Abstract
Astrocytes isolated from individuals with amyotrophic lateral sclerosis (ALS) are toxic towards motor neurons (MNs) and play a non-cell autonomous role in disease pathogenesis. The mechanisms underlying the susceptibility of motor neurons to cell death remains unclear. Here, we report that astrocytes derived from mice bearing ALS mutations and from individuals with ALS reduce expression of major histocompatibility complex class I (MHCI) on MNs. Reduced MHCI expression makes these MNs susceptible to astrocyte-induced cell death. Increasing MHCI expression on MNs increases survival and motor performance in a mouse model of ALS and protects MN against astrocyte toxicity. A single MHCI molecule, HLA-F, protects MNs from ALS astrocyte-mediated toxicity, while knockdown of its receptor, the killer cell immunoglobulin-like receptor KIR3DL2, an inhibitory receptor that recognizes MHCI, on astrocytes results in enhanced MN death. These data indicate that in ALS upon loss of MHCI expression MNs become vulnerable to astrocyte-mediated toxicity.
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal, adult onset neurodegenerative disorder characterized by the progressive loss of motor neurons (MNs) and death due to respiratory failure1. The majority of cases are classified as sporadic ALS (SALS), defined as having no family history of the disease. Despite efforts to identify risk factors and potential susceptibility genes, the etiology of SALS remains largely unknown2. Current understanding of cellular and molecular mechanisms underlying this disease is mostly derived from animals models bearing mutations in genes associated with autosomal dominant familial ALS (FALS)3-7. Despite the difference in etiology, FALS and SALS are phenotypically indistinguishable. Several studies suggest a common disease mechanism underlying FALS and SALS8,9 and therefore knowledge gathered from animal models of FALS can help understand the mechanism behind SALS10.
In addition to cell intrinsic damage occurring within MNs, glial cells such as astrocytes, microglia and oligodendrocytes contribute to MN death in ALS in a non-cell autonomous manner11,12. Astrocytes derived from postmortem biopsies of human ALS individuals induce MN death in vitro, opening the possibility to study these patient specific cells in great detail13,14. However, the understanding of how and why MNs die in this disease remains unknown.
Recent work in development, aging and neurological disorders, including ALS has focused on immune molecules, such as cytokines, proteins of the complement system, and major histocompatibility complex class I (MHCI) proteins and their role in the developing and adult brain15-17. In the central nervous system (CNS), MHCI plays a role in synaptic function15,18-22, and in neurodegenerative processes23,24. In the ALS mouse model, which express a disease-associated variant in superoxide dismutase 1 (SOD1G93A) mice, higher levels of MHCI in MNs were found in animals that presented a slower disease progression25. Furthermore, disease progression is accelerated in SOD1G93A mice lacking beta-2 microglobulin (β2m), an essential component for MHCI presentation26. Taken together these findings suggest that MHCI may have a role in the pathogenesis of ALS.
Results
MHCI expression is reduced on MNs in ALS
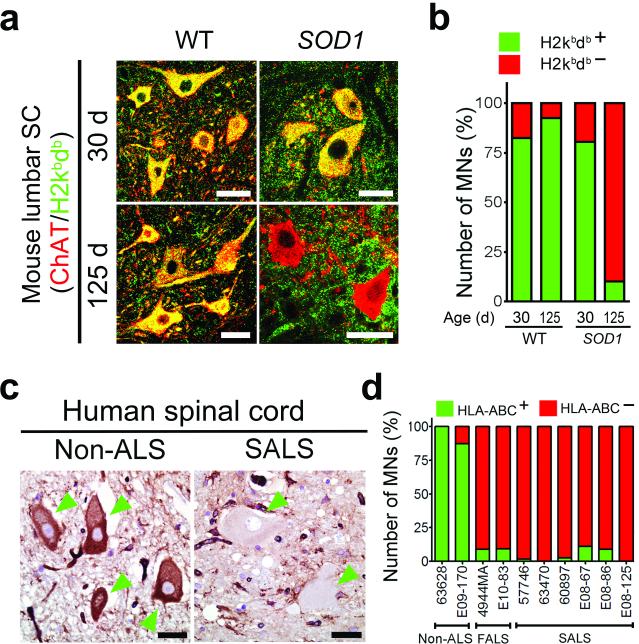
MHCI molecules and β2m are enriched in mouse spinal MNs (Supplementary Fig. 1a,b). In order to examine changes in MHCI expression in MNs in ALS, we analyzed MHCI expression prior to and after disease onset in all segments of the spinal cord of SOD1G93A mice and compared them to wild-type mice. Using an antibody that recognizes subgroups of mouse MHCI (histocompatibility 2 (H2), kb and db) and commonly used for MHCI detection in MNs25,27, we found that MHCI expression was markedly reduced in MN somata but increased in motor axons in SOD1G93A mice after disease onset (Fig. 1a,b and Supplementary Fig. 1c-e). These results are in agreement with previous findings using another rapid progressing SOD1 mouse model (129Sv-SOD1G93A) showing that MHCI protein is transported away from MN cell body and accumulates in peripheral motor axons during disease progression25.
Figure 1. MHCI expression is reduced on spinal MNs in end-stage ALS.
(a) Representative immunofluorescence images (from three images evaluated) showing MHCI (H2db/H2kb) expression in MNs at 30 and 125 days in SOD1G93A mice. (b) Percent of MHCI positive lumbar spinal cord MNs in SOD1G93A and control mice evaluated as shown in (a) For each group, two animals were used and MNs counts were pulled together. 321, 216, 216, 154 MNs were counted in graph columns 1 through 4. (c) Immunohistochemical analysis of MHCI (HLA-ABC) expression in spinal MNs of individuals with ALS post-mortem. Green arrowheads point to MNs. (d) Percent of MHCI positive MNs in the spinal cords of individuals with ALS individuals and controls determined as shown in (c). For analysis two non-ALS, two FALS and 6 SALS individuals were analyzed. 50, 60, 35, 51, 71, 87, 45, 46, 68 and 22 of MNs were counted in graph columns 1 through 10. WT, wild-type mouse. SOD1, SOD1G93A mouse. Scale bars 20 μm.
Next, to determine if loss of MHCI in MNs seen in the mouse model was also seen in human ALS individuals, we evaluated MHCI expression by immunohistochemistry in spinal cord samples from FALS carrying the SOD1A4V mutation and sporadic ALS individuals compared to non-ALS controls. Using an antibody recognizing human leukocyte antigen (HLA)-A, -B, and –C, we observed that MHCI expression in MNs was nearly absent in both FALS and SALS spinal cords as compared to MHCI levels expression in MNs of non-ALS samples (Fig. 1c,d).
Loss of MHCI by MNs after exposure to ALS astrocytes
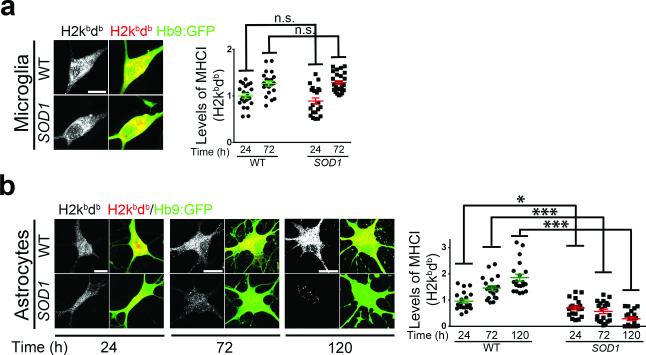
Using our recently described co-culture system of adult CNS derived microglia with MNs28, we evaluated the impact of ALS microglia on the expression of MHCI in MNs. Although SOD1G93A microglia have been shown to be toxic to MNs28, when MNs were co-cultured with SOD1G93A microglia, no overt changes were observed in their MHCI expression (Fig. 2a). By contrast when MNs were co-cultured with ALS astrocytes29, confirmed to be devoid of other glia types or cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells (Supplementary Fig. 2a-g), MHCI expression in MNs was diminished already within 24 hours and steadily declined over the next 96 hours, at which point approximately 73% of MNs completely lost MHCI expression (Fig. 2b). MHCI expression in MNs was slightly increased during the same period when MNs were cultured on top of wild-type astrocytes (Fig. 2b). This could reflect MN maturation in the presence of astrocytes30, which affects MHCI expression patterns in neurons31.
Figure 2. ALS astrocytes induce down-regulation of MHCI expression in MNs.
(a,b) Representative immunofluorescence images (of three independent experiments) showing MHCI (H2db/H2kb) expression in MNs cultured in the presence of either wild type or SOD1G93A microglia for 72 hours (a) and measurement of their MHCI expression relative to levels found in MNs cultured with wild type microglia for 24h (b). (c,d) Representative immunofluorescence images (of three independent experiments) showing MHCI (H2db/H2kb) expression in MNs cultured in the presence of either wild type or SOD1G93A astrocytes during a 120 hours period (c) and measurement of their MHCI expression relative to levels found in MNs cultured with wild type astrocytes for 24h (d). Experiments were made in triplicate. Levels of MHCI are expressed as mean fluorescence intensity (MFI) found in MNs. Error bars represent s.e.m.. Each dot in graphs (b and d) represents MHCI level found per MN (One-Way ANOVA, *P<0.05; ***P<0.001; n.s., non-significant P≥0.05). WT, wild-type mouse. SOD1, SOD1G93A mouse. Scale bars 10 μm.
In order to evaluate if expression of ALS linked-mutant SOD1 protein within MNs could lead to intrinsic down-regulation of MHCI expression, we generated wild-type and SOD1G93A MNs using induced pluripotent stem cell (iPSC) technology32. The iPSCs generated contained the green fluorescent protein (GFP) under the control of the MN specific Hb9 promoter allowing MN enrichment using a fluorescence activated cell sorter upon MN induced cell differentiation (Supplementary Fig. 3a-b)32. Upon confirming that both wild-type and SOD1G93A iPSC showed neuronal morphology and gene expression profiles similar to MNs derived from mouse embryonic stem cells (Supplementary Fig. 3c), MHCI levels were evaluated. During the first 72 hours, no significant change in MHCI expression was found between wild-type and SOD1G93A MNs and only a modest down-regulation of MHCI was observed in SOD1G93A MNs by 120 hour (Supplementary Fig. 4a). A further decrease was not observed if SOD1G93A expressing MNs were culture in the presence of SOD1G93A astrocytes (Supplementary Fig. 4b), suggesting that ALS astrocytes act as a main contributor to the down-regulation of MHCI observed in MNs. Of note, the findings described above were not observed in GABAergic neurons, a neuronal population spared from ALS astrocyte induced toxicity when co-cultured 13,14,33,34, since MHCI expression levels in these cells were found to remained constant throughout the culture period in the presence of SOD1G93A astrocytes (Supplementary Fig. 4c).
Next, in order to determine if cell-cell contact between MNs and astrocytes was required for MHCI loss in MNs to occur, we cultured MNs in medium previously used to culture astrocytes but without physical contact with astrocytes. As observed in the presence SOD1G93A astrocytes by 24 hours majority of MNs lost MHCI expression after culture with SOD1G93A astrocyte conditioned medium a time point were at least 95% of MNs are still alive (Supplementary Fig. 5a), suggesting ALS astrocytes secrete factors that induce MHCI down-regulation in MNs. Based on these findings we then evaluated if we could recapitulate this down-regulation of MHCI in MNs observed in the presence of SOD1G93A astrocyte conditioned medium by using compounds acting on pathways shown to be affected in ALS. Compounds causing endoplasmic reticulum (ER) stress, oxidative stress, and inflammatory response were tested. Thapsigargin, a sarco-endoplasmic reticulum calcium ATPase inhibitor, known to induce ER stress in MNs35, led to loss of MHCI expression in more than 76% of MNs, where menadione, an oxidative stress inducer, and the pro-inflammatory molecules, tumor necrosis factor (TNFα), interferon gamma (IFNγ) and interleukin 2 (IL2) showed no or only moderate effects on MNs MHCI levels (Supplementary Fig. 5b). Taken together, these results suggest that astrocytes may secrete inducers of ER stress resulting in loss of MHCI in MNs.
Levels of MHCI in MN determine susceptibility to ALS astrocyte-induced toxicity
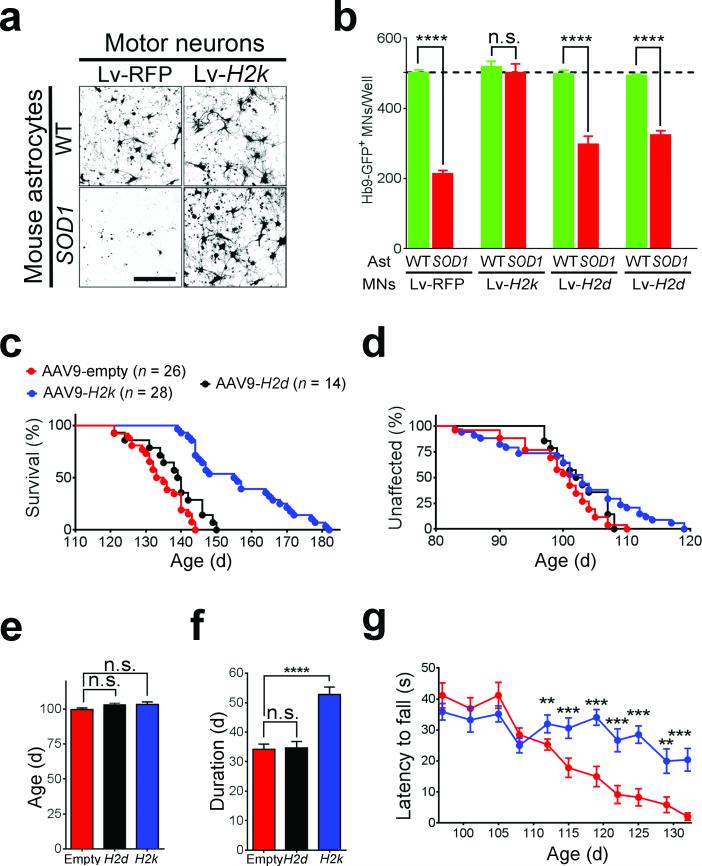
To test the hypothesis that restoring MHCI expression to MNs may prevent astrocyte mediated toxicity, we overexpressed three MHCI molecules in MNs prior to co-culture with mouse ALS astrocytes. Mouse classical MHCI of the subclasses H2db, H2kb or H2ld were overexpressed via lentiviral vectors in Hb9:GFP sorted MNs, resulting in transduction of more than 80% of MNs and rescue of MHCI levels on MNs exposed to SOD1G93A astrocyte (Supplementary Fig. 6a-d)36. While overexpression of H2db or H2ld in MNs resulted in a modest increase in MN survival, overexpression of H2kb fully protected MN from the toxic effects of SOD1G93A astrocytes with no changes in cell morphology (Fig. 3a). This effect was not due to the lentivirus infection per se since expression of RFP did not alter SOD1G93A astrocyte-mediated toxicity, with more than 60% of MNs dying within 120 hours of co-culture (Fig. 3b).
Figure 3. H2kb expression protects MNs from ALS astrocyte-induced toxicity and delays disease progression in SOD1G93A mice.
(a) Representative immunofluorescence images (of three independent experiments) of MNs expressing either RFP or H2k and cultured in the presence of wild type or SOD1G93A astrocytes for 120 hours. Scale bar 100 μm. (b) Survival of MNs expressing either, RFP, H2k, H2k or H2l and cultured in the presence of wild type or SOD1G93A astrocytes for 120 hours. Data shown is a representative of three independent experiments and is displayed as the mean ± s.e.m of counts three replicates (****P<0.0001; n.s., non-significant, one-Way ANOVA). (c) Kaplan-Meier survival curve of female SOD1G93A mice treated with AAV9-H2k (n = 28), AAV9-H2k (n = 14) or AAV9-empty controls (n = 26). Mean survival, AAV9-H2k 156.9 ± 2.6, AAV9-H2k 139.2 ± 1.4, AAV-9-empty 135.5 ± 1.6 days, mean ± s.e.m, P< 0.0001, unpaired t-test. (d,e) Age of onset observed in AAV9 treated SOD1G93A mice and displayed by the Kaplan-Meier curve (d) and the mean age of onset (e). AAV9-H2k 103.3 ± 2.0, AAV9-H2k 103.1 ± 1.2 days, AAV-9-empty 99.73 ± 1.2, mean ± s.e.m, P=0.1, unpaired t- test. (f) Mean disease progression observed in AAV9 treated SOD1G93A mice. AAV9-H2k, 52.7 ± 2.6; AAV9-H2k, 34.62 ± 2.2; AAV9-empty 34.1 ± 1.8 days, mean ± s.e.m, P<0.0001, unpaired t-test. (g) Rotarod performance of AAV9-H2k treated SOD1G93A mice compared with age-matched controls, mean ± s.e.m, *P<0.05; **P<0.01; ***P<0.005, unpaired t- test. WT, wild-type. SOD1, SOD1G93A. Ast, astrocytes.
Knockdown of H2kb expression in MN did not induce MN cell death per se or alter their susceptibility to stress molecules (Supplementary Fig. 7a), but upon co-culture with SOD1G93A astrocytes increased susceptibility to SOD1G93A astrocyte-induced toxicity (Supplementary Fig. 7b). Importantly, reduction of MHC I expression in GABAergic neurons lead to decreased survival upon co-culture with SOD1G93A astrocytes, with an observed 43.7% cell death as compared to scrambled shRNA transduced GABAergic neurons (Supplementary Fig. 7c-e). These findings suggest that MHCI expression by neuronal cells modulates their susceptibility to ALS astrocyte-induced toxicity.
Sustained H2k expression delayed SOD1 disease progression
Since AAV9 readily transduces MNs in the spinal cord when delivered in to the cerebral spinal fluid (CSF)37,38, we constructed AAV9 vectors to target MNs in SOD1G93A mice with H2kb (AAV9-H2k) or H2db (AAV9-H2d). We also used AAV9-GFP to confirm that high level of transduction in spinal cord MNs of SOD1G93A mice could be achieved by our delivery method (Supplementary Fig. 8a). AAV9-H2k (or AAV9-H2k) increased H2kb (or H2db) mRNA expression levels in spinal cords (Supplementary Fig. 8b). Expression of H2kb in MNs via AAV9 delivery starting at post-natal day 1 resulted in a 21-day extension in the mean survival SOD1G93A mice as compared to control (AAV9-empty) littermates (156.9 ± 2.6 days in AAV9-H2k versus 135.5 ± 1.6 days in AAV9-empty (Fig. 3c). Amongst AAV9-H2k injected animals, 39% of the animals survived over 165 days, with the longest living mouse reaching 182 days. When AAV9-H2k was delivered to SOD1G93A mice, no significant (P> 0.05) changes in mean survival were observed (139.2 ± 1.4 days in AAV9-H2k versus 135.5 ± 1.6 days in AAV9-empty). Mean disease onset, as assessed by age at peak body weight did not differ between AAV9-H2k, AAV9-H2k and AAV9-empty groups (103.3 ± 2.0, 103.1 ± 1.2 and 99.73 ± 1.2 days, respectively; Fig. 3d-e). However, disease duration, assessed by disease progression, was increased by 50.3% in AAV9-H2k treated mice, but not in AAV9-H2k treated mice (Fig. 3f). Videos taken of SOD1G93A mice during the disease progression period, show a marked difference between AAV9-H2k injected versus AAV9-empty injected mice (Supplementary Videos 1-4). During this stage, AAV9-H2k injected SOD1G93A mice showed greater ambulatory capacity compared to AAV9-empty injected animals. Motor function was also improved as assessed by rotarod test (Fig. 3g). The delay in disease progression observed when SOD1G93A mice were treated with AAV9-H2k is likely not derived from H2kb expression in astrocytes since the overexpression of H2kb in SOD1G93A astrocytes in vitro did not modify their toxicity towards MNs (Supplementary Fig. 9).
ALS astrocytes express MHCI inhibitory receptors
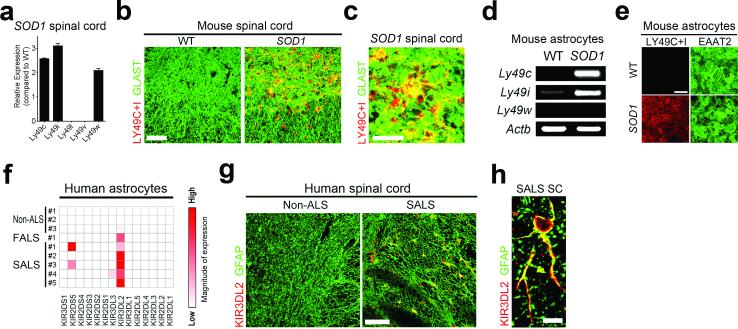
The levels of MHCI expression helps innate immune cells, including natural killer (NK) cells, to distinguish target cells from healthy cells39. Reduced presentation of MHCI antigen on target cells acts as a trigger for cytotoxic T lymphocytes (CTLs) to secrete effector molecules and kill the target cells40. However, when target cells retain MHCI expression, CTLs can sense this MHCI leading to the engagement of signaling cascades that inhibit lymphocytic toxicity and result in the preservation of target cells40,41. To determine if ALS astrocytes have the ability to sense MHCI levels on MNs, we examined expression of MHCI receptors in astrocytes. mRNA expression of H2k inhibitory receptors of the Ly49 family such Ly49c, Ly49i and Ly49w were highly expressed SOD1G93A mice at end stages of disease (Fig. 4a). Immunostaining analysis using antibodies to detect Ly49c and Ly49i (Ly49c/i) receptors confirmed expression of these two receptors in the ventral horn of the lumbar spinal cord of SOD1G93A mice, with little to no expression in age-matched wild-type mice (Fig. 4b-c). 96 ± 2% of Ly49c/i positive cells were astrocytes (defined by immunoreactivity to the astrocyte specific membrane protein GLAST or the cytoplasmic protein GFAP) (Fig. 4c and Supplementary Fig. 10a). Ly49c/i receptors were also expressed in SOD1G93A astrocytes used in our in vitro studies (Fig. 4d-e). These receptors were also detected in infiltrating CTLs found in the spinal cord of SOD1G93A mice42, however CTL numbers were minimal and therefore only accounted for a small fraction of cells expressing Ly49c/i receptors (Supplementary Fig. 10b).
Figure 4. ALS astrocytes express MHCI inhibitory receptors.
(a-e) Evaluation of the expression of MHCI inhibitory receptors found in SOD1G93A mice spinal cord at disease end-stage (a-c) or astrocytes lines (d-e), by RNA analysis (a,d) or immunohistochemistry analysis (b,c,e). (f-h) Expression of the MHCI inhibitory receptor KIR3DL2 in human ALS astrocytes cell lines as determined by RNA analysis (f) and in sections of spinal cord from SALS post–mortem tissue (g-h). Data presented in (a-h) is a representative of finding form three independent experiments. WT, wild-type. SOD1, SOD1G93A. Scale bars 50 μm (b), 200 μm (e, g), 10 μm (c), 5 μm (h).
We extended our analysis to astrocytes derived from individuals with ALS and analyzed mRNA expression of a wide panel of 14 MHCI receptors. Killer cell immunoglobulin-like receptor 3DL2 (KIR3DL2), an inhibitory MHCI receptor, was uniquely expressed in all human ALS astrocyte cell lines tested (Fig. 4f). There was no detectable expression of MHCI inhibitory receptor, including KIR3DL2 or any other KIR in non-ALS control astrocyte cell lines tested. Expression of KIR3DL2 was also confirmed in post-mortem spinal cord samples of SALS individuals, where KIR3DL2 expression was predominantly localized to GFAP positive astrocytes (Fig. 4g-h).
HLA-F protects human MNs from ALS astrocyte toxicity
Major histocompatibility complex, class I, F (HLA-F), a human MHCI molecule, has been identified as a ligand that can physically and functionally interact with the KIR3DL2 receptor43. HLA-F is expressed in MNs of non-ALS spinal cord samples, and its expression is reduced in ALS MNs (Fig. 5a-b). To test whether sustained expression of HLA-F in human MNs protects them from ALS astrocyte induced toxicity, we used an in vitro model system where human MNs and human astrocytes were co-cultured14. MNs were generated from human embryonic stem cells and found to exhibit good neuronal morphology, expression of the MN markers homeobox gene (HB9), neurofilament marker (SMI32) and choline acetyltransferase (ChAT) and minimal to none non-neuronal cell contamination (Fig. 5c). Given that MHCI recognition is species specific, we needed to construct a vector that could deliver HLA-F to human MNs. A lentiviral vector encoding human HLA-F cDNA along with an IRES and eGFP (Lv-HLAF-IRES-eGFP) to track transduced cells was produced. Transgene expression and high levels of MN transduction were confirmed, with a transduction efficiency of over 90% (Fig. 5d), and all cells expressing eGFP were found to express HLA-F (Supplementary Fig. 11). Three-days after the MNs were transduced with Lv-HLAF-IRES-eGFP, astrocytes devoid of contamination by other glia and CTLs, (Supplementary Fig. 2d,e) were added to cultures. After 2 weeks of co-culture we observed that overexpression of HLA-F in human derived MNs increased their survival upon co-culture with either FALS or SALS astrocytes (Fig. 5e-f). No change in MN survival was observed when MNs overexpressing HLA-F were co-cultured with non-ALS astrocytes, suggesting a specific effect of HLA-F in preventing ALS astrocyte mediated toxicity rather than improved general MN survival. To test whether suppression of KIR3DL2 in ALS astrocytes will enhance their toxicity towards MNs, we reduced KIR3DL2 expression in human astrocytes using shRNA directed against KIR3DL2 (Supplementary Fig. 12a). All ALS astrocytes treated with scrambled shRNA were toxic to MNs, with approximate 50% MNs death observed by day 14 (Supplementary Fig. 12b). When the same MNs were co-cultured with ALS astrocytes lacking KIR3DL2 expression, 50% MN death could already be observed by day 7 of co-culture (Supplementary Fig.12b) suggesting an increase cell death activity of ALS astrocytes if they lack the functional interacting domain to HLA-F.
Figure 5. HLA-F expression protects human MNs from ALS astrocyte-induced toxicity.
(a) Representative images (from three independent experiments) of immunohistochemistry analysis performed in human spinal cord tissue probing for HLA-F expression. Green arrowheads point to MNs. HLA-F was visualized by 3,3'-Diaminobenzidine (DAB) staining. (b) Percent of HLA-F positive MNs found in human spinal cords of ALS individuals and control determined as shown in (a). For columns 1 through 5, the total number of MNs was 62, 58, 54, 26 and 42, respectively. Scale bars 20 μm. Number at the bottom of the columns represent subject ID number. (c) Microscope images of human ESC derived MNs used to evaluate morphology and expression of prototypic MN markers. (d) RNA (upper panel) and immunocytochemistry analysis (lower panel) of lentivirus infected cells displaying HLA-F and eGFP expression in human MNs. (e) Representative images (from three independent experiments) of human MNs expressing HLA-F and co-cultured with FALS and SALS astrocyte visualized by ChAT staining. (f) Quantification of the number of surviving MNs as shown in (e). Dotted line represents average MN counts when co-cultured with non-ALS controls. Data shows a representative of three independent experiments and is displayed as the mean ± s.e.m counts of triplicates. (One-Way ANOVA, *P< 0.05; **P< 0.01; ***P<0.001; ns, non-significant P≥0.5). Scale bars 20 μm (a), 50 μm (c-d), 100 μm (e).
Discussion
We and others have recently shown that astrocytes derived from individuals with ALS are toxic to MNs13,14,44. These findings add to the growing evidence that non-cell autonomous mechanisms contribute to MN death in ALS, and point to a crucial role of glia in ALS pathogenesis11,12. However the mechanisms behind ALS astrocyte toxicity and selectivity towards MNs remain unknown. The understanding of these mechanisms is required to develop therapies aiming at delaying or even preventing MN degeneration in ALS. Our findings reported here show that ALS astrocytes recognize MNs as their targets based on the reduced expression of MHCI observed in MNs at later stages of disease in SOD1G93A mouse model as well in ALS autopsy material. The relatively stable levels of MHCI found in GABAergic neurons upon culture with ALS astrocytes helps explain why certain neuronal subtypes other than MNs are spared from the astrocyte toxicity. Interestingly, in ALS microglia do not cause down-regulation of MHCI in MNs, but still induce MN death28, suggesting that the mechanisms for MN recognition by reduced level of MHCI are astrocyte specific within the glial population. Overall, along with previous reports suggesting possible involvement of MHCI in many neurologic disorders23-25,45-47, our findings support the important role of MHCI molecules in the motor neuron degeneration occurring in ALS.
Further studies are necessary to precisely identify signals originating from the astrocytes and the molecular mechanism behind MHCI reduction in MNs upon contact with ALS astrocytes. Here, we showed soluble factors secreted from ALS astrocytes can down-regulate MN MHCI expression, and identify ER stress as a likely mechanism governing this down-regulation. ER dysfunction is a critical contributor to MHCI loss on various cell types48. Indeed, in the SOD1 mouse models, ER stress is one of the first phenotypic alterations detected in vulnerable MNs49,50, and remains a prominent feature of post-mortem ALS subject spinal cord MNs51. Since down-regulation of MHCI may initiate cellular events to trigger ALS astrocyte toxicity, it is imperative to identify how ALS astrocytes cause ER stress in MNs. Among the potential candidates, misfolded SOD1 should be considered due to their known involvements towards ER stress35,52.
Our results are in alignment with the finding that ALS astrocytes might develop aberrant characteristics to convey toxicity to MNs53. Despite this, the observation that FALS and SALS astrocytes express predominantly a single killer cell immunoglobulin-like receptor, KIR3DL2, opens the possibility to find common molecules that have potential therapeutic strategy to ALS despite disease etiology. Due to its ability to interact with KIR3DL2 43, and the low frequency of polymorphisms among different human populations54, HLA-F is a promising candidate. Strikingly, HLA-F protein expression was found to be down regulated in post-mortem samples of ALS spinal cord MNs, and sustained HLA-F expression in human MNs in vitro confers protection from both FALS and SALS astrocyte toxicity. These findings suggest viral vectors, such as AAVs that may be developed to treat CNS disorders55-59 in order to deliver HLA-F to MNs and hamper astrocyte toxicity in the larger body of SALS individuals.
Online Methods
Animals
All procedures were performed in accordance with the NIH Guidelines on the care and use of vertebrate animals and approved by the Institutional Animal Care and Use Committee of the Research Institute at Nationwide Children's Hospital. Transgenic mice that expressed human SOD1 carrying the G93A mutation (B6SJL-TgSOD1G93A), referred to here as SOD1G93A mice, were obtained from Jackson Laboratories and maintained, characterized by the guidelines of Jackson Laboratory for the entire of animal study60 (Bar Harbor, ME). Animals were bred at our facilities and were housed under light/dark (12:12 hour) cycle with food and water ad libitum. At each generation, animals were genotyped, SOD1G93A transgene copy number were verified by quantitative PCR, prior to either the isolation of primary cells or the injection of AAV9. To minimize variability due to gender effects on survival and behavior analysis, only female mice were used for AAV9-H2k injection experiments. After genotyping, SOD1G93A animals were randomly selected for AAV9 injections of control, H2k or H2k. In each litter, half of the animals were treated with AAV9-empty and half with AAV9-H2k or AAV9-H2k. All procedures were performed in accordance with the NIH Guidelines and were approved by the Nationwide Children's Research Institutional Animal Care and Use Committee. The numbers of mice used in each experiment varied are indicated in the figure legends.
SOD1G93A mouse survival and behavioral analysis
Disease stages (previously described28,57) included the following: “Pre-symptomatic stage,” during which mice displayed no disease symptoms and were not yet at peak body weight; “Symptomatic-stage,” during which mice showed overt symptoms characterized by tremors and hindlimb paralysis and showed a 10% or more decrease from the peak of body weight; “End-stage,” during which animals exhibited forelimb and hindlimb paralysis and were unable to right themselves within 30 seconds after being placed on its back. “Disease onset” was defined as the age at which mice reach their peak body weight. “Disease progression” was defined as the time period between disease onset and end stage. Motor coordination was recorded using a rotarod instrument (Columbus Instruments, Columbus, OH), Three trials were performed on accelerating rotarod beginning at 5 rpm per minute twice a week. The time each mouse remained on the rod was recorded. Data collection and analysis was performed by a researcher that was blinded to the treatment of animal groups.
Isolation and culture of mouse glial cells
Astrocytes and microglia were isolated from 110-130 day old SOD1G93A and wild-type B6SJL mice. Astrocyte cultures were prepared as previously described with minor modifications61. Briefly, spinal cords were enzymatically dissociated to single cells with a mixture of Papain (2.5 U/ml; Worthington Biochemical, Lakewood, NJ), Dispase grade II (1 U/ml; Boehringer Mannheim Corporation, Indianapolis, IN) and Dnase I (250 U/ml; Worthington Biochemical) for about 20 minutes. After filtration with a 70 μm nylon mesh, cells were pelleted, and resuspended in DMEM/F12 (Invitrogen, Carlsbad, CA) which was supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 0.2% N2 supplement (Invitrogen). The cells were then plated onto laminin coated 75 cm2 tissue culture flasks. Upon confluence, flasks were shaken overnight in order to remove potential microglial cells and then were treated with cytosine arabinose (20 μM, Sigma-Aldrich, St. Louis, MO). Prior to use, astrocyte preparations were screened for the presence of CTLs and natural killer (NK) cells and were found to be devoid of them.
Microglia were isolated following a protocol previously described28. Briefly, tissues were fragmented with a scalpel and incubated in enzymatic solution containing papain (2.5 U/ml; Worthington Biochemical) for 60 minutes at 37 °C. 20% FBS in Hank's Balanced Salt Solution (HBSS, Invitrogen) was applied to the tissue, and they were then centrifuged at 200×g for 4 minutes. Cell pellets were resuspended in 2 ml of DNase I (0.5 mg/ml, Worthington Biochemical) in HBSS and were incubated for 5 minutes at room temperature. Tissue was gently disrupted with fire-polished Pasteur pipettes, filtered through a 70 micron cell strainer, and centrifuged at 200×g for 4 minutes. Pellet was then resuspended in 20ml of 20% isotonic Percoll (GE healthcare) in HBSS. 20ml of pure HBSS was carefully laid on top the percoll layer and centrifugation was performed at 200×g for 20 minutes with slow acceleration and no brake. The pellet containing the mixed glial cell population was washed once with HBSS and was suspended in Dulbecco's modified Eagle's/F12 medium with GlutaMAX™ (DMEM/F12, Invitrogen) supplemented with 10% heat inactivated FBS, antibiotic-antimycotic (all from Life Technologies) and 5 ng/ml of carrier-free murine recombinant granulocyte and macrophage colony stimulating factor (GM-CSF) (R&D systems, Minneapolis, MN). Cell suspension was then plated on a poly-L-lysine (Sigma) coated plate and maintained at 37°C. The media was replaced every 3 days until the cells reached confluency. Microglia that formed a non-adherent, floating cell layer were collected, replated, and cultured for an extended period of time. Microglia were incubated for 3 days without GM-CSF before being re-plated for co-culture with MNs. Prior to analysis, microglia preparations were tested for the presence of CTLs and NK cells and were found to be devoid of them.
Mouse NPC isolation and differentiation into astrocytes
NPCs were isolated according to methods previously described29,62. Briefly, spinal cords were enzymatically dissociated in the same way as described for astrocytes. The cell suspension obtained was then mixed with an equal volume of isotonic Percoll (GE Healthcare) and was centrifuged at 20,000×g for 30 minutes at room temperature. Cells from the low-buoyancy fraction (5-10 ml above the red blood cell layer) were harvested, washed thoroughly with D-PBS/PSF (Invitrogen) and plated in 60 mm uncoated plates. Cells were grown in growth medium (DMEM/F12, Invitrogen) with 1% N2 supplement (Invitrogen), 20 ng/ml of fibroblast growth factor-2 (FGF-2, Peprotech, Rocky Hill, NJ) and 20 ng/ml of endothelial growth factor (EGF, Peprotech). Cells were first grown as neurospheres and then were placed on a polyornithine-laminin (P/L)-coated plates, in which they grow as monolayer cultures. NPC cultures were found to be devoid of astrocytes, microglia, CTLs and NK cells contaminants. Once cultures were established, NPCs from wild-type and SOD1G93A mice were used to generate astrocytes by withdrawing growth factors and supplementing the medium with 10% FBS (astrocyte media). The media was changed every 2 days thereafter. Astrocytes were allowed to mature for 7 days prior to being used in the experiments described above. Highly enriched astrocyte cultures were obtained with no detectable levels of microglia, CTLs and NK cells.
Human post-mortem NPC derived astrocytes
Post-mortem spinal cords were obtained from the National Disease Research Interchange (NDRI, Philadelphia, PA) and from Dr. Fred Gage (Salk Institute, CA). Informed consent was obtained from all subjects. Receipt of human tissues was granted through Nationwide Children's Hospital Institutional Review Board (IRB08-00402) and all human samples were used in accordance with their approved protocols. Extensive phenotypic characterization of the cell lines used here has been previously described13,44. Cells were grown on laminin-coated plates in astrocyte media supplemented 0.2% N2 supplement (Invitrogen). Media change occurred every 3 days, and cells were passaged when cultures reached 80% confluency. Human astrocyte cultures were found to be devoid of microglia, CTLs and NK cells.
iPSC generation
NPCs, expressing the MN Hb9::GFP reporter, obtained from wild-type and SOD1G93A mice were converted to iPSCs. As previously described, retrovirus encoding OCT3/4 and KLF4 were sufficient to generate iPSC clones63,64. 20 viral particles per cell were needed to efficiently reprogram the cells. Cells were cultured in the presence of NPC media for four days followed by a change to mouse embryonic stem cell (mESC) media with DMEM (Millipore, Billerica, MA), supplemented with 18% ES FBS (Invitrogen), L-glutamine (2mM, Invitrogen), nonessential amino acids (1x, Millipore), antibiotic-antimycotic (Invitrogen), 2-mercaptoethanol (Sigma), and recombinant LIF (100U/ml, Millipore). iPSC clones were morphologically similar to mouse ESCs (HBG3 cells, Thomas Jessell, Columbia University) and were obtained within two weeks. A wide panel of markers was used to compare ESCs with the newly generated iPSC lines.
Mouse MN differentiation
Mouse ESCs or iPSCs expressing Hb9::GFP reporter were cultured on top of inactivated mouse fibroblasts (Millipore). MN differentiation was induced by plating 1-2 × 106 cells per 10 cm dish in the presence of 2 μM retinoic acid (Sigma-Aldrich) and 2 μM purmorphamine (Calbiochem, Billerica, MA). After 5 days of differentiation, embryonic bodies were dissociated and sorted based on levels of GFP using a FACSVantage/DiVa sorter (BD Biosciences, Rockville, MD).
NPC differentiation into GABAergic neurons
Mouse NPCs were induced to differentiate into GABAergic neurons by supplementing growth medium with 0.1% FBS (Invitrogen), retinoic acid (1 μM, Sigma-Aldrich), and forskolin (5 μM, Sigma-Aldrich). Media were changed every day. Cultures were allowed to differentiate for 7 days prior to being used for experiments.
Co-culture of mouse astrocytes with mouse MNs
Astrocytes were plated at the density of 35,000 cells per well in 96-well plates coated with laminin. After 48 hours, FACS sorted GFP+ MNs were plated on top of the astrocyte monolayer at a density of 10,000 cells per well. Co-cultures were performed in MN media composed of DMEM/F12 (Invitrogen) supplemented with 5% horse serum (Equitech Bio, Kerrville, TX), 2% N2 supplement (Invitrogen), 2% B27 supplement (Invitrogen), 10 ng/ml GDNF (Invitrogen), 10 ng/ml BDNF (Invitrogen), 10 ng/ml CNTF (Invitrogen). Half of the media was replaced every other day, with the addition of fresh growth factors.
Sustained expression of MHCI molecules in mouse MNs
To express histocompatibility 2 subclass MHCI in MNs, a previously described protocol was followed, with minor modifications65. Briefly, wild-type astrocytes were plated on a laminin-coated transwell (Corning, Lowell, MA) using MN media. After 24 hours, sorted GFP+ MNs were plated on a separate laminin-coated 96 well plate in media, conditioned by wild-type astrocytes. Four hours later, the transwell containing wild-type astrocytes was transferred into the MN plate, after verification that all MNs were fully attached and were starting to show neuritic extensions. The following day, the transwell of wild-type astrocytes was removed and the MNs were infected with Lv- H2k, H2k or H2l (40 viral particles per MN). Twelve hours post-infection, co-culture with wild-type astrocytes via transwell was resumed. After 72 hours, the transwell was removed and the co-culture experiments with wild-type and SOD1G93A astrocytes were initiated. Experiments were performed independently by two investigators.
Astrocyte conditioned media
Astrocyte conditioned medium was prepared by co-culturing mouse MNs and mouse astrocytes for 120 hours. After removal of cell debris by centrifugation (500 ×g for 10 min), medium was supplemented with GDNF, CNTF and BDNF. This medium was added to MNs cultures and cultures were evaluated after 24 hours.
Co-culture of human astrocytes with human MNs expressing HLA-F
MNs were obtained by differentiating human ES cell-derived MN progenitors (Lonza, Walkersville, MD) following the manufacturer's instructions. MN progenitors were plated at a density of 10,000 cells per well in a laminin coated 96-well plate. 48 hours after plating, the cells were infected with adenovirus encoding NGN2, ISL1, and LHX3 in order to enhance efficiency and shorten the time required for MN differentiation66. After 10 days of MN differentiation, MNs were infected with lentivirus to overexpress HLA-F (20 viral particles per MN). 3 days after, 10,000 human astrocytes were added to each well. Co-cultures were allowed to continue for another 14 days, with half of the media being replaced every other day. Due to the limited number of MNs available at a time of study, astrocyted were randomly choosen and co-culture initiated.
Viral vectors
To knockdown H2kb levels in MNs or GABAergic neurons, sequences from the RNAi Consortium lentiviral shRNA library were screened and the sequence 5’-TAAAGAGAACTGAGGGCTCTG -3’ was used. The sequence 5’-GGCGTAGATGTCCGATAAGAA-3’ was used for the scrambled shRNA control. Sequences were cloned into a lentiviral vectors. H2kb cDNA was purchased from Genecopia (Rockville, MD) and referred to as H2k. H2db cDNA (NM_010380.3) was purchased from Thermoscientific (Pittsburgh, PA) and referred to as H2k. H2-ld cDNA (NM_001267808.1) was synthesized by Genscript (Piscataway, NJ) referred to as H2l. To knockdown KIR3DL2 in human ALS astrocytes, sequences from the RNAi Consortium lentiviral shRNA library were also screened and the sequence 5’-TAAAGGAGAAAGAAGAGGAGG -3’ was used. The sequence 5’-GGGAGAAAGAAGGAGGATAAA-3’ was used for the scrambled shRNA control. The HLA-F cDNA (NM_001098479.1) was purchased from Genecopia (Rockville). The production and purification of the lentivirus were performed as previously reported29.
MN cell viability
At various time points during the co-culture of mouse astrocytes and mouse MNs, cell survival, neuritic length and soma size of MNs were recorded using a fully automated IN CELL 6000 cell imager (GE Healthcare) as previously reported44. Images were processed with the Developer and Analyzer software package (GE Healthcare). Otherwise noted, images shown represent 120 hours post co-culture. All counts were performed in triplicate and repeated at least three times.
AAV9 injection in SOD1G93A mice
H2kb or H2db cDNA sequence used in our in vitro experiments was cloned into a AAV9 vector that has been reported to transduce high levels of MNs in brain and spinal cords56,57. Self-complementary AAV9 encoding no transgene (AAV9-empty), or GFP (AAV9-GFP) or H2db (AAV9-H2k) or H2kb (AAV9-H2k) was produced by transient transfection procedures using a double-stranded AAV2-ITR-based CB vector, with a plasmid encoding Rep2Cap9 sequence as previously described along with an adenoviral helper plasmid pHelper (Stratagene, Santa Clara, CA) in 293 cells. Injections of AAV9 were performed directly to the cerebral spinal fluid (CSF) at postnatal day 1 by direct injection into the lateral ventricles. Injection was performed with laser-pulled borosilicate glass needles (Sutter Instruments, Novato, CA, O.D.: 1.2 mm, I.D.: 0.69 mm 10 cm length) as previously described59. Animals received a total dose of 2.33×1013vg/kg in to 4 μl volume. To validate and minimize variability associated with the injection procedure, at least two fold (n = 24) of the minimum number of animals that the guidelines for preclinical animal research in ALS/MND suggests67 was used for our survival studies.
RNA isolation and RT-PCR
RNA was harvested using the RT2 q-PCR-grade RNA isolation kit (Qiagen, Frederick, MD) and total RNA was reverse transcribed with RT2 First Strand Kit (Qiagen) according to the manufacturer's instructions. After ensuring all cDNAs were devoid of genomic DNA contamination, mouse and human gene transcripts were amplified using gene-specific primers described in Supplementary Table 4. For detection of MHCI inhibitor receptor transcripts from the Ly49 gene and killer-cell immunoglobulin-like receptors (KIRs) families, astrocytes were prepared by co-culturing with mouse MNs and RT-PCR was performed using primer sets previously described68. Real-time quantitative PCR reactions were performed using RT2 Real-Time SYBR Green/Rox Master Mix (Qiagen, Frederick, MD). Each sample was run in triplicate and relative concentration was calculated using the ddCt values normalized to endogenous actin transcript.
In situ hybridization
Spinal cords were removed from 60 day old wild-type mice and frozen in M1 embedding matrix (Shandon, Pittsburgh). The negative control, labeled with H2kbdb KO, was an H2kb−/−db−/− double knockout as previously described22. Twelve μm cryostat sections were obtained, affixed to slides, air-dried, and stored at −80°C. In situ hybridization was performed as previously described22,69. Briefly, slides were thawed and fixed in 4% paraformaldehyde before proteinase K (1 μg/ml) treatment. Slides were then acetylated and dehydrated in an ethanol series (50%, 75%, 2 X 95%, and 2X 100%). Labeled (α-35S-UTP) riboprobe was diluted to 0.75 × 107 cpm/ml in 1X Denhardt's solution with 50% deionized formamide, 10% dextran sulfate, 0.3 M NaCl, 10 mM Tris-HCl pH 8.0, and 1 mM EDTA pH 8.0; applied to sections; and then hybridization took place at 62°C for 12–18 h. After hybridization, coverslips were floated off in 4× SSC, and then treated with 50 μg/ml RNase A for 30 min at 37°C. Slides were washed with a series of SSC solutions, beginning at 2X and concluding with a high-stringency wash of 0.1X SSC (0.15 M sodium chloride/0.015 M sodium citrate, pH 7) at 60°C for 30 min. Finally, sections were dehydrated through an ethanol series and placed on film. After exposure to Kodak XAR-5 film at room temperature, sections were coated with NTB-2 emulsion and developed after 2–4 weeks.
The sequence of the H2db probe was: 3’-AGGTGGGCTACGTGGACGACGAGGAGTTCGTGCGCTTCGACAGCGACGCGGAGA ATCCGAGATATGAGCCGCGGGCGCCGTGGATGGAGCAGGAGGGGCCGGAGTATT GGGAGCGGGAAACACAGAAAGCCAAGGGCCAAGAGCAGTGGTTCCGAGTGAGCC TGAGGAACCTGCTCGGCTACTACAACCAGAGCGCGGGCGGCTCTCACACACTCCA GCAGATGTCTGGCTGTGACTTGGGGTCGGACTGGCGCCTCCTCCGCGGGTACCT GCAGTTCGCCTATGAAGGCCGCGATTACATCGCCCTGAACGAGAACCCAC-5’.
Adjacent sections were hybridized with sense and antisense probes. No specific hybridization was seen using sense probes.
Fixation and immunostaining
Cells were fixed with 4% paraformaldehyde (PFA) for 10 min. Mouse spinal cords were obtained by intracardiac perfusion with 4% PFA followed by 24 hours of post-fixation. Spinal cords were rinsed twice with 0.1 M sodium phosphate buffer and immersed in 30% sucrose for 2 days at 4°C or until the spinal cords sank to the bottom of the 50ml conical. Fixed spinal cords were embedded and sectioned using a vibratome (40 μm). For antigen detection using frozen sections, mouse spinal cord tissues were cut in 5- to 6-mm sections and embedded in Tissue-Tek OCT compound (Sakura Finetek) and frozen with dry ice. Tissues were then sectioned at 10 μm with a cryostat, collected directly on gelatinized objective slides and then stored at −20 °C before immunocytochemical analysis. Paraffin-embedded human spinal cord tissues were obtained from NDRI and from Dr. Jonathan Glass and Dr. Marla Gearing (Emory University, GA), which had obtained informed consent from all prospective donors. Tissues were sectioned at 10 μm and antigen retrieval methods were applied based on manufacturer's suggestions where primary antibodies were purchased. Staining of control and experimental groups was always performed in parallel. Antibodies used are listed in Supplementary Table 1. For most antigens, samples were first incubated for 1 hour in TBS containing 0.1% triton-X and 10% donkey serum, followed by incubation with the primary antibody for 48-72 hours at 4°C. For Ly49C/I staining, frozen sections previously prepared on slides were washed with TBS three times, then incubated for two hours in TBS containing 5% donkey serum, followed by incubation with primary antibody for 72 hours at 4°C. detection with cyanine dyes (Cy2, Cy3, and Cy5) labeled secondary antibodies used at the dilution of 1:250 (Jackson immunoresearch, West Grove, PA) was performed for 2 hours at room temperature.
MHCI staining was performed according to a previously described protocol, with minor modifications25,46. The antibody ER-HR52 recognizes histocompatibility 2 subclasses for mouse classical MHCI molecules (H2kbdb) and the antibody EMR8-5 recognizes all HLA-A, B and C of the human classical MHCI molecules (HLA-ABC), therefore we refer to it in the text as MHCI. HLA-F expression was probed with a rabbit-anti-HLA-F antibody75,76. Membrane and cytoplasmic expression of HLA-F was interpreted as positive. Briefly, for in vitro MHCI labeling, cells on coverslips were fixed, blocked and incubated with primary and secondary antibodies without membrane permeabilization during the staining process. MHCI levels were obtained from randomly selected MNs using confocal microscope. MHCI fluorescence intensity values per MN were automatically measured using Adobe Photoshop CS5 extended version (Adobe, San Jose, CA) and were corrected to soma size and GFP expression of cells analyzed. For in vivo MHCI labelling, cell permeabilization was achieved using 0.05% triton-X for mouse spinal cord samples and 0.1% saponin for human spinal cord samples for 30 minutes at room temperature. Incubation with primary and secondary antibodies was performed in 10% donkey serum without any detergent. Detection of MHCI in paraffin embedded human tissue was achieved with 3,3’-diaminobensidine staining by using the ABC and VectorRed Kit protocols (Vector Laboratories, Burlingame, CA). Tissues were counterstained with Hematoxylin QS solution (Vector Laboratories). Fluorescence images were captured on a laser scanning confocal microscope (Carl Zeiss Microscopy, Thornwood, NY) and 3,3’-diaminobensidine stained images were captured with the Zeiss Axioscope. Evaluation of MHCI levels in MNs were performed by an operator blinded to the identity of sample been evaluated.
Statistical analysis
Statistical analysis was performed using Graph Pad Prism 6 software (La Jolla). Depending on the number of variables and time-points in each experiment, statistical analysis of mean differences between groups was performed by either Student's t-test or multiway ANOVA followed by a Bonferroni post hoc analysis. Kaplan-Meier survival analyses were analyzed by the log-rank test. Comparison of mean survival, disease onset and progression were analyzed by the unpaired t test. Specific statistical tests, P values and sample size are indicated in figure legends.
Supplementary Material
Acknowledgements
We would like to thank C. Shatz (Stanford University) for critical discussions, S. Eckardt for expert editorial assistance, The National Disease Research Institute (NDRI), A. Burghes (The Ohio State University), J. R. Mendell (Nationwide Children's), J. Glass and M. Gearing (Emory University supported by NIH/NINDS P30NS055077) for providing human spinal cord specimens, F. Gage (Salk Institute) for providing human post-mortem NPCs used to generate non-ALS astrocytes, M. Hester for guidance with iPSC generation, and K. Campbell for technical assistance. This work was supported by the US National Institutes of Health (NIH) grant R01-NS644912, RC2-NS69476, and funding from the Robert Packard Center for ALS Research, the Project A.L.S. and the Helping Link Foundation. Authors part of this work also received research fellowships from the Swiss National Science Foundation, the Marie Curie Foundation, NINDS Training in Neuromuscular Disease, The Ohio State University Presidential Fellowship and The Ohio State University and Nationwide Children's Hospital Muscle Group Fellowship.
Footnotes
Author contributions All authors contributed to the design of the experiments. Mouse astrocyte, microglia and NPC isolation was performed by S. Song, C. J. Miranda, L. Braun, A. Frakes and S. Likhite. Human astrocytes cultures were performed by S. Song, C. J. Miranda, K. Meyer, and L. Ferraiuolo. Neuronal cell differentiation and co-culture experiments were performed by S. Song, C. J. Miranda, K. Meyer, A. Frakes and S. Likhite. In situ hybridization of H2db transcripts was performed by M. J. McConnell. RT-PCR and immunocytochemistry were performed by S. Song, C. J. Miranda, L. Braun, and A. K. Bevan. Lentivirus production was performed by S. Song, C. J. Miranda, L. Braun, K. Meyer, A. Frakes, L. Ferraiuolo, and S. Likhite. Data analysis was performed by S. Song, C. J. Miranda, L. Braun, K. Meyer, A. Frakes, L. Ferraiuolo, S. Likhite, A. K. Bevan, K. D. Foust, M. J. McConnell, C. M. Walker, and B. K. Kaspar. The manuscript and figures were prepared by S. Song, C. J. Miranda, and B. K. Kaspar with input from all co-authors.
Supplementary Information is available in the online version of the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Hardiman O, van den Berg LH, Kiernan MC. Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7:639–649. doi: 10.1038/nrneurol.2011.153. [DOI] [PubMed] [Google Scholar]
- 2.Brown RH., Jr. Amyotrophic lateral sclerosis. Insights from genetics. Arch Neurol. 1997;54:1246–1250. doi: 10.1001/archneur.1997.00550220050013. [DOI] [PubMed] [Google Scholar]
- 3.Kwiatkowski TJ, Jr., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 4.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 5.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Deerlin VM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosco DA, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nature neuroscience. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruzman A, et al. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12524–12529. doi: 10.1073/pnas.0705044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurney ME, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 11.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philips T, Rothstein JD. Glial cells in amyotrophic lateral sclerosis. Exp Neurol. 2014 doi: 10.1016/j.expneurol.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haidet-Phillips AM, et al. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat Biotechnol. 2011;29:824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Re DB, et al. Necroptosis drives motor neuron death in models of both sporadic and familial ALS. Neuron. 2014;81:1001–1008. doi: 10.1016/j.neuron.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boulanger LM. Immune proteins in brain development and synaptic plasticity. Neuron. 2009;64:93–109. doi: 10.1016/j.neuron.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Tian L, Ma L, Kaarela T, Li Z. Neuroimmune crosstalk in the central nervous system and its significance for neurological diseases. Journal of neuroinflammation. 2012;9:155. doi: 10.1186/1742-2094-9-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Needleman LA, Liu XB, El-Sabeawy F, Jones EG, McAllister AK. MHC class I molecules are present both pre- and postsynaptically in the visual cortex during postnatal development and in adulthood. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16999–17004. doi: 10.1073/pnas.1006087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fourgeaud L, et al. MHC class I modulates NMDA receptor function and AMPA receptor trafficking. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22278–22283. doi: 10.1073/pnas.0914064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goddard CA, Butts DA, Shatz CJ. Regulation of CNS synapses by neuronal MHC class I. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6828–6833. doi: 10.1073/pnas.0702023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huh GS, et al. Functional requirement for class I MHC in CNS development and plasticity. Science. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H, et al. Synapse elimination and learning rules co-regulated by MHC class I H2-Db. Nature. 2014;509:195–200. doi: 10.1038/nature13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McConnell MJ, Huang YH, Datwani A, Shatz CJ. H2-K(b) and H2-D(b) regulate cerebellar long-term depression and limit motor learning. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6784–6789. doi: 10.1073/pnas.0902018106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freria CM, Zanon RG, Santos LM, Oliveira AL. Major histocompatibility complex class I expression and glial reaction influence spinal motoneuron synaptic plasticity during the course of experimental autoimmune encephalomyelitis. J Comp Neurol. 2010;518:990–1007. doi: 10.1002/cne.22259. [DOI] [PubMed] [Google Scholar]
- 24.Kim T, et al. Human LilrB2 is a beta-amyloid receptor and its murine homolog PirB regulates synaptic plasticity in an Alzheimer's model. Science. 2013;341:1399–1404. doi: 10.1126/science.1242077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nardo G, et al. Transcriptomic indices of fast and slow disease progression in two mouse models of amyotrophic lateral sclerosis. Brain. 2013;136:3305–3332. doi: 10.1093/brain/awt250. [DOI] [PubMed] [Google Scholar]
- 26.Staats KA, et al. Beta-2 microglobulin is important for disease progression in a murine model for amyotrophic lateral sclerosis. Front Cell Neurosci. 2013;7:249. doi: 10.3389/fncel.2013.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linda H, Hammarberg H, Piehl F, Khademi M, Olsson T. Expression of MHC class I heavy chain and beta2-microglobulin in rat brainstem motoneurons and nigral dopaminergic neurons. J Neuroimmunol. 1999;101:76–86. doi: 10.1016/s0165-5728(99)00135-6. [DOI] [PubMed] [Google Scholar]
- 28.Frakes AE, et al. Microglia induce motor neuron death via the classical NF-kappaB pathway in amyotrophic lateral sclerosis. Neuron. 2014;81:1009–1023. doi: 10.1016/j.neuron.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miranda CJ, et al. Aging brain microenvironment decreases hippocampal neurogenesis through Wnt-mediated survivin signaling. Aging Cell. 2012 doi: 10.1111/j.1474-9726.2012.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clarke LE, Barres BA. Emerging roles of astrocytes in neural circuit development. Nature reviews. Neuroscience. 2013;14:311–321. doi: 10.1038/nrn3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, et al. The expression pattern of classical MHC class I molecules in the development of mouse central nervous system. Neurochemical research. 2013;38:290–299. doi: 10.1007/s11064-012-0920-0. [DOI] [PubMed] [Google Scholar]
- 32.Israelson A, et al. Macrophage Migration Inhibitory Factor as a Chaperone Inhibiting Accumulation of Misfolded SOD1. Neuron. 2015;86:218–232. doi: 10.1016/j.neuron.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchetto MC, et al. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Nagai M, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nature neuroscience. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishitoh H, et al. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes & development. 2008;22:1451–1464. doi: 10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dodge JC, et al. Delivery of AAV-IGF-1 to the CNS extends survival in ALS mice through modification of aberrant glial cell activity. Molecular therapy : the journal of the American Society of Gene Therapy. 2008;16:1056–1064. doi: 10.1038/mt.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakrabarty P, et al. Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS One. 2013;8:e67680. doi: 10.1371/journal.pone.0067680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robbins KL, Glascock JJ, Osman EY, Miller MR, Lorson CL. Defining the therapeutic window in a severe animal model of spinal muscular atrophy. Hum Mol Genet. 2014;23:4559–4568. doi: 10.1093/hmg/ddu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tay CH, Szomolanyi-Tsuda E, Welsh RM. Control of infections by NK cells. Current topics in microbiology and immunology. 1998;230:193–220. doi: 10.1007/978-3-642-46859-9_12. [DOI] [PubMed] [Google Scholar]
- 40.Lanier LL. NK cell recognition. Annual review of immunology. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 41.Long EO. Regulation of immune responses through inhibitory receptors. Annual review of immunology. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 42.Chiu IM, et al. T lymphocytes potentiate endogenous neuroprotective inflammation in a mouse model of ALS. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17913–17918. doi: 10.1073/pnas.0804610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodridge JP, Burian A, Lee N, Geraghty DE. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J Immunol. 2013;191:3553–3562. doi: 10.4049/jimmunol.1300081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer K, et al. Direct conversion of patient fibroblasts demonstrates non-cell autonomous toxicity of astrocytes to motor neurons in familial and sporadic ALS. Proc Natl Acad Sci U S A. 2014;111:829–832. doi: 10.1073/pnas.1314085111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oliveira AL, et al. A role for MHC class I molecules in synaptic plasticity and regeneration of neurons after axotomy. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17843–17848. doi: 10.1073/pnas.0408154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thams S, et al. Classical major histocompatibility complex class I molecules in motoneurons: new actors at the neuromuscular junction. J Neurosci. 2009;29:13503–13515. doi: 10.1523/JNEUROSCI.0981-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rivera-Quinones C, et al. Absence of neurological deficits following extensive demyelination in a class I-deficient murine model of multiple sclerosis. Nat Med. 1998;4:187–193. doi: 10.1038/nm0298-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen TH, Bouvier M. MHC class I antigen presentation: learning from viral evasion strategies. Nat Rev Immunol. 2009;9:503–513. doi: 10.1038/nri2575. [DOI] [PubMed] [Google Scholar]
- 49.Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nature neuroscience. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 50.Filezac de L'Etang A, et al. Marinesco-Sjogren syndrome protein SIL1 regulates motor neuron subtype-selective ER stress in ALS. Nature neuroscience. 2015;18:227–238. doi: 10.1038/nn.3903. [DOI] [PubMed] [Google Scholar]
- 51.Lautenschlaeger J, Prell T, Grosskreutz J. Endoplasmic reticulum stress and the ER mitochondrial calcium cycle in amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2012;13:166–177. doi: 10.3109/17482968.2011.641569. [DOI] [PubMed] [Google Scholar]
- 52.Kikuchi H, et al. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6025–6030. doi: 10.1073/pnas.0509227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diaz-Amarilla P, et al. Phenotypically aberrant astrocytes that promote motoneuron damage in a model of inherited amyotrophic lateral sclerosis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18126–18131. doi: 10.1073/pnas.1110689108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moscoso J, Serrano-Vela JI, Pacheco R, Arnaiz-Villena A. HLA-G, -E and -F: allelism, function and evolution. Transpl Immunol. 2006;17:61–64. doi: 10.1016/j.trim.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 55.Bevan AK, et al. Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:1971–1980. doi: 10.1038/mt.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foust KD, et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foust KD, et al. Therapeutic AAV9-mediated suppression of mutant SOD1 slows disease progression and extends survival in models of inherited ALS. Mol Ther. 2013;21:2148–2159. doi: 10.1038/mt.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyazaki Y, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 2012;18:1136–1141. doi: 10.1038/nm.2791. [DOI] [PubMed] [Google Scholar]
- 59.Meyer K, et al. Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA: a dose-response study in mice and nonhuman primates. Molecular therapy : the journal of the American Society of Gene Therapy. 2015;23:477–487. doi: 10.1038/mt.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melanie Leitner SM. Cathleen Lutz. Working with ALS Mice (Guidelines for preclinical testing & colony management). Manual in Jackson Laboratory. 2009 [Google Scholar]
- 61.Noble M, Mayer-Proschel M. Culture of astrocytes, oligodendrocytes, and O-2A progenitor cells. MIT press; Cambridge: 1998. [Google Scholar]
- 62.Ray J, Gage FH. Differential properties of adult rat and mouse brain-derived neural stem/progenitor cells. Mol Cell Neurosci. 2006;31:560–573. doi: 10.1016/j.mcn.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 63.Hester ME, et al. Two factor reprogramming of human neural stem cells into pluripotency. PLoS One. 2009;4:e7044. doi: 10.1371/journal.pone.0007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim JB, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 65.Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 66.Hester ME, et al. Rapid and efficient generation of functional motor neurons from human pluripotent stem cells using gene delivered transcription factor codes. Mol Ther. 2011;19:1905–1912. doi: 10.1038/mt.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ludolph AC, et al. Guidelines for preclinical animal research in ALS/MND: A consensus meeting. Amyotrophic lateral sclerosis : official publication of the World Federation of Neurology Research Group on Motor Neuron Diseases. 2010;11:38–45. doi: 10.3109/17482960903545334. [DOI] [PubMed] [Google Scholar]
- 68.Thompson A, van der Slik AR, Koning F, van Bergen J. An improved RTPCR method for the detection of killer-cell immunoglobulin-like receptor (KIR) transcripts. Immunogenetics. 2006;58:865–872. doi: 10.1007/s00251-006-0163-9. [DOI] [PubMed] [Google Scholar]
- 69.Syken J, Shatz CJ. Expression of T cell receptor beta locus in central nervous system neurons. Proc Natl Acad Sci U S A. 2003;100:13048–13053. doi: 10.1073/pnas.1735415100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.