Abstract
The transforming growth factor beta (TGFβ) superfamily member Nodal is an established regulator of early embryonic development, with primary roles in endoderm induction, left-right asymmetry and primitive streak formation. Nodal signals through TGFβ family receptors at the plasma membrane and induces signaling cascades leading to diverse transcriptional regulation. While conceptually simple, the regulation of Nodal and its molecular effects are profoundly complex and context dependent. Pioneering work by developmental biologists has characterized the signaling pathways, regulatory components, and provided detailed insight into the mechanisms by which Nodal mediates changes at the cellular and organismal levels. Nodal is also an important factor in maintaining pluripotency of embryonic stem cells through regulation of core transcriptional programs. Collectively, this work has led to an appreciation for Nodal as a powerful morphogen capable of orchestrating multiple cellular phenotypes. Although Nodal is not active in most adult tissues, its re-expression and signaling have been linked to multiple types of human cancer, and Nodal has emerged as a driver of tumor growth and cellular plasticity. In vitro and in vivo experimental evidence has demonstrated that inhibition of Nodal signaling reduces cancer cell aggressive characteristics, while clinical data have established associations with Nodal expression and patient outcomes. As a result, there is great interest in the potential targeting of Nodal activity in a therapeutic setting for cancer patients that may provide new avenues for suppressing tumor growth and metastasis. In this review, we evaluate our current understanding of the complexities of Nodal function in cancer and highlight recent experimental evidence that sheds light on the therapeutic potential of its inhibition.
Keywords: Nodal, embryogenesis, cancer stem cells, pluripotency
1. Introduction
Nodal is a member of the transforming growth factor β (TGFβ) superfamily of embryonic morphogens that coordinate tissue specificity during development. This family, which includes TGFβs, bone morphogenetic proteins (BMPs), and growth differentiation factors (GDFs), regulates a diverse array of functions that are cell and tissue dependent [1, 2]. Ligands of the TGFβ superfamily form dimers via disulfide bonds with pro-domains that are cleaved during activation to yield mature protein. Processed ligands bind to TGFβ superfamily receptors consisting of type I and type II serine/threonine kinases. Binding of ligand induces receptor complex formation and phosphorylation of type I receptor by type II. Type I receptors in turn transmit downstream phosphorylation signals to the Smad family of signaling proteins. Receptor-regulated Smad proteins (R-Smads 1–3, 5, 8) are directly phosphorylated by active receptors and bind a common intermediate, Smad4 (Co-Smad), leading to translocation of this complex into the nucleus where it cooperates with transcription factors to regulate gene expression and cellular function. Smad signaling is highly dynamic and regulated by functional domains that collectively mediate DNA binding, co-factor interactions, nuclear import/export and are subject to ubiquitin and phosphorylation events by a multitude of upstream signaling activities that impart molecular control and connection to other pathways (reviewed in [3–5]). Inhibitory Smads (Smad6, Smad7) antagonize receptor activation of R-Smads and R-Smad/Co-Smad complex formation while phosphatases and nuclear export contribute to the attenuation of Smad signals. In addition, the complexity of TGFβ superfamily signaling is further expanded by the involvement of co-receptors, soluble inhibitors, heterodimerization of ligands, and numerous receptor combinations. As the TGFβ superfamily pathways are crucial to embryonic development, defects can lead to clinical manifestation of disease, while aberrant signaling drives neoplasia [6–8].
Originally discovered in mouse, Nodal plays a crucial role in mesoderm and endoderm formation, left-right patterning, neurogenesis, and maintenance of stem cell pluripotency [9, 10]. The human genome encodes one Nodal gene located on chromosome 10q22.1 that contains three exons. The translated protein constitutes a 26 amino acid signal sequence, 211 amino acid pro-domain, and a 110 amino acid mature protein. The mouse and chick genomes also encode a single Nodal gene, while multiple Nodal genes have been characterized in Zebrafish (cyclops, squint, southpaw) and Xenopus (Xnr 1,2,4–6) [11]. The complex roles of Nodal in embryogenesis and pluripotency have been reviewed in detail in excellent comprehensive reviews to which the reader is referred for these topics [10, 12–17]. This review will summarize components of the Nodal signaling pathway and its effects in developmental biology and stem cell maintenance followed by a more detailed translational review of the role Nodal and its signaling pathway play in cancer.
2. Canonical Nodal signaling and biological roles
Nodal exhibits processing and signaling typical for ligands of the TGFβ superfamily (Figure 1). Nodal is translated as a pro-protein that is cleaved to bind membrane receptors. Specifically, Nodal precursor protein is processed by pro-protein convertases Furin and Pace4 (PCSK6) to yield the mature Nodal protein [18]. Mature Nodal propagates its signal by binding the type I Activin receptors Alk4/7 (ACVR1B/ACVR1C) and type II ActRIIA/ActRIIB (ACVR2A/ACVR2B) in cooperation with its co-receptors Cripto-1 (FDGF1) or Cryptic (CFC1). Cripto-1 and Cryptic are GPI-linked members of the EGF-CFC family that contain an epidermal growth factor motif (EGF) and a cysteine-rich Cripto-1/FLR1/cryptic (CFC) region that binds Alk4 and promotes the recruitment of ActRIIA/B, facilitating formation of the receptor signaling complex [19, 20]. Cripto-1 can also directly bind Furin and Pace4 acting as an organizing center for processing and facilitation of signaling [21]. Cripto-1 and Cryptic mediate the binding of GDF1 and GDF3 to the same receptors as Nodal, whereas other TGFβ superfamily ligands such as Activin-βA and –βB can signal through Alk4 and ActRIIA or ActRIIB without co-receptor requirement but are inhibited by the presence of Cripto-1 [7, 22–26]. Therefore, Cripto-1/Cryptic mediate signaling specificity among TGFβ superfamily ligands and Cripto-1 is also capable of inducing signaling of other pathways [27–32]. Of note however, although Cripto-1 involvement in Nodal signaling is well established, Cripto-1 independent signaling by Nodal has also been demonstrated. Pro-Nodal secreted from the mouse epiblast signals within the extra-embryonic ectoderm and in cell culture experiments binds Alk4 and ActRIIB [33]. Nodal induces Alk7 signaling by direct binding, although at lower levels than in the presence of Cripto-1 [34]. Additionally, Nodal can induce anterior-posterior axis formation in the mouse in the absence of Cripto-1 [35]. However, unlike Cripto-1 or Cryptic mutant mice, Cripto-1/Cryptic double mutant mice phenocopy the developmental defects of Nodal knockout embryos, indicating functional redundancy of the EGF-CFC proteins and highlighting co-receptor dependency during mouse embryogenesis, suggesting that EGF-CFC co-receptor requirement is necessary for the majority of Nodal functions in normal biology [36].
Figure 1.
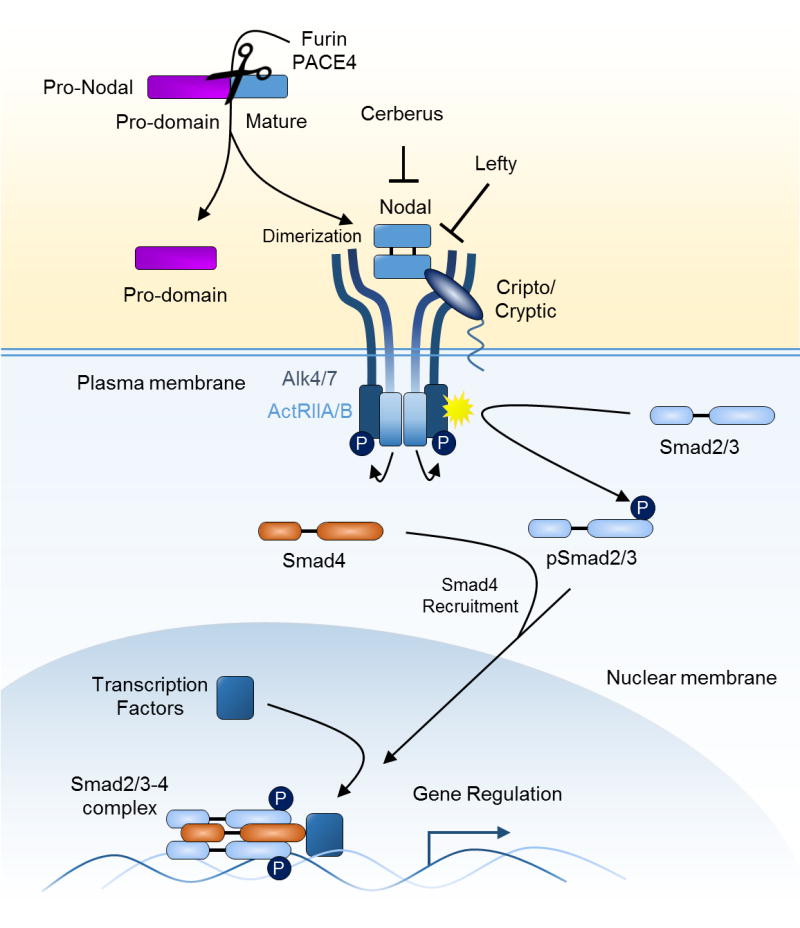
Summary of Nodal processing and canonical signaling. The pro-form of Nodal is cleaved by pro-protein convertases Furin or Pace4 to generate mature Nodal. Mature Nodal forms a dimer via disulfide bonds and binds to type I (Alk4/7) and type II (ActRIIA/B) Activin receptors in combination with its co-receptor Cripto-1 or Cryptic. Extracellular inhibitors, such as Lefty, disrupt the interaction of Nodal with Cripto-1/Cryptic while inhibitors such as Cerberus inhibit signaling by directly binding Nodal. Upon receptor complex formation, constitutively active ActRIIA/B phosphorylates and activates Alk4/7 leading to phosphorylation of Smad2 and/or Smad3. Smad2/3 binds Smad4 and translocates to the nucleus as a trimer of two Smad2/3 and one Smad4 molecules. Within the nucleus, the Smad2/3–4 complex interacts with numerous transcription factors and binds DNA to regulate a diverse array of gene expression that coordinates embryogenesis and maintains pluripotency.
Stimulation of the Nodal receptor complex induces receptor-mediated phosphorylation of Smad2 and/or Smad3 (Smad2/3) which binds Smad4 and translocates to the nucleus. Smad2/3-Smad4 complexes associate with binding partners such as forkhead box H1 (FoxH1) leading to transcriptional regulation of developmental genes such as Goosecoid, Pitx2 and Mixl1 [11]. Nodal is inhibited by soluble extracellular proteins of the Dan family such as Cerberus (Cer1) which bind Nodal and disrupt interaction with receptors, and Lefty (and species specific orthologues), divergent members of the TGFβ superfamily which antagonize Nodal signaling through disruption of interactions with Nodal and EGF-CFC proteins (Cripto-1/Cryptic) but do not interact with Activin receptors and are therefore not competitive inhibitors [37]. Nodal upregulates its own expression through an internal enhancer element responsive to Smad2/FoxH1 binding located within intron 1, as well as the expression of Lefty [38]. During embryogenesis, in vivo models and recent biophysical data indicate that the diffusion of Lefty surpasses that of Nodal, indicating a reaction-diffusion system that causes Nodal to signal locally while inducing inhibition at distant sites [39–41]. Through these actions, Nodal activates a tight auto-regulatory circuit of activity that involves both induction and control of its signaling.
The mature cleaved form of Nodal is highly susceptible to rapid degradation and clearance, supporting the concept that Nodal signaling is a transient and context-dependent process, while its stability is maintained during secretion by a glycosylated pro-domain [42]. In the case of Zebrafish, cyclops has a significantly shorter range of signaling than squint, and these ranges are imparted by differences in the pro-domain of each protein [43–45]. The ability of the pro-domain to stabilize Nodal and restrict its activity is important for the diffusion of Nodal to target cells located at a distance from its source. In cell culture experiments using mouse cells, Nodal is internalized in complex with its receptors and Cripto-1 by endocytosis with markers that overlap with flotilin and sorted to the limiting membrane of endosomes [21, 46]. Association with smad anchor for receptor activation (SARA; ZFYVE9) couples Nodal signaling to the endosomal membrane where Activin receptors phosphorylate Smads on the cytoplastmic side. The EGF domain of Cripto-1 is essential for this trafficking function and in the absence of Cripto-1, mature Nodal is localized to the lumen of endosomes and subject to lysosomal targeting. These discoveries provide a better understanding of Nodal signaling at the subcellular level and are in line with other members of the TGFβ superfamily that signal through similar endocytosis/SARA mediated mechanisms [47, 48]. Nodal activity can also be regulated by association with other members of the TGFβ superfamily, such as BMPs and GDFs, which alter Nodal signaling potency. For example, association with BMP7 results in inactivation of the mature Nodal protein, while association with GDF1 greatly increases the stimulation of Smad phosphorylation [49, 50]. Collectively, the highly regulated and dynamic nature of Nodal activity can be appreciated by its precise and elaborate roles in development.
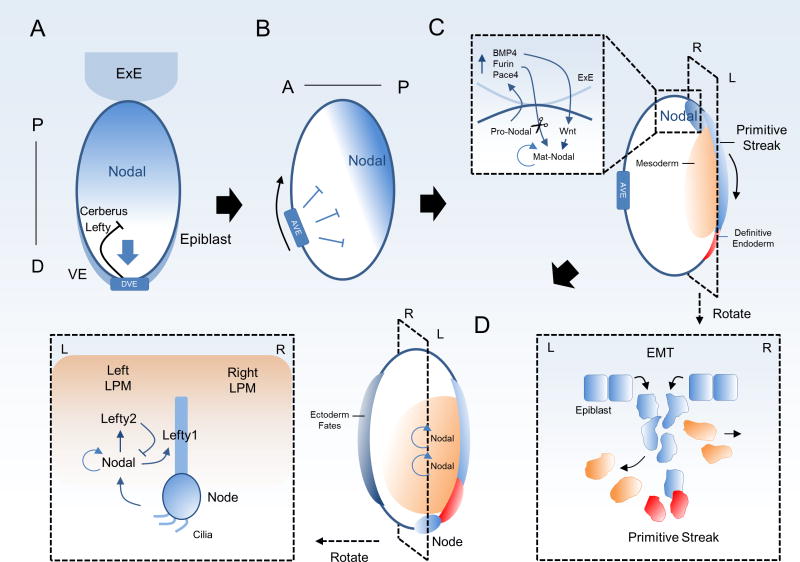
Nodal is involved in the induction of gastrulation, organization of embryonic axes and specification of endoderm and mesoderm lineages in multiple vertebrate species [12, 14, 51–55]. Nodal fulfills these functions, in part, by mediating spatio-temporal signaling and providing positional information for differentiating cells that respond to both dose and duration of Nodal signals [56]. As an example, in the early mouse embryo Nodal cooperates with BMP and Wnt signaling to coordinate proximal-distal and anterior-posterior axis patterning and is required for formation of the primitive streak, a process in which epiblast cells undergo an epithelial-to-mesenchymal transition (EMT) to migrate and give rise to mesoderm and endoderm lineages. These early processes are important for the coordinated movement of embryonic cells to properly initiate gastrulation and form the primary germ layers. During these processes, Nodal mediates signaling within the epiblast, which will form the mature embryo, as well as within extra-embryonic tissues that support embryonic development (for review [14, 15]). Briefly, in the post-implantation embryo, Nodal is expressed throughout the epiblast and promotes posterior cell fates as well as maintenance of the extra-embryonic ectoderm [57]. Nodal also induces Smad2 signaling in the underlying visceral endoderm (VE) to pattern a specialized region known as the distal visceral endoderm (DVE) [15, 57]. Cells of the DVE secrete Nodal inhibitors Cerberus and Lefty which establish a proximal gradient of Nodal signaling in the epiblast, forming one of the earliest embryonic patterns in the mouse (Figure 2A). In a Nodal-dependent process, the DVE migrates anteriorly to become the anterior visceral endoderm (AVE) and localizes Nodal to the posterior side of the developing anterior-posterior axis through continued secretion of inhibitors (Figure 2B). Nodal signaling is concentrated in the proximal posterior epiblast prior to primitive streak formation in a process that involves the activity of pro-Nodal produced in the epiblast to signal in the ExE and induce the production of Furin, Pace4 and BMP4 [33]. Furin and Pace4 in turn process Nodal which amplifies its own signal through its autoregulatory enhancer region, while BMP4 signals back to the posterior epiblast to upregulate Wnt3/β-catenin which activates the Nodal proximal epiblast enhancer region, collectively increasing Nodal expression (Figure 2C inset). High levels of Nodal within this region are necessary for induction of the primitive streak and Nodal deficient embryos are not capable of initiating this process [58]. Epiblast cells entering the streak migrate anteriorly and laterally and undergo complex genetic and transcriptional changes to develop the endoderm and mesoderm lineages (Figure 2C). High levels of Nodal specify endoderm, while cells on the anterior side of the streak take on neuroectoderm characteristics. Following the induction of gastrulation, the primitive node forms near the anterior end of the streak and Nodal is expressed at the periphery of this structure through a node-dependent enhancer element [59–64]. The node acts as an organizing center to orchestrate left-right (LR) asymmetry and organ development in vertebrates [65]. Upregulation of Nodal on the left side of the node is a conserved feature in all examined animal species for asymmetric expression during embryogenesis [66, 67]. This localization of Nodal expression in the left lateral plate mesoderm results from multiple mechanisms including a leftward flow of fluid from active cilia within the node and localization of Nodal inhibitors on the right side [68]. Nodal on the left side upregulates its own expression, as well as that of Lefty1/2 [69, 70]. Specifically, expression of Lefty1 at the dorsal midline barrier and Lefty 2 on the right side restrict Nodal’s range and prevents Nodal signaling from spreading from the left side of the midline to the right (Figure 2D) [68, 70]. Thus, Nodal serves to pattern the asymmetric features of the embryo and defects in Nodal signaling during these processes lead to significant disruption of body axis formation and developmental impairments. Following gastrulation, Nodal is downregulated during somitogenesis, completing its role in organization of the early mouse embryo. Similar gradients and transitory patterns of Nodal signaling have been demonstrated in other vertebrate species, highlighting the powerful ability of Nodal to coordinate morphogenesis and cellular movements during development [71, 72].
Figure 2.
Role of Nodal in the early mouse embryo. A, following implantation, Nodal is expressed throughout the epiblast and plays a role in specifying the underlying distal visceral endoderm (DVE). The DVE in response secretes Nodal inhibitors Cerberus and Lefty, generating a posterior-distal gradient of Nodal and forming one of the earliest patterns in the embryo. ExE, extra-embryonic ectoderm; VE, visceral endoderm; P-D, proximal-distal. B, the DVE migrates anteriorly to become the anterior visceral endoderm (AVE) concentrating Nodal further to the proximal and newly formed posterior axis by continued secretion of Nodal inhibitors. A-P, anterior-posterior. C, in a process that requires high levels of Nodal, gastrulation begins with formation of the early primitive streak. Epiblast cells undergo EMT and migrate with subsequent differentiation into mesoderm and endoderm lineages. Inset: Pro-Nodal expressed in the epiblast upregulates Furin, Pace4 and BMP4 within the ExE. Furin and Pace4 subsequently process pro-Nodal and BMP4 stimulates Wnt3 signaling in the epiblast. Wnt3 acts on a Nodal proximal epiblast enhancer to increase Nodal expression. Nodal also amplifies its own signal through an enhancer region in intron 1 stimulated by Smad2/3-FoxH1, collectively increasing levels of Nodal in the proximal epiblast. D, formation of the primitive node organizes left-right asymmetry. Nodal expressed within the node is concentrated to the left side of the embryo by cooperation of inhibitors and a leftward fluid flow from the Node. Nodal expression is increased through autoregulatory mechanisms and patterns the left-lateral plate mesoderm (LPM) through regulation of morphogenetic genes while activity on the right side is suppressed.
Crucial to proper embryonic development is the sustainment of stem cells that give rise to somatic lineages. Embryonic stem cells regulate a transcriptional profile that involves the expression of core pluripotency genes such as Nanog, Sox2 and Oct4. These genes maintain a stem cell state by activating genes which maintain pluripotency and prepare lineage genes for differentiation through chromatin modifications. “Poised” genes contain transcriptional activating marks such as tri-methylation of histone 3 at lysine 4 (H3K4me3) as well as inhibitory chromatin modifications (e.g. H3K9me3, H3K27me3) that prevent or pause transcription, leaving genes silent but priming them for prompt response to differentiation signals [73]. This allows for both maintenance of the stem cell as well as the ability to rapidly produce differentiated progeny. Numerous reports have demonstrated that Nodal is required for the pluripotency of human embryonic stem cells (hESCs) and prevents differentiation into neuroectoderm lineages [74–80]. Nodal−/− mouse epiblast stem cells exhibit neural differentiation following explant culture in vitro, and Nodal is required for the maintenance of Nanog and Oct4 expression in early mouse epiblast cells for formation of the embryonic visceral endoderm [18, 81, 82]. In line with this, Nodal is required to sustain in vitro pluripotency of mouse and rat epiblast stem cells isolated from post-implantation embryos (which closely resemble hESCs) [83]. Other studies have shown the requirement for Nodal to drive differentiation of mouse and hESC cultures to endoderm and mesoderm lineages [84–87]. Thus, while Nodal is involved in lineage specification, it also maintains the pluripotency of embryonic stem cells. Recent work has begun to elucidate the mechanisms by which Nodal mediates these seemingly contradictory effects. Modulation of transcription by chromatin remodeling and switching of Smad2/3 binding partners during differentiation are key mediators of the process, as discussed below.
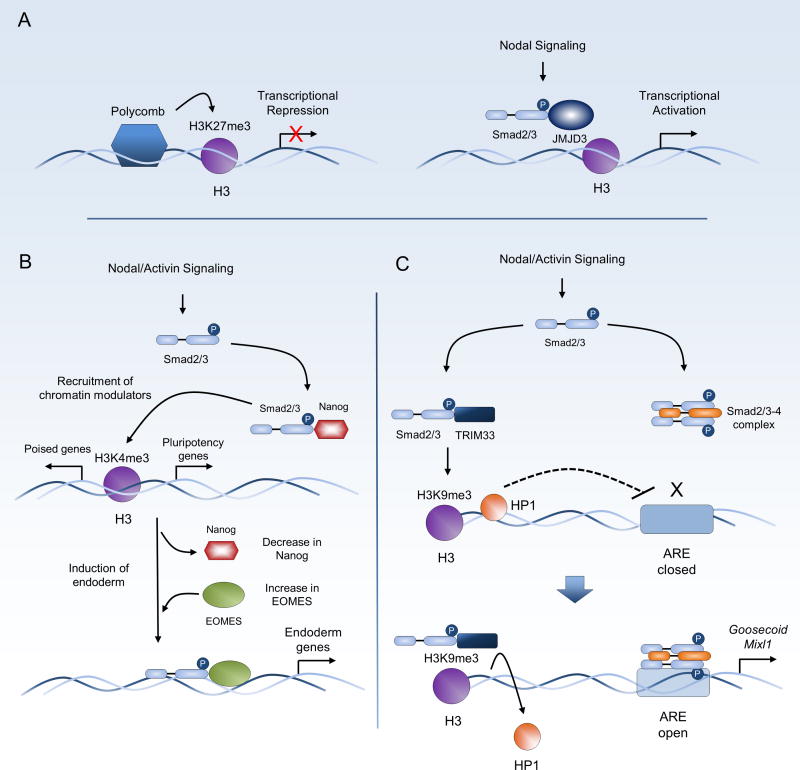
The necessity of chromatin remodeling in response to Nodal has been demonstrated for the induction of Nodal target genes. For instance, activated Smad2/3 in response to Nodal signaling binds the DNA demethylase JMJD3 and relieves Polycomb H3K27me3 mediated chromatin condensation to allow for transcriptional activation of Brachyury and Nodal itself [88]. Furthermore, in hESC, Nanog expression is maintained by Activin/Nodal, and Nanog in turn regulates Nodal signaling intensity by binding Smad2/3 to modulate activity and prevent differentiation to endoderm. Furthermore, Smad2/3-Nanog complexes recruit Compass and DPY30 proteins to deposit transcriptional activating H3K4me3 marks on genes required to maintain pluriopotency, as well as lineage genes that also contain inhibitory H3K27me3 modification. Upon induction of endoderm, Eomedosermin (EOMES), a master regulator of endoderm and mesoderm formation, is upregulated with a progressive decrease in Nanog expression [89]. This leads to a switch from Smad2/3-Nanog complexes to Smad2/3-Eomes which drive the activation of endoderm specification genes such as Sox17 and FoxA2. In another example, Nodal signaling regulates poised chromatin through association of activated Smad2/3 with TRIM33, a Smad binding protein and regulator of transcription [90]. Smad2/3-TRIM33 complexes bind chromatin at regions containing H3K9me3 and H3K18ac and relieve transcriptional repression in nearby loci for the mesendoderm genes Goosecoid and Mixl1 by displacing chromatin condensing protein heterochromatin protein 1 and allowing for binding of Smad2/3-Smad4 complexes. These examples highlight direct chromatin modifications downstream of Nodal that act as molecular switches mediating pluripotency and differentiation (summarized in Figure 3). These studies are further supported by evidence of changes to the Smad2/3 chromatin binding landscape during the switch from pluripotency to differentiation in embryonic stem cells [91, 92].
Figure 3.
Nodal signaling maintains pluripotency and prepares stem cells for differentiation through chromatin modifications and Smad2 binding partners. A, Polycomb tri-methylation of lysine 27 at histone 3 (H3K27me3) represses transcriptional activation. Nodal signaling induces pSmad2 binding of JMJD3 demethylase that removes Polycomb methylation and allows for transcriptional activation [88]. B, Nodal signaling is required for the maintenance of pluripotency through cooperation with Nanog and recruitment of chromatin modifying protein complexes to sustain a stem cell phenotype, in part through the deposition of H3K4me3 marks on active promoters and genes poised for lineage fate determination. Following the induction of differentiation signals, Smad2 complexes associate with EOMES, a master regulator of endoderm formation, to regulate numerous genes involved in endoderm differentiation while expression of Nanog is reduced [89, 170, 171]. C, activated Smad2 binds to TRIM33 and recognizes the inhibitory H3K9me3 mark, displacing heterochromatin protein 1 (HP1). This relieves chromatin condensation at nearby sites and allows for binding of Smad2/3–4 complexes to activin response elements (ARE) to upregulate endodermal differentiation genes such as Goosecoid and Mixer [90].
A recently characterized Nodal enhancer, highly bound element (HBE), provides additional insight into Nodal regulation in embryonic stem cell pluripotency. HBE is activated in the early pre-implantation mouse epiblast and is dependent upon pluripotency factors such as Oct4, Nanog, Klf4 and Sox2 [93]. This regulates Nodal expression prior to implantation and activates the autoregulatory Nodal enhancer element which subsequently drives Nodal expression in the post-implantation epiblast. Removal of HBE in mESC leads to a lack of Nodal expression and increased inhibitory transcriptional histone mark H3K27me3 near the Smad2/FoxH1 responsive element (ASE), suggesting that HBE regulated Nodal expression leads to chromatin changes that relieve the inhibition of the ASE element. Characterization of this novel enhancer provides further molecular detail underlying the regulation of Nodal in stem cells. How chromatin modification downstream of Nodal signaling coupled with enhancer activation collectively unite in dynamic stem cell signaling contexts remain to be fully elucidated. Thus, the continued study of the pathways by which Nodal modifies chromatin to regulate pluripotency and differentiation in stem cells is an area of exceptional importance. Of note, the establishment of Nodal as a key regulator of pluripotency draws attention to its dysregulation in neoplastic progression where cancer stem cells may exploit these authoritative genetic controls for survival and proliferation, as discussed below.
3. Nodal and Cancer
Since the initial demonstration of Nodal’s influence on metastatic melanoma aggressiveness and correlation to melanoma progression in clinical samples, Nodal re-expression in neoplasia has been linked to aggressiveness in numerous cancer types, frequently in the absence of its endogenous inhibitors (e.g. Lefty) [94]. The pro-tumorigenic effects of the re-expression of Nodal protein and its signaling pathway in cancer cells have been well documented and correlated with observations from experiments that have inhibited Nodal by different mechanisms (e.g. chemical inhibitors, Morpholinos, shRNA, neutralizing antibody). For example, reduction of Nodal activity in aggressive breast cancer cells has been shown to decrease their proliferative capacity, clonogenicity and ability to engage in vasculogenic mimicry while increasing basal rates of apoptosis. In addition, the level of Nodal expression has been shown to correlate with aggressiveness in different human breast cancer cell lines while increasing Nodal expression in poorly aggressive breast cancer cells resulted in increased proliferation [95]. Furthermore, reduction of Nodal expression decreased tumorigencity and metastasis of breast cancer cells in multiple in vivo models [95, 96]. Similar associations have been reported for glioma cells [97, 98]. For instance, in glioma cell lines, Nodal expression was shown to correlate with invasive potential and treatment of glioma cells with rNodal increased invasion, proliferation, in vivo tumor growth and production of MMP-2, while knockdown decreased these effects and induced a differentiated phenotype [97]. A multitude of similar in vitro and in vivo work demonstrating reductions in aspects such as invasion, tumor growth, proliferative capacity and promotion of an overall less aggressive phenotype have been described for Nodal in various cancers including melanoma, prostate, pancreatic and ovarian carcinoma (reviewed in [94, 99, 100]).
Clinical correlations with Nodal expression and disease progression continue to be established [99, 100]. Thus, the detrimental effects related to the re-emergence of Nodal in the absence of its endogenous inhibitors, proper transcriptional regulation, and appropriate microenvironmental and embryological contexts, have been well validated. While the mechanistic complexities of Nodal signaling in cancer are being resolved, it remains a difficult problem to fully solve due to the heterogeneous nature of tumors [101, 102].
3.1 Pluripotency in cancer stem cells
Given the role of Nodal in stem cell pluripotency and the influence of cancer stem cells on neoplastic progression, several lines of evidence strongly suggest the presence of stem cell features and cellular plasticity in Nodal positive cancers including the maintenance of self-renewal and association with stem cell markers. In one report examining primary pancreatic cancer stem cells, the Nodal pathway was found to be upregulated in the CD133+ stem-cell like population when compared to more differentiated pancreatic cancer cells [103]. The Nodal signaling pathway in these cells was competent and responded to both recombinant Nodal (rNodal) and Activin, demonstrating strong phosphorylation of Smad2/3, but did not respond to stimulation with TGF-β1. Moreover, inhibition of Nodal in these stem cells inhibited their ability to self-renew, causing a reduction in soft agar colony formation as well as invasive capacity. In a subsequent study, Nodal produced by stromal pancreatic stellate cells promoted the invasive qualities and sphere formation of pancreatic cancer stem cells [104]. Thus, paracrine Nodal activity may act to provide a niche for pancreatic cancer stem cells. In another report, Nodal associated with colon cancer stem cell markers (CD44+/CD24+) in colon cancer cell lines and promoted their self-renewal capacity. This effect was reduced with treatment of SB431542, an Alk4/5/7 inhibitor, suggesting a role for Nodal in the maintenance of colon cancer stem cells. In support of this, higher levels of Nodal and CD44+/CD24+ were observed in colon cancer patient tissues when compared with adjacent non-cancerous tissues [105].
Evidence in testicular germ cell cancers (TGCC) has also provided evidence for the role of Nodal on stem cell phenotypes and neoplastic transitions. Type II TGCC are divided into two categories: seminomas which resemble a less aggressive carcinoma in-situ, and non-seminomas which display a high degree of pluripotency factors [106]. Non-seminomas form highly heterogeneous tumors with both differentiated and undifferentiated cells forming teratomas and choriocarcinomas and display more embryonal-like features. Using a seminoma xenograft model in which seminoma cells are reprogrammed to form carcinomas with more embryonal-like features, expression of key pluripotency and chromatin remodeling genes was examined [107]. Nodal was upregulated during this transition by demethylation of the Nodal promoter. Interestingly, in this model, inhibition of BMP signaling by the microenvironment was linked to the induction of Nodal expression. This study supports previous evidence which demonstrated that Nodal is expressed during mouse germ cell development and regulates pluripotency [108]. Interestingly, Nodal, Cripto-1 and Lefty1 were found to be expressed in human testis cancers that contained carcinoma in-situ, but Lefty1 was absent following progression to malignant embryonal carcinoma, suggesting unabated Nodal signaling during this transition.
3.2 Alternative Signaling Pathways and EMT
TGFβ family signaling through non-Smad proteins (non-canonical signaling) has been demonstrated, although remains poorly understood. This signaling can be mediated by both type I and type II receptors and involves activation of pathways such as p38, JNK, ERK, PI3K and Rho. These signaling pathways can act independently, or in cooperation with Smads [109]. In particular, the ERK and PI3K pathways have been highly implicated in cancer. For example, ERK and TGFβ1 have been shown to act in concert with other factors to induce pathways such as the epithelial-to-mesenchymal transition (EMT), one of the most intensely studied processes in cancer biology [110, 111]. Traits of EMT such as the acquisition of motile phenotypes, loss of cell-cell contacts including down-regulation of E-cadherin protein and gain of a genetic program that induces a mesenchymal phenotype, contribute to the process of metastasis and have been implicated in numerous cancers [112]. The relevance of signaling pathways involved with EMT resides in the notion that many cancer cells are physically and biochemically changing in order to interact with and migrate through the surrounding microenvironment, much like mesenchymal cells [113, 114]. It is interesting to note that the first instance of EMT in mammalian organisms occurs during the formation of the primitive streak, and as previously discussed above, high levels of Nodal are a requirement for this process.
Two recent studies using breast cancer cells have demonstrated a link between the pro-tumorigenic effects of Nodal and the ERK pathway [96, 115]. Knockdown of Nodal in metastatic breast cancer cells led to a reduced aggressive phenotype with decreases in viability, invasiveness and proliferation, as well as reduced clonogenicity, indicating a loss of self-renewal capacity, consistent with a decrease in phosphorylation of Smad3. Nodal knockdown also inhibited tumor formation and growth of these cells in orthotopic murine xenografts. Molecular analysis revealed an impairment to cell cycle progression with reduction of cyclin B1 and upregulation and increased nuclear localization of the cell cycle inhibitor p27. Especially noteworthy in this study was the down-regulation of the proto-oncogene c-myc and global down-regulation of H3K4Me3 chromatin marks (active promoters) with an increase in H3K27Me3 (transcriptional inhibition). A key upstream regulator of both c-myc and p27 is the ERK signaling pathway. It was shown in this study that chemical inhibition of ERK reduced the ability of Nodal to increase c-myc and downregulate p27, suggesting that Nodal was acting to regulate aspects of the cell cycle through ERK, independent of Smad signaling. In support of this, ERK activity localized with Nodal expression by immunohistochemistry in aggressive breast cancer samples, indicating potential clinical connections. In another study, overexpression of Nodal or treatment with rNodal in non-aggressive estrogen receptor (ER) positive breast cancer cell lines reduced ER and caused a loss of E-cadherin localization from the plasma membrane [115]. Overexpression of Nodal also upregulated mesenchymal markers including Vimentin, N-cadherin and Twist1, inducing an EMT phenotype that was mediated by signaling through the ERK pathway. While rNodal stimulation increased the invasive abilities of these cell lines, the presence of an ERK inhibitor disrupted the inductive effect of rNodal on Twist1 and Vimentin and E-cadherin re-localized to the membrane. Reciprocal experiments in aggressive breast cancer cells expressing high activity of the Nodal pathway by knocking down Nodal with shRNA reduced Twist1 and Vimentin and inhibited cellular invasion and migration. Collectively, these studies demonstrate the ability of Nodal to induce cell cycle changes and promote EMT by signaling through the ERK pathway. In support of these studies in breast cancer, overexpression of Lefty2 in glioma cells is capable of reducing signaling through ERK [116]. Additional studies addressing the upstream molecular detail of Nodal-dependent ERK activation are of interest, but the downstream molecular and functional effects of this stimulation are readily apparent.
Studies in other cancer types have also reported induction of alternative signaling and/or EMT phenotypes related to Nodal signaling. Treatment of B16 murine melanoma cells with rNodal induced EMT characterized by an increase in migratory capabilities and increase in Vimentin, Fibronectin and a decrease in E-cadherin, while blocking Nodal signaling inhibited these effects [117]. Additionally, overexpression of endogenous Nodal also induced EMT in these cells while knock-down inhibited the process [118]. Specifically, Nodal stimulation led to expression of Snail and Slug, key transcription factors in the EMT process that mediate their effects, in part, through transcriptional downregulation of E-cadherin and upregulation of Vimentin. Furthermore, induction of Snail and Slug were partially regulated by non-canonical Nodal signaling through the PI3K/AKT pathway [117]. In both studies, inhibiting Snail reduced the induction of EMT in response to increased Nodal levels. Similar to these reports, an increase in Snail was observed following rNodal treatment of pancreatic cancer cell lines. This stimulation led to increases in the mesenchymal markers N-cadherin and Vimentin and down-regulation of E-cadherin [119]. Nodal treatment also induced expression of the malignancy related extracellular protease MMP-2, implicated in the breakdown of extracellular matrix, and CXCR4, a chemokine receptor involved in metastasis, collectively leading to a more aggressive phenotype.
Hypoxia is an effect commonly encountered within solid tumors due to decreased capillary penetration of growing tumors and induces a shift towards the use of glycolytic pathways by cancer cells. High glycolytic activity is characteristic of cancer stem cells and activates proteins such as hypoxia inducible factors (HIFs) that respond to low oxygen levels. HIFs induce the transcription of genes needed for the induction of new blood vessels and promote anaerobic metabolism. A state of hypoxia has been shown to support cancer stem cell growth, as well as maintain self-renewal capacity through genes such as Oct4 and Nanog. Interestingly, in breast cancer and melanoma cells, hypoxia upregulated Nodal protein levels [120]. This induction was shown to be mediated by HIFs enhancing Notch-mediated transcription of Nodal through the node dependent enhancer which is normally active within the primitive node during left-right asymmetry in embryogenesis. This Nodal upregulation led to increases in migration, invasion and induction of endothelial tube formation in response to conditioned media from breast cancer cells grown in hypoxia and blocked by shRNA to Nodal. A reciprocal but mutual connection between HIF1α and Nodal seems to be present in glioma. Using human glioma cells, hypoxic conditions led to Nodal-dependent induction of HIF1α expression. The increase in HIF1α transitioned cells to a glycolytic state and decreased mitochondrial energy production. Consistent with these results, increased HIF1α upregulated numerous genes involved in glycolysis including glucose transporter-1 and pyruvate dehydrogenase-1 with a corresponding increase in glucose uptake and lactate production [121]. Moreover, Nodal mediates HIF1α in part through ERK1/2 signaling in glioma cells. Thus, while the induction and feedback of this connection may be cell type specific, it appears that HIFs and Nodal may cooperate to induce a shift to glycolysis and maintain the pluripotency and survival of cancer stem cells in response to hypoxia.
In addition to diverse activation of multiple downstream pathways, further complexities in Nodal signaling have recently been demonstrated at the receptor level in metastatic melanoma cells. When hESC receptor profiles were compared to metastatic melanoma cells, both cell types expressed Alk4, but metastatic melanoma cells exhibited little to no expression of ActRIIA or ActRIIB, whereas TGFβRII was present at high levels in melanoma cells, with little expression in hESCs. Thus, these cell types appeared to diverge in their TGFβ family type II receptors. In vitro binding assays and receptor/ligand crosslinking experiments with melanoma cells demonstrated the binding of Nodal to TGFβRII. Interestingly, antibody neutralization of either Nodal or TGFβRII led to attenuation of each protein, suggesting potential feedback loops between these two proteins. Consistent with this finding, transfection of a dominant negative TGFβRII construct decreased Nodal levels and phosphorylation of Smad2/3. Thus, these data suggest that in metastatic melanoma cells, a lack of expression of ActRIIA/B causes a shift in Nodal to TGFβRII and TGFβRI (Alk5) binding. This is the first report to demonstrate the utilization of a TGFβRII/TGFβRI complex by Nodal and adds to the intricacies of alternative signaling pathways of Nodal in cancer [122]. This work also highlights potential mechanisms by which therapeutics that target members of the Nodal signaling pathway, rather than Nodal itself, may be bypassed.
3.3 Epigenetic influence of an embryonic microenvironment on Nodal signaling in metastatic tumor cells
One of the most striking features of Nodal in metastatic cells is the influential relationship between Nodal positive tumors and the embryonic microenvironment, as discussed below. The multipotent tumor cell in cancer and pluripotent embryonic stem cell demonstrate levels of plasticity that can lead to their differentiation into various cell types, and the bi-directional communication between cells and their microenvironment can modulate and determine both the fate and behavior of these cells [123–141]. While these observations suggest a commonality in processes associated with cell signaling and fate determination between the areas of developmental biology and cancer biology, this relationship was strengthened by the identification of tumor specific stem cell populations in some cancers [124–128]. This concept led to studies focused on the bi-directional communication between cancer cells with an embryonic microenvironment and has yielded significant insight into mechanisms associated with normal versus aberrant cell behavior and cell fate determination in cancer. For example, an early study showed that the tumorigenic phenotype of teratocarcinoma cells was suppressed when introduced into a mouse embryonic blastocyst microenvironment while the tumor cells themselves contributed to the formation of normal tissue, which was interpreted as a demonstration of the cells’ developmental plasticity [142]. In addition, the concept that an embryonic microenvironment that can differentiate a stem cell through a particular lineage should be able to reprogram cancer cells derived from that lineage was proposed [143]. This concept was supported by experiments showing an inhibition of tumor formation by B16 murine melanoma cells after they were exposed to microenvironmental factors derived from the embryonic skin of a developing mouse [144], and in a chick model, where Rous sarcoma virus failed to induce sarcomas in embryos [145]. More recent studies have extended these observations and gained insights into the molecular reprogramming of metastatic melanoma cells exposed to the embryonic microenvironments of hESCs [123, 146], zebrafish [147–149] and chick embryo [150]. This work has led to the discovery of novel signaling pathways in melanoma that underlie their stem cell-like plasticity and provide potential targets for therapeutic intervention. Studies concerned with the interactions of cells with their microenvironments, whether stem cells or metastatic tumor cells, are focused on how these interactions regulate not only the cells’ behavior, but determine or modulate the cells’ fate. In this regard, identifying the molecular pathways that regulate the plasticity of the cells is a principle concern and involves examining the epigenetic influences of the microenvironment that contribute to regulating the cell phenotype [123, 126, 138, 151]. Given the same DNA code in every cell in an organism, differences in gene expression during differentiation, or in response to microenvironmental influences, are due to epigenetic effects on specific cellular processes. Unlike genetic modifications that alter the DNA sequence, epigenetic influences affect reversible processes, such as cell signaling or DNA modifications [123, 151]. The three interacting events known to occur in epigenetic regulation involve DNA methylation, histone modification and nucleosome remodeling. In the case of cancer cells, the initiation, promotion and maintenance of the tumorigenic phenotype is associated with DNA methylation in the promoter regions and the first exons of certain genes [151]. Epigenetic modifications can also result from changes in ion flow that alter cell signaling events changing cellular functions at the post-translational level that affect complex signaling pathways, which ultimately regulate gene transcription and subsequent level of protein expression [151].
Given the similarities between the plastic phenotype of hESCs and multipotent tumor cells, coupled with the observation that hESCs do not form tumors during blastocyst implantation (though do form teratomas when implanted into immunodeficient mice) [142], prompted the notion that the embryonic microenvironment exerts a dominant role over stem cell fate [123]. This concept was examined using a model where a three-dimensional matrix was conditioned by hESCs, and after the hESCs were removed, multipotent metastatic melanoma cells were plated onto this hESC-conditioned matrix [123, 146]. Free of potential epigenetic influences associated with cell-cell interactions, this work demonstrated that factors secreted by hESCs into their microenvironment dramatically altered the subsequent behavior of the metastatic melanoma cells (Figure 4). Melanoma cells formed spheroids in areas of the matrix that had previously contained hESCs colony clusters during their conditioning of the matrix, while parallel experiments showed that the conditioned medium from hESCs did not induce melanoma spheroid formation. Biochemical and molecular analyses of the melanoma cells (which were phenotypically amelanotic) recovered from the conditioned matrix showed an epigenetic induction of MLANA (a melanocyte-specific phenotype marker), which did not occur when the melanoma cells were exposed to a melanocyte-conditioned matrix. After exposure to the hESC conditioned matrix, the entire population of melanoma cells was less invasive in vitro and less tumorigenic in vivo, though neither their invasiveness nor tumorigenicity was completely abrogated. This failure of the hESC-conditioned matrix to completely inhibit the invasive and tumorigenic potential of the melanoma cell population was presumed to be the result of the heterogeneous nature of both the hESC-conditioned matrix (as demonstrated by regions of the conditioned matrix that promoted melanoma spheroid formation versus regions that did not), and the melanoma cell line(s) themselves [146]. Similar results were subsequently demonstrated for metastatic breast and prostate cancer cells cultured on an hESC conditioned matrix [123].
Figure 4.
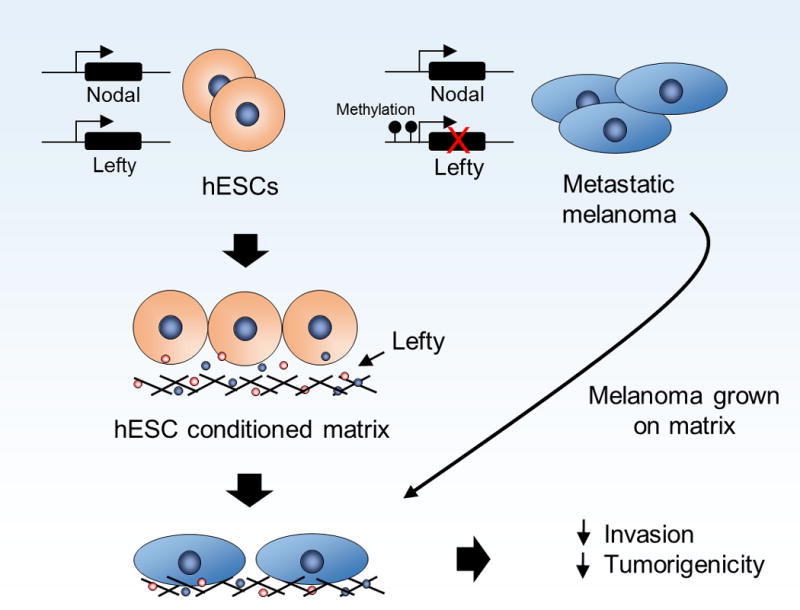
Crosstalk between human embryonic stem cells and multi-potent tumor cells. hESCs express both Nodal and Lefty while in contrast, metastatic melanoma cells fail to express Lefty due to epigenetic changes that involve methylation of the Lefty promoter, leading to aberrant Nodal signaling. hESCs condition growth matrix including the deposition of Lefty. Metastatic melanoma cells grown on this matrix exhibit decreased invasive capabilities and in vivo tumorigenicity, demonstrating an example of the influence hESCs and an embryonic microenvironment on cancer cell phenotypes [123, 146, 151].
The concept of the commonality in plasticity of hESCs and cancer cells was further examined using an embryonic zebrafish model to investigate the potential bi-directional cellular communication that exists between cells and an embryonic microenvironment [123, 147–149]. Of particular interest were common molecular messengers used by both hESCs and metastatic tumor cells. These include Wnt, Notch and TGF-β superfamilies, which are classically associated with developmental processes and are now recognized as important modulators of tumor progression [123, 129, 131, 147, 152, 153]. A major concern of these studies was to understand which embryonic stem cell signaling pathways could be exploited by cancer cells to sustain their plasticity [123]. These studies found that when multipotent, metastatic melanoma cells were injected into the blastula-stage of zebrafish embryos towards the animal pole, the embryos developed an abnormal anterior outgrowth, and when injected near the yolk margin, a duplication of the body axis occurred [147]. The induced ectopic growths did not occur when non-aggressive melanoma cells were injected. It was also found that the melanoma cells did not directly form these structures but recruited and instructed the host zebrafish cells. These results suggested that the metastatic melanoma cells were secreting a potent embryonic morphogen capable of initiating embryonic axis formation and Nodal, a member of the TGF-β superfamily, was identified as this signaling molecule. The identification of Nodal secretion by the metastatic melanoma cells was supported by the observation that goosecoid (a Nodal-responsive gene) was induced in zebrafish cells surrounding the melanoma cells in as little as one hour after engraftment. Furthermore, the induction of ectopic outgrowths by the melanoma cells was abrogated by either inhibiting the expression of Nodal in the cancer cells, or overexpressing Lefty, the endogenous inhibitor of Nodal, in the zebrafish embryo. Western blot analysis confirmed the expression and secretion of Nodal protein by the metastatic, but not non-aggressive melanoma cells [147]. While these zebrafish embryo experiments showed how metastatic melanoma cells can affect and alter the embryonic environment, other work has demonstrated that extracts from zebrafish embryos can inhibit proliferation and induce apoptosis in several cancer cell types, and exposure of metastatic melanoma cells to the zebrafish embryonic microenvironment can reprogram the cells to a non-tumorigenic phenotype [149].
When metastatic melanoma cells were transplanted into the neural crest region of a chick embryonic microenvironment, the melanoma cells invaded host neural crest targets and did not form tumors [150]. While a subset of the tumor cells were reprogrammed to a neural crest cell-like phenotype, melanoma cells were also seen to populate host peripheral structures in a programmed manner, including branchial arches, dorsal root and sympathetic ganglia. In addition, a subpopulation of melanoma cells that invaded the chick periphery was reprogrammed to express the melanocyte-associated protein MLANA, and the neuronal marker TUJ1. Together, these observations suggested that metastatic melanoma cells can respond to developmental cues and that factors unique to the neural crest embryonic milieu may reprogram melanoma cells to a more benign melanocytic cell type.
The embryonic zebrafish model identified Nodal as a crucial component of the signaling pathway between human metastatic melanoma cells and zebrafish stem cells and prompted additional studies into the molecular mechanisms that underlie this communication. The key and defining differences in the Nodal signaling pathway between hESCs and metastatic melanoma cells were the observations that: 1) unlike hESCs, metastatic melanoma cells only express Nodal and not its primary inhibitor, Lefty, and 2) only subpopulations of the metastatic melanoma cells express Cripto-1 [123, 153]. This suggested that Nodal signaling occurs in an unregulated manner and without control in the metastatic melanoma cells. To further explore this premise, two approaches were used to block Nodal signaling in metastatic melanoma cells to examine what happens to the cells when Nodal signaling is inhibited. These approaches used a small-molecule inhibitor of the activin-like-kinase (Alk) activity and Nodal specific morpholinos (MO) [153]. Treating metastatic melanoma cells with the Alk4/5/7 small-molecule inhibitor (SB431542) resulted in an inhibition of Nodal signaling measured as a reduction in Smad2 phosphorylation and a decrease in Nodal protein expression. These results suggested that Nodal expression is regulated, at least in part, by a Smad2-dependent positive-feedback loop. The SB431542-treated metastatic melanoma cells appeared to revert to a less plastic, melanocyte-like phenotype as demonstrated by being less invasive in vitro, less able to engage in vasculogenic mimicry, and the re-expression of Tyrosinase (a melanocytic marker). The anchorage-independent growth of the metastatic melanoma cells was significantly reduced by treating them with Nodal MOs, and this anchorage-independent growth could be rescued by adding rNodal to the cells. Orthotopic transplantation of Nodal MO-treated metastatic melanoma cells in a nude mouse model resulted in a significant decrease in tumor incidence [153]. Additional studies revealed that when Nodal-deficient metastatic melanoma cells were transplanted into the chick embryonic neural crest microenvironment, they failed to migrate and populate neural crest-cell migratory pathways and peripheral targets [123]. Furthermore, analysis of the conditioned matrix of the hESCs model demonstrated that the Lefty protein that was deposited into the microenvironment by the hESCs resulted in the decrease in Nodal expression by the metastatic melanoma cells that were subsequently cultured on this conditioned matrix [123, 146].
Additional studies examined how the embryonic microenvironment could epigenetically reprogram metastatic tumor cells via the Nodal signaling pathway based on the observation that epigenetic regulation of genes can also occur via miRNAs, which are a large group of noncoding RNAs that can block mRNA translation and affect mRNA stability. These studies showed that metastatic melanoma cells exposed to an hESC conditioned microenvironment expressed a higher level of miR-302a and lower level of miR-27b and that Notch4, an upstream regulator of Nodal expression, is one of the targets for miR-302a, while DNA methylation was found to be associated with the down-regulation of Lefty, the primary inhibitor of Nodal [151].
During early vertebrate development, Nodal expression is regulated by the Notch signaling pathway and is involved in the establishment of left-right asymmetry [11, 51]. Given the reestablishment of Nodal expression in metastatic melanoma associated with the aggressive phenotype, additional studies were initiated to examine the potential role for the Notch signaling pathway in Nodal expression in metastatic melanoma. It was found that there was a molecular link between Notch and Nodal signaling in metastatic melanoma cells that occurred via an RBPJ-dependent Nodal enhancer element [154]. Furthermore, the specific correlation between Notch4 and Nodal expression was shown in a number of aggressive but not non-aggressive melanoma cell lines. It was also shown that Notch4 is required for the expression of Nodal, which not only regulates the aggressive phenotype, but also contributes to the regulation of vasculogenic mimicry and anchorage-independent growth in vitro. This work, therefore, identified significant molecular cross talk between Notch4 and Nodal as a key element in metastatic melanoma and placed Notch4 signaling upstream in regulating Nodal expression [154].
It has become apparent that the complexities of the Nodal signaling pathway associated with stem cell fate are relevant to and involved in the aggressive, metastatic phenotype. As such, the Nodal signaling pathway, as well as potential intersecting signaling pathways known from developmental studies, may provide unique and specific therapeutic targets for treating metastatic melanoma. In this regard, and as previously presented, the cross talk between the Notch4 signaling pathway and Nodal expression is a key factor in the aggressiveness of metastatic melanoma and suggests additional therapeutic targets for treatment. Ongoing studies are examining in greater detail how the embryonic microenvironment epigenetically reprograms multipotent, metastatic tumor cells and are focused on the potential downregulation and or silencing of the aberrantly expressed Nodal plasticity gene.
3.4 Clinical significance of Nodal in cancer
In support of the strong experimental evidence linking Nodal to cancer cell aggressiveness, clinical evidence has documented correlations between Nodal and advanced malignancies and suggests the potential therapeutic benefits of targeting Nodal with novel therapies. In melanoma, Nodal was correlated with disease progression in which expression was significantly higher in cutaneous metastases compared to primary tumors of benign skin samples and was not detected in normal skin or melanocytes [147]. These initial observations were strengthened in a large study which examined Nodal in 269 melanocytic lesions, including 109 melanoma samples. Results from this study confirmed an increase in Nodal expression during melanoma progression with low levels noted in some benign nevi [155]. In particular, Nodal expression was highest in deep malignant melanomas and metastatic melanomas. This study constitutes the largest clinical analysis of Nodal in melanoma to date. However, in another study, Nodal was found to be expressed in over half of stage III and IV melanoma samples, although it did not specifically correlate with disease progression in this analysis [156]. Ongoing work focuses on the expression of Nodal in nevi from patients with a history of melanoma, and suggests a potential, prognostic value in this association.
Examining Nodal expression within the endometrium revealed an upregulation during the proliferative phase of the menstrual cycle while Lefty was upregulated at later stages and coincided with Nodal [157]. These observations suggested a proliferation inducing role of Nodal that is controlled in later stages by the subsequent activation of Lefty. When Nodal was examined in endometrial cancer samples, both Nodal and Cripto-1 increased with tumor grade. Remarkably, Lefty levels were low in all cancer samples, indicating unregulated Nodal signaling during endometrial cancer progression. Analysis in pancreatic cancer revealed a lack of Nodal detection at the message and protein levels in normal tissue, whereas high expression levels were noted in pancreatic adenocarcinoma [103]. This work has been further confirmed by recent studies in which Nodal was significantly expressed at higher levels in pancreatic cancer samples compared to normal tissue and associated with poor differentiation [119]. Additionally, high levels of Nodal expression correlated with reduced survival of patients with pancreatic ductal adenocarcinoma [158]. In hepatocellular carcinoma, Nodal was detected in 70 of 96 samples and was absent in normal liver tissue [159]. Nodal associated with numerous parameters correlating to disease progression such as tumor size, metastasis status, microvascular density and vascular invasion. Nodal was found to be an independent prognostic indicator of overall survival in this study in patients following a curative hepatectomy with three-year survival rates dropping from 59.8% in low Nodal expressing samples to 9.7% in the high Nodal expressing group. Still other studies have reported associations with Nodal and neoplasia. In thyroid cancer, Nodal was strongly correlated with papillary and follicular carcinomas while Nodal was detected at higher levels in colon cancer tissue compared to adjacent non-cancerous tissue [105, 160].
The largest clinical analysis to date on the expression of Nodal in cancer was performed in breast cancer [99]. Specifically, 431 therapy naïve breast cancer patient samples were examined for Nodal expression by immunohistochemistry and stratified by clinical-pathological features. Increasing Nodal scores correlated with malignancy, stage, lymph node involvement and lower degree of differentiation. Of particular note in this study, the expression of Nodal did not correlate with any of the established markers for breast cancer (i.e. ER, PR, Her-2). This later result suggests that Nodal positive tumors may represent a novel subtype of breast cancer with independent prognostic value. It also highlights the potential for targeting Nodal in breast cancer. Adding to this information, another study examined vascularization in breast cancer samples and found that levels of Nodal protein within tumors correlated with microvascular density as assessed by CD31 staining [161]. In vitro analysis in this study revealed that Nodal regulated factors associated with angiogenesis such as VEGF and PDGF in breast cancer cells. In line with this association to vascularization, Nodal mRNA in human glioma samples corresponded with VEGF in human cDNA arrays and Nodal protein correlated to tumor grade in human glioma samples with highest Nodal expression in grade IV glioblastoma compared to grade III astrocytoma with minimal detection in non-tumor controls as assessed by immunohistochemistry [162].
While additional clinical analysis is needed for interpretation in other types of cancers, observations from reports such as these indicate potential prognostic implications for the significance of Nodal in cancer. The clinical results are in agreement with numerous findings in experimental models that have demonstrated the ability of Nodal to mediate many aspects of tumorigenesis and the metastatic cascade.
3.5 Targeting Nodal in metastatic tumor cells
The strong connections between Nodal and cancer, validated by molecular mechanistic data, have generated great interest in reducing the influence of Nodal on neoplastic progression in cancer. While reports of Nodal being detected in adult tissue(s) have been described; the majority of studies to date have noted low, or complete absence of Nodal in adult tissue samples and how Nodal might function or signal in somatic cells have not been identified. Furthermore, tissues that require frequent remodeling and undergo dramatic changes such as the endometrium, placenta, and mammary gland transiently express various levels of Nodal during adulthood [reviewed in 94]. Thus, our understanding of Nodal expression and function in adult tissue remains to be further elucidated. Moreover, the ephemeral nature of Nodal combined with its labile stability (in its mature form) and challenging detection methods complicate interpretations, emphasizing validation by multiple experimental methods as a requirement for appropriate analysis and conclusions. However, the compelling burden of evidence that exists within the literature and outlined in this review strongly suggest significant therapeutic potential for the careful development of approaches that aim to inhibit Nodal signaling in cancer cells.
While the heterogeneous and dynamic nature of cancer complicates therapeutic approaches, the development of targeted therapies, specifically against secreted or membrane bound molecules, has been met with success. For example, breast cancers with human epidermal growth factor 2 (Her2) amplification previously carried a poor prognosis; however, since the development of Herceptin (Trastazumab), a monoclonal antibody targeting the Her2 receptor, prognosis for these patients has greatly improved [163, 164]. Conversely, in diseases such as triple-negative breast cancer, in which no established targeted therapies exist, patients continue to face poor survival rates as well as the therapeutic consequences of non-specific therapies with detrimental side effects. Targeted therapies are often used in combination with broad spectrum chemotherapeutics and have shown therapeutic synergy [165]. Thus, studies examining combinatorial approaches to therapy in the context of Nodal inhibition may hold the greatest potential. As noted above, the inhibition of Nodal in the laboratory setting can be achieved through various means, but translating experimental manipulation into clinical application remains challenging. Since Nodal functions as a secreted protein, the potential for therapeutic targeting exists, especially in the development and application of monoclonal antibodies.
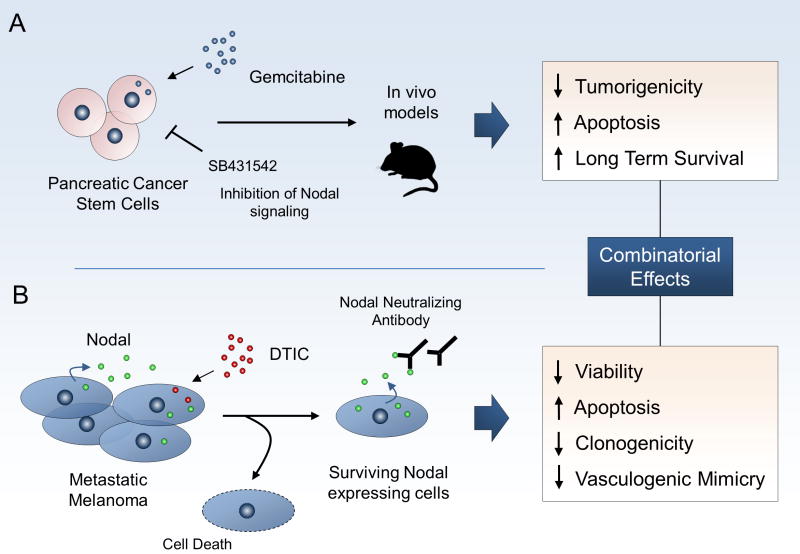
A previous study in pancreatic carcinoma cells demonstrated synergistic effects of targeting Nodal in combination with chemotherapy [103]. Chemical inhibition of Nodal signaling in pancreatic cancer stem cells reduced CD133 expression indicating a greater degree of differentiation and upon withdrawal of Nodal inhibition, CD133 levels were restored. If, however, cells were treated with gemcitabine, a DNA synthesis inhibiting chemotherapy, in the context of Nodal inhibition, in vivo tumorigenicity was completely abolished and survival was significantly increased in orthotopic mouse models. These results were further validated using a pre-clinical model involving xenografted human pancreatic tissue. Emerging evidence suggests that Nodal signaling in these cells is regulated by the miR-17-92 cluster which may impact their response to chemotherapy and maintenance of stem cell like features [166]. A recent study has also supported a link between Nodal inhibition and sensitivity to chemotherapy in the context of melanoma [167]. Metastatic melanoma is a disease with a poor prognosis and short five-year survival period with few effective therapies available. The alkylating agent dacarbazine (DTIC) has been a mainstay of therapy, although its efficacy remains low. Treatment of metastatic melanoma cell lines with DTIC resulted in residual surviving cells that retained Nodal expression and displayed more aggressive qualities. In the remaining DTIC treated cells, using neutralizing antibodies against Nodal reduced their proliferative capacity and increased cell death. Intriguingly, the combinatorial treatment of DTIC with subsequent targeting of Nodal produced synergistic effects including a decrease in cellular viability and invasive capacity that was greater than either approach alone. The observed functional changes were validated by molecular analysis of proliferation and apoptosis markers. Additionally, in human melanoma samples refractory to DTIC, Nodal was strongly expressed within the tumors before and after therapy, highlighting the relevancy of the therapeutic potential obtained in vitro (summarized in Figure 5). Thus, given the ability of Nodal to maintain self-renewal and promote tumorigenic effects, Nodal may contribute to the survival of cancer cells through as yet identified mediators of response to therapies. It is not entirely surprising that the inhibition of Nodal leads to changes in sensitivity to chemotherapy due to the stem cell like features that Nodal imparts on cancer cells and the intrinsic resistance of cancer stem cells to therapeutics.
Figure 5.
Inhibition of Nodal modulates response to chemotherapy. A, pancreatic carcinoma cells were treated with gemcitabine, SB431542, or both molecules. In combination treatment, in vivo tumorigenicity was abolished compared to the effects of SB431542 or gemcitabine alone. Combination treatment also led to enhanced survival of mice xenografted with human pancreatic cancer cells and reductions in pancreatic stem cell markers [103]. B, when metastatic melanoma cells were treated with dacarbazine (DTIC), an increase in the Nodal expressing cell population was observed in surviving cells. Subsequent treatment of this population with anti-Nodal neutralizing antibodies decreased aggressive qualities of the cells, including a reduction in viability and increases in cell death when compared to IgG controls, demonstrating greater effects than DTIC or neutralizing antibody alone [167].
Although therapeutic agents have been in development that target aspects of the Nodal signaling pathway (BIIB015, Biogen-Idec, monoclonal antibody targeting Cripto-1 [168]; LY2157299, Lilly, TGFβ kinase inhibitor with effects on Alk4/7), no developmental therapeutics directly targeting Nodal have been previously described. Recently, a novel Nodal-targeting monoclonal antibody has been developed [169, (Strizzi et al. in press)]. This monoclonal antibody (3D1), was generated against human Nodal amino acids 44–67, targeting the Cripto-1 binding site. 3D1 binds Nodal with a dissociation constant of 1.4 nM KD (rNodal) and disrupts interaction with the Cripto-1 co-receptor complex. In vitro analysis of cancer cells following treatment with 3D1 reproduced many of the previously reported effects of Nodal inhibition in cancer cells including decreases in anchorage independent growth and vasculogenic mimicry. Reductions in downstream mediators of the canonical and non-canonical Nodal signaling pathway were also observed and included reduced phosphorylation of Smad2 and ERK. Additionally, changes in cell cycle markers such as increased p27, decreased cyclin B1 and decreased serine 10 phosphorylation of Histone 3 were observed. In vivo, treatment of C8161 metastatic melanoma cells with 3D1 inhibited cancer cell growth in multiple xenograft models. When cells were injected orthotopically in a cutaneous model, tumor volumes were decreased in 3D1 treated tumors using direct intra-tumoral injection. Additionally, in a metastatic model characterized by lung metastases, 3D1 was administered via intra-peritoneal injections, and lung tumor burden was reduced in the 3D1 treated group compared with control. In each case, immunohistochemistry of the tumors revealed a marked decrease of phosphorylated Smad2 within the nucleus of the 3D1 treatment groups. Analysis of lung sections also revealed a decrease in cyclin B1 staining and an increase in p27 compared to IgG controls. This study represents the first report of an effective monoclonal antibody targeting Nodal that is in development for translational application.
4. Conclusion and Future Perspectives
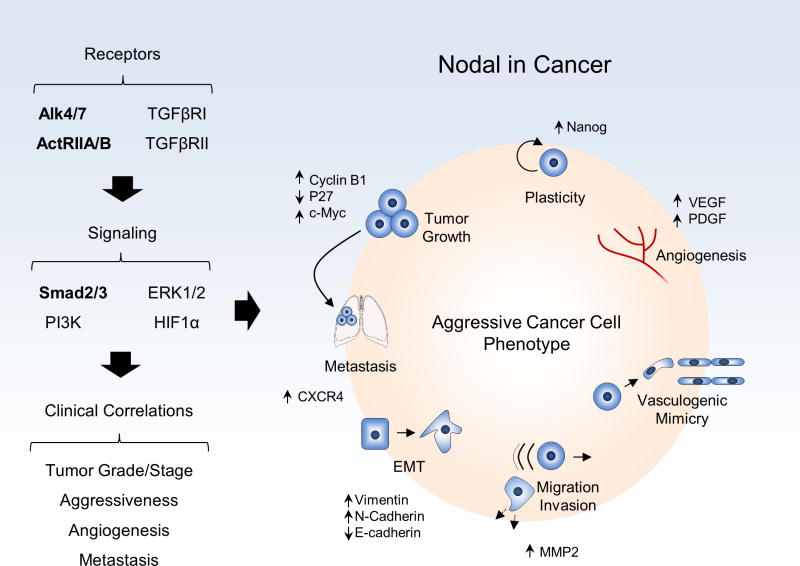
The body of evidence presented in this review provides new insights into the cancer stem cell phenotype expressed by aggressive tumor cells, with particular focus on the reactivation of the embryonic Nodal signaling pathway (Figure 6). Although cancer stem cells form the minority subpopulation(s) within heterogeneous tumors, they commonly express drug resistance markers and are therefore challenging to target. In a similar manner, as Nodal is reactivated in aggressive tumor cells while its inhibitor Lefty is mostly silenced, it is now apparent that these Nodal-expressing cells are resistant as well to front-line therapies. In humans, Nodal expression is largely restricted to embryonic tissues, and is lost in most normal adult tissues. This selective expression pattern suggests a strong rationale for targeting Nodal in cancer. Equally noteworthy are the recent findings generated by immunohistochemistry and molecular analyses of Nodal in patient tumors, together with experimental animal studies, showing the significance of Nodal as a valuable prognostic biomarker in many different cancers and its quintessential role underlying tumor growth and metastasis. As more evidence accumulates demonstrating the inability of conventional therapy to target cancer stem cells, the notion of combinatorial approaches is gaining prestige. Indeed, recent studies in various tumor types support the validity of this strategy, including the targeting of Nodal together with front-line therapy. As additional cancer targets emerge from sophisticated molecular analyses, precision, personalized therapies will become more prevalent -- bolstered by our new found knowledge of cancer stem cells.
Figure 6.
Summary of Nodal signaling and its effects in cancer (canonical pathways listed in bold).
Acknowledgments
This work was supported in part by NIH R37CA59702, R01CA121205, Susan G. Komen, H Foundation, Dixon Translational Grants Initiative, and a kind gift from the Robert Kriss Family (to MJCH), and a Brinson Foundation Junior Investigator Award (to TMB). We thank Zhila Khalkhali-Ellis and Luigi Strizzi for critical reading of the manuscript.
References
- 1.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss A, Attisano L. The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol. 2013;2:47–63. doi: 10.1002/wdev.86. [DOI] [PubMed] [Google Scholar]
- 3.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes and Development. 2005;19:2783–810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 4.Xu P, Liu J, Derynck R. Post-translational regulation of TGF-beta receptor and Smad signaling. FEBS Letters. 2012;586:1871–84. doi: 10.1016/j.febslet.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imamura T, Oshima Y, Hikita A. Regulation of TGF-beta family signalling by ubiquitination and deubiquitination. Journal of Biochemistry. 2013;154:481–9. doi: 10.1093/jb/mvt097. [DOI] [PubMed] [Google Scholar]
- 6.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wakefield LM, Hill CS. Beyond TGFbeta: roles of other TGFbeta superfamily members in cancer. Nature Reviews Cancer. 2013;13:328–41. doi: 10.1038/nrc3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derynck R, Akhurst RJ. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nature Cell Biology. 2007;9:1000–4. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 9.Robertson E, Bradley A, Kuehn M, et al. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323:445–8. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 10.Schier AF. Nodal morphogens. Cold Spring Harb Perspect Biol. 2009;1:a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schier AF, Shen MM. Nodal signaling in vertebrate development. Nature. 2000;403:385–9. doi: 10.1038/35000126. [DOI] [PubMed] [Google Scholar]
- 12.Whitman M. Nodal signaling in early vertebrate embryos: themes and variations. Developmental Cell. 2001;1:605–17. doi: 10.1016/s1534-5807(01)00076-4. [DOI] [PubMed] [Google Scholar]
- 13.Shen MM. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–34. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- 14.Tam PP, Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nature Reviews Genetics. 2007;8:368–81. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- 15.Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nature Reviews Molecular Cell Biology. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 16.Constam DB. Running the gauntlet: an overview of the modalities of travel employed by the putative morphogen Nodal. Current Opinion in Genetics and Development. 2009;19:302–7. doi: 10.1016/j.gde.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Pauklin S, Vallier L. Activin/Nodal signalling in stem cells. Development. 2015;142:607–19. doi: 10.1242/dev.091769. [DOI] [PubMed] [Google Scholar]
- 18.Beck S, Le Good JA, Guzman M, et al. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nature Cell Biology. 2002;4:981–5. doi: 10.1038/ncb890. [DOI] [PubMed] [Google Scholar]
- 19.Salomon DS, Bianco C, Ebert AD, et al. The EGF-CFC family: novel epidermal growth factor-related proteins in development and cancer. Endocrine Related Cancer. 2000;7:199–226. doi: 10.1677/erc.0.0070199. [DOI] [PubMed] [Google Scholar]
- 20.Shen MM, Schier AF. The EGF-CFC gene family in vertebrate development. Trends in Genetics. 2000;16:303–9. doi: 10.1016/s0168-9525(00)02006-0. [DOI] [PubMed] [Google Scholar]
- 21.Blanchet MH, Le Good JA, Mesnard D, et al. Cripto recruits Furin and PACE4 and controls Nodal trafficking during proteolytic maturation. EMBO Journal. 2008;27:2580–91. doi: 10.1038/emboj.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng SK, Olale F, Bennett JT, et al. EGF-CFC proteins are essential coreceptors for the TGF-beta signals Vg1 and GDF1. Genes and Development. 2003;17:31–6. doi: 10.1101/gad.1041203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, Ware SM, Sato A, et al. The Vg1-related protein Gdf3 acts in a Nodal signaling pathway in the pre-gastrulation mouse embryo. Development. 2006;133:319–29. doi: 10.1242/dev.02210. [DOI] [PubMed] [Google Scholar]
- 24.Mathews LS, Vale WW. Expression cloning of an activin receptor, a predicted transmembrane serine kinase. Cell. 1991;65:973–82. doi: 10.1016/0092-8674(91)90549-e. [DOI] [PubMed] [Google Scholar]
- 25.Attisano L, Wrana JL, Cheifetz S, et al. Novel activin receptors: distinct genes and alternative mRNA splicing generate a repertoire of serine/threonine kinase receptors. Cell. 1992;68:97–108. doi: 10.1016/0092-8674(92)90209-u. [DOI] [PubMed] [Google Scholar]
- 26.Gray PC, Harrison CA, Vale W. Cripto forms a complex with activin and type II activin receptors and can block activin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5193–8. doi: 10.1073/pnas.0531290100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe K, Nagaoka T, Strizzi L, et al. Characterization of the glycosylphosphatidylinositol-anchor signal sequence of human Cryptic with a hydrophilic extension. Biochimica et Biophysica Acta. 2008;1778:2671–81. doi: 10.1016/j.bbamem.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bianco C, Adkins HB, Wechselberger C, et al. Cripto-1 activates nodal- and ALK4-dependent and -independent signaling pathways in mammary epithelial Cells. Molecular and Cellular Biology. 2002;22:2586–97. doi: 10.1128/MCB.22.8.2586-2597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bianco C, Mysliwiec M, Watanabe K, et al. Activation of a Nodal-independent signaling pathway by Cripto-1 mutants with impaired activation of a Nodal-dependent signaling pathway. FEBS Letters. 2008;582:3997–4002. doi: 10.1016/j.febslet.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Andrea D, Liguori GL, Le Good JA, et al. Cripto promotes A-P axis specification independently of its stimulatory effect on Nodal autoinduction. Journal of Cell Biology. 2008;180:597–605. doi: 10.1083/jcb.200709090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray PC, Vale W. Cripto/GRP78 modulation of the TGF-beta pathway in development and oncogenesis. FEBS Letters. 2012;586:1836–45. doi: 10.1016/j.febslet.2012.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klauzinska M, Bertolette D, Tippireddy S, et al. Cripto-1: an extracellular protein - connecting the sequestered biological dots. Connective Tissue Research. 2015:1–17. doi: 10.3109/03008207.2015.1077239. [DOI] [PubMed] [Google Scholar]
- 33.Ben-Haim N, Lu C, Guzman-Ayala M, et al. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Developmental Cell. 2006;11:313–23. doi: 10.1016/j.devcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 34.Reissmann E, Jornvall H, Blokzijl A, et al. The orphan receptor ALK7 and the Activin receptor ALK4 mediate signaling by Nodal proteins during vertebrate development. Genes and Development. 2001;15:2010–22. doi: 10.1101/gad.201801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liguori GL, Borges AC, D’Andrea D, et al. Cripto-independent Nodal signaling promotes positioning of the A-P axis in the early mouse embryo. Developmental Biology. 2008;315:280–9. doi: 10.1016/j.ydbio.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 36.Chu J, Shen MM. Functional redundancy of EGF-CFC genes in epiblast and extraembryonic patterning during early mouse embryogenesis. Developmental Biology. 2010;342:63–73. doi: 10.1016/j.ydbio.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C, Shen MM. Two modes by which Lefty proteins inhibit nodal signaling. Current Biology. 2004;14:618–24. doi: 10.1016/j.cub.2004.02.042. [DOI] [PubMed] [Google Scholar]
- 38.Norris DP, Brennan J, Bikoff EK, et al. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 2002;129:3455–68. doi: 10.1242/dev.129.14.3455. [DOI] [PubMed] [Google Scholar]
- 39.Muller P, Rogers KW, Jordan BM, et al. Differential diffusivity of Nodal and Lefty underlies a reaction-diffusion patterning system. Science. 2012;336:721–4. doi: 10.1126/science.1221920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakuma R, Ohnishi Yi Y, Meno C, et al. Inhibition of Nodal signalling by Lefty mediated through interaction with common receptors and efficient diffusion. Genes to Cells. 2002;7:401–12. doi: 10.1046/j.1365-2443.2002.00528.x. [DOI] [PubMed] [Google Scholar]
- 41.Juan H, Hamada H. Roles of nodal-lefty regulatory loops in embryonic patterning of vertebrates. Genes to Cells. 2001;6:923–30. doi: 10.1046/j.1365-2443.2001.00481.x. [DOI] [PubMed] [Google Scholar]
- 42.Le Good JA, Joubin K, Giraldez AJ, et al. Nodal stability determines signaling range. Current Biology. 2005;15:31–6. doi: 10.1016/j.cub.2004.12.062. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Schier AF. The zebrafish Nodal signal Squint functions as a morphogen. Nature. 2001;411:607–10. doi: 10.1038/35079121. [DOI] [PubMed] [Google Scholar]
- 44.Jing XH, Zhou SM, Wang WQ, et al. Mechanisms underlying long- and short-range nodal signaling in Zebrafish. Mechanisms of Development. 2006;123:388–94. doi: 10.1016/j.mod.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 45.Tian J, Andree B, Jones CM, et al. The pro-domain of the zebrafish Nodal-related protein Cyclops regulates its signaling activities. Development. 2008;135:2649–58. doi: 10.1242/dev.019794. [DOI] [PubMed] [Google Scholar]
- 46.Blanchet MH, Le Good JA, Oorschot V, et al. Cripto localizes Nodal at the limiting membrane of early endosomes. Science Signaling. 2008;1:ra13. doi: 10.1126/scisignal.1165027. [DOI] [PubMed] [Google Scholar]
- 47.Tsukazaki T, Chiang TA, Davison AF, et al. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–91. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 48.Chen YG. Endocytic regulation of TGF-beta signaling. Cell Research. 2009;19:58–70. doi: 10.1038/cr.2008.315. [DOI] [PubMed] [Google Scholar]
- 49.Yeo C, Whitman M. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Molecular Cell. 2001;7:949–57. doi: 10.1016/s1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka C, Sakuma R, Nakamura T, et al. Long-range action of Nodal requires interaction with GDF1. Genes and Development. 2007;21:3272–82. doi: 10.1101/gad.1623907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schier AF. Nodal signaling in vertebrate development. Annual Review of Cell and Developmental Biology. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- 52.Feldman B, Gates MA, Egan ES, et al. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature. 1998;395:181–5. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- 53.Rebagliati MR, Toyama R, Fricke C, et al. Zebrafish nodal-related genes are implicated in axial patterning and establishing left-right asymmetry. Developmental Biology. 1998;199:261–72. doi: 10.1006/dbio.1998.8935. [DOI] [PubMed] [Google Scholar]
- 54.Dougan ST, Warga RM, Kane DA, et al. The role of the zebrafish nodal-related genes squint and cyclops in patterning of mesendoderm. Development. 2003;130:1837–51. doi: 10.1242/dev.00400. [DOI] [PubMed] [Google Scholar]
- 55.Blum M, Feistel K, Thumberger T, et al. The evolution and conservation of left-right patterning mechanisms. Development. 2014;141:1603–13. doi: 10.1242/dev.100560. [DOI] [PubMed] [Google Scholar]
- 56.Robertson EJ. Dose-dependent Nodal/Smad signals pattern the early mouse embryo. Seminars in Cell and Developmental Biology. 2014;32:73–9. doi: 10.1016/j.semcdb.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 57.Brennan J, Lu CC, Norris DP, et al. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–9. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- 58.Conlon FL, Lyons KM, Takaesu N, et al. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development. 1994;120:1919–28. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- 59.Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes and Development. 2002;16:2339–44. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Norris DP, Robertson EJ. Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements. Genes and Development. 1999;13:1575–88. doi: 10.1101/gad.13.12.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krebs LT, Iwai N, Nonaka S, et al. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes and Development. 2003;17:1207–12. doi: 10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raya A, Kawakami Y, Rodriguez-Esteban C, et al. Notch activity induces Nodal expression and mediates the establishment of left-right asymmetry in vertebrate embryos. Genes and Development. 2003;17:1213–8. doi: 10.1101/gad.1084403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adachi H, Saijoh Y, Mochida K, et al. Determination of left/right asymmetric expression of nodal by a left side-specific enhancer with sequence similarity to a lefty-2 enhancer. Genes and Development. 1999;13:1589–600. doi: 10.1101/gad.13.12.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto M, Meno C, Sakai Y, et al. The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes and Development. 2001;15:1242–56. doi: 10.1101/gad.883901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamada H, Tam PP. Mechanisms of left-right asymmetry and patterning: driver, mediator and responder. F1000 Faculty Reviews. 2014;6:110. doi: 10.12703/P6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Komatsu Y, Mishina Y. Establishment of left-right asymmetry in vertebrate development: the node in mouse embryos. Cellular and Molecular Life Sciences. 2013;70:4659–66. doi: 10.1007/s00018-013-1399-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ermakov AS. Establishment of visceral left-right asymmetry in mammals: the role of ciliary action and leftward fluid flow in the region of Hensen’s node. Ontogenez. 2013;44:341–56. [PubMed] [Google Scholar]
- 68.Mercola M. Left-right asymmetry: nodal points. Journal of Cell Science. 2003;116:3251–7. doi: 10.1242/jcs.00668. [DOI] [PubMed] [Google Scholar]
- 69.Saijoh Y, Adachi H, Sakuma R, et al. Left-right asymmetric expression of lefty2 and nodal is induced by a signaling pathway that includes the transcription factor FAST2. Molecular Cell. 2000;5:35–47. doi: 10.1016/s1097-2765(00)80401-3. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto M, Mine N, Mochida K, et al. Nodal signaling induces the midline barrier by activating Nodal expression in the lateral plate. Development. 2003;130:1795–804. doi: 10.1242/dev.00408. [DOI] [PubMed] [Google Scholar]
- 71.Schier AF, Talbot WS. Molecular genetics of axis formation in zebrafish. Annual Review of Genetics. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- 72.Blum M, Beyer T, Weber T, et al. Xenopus, an ideal model system to study vertebrate left-right asymmetry. Developmental Dynamics. 2009;238:1215–25. doi: 10.1002/dvdy.21855. [DOI] [PubMed] [Google Scholar]
- 73.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–54. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parisi S, D’Andrea D, Lago CT, et al. Nodal-dependent Cripto signaling promotes cardiomyogenesis and redirects the neural fate of embryonic stem cells. Journal of Cell Biology. 2003;163:303–14. doi: 10.1083/jcb.200303010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Besser D. Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. Journal of Biological Chemistry. 2004;279:45076–84. doi: 10.1074/jbc.M404979200. [DOI] [PubMed] [Google Scholar]
- 76.Brandenberger R, Wei H, Zhang S, et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nature Biotechnology. 2004;22:707–16. doi: 10.1038/nbt971. [DOI] [PubMed] [Google Scholar]
- 77.Vallier L, Reynolds D, Pedersen RA. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Developmental Biology. 2004;275:403–21. doi: 10.1016/j.ydbio.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 78.James D, Levine AJ, Besser D, et al. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–82. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 79.Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. Journal of Cell Science. 2005;118:4495–509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 80.Smith JR, Vallier L, Lupo G, et al. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Developmental Biology. 2008;313:107–17. doi: 10.1016/j.ydbio.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 81.Mesnard D, Guzman-Ayala M, Constam DB. Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development. 2006;133:2497–505. doi: 10.1242/dev.02413. [DOI] [PubMed] [Google Scholar]
- 82.Camus A, Perea-Gomez A, Moreau A, et al. Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Developmental Biology. 2006;295:743–55. doi: 10.1016/j.ydbio.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 83.Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–5. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 84.Kubo A, Shinozaki K, Shannon JM, et al. Development of definitive endoderm from embryonic stem cells in culture. Development. 2004;131:1651–62. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- 85.D’Amour KA, Agulnick AD, Eliazer S, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nature Biotechnology. 2005;23:1534–41. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 86.Pfendler KC, Catuar CS, Meneses JJ, et al. Overexpression of Nodal promotes differentiation of mouse embryonic stem cells into mesoderm and endoderm at the expense of neuroectoderm formation. Stem Cells and Development. 2005;14:162–72. doi: 10.1089/scd.2005.14.162. [DOI] [PubMed] [Google Scholar]
- 87.Takenaga M, Fukumoto M, Hori Y. Regulated Nodal signaling promotes differentiation of the definitive endoderm and mesoderm from ES cells. Journal of Cell Science. 2007;120:2078–90. doi: 10.1242/jcs.004127. [DOI] [PubMed] [Google Scholar]
- 88.Dahle O, Kumar A, Kuehn MR. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Science Signaling. 2010;3:ra48. doi: 10.1126/scisignal.2000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teo AK, Arnold SJ, Trotter MW, et al. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes and Development. 2011;25:238–50. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xi Q, Wang Z, Zaromytidou AI, et al. A poised chromatin platform for TGF-beta access to master regulators. Cell. 2011;147:1511–24. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim SW, Yoon SJ, Chuong E, et al. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Developmental Biology. 2011;357:492–504. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 92.Brown S, Teo A, Pauklin S, et al. Activin/Nodal signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells. 2011;29:1176–85. doi: 10.1002/stem.666. [DOI] [PubMed] [Google Scholar]
- 93.Papanayotou C, Benhaddou A, Camus A, et al. A novel nodal enhancer dependent on pluripotency factors and smad2/3 signaling conditions a regulatory switch during epiblast maturation. PLoS Biology. 2014;12:e1001890. doi: 10.1371/journal.pbio.1001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Quail DF, Siegers GM, Jewer M, et al. Nodal signalling in embryogenesis and tumourigenesis. International Journal of Biochemistry and Cell Biology. 2013;45:885–98. doi: 10.1016/j.biocel.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 95.Quail DF, Zhang G, Walsh LA, et al. Embryonic morphogen nodal promotes breast cancer growth and progression. PLoS One. 2012;7:e48237. doi: 10.1371/journal.pone.0048237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kirsammer G, Strizzi L, Margaryan NV, et al. Nodal signaling promotes a tumorigenic phenotype in human breast cancer. Seminars in Cancer Biology. 2014;29:40–50. doi: 10.1016/j.semcancer.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee CC, Jan HJ, Lai JH, et al. Nodal promotes growth and invasion in human gliomas. Oncogene. 2010;29:3110–23. doi: 10.1038/onc.2010.55. [DOI] [PubMed] [Google Scholar]
- 98.De Silva T, Ye G, Liang YY, et al. Nodal promotes glioblastoma cell growth. Frontiers Endocrinology. 2012;3:59. doi: 10.3389/fendo.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strizzi L, Hardy KM, Margaryan NV, et al. Potential for the embryonic morphogen Nodal as a prognostic and predictive biomarker in breast cancer. Breast Cancer Res. 2012;14:R75. doi: 10.1186/bcr3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Strizzi L, Hardy KM, Kirsammer GT, et al. Embryonic signaling in melanoma: potential for diagnosis and therapy. Laboratory Investigation. 2011;91:819–24. doi: 10.1038/labinvest.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Strizzi L, Hardy KM, Kirschmann DA, et al. Nodal expression and detection in cancer: experience and challenges. Cancer Research. 2012;72:1915–20. doi: 10.1158/0008-5472.CAN-11-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seftor EA, Seftor RE, Weldon DS, et al. Melanoma tumor cell heterogeneity: a molecular approach to study subpopulations expressing the embryonic morphogen nodal. Seminars in Oncology. 2014;41:259–66. doi: 10.1053/j.seminoncol.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lonardo E, Hermann PC, Mueller MT, et al. Nodal/Activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell. 2011;9:433–46. doi: 10.1016/j.stem.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 104.Lonardo E, Frias-Aldeguer J, Hermann PC, et al. Pancreatic stellate cells form a niche for cancer stem cells and promote their self-renewal and invasiveness. Cell Cycle. 2012;11:1282–90. doi: 10.4161/cc.19679. [DOI] [PubMed] [Google Scholar]
- 105.Gong Y, Guo Y, Hai Y, et al. Nodal promotes the self-renewal of human colon cancer stem cells via an autocrine manner through Smad2/3 signaling pathway. Biomedical Research International. 2014;2014:364134. doi: 10.1155/2014/364134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spiller CM, Bowles J, Koopman P. Nodal/Cripto signaling in fetal male germ cell development: implications for testicular germ cell tumors. International Journal of Developmental Biology. 2013;57:211–9. doi: 10.1387/ijdb.130028pk. [DOI] [PubMed] [Google Scholar]
- 107.Nettersheim D, Jostes S, Sharma R, et al. BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma. PLoS Genetics. 2015;11:e1005415. doi: 10.1371/journal.pgen.1005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Spiller CM, Feng CW, Jackson A, et al. Endogenous Nodal signaling regulates germ cell potency during mammalian testis development. Development. 2012;139:4123–32. doi: 10.1242/dev.083006. [DOI] [PubMed] [Google Scholar]
- 109.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Research. 2009;19:128–39. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Davies M, Robinson M, Smith E, et al. Induction of an epithelial to mesenchymal transition in human immortal and malignant keratinocytes by TGF-beta1 involves MAPK, Smad and AP-1 signalling pathways. Journal of Cellular Biochemistry. 2005;95:918–31. doi: 10.1002/jcb.20458. [DOI] [PubMed] [Google Scholar]
- 111.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes and Development. 2013;27:2192–206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nature Reviews Molecular Cell Biology. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Micalizzi DS, Ford HL. Epithelial-mesenchymal transition in development and cancer. Future Oncology. 2009;5:1129–43. doi: 10.2217/fon.09.94. [DOI] [PubMed] [Google Scholar]
- 114.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Quail DF, Zhang G, Findlay SD, et al. Nodal promotes invasive phenotypes via a mitogen-activated protein kinase-dependent pathway. Oncogene. 2014;33:461–73. doi: 10.1038/onc.2012.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun G, Shi L, Li M, et al. Lefty inhibits glioma growth by suppressing Nodal-activated Smad and ERK1/2 pathways. Journal of the Neurological Sciences. 2014;347:137–42. doi: 10.1016/j.jns.2014.09.034. [DOI] [PubMed] [Google Scholar]
- 117.Fang R, Zhang G, Guo Q, et al. Nodal promotes aggressive phenotype via Snail-mediated epithelial-mesenchymal transition in murine melanoma. Cancer Letters. 2013;333:66–75. doi: 10.1016/j.canlet.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 118.Guo Q, Ning F, Fang R, et al. Endogenous Nodal promotes melanoma undergoing epithelial-mesenchymal transition via Snail and Slug in vitro and in vivo. American Journal of Cancer Research. 2015;5:2098–112. [PMC free article] [PubMed] [Google Scholar]
- 119.Duan W, Li R, Ma J, et al. Overexpression of Nodal induces a metastatic phenotype in pancreatic cancer cells via the Smad2/3 pathway. Oncotarget. 2015;6:1490–506. doi: 10.18632/oncotarget.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Quail DF, Taylor MJ, Walsh LA, et al. Low oxygen levels induce the expression of the embryonic morphogen Nodal. Molecular Biology of the Cell. 2011;22:4809–21. doi: 10.1091/mbc.E11-03-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lai JH, Jan HJ, Liu LW, et al. Nodal regulates energy metabolism in glioma cells by inducing expression of hypoxia-inducible factor 1alpha. Neuro-Oncology. 2013;15:1330–41. doi: 10.1093/neuonc/not086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Khalkhali-Ellis Z, Kirschmann DA, Seftor EA, et al. Divergence(s) in nodal signaling between aggressive melanoma and embryonic stem cells. International Journal of Cancer. 2015;136:E242–51. doi: 10.1002/ijc.29198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hendrix MJ, Seftor EA, Seftor RE, et al. Reprogramming metastatic tumour cells with embryonic microenvironments. Nature Reviews Cancer. 2007;7:246–55. doi: 10.1038/nrc2108. [DOI] [PubMed] [Google Scholar]
- 124.Sell S. Stem cell origin of cancer and differentiation therapy. Critical Reviews in Oncology/Hematology. 2004;51:1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 125.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Research. 2006;66:9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 126.Li L, Neaves WB. Normal stem cells and cancer stem cells: the niche matters. Cancer Research. 2006;66:4553–7. doi: 10.1158/0008-5472.CAN-05-3986. [DOI] [PubMed] [Google Scholar]
- 127.Tan BT, Park CY, Ailles LE, et al. The cancer stem cell hypothesis: a work in progress. Laboratory Investigation. 2006;86:1203–7. doi: 10.1038/labinvest.3700488. [DOI] [PubMed] [Google Scholar]
- 128.Monk M, Holding C. Human embryonic genes re-expressed in cancer cells. Oncogene. 2001;20:8085–91. doi: 10.1038/sj.onc.1205088. [DOI] [PubMed] [Google Scholar]
- 129.Bittner M, Meltzer P, Chen Y, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–40. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 130.Carr KM, Bittner M, Trent JM. Gene-expression profiling in human cutaneous melanoma. Oncogene. 2003;22:3076–80. doi: 10.1038/sj.onc.1206448. [DOI] [PubMed] [Google Scholar]
- 131.Hoek K, Rimm DL, Williams KR, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Research. 2004;64:5270–82. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 132.Gao CF, Xie Q, Su YL, et al. Proliferation and invasion: plasticity in tumor cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10528–33. doi: 10.1073/pnas.0504367102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Luo J, Duggan DJ, Chen Y, et al. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Research. 2001;61:4683–8. [PubMed] [Google Scholar]
- 134.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 136.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 138.Lotem J, Sachs L. Epigenetics and the plasticity of differentiation in normal and cancer stem cells. Oncogene. 2006;25:7663–72. doi: 10.1038/sj.onc.1209816. [DOI] [PubMed] [Google Scholar]
- 139.Ivanova NB, Dimos JT, Schaniel C, et al. A stem cell molecular signature. Science. 2002;298:601–4. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 140.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 142.Mintz B, Illmensee K. Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:3585–9. doi: 10.1073/pnas.72.9.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pierce GB, Pantazis CG, Caldwell JE, et al. Specificity of the control of tumor formation by the blastocyst. Cancer Research. 1982;42:1082–7. [PubMed] [Google Scholar]
- 144.Gerschenson M, Graves K, Carson SD, et al. Regulation of melanoma by the embryonic skin. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:7307–10. doi: 10.1073/pnas.83.19.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dolberg DS, Bissell MJ. Inability of Rous sarcoma virus to cause sarcomas in the avian embryo. Nature. 1984;309:552–6. doi: 10.1038/309552a0. [DOI] [PubMed] [Google Scholar]
- 146.Postovit LM, Seftor EA, Seftor RE, et al. A three-dimensional model to study the epigenetic effects induced by the microenvironment of human embryonic stem cells. Stem Cells. 2006;24:501–5. doi: 10.1634/stemcells.2005-0459. [DOI] [PubMed] [Google Scholar]
- 147.Topczewska JM, Postovit LM, Margaryan NV, et al. Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nature Medicine. 2006;12:925–32. doi: 10.1038/nm1448. [DOI] [PubMed] [Google Scholar]
- 148.Cucina A, Biava PM, D’Anselmi F, et al. Zebrafish embryo proteins induce apoptosis in human colon cancer cells (Caco2) Apoptosis. 2006;11:1617–28. doi: 10.1007/s10495-006-8895-4. [DOI] [PubMed] [Google Scholar]
- 149.Lee LM, Seftor EA, Bonde G, et al. The fate of human malignant melanoma cells transplanted into zebrafish embryos: assessment of migration and cell division in the absence of tumor formation. Developmental Dynamics. 2005;233:1560–70. doi: 10.1002/dvdy.20471. [DOI] [PubMed] [Google Scholar]
- 150.Kulesa PM, Kasemeier-Kulesa JC, Teddy JM, et al. Reprogramming metastatic melanoma cells to assume a neural crest cell-like phenotype in an embryonic microenvironment. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3752–7. doi: 10.1073/pnas.0506977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Costa FF, Seftor EA, Bischof JM, et al. Epigenetically reprogramming metastatic tumor cells with an embryonic microenvironment. Epigenomics. 2009;1:387–98. doi: 10.2217/epi.09.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hendrix MJ, Seftor EA, Hess AR, et al. Molecular plasticity of human melanoma cells. Oncogene. 2003;22:3070–5. doi: 10.1038/sj.onc.1206447. [DOI] [PubMed] [Google Scholar]
- 153.Postovit LM, Costa FF, Bischof JM, et al. The commonality of plasticity underlying multipotent tumor cells and embryonic stem cells. Journal of Cellular Biochemistry. 2007;101:908–17. doi: 10.1002/jcb.21227. [DOI] [PubMed] [Google Scholar]
- 154.Hardy KM, Kirschmann DA, Seftor EA, et al. Regulation of the embryonic morphogen Nodal by Notch4 facilitates manifestation of the aggressive melanoma phenotype. Cancer Research. 2010;70:10340–50. doi: 10.1158/0008-5472.CAN-10-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yu L, Harms PW, Pouryazdanparast P, et al. Expression of the embryonic morphogen Nodal in cutaneous melanocytic lesions. Modern Pathology. 2010;23:1209–14. doi: 10.1038/modpathol.2010.101. [DOI] [PubMed] [Google Scholar]
- 156.Hooijkaas AI, Gadiot J, van Boven H, et al. Expression of the embryological morphogen Nodal in stage III/IV melanoma. Melanoma Research. 2011;21:491–501. doi: 10.1097/CMR.0b013e32834bf37b. [DOI] [PubMed] [Google Scholar]
- 157.Papageorgiou I, Nicholls PK, Wang F, et al. Expression of nodal signalling components in cycling human endometrium and in endometrial cancer. Reproductive Biology and Endocrinology. 2009;7:122. doi: 10.1186/1477-7827-7-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kong B, Wang W, Esposito I, et al. Increased expression of Nodal correlates with reduced patient survival in pancreatic cancer. Pancreatology. 2015;15:156–61. doi: 10.1016/j.pan.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 159.Chen J, Liu WB, Jia WD, et al. Embryonic morphogen nodal is associated with progression and poor prognosis of hepatocellular carcinoma. PLoS One. 2014;9:e85840. doi: 10.1371/journal.pone.0085840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chai YJ, Kim YA, Jee HG, et al. Expression of the embryonic morphogen Nodal in differentiated thyroid carcinomas: Immunohistochemistry assay in tissue microarray and The Cancer Genome Atlas data analysis. Surgery. 2014;156:1559–67. doi: 10.1016/j.surg.2014.08.050. discussion 1567–8. [DOI] [PubMed] [Google Scholar]
- 161.Quail DF, Walsh LA, Zhang G, et al. Embryonic protein nodal promotes breast cancer vascularization. Cancer Research. 2012;72:3851–63. doi: 10.1158/0008-5472.CAN-11-3951. [DOI] [PubMed] [Google Scholar]
- 162.Hueng DY, Lin GJ, Huang SH, et al. Inhibition of Nodal suppresses angiogenesis and growth of human gliomas. Journal of Neuro-Oncology. 2011;104:21–31. doi: 10.1007/s11060-010-0467-3. [DOI] [PubMed] [Google Scholar]
- 163.Hudis CA. Trastuzumab--mechanism of action and use in clinical practice. New England Journal of Medicine. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 164.Pinto AC, Ades F, de Azambuja E, et al. Trastuzumab for patients with HER2 positive breast cancer: delivery, duration and combination therapies. Breast. 2013;22(Suppl 2):S152–5. doi: 10.1016/j.breast.2013.07.029. [DOI] [PubMed] [Google Scholar]
- 165.Li F, Zhao C, Wang L. Molecular-targeted agents combination therapy for cancer: developments and potentials. International Journal of Cancer. 2014;134:1257–69. doi: 10.1002/ijc.28261. [DOI] [PubMed] [Google Scholar]
- 166.Cioffi M, Trabulo SM, Sanchez-Ripoll Y, et al. The miR-17-92 cluster counteracts quiescence and chemoresistance in a distinct subpopulation of pancreatic cancer stem cells. Gut. 2015;64:1936–48. doi: 10.1136/gutjnl-2014-308470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hardy KM, Strizzi L, Margaryan NV, et al. Targeting nodal in conjunction with dacarbazine induces synergistic anticancer effects in metastatic melanoma. Molecular Cancer Research. 2015;13:670–80. doi: 10.1158/1541-7786.MCR-14-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kelly RK, Olson DL, Sun Y, et al. An antibody-cytotoxic conjugate, BIIB015, is a new targeted therapy for Cripto positive tumours. European Journal of Cancer. 2011;47:1736–46. doi: 10.1016/j.ejca.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 169.Foca A, Sanguigno L, Foca G, et al. New Anti-Nodal Monoclonal Antibodies Targeting the Nodal Pre-Helix Loop Involved in Cripto-1 Binding. Int J Mol Sci. 2015;16:21342–62. doi: 10.3390/ijms160921342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Vallier L, Mendjan S, Brown S, et al. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–49. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bertero A, Madrigal P, Galli A, et al. Activin/nodal signaling and NANOG orchestrate human embryonic stem cell fate decisions by controlling the H3K4me3 chromatin mark. Genes and Development. 2015;29:702–17. doi: 10.1101/gad.255984.114. [DOI] [PMC free article] [PubMed] [Google Scholar]