Abstract
Background & Aims
The embryonic small intestinal epithelium is highly proliferative, and although much is known about mechanisms regulating proliferation in the adult intestine, the mechanisms controlling epithelial cell proliferation in the developing intestine are less clear. GATA4, a transcription factor that regulates proliferation in other developing tissues, is first expressed early in the developing gut in midgut endoderm. GATA4 function within midgut endoderm and the early intestinal epithelium is unknown.
Methods
By using Sonic Hedgehog Cre to eliminate GATA4 in the midgut endoderm of mouse embryos, we determined the impact of loss of GATA4 on intestinal development, including epithelial cell proliferation, between embryonic day (E)9.5 and E18.5.
Results
We found that intestinal length and width were decreased in GATA4 mutants compared with controls. GATA4-deficient intestinal epithelium contained fewer cells, and epithelial girth was decreased. We further observed a decreased proportion of proliferating epithelial cells at E10.5 and E11.5 in GATA4 mutants. We showed that GATA4 binds to chromatin containing GATA4 consensus binding sites within cyclin D2 (Ccnd2), cyclin-dependent kinase 6 (Cdk6), and frizzled 5 (Fzd5). Moreover, Ccnd2, Cdk6, and Fzd5 transcripts were reduced at E11.5 in GATA4 mutant tissue. Villus morphogenesis was delayed, and villus structure was abnormal in GATA4 mutant intestine.
Conclusions
Our data identify GATA4 as an essential regulator of early intestinal epithelial cell proliferation. We propose that GATA4 controls proliferation in part by directly regulating transcription of cell-cycle mediators. Our data further suggest that GATA4 affects proliferation through transcriptional regulation of Fzd5, perhaps by influencing the response of the epithelium to WNT signaling.
Keywords: Transcriptional Regulation, WNT Signaling, Villus Morphogenesis
Abbreviations used in this paper: Atoh, atonal homolog 1; Bio-ChIP-Seq, biotin-mediated chromatin immunoprecipitation with high-throughput sequencing; Ccnd1/2, cyclin D1/2; Cdk6, cyclin-dependent kinase 6; cKO, conditional knockout; EdU, 5-ethynyl-2'deoxyuridine; E, embryonic day; Fzd5, frizzled 5; Hes1/6, hairy and enhancer of split-1/6; PCR, polymerase chain reaction; Pdgfa, platelet-derived growth factor α; PDGFRA, platelet-derived growth factor receptor α; Prdm1, PR domain containing 1; Olfm4, olfactomedin 4; SBS, short-bowel syndrome; Shh, sonic hedgehog; Wnt, wingless type MMTV integration site family
Summary.
Gata4 deletion from the small intestinal epithelium beginning at embryonic day 9.5 disrupts epithelial cell proliferation. GATA4 regulates proliferation in the early developing intestine by directly regulating Ccnd2, Cdk6, and Fzd5. Villus morphogenesis is delayed in GATA4 mutant intestine.
The small intestine has one of the highest surface area–to–volume ratios in the body, which is critical for nutrient absorption. When intestinal mass is lost, decreased nutrient, vitamin, and fluid absorption can restrict growth and disrupt hydration and electrolyte balance.1 This type of intestinal failure, termed short-bowel syndrome (SBS), occurs in neonates and infants as a consequence of necrotizing enterocolitis or congenital defects such as reduced intestinal growth, intestinal atresia, midgut volvulus, and gastroschisis.2, 3 High mortality and morbidity are associated with SBS, and SBS-related economic and quality of life costs are high.2 One strategy to gain insights into congenital SBS causes, which are poorly understood, and to discover novel SBS therapies, including those based in regenerative medicine, is to uncover the cellular and molecular mechanisms of intestinal development, particularly those relevant to intestinal growth and elongation.
Distinct epithelial morphogenetic processes occurring during development contribute to the establishment of an intestine of requisite length and absorptive area.1 At embryonic day (E)9.0–E9.5 in the mouse, the primitive small intestine is lined by a polarized epithelium derived from midgut endoderm.1 Between E9.5 and E14.5, the gut rapidly expands. During this time, proliferating epithelial cells elongate and densely pack, and the simple epithelium transitions to pseudostratified.4 Epithelial pseudostratification often occurs in highly proliferative tissues, allowing for greater cell density compared with simple epithelia.4, 5 Increased intestinal epithelial girth coincides with pseudostratification, and actomyosin-dependent and microtubule-dependent signaling events drive changes in cell shape that increase girth.4 Between E14.5 and E18.5, villus morphogenesis ensues, converting the pseudostratified epithelium to simple columnar, and cytodifferentiation generates absorptive and secretory epithelial cell populations.1
Although the establishment of a highly proliferative pseudostratified epithelium is central for gut expansion, the mechanisms controlling proliferation within the epithelium during early gut development are largely unknown. Hedgehog, wingless type MMTV integration site family (Wnt5a), receptor tyrosine kinase-like orphan receptor 2 (Ror2), secreted frizzled-related protein, fibroblast growth factor 9, fibroblast growth factor receptor, and caudal type homeobox 2 (Cdx2) mutants all have shorter guts, but only WNT5A has been shown to play an essential role in regulating epithelial cell proliferation during this developmental window.6, 7, 8, 9, 10, 11, 12 Secreted by the mesenchyme, WNT5A influences epithelial cell proliferation in a paracrine manner.7 The factors functioning within the nascent epithelium itself to regulate proliferation, including those downstream of WNT5A, are unknown. Our laboratory previously studied the zinc-finger DNA binding transcription factor GATA4 in the context of intestinal development and showed that elimination of GATA4 from the small intestinal epithelium after E14.5 only subtly affects the embryonic intestine.13 GATA4, however, is expressed in the developing intestine before E14.5. It is present in the midgut endoderm fated to form the small intestinal epithelium.14 GATA4 function within the midgut endoderm and the early intestinal epithelium has not been investigated. Therefore, to determine GATA4 function during early intestinal development (before E14.5), and particularly the role of GATA4 in epithelial cell proliferation, we eliminated GATA4 in the midgut endoderm using sonic hedgehog Cre (ShhCre).15 Small intestine lacking GATA4 was shorter than control intestine. Furthermore, we observed a decreased proportion of proliferating epithelial cells at E10.5 and E11.5 in GATA4 mutants compared with controls. We found that GATA4 binds to chromatin within cyclin D2 (Ccnd2), cyclin-dependent kinase 6 (Cdk6), and frizzled 5 (Fzd5), suggesting that GATA4 directly regulates proliferation within the early gut by controlling transcription of cell-cycle regulators and WNT receptors. Villus morphogenesis also was delayed in GATA4 mutant intestine. Collectively, these data show GATA4 as a key regulator of early intestinal epithelial cell proliferation and suggest novel relationships between GATA4 and WNT signaling and between proliferation and villus morphogenesis.
Materials and Methods
Animals
Gata4loxP(Gata4tm1.1Sad), Gata4-(Gata4tmo1Eno), ShhCre(Shhtm1(EGFP/cre)Cjt/J), Gata4flbio/flbio(Gata4tm3.1Wtp), Rosa26BirA(Gt(ROSA)26SorTm1(birA)Mejr), and CD-1 (Charles River Laboratories, Wilmington, MA) mice were used.15, 16, 17, 18, 19 Embryos were obtained by timed mating with a vaginal plug at noon considered E0.5. Genotypes were determined by polymerase chain reaction (PCR) with tail tip, ear punch, or yolk sac genomic DNA (Table 1, primers). For proliferation studies, 200 μg of 5-ethynyl-2’deoxyuridine (EdU) (Invitrogen, Carlsbad, CA) was administered to pregnant mice by intraperitoneal injection 30 minutes before euthanizing. EdU incorporation was detected with the Click-it EdU Alexa-Fluor kit (Invitrogen). The Medical College of Wisconsin’s Animal Care Committee approved all animal procedures.
Table 1.
Primers Used for PCR Genotyping and Bio-ChIP-PCR
| Gene | Forward | Reverse | Size, bp |
|---|---|---|---|
| Genotyping | |||
| Gata4loxP | CCCAGTAAAGAAGTCAGCACAAGGAAAC | AGACTATTGATCCCGGAGTGAACATT | 355, wt 455, loxP |
| Gata4null | GGGGCAGGACAGCAAGGGGGAGGATT | GTGAGACCTGCAGAATGGGAGTGGAGAATG | 207 |
| ShhCre | GGGACAGCTCACAAGTCCTC | GGTGCGCTCCTGGACGTA | 350 |
| Bio-ChIP | |||
| Ccnd2 | ATCTTTTGAGGAAAAGGACGGCTTG | TCCTCTCCAGAGCTCAGAATAAACG | 229 |
| Cdk6 | TTTGAGTTCAGAAGAGGGCACAGTT | ACAGGCCTGGGTTTTACAGTTCAGT | 251 |
| Negative control (Dll1 upstream) | TGGCAAGGGAGAGAAGGAAGCA | ACAGCCTTGGTCTGTTCAGTG T | 158 |
| Fzd5 | GCTGTTGATCAATGTGGGAAAGATT | CTGCTTTCCGATGAGAGACAGAGAC | 233 |
| Pdgfa | GTGGAGAGGCCTGAACTGTTTTTGT | CTCTGGGAGTATTTCCCCAAGTGAC | 233 |
Histochemistry, Immunohistochemistry, and Immunofluorescence
Histochemistry, immunohistochemistry, and immunofluorescence were performed as previously described.20 At E9.5–E10.5, whole embryos were used. At E11.5–E18.5, dissected intestines were used. The entire intestine was examined at E9.5–E12.5. At E14.5–E18.5, jejunal tissue (0.5 cm) collected from the intestinal midpoint was assayed unless otherwise noted (Table 2 lists antibodies). Measurements and cell counts were analyzed using ImageJ software (National Institutes of Health, Bethesda, MA). Fluorescent images were captured on a Nikon Eclipse 80i microscope (Melville, NY) using a Nikon digital smart DS-QiMc camera. Histochemistry and immunohistochemistry slides were scanned using a NanoZoomer slide scanner (Hamamatsu, Bridgewater, NJ), and images from scanned slides were captured using NDP.view 2 software (Hamamatsu).
Table 2.
List of Antibodies Used in Immunohistochemistry and Immunofluorescence
| Antibody | Dilution | Manufacturer | Catalog Number |
|---|---|---|---|
| CDX2 (rabbit polyclonal) | 1:250 | Bethyl Laboratories (Montgomery, TX) | A300-692A |
| Cleaved Casp-3 (Asp175) (5A1E) (rabbit monoclonal) | 1:1000 | Cell Signaling Technologies (Danvers, MA) | 9644 |
| E-cadherin (mouse monoclonal) | 1:2000 | BD Biosciences (San Jose, CA) | 610181 |
| GATA4 C-20 (goat polyclonal) | 1:250 | Santa Cruz Biotechnology (Santa Cruz, CA) | sc-1237 |
| HNF4A C-19 (goat polyclonal) | 1:500 | Santa Cruz Biotechnology | sc-6556 |
| Laminin (rabbit polyclonal) | 1:500 | Sigma-Aldrich (St. Louis, MO) | L9393 |
| MUCIN 2 (rabbit polyclonal) | 1:250 | Santa Cruz Biotechnology | sc-15334 |
| PDGFRA (rabbit polyclonal) | 1:500 | Santa Cruz Biotechnology | sc-338 |
| Atypical protein kinase C zeta (rabbit polyclonal) | 1:500 | Santa Cruz Biotechnology | sc-216 |
| Alexa-Fluor 488 donkey anti-rabbit | 1:500 | Life Technologies (Grand Island, NY) | A-21206 |
| Alexa-Fluor 488 donkey anti-mouse | 1:500 | Life Technologies | A-21202 |
| Alexa-Fluor 488 donkey anti-goat | 1:500 | Life Technologies | A-11055 |
| Alexa-Fluor 594 donkey anti-rabbit | 1:500 | Life Technologies | A-21207 |
| Alexa-Fluor 594 donkey anti-mouse | 1:500 | Life Technologies | A-21203 |
| Alexa-Fluor 594 donkey anti-goat | 1:500 | Life Technologies | A-11058 |
| Biotinylated goat anti-rabbit (heavy and light chains) IgG | 15 μL/mL | Vector Labs (Burlingame, CA) | BA-1000 |
| Biotinylated goat anti–guinea pig IgG | 15 μL/mL | Vector Labs | BA-7000 |
| Biotinylated rabbit anti-goat IgG | 15 μL/mL | Vector Labs | BA-5000 |
| Phalloidin-488 | 1:100 | Life Technologies | A12379 |
Quantitative Reverse-Transcription PCR
DNase-treated RNA from whole intestine (E11.5, E12.5, E14.5, E18.5) or from epithelial cells (E16.5) was used for quantitative reverse-transcription (qRT)-PCR13 (Table 3 lists TaqMan assays [Life Technologies, Carlsbad, CA] used). Epithelial cells were isolated as previously described.20 Epithelial cell preparations excluded the distal 10% of the intestine. For each gene assayed, 3 technical replicates were performed with at least 3 biological samples per genotype. Glyceraldehyde-3-phosphate dehydrogenase was used for normalization. Data were analyzed as previously described.13
Table 3.
TaqMan Assay Identifiers Used for Quantitative Reverse-Transcription PCR
| Gene | TaqMan ID | Gene | TaqMan ID |
|---|---|---|---|
| Abcg5 | Mm00433937_m1 | Gfi1 | Mm00515855_m1 |
| Abcg8 | Mm00433937_m1 | Gip | Mm00433601_m1 |
| Apoa4 | Mm00431814_m1 | Hes1 | Mm01342805_m1 |
| Apob | Mm01545156_m1 | Hes6 | Mm00517097_g1 |
| Apoc2 | Mm00433937_m1 | Klf4 | Mm00516104_m1 |
| Apoc3 | Mm00433937_m1 | Lct | Mm01285112_m1 |
| Atoh1 | Mm00476035_s1 | Muc1 | Mm00449604_m1 |
| Axin2 | Mm00443610_m1 | Muc2 | Mm01276696_m1 |
| Cck | Mm00446170_m1 | Ngn3 | Mm00437606_s1 |
| Ccnd1 | Mm00432359_m1 | Olfm4 | Mm01320260_m1 |
| Ccnd2 | Mm00438070_m1 | Pdgfa | Mm01205760_m1 |
| Cd44 | Mm01277161_m1 | Prdm1 | Mm00476128_m1 |
| Cdk6 | Mm01311342_m1 | Pyy | Mm00520716_g1 |
| Chga | Mm00514341_m1 | Shh | Mm00436528_m1 |
| Cldn8 | Mm00516972_s1 | Slc2a2 | Mm00433937_m1 |
| Fabp1 | Mm00444340_m1 | Slc2a5 | Mm00600311_m1 |
| Fabp2 | Mm00433188_m1 | Slc5a11 | Mm00433937_m1 |
| Fabp6 | Mm00434316_m1 | Slc10a2 | Mm00488258_m1 |
| Fgf15 | Mm00433278_m1 | Sox9 | Mm00448840_m1 |
| Fzd5 | Mm00445623_s1 | Spdef | Mm00600221_m1 |
| Gapdh | Mm99999915_g1 |
Biotin-Mediated Chromatin Immunoprecipitation
Previously published publically available BAM files containing alignment data from GATA4 biotin-mediated chromatin immunoprecipitation sequencing (Bio-ChIP-Seq) of mouse jejunal epithelial cells mapped to Mus musculus build 9 (GSE68957) were imported into Partek Genomics Suite (PGS) software version 6.6 (Partek, Inc, St. Louis, MO) for analysis using the ChIP-Seq workflow.21 Peaks were detected with a false discovery rate (FDR) threshold of 0.001. Peaks enriched in GATA4 Bio-ChIP compared with input were identified using a threshold of P < .05. Peaks were annotated to genes by PGS software by considering a gene to include all exons and introns as well as 10 Kb upstream of the transcriptional start site. Supplementary Table 1 contains a list of all annotated peaks. We performed Bio-ChIP-PCR as previously described.22, 23 Briefly, jejunal epithelial cells from adult Gata4flbio/flbio::Rosa26BirA/BirA (n = 6) and Gata4wt/wt::Rosa26BirA/BirA (n = 6) mice were used. Endogenous GATA4 protein is biotinylated in Gata4flbio/flbio::Rosa26BirA/BirA mice, allowing for precipitation of GATA4–chromatin complexes with streptavidin. GATA4 occupancy was detected by PCR, with primers (Table 1) flanking predicting GATA4 binding sites and α32P-2'-deoxyadenosine 5'-triphosphate (dATP). As a negative control, PCR was performed using primers amplifying an upstream region of Dll1.23 PCR products were separated by 4% polyacrylamide gels and visualized by autoradiography. For quantitation, PCR products were visualized using a Storm820 Phosphor Imager (Amersham Biosciences, Pittsburgh, PA). Band intensity was measured using ImageQuant 5.2 (Molecular Dynamics, Sunnyvale, CA). The percentage of enrichment was calculated by dividing the band intensity for the ChIP sample by 10× the band intensity for the input sample to account for the 1:10 dilution of input sample used in the PCR reaction. A 2-sample Student t test was used to compare means.
All authors had access to the study data. All authors reviewed and approved the final manuscript.
Results
Small Intestinal Length and Width Are Decreased in Intestine of Gata4 Conditional Knockout Embryos
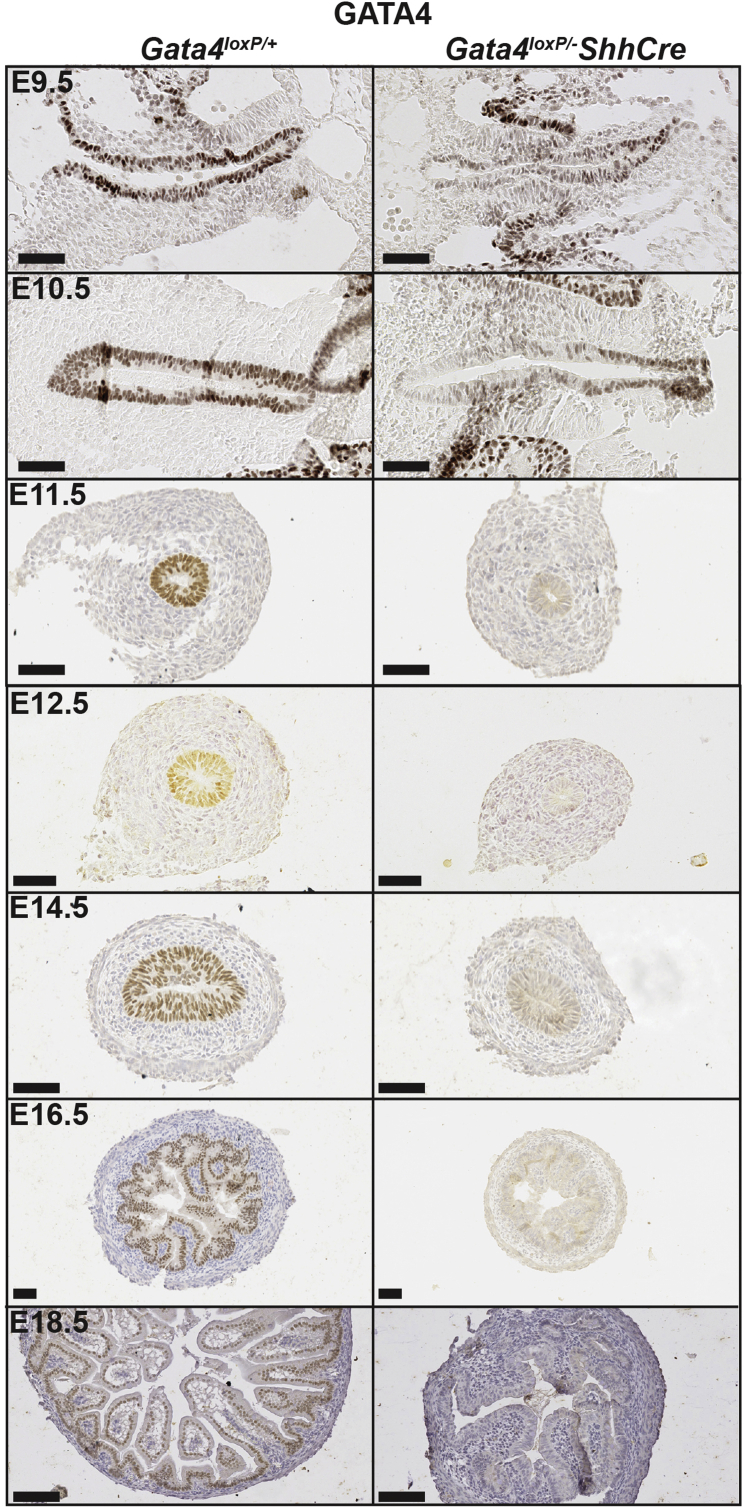
To determine GATA4 function during early intestinal development, we eliminated GATA4 from the midgut endoderm using ShhCre, which has robust activity in midgut endoderm by E9.5.15, 24 To identify when GATA4 first was deleted efficiently in mutant intestine, we isolated Gata4loxP/+ and Gata4loxP/-ShhCre (Gata4 conditional knockout [cKO]) embryos beginning at E9.5 and stained for GATA4. At E9.5, all Gata4 cKO embryos examined contained reduced GATA4 in midgut endoderm (Figure 1) (n = 10 embryos). GATA4 was reduced further in E10.5 mutant embryos with a small subset showing low to no GATA4 protein (Figure 1) (n = 8 embryos: 6 of 8 with reduced GATA4 and 2 of 8 with low to no GATA4). By E11.5, all mutant embryos examined had little to no detectable GATA4 (Figure 1) (n = 13 embryos: 12 of 13 with no GATA4 and 1 of 13 with sparse cells with reduced GATA4). As expected, GATA4 protein was undetectable in mutant intestinal epithelium at E12.5, E14.5, E16.5, and E18.5 (Figure 1). Therefore, we concluded that GATA4 was depleted efficiently in mutant intestine by E11.5.
Figure 1.
GATA4 protein was eliminated from the small intestine of Gata4loxP/-ShhCre cKO embryos by E11.5. Immunohistochemistry was used to detect GATA4 protein over a time course during development (n ≥ 3 controls and mutants per stage). Hematoxylin was used to counterstain. GATA4 protein (brown nuclear staining) was reduced at E9.5 and E10.5. GATA4 protein was undetectable in intestinal epithelium of Gata4loxP/-ShhCre cKO embryos at E11.5, E12.5, E14.5, E16.5, and E18.5. Scale bars: 50 μm.
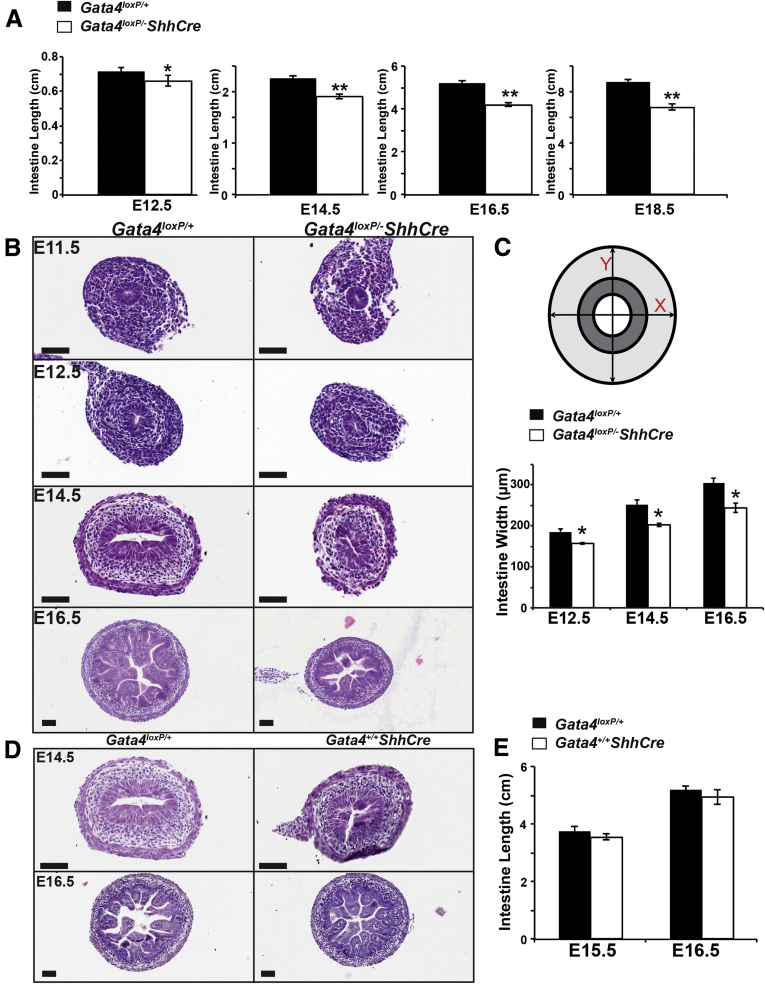
To determine if GATA4 loss affected gut elongation, we measured small intestinal length at E12.5, E14.5, E16.5, and E18.5. We found that GATA4-deficient small intestine was shorter than control intestine by 8% at E12.5, by 16% at E14.5, by 19% at E16.5, and by 22% at E18.5, suggesting that GATA4 is required for gut elongation (Figure 2A). Moreover, the total width of GATA4 mutant intestine was smaller than controls (Figure 2B and C). We determined total small intestinal girth in control and mutant intestine at E12.5, E14.5, and E16.5 by measuring the total cross-sectional width at 2 points, as shown in Figure 2C, and averaging these values. We found that the average width of GATA4-deficient small intestine was decreased by 15% at E12.5, by 19% at E14.5, and by 20% at E16.5 (Figure 2C). To confirm that changes observed were not related to ShhCre, we compared H&E sections and overall length of small intestine between Gata4+/+ShhCre and Gata4loxP/+ embryos and found no differences (Figure 2D and E).
Figure 2.
Small intestinal length and width were decreased in GATA4 mutants. (A) Small intestine of Gata4loxP/-ShhCre cKO embryos was shorter than that of Gata4loxP/+ embryos (n ≥ 15 controls and mutants per stage). (B) H&E staining suggested that intestine of Gata4loxP/-ShhCre embryos was smaller than that of controls. (C) Total intestinal width was measured across the x- and y-axes as shown. Gata4loxP/-ShhCre cKO intestinal width was decreased compared with controls starting at E12.5 (n ≥ 4 sections, ≥3 controls and mutants per stage). (D) H&E staining showed comparable structure between control and Gata4+/+ShhCre intestines at E14.5 and E16.5. (E) Small intestine length was measured in control and Gata4+/+ShhCre embryos at E15.5 and E16.5 and no difference was observed (n ≥ 4 per genotype, per stage). Error bars show SEM. P values were determined using a 2-sample Student t test: *P ≤ .05, **P ≤ .01. Scale bars: 50 μm.
Epithelial Girth Is Decreased in Intestine of Gata4 cKO Embryos
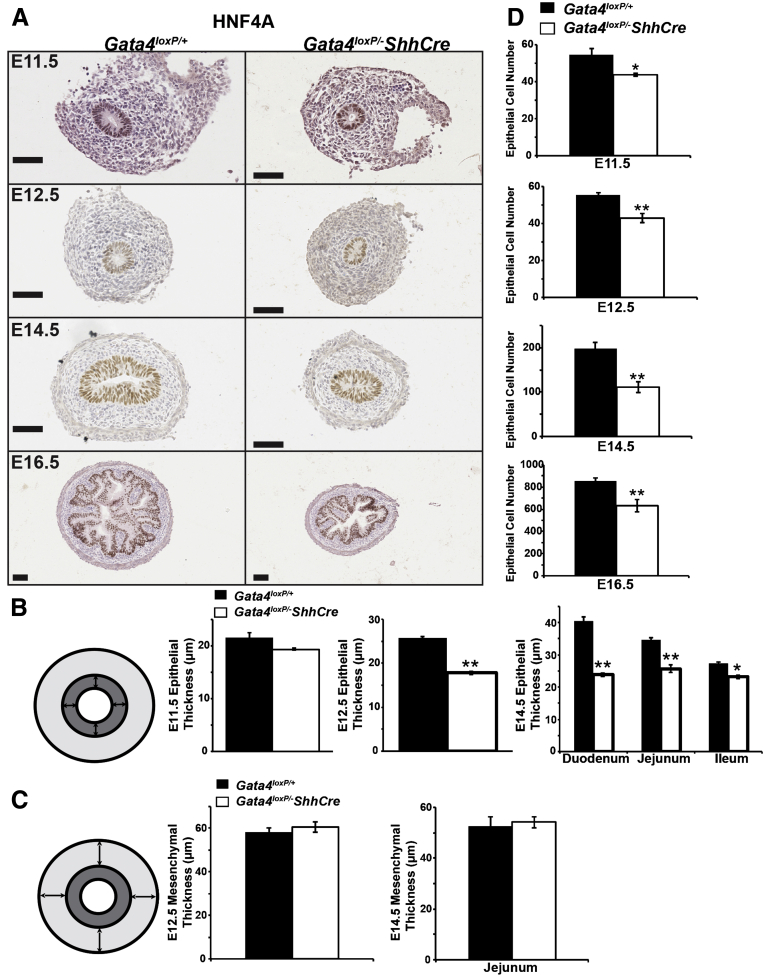
Closer examination of H&E-stained tissues (Figure 2B) suggested that reduced total intestinal width observed in GATA4 mutants reflected a change in epithelial thickness rather than a change in mesenchymal thickness. To mark the epithelium, we stained for HNF4A, a transcription factor exclusively expressed in epithelium (Figure 3A). We determined epithelial girth by measuring thickness of HNF4A+ tissue at 4 sites per cross-section and averaging these values to obtain an average epithelial thickness per section (Figure 3B). We observed a trend toward a thinner epithelium at E11.5 (10% reduction; P = .140) (Figure 3B). At E12.5, we observed a statistically significant 28% decrease in average epithelial thickness in GATA4 mutants compared with controls (Figure 3B). At E14.5, we quantified epithelial girth in duodenum, jejunum, and ileum and found it to be reduced in GATA4 mutants in all regions (reduced by 42% in duodenum, by 25% in jejunum, and by 15% in ileum) (Figure 3B). We used the same strategy to determine the average mesenchymal thickness in controls and mutants at E12.5 and E14.5 (jejunum) and found no difference (Figure 3C), supporting the conclusion that reduced intestinal girth reflects a change in epithelial thickness rather than mesenchymal thickness.
Figure 3.
Epithelial girth and cell number were decreased in intestine lacking GATA4. (A) HNF4A immunohistochemistry (brown nuclear stain) marked intestinal epithelial cells of Gata4loxP/+ control and Gata4loxP/-ShhCre cKO embryos. Hematoxylin was used to counterstain. Epithelial thickness appeared decreased in GATA4 mutants. (B) Epithelial thickness was decreased in GATA4 mutants. Per HNF4A stained section, 4 measurements were taken (diagram, arrows) and averaged (n ≥ 5 sections, ≥3 controls and mutants per stage). At E14.5, measurements were made in the duodenum, jejunum, and ileum. (C) Mesenchymal thickness was equivalent between GATA4 mutants and controls. Per HNF4A stained section, 4 measurements were taken (diagram, arrows) and averaged (n ≥ 5 sections, ≥5 controls and mutants per stage). At E14.5, jejunum was measured. (D) GATA4 mutant epithelium contained fewer HNF4+ epithelial cells compared with controls (n ≥ 5 sections, ≥4 controls and mutants per stage; jejunal sections counted at E14.5 and E16.5). Error bars show SEM. P values were determined by 2-sample Student t test: *P ≤ .05, **P ≤ .01. Scale bars: 50 μm.
Gata4 Is Required for Epithelial Cell Proliferation at E10.5 and E11.5
It has been argued that epithelial girth is related to cell number.4, 5 Because cellular packing density increases with pseudostratification, individual cells become longer and thinner, thereby increasing epithelial thickness. Consequently, a thinner epithelium in GATA4 mutants may reflect a change in epithelial cell number. Therefore, we determined epithelial cell number in control and mutant intestine at E11.5, E12.5, E14.5, and E16.5 by counting HNF4A+ cells/section. At E14.5 and E16.5, we quantified HNF4A+ cells in jejunum. At all stages, we found fewer epithelial cells in GATA4-deficient small intestine compared with controls (Figure 3D) (reduced by 19% at E11.5, by 23% at E12.5, by 44% at E14.5, and by 26% at E16.5).
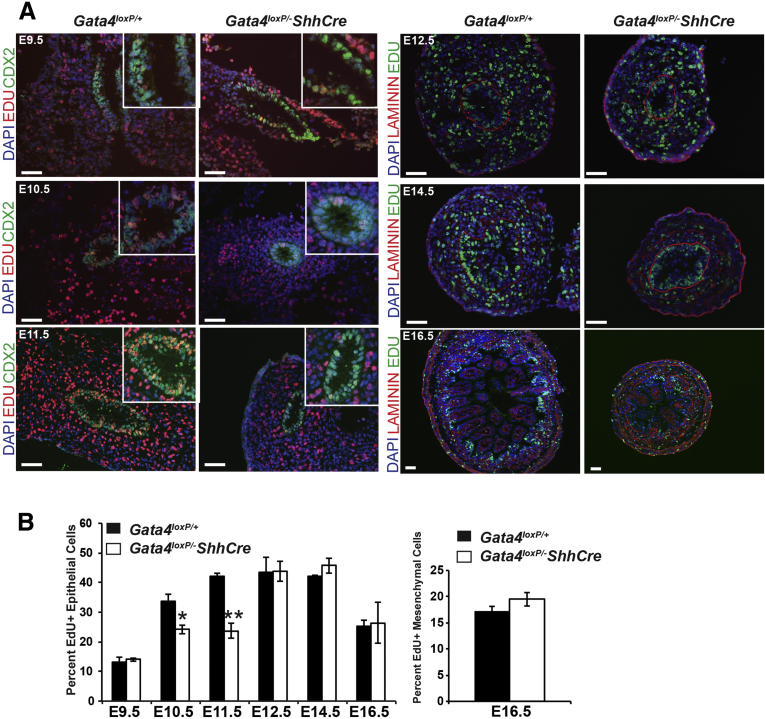
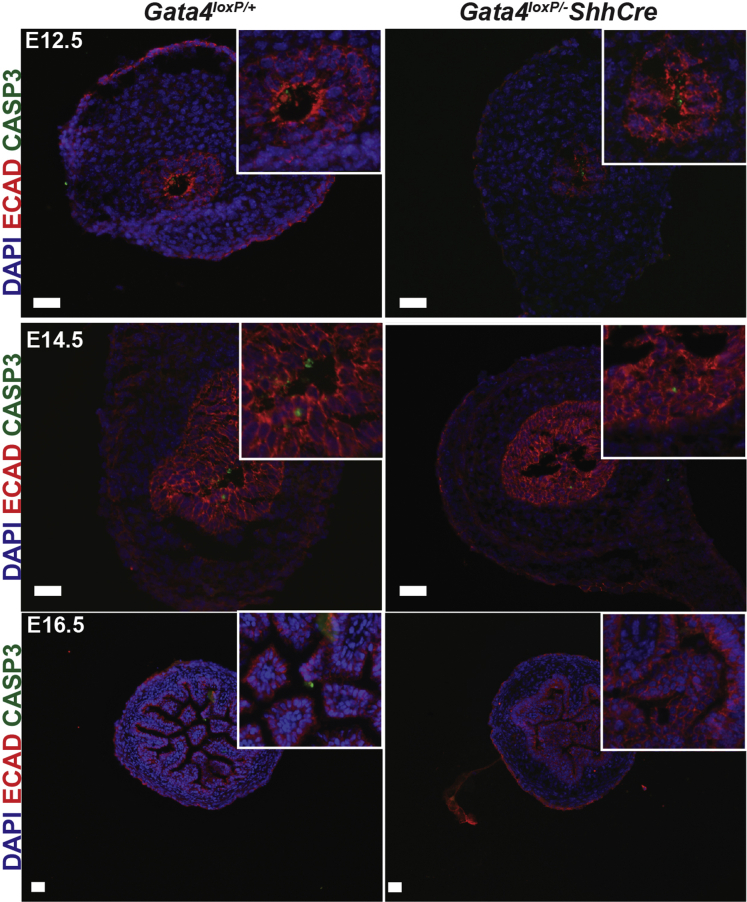
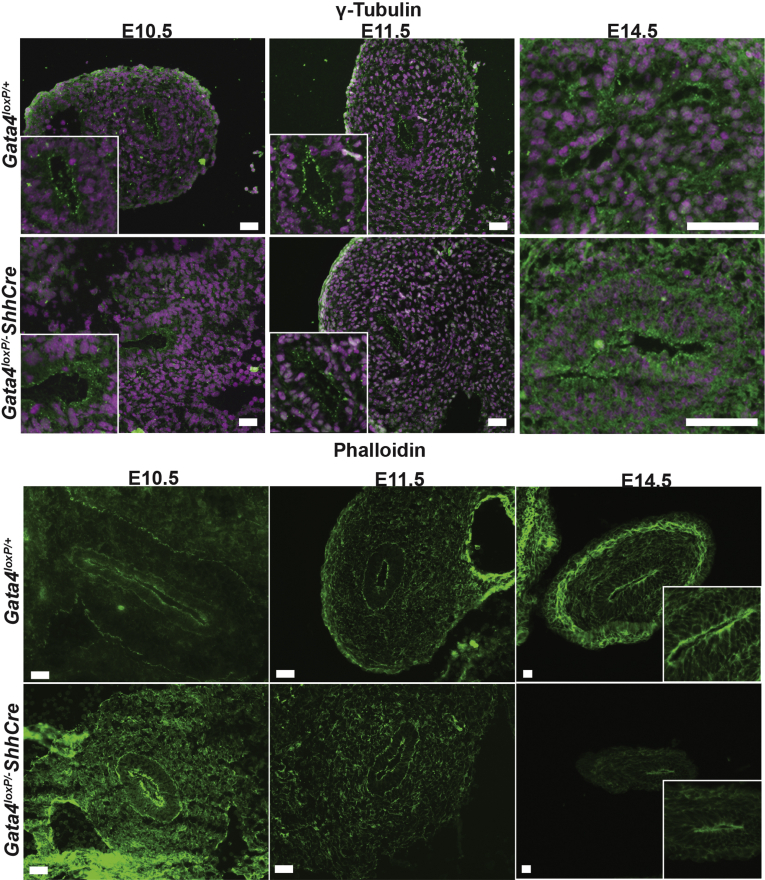
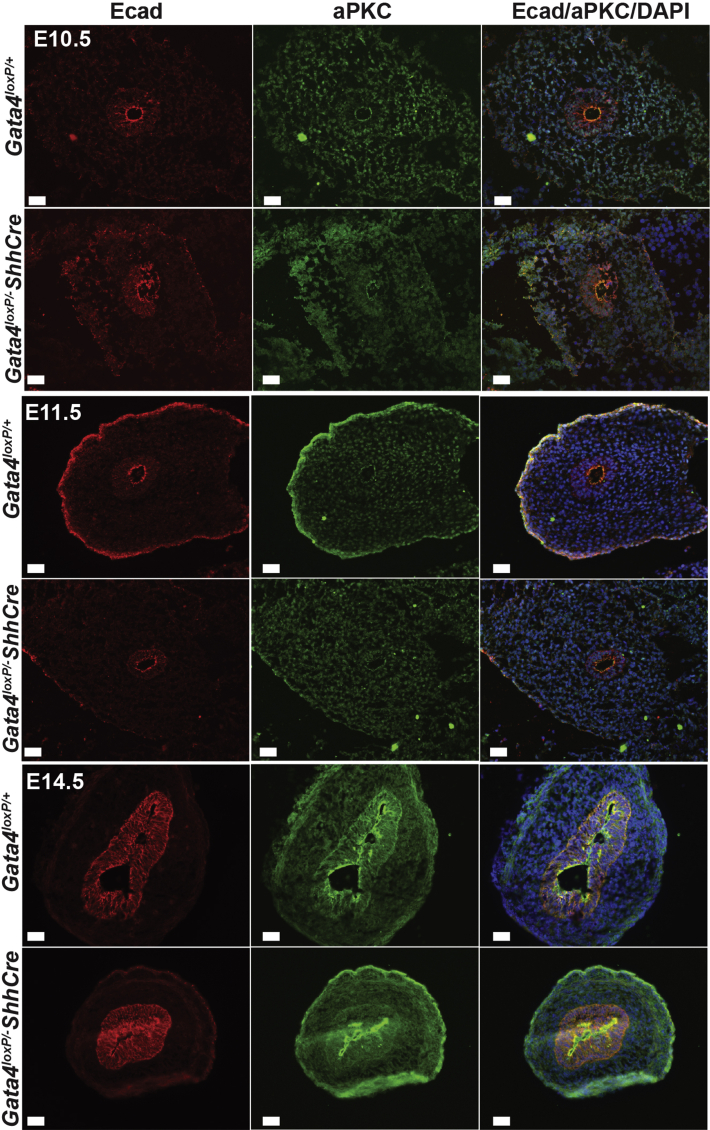
One explanation for fewer epithelial cells is that proliferation is reduced without GATA4. Therefore, to quantify proliferation in the developing small intestinal epithelium, we measured EdU incorporation at E9.5, E10.5, E11.5, E12.5, E14.5, and E16.5. Between E9.5 and E11.5, we identified proliferating epithelial cells as CDX2+/EdU+ cells (Figure 4A). At E12.5, E14.5, and E16.5, we identified proliferating epithelial cells as EdU+ cells located on the luminal side of the laminin+ basement membrane (Figure 4A). At E9.5, we observed no change between controls and mutants (Figure 4B). By E10.5 and E11.5, however, we found a reduced proportion of proliferating epithelial cells in mutants compared with controls (Figure 4B) (decreased by 28% at E10.5 and by 44% at E11.5). By E12.5 and beyond, the proportion of proliferating cells recovered in GATA4 mutants matching that of control tissue (Figure 4B). From these data, we conclude that GATA4 is essential for epithelial cell proliferation during the early stages (E10.5–E11.5), but not the later stages (E12.5–E16.5), of intestinal development. Studies using VillinCre to eliminate GATA4 from the intestinal epithelium after E14.5 concur with this finding because proliferation is normal in Gata4VillinCre mutants.13, 25 Although the proportion of proliferating cells returned to control levels at E12.5 and beyond, this was insufficient to rescue epithelial cell number (Figure 3D). We assayed cell death in control and mutant intestine at E12.5, E14.5, and E16.5 by active caspase 3 staining and observed no difference between mutant and control groups (Figure 5). We also found no difference in epithelial cell polarity because E-cadherin, atypical protein kinase C, phalloidin, and γ-tubulin staining were comparably localized at the apical cell surface in cells of both control and mutant epithelium at E10.5, E11.5, and E14.5 (Figures 6 and 7).
Figure 4.
Proliferation was decreased at E10.5 and E11.5 in intestine lacking GATA4. (A) EdU incorporation (E9.5–E11.5, red; E12.5–E16.5, green) was used to measure proliferation within the intestinal epithelium of Gata4loxP/+ and Gata4loxP/-ShhCre cKO embryos. At E9.5–E11.5, CDX2 (green) marked epithelial cells. At E12.5, E14.5, and E16.5, laminin (red), which marks the basement membrane separating epithelium and mesenchyme, distinguished between epithelium and mesenchyme. At all stages, 4',6-diamidino-2-phenylindole (DAPI) (blue) labeled nuclei. (B) The proportion of proliferating epithelial cells was decreased in GATA4 mutant intestine compared with controls at E10.5 and E11.5 (left). The percentage of proliferating epithelial cells was determined at E9.5–E11.5 by dividing EdU+, CDX2+ cells by total CDX2+ cells and at E12.5, E14.5, and E16.5 by dividing EdU+ cells by total DAPI+ cells located on the luminal side of laminin staining (n ≥ 5 sections, ≥3 controls and mutants per stage). The proportion of proliferating mesenchymal cells was not changed in GATA4 mutants compared with controls (right). The percentage of proliferating mesenchymal cells was determined at E16.5 by dividing EdU+ cells by the total DAPI+ cells located on the basal side of the ring of laminin staining demarcating the epithelial–mesenchymal boundary. Error bars show SEM. P values were determined by 2-sample Student t test: *P ≤ .05, **P ≤ .01. Scale bars: 50 μm.
Figure 5.
Apoptotic cell death was not increased in intestine lacking GATA4. Immunofluorescent staining with an antibody against cleaved caspase 3 (green) was used to assess apoptotic cell death in intestine from control and Gata4loxP/-ShhCre embryos at E12.5, E14.5, and E16.5 (n = 3 controls and mutants per stage). E-cadherin staining (red) was used to mark the epithelium. 4',6-diamidino-2-phenylindole (DAPI) (blue) was used to mark nuclei. No differences were observed between groups. Scale bars: 50 μm.
Figure 6.
Cell polarity appears normal in GATA4 mutant intestine. Immunofluorescent staining with an antibody against γ-tubulin (red) or with phalloidin (green) was used to assess cell polarity. Either sections from control and Gata4loxP/-ShhCre embryos containing midgut endoderm (E10.5) or sections of intestine dissected from control and Gata4loxP/-ShhCre embryos (E11.5, E14.5) were stained (n = 3 controls and mutants per stage). 4',6-diamidino-2-phenylindole (DAPI) (blue) was used to visualize nuclei. Scale bars: 50 μm.
Figure 7.
Cell polarity was not affected by loss of GATA4. Immunofluorescent staining with antibodies against E-cadherin (red) and atypical PKC zeta (green) was used to assess cell polarity. Either sections from control and Gata4loxP/-ShhCre embryos containing midgut endoderm (E10.5) or sections of intestine dissected from control and Gata4loxP/-ShhCre embryos (E11.5, E14.5) were stained (n = 3 controls and mutants per stage). 4',6-diamidino-2-phenylindole (DAPI) (blue) was used to visualize nuclei. Scale bars: 50 μm.
GATA4 Regulates Cyclin D2, Cyclin-Dependent Kinase 6, and Frizzled 5 in the Developing Small Intestine
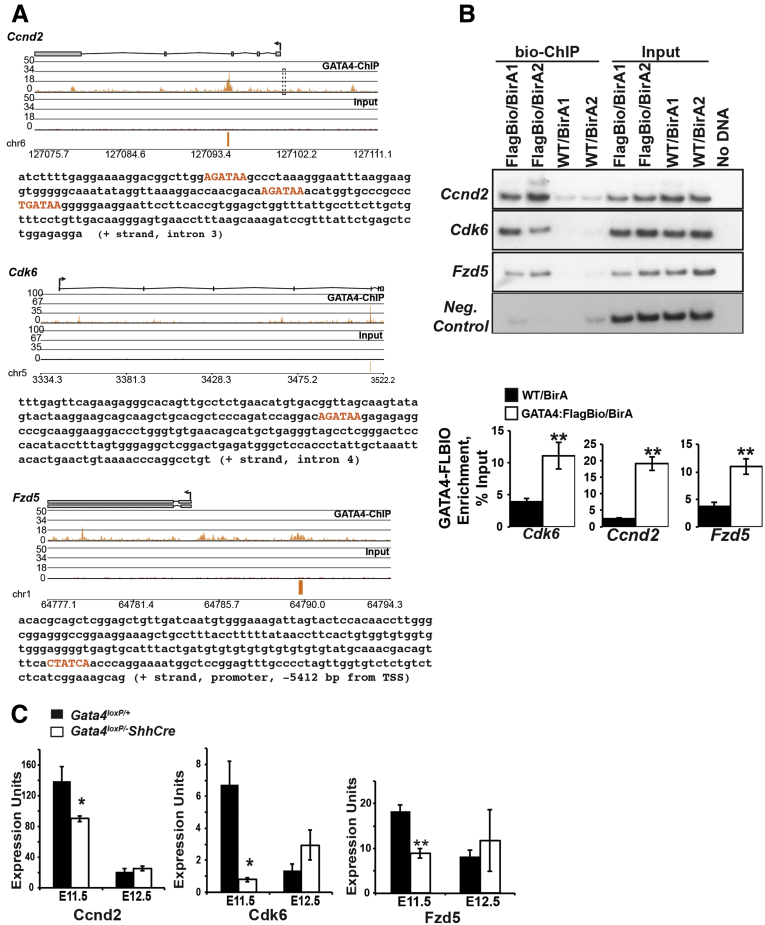
One mechanism through which GATA4 could control proliferation is by transcriptional regulation of cell-cycle mediators. Studies in the heart have shown that GATA4 directly regulates Ccnd2 and Cdk4, suggesting these as candidates for GATA4 regulation in the intestine.26 We obtained published GATA4 Bio-ChIP-Seq data generated from mouse jejunal small intestinal epithelium (GSE68957)21 and analyzed these data with Partek Genomics Suite software to identify GATA4 enrichment sites within Ccnd2 or Cdk4. We observed one peak in Ccnd2 intron 3 (Figure 8A), and no peaks within Cdk4. The region identified in Ccnd2 represented a novel GATA4 regulatory element because sites identified as GATA4-bound in cardiac tissue reside in the promoter 558 bp upstream of the transcriptional start site.26 No GATA4 enrichment was observed in this region of Ccnd2 in GATA4 Bio-ChIP-Seq in intestine (Figure 8A, dotted box). Although no GATA4 enrichment peaks were found in Cdk4, we did identify a peak in Cdk6 intron 4 (Figure 8A). CCND2 complexes with either CDK4 or CDK6 to regulate the cell cycle.27 Furthermore, the stringent GATA4 consensus sequence WGATAR was found within Ccnd2 and Cdk6 regions identified by ChIP-Seq (Figure 8A, orange text). To verify GATA4 occupancy of novel GATA4 sites within Ccnd2 and Cdk6, we performed Bio-ChIP-PCR using small intestinal epithelial cell chromatin from adult Gata4flbio/flbio::Rosa26BirA/BirA and Rosa26BirA/BirA mice.16, 17 We conducted 3 independent Bio-ChIP–PCR experiments, and each experiment assayed chromatin from 2 Gata4flbio/flbio::Rosa26BirA/BirA and 2 Rosa26BirA/BirA mice (n = 6 mice/genotype). By using PCR with primers flanking putative GATA4 binding sites in Ccnd2 and Cdk6, we found GATA4 enriched at these loci in chromatin from Gata4flbio/flbio::Rosa26BirA/BirA intestine compared with chromatin from Rosa26BirA/BirA intestine (Figure 8B). Therefore, we conclude that GATA4 binds to Ccnd2 and Cdk6 in the intestine.
Figure 8.
GATA4 regulates Ccnd2, Cdk6, and Fzd5. (A) GATA4 Bio-ChIP-Seq data were analyzed using PGS software. Enrichment of sequencing tags (orange) was identified within Ccnd2, Cdk6, and Fzd5 in the GATA4-Bio-ChIP sample compared with input. Consensus GATA4 binding sites (WGATAR) were identified in Ccnd2 (3 binding sites), Cdk6 (1 binding site), and Fzd5 (1 binding site) and are shown in orange in sequence text. Arrows show the transcriptional start sites. The dotted box on the Ccnd2 plot indicates a GATA4 binding site shown to bind GATA4 in cardiac tissue but not identified as GATA4 occupied in intestine. Each diagram contains the complete genomic region that was analyzed for the presence of GATA4 binding sites, which includes the entire gene and 10 Kb upstream of the transcriptional start site. Scales for each diagram differ because the size of each gene differs. (B) Bio-ChIP-PCR showed GATA4 enrichment at predicted binding sites in Ccnd2, Cdk6, and Fzd5 in GATA4FlagBio/BirA (FlagBio/BirA) chromatin compared with GATA4WT/BirA (WT/BirA) chromatin. PCR using chromatin from 2 mice per genotype (Gata4flbio/flbio::Rosa26BirA/BirAand Rosa26BirA/BirA) shown; a total of 6 mice per genotype were assayed. No enrichment was detected within a negative control region (upstream sequence of Dll1).23 Input showed equivalent chromatin amounts were used. Graphs compare average enrichment of GATA4 binding to Ccnd2, Cdk6, and Fzd5 in jejunal intestinal epithelium from GATA4FlagBio/BirA and GATA4WT/BirA mice (n = 6 mice per genotype). (C) qRT-PCR showed that Ccnd2, Cdk6, and Fzd5 transcripts were decreased in GATA4 mutants at E11.5 and comparable with controls at E12.5 (n = 3 intestine per genotype per age; experiments performed in triplicate). All error bars show SEM. All P values were determined by 2-sample Student t test: *P ≤ .05, **P ≤ .01. Neg, negative.
If GATA4 controls proliferation by directly regulating Ccnd2 and Cdk6, we expected these transcripts to be decreased in Gata4 cKO intestine compared with control intestine. We evaluated Ccnd2 and Cdk6 levels by qRT-PCR and found that both transcripts were reduced in GATA4-deficient small intestine compared with control intestine at E11.5, when the proportion of proliferating cells was lower in mutants compared with controls. At E12.5, when proliferation was comparable between mutants and controls, we detected no difference in Ccnd2 or Cdk6 levels (Figure 8C).
It also was striking that the magnitude and timing of the proliferation decrease that we observed in GATA4 mutants mirrored that observed in WNT5A mutants, suggesting a relationship between GATA4 and WNT5A. At E11.5, epithelial cell proliferation was reduced by 44% in GATA4 mutants and by 37% in WNT5A mutants.7 The mechanism through which WNT5A acts on epithelial cells to influence proliferation is unknown. No change in epithelial proliferation was observed in WNT5A-receptor ROR2 mutants, suggesting an ROR2-independent mechanism.12 FZD5, which is expressed by intestinal epithelial cells, is another WNT5A receptor.28, 29 Although FZD5 function has not been investigated during early intestinal development because Fzd5-/- embryos die at approximately E10.75, endothelial cell proliferation was reduced in the FZD5-deficient yolk sac.28 Because studies of yolk sac development suggest a role for FZD5 in regulating proliferation, we queried ChIP-Seq data to determine if GATA4 enrichment was present in Fzd5 and found a peak within the Fzd5 promoter containing a single WGATAR consensus sequence (Figure 8A). We confirmed GATA4 occupancy within this region by Bio-ChIP-PCR and found that Fzd5 transcript was decreased in GATA4 mutant E11.5 intestine (Figure 8B and C). Similar to Ccnd2 and Cdk6, Fzd5 levels were unchanged at E12.5 (Figure 8C). We infer from Bio-ChIP and expression data that GATA4 controls proliferation at E11.5 by not only directly regulating transcription of cell-cycle regulators themselves (Ccnd2 and Cdk6), but also by directly regulating transcription of Fzd5, thereby potentially impacting the magnitude of WNT5A signaling received by the epithelium.
Villus Morphogenesis Is Delayed in Gata4 cKO Intestine
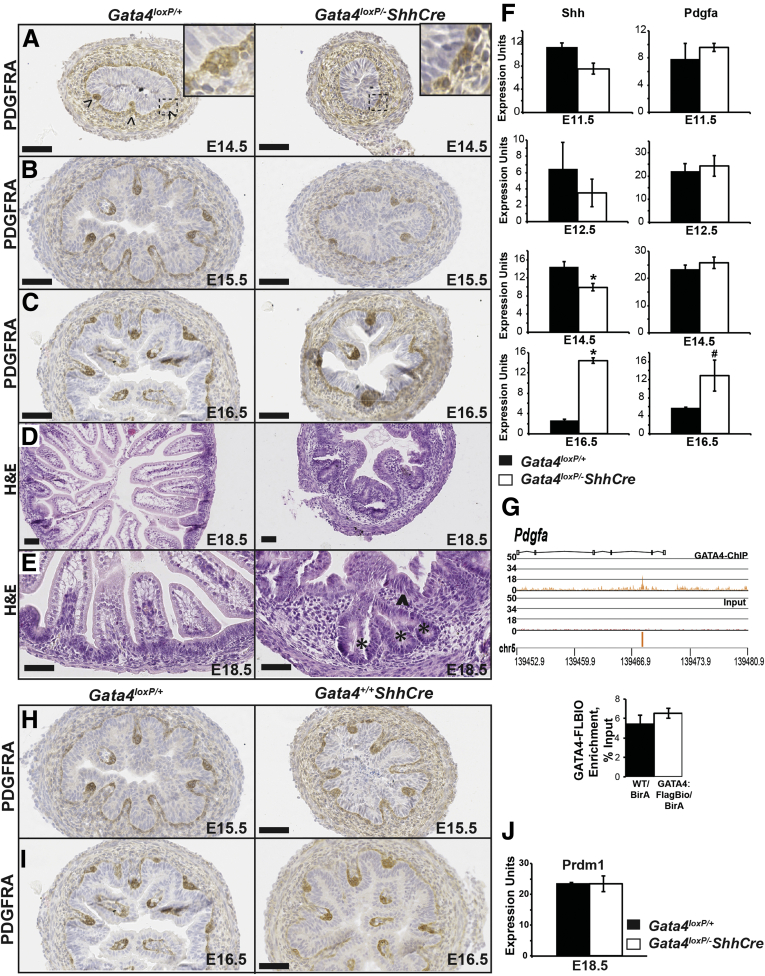
We noted that villi appeared to be abnormal in GATA4-deficient intestine compared with control tissue (Figures 1, 2B, and 3A). Therefore, we evaluated villus morphogenesis in controls and mutants between E14.5 and E16.5 by platelet-derived growth factor receptor α (PDGFRA) immunohistochemistry, which marks mesenchymal cell clusters where presumptive villi will emerge.30 In E14.5 controls, PDGFRA+ mesenchymal clusters were readily observed (Figure 9A, arrowheads). Although PDGFRA+ cells were present throughout the mesenchyme of GATA4 mutants, no clusters comparable with those in controls were evident (Figure 9A). By E15.5, control tissue contained numerous villi. PDGFRA+ clusters populated nascent villi tips and marked sites of new villi (Figure 9B). GATA4 E15.5 mutant tissue, however, resembled control tissue at E14.5 with sparse PDGFRA+ clusters (Figure 9B). Although villi were present in E16.5 mutant tissue, they appeared to be fewer in number compared with controls (Figure 9C). By E18.5, the intestinal epithelium of controls was organized as long, slender villi with columnar epithelial cells (Figure 9D and E). In contrast, GATA4-deficient epithelium appeared disorganized with fewer shorter, wider villi (Figure 9D and E). Regions resembling pseudostratified epithelium also persisted (Figure 9E, arrowhead). Taken together, we conclude that villus morphogenesis is delayed and that villus structure is abnormal in GATA4 mutants.
Figure 9.
Villus morphogenesis was delayed and villus structure was altered in the absence of GATA4. (A–C) PDGFRA immunohistochemistry was used to evaluate villus morphogenesis from E14.5 to E16.5 in intestine from Gata4loxP/+ and Gata4loxP/-ShhCre cKO embryos (n ≥ 4 controls and mutants per stage). PDGFRA+ mesenchymal cell clusters associated with the epithelium were observed by E14.5 in controls (A, arrowheads; dotted box, higher-magnification inset). Although PDGFRA+ cells were observed in E14.5 GATA4-deficient intestine, clusters were not apparent (A, dotted box, higher-magnification inset). At (B) E15.5 and (C) E16.5, PDGFRA+ cells populated villus tips and marked sites where new villi would emerge in controls. E15.5 GATA4 mutant tissue resembled E14.5 control tissue (compare the left side of panel A with the right side of panel B). (B) PDGFRA+ clusters became apparent in GATA4 mutant tissue at E15.5. (C) At E16.5, fewer villi were observed in GATA4 mutants compared with controls. (D and E) H&E staining of intestine from E18.5 Gata4loxP/+ and Gata4loxP/-ShhCre cKO embryos showed abnormal villus structure in mutants. Controls contained long, slender villi with columnar epithelial cells. GATA4-deficient epithelium appeared disorganized, with fewer villi that were shorter and wider than controls. (E) Regions resembling pseudostratified epithelium persisted (right panel, arrowhead). Crypt-like structures also were observed (right, stars). (F) Quantitative reverse-transcription PCR was used to determine expression of Shh and Pdgfa transcripts in RNA from whole intestine at E11.5 (n = 4 controls, 3 mutants), E12.5 (n = 3 controls, 3 mutants), and E14.5 (n = 3 controls, 3 mutants), and in RNA from intestinal epithelial cells at E16.5 (n = 3 controls, 3 mutants). (G) GATA4 Bio-ChIP-Seq data were analyzed using PGS software, and enrichment of sequencing tags (orange) was identified within Pdgfa in the GATA4-Bio-ChIP sample compared with input. The diagram contains the complete genomic region analyzed for the presence of GATA4 binding sites, which includes the entire gene and 10 Kb upstream of the transcriptional start site. Bio-ChIP-PCR showed no GATA4 enrichment at the predicted binding site in Pdgfa in GATA4FlagBio/BirA chromatin compared with GATA4WT/BirA chromatin. Graph compares average enrichment of GATA4 binding with Pdgfa in jejunal intestinal epithelium from GATA4FlagBio/BirA and GATA4WT/BirA mice (n = 6 mice per genotype [Gata4flbio/flbio::Rosa26BirA/BirAand Rosa26BirA/BirA]). (H and I) Villus morphogenesis was assessed by PDGFRA staining of E15.5 embryonic intestine from control and Gata4+/+ShhCre embryos (n = sections from 2 Gata4+/+ShhCre embryos at E15.5 and from 3 Gata4+/+ShhCre embryos at E16.5). No difference was observed. (J) To determine if premature crypt formation occurred in GATA4 mutant intestine, quantitative reverse-transcription PCR was used to determine Prdm1 expression in intestine at E18.5 (n = 3 controls, 3 mutants). All error bars show SEM. All P values were determined by 2-sample Student t test: *P ≤ .05, #P = .053. Scale bars: 50 μm.
Two important morphogens secreted by epithelium known to regulate villus morphogenesis are SHH and PDGFA.30, 31 To assess Shh and platelet-derived growth factor α (Pdgfa) expression in GATA4-deficient intestine, we used qRT-PCR to measure transcript abundance at E11.5, E12.5, E14.5, and E16.5. We found that at early stages (E11.5–E14.5), when villus morphogenesis was delayed in GATA4 mutant embryos, Shh transcript was less abundant in GATA4 mutant intestine compared with control intestine; Pdgfa levels were unchanged (Figure 9F). At E16.5, when villus morphogenesis was underway in GATA4 mutant intestine, both Shh and Pdgfa increased in mutant epithelium (Figure 9F). Analysis of GATA4 Bio-ChIP-Seq data showed a GATA4 binding site within the Pdgfa gene but not within the Shh gene (Figure 9G). We performed Bio-ChIP-PCR using small intestinal epithelial cell chromatin from Gata4flbio/flbio::Rosa26BirA/BirA and Rosa26BirA/BirA mice as described earlier and found no enrichment of GATA4 at this binding site within Pdgfa (Figure 9G). Together, these data suggest that changes in Shh and Pdgfa abundance observed in GATA4 mutant tissue were not related directly to GATA4 transcriptional control. Because changes in Pdgfa could affect proliferation in the mesenchymal compartment,30 we compared the proportion of EdU+ proliferating mesenchymal cells between control and GATA4 mutant intestine at E16.5 and found no difference (Figure 4B, right panel).
Because we used ShhCre for these studies and SHH itself is an important regulator of villus morphogenesis, we verified that defects in villus morphogenesis observed in GATA4 mutants were specific to loss of GATA4 by assessing villus morphogenesis in ShhCre control embryos. We performed PDGFRA staining with tissue from Gata4+/+ShhCre embryos and found no change in villus morphogenesis (Figure 9H and I), showing that the phenotype is specific to loss of GATA4.
Finally, we observed potential premature crypt formation in GATA4 mutant tissue (Figure 9E, stars). To determine if premature crypt formation occurred in the absence of GATA4, we measured PR domain containing 1 (Prdm1) expression in control and GATA4 mutant intestine at E18.5. Prdm1 levels normally decrease as crypts form, and precocious crypt development occurs in Prdm1 null mice.32 We observed no change in Prdm1 expression between control and mutant tissue, suggesting that premature crypt formation did not occur in GATA4 mutant intestine (Figure 9J).
Epithelial Cell Populations Are Altered in GATA4-Deficient Intestine
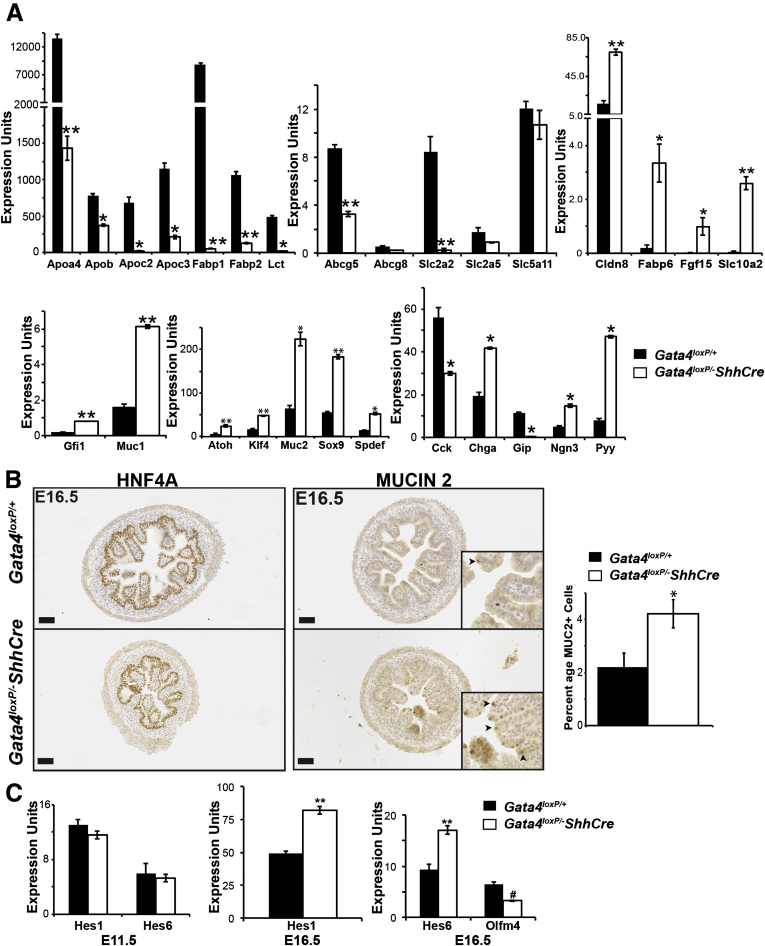
When GATA4 is deleted with VillinCre, differentiated epithelial cell marker gene expression is altered, and goblet cells increase.13, 23, 25, 33 Because earlier loss of GATA4 could influence this phenotype, we compared enterocyte and secretory cell lineage gene expression between control and Gata4–ShhCre mutants. Similar to Gata4–VillinCre mutants, jejunal-enriched enterocyte transcripts were reduced, ileal-enriched enterocyte transcripts were increased, and goblet cell transcripts were increased in Gata4–ShhCre mutants compared with controls (Figure 10A). Furthermore, the percentage of MUC2+ goblet cells was increased by 2-fold in Gata4–ShhCre mutants, similar to the 1.5-fold increase observed in Gata4–VillinCre mutants (Figure 10B).13 Increased neurogenin 3 and chromogranin A levels implied that Gata4–ShhCre mutant jejunum contained more enteroendocrine cells (Figure 10A). This reflects a difference between Gata4–VillinCre and Gata4–ShhCre mutants because the expression of neurogenin 3 and chromogranin A, and by extension enteroendocrine cell number, was unchanged in Gata4–VillinCre mutants.13
Figure 10.
Epithelial cell populations were altered in GATA4-deficient small intestine. (A) Quantitative reverse-transcription PCR determined enterocyte, goblet cell, and enteroendocrine cell transcript levels in intestinal epithelium of Gata4loxP/+ and Gata4loxP/-ShhCre cKO E16.5 embryos. Enterocyte gene expression was altered with jejunal-enriched transcripts decreasing (Apoa4, Apob, Apoc2, Apoc3, Fabp1, Lct, Slc2a2), pan-enterocyte transcripts decreasing (Fabp2, Abcg5), and ileal-enriched transcripts increasing (Cldn8, Fabp6, Fgf15, Slc10a2). Goblet cell transcripts were increased in mutants (Gfi1, Muc1, Muc2, Spdef, Atoh, Klf4). The Paneth cell marker sry-box 9 (Sox9) also was increased in mutants. Pan-enteroendocrine cell transcripts were increased in mutants (Ngn3, Chga). Markers of proximal-enriched enteroendocrine cell types (Cck, Gip) were decreased in mutants. Pyy, a distally enriched enteroendocrine cell marker, was increased in mutants. (B) MUC2+ goblet cells were increased in mutants. MUC2+ and HNF4A+ cells were counted in serial jejunal sections to determine goblet cell number and total epithelial cell number, respectively (n = 5 sections, 4 embryos/genotype). (C) Quantitative reverse-transcription PCR was used to determine the expression of Notch targets Hes1, Hes6, and Olfm4 in RNA from whole intestine at E11.5 (n = 4 controls, 3 mutants) and in RNA from intestinal epithelial cells at E16.5 (n = 3 controls, 3 mutants). Olfm4 was undetectable at E11.5. All error bars show SEM. All P values were determined by 2-sample Student t test: *P ≤ .05, **P ≤ .01, #P = .12. Scale bars: 50 μm.
Increased numbers of secretory cells and a 3.5-fold atonal homolog 1 (Atoh1) transcript increase in GATA4 mutant intestine suggested Notch signaling was decreased in mutants. Therefore, we examined the expression of the Notch transcriptional targets hairy and enhancer of split (Hes)1, Hes6, and olfactomedin 4 (Olfm4) at E11.5 and at E16.5.34 At E11.5, we found no change in Hes1 or Hes6 abundance between groups, and Olfm4 was undetectable in both control and GATA4 mutant intestine (Figure 10C). At E16.5, we observed slight increases in Hes1 and Hes6 abundance in GATA4 mutant epithelium (1.7-fold and 1.8-fold, respectively), but Olfm4 transcript was decreased 2-fold (P = .12) (Figure 10C). Although we observed nonuniform changes in levels of Notch target transcripts, together reduced Olfm4, increased Atoh1, and increased goblet cell number suggest that Notch signaling is abnormal in GATA4 mutant intestine. Cumulatively, the data indicate that Notch signaling likely is decreased in GATA4 mutant intestine.
Discussion
During embryonic development, the intestine grows dramatically, and epithelial proliferation is an important driver of organ growth. However, mechanisms through which epithelial cells regulate proliferation during this period are poorly understood. Our data establish GATA4 as an epithelial cell–specific factor that controls proliferation during transition of the midgut endodermal epithelium from simple to pseudostratified. We propose that GATA4 controls proliferation through transcriptional regulation of the cell-cycle regulators Ccnd2 and Cdk6 and the WNT receptor Fzd5. GATA4 has been shown previously to regulate Ccnd2 in the developing heart.26 Our data indicate that GATA4 activates intestinal Ccnd2 expression through a tissue-specific mechanism using a regulatory element in Ccnd2 that is distinct from that used in the heart. Moreover, GATA4 regulates an alternative CCND2 binding partner in the intestine (Cdk6) compared with the heart (Cdk4). Interactions with tissue-restricted factors likely contribute to the tissue specificity of GATA4 function. Although reductions in Ccnd2, Cdk6, or Fzd5 transcripts may be modest individually, we argue that the cumulative change in expression of these genes translates into decreased proliferation. Furthermore, our studies do not exclude the possibility that GATA4 regulates additional targets involved in controlling proliferation to regulate expansion of the early gut epithelium.
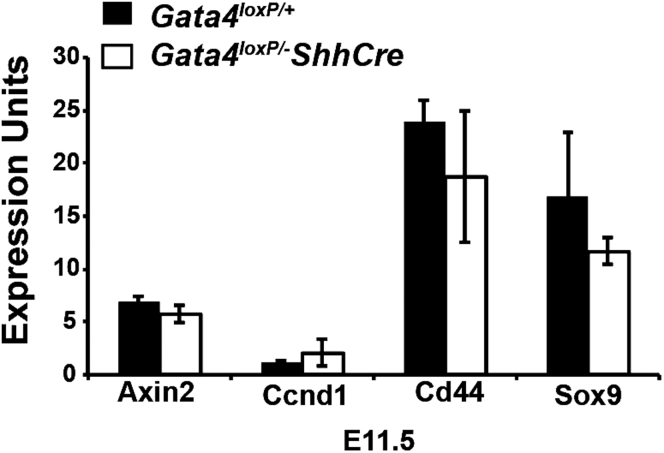
The overlap between GATA4 and WNT5A mutant phenotypes suggested that there could be interplay between these molecules/pathways with respect to epithelial proliferation. Our finding that the WNT5A receptor Fzd5 is a GATA4 transcriptional target provides a possible link. The mechanism through which WNT5A acts on epithelial cells to influence proliferation is unknown, but studies of ROR2 mutants indicate that ROR2 is not required, leading us to focus on FZD5.12 Our data suggest that WNT5A could function through FZD5 to regulate proliferation in the early intestine. Supporting a role for FZD5 in regulating proliferation, endothelial cell proliferation is reduced in the Fzd5 null yolk sac.28 In adult FZD5 mutant intestine, Paneth cell localization is altered.29 FZD5 function has not yet been evaluated during early intestinal development. We propose that in GATA4 mutants, reduced levels of Fzd5 affect WNT5A signaling, thereby providing another possible mechanism through which GATA4 regulates proliferation. Whether WNT5A–FZD5 signaling in the early intestine occurs through canonical or noncanonical pathways to affect proliferation is unclear. Wnt5a null intestine shows no change in activity of a canonical WNT reporter compared with controls, and the abundance of canonical WNT targets Axin2, Lef1, and Tcf1 is unchanged in WNT5A mutants.7 We investigated canonical WNT signaling in GATA4 mutant intestine at E11.5 by examining levels of canonical target transcripts and found no change in Axin2, Cd44, Ccnd1, or sry-box 9 levels in GATA4 mutants compared with controls (Figure 11). Together, these data suggest that WNT5A-FZD5 does not affect proliferation in the early intestine via canonical WNT signaling. Our studies provide a rationale for future studies to investigate the relationship between FZD5 and WNT5A and the mechanisms through which FZD5 affects epithelial proliferation during intestinal development.
Figure 11.
Expression of canonical WNT targets was normal in GATA4 mutant intestine at E11.5. Quantitative reverse-transcription PCR was used to determine expression of Axin2, Ccnd1, Cd44, and sry-box 9 (Sox9) in RNA from whole intestine at E11.5 (n = 4 controls, 3 mutants). Error bars show SEM. P values were determined by 2-sample Student t test. All P values computed were P > .05.
Between E9.5 and E11.5, we observed a dramatic increase in the percentage of proliferating cells in control epithelium, which was maintained through E14.5 (Figure 4). During this time, the epithelium transforms from simple to pseudostratified.4 Epithelial pseudostratification likely occurs in highly proliferative tissues as an adaptation to accommodate greater cell number by densely packing elongated cells.4, 5 Correspondingly, intestinal epithelial girth increases.4 In addition to actomyosin-dependent and microtubule-dependent signaling events that reorganize cell shape to alter girth,4 we propose that the number of epithelial cells present in a given space necessarily influences epithelial girth. In an environment with fewer cells, such as GATA4 mutant intestine, space is less constrained, suggesting that cells do not pack as densely, thereby affecting girth.
Between E14.5 and E16.5, the proportion of proliferating cells decreases (Figure 4). During this period, the epithelium undergoes villus morphogenesis, transitioning from pseudostratified to simple columnar. Cytodifferentiation also ensues. Epithelial remodeling may provide a quick and efficient mechanism to extend gut length.1, 4, 5 In GATA4 mutants, decreased proliferation resulted in fewer epithelial cells. Consequently, fewer epithelial cells were available to lengthen the gut, and GATA4 mutant intestine was shorter than control intestine. Villus morphogenesis also was delayed in GATA4 mutants. The events driving initiation of villus morphogenesis are unclear. One explanation for the GATA4 mutant phenotype is that proliferation and villus morphogenesis are interdependent. We could interpret our data to indicate that a critical number of epithelial cells is required to initiate villus morphogenesis, possibly reflecting a requirement for threshold levels of epithelial-derived morphogens essential for villus morphogenesis. GATA4 thereby indirectly would influence villus morphogenesis indirectly by regulating proliferation. Our finding that Shh levels were decreased between E11.5 and E14.5, without evidence for GATA4 direct transcriptional regulation of Shh, supports this interpretation. However, if delayed villus morphogenesis was simply a consequence of cell numbers and morphogen thresholds in GATA4 mutants, we would predict that Pdgfa levels also would have been lower during this period, which we did not observe. A caveat to the interpretation of Pdgfa expression data is that Pdgfa, unlike Shh, also is expressed in nonepithelial cell types. The developing muscle layer expresses Pdgfa.30, 35 Because we used total intestinal RNA to analyze Pdgfa expression between E11.5 and E14.5, it is possible that any decrease in epithelial expression of Pdgfa was masked by its expression in the muscle layer. We propose that compensatory mechanisms likely explain the Shh and Pdgfa increases at E16.5.
Together, studies of GATA4 in ShhCre and VillinCre conditional knockout mice indicate that the requirement for GATA4 during intestinal development changes over time.13, 25, 33 Gata4 deletion from the midgut endoderm caused a more severe intestinal phenotype compared with Gata4 deletion from the epithelium after E14.5.13, 25, 33 Proliferation was uniquely altered in Gata4–ShhCre mutant intestine, suggesting that it is controlled differently during early and late intestinal development. Overlapping function between GATA4 and GATA6 or GATA4 and CDX2 could explain why loss of GATA4 at later stages does not alter proliferation, and compound GATA4–GATA6 and GATA4–CDX2 mutants each show proliferation defects.23, 33, 36 When both GATA4 and GATA6 are deleted in the developing intestinal epithelium with VillinCre, proliferation is increased.23 Deletion of GATA4-GATA6 or GATA4-CDX2 in the adult intestine using tamoxifen-inducible VillinCre reduces crypt cell proliferation.36, 37 It is unclear why GATA6 and CDX2 compensate for GATA4 function in late embryonic and adult intestine, but not in early intestine. One interpretation is that GATA4 uniquely regulates proliferation during this discrete developmental window.
Our previous study of intestine from Gata4Gata6–VillinCre double conditional knockout embryos provides evidence that GATA factors influence Notch signaling and epithelial cytodifferentiation because we identified GATA factors as direct regulators of the Notch ligand Dll1.23 In GATA4–GATA6–deficient intestine at E18.5, Dll1 expression was reduced, Olfm4 expression was decreased 8.7-fold, and goblet cells were increased 4.3-fold.23 Hes1 and Hes6 levels, however, remained normal in double mutants.23 Dll1 knockout intestine also contains increased goblet cells, and expression of at least one Notch target, Hes5, remained unchanged.38 Similarly, in Gata4–ShhCre mutant intestine, we observed variable outcomes in Notch target gene expression with Olfm4 decreased and Hes1 and Hes6 increased. Downstream of Notch signaling is Atoh1.1 When Notch signaling is high, Atoh1 is repressed.1 Conversely, low Notch signaling is permissive for Atoh1 expression, and secretory cell differentiation occurs.1 Both Atoh1 and goblet cell number were increased in GATA4 mutant intestine, suggesting that despite discordant changes in Notch target gene expression, Notch signaling was lower in GATA4 mutant tissue. It is clear from these studies that Notch signaling in the developing intestine is complex. Future studies should aim to precisely map the molecular mechanisms of Notch signaling in the developing intestine so that the significance of changes in expression of Notch targets including Hes1, Hes6, and Olfm4 can be better interpreted. Finally, because we identified Dll1 as a GATA target gene in intestine of Gata4Gata6–VillinCre double conditional knockout embryos, we examined Dll1 expression in E16.5 Gata4–ShhCre mutant intestine. We found its expression to be unchanged between controls and mutants (data not shown). It is possible that GATA6 compensates for loss of GATA4 in regulating Dll1 expression in Gata4–ShhCre mutant intestine. This could provide an explanation for the less severe secretory cell phenotype observed in single GATA4 knockouts compared with double GATA4–GATA6 knockouts.
One strategy to increase our understanding of the etiology of congenital forms of SBS and to discover novel therapies for SBS is to determine the cellular and molecular mechanisms of intestinal development, particularly those relevant to intestinal growth and elongation. Through a better understanding of the fundamental mechanisms of intestinal development, it may be possible to develop methods to restore intestinal tissue in patients with SBS or to develop methods to generate much needed intestinal tissue in vitro that ultimately could be used for transplantation. Our work shows that GATA4 is an essential regulator of intestinal development. We provide insight into mechanisms controlling intestinal epithelial cell proliferation during early development and the initiation of villus morphogenesis. We conclude that GATA4 regulates proliferation, at least in part, by regulating transcription of the Ccnd2, Cdk6, and Fzd5 genes. We identified novel tissue-specific GATA4 regulation of cell-cycle mediators and suggest that GATA4 affects WNT signaling by contributing to transcriptional regulation of Fzd5. The relationship between GATA4, FZD5, and WNT5A will be explored further in future studies. Moreover, we propose that by regulating proliferation, GATA4 influences the onset of villus morphogenesis, suggesting a novel relationship between proliferation and villus morphogenesis. We also detected abnormal Notch signaling in GATA4 mutants, supporting the proposal that GATA4 and Notch signaling interact during intestinal development. Finally, our studies of GATA4 using ShhCre and VillinCre show that GATA4 function changes during the course of intestinal development.
Acknowledgments
The authors thank Drs Stephen Krasinski and Boaz Aronson (Division of Gastroenterology and Nutrition, Department of Medicine, Children’s Hospital Boston, and Harvard Medical School, Boston, MA) for providing Gata4Flag-biotin and Wt/BirA mouse lines and bio-ChIP-Seq data. The authors also thank Dr Stephen Duncan (Medical University of South Carolina, Charleston, SC), Dr Brian Link (Medical College of Wisconsin, Milwaukee, WI), Dr Sid Rao (Medical College of Wisconsin, Milwaukee, WI), and Dr Jason Spence (University of Michigan, Ann Arbor, MI) for helpful discussions and input into the manuscript. The authors thank Dr Meijun Du (Medical College of Wisconsin, Milwaukee, WI) and Dr Liang Wang (Medical College of Wisconsin, Milwaukee, WI) for expert advice and technical assistance.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by grants from the US National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (DK087873), the American Gastroenterological Association Foundation Research Scholar Award, and Advancing a Healthier Wisconsin (M.A.B.).
Supplementary Material
References
- 1.Thompson C.A., Battle M.A. Basic science of small intestinal development. In: Gumucio D.L., Samuelson L.C., Spence J.R., editors. Translational gastroenterology: organogenesis to disease. John Wiley & Sons; Hoboken, NJ: 2014. p. 85. [Google Scholar]
- 2.Gutierrez I.M., Kang K.H., Jaksic T. Neonatal short bowel syndrome. Semin Fetal Neonatal Med. 2011;16:157–163. doi: 10.1016/j.siny.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Helmrath M.A., Deonarine K. Clincal small intestine. In: Gumucio D.L., Samuelson L.C., Spence J.R., editors. Translational gastroenterology: organogenesis to disease. John Wiley & Sons; Hoboken, NJ: 2014. p. 99. [Google Scholar]
- 4.Grosse A.S., Pressprich M.F., Curley L.B. Cell dynamics in fetal intestinal epithelium: implications for intestinal growth and morphogenesis. Development. 2011;138:4423–4432. doi: 10.1242/dev.065789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee H.O., Norden C. Mechanisms controlling arrangements and movements of nuclei in pseudostratified epithelia. Trends Cell Biol. 2013;23:141–150. doi: 10.1016/j.tcb.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Bakker E.R., Raghoebir L., Franken P.F. Induced Wnt5a expression perturbs embryonic outgrowth and intestinal elongation, but is well-tolerated in adult mice. Dev Biol. 2012;369:91–100. doi: 10.1016/j.ydbio.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Cervantes S., Yamaguchi T.P., Hebrok M. Wnt5a is essential for intestinal elongation in mice. Dev Biol. 2009;326:285–294. doi: 10.1016/j.ydbio.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao N., White P., Kaestner K.H. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geske M.J., Zhang X., Patel K.K. Fgf9 signaling regulates small intestinal elongation and mesenchymal development. Development. 2008;135:2959–2968. doi: 10.1242/dev.020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao J., Kim B.M., Rajurkar M. Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development. 2010;137:1721–1729. doi: 10.1242/dev.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama M., Aizawa S., Shimono A. Sfrp controls apicobasal polarity and oriented cell division in developing gut epithelium. PLoS Genet. 2009;5:e1000427. doi: 10.1371/journal.pgen.1000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada M., Udagawa J., Matsumoto A. Ror2 is required for midgut elongation during mouse development. Dev Dyn. 2010;239:941–953. doi: 10.1002/dvdy.22212. [DOI] [PubMed] [Google Scholar]
- 13.Walker E.M., Thompson C.A., Kohlnhofer B.M. Characterization of the developing small intestine in the absence of either GATA4 or GATA6. BMC Res Notes. 2014;7:902. doi: 10.1186/1756-0500-7-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watt A.J., Zhao R., Li J. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev Biol. 2007;7:37. doi: 10.1186/1471-213X-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harfe B.D., Scherz P.J., Nissim S. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Driegen S., Ferreira R., van Zon A. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res. 2005;14:477–482. doi: 10.1007/s11248-005-7220-2. [DOI] [PubMed] [Google Scholar]
- 17.He A., Shen X., Ma Q. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molkentin J.D., Lin Q., Duncan S.A. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 19.Watt A.J., Battle M.A., Li J. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bondow B.J., Faber M.L., Wojta K.J. E-cadherin is required for intestinal morphogenesis in the mouse. Dev Biol. 2012;371:1–12. doi: 10.1016/j.ydbio.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aronson B.E., Rabello Aronson S., Berkhout R.P. GATA4 represses an ileal program of gene expression in the proximal small intestine by inhibiting the acetylation of histone H3, lysine 27. Biochim Biophys Acta. 2014;1839:1273–1282. doi: 10.1016/j.bbagrm.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He A., Pu W.T. Genome-wide location analysis by pull down of in vivo biotinylated transcription factors. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb2120s92. Chapter 21:Unit 21.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker E.M., Thompson C.A., Battle M.A. GATA4 and GATA6 regulate intestinal epithelial cytodifferentiation during development. Dev Biol. 2014;392:283–294. doi: 10.1016/j.ydbio.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris K.S., Zhang Z., McManus M.T. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Battle M.A., Bondow B.J., Iverson M.A. GATA4 is essential for jejunal function in mice. Gastroenterology. 2008;135:1676–1686.e1. doi: 10.1053/j.gastro.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rojas A., Kong S.W., Agarwal P. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherr C.J. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa T., Tamai Y., Zorn A.M. Mouse Wnt receptor gene Fzd5 is essential for yolk sac and placental angiogenesis. Development. 2001;128:25–33. doi: 10.1242/dev.128.1.25. [DOI] [PubMed] [Google Scholar]
- 29.van Es J.H., Jay P., Gregorieff A. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 30.Karlsson L., Lindahl P., Heath J.K. Abnormal gastrointestinal development in PDGF-A and PDGFR-(alpha) deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development. 2000;127:3457–3466. doi: 10.1242/dev.127.16.3457. [DOI] [PubMed] [Google Scholar]
- 31.Walton K.D., Kolterud A., Czerwinski M.J. Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc Natl Acad Sci U S A. 2012;109:15817–15822. doi: 10.1073/pnas.1205669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muncan V., Heijmans J., Krasinski S.D. Blimp1 regulates the transition of neonatal to adult intestinal epithelium. Nat Commun. 2011;2:452. doi: 10.1038/ncomms1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosse T., Piaseckyj C.M., Burghard E. Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol Cell Biol. 2006;26:9060–9070. doi: 10.1128/MCB.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.VanDussen K.L., Carulli A.J., Keeley T.M. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrae J., Gouveia L., He L. Characterization of platelet-derived growth factor-A expression in mouse tissues using a lacZ knock-in approach. PLoS One. 2014;9:e105477. doi: 10.1371/journal.pone.0105477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beuling E., Baffour-Awuah N.Y., Stapleton K.A. GATA factors regulate proliferation, differentiation, and gene expression in small intestine of mature mice. Gastroenterology. 2011;140:1219–1229.e1. doi: 10.1053/j.gastro.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.San Roman A.K., Aronson B.E., Krasinski S.D. Transcription factors GATA4 and HNF4A control distinct aspects of intestinal homeostasis in conjunction with transcription factor CDX2. J Biol Chem. 2015;290:1850–1860. doi: 10.1074/jbc.M114.620211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamataki D., Holder M., Hodgetts C. Delta1 expression, cell cycle exit, and commitment to a specific secretory fate coincide within a few hours in the mouse intestinal stem cell system. PLoS One. 2011;6:e24484. doi: 10.1371/journal.pone.0024484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.