Summary
Chlamydiae and Chlamydiae-related organisms are obligate intracellular bacterial pathogens. They reside in a membrane-bound compartment termed the inclusion and have evolved sophisticated mechanisms to interact with cellular organelles. This review focuses on the nature, the function(s) and the consequences of Chlamydiae inclusions interaction with the endoplasmic reticulum (ER). The inclusion membrane establishes very close contact with the ER at specific sites termed ER-Inclusion membrane contact sites (MCSs). These MCSs are constituted of a specific set of factors, including the C. trachomatis effector protein IncD and the host cell proteins CERT and VAPA/B. Because CERT and VAPA/B have a demonstrated role in the non-vesicular trafficking of lipids between the ER and the Golgi, it was proposed that Chlamydia establish MCSs with the ER to acquire host lipids. However, the recruitment of additional factors to ER-Inclusion MCSs, such as the ER calcium sensor STIM1, may suggest additional functions un-related to lipid acquisition. Finally, Chlamydiae interaction with the ER appears to induce the ER stress response, but this response is quickly dampened by Chlamydiae to promote host cell survival.
Introduction
The Chlamydiae phylum regroups gram-negative obligate intracellular bacterial pathogens of humans and animals. This phylum is sub-divided into three families. The Chlamydiacea family includes the major human pathogens species Chlamydia trachomatis and Chlamydia pneumoniae, which are respectively responsible for ocular and genital infections (Schachter, 1999) and pneumonia (Kuo et al., 1995). The Simkaniaceae and the Waddliaceae families respectively include the Chlamydia-related species Simkania negevensis and Waddlia chondrophila. These emerging pathogens infect humans, animals and amoebae and are respectively linked to human pulmonary infections (Friedman et al., 2003) and miscarriages in humans and abortions in ruminants (Dilbeck et al., 1990, Henning et al., 2002).
Chlamydiae and Chlamydiae-related organisms share a strictly intracellular bi-phasic developmental cycle. In the early stages, infectious bacteria (EB, Elementary Body) enter the cells and reside in a membrane bound compartment, named the inclusion. EBs then differentiate into non-infectious bacteria (RB, Reticulate Body) and undergo several rounds of replication. In the mid-stages, the cycle becomes asynchronous and some RBs start to differentiate back into EBs. In the late stages, EBs are released to allow a second round of infection (Moulder, 1991). Depending on the species, the cycle last 2-3 days (C. trachomatis, C. pneumonia and W. chondrophila) and up to 10 days (S. negevensis). To establish an intracellular niche permissive to bacterial replication, Chlamydia species rely on their ability to manipulate the host cellular environment. The bacterial type III secretion system (T3SS) and T3SS effectors proteins are key to this process (reviewed by (Mueller et al., 2014)). Some effectors are released into the cytosol, while others, belonging to the family of Inc proteins, are inserted into the inclusion membrane (Dehoux et al., 2011, Lutter et al., 2012). The effector-dependent subversion of various cellular pathways allows the Chlamydia inclusion to rapidly evades the endocytic pathway, while interacting with various organelles such as mitochondria, lipid droplets, multi-vesicular bodies, the Golgi and the endoplasmic reticulum (ER) (Reviewed by (Bastidas et al., 2013). Moreover, the interaction of C. pneumoniae with the ER was recently shown (Shima et al., 2015). W. chondrophila also encode a T3SS (Bertelli et al., 2010) and although T3SS effector proteins have yet to be identified, evasion of the endocytic pathway and interaction with mitochondria and the ER have been described (Croxatto et al., 2010, Kebbi-Beghdadi et al., 2011, de Barsy et al., 2013). S. negevensis has been less studied, but the genome sequence revealed the presence of both a T3SS and a type IV secretion system (T4SS) (Collingro et al., 2011) and interaction with the ER has been recently reported (Mehlitz et al., 2014, Pilhofer et al., 2014).
Over the past five years, studies of Chlamydia, Waddlia and Simkania have brought to light the intimate association of their inclusion with the ER. This review focuses on the current knowledge of the nature of this interaction, the bacterial and host factors involved and the biological consequences for the bacteria and the host.
DIRECT CONTACT BETWEEN THE ER AND THE INCLUSION MEMBRANE
C. trachomatis and C. pneumoniae ER-inclusion Membrane Contact Sites (MCSs)
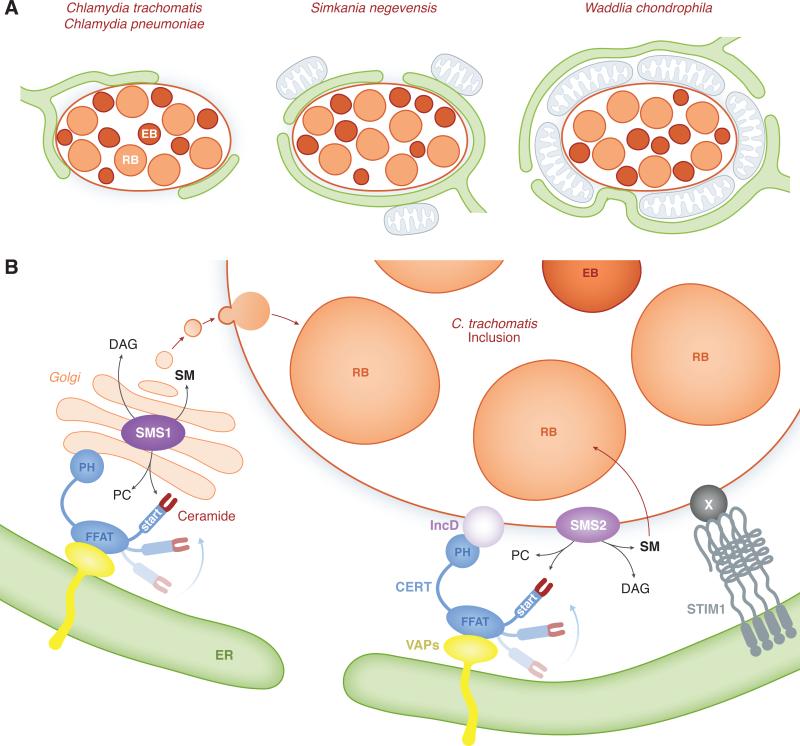
Electron micrographs by Giles et. al. provided the first evidence that ER tubules were present in the close proximity of the C. trachomatis inclusion (Giles et al., 2008). This observation was later on confirmed by independent studies that further characterized the connection between the C. trachomatis inclusion membrane and the ER (Derré et al., 2011, Dumoux et al., 2012). Electron micrographs of C. trachomatis infected cells revealed that the inclusion was covered with several patches of ER located 10-20nm away from the inclusion membrane (Figure 1A, left cartoon). In eukaryotic cells, zones of close apposition (10-50nm) between two organelles are defined as Membrane Contact Sites (MCSs). The ER is often one of the partnering organelle contacting mitochondria, endosomes, the Golgi or the plasma membrane (PM) (Levine et al., 2006, Lebiedzinska et al., 2009, Prinz, 2014). Based on this knowledge, the points of contact between the ER and C. trachomatis inclusion membrane were proposed to represent a novel type of MCSs, named ER-Inclusion MCSs (Derré et al., 2011). These structures have since been recognized as bona fide MCSs (Prinz, 2014) and were also detected during C. pneumoniae productive or persistent infection (Shima et al., 2015) (Figure 1A, left cartoon).
Figure 1. Nature, composition and function of membrane contact sites between Chlamydiae inclusion and the ER.
A. Nature of the interaction between Chlamydiae inclusion and the ER. Schematic representation of C. trachomatis, C. pneumoniae, S. negevensis and W. chondrophila inclusions filled with RBs (light orange) and EBs (dark orange). The ER (green) is found in the vicinity of the inclusion membrane for each species. C. trachomatis and C. pneumoniae inclusion membrane is in close contact with and partially covered by the ER (left cartoon). S. negevensis inclusion is almost entirely covered by and tightly associated with the ER and ER-associated mitochondria (blue) (middle cartoon). W. chondrophila inclusion is surrounded by an inner layer of tightly associated mitochondria, which interact with a dense ER network (right cartoon).
B. Molecular composition of ER-Golgi and ER-Inclusion MCSs and their role in the vesicular and non-vesicular trafficking of lipids to C. trachomatis inclusion. CERT (blue) localize to ER-Golgi MCSs where it participates to the transfer of ceramide from the ER to the Golgi, via the START domain. The FFAT motif of CERT binds to the ER-resident proteins VAPA/B (yellow) and the PH domain of CERT binds to the Golgi. CERT and VAPA/B also localize to ER-Inclusion MCSs. The PH domain of CERT binds to the C. trachomatis inclusion membrane protein IncD (light purple) and the FFAT motif of CERT allows CERT association with VAPA/B on the ER. In addition to CERT, VAPA/B and IncD, the ER Calcium sensor STIM1 (grey) also localize to ER-Inclusion MCSs. A Chlamydia factor X (dark grey) may be involved in this process. At ER-Golgi MCSs, the CERT-dependent transfer of ceramide from the ER to the Golgi-localized Sphingomyelin synthase SMS1 (dark purple) supports Sphingomyelin (SM) synthesis at the Golgi. In the context of Chlamydia infection, Golgi-derived vesicles traffic to the inclusion and SM is incorporated into the inclusion membrane and the cell wall of the bacteria. It is proposed that at ER-Inclusion MCSs, the CERT-dependent transfer of ceramide from the ER to the inclusion membrane-localized Sphingomyelin synthase SMS2 (purple) supports SM synthesis directly at the inclusion membrane. SM would then be incorporated into the bacteria.
C. trachomatis pathogen synapse
Electron tomography confirmed the presence of ER patches in close apposition with C. trachomatis inclusion, but also revealed the presence of host ribosomes onto the cytosolic side of the ER (Dumoux et al., 2012). Moreover, structures similar to ER-Inclusion MCSs, but where ordered arrays of Chlamydia T3SS connected luminal RBs with the inclusion membrane and the apposed rough ER (rER) were also identified and named pathogen synapses (Dumoux et al., 2012).
S. negevensis ER-inclusion and ER-mitochondria MCSs
S. negevensis was observed in a vacuole that is tightly associated with the ER in both amoebae and human cells (Mehlitz et al., 2014, Pilhofer et al., 2014). Electron-cryotomography of infected amoeba revealed that S. negevensis inclusions were almost entirely enveloped by the rER leaving only very small areas of inclusion membrane directly in contact with the cytosol (Pilhofer et al., 2014) (Figure 1A, middle cartoon). It was noted that despite the very tight association of the inclusion with the ER, fusion of the respective membranes were not observed. In human cells, an extensive network of ribosomes studded membranes was also observed in the close proximity of the S. negevensis inclusion (S. negevensis inclusion was referred to by the authors as the Simkania containing vacuole (SCV) because of the different morphology compare to other Chlamydia inclusions) (Mehlitz et al., 2014). Moreover, the association with the ER correlated with a recruitment of mitochondria that were directly in contact with the ER covering the S. negevensis inclusion (Figure 1A, middle cartoon). These structures resemble ER-mitochondria MCSs (Kornmann, 2013) and were observed in infected human cells but not in amoeba.
W. chondrophila mitochondria-inclusion and ER-mitochondria MCSs
Immunofluorescence studies of W. chondrophila infected monocyte-derived human macrophages revealed the recruitment of mitochondria and the ER to the inclusion within the first 8 hours of infection. The mitochondria were recruited first and the association with the ER followed. Ultra-structural analysis revealed that W. chondrophila inclusions were surrounded by an inner layer of mitochondria that directly contacted the inclusion membrane and an outer layer composed of a dense ER network that was closely apposed to the mitochondria and occasionally contacted the inclusion membrane in small areas (Croxatto et al., 2010) (Figure 1A, right cartoon).
Molecular composition of ER-Inclusion MCSs
While some ER resident proteins have been observed in the vicinity of the Chlamydia, Simkania and Waddlia inclusion, studies of C. trachomatis have led to the identification of proteins that are specifically enriched at ER-Inclusion MCSs.
CERT/VAPs/IncD
CERT is a functional component of ER-Golgi MCSs involved in the non-vesicular transfer of ceramide from the ER to the Golgi (Hanada et al., 2003). In addition to the carboxy-terminal START domain (Ponting et al., 1999) that binds ceramide, the ER-to-Golgi transfer requires a central FFAT domain (Loewen et al., 2003) which binds the ER resident proteins VAPA and VAPB (Vesicle-associated membrane protein-associated protein) (Lev et al., 2008) and an amino-terminal PH domain (Lemmon, 2008) which binds PtdIns(4)P and potentially Arf1 at the Golgi membrane (Balla et al., 2006, Hanada et al., 2009) (Figure 1B).
Immuno-fluorescence microscopy revealed that CERT and VAPA/B localized to patches associated with C. trachomatis inclusion membrane as early as 2h post infection (Derré et al., 2011, Elwell et al., 2011). Cryo-electron microscopy and immunogold-labeling confirmed their localization to ER-Inclusion MCSs, with CERT localizing at the interface between the inclusion membrane and the ER tubules and VAPB localizing to the ER (Derré et al., 2011) (Figure 1B).
The CERT/VAPB localization at ER-Inclusion MCSs was therefore reminiscent of the one observed at ER-Golgi MCSs, except that the Golgi membrane was substituted with the inclusion membrane. The molecular mechanism(s) involved in CERT recruitment to C. trachomatis inclusion membrane were further characterized as follows.
Elwell et. al. investigated if CERT association with the inclusion involved PtdIns(4)P, VAP, ARF1 or the binding to ceramide. They showed that single amino acid point mutation preventing CERT ability to bind PtdIns(4)P (G67E) or VAPA/B (D324A) did not affect CERT recruitment to the inclusion. A similar result was observed upon Arf1 inhibition using Exo1. On the contrary, treatment of infected cells with HPA-12, a synthetic analogue of ceramide that inhibits CERT-mediated transfer of ceramide, prevented CERT association to the inclusion membrane (Elwell et al., 2011). The authors concluded that PtdIns(4)P, VAP and Arf1 were not essential for CERT association with the inclusion, while CERT ceramide binding and/or transfer activity was.
Derré et. al. also showed that CERT associated with the inclusion in ARF1-depleted cells, confirming that CERT association with the inclusion was ARF1 independent. They also showed that the PH domain of CERT was necessary and sufficient for CERT association with the inclusion (Derré et al., 2011). Although the PH domain of CERT is involved in PtdIns(4)P binding at the Golgi, the data obtained by Elwell et. al. with CERT G67E mutant and data collected by Derré et. al. demonstrated that in C. trachomatis infected cells, the PH domain of CERT is most likely not required for binding PtdIns(4)P onto the inclusion membrane. In order to identify a potential binding partner for CERT on the inclusion, Derré et. al. conducted pull down experiments of 3xFLAG-CERT from infected cell lysates followed by mass spectrometry analysis. This approach led to the identification of the C. trachomatis inclusion membrane protein IncD. The endogenous IncD protein localized to CERT-positive patches onto the inclusion membrane and co-immunoprecipitation experiments showed that 3xFLAG-IncD and GFP-CERT interacted when co-expressed in eukaryotic cells. Moreover, the PH domain of CERT was identified as the domain mediating IncD/CERT interaction. The direct interaction between IncD and the PH domain of CERT was also shown using purified proteins in vitro (Derré et al., 2011).
The advances in Chlamydia genetics (Wang et al., 2011) allowed for probing the IncD/CERT/VAPB interaction during infection, when IncD-3xFLAG was expressed from C. trachomatis under the control of an anhydrotetracyclin inducible promoter and inserted into the inclusion membrane (Agaisse et al., 2014). In the absence of IncD-3xFLAG expression, CERT localized to discrete patches onto the inclusion membrane, however CERT was massively associated with the inclusion membrane upon (over)-expression of IncD-3xFLAG. This association was dependent on the PH domain of CERT. In addition, the IncD-dependent massive recruitment of CERT to the inclusion correlated with a massive association of VAPB that was dependent on the FFAT domain of CERT (Agaisse et al., 2014). Altogether these results experimentally validated a model in which, at ER-Inclusion MCSs, the C. trachomatis effector protein IncD recruits CERT to the inclusion membrane by direct interaction with CERT PH domain, which mediates the FFAT motif-dependent recruitment of the ER-resident protein VAPB to the inclusion (Figure 1B). It is unclear at this point whether the IncD-CERT-VAP interaction is sufficient to bring the ER in close apposition to the inclusion membrane or if additional factors are required to establish the ER-Inclusion MCSs.
STIM1
The ER calcium sensor, STIM1, and the plasma membrane localized calcium channel, Orai1, play a central role at ER-PM MCSs during store-operated calcium entry (SOCE) (Soboloff et al., 2012, Srikanth et al., 2012, Prakriya, 2013). In resting cells, STIM1 is bound to Ca2+ and localized to the bulk of the ER. Upon ER Ca2+ store depletion, the unbinding of Ca2+ triggers STIM1 oligomerization and redistribution to ER-PM MCSs, where the CAD domain of STIM1 interacts with and activates the Orai1 Ca2+ channel leading to Ca2+ influx and replenishment of the ER store.
Agaisse et. al. showed that, compared to general ER markers, such as Rtn3C and Sec61ß, that appeared evenly distributed throughout the bulk of the ER and in the vicinity of the inclusion, STIM1 was highly enriched in patches at the inclusion membrane (Agaisse et al., 2015). Ultra-structural studies indicated that STIM1 localized to ER structures closely apposed to the inclusion membrane confirming its localization to ER-Inclusion MCSs. A time course of infection revealed that STIM1 associated with the inclusion throughout the developmental cycle. The Ca2+ channel Orai1 was never found in contact with the inclusion, however CERT co-localized with STIM1 at all stages of the developmental cycle (Agaisse et al., 2015). It was therefore suggested that the ER-inclusion MCSs formed during C. trachomatis infection are hybrid MCSs composed of proteins usually found at ER-Golgi or ER-PM MCSs (e.g. CERT/VAP and STIM1 respectively) (Figure 1B).
The CAD domain of STIM1, which mediates the interaction with Orai1, was identified as the minimal domain required for STIM1 association with the inclusion (Agaisse et al., 2015). It was proposed that, as shown for CERT, a Chlamydia effector protein might be involved in the CAD-domain-dependent recruitment of STIM1 to the inclusion.
It was shown that, similar to the situation observed in non-infected cells, STIM1 molecules localized to ER-PM MCSs upon Ca2+ store depletion of C. trachomatis infected cells. Moreover, a pool of STIM1 molecules remained associated with the inclusion after Ca2+ store depletion, suggesting that de novo formation of ER-PM MCSs did not disengage STIM1 from ER-Inclusion MCSs (Agaisse et al., 2015).
Finally, while CERT or VAPA/B depletion have an adverse effect on C. trachomatis intracellular replication (Derré et al., 2011, Elwell et al., 2011), STIM1 depletion was not associated with a growth defect which could suggest redundancy and leaves the role of STIM1 during C. trachomatis infection unanswered (Agaisse et al., 2015).
Chlamydia interaction with the ER and potential connection with lipid transfer and acquisition
Host lipids, such as sphingomyelin (SM) and cholesterol, are essential for Chlamydia development (original observation by (Hackstadt et al., 1996) and reviewed by (Elwell et al., 2012)). The bulk of SM is synthetized at the Golgi by the sphingomyelin synthase SMS1, while some SM synthesis also occurs at the PM via SMS2 (Huitema et al., 2004, Tafesse et al., 2007). At the Golgi, SM synthesis results from the CERT/VAP-dependent non-vesicular transfer of the SM precursor, ceramide, from the ER to the Golgi at ER-Golgi MCSs (Figure 1B). In the context of C. trachomatis infection, it was shown by labeling infected cells with fluorescent ceramide, which was subsequently metabolized into fluorescent SM in the Golgi, that SM-containing vesicles trafficked from the Golgi to the inclusion, where SM was transiently incorporated into the inclusion membrane before accumulating in the cell wall of the Chlamydia. These results highlighted the existence of a vesicular-dependent pathway of SM acquisition by C. trachomatis (Hackstadt et al., 1996, Heuer et al., 2009) (Figure 1B).
Three complementary approaches (the pharmacological inhibitor HAP-12, down-regulation of CERT activity through phosphorylation and protein depletion) were used by Elwell et. al. and Derré et. al. to show that CERT inhibition resulted in a decrease in the size of C. trachomatis inclusion and in the numbers of infectious bacteria recovered at the end of the developmental cycle (Derré et al., 2011, Elwell et al., 2011). A similar phenotype was observed upon VAPA/B depletion (Derré et al., 2011) and SMS1 or SMS2 depletion (Elwell et al., 2011). Moreover, while CERT depletion did not seem to affect the trafficking of fluorescently labeled SM to the inclusion (Derré et al., 2011), HAP-12 treatment had a strong inhibitory effect (Elwell et al., 2011). The decrease of C. trachomatis replication and lipid trafficking to the inclusion described above could originated from interference with the CERT/VAP-dependent transfer of ceramide at ER-Golgi MCSs or SM synthesis at the Golgi, which would in turn affect the vesicular pathway of SM acquisition by C. trachomatis (Figure 1B). However, the fact that anti-Ceramide antibodies partially labeled the inclusion membrane (Elwell et al., 2011) and the fact that SMS2, but not SMS1, was recruited to the inclusion membrane ((Elwell et al., 2011), Agaisse and Derré Unpublished), led to propose that, in addition to the vesicular-dependent pathway of SM acquisition, ER-Inclusion MCSs might play a role in the non-vesicular transfer of lipids to C. trachomatis inclusion (Derré et al., 2011, Elwell et al., 2011). In this model (Figure 1B), CERT and VAPA/B would participate to the non-vesicular trafficking of ceramide from the ER to the inclusion membrane, where ceramide would be further synthetized into SM by SMS2 and then transferred to the Chlamydia.
Altogether, these data suggest that C. trachomatis interaction with the ER could facilitate the transfer of lipids from the ER to the inclusion membrane and/or to the bacteria. These lipids could serve as a source of nutrient during bacterial replication. In addition, these lipids could be used to change the lipid composition of the inclusion membrane and avoid immune recognition and/or facilitate/prevent interaction with certain organelles or proteins. It would be interesting to investigate whether additional lipid transfer proteins are recruited to ER-Inclusion MCSs to increase the panel of host lipids that are scavenged during C. trachomatis infection. Finally, while the IncD-dependent recruitment of CERT to the inclusion membrane has been established, how SMS2 associates with the inclusion membrane and how the SM synthetized at the inclusion membrane would be transferred to the bacteria remain to be explored.
CONSEQUENCES OF THE INTERACTION WITH THE ER: MODULATION OF THE ER STRESS RESPONSE
The ER plays a major role in controlling cellular homeostasis and perturbations of ER functions lead the activation of a process called the unfolded protein response (UPR) (reviewed by (Celli et al., 2015) in the context of host-pathogens interaction). UPR relies on the ER membrane localized sensors activating transcription factor 6 (ATF6), inositol requiring enzyme 1 (IRE1) and double-stranded RNA-dependent protein kinase R (PKR)-like ER kinase (PERK) which at steady state are bound to the ER chaperone immunoglobulin protein (BiP), also known as 78kDa glucose related protein (GRP78). If unfolded proteins accumulate within the ER, BiP/GRP78 associates with these unfolded proteins and dissociates from ATF6, IRE1 and PERK leading to their activation and the initiation of the UPR signaling cascades. These signaling events are complex and lead to the increased production of chaperones such as BiP/GRP78, to the phosphorylation of the eukaryotic translation initiation factor-a (elF2a) and to the C/EBP-homologous protein (CHOP) translocation to the nucleus and activation of apoptotic cell death, to the proteolytic cleavage of ATF6 and to the splicing of X-box binding protein 1 (XBP1) mRNA.
The association of pathogens with the ER could have adverse effects on ER functions, especially when, as observed for S. negevensis, the inclusion is massively and tightly associated with the ER. This idea prompt Mehlitz et. al. to investigate the effect of S. negevensis infection on the UPR (Mehlitz et al., 2014). During the early stages of S. negevensis infection, the increase of in BiP/GRP78 mRNA levels was indicative of an initial pulse of UPR, which was however not sustained during the rest of the developmental cycle. Accordingly, in comparison to un-infected cells, S. negevensis infected cells treated with the ER-stress inducers Thapsigargin or Tunicamycin displayed reduced levels of BiP/GRP78 and ERp72 proteins, reduced elF2a phosphorylation and decreased nuclear translocation of CHOP. Finally, S. negevensis replication was affected at high dose of ER-stress inducer. It was therefore proposed that although ER-stress is initially induced upon S. negevensis interaction with the ER, the bacteria is able to down-regulate this response by inhibiting the UPR signaling cascades.
Shima et. al. also investigated the effect of C. pneumoniae infection on the UPR in the context of persistent infection (Shima et al., 2015). Persistent Chlamydia infections are characterized by the formation of small inclusions containing non-infectious but viable enlarged RBs and can be induced by stimuli such as INFγ, antimicrobial treatment or nutrient starvation (Hogan et al., 2004, Wyrick, 2010, Lewis et al., 2014). As observed during S. negevensis infection, BiP/GRP78 expression was also induced during the early phase, but not in the late phase of C. pneumoniae INFγ-induced persistent infection. Interestingly, this effect was not observed in penicillin-induced persistent infection, suggesting that in the case of C. pneumonia, interaction with the ER alone was not sufficient to induce BiP/GRP78 expression. The analysis of UPR-mediating signaling pathways revealed that elF2a was phosphorylated during C. pneumoniae INFγ-induced persistent infection, however ATF6 proteolytic cleavage and XBP1 splicing did not occur. Moreover, the analysis of the UPR induced by the exogenous ER-stress inducer dithiothreitol (DTT) revealed that DTT-mediated phosphorylation of elF2a was attenuated in C. pneumoniae INFγ-induced persistent infection. However in BiP/GRP78 depleted cells, elF2a phosphorylation was increased and an increase in the percentage of apoptotic cells was observed. It was therefore proposed that during the early stages of C. pneumoniae INFγ-induced persistent infection the increase in BiP/GFP78 expression level activates the UPR signaling cascade and that failure to do so result in host cell death.
Conclusion and Perspectives
Over the past five years, it has become clear that Chlamydiae and Chlamydiae-related organisms interact with the ER. The Chlamydiae-containing inclusions do not appear to fuse with the ER, but instead very close contacts, the ER-Inclusion MCSs, are established between the membrane of the inclusion and the ER. How MCSs are established and maintained are open questions. It is possible that as shown for ER-mitochondria MCSs (Kornmann, 2013), special structural components of host and/or bacterial origin play a role in this process. In addition, the bacteria recruit specific host factors. The role of some of these factors, such as CERT and VAPA/B, in the non-vesicular trafficking of lipids suggests that Chlamydia could target the ER to directly harvest host lipids. Several lipid transfer proteins localize to MCSs between the ER and various organelles (Prinz, 2014) and it would be interesting to investigate whether any of them are also recruited to ER-Inclusion MCSs. In addition, the identification of additional factors, such as STIM1, that are recruited at point of contact between the ER and the inclusion, may uncover functions un-related to lipid acquisition. Finally, it appears that Chlamydiae interaction with the ER may have an adverse effect on the host cell by inducing the ER stress response. However, this response is quickly dampened, suggesting that Chlamydiae have evolved specific mechanisms to inhibit the ER stress response. It would be interesting to investigate whether any of the type III secretion effectors are involved in this process.
Altogether the discovery that Chlamydiae interacts with the ER has opened a novel and exciting area of research which will lead to a better understanding of the mechanisms involved in the subversion of cellular organelles by these obligate intracellular bacteria.
Acknowledgements
I thank Hervé Agaisse for critical reading of this manuscript. I.D. is supported by NIH grant R01-AI101441.
Footnotes
I have no conflict of interest to declare.
References
- Agaisse H, Derré I. Expression of the effector protein IncD in Chlamydia trachomatis mediates recruitment of the lipid transfer protein CERT and the endoplasmic reticulum-resident protein VAPB to the inclusion membrane. Infection and immunity. 2014;82:2037–2047. doi: 10.1128/IAI.01530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaisse H, Derré I. STIM1 is a novel component of ER-Chlamydia trachomatis inclusion membrane contact sites. PloS one. 2015 doi: 10.1371/journal.pone.0125671. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends in cell biology. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Bastidas RJ, Elwell CA, Engel JN, Valdivia RH. Chlamydial intracellular survival strategies. Cold Spring Harbor perspectives in medicine. 2013;3:a010256. doi: 10.1101/cshperspect.a010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C, Collyn F, Croxatto A, Ruckert C, Polkinghorne A, Kebbi-Beghdadi C, et al. The Waddlia genome: a window into chlamydial biology. PloS one. 2010;5:e10890. doi: 10.1371/journal.pone.0010890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Tsolis RM. Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes? Nature reviews. Microbiology. 2015;13:71–82. doi: 10.1038/nrmicro3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro A, Tischler P, Weinmaier T, Penz T, Heinz E, Brunham RC, et al. Unity in variety--the pan-genome of the Chlamydiae. Molecular biology and evolution. 2011;28:3253–3270. doi: 10.1093/molbev/msr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxatto A, Greub G. Early intracellular trafficking of Waddlia chondrophila in human macrophages. Microbiology. 2010;156:340–355. doi: 10.1099/mic.0.034546-0. [DOI] [PubMed] [Google Scholar]
- de Barsy M, Greub G. Waddlia chondrophila: from biology to pathogenicity. Microbes and Infection. 2013;15:1033–1041. doi: 10.1016/j.micinf.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Dehoux P, Flores R, Dauga C, Zhong G, Subtil A. Multi-genome identification and characterization of chlamydiae-specific type III secretion substrates: the Inc proteins. BMC genomics. 2011;12:109. doi: 10.1186/1471-2164-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derré I, Swiss R, Agaisse H. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS pathogens. 2011;7:e1002092. doi: 10.1371/journal.ppat.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilbeck PM, Evermann JF, Crawford TB, Ward AC, Leathers CW, Holland CJ, et al. Isolation of a previously undescribed rickettsia from an aborted bovine fetus. Journal of clinical microbiology. 1990;28:814–816. doi: 10.1128/jcm.28.4.814-816.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoux M, Clare DK, Saibil HR, Hayward RD. Chlamydiae assemble a pathogen synapse to hijack the host endoplasmic reticulum. Traffic. 2012;13:1612–1627. doi: 10.1111/tra.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell CA, Engel JN. Lipid acquisition by intracellular Chlamydiae. Cell Microbiol. 2012;14:1010–1018. doi: 10.1111/j.1462-5822.2012.01794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, et al. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS pathogens. 2011;7:e1002198. doi: 10.1371/journal.ppat.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman MG, Dvoskin B, Kahane S. Infections with the chlamydia-like microorganism Simkania negevensis, a possible emerging pathogen. Microbes Infect. 2003;5:1013–1021. doi: 10.1016/s1286-4579(03)00188-6. [DOI] [PubMed] [Google Scholar]
- Giles DK, Wyrick PB. Trafficking of chlamydial antigens to the endoplasmic reticulum of infected epithelial cells. Microbes Infect. 2008;10:1494–1503. doi: 10.1016/j.micinf.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. The EMBO journal. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Tomishige N, Yamaji T. CERT-mediated trafficking of ceramide. Biochimica et biophysica acta. 2009;1791:684–691. doi: 10.1016/j.bbalip.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- Henning K, Schares G, Granzow H, Polster U, Hartmann M, Hotzel H, et al. Neospora caninum and Waddlia chondrophila strain 2032/99 in a septic stillborn calf. Veterinary microbiology. 2002;85:285–292. doi: 10.1016/s0378-1135(01)00510-7. [DOI] [PubMed] [Google Scholar]
- Heuer D, Rejman Lipinski A, Machuy N, Karlas A, Wehrens A, Siedler F, et al. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature. 2009;457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. Chlamydial persistence: beyond the biphasic paradigm. Infection and immunity. 2004;72:1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitema K, van den Dikkenberg J, Brouwers JF, Holthuis JC. Identification of a family of animal sphingomyelin synthases. The EMBO journal. 2004;23:33–44. doi: 10.1038/sj.emboj.7600034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebbi-Beghdadi C, Cisse O, Greub G. Permissivity of Vero cells, human pneumocytes and human endometrial cells to Waddlia chondrophila. Microbes Infect. 2011;13:566–574. doi: 10.1016/j.micinf.2011.01.020. [DOI] [PubMed] [Google Scholar]
- Kornmann B. The molecular hug between the ER and the mitochondria. Current opinion in cell biology. 2013;25:443–448. doi: 10.1016/j.ceb.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Kuo CC, Jackson LA, Campbell LA, Grayston JT. Chlamydia pneumoniae (TWAR). Clinical microbiology reviews. 1995;8:451–461. doi: 10.1128/cmr.8.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebiedzinska M, Szabadkai G, Jones AW, Duszynski J, Wieckowski MR. Interactions between the endoplasmic reticulum, mitochondria, plasma membrane and other subcellular organelles. The international journal of biochemistry & cell biology. 2009;41:1805–1816. doi: 10.1016/j.biocel.2009.02.017. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nature reviews. Molecular cell biology. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lev S, Ben Halevy D, Peretti D, Dahan N. The VAP protein family: from cellular functions to motor neuron disease. Trends in cell biology. 2008;18:282–290. doi: 10.1016/j.tcb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Levine T, Loewen C. Inter-organelle membrane contact sites: through a glass, darkly. Current opinion in cell biology. 2006;18:371–378. doi: 10.1016/j.ceb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Lewis ME, Belland RJ, AbdelRahman YM, Beatty WL, Aiyar AA, Zea AH, et al. Morphologic and molecular evaluation of Chlamydia trachomatis growth in human endocervix reveals distinct growth patterns. Frontiers in cellular and infection microbiology. 2014;4:71. doi: 10.3389/fcimb.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. The EMBO journal. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter EI, Martens C, Hackstadt T. Evolution and conservation of predicted inclusion membrane proteins in chlamydiae. Comparative and functional genomics. 2012;2012:362104. doi: 10.1155/2012/362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlitz A, Karunakaran K, Herweg JA, Krohne G, van de Linde S, Rieck E, et al. The chlamydial organism Simkania negevensis forms ER vacuole contact sites and inhibits ER-stress. Cell Microbiol. 2014 doi: 10.1111/cmi.12278. [DOI] [PubMed] [Google Scholar]
- Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KE, Plano GV, Fields KA. New frontiers in type III secretion biology: the Chlamydia perspective. Infection and immunity. 2014;82:2–9. doi: 10.1128/IAI.00917-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M, Aistleitner K, Ladinsky MS, Konig L, Horn M, Jensen GJ. Architecture and host interface of environmental chlamydiae revealed by electron cryotomography. Environ Microbiol. 2014;16:417–429. doi: 10.1111/1462-2920.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends in biochemical sciences. 1999;24:130–132. doi: 10.1016/s0968-0004(99)01362-6. [DOI] [PubMed] [Google Scholar]
- Prakriya M. Store-operated Orai channels: structure and function. Current topics in membranes. 2013;71:1–32. doi: 10.1016/B978-0-12-407870-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. The Journal of cell biology. 2014;205:759–769. doi: 10.1083/jcb.201401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter J. Infection and disease epidemiology. In: Stephens RS, editor. Chlamydia: Intracellular biology, pathogenesis, and immunity. American Society for Microbiology; Wahsington, DC: 1999. pp. 139–170. [Google Scholar]
- Shima K, Klinger M, Schutze S, Kaufhold I, Solbach W, Reiling N, Rupp J. The role of ER-related BiP/GRP78 in IFN-gamma induced persistent Chlamydia pneumoniae infection. Cell Microbiol. 2015 doi: 10.1111/cmi.12416. [DOI] [PubMed] [Google Scholar]
- Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nature reviews. Molecular cell biology. 2012;13:549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S, Gwack Y. Orai1, STIM1, and their associating partners. The Journal of physiology. 2012;590:4169–4177. doi: 10.1113/jphysiol.2012.231522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafesse FG, Huitema K, Hermansson M, van der Poel S, van den Dikkenberg J, Uphoff A, et al. Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. The Journal of biological chemistry. 2007;282:17537–17547. doi: 10.1074/jbc.M702423200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS pathogens. 2011;7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick PB. Chlamydia trachomatis persistence in vitro: an overview. The Journal of infectious diseases. 2010;201(Suppl 2):S88–95. doi: 10.1086/652394. [DOI] [PMC free article] [PubMed] [Google Scholar]