. In the fungal laccase Tvl from Trametes versicolor (Piontek et al., 2002 ▸) there was a 40-residue extended loop region between domains 2 and 3, which is approximately ten residues longer than that in the bacterial laccase. While the 40-residue loop in Tvl laccase wound from the side of the β-barrel domains via an internal connection, this connection did not result in the formation of a hole in the fungal laccase. This loop was clearly distant from the 310-helical fragment between domains 1 and 2 (Fig. 2 ▸ a). However, the equivalent loop in the bacterial laccase spanned over the β-barrel by an external connection (Figs. 2 ▸ b–2 ▸ e), which may be a characteristic of prokaryotic enzymes.
Figure 2.
Crystal structures of laccases. (a) Tvl from T. versicolor (PDB entry 1gyc; Piontek et al., 2002 ▸), (b) CueO from E. coli (PDB entry 1n68; Roberts et al., 2003 ▸), (c) Thhb27 from Thermus thermophilus (PDB entry 2xu9; Serrano-Posada et al., 2015 ▸), (d) McoC from Campylobacter jejuni CGUG11284 (PDB entry 3zx1; Silva, Durão et al., 2012 ▸), (e) CotA from B. subtilis. Domains 1, 2 and 3 are coloured green, blue and yellow, respectively. Cu atoms are represented as spheres and are coloured orange. The linking loop between domains 2 and 3 is coloured red and the peptide connection between domains 1 and 2 is coloured magenta.
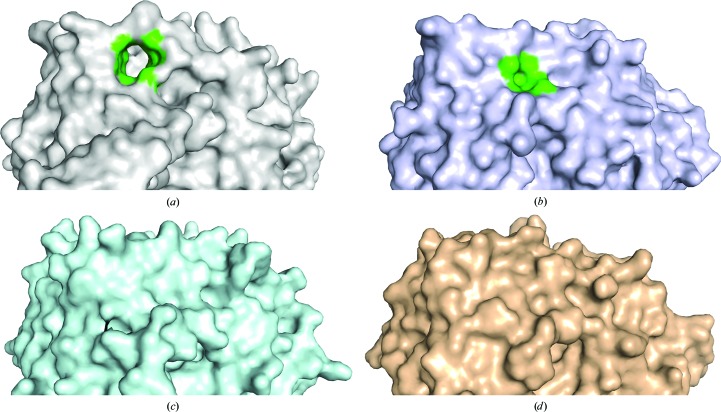
Moreover, cavity analysis was performed on CotA and another three bacterial laccases (Fig. 3 ▸). In CotA, the predicted hole was exactly the same as that in the crystal structure. In CueO (Roberts et al., 2003 ▸) two cavities were found adjacent to the linking loop; however, the cavities were only open on one side and the surface areas of the two cavities were 11 and 4 Å2, respectively. In both McoC and Thhb27 (Silva, Durão et al., 2012 ▸; Serrano-Posada et al., 2011 ▸) no obvious cavity was formed by the corresponding loop. The loop linking domains 2 and 3 in CotA was 27 residues in length, which is longer than those in CueO (20 amino acids), McoC (19 amino acids) and Thhb27 (22 amino acids). This may lead to the loose connection between domains 2 and 3 in CotA and favour formation of the hole.
Figure 3.
Molecular-surface representation and the putative cavity adjacent to the linking loop between domains 2 and 3. (a) CotA, (b) CueO, (c) McoC, (d) Thhb27. The surface is coloured grey for CotA, blue-white for CueO, cyan for McoC and orange for Thhb27. All molecular-surface representations are in the same orientation. The cavity is coloured green; the other cavity of CueO is on the opposite side and is not visible in this orientation.
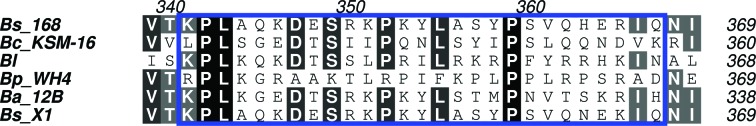
Laccases are multi-copper oxidases that are widely distributed in nature. Structural studies revealed that laccases possess conserved copper-ion centres and overall topology. In contrast, it was reported that the laccases from various species contained specific features. A conserved C-terminal end (DSGL) was observed in the laccase from Melanocarpus albomyces, which penetrated the tunnel leading from the protein surface to the TNC (Andberg et al., 2009 ▸). A laccase from silkworm (Bombyx mori) was identified as a dimer, containing an extra domain of approximately 200 amino acids that was not present in laccases from plants and fungi (Dittmer et al., 2004 ▸). CueO from Escherichia coli contained a fifth labile copper bound to a methionine-rich motif. This labile copper could mediate electron transfer from the substrate to the T1 copper (Roberts et al., 2002 ▸). As a component of the spore-coat outer layer from B. subtilis, CotA may also exhibit specific features (McKenney et al., 2013 ▸). The sequences of CotA from strains of Bacillus are highly conserved (Xie et al., 2015 ▸). Therefore, all of the CotA enzymes could have a similar structure. Multiple sequence alignment shows that the lengths of the loops linking domains 2 and 3 in the CotA proteins are similar to each other (Fig. 4 ▸) and, moreover, the hole structure of CotA was not found in the fungal laccase or other bacterial laccases; therefore, this hole structure could be a specific feature of the CotA protein from strains of Bacillus.
Figure 4.
Multiple sequence alignment of the loop region linking domains 2 and 3 of CotA laccase. The linking loop is boxed in blue. Bs_168, B. subtilis 168; Bc_KSM-K16, B. clausii KSM-K16; Bl, B. licheniformis; Bp-WH4, B. pumilus WH-4; Ba_12B, B. amyloliquefaciens 12B; Bs_X1, B. subtilis X1.
3.2. Novel binding site and conformational changes induced by the binding of ABTS
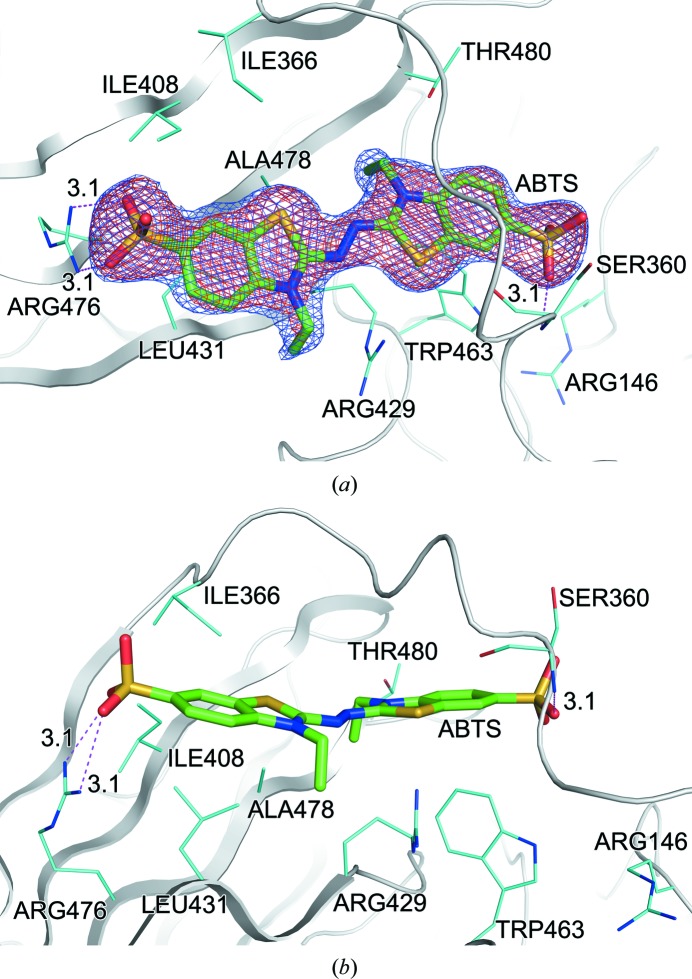
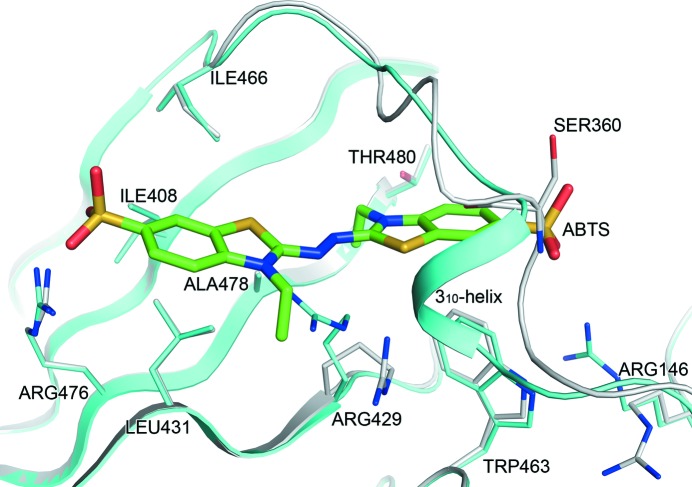
The ABTS molecule was identified from an interpretable electron density wrapped by the loop region of residues 359–365 (Fig. 5 ▸). This novel binding site was mainly formed by the extruding loop segment and four β-stands of domain 3. The bottom of the hole was positively charged owing to residues Arg146, Arg429 and Arg476. The ABTS molecule passed through the hole in a linear shape and interacted with adjacent residues by hydrophobic contacts: Ile408, Leu431, Ile366, Ala478 and Trp463. Moreover, the ABTS was directly bound to Arg476 and Ser360. The sulfonate moiety at one end of ABTS formed two hydrogen bonds to the guanidyl group of Arg476 (Arg476 NH1–O48 = 3.1 Å, Arg476 NH2–O49 = 3.1 Å), and the sulfonate moiety at the other end of ABTS formed a hydrogen bond to Ser360 (Ser360 N–O45 = 3.1 Å). The thiazoline ring of ABTS was almost buried in the hole, and the two sulfonate moieties of ABTS were exposed to the external environment. In the CotA–ABTS complex, conformational changes were observed upon substrate binding. A 310-helical fragment composed of residues 359–362 was dissolved and the side chains of Arg429 and Arg146 were rotated by about 40 and 120°, respectively, providing more space for ABTS binding (Fig. 6 ▸).
Figure 5.
CotA–ABTS interactions. Residues in close proximity to ABTS are labelled and represented as cyan stick models showing (a) the top view and (b) the side view. The 2F o − F c electron-density map (1σ level) around ABTS is represented in blue and the OMIT F o − F c map (4σ level) around ABTS is represented in red. The ABTS molecule is shown as a green stick model.
Figure 6.
Superposition of the CotA–ABTS complex (grey) onto native CotA (cyan). Residues in close proximity to ABTS are labelled and represented as stick models. The ABTS molecule is represented as a green stick model.
In a previous study, ABTS bound in the T1 binding pocket as a shallow U-shaped molecule with an overall B factor of 55.1 Å2 and 40% occupancy. Only one half of the ABTS molecule was partially buried in the CotA structure (Enguita et al., 2003 ▸, 2004 ▸). In our study, the ABTS passed through the hole in a linear shape; the B factor of ABTS was 35.6 Å2 and it had full occupancy. Compared with the U-shaped binding state, the binding of ABTS to CotA described here was more stable.
In this work, the CotA crystal was soaked in 5 mM ABTS for 20 s. The use of a high concentration of ABTS meant that the molecule was more easily able to access the hole motif. In addition, the interactions between ABTS and the adjacent residues in the hole stabilized the binding of ABTS without any oxidation occurring. However, in the T1 binding pocket oxidation proceeded rapidly, which resulted in the product being released. It is noted that the shape of the T1 binding pocket continuously changed, making it hard to form a stable complex. In the previous report, the CotA crystal was soaked in 0.5 mM ABTS for 1 h. The lower substrate concentration and longer soaking time used in that research may account for the difference, as after 1 h of soaking the ABTS was already oxidized, meaning that the ABTS was bound in its cationic state (Enguita et al., 2004 ▸).
3.3. Kinetic characterization of CotA and mutants
A group of residues (Arg146, Arg429, Arg476, Leu431, Ala478 and Thr480) in the novel binding site have been mutated. The substrates ABTS and SGZ were used to study the catalytic properties of the mutants. The data for the kinetic parameters are shown in Table 4 ▸. The copper contents of wild-type CotA and five mutants, R146K, R429K, A478F, T480A and T480F, were also measured, as shown in Table 5 ▸. A stoichiometry of close to four coppers per protein was calculated for both mutant and wild-type CotA, showing that all four copper ions were incorporated into the active site. Since the mutation sites were distant from the copper-coordinating sites, the mutation may not affect the stability of the Cu atoms; this may also be the case for the L431F and R476K mutants.
Table 4. Kinetic constants of CotA and its mutants for ABTS and SGZ.
| ABTS | SGZ | ||||||
|---|---|---|---|---|---|---|---|
| Enzyme | K m (µM) | k cat (s−1) | k cat/K m (µM −1 s−1) | K m (µM) | k cat (s−1) | k cat/K m (µM −1 s−1) | (k cat/K m)ABTS/(k cat/K m)SGZ |
| WT | 105.51 ± 19.32 | 51.82 ± 3.85 | 0.50 ± 0.05 | 25.78 ± 4.54 | 15.48 ± 1.41 | 0.61 ± 0.05 | 0.82 ± 0.03 |
| R146K | 257.66 ± 3.8 | 0.36 ± 0.01 | 0.0014 ± 0.0008 | 13.45 ± 3.43 | 0.05 ± 0.005 | 0.004 ± 0.001 | 0.35 ± 0.09 |
| R429K | 90.61 ± 16.8 | 1.71 ± 0.23 | 0.02 ± 0.001 | 7.44 ± 1.67 | 0.14 ± 0.01 | 0.02 ± 0.003 | 1.00 ± 0.07 |
| R476K | 101.03 ± 19.03 | 11.31 ± 2.49 | 0.11 ± 0.003 | 29.32 ± 6.21 | 4.27 ± 0.68 | 0.15 ± 0.001 | 0.73 ± 0.01 |
| L431F | 77.68 ± 9.34 | 20.93 ± 2.91 | 0.27 ± 0.01 | 32.93 ± 3.49 | 8.08 ± 1.87 | 0.25 ± 0.08 | 1.08 ± 0.03 |
| A478F | 109.88 ± 15.24 | 40.22 ± 4.67 | 0.37 ± 0.01 | 32.71 ± 7.20 | 12.53 ± 3.07 | 0.38 ± 0.05 | 0.97 ± 0.03 |
| T480A | 110.09 ± 22.48 | 35.88 ± 6.18 | 0.33 ± 0.03 | 37.40 ± 6.37 | 12.92 ± 2.96 | 0.34 ± 0.04 | 0.97 ± 0.05 |
| T480F | 52.95 ± 8.56 | 29.33 ± 5.89 | 0.53 ± 0.08 | 40.03 ± 6.76 | 7.36 ± 2.00 | 0.18 ± 0.03 | 2.90 ± 0.09 |
Table 5. Copper content in wild-type and mutant CotA.
| Protein | Copper content (moles of Cu/moles of protein) |
|---|---|
| WT | 4.51 ± 0.02 |
| R146K | 3.71 ± 0.03 |
| R429K | 4.52 ± 0.02 |
| A478F | 3.78 ± 0.06 |
| T480A | 4.19 ± 0.05 |
| T480F | 3.83 ± 0.06 |
In our work, substitution of arginine by lysine in the novel binding cavity dramatically affected the activity of the CotA enzyme, and the decrease in activity was not substrate-dependent. The activity of the R146K and R429K mutants decreased 30–100-fold in efficiency (k cat/K m) compared with the wild type (WT). The R476K mutant retained approximately 20% of the WT activity with both ABTS and SGZ. The L431F, T480A and A478F mutants displayed approximately 50, 60 and 70% of the activity of the WT, respectively. Moreover, the T480F mutant exhibited different effects in the oxidation of ABTS and SGZ. The T480F mutant showed equivalent activity compared with the WT in the oxidation of ABTS, whereas only about 30% of the activity of the WT was retained in the oxidation of SGZ. To assess substrate specificity, (k cat/K m)ABTS/(k cat/K m)SGZ was calculated. The ratios for wild-type CotA and the T480F mutant were approximately 0.82 ± 0.03 and 2.90 ± 0.09, respectively. Therefore, the T480F mutant was almost 3.5 times more specific for ABTS than for SGZ compared with the WT. It was reported that a T480A mutant of CotA displayed on the surface of the bacterium spore was 2.38-fold more active compared with WT CotA in organic solvent (Jia et al., 2014 ▸). In our work, the T480A mutant showed 60% of the activity compared with the WT in a buffer solution. This may be because of structural differences between the enzyme in solution and the enzyme displayed on the spore surface.
All of these residues except Arg146 are located in domain 3 of CotA. This domain not only contains the mononuclear copper centre but also contributes to the formation of the binding site of the TNC. Mutations in the specific residues that compose the novel binding cavity may contribute to changes at the catalytic centre of CotA; this was also reported for various mutant sites located in domain 3, although these sites were distant from the copper centres (Gupta & Farinas, 2010 ▸; Bao et al., 2013 ▸; Kenzom et al., 2014 ▸; Jia et al., 2014 ▸).
Over the years, laccases have become attractive candidates for protein engineering. Various CotA mutants have been prepared by rational, semi-rational or directed evolution approaches, and biocatalysts with improved features or new functions were obtained. A mutant of CotA from Bacillus sp. HR03 exhibited higher thermal stability (Mollania et al., 2011 ▸), and the substrate specificity of CotA was narrowed by saturation mutagenesis (Gupta & Farinas, 2009 ▸, 2010 ▸). In the B. subtilis spore, the CotA-ABTS-SD1 mutant (T260L) exhibited 120-fold higher specificity for ABTS compared with the WT (Gupta & Farinas, 2010 ▸). In addition, the double mutant K316N/D500G of CotA from B. licheniformis displayed a 11.4-fold higher expression level in E. coli compared with the WT (Koschorreck et al., 2009 ▸). Here, a novel binding site in CotA was revealed, with the CotA mutants described above showing different effects on the oxidation of ABTS and SGZ. Specially, the T480F mutant was found to be almost 3.5 times more specific for ABTS than for SGZ compared with the WT. This implies that rational design could be performed on the novel binding site to improve some of the properties of CotA proteins.
4. Conclusions
In summary, the hole motif described in this paper is a specific feature of CotA proteins into which the substrate ABTS can bind. This novel binding site could be a potential structural target for protein engineering of CotA laccases.
Supplementary Material
PDB reference: CotA, wild type, 4yvu
PDB reference: complex with ABTS, 4yvn
Acknowledgments
Professor Zihe Rao, Professor Fei Sun and Professor Zhiyong Lou are gratefully acknowledged for their support and helpful discussions. This work was supported by initial grants from The 100 Talents Program of the Chinese Academy of Sciences, the Western Light Talent Culture Project of the Chinese Academy of Sciences and the open fund of the Key Laboratory of Environmental and Applied Microbiology, Chinese Academy of Sciences (No. KLCAS-2014-06). The Beijing Synchrotron Radiation Facility is gratefully acknowledged for the provision of synchrotron-radiation facilities and support.
References
- Andberg, M., Hakulinen, N., Auer, S., Saloheimo, M., Koivula, A., Rouvinen, J. & Kruus, K. (2009). FEBS J. 276, 6285–6300. [DOI] [PubMed]
- Bao, S., Teng, Z. & Ding, S. (2013). Mol. Biol. Rep. 40, 1927–1936. [DOI] [PubMed]
- Bauer, T., Little, S., Stöver, A. G. & Driks, A. (1999). J. Bacteriol. 181, 7043–7051. [DOI] [PMC free article] [PubMed]
- Bento, I., Martins, L. O., Lopes, G. G., Carrondo, M. A. & Lindley, P. F. (2005). Dalton Trans., pp. 3507–3513. [DOI] [PubMed]
- Bento, I., Silva, C. S., Chen, Z., Martins, L. O., Lindley, P. F. & Soares, C. M. (2010). BMC Struct. Biol. 10, 28. [DOI] [PMC free article] [PubMed]
- Bertrand, T., Jolivalt, C., Briozzo, P., Caminade, E., Joly, N., Madzak, C. & Mougin, C. (2002). Biochemistry, 41, 7325–7333. [DOI] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Chen, Z., Durão, P., Silva, C. S., Pereira, M. M., Todorovic, S., Hildebrandt, P., Bento, I., Lindley, P. F. & Martins, L. O. (2010). Dalton Trans. 39, 2875–2882. [DOI] [PubMed]
- Dittmer, N. T., Suderman, R. J., Jiang, H., Zhu, Y.-C., Gorman, M. J., Kramer, K. J. & Kanost, M. R. (2004). Insect Biochem. Mol. Biol. 34, 29–41. [DOI] [PubMed]
- Dundas, J., Ouyang, Z., Tseng, J., Binkowski, A., Turpaz, Y. & Liang, J. (2006). Nucleic Acids Res. 34, W116–W118. [DOI] [PMC free article] [PubMed]
- Durão, P., Chen, Z., Silva, C. S., Soares, C. M., Pereira, M. M., Todorovic, S., Hildebrandt, P., Bento, I., Lindley, P. F. & Martins, L. O. (2008). Biochem. J. 412, 339–346. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Enguita, F. J., Marçal, D., Martins, L. O., Grenha, R., Henriques, A. O., Lindley, P. F. & Carrondo, M. A. (2004). J. Biol. Chem. 279, 23472–23476. [DOI] [PubMed]
- Enguita, F. J., Martins, L. O., Henriques, A. O. & Carrondo, M. A. (2003). J. Biol. Chem. 278, 19416–19425. [DOI] [PubMed]
- Fernandes, A. T., Pereira, M. M., Silva, C. S., Lindley, P. F., Bento, I., Melo, E. P. & Martins, L. O. (2011). J. Biol. Inorg. Chem. 16, 641–651. [DOI] [PubMed]
- Garavaglia, S., Cambria, M. T., Miglio, M., Ragusa, S., Iacobazzi, V., Palmieri, F., D’Ambrosio, C., Scaloni, A. & Rizzi, M. (2004). J. Mol. Biol. 342, 1519–1531. [DOI] [PubMed]
- Gupta, N. & Farinas, E. T. (2009). Comb. Chem. High. Throughput Screen. 12, 269–274. [DOI] [PubMed]
- Gupta, N. & Farinas, E. T. (2010). Protein Eng. Des. Sel. 23, 679–682. [DOI] [PubMed]
- Jia, H., Lee, F. S. & Farinas, E. T. (2014). ACS Comb. Sci. 16, 665–669. [DOI] [PubMed]
- Kenzom, T., Srivastava, P. & Mishra, S. (2014). Appl. Environ. Microbiol. 80, 7484–7495. [DOI] [PMC free article] [PubMed]
- Koschorreck, K., Schmid, R. D. & Urlacher, V. B. (2009). BMC Biotechnol. 9, 12. [DOI] [PMC free article] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Martins, L. O., Soares, C. M., Pereira, M. M., Teixeira, M., Costa, T., Jones, G. H. & Henriques, A. O. (2002). J. Biol. Chem. 277, 18849–18859. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McKenney, P. T., Driks, A. & Eichenberger, P. (2013). Nature Rev. Microbiol. 11, 33–44. [DOI] [PMC free article] [PubMed]
- Messerschmidt, A. & Huber, R. (1990). Eur. J. Biochem. 187, 341–352. [DOI] [PubMed]
- Messerschmidt, A., Ladenstein, R., Huber, R., Bolognesi, M., Avigliano, L., Petruzzelli, R., Rossi, A. & Finazzi-Agró, A. (1992). J. Mol. Biol. 224, 179–205. [DOI] [PubMed]
- Mollania, N., Khajeh, K., Ranjbar, B. & Hosseinkhani, S. (2011). Enzyme Microb. Technol. 49, 446–452. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Piontek, K., Antorini, M. & Choinowski, T. (2002). J. Biol. Chem. 277, 37663–37669. [DOI] [PubMed]
- Roberts, S. A., Weichsel, A., Grass, G., Thakali, K., Hazzard, J. T., Tollin, G., Rensing, C. & Montfort, W. R. (2002). Proc. Natl Acad. Sci. USA, 99, 2766–2771. [DOI] [PMC free article] [PubMed]
- Roberts, S. A., Wildner, G. F., Grass, G., Weichsel, A., Ambrus, A., Rensing, C. & Montfort, W. R. (2003). J. Biol. Chem. 278, 31958–31963. [DOI] [PubMed]
- Serrano-Posada, H., Centeno-Leija, S., Rojas-Trejo, S. P., Rodríguez-Almazán, C., Stojanoff, V. & Rudiño-Piñera, E. (2015). Acta Cryst. D71, 2396–2411. [DOI] [PMC free article] [PubMed]
- Serrano-Posada, H., Valderrama, B., Stojanoff, V. & Rudiño-Piñera, E. (2011). Acta Cryst. F67, 1595–1598. [DOI] [PMC free article] [PubMed]
- Silva, C. S., Damas, J. M., Chen, Z., Brissos, V., Martins, L. O., Soares, C. M., Lindley, P. F. & Bento, I. (2012). Acta Cryst. D68, 186–193. [DOI] [PubMed]
- Silva, C. S., Durão, P., Fillat, A., Lindley, P. F., Martins, L. O. & Bento, I. (2012). Metallomics, 4, 37–47. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Xie, T., Liu, Z., Liu, Q. & Wang, G. (2015). J. Struct. Biol. 190, 155–161. [DOI] [PubMed]
- Zaitseva, I., Zaitsev, V., Card, G., Moshkov, K., Bax, B., Ralph, A. & Lindley, P. (1996). J. Biol. Inorg. Chem. 1, 15–23.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: CotA, wild type, 4yvu
PDB reference: complex with ABTS, 4yvn