Abstract
The family with sequence similarity 83, member D (FAM83D) gene has been proposed as a new prognostic marker for breast cancer. Here we further evaluate the prognostic significance of FAM83D expression in different breast cancer subtypes using a meta-analysis. Patients with higher FAM83D mRNA levels have significantly decreased overall and metastatic relapse-free survival, particularly in the group of patients with ER-positive, or luminal subtype tumors. We also assessed FAM83D alterations and its prognostic significance across 22 human cancer types using The Cancer Genome Atlas (TCGA). FAM83D is frequently gained in the majority of human cancer types, resulting in the elevated expression of FAM83D. Higher levels of FAM83D mRNA expression are significantly associated with decreased overall survival in several cancer types. Finally, we demonstrate that TP53 mutation in human cancers is coupled to a significant increase in the expression of FAM83D, and that a higher level of FAM83D expression is positively correlated with an increase in genome instability in many cancer types. These results identify FAM83D as a potential novel oncogene across multiple human cancer types.
Keywords: FAM83D, prognosis, human cancer, genetic instability
INTRODUCTION
Cancer is a complex and intrinsically heterogeneous disease in which patients may exhibit similar symptoms, and appear to have the same disease, for entirely different genetic reasons [1, 2, 3]. Microarray and next generation sequencing technologies have been invaluable tools for deconvoluting the heterogeneity and complexity of somatic cancer genetics. These technologies are also facilitating development of a catalogue of genomic changes with which to identify new biomarkers for the diagnosis, prognosis, and prediction of therapeutic response, and the discovery of new therapeutic targets. However, while improvements have been made in the diagnosis and treatment of some cancers, the prognosis and survival for most patients, especially those with metastasis, have not dramatically changed. Therefore, an urgent need exists for new cellular and mechanistic insights into why tumor metastases occur, and for the development of new therapies to improve patient survival and overall quality of life.
The family with sequence similarity 83, member D (FAM83D) gene is located on chromosome 20q, a region that is frequently amplified in various types of human cancer. We recently identified FAM83D as a novel oncogene for breast cancer (BC) by demonstrating that higher FAM83D expression is significantly correlated with shorter disease- and distant metastasis-free surivival in BC patients, and that forced expression of FAM83D in vitro promotes BC cell proliferation, migration and invasion, while FAM83D depletion by shRNA leads to cell death [4]. FAM83D excutes these biological functions at least in part through regulation of the tumor suppressor gene FBXW7 [4]. The fact that FAM83D expression is elevated in hepatocellular carcinoma [5, 6], ovarian cancer [7] and metastatic lung adenocarcinomas [8] suggests the possibility that FAM83D is an oncogene for additional cancer types. In this study, using recently available cancer genomic data from ‘The Cancer Genome Atlas' (TCGA), we investigated the prognostic significance of FAM83D expression across 22 human cancer types.
Genetic instability is a characteristic of most human cancers and is believed to enable acquisition of other cancer hallmarks [9]. Genetic instability in tumors has been characterized at the level of single nucleotide (mutation), small microsatellite sequences and whole chromosomes (aneuploidy) [10, 11]. Although the mechanisms that control small scale changes (mismatch repair mechanisms) [12, 13, 14] and large scale chromosomal changes [10, 11] have been widely studied, much less is known about the mechanisms that control structural defects, deletions, and amplifications. FAM83D interacts with the chromokinesin KID22 and is required for correct chromosome congression in metaphase [15]. In this study, we utilized the TCGA database to assess whether overexpression of FAM83D correlates with genetic instability (e.g. the fraction of cancer genomes with copy number alteration and mutation frequencies).
RESULT
FAM83D expression and overall and metastatic relapse-free survival in breast cancers
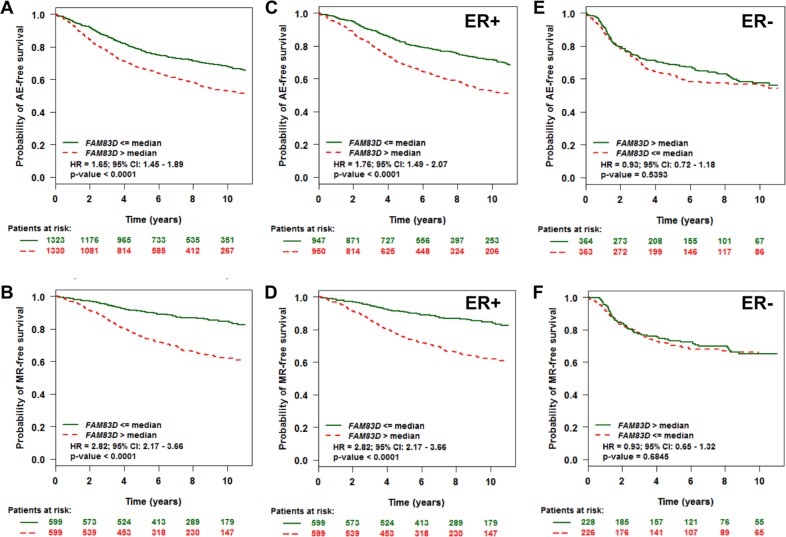
We conducted a meta-analysis of the prognostic significance of FAM83D expression in human BC patients using the Breast Cancer Gene-Expression Miner v3.1 (bc-GenExMiner v3.1) software program [16]. Consistent with a previous report [4], patients with high FAM83D mRNA expression levels (greater than median expression) had significantly decreased overall survival (OS, any event [AE]) in comparison to those with low FAM83D mRNA levels (less than median expression) (Figure 1A, Supplementary Figure S1A). Additionally, high levels of FAM83D mRNA were significantly correlated with decreased metastatic relapse (MR)-free survival (MRFS) (Figure 1B, Supplementary Figure S1B). These results indicate that FAM83D is a statistically significant biomarker for OS and MRFS.
Figure 1. Evaluation of the prognostic impact of FAM83D mRNA expression on any event (AE) - and metastatic relapse (MR)-free survival.
A. Association of FAM83D expression with AE-free survival. B. Association of FAM83D expression with MR-free survival. C–F. Effect of FAM83D expression levels on AE- and MR-free survival according to ER status. “Patients at risk” refers to patients that are at risk of the event occurrence, such as death or metastatic relapse. Kaplan-Meier survival curves for breast cancer patients according to tumor expression of FAM83D are presented. The p values were obtained from a log-rank test among two groups.
FAM83D is an independent marker of disease outcome in ER-positive patients
Estrogen receptor (ER) and nodal status in BC are important predictors of recurrence and greatly influence treatment regimens. We therefore performed univariate Cox proportional hazards model analysis on each of the 18 possible pools corresponding to every combination of population (nodal and ER status) and event criteria (MR or any event [AE]) to assess the prognostic impact of FAM83D expression on patients with different ER and nodal statuses. As summarized in Table 1, we found that high FAM83D expression levels shortened both AE- and MR-free survival only within the groups of ER-positive (ER+) or mixed (ERm) patients, and not within the group of ER-negative (ER−) patients. To further clarify these results, we performed a subset analysis of FAM83D in ER+ and ER- tumors. High levels of FAM83D expression were significantly associated with shorter AE- and MR-free survival in patients with ER+, but not ER- tumors (Figure 1C–1F).
Table 1. Prognostic impact of FAM83D expression level in 18 possible pools corresponding to every combination of populations (nodal and ER status).
| LN | ER | Event status | p-value | Hazard ratio | 95% CI | No patients | No events |
|---|---|---|---|---|---|---|---|
| Nm | Erm | AE | < 0.0001 | 1.34 | 1.26 - 1.43 | 2653 | 909 |
| Nm | ER+ | MR | < 0.0001 | 1.56 | 1.41 - 1.71 | 1198 | 282 |
| Nm | ERm | MR | < 0.0001 | 1.44 | 1.33 - 1.56 | 1672 | 425 |
| Nm | ER+ | AE | < 0.0001 | 1.42 | 1.31 - 1.54 | 1897 | 624 |
| N+ | ERm | MR | < 0.0001 | 1.56 | 1.36 - 1.80 | 439 | 166 |
| N+ | ER+ | MR | < 0.0001 | 1.63 | 1.38 - 1.92 | 343 | 118 |
| N- | ERm | AE | < 0.0001 | 1.38 | 1.23 - 1.55 | 1048 | 299 |
| N- | ER+ | AE | < 0.0001 | 1.5 | 1.29 - 1.75 | 743 | 213 |
| N+ | ERm | AE | < 0.0001 | 1.29 | 1.17 - 1.43 | 911 | 406 |
| N- | ER+ | MR | < 0.0001 | 1.78 | 1.41 - 2.27 | 422 | 78 |
| N+ | ER+ | AE | < 0.0001 | 1.33 | 1.18 - 1.51 | 685 | 284 |
| N- | ERm | MR | 0.0001 | 1.42 | 1.20 - 1.69 | 590 | 111 |
| Nm | ER- | MR | 0.0681 | 1.17 | 0.99 - 1.38 | 454 | 140 |
| Nm | ER- | AE | 0.0878 | 1.12 | 0.98 - 1.27 | 727 | 278 |
| N- | ER- | AE | 0.0909 | 1.23 | 0.97 - 1.56 | 285 | 82 |
| N+ | ER- | MR | 0.3033 | 1.16 | 0.87 - 1.55 | 94 | 48 |
| N- | ER- | MR | 0.7622 | 1.06 | 0.74 - 1.51 | 153 | 32 |
| N+ | ER- | AE | 0.7638 | 1.03 | 0.84 - 1.27 | 223 | 122 |
Node or ER status (+: positive, −: negative, m: mixed); AE: any event; MR: metastatic relapse; HR: hazards ratio.
FAM83D is an independent marker of disease outcome in luminal patients
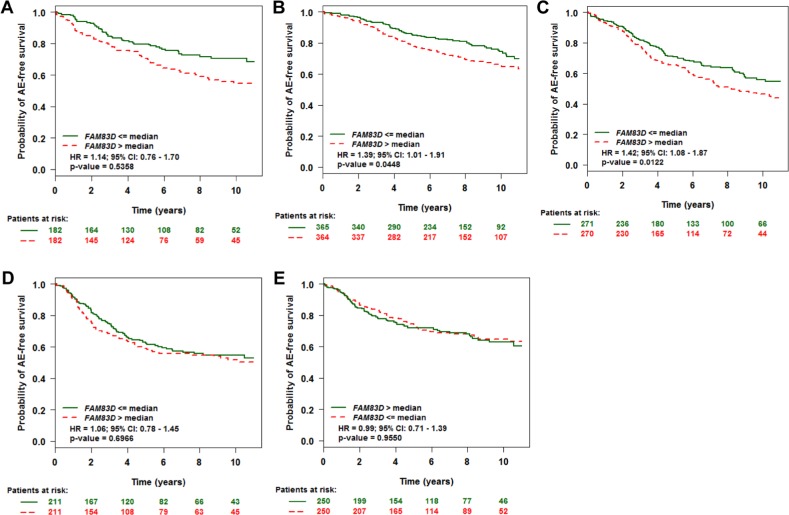
We investigated whether FAM83D could predict disease outcome within the individual molecular subtypes. Tumors were classified into normal-like, luminal A, luminal B, HER2+, and basal-like subtypes based on criteria described by PAM50 [17]. This resulted in samples assigned as normal-like (n = 364), luminal A (n = 729), luminal B (n = 541), HER2+ (n = 422), or basal-like (n = 500). Overall, expression levels of FAM83D were relatively high in luminal B, HER2+ and basal-like tumors, and low in luminal A and normal-like tumors (Supplementary Figure S2). In luminal A and B subtypes, patients with high expression levels of FAM83D presented with significantly decreased AE-free survival (Figure 2B and 2C). Conversely, there was no significant effect of FAM83D expression levels on AE-free survival in the normal-like, HER2+ and basal-like subtype groups (Figure 2A, 2D and 2E).
Figure 2. Effect of FAM83D expression levels on any event (AE)-free survival according to molecular subtypes.
A. Normal-like, B. Luminal A, C. Luminal B, D. ERBB2, and E. Basal subtype. “Patients at risk” refers to patients that are at risk of the event occurrence, such as death or metastatic relapse. Kaplan-Meier estimates of AE-free survival according to the FAM83D expression are presented. The p values were obtained from a log-rank test among two groups.
FAM83D is an independent marker of disease outcome in several human cancer types
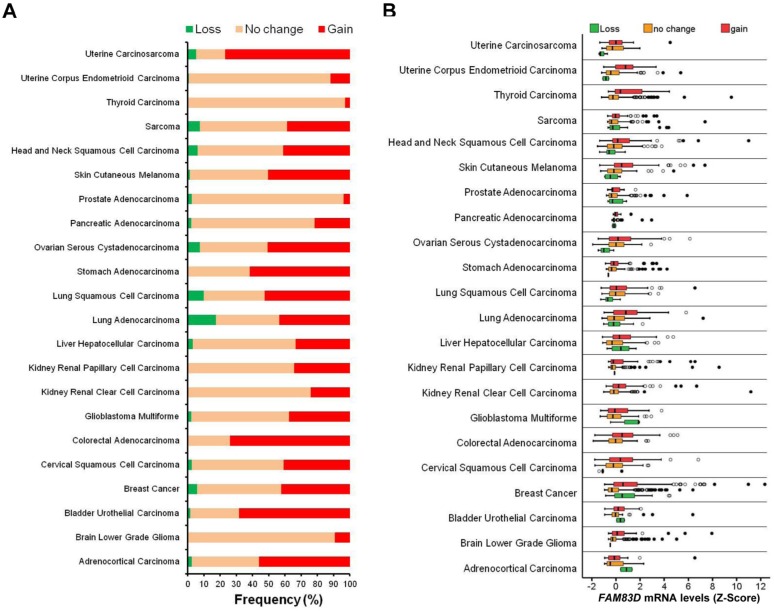
TCGA data was analyzed to determine whether FAM83D might be involved in other cancers (Supplementary Table S1). We found that FAM83D was gained in more than 20% of cases in 18 of 22 human cancer types analyzed (Figure 3A). Expression of FAM83D is significantly higher in tumors with a gain of FAM83D in comparison to those without such changes, suggesting that the gain of FAM83D results in elevated expression of FAM83D (Figure 3B, Supplementary Table S2). In a survey of the data available for BC, lung adenocarcinomas and squamous cell carcinomas, ovarian serous cystadenocarcinoma, head and neck squamous cell carcinoma, and sarcoma, more than 10 cases in which there was a loss of FAM83D were identified (Supplementary Table S2). In lung squamous cell carcinoma and ovarian serous cystadenocarcinoma, FAM83D loss was found to be associated with a significant reduction in FAM83D expression (Figure 3B, Supplementary Table S2). Notably, in the case of BC, FAM83D was expressed at significantly higher levels in cases with FAM83D loss (Figure 3B, Supplementary Table S2), suggesting that additional mechanisms may lead to elevated expression of FAM83D.
Figure 3. Alteration of FAM83D across human cancer types.
A. Frequencies of FAM83D genomic alterations in different human cancer types. B. Relationship between FAM83D mRNA expression levels and genomic alterations.
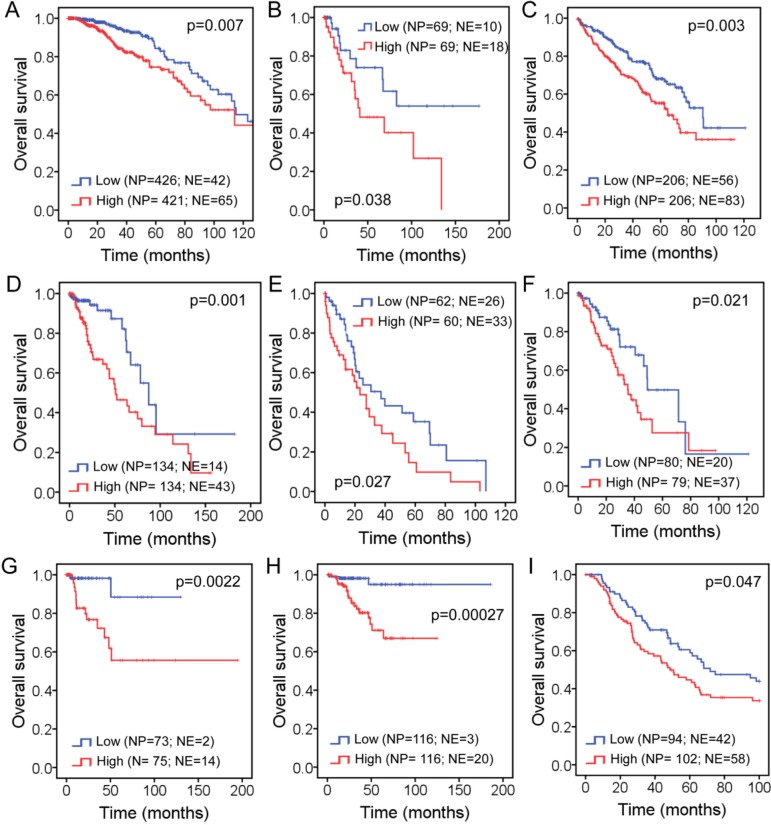
Kaplan-Meier analyses of the TCGA cohort were used to assess the prognostic value of FAM83D expression in different human cancer types. We found that BC patients with higher levels of FAM83D mRNA have significantly shorter overall survival (Figure 4A), which is consistent with the results of a meta-analysis of microarray datasets. In addition, we discovered that higher levels of FAM83D mRNA are also correlated with shorter overall survival in 8 of 16 other human cancer types (Figure 4B to 4I, Supplementary Figure S3). Moreover, it appears that the cervical (Figure 4B), kidney (Figure 4G) and uterine (Figure 4H) carcinomas show a larger difference in overall survival between high and low levels of FAM83D expression in comparison to other types of cancer. Taken together with the results from earlier studies of breast [4] and liver [5] cancers, these latest findings strengthen the proposal that FAM83D is a general oncogene in many different human cancer types.
Figure 4. Impact of FAM83D expression level on overall survival in patients with: A. Breast cancer, B. Cervical squamous cell carcinoma, C. Kidney renal clear cell carcinoma, D. Brain lower grade glioma, E. Liver hepatocellular carcinoma, F. Lung adenocarcinoma, G. Kidney renal papillary cell carcinoma, H. Uterine corpus endometrioid carcinoma, and I. Skin cutaneous melanoma.
“NP” refers to “Number of Patients”, whereas “NE” refers to “Number of Events”. Kaplan-Meier estimates of overall survival according to the FAM83D expression are presented. The p values were obtained from a log-rank test among two groups.
Mutation of TP53 is coincident with increased FAM83D expression in many human cancer types
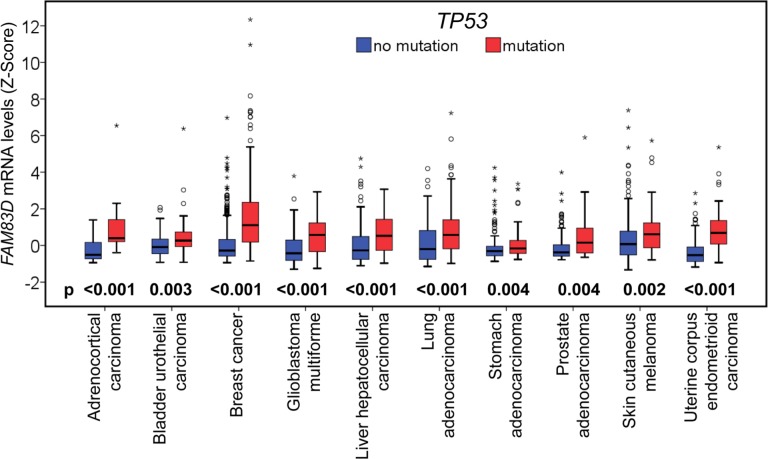
As TP53 is situated at the crossroads of a network of signaling pathways that suppresses cancer development, we sought to investigate the potential relationship between TP53 and FAM83D. 16 of 22 cancer types were used to compare FAM83D expression in TP53 wild-type and mutated cases (Supplementary Table S3). In 10 of 16 cancer types, TP53 mutations coincided with a significant increase in FAM83D expression (Figure 5, Supplementary Table S3), suggesting that TP53 may be a factor in the regulation of FAM83D expression.
Figure 5. Relationship between FAM83D mRNA expression and TP53 mutations.
FAM83D mRNA expression levels were compared between samples with TP53 wildtype (no mutation) and mutant forms. The p-values were obtained from Mann-Whitney U test. “O” signifies an outlier; and “*” an extreme outlier.
FAM83D expression correlates with the level of genomic instability in many human cancer types
FAM83D interacts with the chromokinesin KID22 and is required for correct chromosome congression in metaphase. We therefore investigated whether higher levels of FAM83D expression is correlated with the extent of genome instability in cancer samples. TCGA datasets were used to compare FAM83D expression to the fraction of cancer genomes with copy number alteration (CNA) and mutation frequencies (Table 2). For 8 cancer types, including breast, low-grade brain gliomas and lung adenocarcinomas, we detected a significant positive correlation between both CNA fractions and mutation frequencies, and FAM83D expression level (Spearman's rho, p < 0.05) (Table 2). For some other cancers, such as colorectal carcinoma, kidney renal papillary cell carcinoma, pancreatic and prostate adenocarcinoma, a significant correlation was only detected between FAM83D expression and either CNA fraction or mutation frequency (Table 2). No correlation was detected for cancers, such as kidney renal clear cell carcinoma, cervical, stomach adenocarcinoma, and ovarian serous cystadenocarcinoma (Table 2). Overall, we conclude that there is a significant correlation between FAM83D expression and the level of genome instability in multiple cancer types in the TCGA datasets.
Table 2. Association of FAM83D expression level with the fraction of cancer genomes with copy number alteration and mutation frequencies (number of mutated genes in each sample).
| Cancer types | Mutation frequency | Fraction of copy number altered genome | ||
|---|---|---|---|---|
| Spearman's rho | p-value | Spearman's rho | p-value | |
| Adrenocortical Carcinoma | 0.444 | 6.63E-05 | 0.223 | 0.055 |
| Brain Lower Grade Glioma | 0.197 | 0.00098 | 0.391 | 6.74E-12 |
| Bladder Urothelial Carcinoma | 0.129 | 0.15 | 0.035 | 0.7 |
| Breast Cancer | 0.413 | 1.19E-40 | 0.56 | 2.39E-80 |
| Cervical Squamous Cell Carcinoma | 0.114 | 0.12 | 0.167 | 0.022 |
| Colorectal Adenocarcinoma | −0.071 | 0.34 | 0.166 | 0.026 |
| Glioblastoma Multiforme | 0.013 | 0.88 | 0.237 | 0.0055 |
| Kidney Renal Clear Cell Carcinoma | −0.096 | 0.051 | −0.049 | 0.32 |
| Kidney Renal Papillary Cell Carcinoma | 0.022 | 0.78 | 0.172 | 0.029 |
| Liver Hepatocellular Carcinoma | 0.192 | 0.0078 | 0.319 | 7.42E-06 |
| Lung Adenocarcinoma | 0.336 | 7.88E-06 | 0.259 | 0.00062 |
| Lung Squamous Cell Carcinoma | 0.036 | 0.63 | 0.32 | 0.000013 |
| Stomach Adenocarcinoma | −0.045 | 0.53 | 0.057 | 0.36 |
| Ovarian Serous Cystadenocarcinoma | 0.012 | 0.88 | 0.099 | 0.22 |
| Pancreatic Adenocarcinoma | −0.061 | 0.59 | 0.341 | 0.0016 |
| Prostate Adenocarcinoma | 0.063 | 0.31 | 0.135 | 0.031 |
| Skin Cutaneous Melanoma | 0.146 | 0.015 | 0.219 | 0.00024 |
| Head and Neck Squamous Cell Carcinoma | 0.303 | 2.39E-07 | 0.255 | 0.000016 |
| Sarcoma | NA | NA | 0.419 | 3.04E-12 |
| Thyroid Carcinoma | −0.00045 | 0.99 | 0.108 | 0.031 |
| Uterine Corpus Endometrioid Carcinoma | 0.16 | 0.015 | 0.357 | 2.13E-08 |
| Uterine Carcinosarcoma | 0.294 | 0.028 | 0.162 | 0.23 |
DISCUSSION
In this study, we initially performed a meta-analysis of the public microarray profiles to evaluate the prognostic value of FAM83D expression in BC. Consistent with the results of the previous study [4], we found that higher levels of FAM83D were significantly associated with shorter overall and metastatic relapse-free survival, particularly in patients with ER+ and luminal subtype tumors. As no correlation was found in samples from patients with ER− and other subtype tumors, we conclude that the association between FAM83D expression and overall and metastatic relapse-free survival in the complete patient sample set, was driven by the ER+ and luminal subtype tumors. Using TCGA breast cancer data, we further confirmed that higher levels of FAM83D expression significantly reduce overall survival. These results indicate that FAM83D is a prognostic biomarker for BC.
Three genetic mechanisms activate oncogenes in human cancers: (1) mutation, (2) gene amplification, and (3) chromosome rearrangements. How is the FAM83D oncogene activated? Using TCGA data, we found that FAM83D is rarely mutated, but is frequently amplified in the majority of human cancer types. FAM83D amplification is strongly correlated with an increase in its expression. In addition, we found that TP53 mutations coincide with an increase in the expression of FAM83D. TP53 is widely mutated in various human cancers. Therefore, not surprisingly, it has been reported that FAM83D expression is elevated in various cancers [18]. Moreover, higher levels of FAM83D expression positively correlate with a poor prognosis in many cancer types, including liver hepatocellular carcinoma. Two recent studies have identified FAM83D as a prognostic marker for hepatocellular carcinoma [5, 6].
FAM83D was first identified as a spindle protein localizing with the spindle apparatus during mitosis [15]. FAM83D interacts with chromokinesin KID22 and is required for correct chromosome congression during metaphase [15]. The mitotic spindle is responsible for the accurate distribution of sister chromatids during cell division. Functional aberration of the mitotic spindle can lead to errors in chromosome separation and subsequent aneuploidy as often seen in advanced human cancers. In this study, we have found that high levels of FAM83D expression are strongly correlated with an increase in genomic instability in the cells of multiple cancer types (Table 2). The findings reported here identify FAM83D as a potential oncogene for many human cancer types, and highlight the prognostic value of FAM83D expression in cancer outcomes. Further studies, however, will be needed to develop a deeper understanding of the mechanistic roles of FAM83D in the development and progression of cancer.
MATERIALS AND METHODS
Datasets used in this study
FAM83D genomic alterations and mRNA expression levels, TP53 mutations, fraction of copy number alteration, frequency of gene mutations and clinical information for the set of samples in each TCGA study were obtained from cBioPortal [19, 20]. All samples associated with the datasets analyzed have been included in this study. Further details can be found in Supplementary Tables S1, S2 and S3. Clinical information concerning these samples can be downloaded from cBioPortal (http://www.cbioportal.org/data_sets.jsp).
Statistical analysis
We performed meta-analysis for breast cancer AE-survival, MR-free survival, breast cancer subtype, and breast cancers with clinicopathological information on 36 breast cancer datasets using bc-GenExMiner v3.1 (the website is: http://bcgenex.centregauducheau.fr/BC-GEM/GEM_Requete.php?mode=1) [16]. The analytical tools available for prognostic gene expression analysis in bc-GenExMiner were used to generate Figure 1 and 2 and Table 1, which include (1) Targeted analysis with N and ER subtyping; (2) Exhaustive analysis with N and ER subtyping; and (3) Analysis by molecular subtype. The difference in FAM83D mRNA expression levels between different statuses of FAM83D genomic alteration and TP53 mutation was analyzed by Mann-Whitney U. Kaplan-Meier plots were constructed and a log-rank test was used to determine differences among overall survival according to FAM83D mRNA levels in different cancer types. Spearman correlation was used to assess the association of FAM83D mRNA levels with CNA and mutation frequencies. All analyses were performed by SPSS 11.5.0 for Windows. A two-tailed p-value of less than 0.05 was considered to indicate statistical significance.
SUPPLEMENTARY FIGURES AND TABLES
Acknowledgments
This work was supported by the NIH, National Cancer Institute grant R01 CA116481, and Low Dose Scientific Focus Area, Office of Biological and Environmental Research, U.S. Department of Energy under Contract No. DE AC02-05CH11231.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no competing interests.
REFERENCES
- 1.Allison KH, Sledge GW. Heterogeneity and cancer. Oncology (Williston Park) 2014;28:772–778. [PubMed] [Google Scholar]
- 2.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 3.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Liu Y, Zhang P, Zhang W, Wang W, Curr K, Wei G, Mao JH. FAM83D promotes cell proliferation and motility by downregulating tumor suppressor gene FBXW7. Oncotarget. 2013;4:2476–2486. doi: 10.18632/oncotarget.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao W, Liu W, Liu X, Yuan Q, Ou Y, Qi Y, Huang W, Wang Y, Huang J. Upregulation of FAM83D affects the proliferation and invasion of hepatocellular carcinoma. Oncotarget. 2015;6:24132–24147. doi: 10.18632/oncotarget.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D, Han S, Peng R, Wang X, Yang XX, Yang RJ, Jiao CY, Ding D, Ji GW, Li XC. FAM83D activates the MEK/ERK signaling pathway and promotes cell proliferation in hepatocellular carcinoma. Biochem Biophys Res Commun. 2015;458:313–320. doi: 10.1016/j.bbrc.2015.01.108. [DOI] [PubMed] [Google Scholar]
- 7.Ramakrishna M, Williams LH, Boyle SE, Bearfoot JL, Sridhar A, Speed TP, Gorringe KL, Campbell IG. Identification of candidate growth promoting genes in ovarian cancer through integrated copy number and expression analysis. PLoS One. 2010;5:e9983. doi: 10.1371/journal.pone.0009983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inamura K, Shimoji T, Ninomiya H, Hiramatsu M, Okui M, Satoh Y, Okumura S, Nakagawa K, Noda T, Fukayama M, Ishikawa Y. A metastatic signature in entire lung adenocarcinomas irrespective of morphological heterogeneity. Hum Pathol. 2007;38:702–709. doi: 10.1016/j.humpath.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Giam M, Rancati G. Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Div. 2015;10:3. doi: 10.1186/s13008-015-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer. 2001;1:109–117. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- 11.Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer. 2003;3:695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 12.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 13.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 14.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 15.Santamaria A, Nagel S, Sillje HH, Nigg EA. The spindle protein CHICA mediates localization of the chromokinesin Kid to the mitotic spindle. Curr Biol. 2008;18:723–729. doi: 10.1016/j.cub.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 16.Jezequel P, Frenel JS, Campion L, Guerin-Charbonnel C, Gouraud W, Ricolleau G, Campone M. bc-GenExMiner 3. 0: new mining module computes breast cancer gene expression correlation analyses. Database (Oxford) 2013;2013:bas060. doi: 10.1093/database/bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varisli L. Meta-analysis of the expression of the mitosis-related gene Fam83D. Oncol Lett. 2012;4:1335–1340. doi: 10.3892/ol.2012.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.