Abstract
Most membrane attack complex-perforin/cholesterol-dependent cytolysin (MACPF/CDC) proteins are thought to form pores in target membranes by assembling into pre-pore oligomers before undergoing a pre-pore to pore transition. Assembly during pore formation is into both full rings of subunits and incomplete rings (arcs). The balance between arcs and full rings is determined by a mechanism dependent on protein concentration in which arc pores arise due to kinetic trapping of the pre-pore forms by the depletion of free protein subunits during oligomerization. Here we describe the use of a kinetic assay to study pore formation in red blood cells by the MACPF/CDC pneumolysin from Streptococcus pneumoniae. We show that cell lysis displays two kinds of dependence on protein concentration. At lower concentrations, it is dependent on the pre-pore to pore transition of arc oligomers, which we show to be a cooperative process. At higher concentrations, it is dependent on the amount of pneumolysin bound to the membrane and reflects the affinity of the protein for its receptor, cholesterol. A lag occurs before cell lysis begins; this is dependent on oligomerization of pneumolysin. Kinetic dissection of cell lysis by pneumolysin demonstrates the capacity of MACPF/CDCs to generate pore-forming oligomeric structures of variable size with, most likely, different functional roles in biology.
Keywords: Pore formation, Kinetics, MACPF/CDC, Toroidal pore, Oligomerization, Membrane structure
Introduction
The membrane attack complex-perforin/cholesterol-dependent cytolysin (MACPF/CDC) family is the largest-known group of pore-forming proteins (Anderluh and Gilbert 2014; Gilbert et al. 2013). Members have been identified in every kind of cellular life form apart from the Archaebacteria, and within their producing organisms they enact a plethora of different biological functions (Anderluh et al. 2014). The two family names combined within the current denomination for this group of proteins were initially identified separately. Although their pore structures are superficially similar in ultrastructural appearance (Bhakdi and Tranum-Jensen 1991; Bhakdi et al. 1985; Gilbert et al. 2013; Morgan et al. 1994; Young et al. 1986a), they display no directly detectable sequence homology. The cholesterol-dependent cytolysins (CDCs) of Gram-positive bacteria have long been recognized as pathogenicity determinants of their producing organisms such as Streptococcus pneumoniae (producing pneumolysin) (Hirst et al. 2008), Clostridium perfringens (perfringolysin) (Awad et al. 2001) and Listeria monocytogenes (listeriolysin) (Birmingham et al. 2008; Czuczman et al. 2014). CDCs are adapted to their different producing organisms and to the hosts they, in turn, infect. This was made clear from phylogenetic analysis of their sequences, which displays a clustering mapping onto bacterial genus and bacterial environment (Anderluh et al. 2014). The other branch defining this family of proteins, the ‘perforins’ or ‘MACPFs’ (Gilbert 2015), was first identified in the form of the serum complement membrane attack complex (Borsos et al. 1964; Tschopp 1984) and then in mammalian perforin-1 (Podack and Dennert 1983; Young et al. 1986a), which cytotoxic T lymphocytes and natural killer cells use to deliver a lethal hit to target antigen-presenting cells (Metkar et al. 2015; Voskoboinik et al. 2006). It was, however, only the solution of 3D atomic structures for two MACPF proteins, in 2007, which revealed that MACPFs and CDCs clearly are structurally homologous proteins with a common evolutionary ancestor (Hadders et al. 2007; Rosado et al. 2007). Several structures of MACPF proteins are now known, and these are allowing structurally based phylogenetic studies to be carried out with increasing accuracy (Gilbert et al. 2013, 2014; Gilbert 2014, 2015). However, the separate naming of CDCs and MACPFs must now be seen to be a ‘founder effect’ of the initial identification of two clusters of sequentially similar polypeptides from this very large family of proteins, and not a genuine segmentation into two quite distinct groupings. In reality, there is not only a continuity of structure among the family members (Gilbert et al. 2013, 2014; Gilbert 2014) but also of sequence variation.
All MACPF/CDC proteins that have been well characterized to date are, broadly speaking, pore-forming proteins. The mechanism of pore formation has been mostly mapped out with CDC family members (Gilbert 2005; Tilley et al. 2005; Tweten 2005), and the available evidence suggests that perforin (Gilbert et al. 2013; Praper et al. 2011) and the perforin-like fungal protein pleurotolysin (Lukoyanova et al. 2015; Ota et al. 2013) share modes of action with them. In this mechanism, monomeric protein binds to a membrane surface and oligomerizes upon it to generate ring-shaped structures, which constitute pre-pore assemblies (Gilbert et al. 1999b; Hotze et al. 2001; Tilley et al. 2005). Pore formation itself ensues when a large conformational change occurs in which a pair of clusters of α-helices refold into a pair of β-sheet hairpins, which then insert into the membrane to form a pore (Czajkowsky et al. 2004; Reboul et al. 2014; Shatursky et al. 1999; Shepard et al. 1998; Tilley et al. 2005) (Fig. 1a). The initial binding to the membrane is, with CDCs, in most cases thought to be based on a direct interaction with cholesterol, although intermedilysin (from the human-specific bacterium Streptococcus intermedius) uses CD59 to make initial surface contact (Giddings et al. 2004). Perforin’s contact with membranes depends on calcium activation of its C2-like C-terminal membrane-binding domain but is not thought to be lipid specific. The basis for its failure to also damage the cells that release it is still unknown (Lopez et al. 2013; Metkar et al. 2011, 2015). The complement membrane attack complex is however different from the CDCs and perforin in that its membrane binding depends on the activation and formation of a nucleating C5b-8 complex of proteins. This complex then attracts multiple copies of the principal pore-forming subunit, C9 (Aleshin et al. 2012; Sonnen and Henneke 2014). There may not be a pre-pore oligomeric state for the MAC, in contrast to the CDCs and perforin (Aleshin et al. 2012; Sonnen and Henneke 2014).
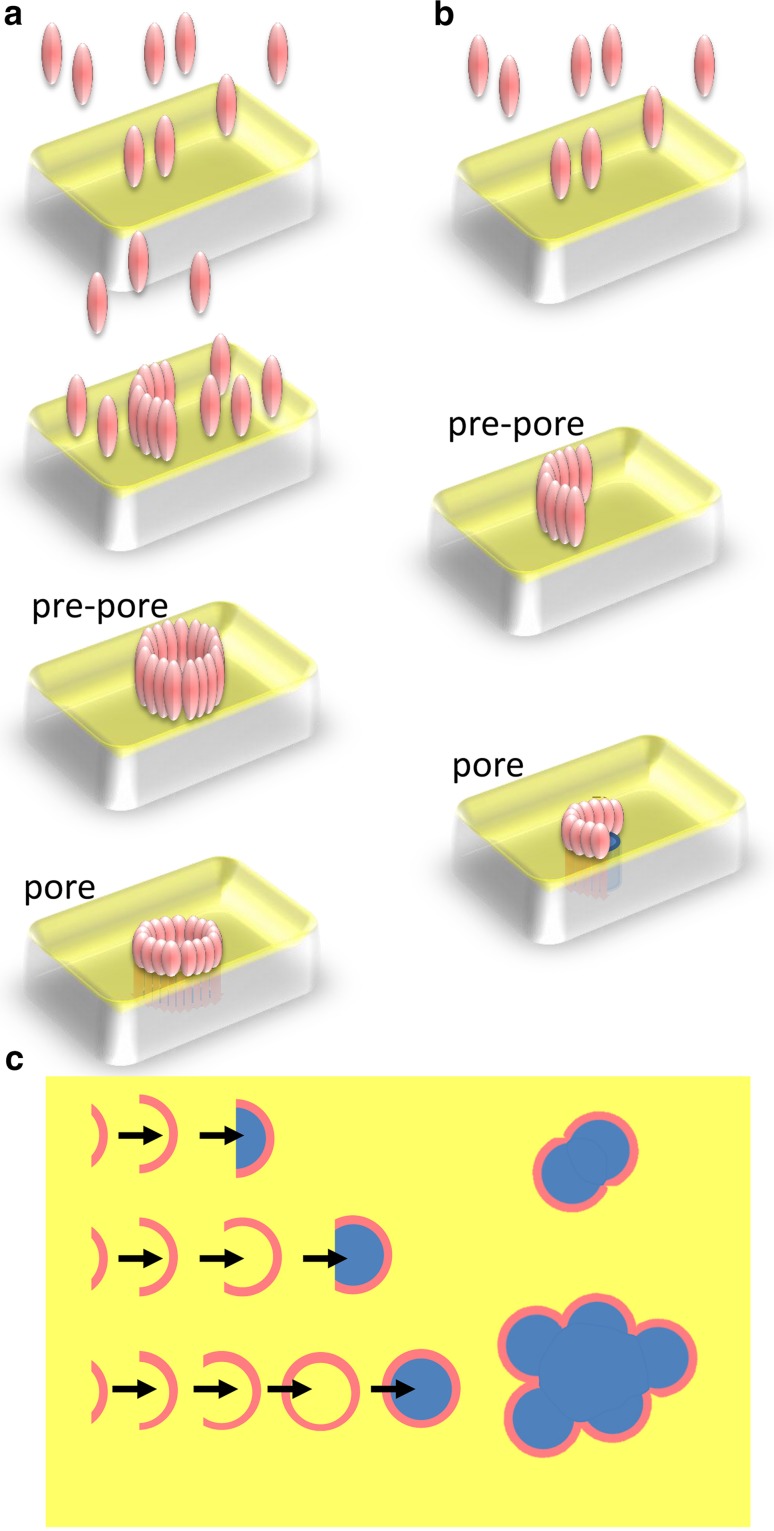
Fig. 1.
Models of pore formation by MACPF/CDC family proteins. a Pore formation by a complete ring of protein subunits—monomers bind to a membrane, assemble on it into a pre-pore, and then undergo a pre-pore to pore transition. b Pore formation by an incomplete ring of protein subunits, or arc, as in a. c Overview of the kinetically governed determination of MACPF/CDC pore size. The size of oligomer formed before the pre-pore to pore transition caps further assembly and is determined by the concentration of protein available (Gilbert 2002, 2005, 2010; Leung et al. 2014). Once formed, arc pores can associate with each other to form larger lesions (Mulvihill et al. 2015; Podobnik et al. 2015; Praper et al. 2011)
In addition to complete rings of subunits forming pores in target membranes, since the earliest days of work on MACPF and CDC proteins there has been a suspicion that incomplete rings, or arcs of subunits, are capable of the same feat (Bhakdi and Tranum-Jensen 1991; Bhakdi et al. 1985; Borsos et al. 1964; Podack and Dennert 1983; Tschopp 1984) (Fig. 1b). This view has continued to be strongly argued (Gilbert 2002, 2005, 2010; Gilbert et al. 2013; Gilbert 2015) but has mostly been neglected (Dunstone and Tweten 2012; Lukoyanova and Saibil 2008) or argued against (Tweten et al. 2015) though the tone of the discussion is changing (Reboul et al. 2016). Among the evidence in favor of the argument that arcs of subunits form functional membrane pores are negative-stain electron microscopy of membranes after MACPF/CDC attack, which have repeatedly shown the presence of protein arcs apparently partly enclosing transmembrane pores with the perimeter completed by the lipid membrane itself (Bhakdi and Tranum-Jensen 1991; Bhakdi et al. 1985; Borsos et al. 1964; Podack and Dennert 1983; Tschopp 1984). In addition, a number of functional studies have also supported pore formation by protein arcs, including the formation of pores with a wide range of conductance values and whose conductance traces are affected by the lipid composition of targeted membranes; the enhancement of lipid “flip-flop” between inner and outer membrane leaflets when pores form; and the imaging of apparent arc-formed pores in situ in tissue sections and on whole cells, which strongly supports their biological relevance (Benz et al. 1986; Felzen et al. 1994; Gilbert 2015; Korchev et al. 1998; Marchioretto et al. 2013; Menestrina et al. 1990; Metkar et al. 2011, 2015; Palmer et al. 1998; Praper et al. 2011; Young et al. 1986a, b, c). Although it might be felt that pores formed at the interface between arcs of protein and membrane bilayer components would be energetically unfeasible, the formation of a partially toroidal structure by the lipids is thought to reduce the line tension and so stabilize the structures, in a way analogous to the formation of purely lipidic toroidal pores during electroporation (Weaver 1994). See elsewhere for further discussion (Gilbert et al. 2013; Gilbert 2015, 2016).
The key contribution of membrane binding to the assembly of MACPF/CDC proteins into pore-forming oligomers is simply the concentration of protein monomers on a plane whereby sufficient interactions are undergone for self-assembly to occur. A variety of data show this, including the demonstration that oligomerization induced in solution is concentration-dependent and can continue indefinitely (leading to helical oligomers rather than the rings found on membranes due to planar constraint) (Gilbert et al. 1998, 1999a, b), that perforin oligomerization can simply be induced by calcium in solution (Metkar et al. 2011), that the size of oligomers can be capped with mutant protein (Palmer et al. 1998), and that disulphide locking of oligomers, which prevents pore formation, affects the distribution of oligomer size (Czajkowsky et al. 2004) (though that was not the inference originally drawn). The observation that oligomer size is affected by the prevention of pore formation has been discussed further elsewhere (Gilbert 2005). This key insight supports a model for the kinetic determination of pore structure whereby the size of oligomeric assembly is determined by the availability of subunits for recruitment into the growing assembly (Gilbert 2002). When an assembly of oligomers terminates with an arc rather than a full ring, it is due to the kinetic trapping of that assembly by a pre-pore to pore transition (Gilbert 2005, 2010). This model has recently been re-proposed and quantified (Leung et al. 2014) and is now considered to be accepted (Mulvihill et al. 2015; Podobnik et al. 2015).
Ultimately, a transition to 3D imaging provided the widely convincing evidence needed that MACPF/CDC proteins form pores using diverse structures—both arcs and rings of subunits. Firstly, cryo-electron tomography and sub-volume averaging allowed the determination of a set of reconstructions of arc and ring assemblies of subunits in pre-pore and pore-forming states (Sonnen et al. 2014). Later, three studies using AFM could image the same assemblies (Leung et al. 2014; Mulvihill et al. 2015; Podobnik et al. 2015), and in the case of the studies on listeriolysin greater ranges of different kinds of assembly to those observed before at this level of detail (Mulvihill et al. 2015; Podobnik et al. 2015) (Fig. 1c). A more recent study of pore formation by streptolysin O using quartz crystal microbalance with dissipation monitoring (QCM-D) has also supported the conclusion that variably sized oligomers (incomplete and complete rings) form membrane pores (Stewart et al. 2015).
In this paper, we describe kinetic data that provide further support for the model of pore formation by rings or kinetically trapped arcs of MACPF/CDC subunits and map out factors determining pore size (Gilbert 2002; Leung et al. 2014; Mulvihill et al. 2015; Podobnik et al. 2015). Although erythrocytes are not thought to be biological targets of MACPF/CDC proteins, saving malaria-associated members of the family (Garg et al. 2013), the red blood cell membrane has long provided a valuable model system for the analysis of pore formation. Bernheimer followed lysis of erythrocytes by a variety of pore-forming agents, observing the pattern now long known for the membrane action of protein toxins: a lag period when lysis did not occur, followed by a rapid lytic event (Bernheimer 1947). In another report, Oberley and Duncan showed how an initial temperature-independent membrane-binding event is followed by lysis, which is prevented at 4 °C because oligomerization and/or pore formation are retarded (Oberley and Duncan 1971). The real-time monitoring of colloid osmotic lysis due to MACPF/CDC pore formation was then established as a way of monitoring the activity of a MACPF/CDC, in particular perfringolysin (Harris et al. 1991). The rapid influx of water into the erythrocyte interior to provide an osmotic balance bursts the red cell membrane and the lysis rate is dependent on the concentration of the pore-forming protein involved and the temperature of the sample. Building on these observations, a variety of kinetic approaches to study MACPF/CDC proteins have been used over the years. Relatively early on, it was shown that the lag period consists of the self-association of subunits to form oligomeric complexes (Harris et al. 1991). The different stages in pore formation were then further defined using mutant forms of perfringolysin in which the pore-forming regions are locked with a disulphide bond so that only pre-pore structures could form (Heuck et al. 2000; Hotze et al. 2001). Binding could be distinguished from oligomerization by their relative timeframes, but both were unaffected by the presence of a disulphide lock and this enabled it to be shown that oligomerization of protein determines the rate of pore formation overall, rather than the pre-pore to pore transition (Heuck et al. 2000; Hotze et al. 2001). The use of liposomes loaded with fluorescent dye-labeled proteins also allowed the real-time monitoring of leakage caused by the activity of perfringolysin, giving a comparative measure of pore formation kinetics (Heuck et al. 2000; Hotze et al. 2002).
Pore formation—or rather, oligomerization—by streptolysin has also been subjected to kinetic studies (Palmer et al. 1995). In a paper in which the sizes of oligomers were quantified using 125I-labeled protein, the temperature dependence of oligomerization was made use of to distinguish a first-order binding event [also reflecting the temperature-independent (Oberley and Duncan 1971) 1:1 interaction of CDCs and cholesterol; (Nollmann et al. 2004)] from a second-order process of oligomerization whose kinetic pattern changes with increasing CDC concentration (Palmer et al. 1995). The conclusion made was that arcs of streptolysin subunits are kinetically significant intermediates; this directly relates to the model of pore formation by kinetically trapped arcs of subunits proposed since then (Gilbert 2002, 2005, 2010; Leung et al. 2014). In work using QCM-D, it has also been shown that streptolysin, as a model CDC, binds non-cooperatively to a lipid membrane (Stewart et al. 2015). The use of mutants alongside wild-type protein then demonstrated that arcs of streptolysin were forming and inserting into membranes and were most likely forming pores (Stewart et al. 2015). These data were interpreted with respect to atomic force microscopy images/measurements generated as part of the same study (Stewart et al. 2015) and also using previously published images (Czajkowsky et al. 2004), which were interpreted in exactly the same way as reported earlier (Gilbert 2005). The QCM-D experiments showed that membrane binding, oligomerization, and insertion to form a pore can be kinetically distinguished, and that the lipids within the membrane are, most likely, not lost from it during pore formation (Stewart et al. 2015). This agreed with an insight from cryo-electron tomography and sub-tomogram averaging that lipids flow out from under an inserting CDC oligomer (Gilbert 2016; Sonnen et al. 2014) but contrasts with another study using AFM that suggested lipids are ejected out of membranes on pore formation (Leung et al. 2014).
In the work reported in this paper, a kinetic assay is used to provide data supporting the model of pore formation by MACPF/CDCs previously set out (Gilbert 2002, 2005, 2010) and with which the work of Stewart et al. (2015), as well as that of others (Leung et al. 2014), agrees. Following the dependence of rate of lysis and length of lag before lysis occurs, on concentration of pneumolysin, indicates that the rate is governed by two different kinds of rate-determining processes, whereas the lag length is governed by the same rate-limiting factors at all concentrations of pneumolysin tested. We show that at low toxin concentrations, the rate of lysis is governed by the pre-pore to pore transition of incomplete rings of subunits, while at higher toxin concentrations the rate of lysis is governed simply by the affinity of protein for the membrane. At all concentrations tested, the lag length is governed by the time taken for oligomerization to occur, in agreement with previous findings (Harris et al. 1991).
Materials and methods
Pneumolysin was expressed and purified exactly as previously described (Gilbert et al. 1999a).
Sheep erythrocytes (in Alsever’s solution; TCS Microbiology Ltd, Botolph Claydon, Buckingham, UK) were centrifuged at 5000 rpm for 1 min in a benchtop centrifuge and the pellet resuspended in 125 mM PBS (8 mM Na2HPO4, 1.5 mM KH2PO4, 2.5 mM KCl, 250 mM NaCl, 125 mM NaCl, pH 7.48) to a concentration of 4 % v/v pellet/buffer. Because of imprecision in pipetting a dense solution such as pelleted cells, the 700-nm scatter of the erythrocyte solution was adjusted so that it was always 2.00 ± 0.05. Then, 40 μl pneumolysin at a known concentration, also in 125 mM PBS, was added to a 5-mm pathlength cuvette held in a CE5500 double-beam spectrophotometer (Cecil Instruments, Cambridge, UK) fitted with a real-time printer-plotter, and with the cuvette housing maintained at constant temperature by a circulating heated water bath; 460 μl of 4 % v/v sheep blood at the required temperature were then added to the pneumolysin solution and the resulting cell lysis followed by the decrease in scattered light. Multiple concentrations of pneumolysin were used, at temperatures ranging from 30 to 38.5 °C, and the effect of cell density was also tested with cell suspensions ranging from 1.5 to 2 OD700. The experiments were repeated several times, using different pneumolysin preparations, and although there was some variation in the absolute values of rate of lysis, the patterns seen within each set of experiments were constant. The conclusions drawn in this paper relate to the patterns of variation of lysis rate, lag length, and so on, and not to their absolute values. They are thus validated by repeated observation. Data were analyzed using equations described in the Results and Discussion section. All plots were made using the curve-fitting program ProFit (QuantumSoft, Ütikon-am-See, Switzerland).
Results and discussion
Primary plots
The kinetic experiments described in this paper constitute a more complete analysis of the concentration dependence of MACPF/CDC pore activity than previously performed, mapping not only the rate of real-time lysis alone (Heuck et al. 2000; Hotze et al. 2002), but also the variation of distinctive features of the real-time lysis curve with protein concentration. It also contrasts with studies that have investigated the MACPF/CDC oligomerization process but not at the same time as the actual functioning of pores (Palmer et al. 1995; Stewart et al. 2015). Our work makes use exclusively of wild-type pneumolysin and relies on the comparison of single-temperature experiments rather than making use of temperature jump approaches. Well-controlled experimental temperatures were ensured by constant maintenance of the reagents at the required value both prior to and after mixing. Our work is therefore distinct from approaches that have used temperature jumps or locking mutants to separate events in cell lysis (Harris et al. 1991; Heuck et al. 2000; Hotze et al. 2001; Oberley and Duncan 1971; Palmer et al. 1995) and complementary to them.
To analyze our data, we sought to apply models for enzymatic activity. However, it is not obvious that cell lysis can appropriately be modeled analogous to enzymatic mechanisms. For example, the standard Michaelis–Menten steady-state model for enzyme activity involves a reversible step in which a metastable transition state, i.e., the enzyme substrate complex, is established, before an irreversible step in which products are formed ensues.
| 1 |
It is not obvious that the action of pore-forming proteins involves any reversible steps once membrane binding has occurred (Ohno-Iwashita et al. 1988; Palmer et al. 1995), but the pre-pore could be argued to resemble a transition state. We therefore attempted to study erythrocyte lysis by pneumolysin within a steady-state kinetic model, and found that, in fact, such a model describes the lytic process very well. Using a spectrophotometer to measure the scatter of light by red blood cells, we observed its subsequent decline in the presence of pneumolysin, indicating cell lysis. The decline in scatter was measured in real time (Fig. 2a), and showed two obvious characteristics. Firstly, a lag period during which the cell density remained constant, and secondly a period of rapid cell disappearance. This pattern replicates that seen previously for CDCs (Bernheimer 1947; Harris et al. 1991), but which has not yet been quantified in terms of toxin concentration, temperature, and cell density. We measured a single value for the rate of lysis by taking the inflection point of the fall in cell density, and from this a lag length by taking the time between the origin and the tangent to the inflection point (Fig. 2a). We then plotted the rate of lysis against the concentration of pneumolysin to obtain a curve that could be well fitted by a rectangular hyperbola using a variation of the Michaelis–Menten equation
| 2 |
where vi is the rate of lysis, p is concentration of pneumolysin, Vmax is the theoretical maximal rate of lysis, and k0.5 is the concentration at half that rate. We repeated this for a series of temperatures, between 30 and 38.5 °C, as shown in Fig. 2b (output values given in Table 1), and it is apparent that Eq. (2) does model the data well. We also plotted the inverse of the lag length against concentration, on the basis that this would allow us to compare directly rate and lag data, using Eq. (3), a simple variant of Eq. (2)
| 3 |
where l is the lag, Lmin the minimal lag length at a particular temperature, and k0.5 the concentration at twice that value. The lag length data, plotted reciprocally, could be fitted well with Eq. (3) at all temperatures (Fig. 2c), which suggests again that the kinetic model we are using is appropriate for measuring cell lysis in our system (output parameters in Table 1).
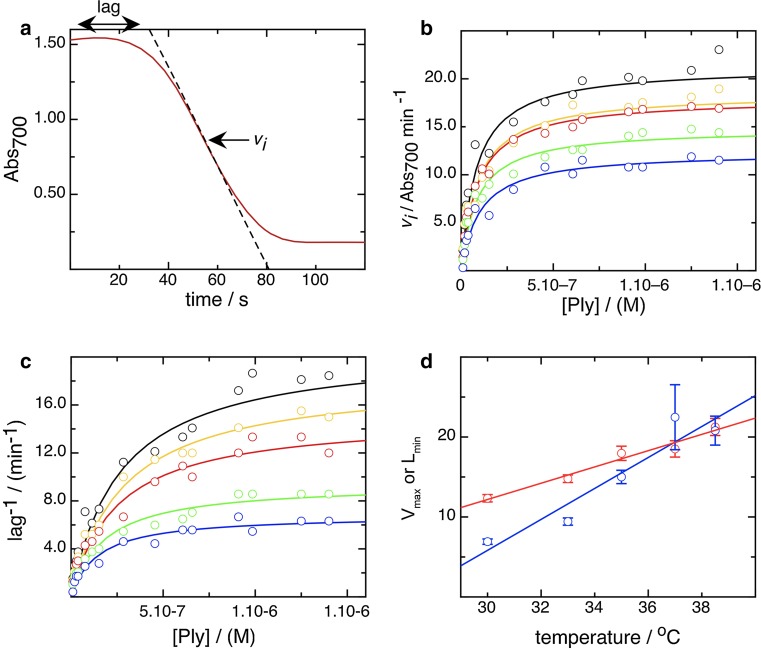
Fig. 2.
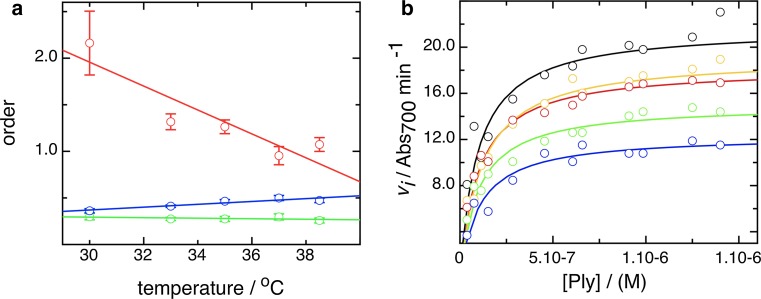
A steady-state kinetic model can be used to model cell lysis. a Typical data plot showing constant absorbance (scatter at 700 nm) immediately after mixing toxin and erythrocyte suspension, and then a hyperbolic decline as the cells lyse. The tangent to the point of inflection defines a rate of lysis, v i and this allows definition of a length of lag, the time from the origin to the tangent, as shown. b Plot of rate of lysis, v i, against toxin concentration at 30 °C (blue symbols/line), 33 °C (green), 35 °C (red), 37 °C (orange), and 38.5 °C (black), fitted using Eq. (2), a modification of the Michaelis–Menten equation. c Plot of reciprocal lag length against toxin concentration for the same five temperatures as in b, with the same color code. These distributions have again been fitted with hyperbolic curves, using Eq. (3). d Plot of dependence of maximum v i (V max) (blue) and minimum lag (red; expressed reciprocally) on temperature, as computed using the fits shown in b and c, respectively
Table 1.
Fit parameters from application of steady-state kinetics to cell lysis
| Temperature (°C) | k 0.5 (nM) | V max (A700 min−1) |
|---|---|---|
| 30.0 | 102 ± 17 | 12.3 ± 0.46 |
| 33.0 | 86 ± 12 | 14.8 ± 0.47 |
| 35.0 | 88 ± 19 | 18.0 ± 0.89 |
| 37.0 | 91 ± 22 | 18.5 ± 1.02 |
| 38.5 | 82 ± 19 | 21.3 ± 1.07 |
| k 0.5 (nM) | L min (minutes) | |
|---|---|---|
| 30.0 | 143 ± 29 | 6.9 ± 0.33 |
| 33.0 | 176 ± 31 | 9.4 ± 0.45 |
| 35.0 | 243 ± 44 | 15.0 ± 0.83 |
| 37.0 | 405 ± 196 | 22.5 ± 4.06 |
| 38.5 | 266 ± 7.3 | 20.8 ± 1.81 |
The values reported arise from a single set of five experiments performed using the same preparation of pneumolysin on a single day. See “Materials and methods” for discussion of the reproducibility of the assay
Secondary and tertiary plots
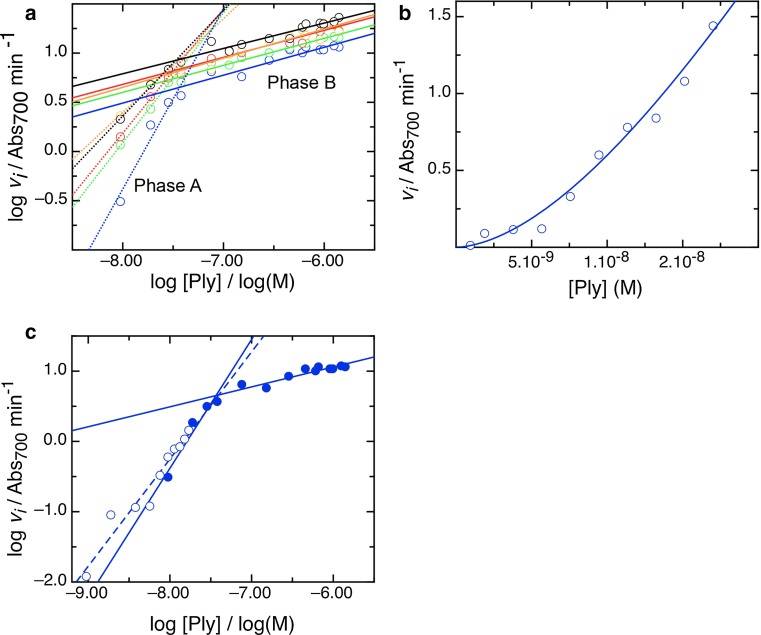
Secondary plots of parameters derived from the primary fits shown in Fig. 2b, 2c allow us further to define the mechanistic stages of cell lysis by pneumolysin. In Fig. 2d we show how the Vmax and Lmin vary with temperature, and that they are clearly distinct from each other in the way that a rise in temperature either increases the rate of cell lysis or decreases the lag in time before it occurs. The difference in the kinetic parameters derived strongly indicates that the phases involve different processes governed by separate rate-determining steps, because the temperature-dependent effect differs in degree. Furthermore, plotting the rate and the lag logarithmically, to obtain a measure of reaction order, also shows a difference between them. The rate data (Fig. 3a) show a biphasic order pattern, with a value of 1–2 at lower concentration (“phase A”) and one of ~0.3 at higher concentration (“phase B”) (see Table 2 for order values). An order >1 suggests the possibility of cooperativity which can be simply described by an alternative model to that described by Eq. (2), incorporating an index of toxin concentration, the Hill coefficient, h
| 4 |
To ascertain if this applies to rate order A, we measured the kinetics of cell lysis by pneumolysin at a series of lower concentrations and at 30 °C to accentuate any effect. As shown in Fig. 3b, a cooperative trend in lysis is apparent, with a cooperativity coefficient h of 1.7. Plotting logarithmically indicates that these data recapitulate the dependence of lysis on concentration already identified as phase A, which is thereby shown to be cooperative (Fig. 3c).
Fig. 3.
Rate of lysis displays two kinds of concentration dependence, and at lower pneumolysin concentrations is cooperative. a Dependence of rate, v i, on concentration at a range of temperatures, displayed by plotting logarithms to calculate the order of the reaction. As in Fig. 2, 30 °C data are colored blue, 33 °C green, 35 °C red, 37 °C orange, and 38.5 °C black. A dotted line fits rate order phase A, a solid line rate order phase b, and their gradients give the actual orders of the reaction. b Measurement of v i over the lower concentration range (rate phase A), at 30 °C, to pick out the cooperative kinetics in rate phase A. Hill coefficient, h = 1.7. c Logarithmic plot of the data shown in b demonstrating it is within rate phase A
Table 2.
Dependence of kinetic order on temperature for cell lysis and lag length
| Temperature (°C) | Order, rate phase A | Order, rate phase B | Order, lag |
|---|---|---|---|
| 30.0 | 1.42 | 0.28 | 0.84 |
| 33.0 | 1.13 | 0.24 | 0.41 |
| 35.0 | 1.10 | 0.24 | 0.46 |
| 37.0 | 0.80 | 0.30 | 0.60 |
| 38.5 | 0.87 | 0.30 | 0.47 |
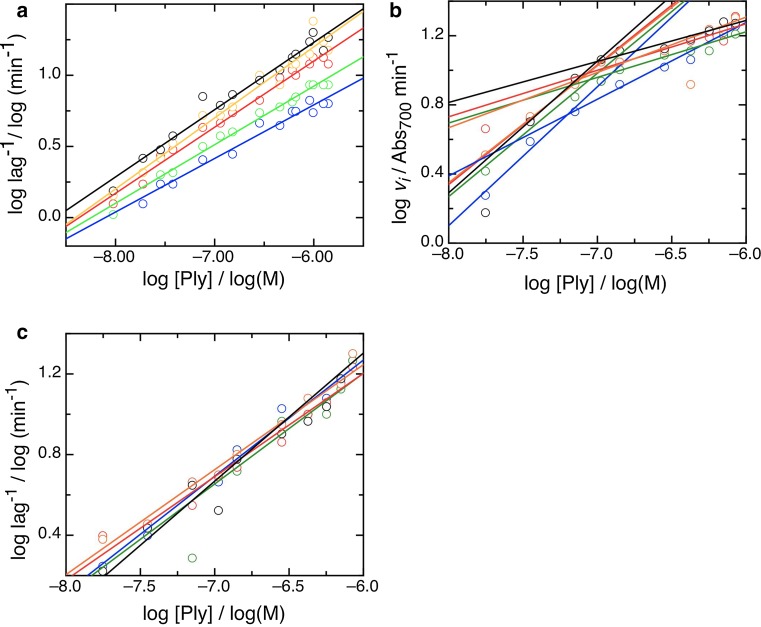
In contrast to the rate data, logarithmic plots of the lag data show a single phase (Fig. 4a), with an order of ~0.5. Thus, the lag length is dependent on pneumolysin to the same extent at all concentrations, whereas the dependence of lysis on concentration undergoes a state change at a critical pneumolysin concentration, which we call pcrit, and which does not vary with temperature; for the data shown in Fig. 3a, pcrit = 60 ± 10.0 nM. A possible rate-determining factor in the rate of cell lysis is the availability of erythrocytes, however as shown in Table 3 and Fig. 4b, pcrit does not vary significantly with cell density, either (pcrit = 74.25 ± 9.0 nM), and the lag order maintains a constant value at all cell densities used (Fig. 4c). This means that length of lag and rate of cell lysis is zero order with respect to the availability of membrane surface at the concentrations of pneumolysin measured, and that we can interpret the kinetics we observe in terms of the action of pneumolysin alone, in the manner of a pseudo-first-order reaction.
Fig. 4.
The lag in lysis has constant concentration dependence and the lysis measured is zero order with respect to cell density. a Dependence of lag length, expressed reciprocally, on concentration at a range of temperatures, colored as previously with data at 30 °C (blue symbols/line), 33 °C (green), 35 °C (red), 37 °C (orange), and 38.5 °C (black). b Dependence of rate, v i, on concentration at a range of temperatures, displayed by plotting logarithms to calculate the order of the reaction. Blue at A700 = 1.52, green at A700 = 1.67, red at A700 = 1.80, orange at A700 = 1.90 and black at A700 = 2.00. In each case, rate phases A and B are fit with solid lines; the switch point, p crit does not vary systematically with cell density. c As b for the lag length, expressed reciprocally. The dependence of lag length on concentration of pneumolysin does not alter with cell density in the range tested
Table 3.
Kinetic measurements are zero order with respect to cell density and rate phase B captures cell binding
| Cell density, 700 nm | Point of inflection (log M) | Critical concentration, p crit (nM) |
|---|---|---|
| 2.00 | −7.15 | 70.8 |
| 1.90 | −7.11 | 77.6 |
| 1.80 | −7.09 | 81.3 |
| 1.67 | −7.09 | 81.3 |
| 1.52 | −7.22 | 60.3 |
| Temperature (°C) | K d (nM) | |
|---|---|---|
| 2.00 | 30.0 | 100 ± 20 |
| 2.00 | 33.0 | 97 ± 15 |
| 2.00 | 35.0 | 98 ± 27 |
| 2.00 | 37.0 | 110 ± 30 |
| 2.00 | 38.5 | 95 ± 27 |
A plot of the kinetic orders of the two phases in lysis rate and of the lag length with temperature is shown in Fig. 5a. It indicates that the order in rate phase A falls markedly as temperature is raised, while that in rate phase B does not vary significantly with temperature. The order of the lag rises slightly with temperature, which indicates that initiation of lysis by pore formation is actually more concentration dependent at higher temperatures.
Fig. 5.
Concentration dependence as a function of temperature and rate phase B K d. a Dependence of order on temperature for rate phase A (red), rate phase B (green), and lag (blue). b Plot to determine the k 0.5 for rate phase B, and thus the K d of pneumolysin binding the membrane. The values computed are listed in Table 3. Colors of data and fits are as follows: 30 °C blue (symbols/line), 33 °C green, 35 °C red, 37 °C orange, and 38.5 °C black
We will now seek to identify the molecular processes underlying each of the steps in cell lysis by pneumolysin that we have shown to be distinguishable on the basis of their dependence on toxin concentration and temperature. The known steps in pore formation are (1) binding to the membrane, (2) self-association to form a pre-pore oligomer, (3) pre-pore to pore transition (Gilbert 2005; Hotze et al. 2001; Tilley et al. 2005). Although a pre-pore assembly must form prior to pore formation, it can be an arc of subunits or a complete ring (Gilbert 2002, 2005; Gilbert et al. 2014; Sonnen et al. 2014), with a recent study indicating a minimal arc size of five subunits within the sensitivity of detection (Leung et al. 2014). We will first consider rate phase A, which is cooperative (Fig. 3b, c) and shows a reduction in dependence on pneumolysin concentration with temperature (Fig. 5a). Firstly, binding of CDC molecules to cholesterol is not cooperative (Oberley and Duncan 1971; Stewart et al. 2015) since each protein molecule binds one cholesterol (Nollmann et al. 2004), but reversibly (Ohno-Iwashita et al. 1988; Palmer et al. 1995). Secondly, other studies have suggested that oligomerization itself is not a cooperative process, since the range of oligomeric sizes seen by electron and atomic force microscopy (Bhakdi et al. 1985; Czajkowsky et al. 2004; Leung et al. 2014; Palmer et al. 1998; Podobnik et al. 2015) and that shown by analytical ultracentrifugation (Solovyova et al. 2004) is continuous, and not skewed as would be expected if oligomerization were cooperative. This contention is also supported by the absence of large conformational changes upon oligomerization to a pre-pore (Czajkowsky et al. 2004; Tilley et al. 2005). By exclusion, this suggests that the pre-pore to pore transition is the rate determining process governing rate phase A, and this makes excellent sense in molecular terms since it involves large conformational changes which might be expected to show cooperativity. The cooperative element to the process indicates that the pre-pore to pore transition becomes easier when more toxin is present, i.e., with increasing oligomeric size (increasing arc length). Ring-shaped pneumolysin oligomers have been reported to vary in size (30–40 nm, 38–44 subunits (Tilley et al. 2005)), but the low level of variability observed suggests that oligomerization to complete rings cannot give rise to the cooperativity observed, and we conclude that the pores present at lower toxin concentrations, in particular, include arcs (Fig. 1b, c). It also makes sense in rather simple molecular terms that oligomers will be smaller in size at lower concentrations. Further support for our interpretation is provided by the observation that the dependence of rate phase A on pneumolysin concentration decreases with rising temperature. This indicates that larger pores tend to form at higher temperatures, in line with the kinetic model for pore formation previously developed (Gilbert 2002, 2005, 2010) and now confirmed (Leung et al. 2014) in which the conversion of pre-pore to pore is delayed by the continued accumulation of new subunits to nascent oligomers but can also be undergone by kinetically trapped oligomeric arcs. Thus, more rapid accumulation of subunits allows for larger pores to form, generating larger pre-pores showing greater cooperativity (i.e., less per-molecule dependence) at pre-pore to pore transition. The insight that at higher temperatures larger oligomers form is discussed further below with respect to the lag time before lysis begins.
If the pre-pore to pore transition governs rate phase A, what governs phase B? The order of phase B shows no apparent variation with temperature (Fig. 5a), which suggests that it does not relate to lipid fluidity or the rate of oligomerization on the membrane surface. It has long been understood that MACPF/CDC proteins bind to membranes in a temperature-independent manner (Oberley and Duncan 1971). We suggest therefore that toxin binding to the membrane surface governs rate phase B, and that at higher concentrations of pneumolysin the rate of lysis is dependent on rate of binding (read out as the amount of toxin bound and therefore able to oligomerize and cause lysis). If this is so, then the equilibrium constant (k0.5 in Eq. 1) of the trend of lysis rate with concentration in this range should give a measure of the Kd of binding. Figure 5b shows a plot from which values of Kd were calculated, as listed in Table 3, showing a value of ~10−7 M. This is the same as the previously calculated affinity of perfringolysin for cholesterol in membranes (Ohno-Iwashita et al. 1988), further supporting the contention that the rate of lysis in phase B is determined by the affinity of toxin for the membrane.
If rate phase A is governed by pre-pore to pore transition, and phase B by binding to the membrane, what governs the dependence of lag on toxin concentration? We have shown that it must involve distinct molecular processes because of the different orders, dependences on temperature, and pattern of order transitions among the three parameters describing lysis that we have measured. It could be argued a priori that the lag is the time taken for sufficient pores to form to initiate cell lysis, and this would agree with earlier observations that oligomerization is essentially complete before lysis begins (Harris et al. 1991). Due to the existence of a pre-pore stage which can persist for significant periods of time (Gilbert et al. 1999b; Leung et al. 2014; Podobnik et al. 2015; Tilley et al. 2005), actual pore formation (i.e. pre-pore to pore transition) is decoupled from the question of how many pores of what size form, in other words from the formation of sufficient pores. The formation of sufficient pores is governed by oligomerization, the stage in the three-step mechanism of lysis by CDCs described above but not yet accounted for, and we conclude that the lag period is oligomerization-dependent. In line with this, and the non-cooperative nature of oligomerization to a pre-pore (see above), lag length shows the same dependency on toxin at all concentrations tested. It also shows a slight increase in dependency on pneumolysin with temperature, suggesting that larger pores are formed at higher temperatures, in agreement again with the identification of rate phase A as governed by the pre-pore to pore transition and as being less dependent on toxin at higher temperatures. Forming a larger pore will take more toxin at higher temperatures but be a necessary consequence of a more rapid migration on the surface of a more energetically dynamic membrane, given a kinetic mechanism for pore formation in which incomplete oligomers (arcs) can get trapped and undergo pre-pore to pore transition (Gilbert 2002, 2005, 2010; Leung et al. 2014; Podobnik et al. 2015). Once formed, however, lysis will be then more rapid because of the increased size of the lesions generated, and the pre-pore to pore transition will itself be less concentration dependent because of its cooperativity (Fig. 3).
In summary, we have shown using pneumolysin as a model system that the dependence of CDC activity on concentration, cell substrate density, and temperature, indicates that all three stages of action (binding, pre-pore oligomerization, and pre-pore to pore transition) are rate limiting at different toxin concentrations, or affect the lag time before lysis rather than the rate of lysis. Taken together, this supports the idea that pore formation by CDC proteins is kinetically governed and that kinetically trapped arcs of subunits form functional pores (Gilbert 2002, 2005, 2010; Leung et al. 2014; Podobnik et al. 2015). Arc pores will form more easily than rings at low toxin concentrations, as supported by our data, and it is likely that the effects of toxins on cells, especially during the early stages of infection, are brought about more by these than by complete rings. Given the similarities between CDCs and MACPF proteins reviewed above, we also hypothesize that our arguments in favor of arciform pores strengthen the case that in perforins, too, unconventional arc-shaped lesion-forming assemblies (Gilbert 2015) as observed by microscopy (Metkar et al. 2015; Young et al. 1986a, 1990) are genuine pores and of physiological significance. This is in strong agreement with single-channel conductance, cell imaging, and cell-labeling experiments (Gilbert 2015; Marchioretto et al. 2013; Metkar et al. 2011, 2015).
Acknowledgments
The Oxford Division of Structural Biology is part of the Wellcome Trust Centre for Human Genetics, Wellcome Trust Core Award Grant Number 090532/Z/09/Z.
References
- Aleshin AE, Schraufstatter IU, Stec B, Bankston LA, Liddington RC, Discipio RG. Structure of complement C6 suggests a mechanism for initiation and unidirectional, sequential assembly of the Membrane Attack Complex (MAC) J Biol Chem. 2012;287:10210–10222. doi: 10.1074/jbc.M111.327809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderluh G, Gilbert RJC. MACPF/CDC proteins—agents of defence, attack and invasion. Dordrecht, NL: Springer; 2014. [Google Scholar]
- Anderluh G, Kisovec M, Krasevec N, Gilbert RJ. Distribution of MACPF/CDC proteins. Sub Cell Biochem. 2014;80:7–30. doi: 10.1007/978-94-017-8881-6_2. [DOI] [PubMed] [Google Scholar]
- Awad MM, Ellemor DM, Boyd RL, Emmins JJ, Rood JI. Synergistic effects of alpha-toxin and perfringolysin O in Clostridium perfringens-mediated gas gangrene. Infect Immun. 2001;69:7904–7910. doi: 10.1128/IAI.69.12.7904-7910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz R, Schmid A, Wiedmer T, Sims PJ. Single-channel analysis of the conductance fluctuations induced in lipid bilayer membranes by complement proteins C5b-9. J Membr Biol. 1986;94:37–45. doi: 10.1007/BF01901011. [DOI] [PubMed] [Google Scholar]
- Bernheimer AW. Comparative kinetics of hemolysis induced by bacterial and other hemolysins. J Gen Physiol. 1947;30:337–353. doi: 10.1085/jgp.30.4.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakdi S, Tranum-Jensen J (1991) Complement lysis: a hole is a hole. Immunol Today 12:318–320; discussion 321 [DOI] [PubMed]
- Bhakdi S, Tranum-Jensen J, Sziegoleit A. Mechanism of membrane damage by streptolysin-O. Infect Immun. 1985;47:52–60. doi: 10.1128/iai.47.1.52-60.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451:350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- Borsos T, Dourmashkin RR, Humphrey JH. Lesions in erythrocyte membranes caused by immune haemolysis. Nature. 1964;202:251–252. doi: 10.1038/202251a0. [DOI] [PubMed] [Google Scholar]
- Czajkowsky DM, Hotze EM, Shao Z, Tweten RK. Vertical collapse of a cytolysin prepore moves its transmembrane beta-hairpins to the membrane. EMBO J. 2004;23:3206–3215. doi: 10.1038/sj.emboj.7600350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czuczman MA, Fattouh R, van Rijn JM, Canadien V, Osborne S, Muise AM, Kuchroo VK, Higgins DE, Brumell JH. Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature. 2014;509:230–234. doi: 10.1038/nature13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstone MA, Tweten RK. Packing a punch: the mechanism of pore formation by cholesterol dependent cytolysins and membrane attack complex/perforin-like proteins. Curr Op Struct Biol. 2012;22:342–349. doi: 10.1016/j.sbi.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felzen B, Berke G, Rosen D, Coleman R, Tschopp J, Young JD, Binah O. Effects of purified perforin and granzyme A from cytotoxic T lymphocytes on guinea pig ventricular myocytes. Cardiovascular Res. 1994;28:643–649. doi: 10.1093/cvr/28.5.643. [DOI] [PubMed] [Google Scholar]
- Garg S, Agarwal S, Kumar S, Yazdani SS, Chitnis CE, Singh S. Calcium-dependent permeabilization of erythrocytes by a perforin-like protein during egress of malaria parasites. Nat Commun. 2013;4:1736. doi: 10.1038/ncomms2725. [DOI] [PubMed] [Google Scholar]
- Giddings KS, Zhao J, Sims PJ, Tweten RK. Human CD59 is a receptor for the cholesterol-dependent cytolysin intermedilysin. Nat Struct Mol Biol. 2004;11:1173–1178. doi: 10.1038/nsmb862. [DOI] [PubMed] [Google Scholar]
- Gilbert RJ. Pore-forming toxins. Cell Mol Life Sci. 2002;59:832–844. doi: 10.1007/s00018-002-8471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert RJ. Inactivation and activity of cholesterol-dependent cytolysins: what structural studies tell us. Structure. 2005;13:1097–1106. doi: 10.1016/j.str.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Gilbert RJ. Cholesterol-dependent cytolysins. Adv Exp Med Biol. 2010;677:56–66. doi: 10.1007/978-1-4419-6327-7_5. [DOI] [PubMed] [Google Scholar]
- Gilbert RJC. Structural features of cholesterol dependent cytolysins and comparison to other MACPF-domain containing proteins. In: Anderluh G, Gilbert RJC, editors. MACPF/CDC proteins—agents of defence, attack and invasion. Dordrecht/NL: Springer; 2014. [DOI] [PubMed] [Google Scholar]
- Gilbert RJC. Perforins. In: Delcour A, editor. Electrophysiology of unconventional channels and pores. Dordrecht/NL: Springer; 2015. pp. 289–312. [Google Scholar]
- Gilbert RJC. Protein-lipid interactions and non-lamellar lipidic structures in membrane pore formation and membrane fusion. Biochimica Biophys Acta. 2016;1858:487–499. doi: 10.1016/j.bbamem.2015.11.026. [DOI] [PubMed] [Google Scholar]
- Gilbert RJ, Rossjohn J, Parker MW, Tweten RK, Morgan PJ, Mitchell TJ, Errington N, Rowe AJ, Andrew PW, Byron O. Self-interaction of pneumolysin, the pore-forming protein toxin of Streptococcus pneumoniae. J Mol Biol. 1998;284:1223–1237. doi: 10.1006/jmbi.1998.2258. [DOI] [PubMed] [Google Scholar]
- Gilbert RJ, Heenan RK, Timmins PA, Gingles NA, Mitchell TJ, Rowe AJ, Rossjohn J, Parker MW, Andrew PW, Byron O. Studies on the structure and mechanism of a bacterial protein toxin by analytical ultracentrifugation and small-angle neutron scattering. J Mol Biol. 1999;293:1145–1160. doi: 10.1006/jmbi.1999.3210. [DOI] [PubMed] [Google Scholar]
- Gilbert RJ, Jimenez JL, Chen S, Tickle IJ, Rossjohn J, Parker M, Andrew PW, Saibil HR. Two structural transitions in membrane pore formation by pneumolysin, the pore-forming toxin of Streptococcus pneumoniae. Cell. 1999;97:647–655. doi: 10.1016/S0092-8674(00)80775-8. [DOI] [PubMed] [Google Scholar]
- Gilbert RJ, Mikelj M, Dalla Serra M, Froelich CJ, Anderluh G. Effects of MACPF/CDC proteins on lipid membranes. Cell Mol Life Sci. 2013;70:2083–2098. doi: 10.1007/s00018-012-1153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert RJ, Dalla Serra M, Froelich CJ, Wallace MI, Anderluh G. Membrane pore formation at protein-lipid interfaces. Trends Biochem Sci. 2014;39:510–516. doi: 10.1016/j.tibs.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Hadders MA, Beringer DX, Gros P. Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense. Science. 2007;317:1552–1554. doi: 10.1126/science.1147103. [DOI] [PubMed] [Google Scholar]
- Harris RW, Sims PJ, Tweten RK. Kinetic aspects of the aggregation of Clostridium perfringens theta-toxin on erythrocyte membranes. A fluorescence energy transfer study. J Biol Chem. 1991;266:6936–6941. [PubMed] [Google Scholar]
- Heuck AP, Hotze EM, Tweten RK, Johnson AE. Mechanism of membrane insertion of a multimeric beta-barrel protein: perfringolysin O creates a pore using ordered and coupled conformational changes. Mol Cell. 2000;6:1233–1242. doi: 10.1016/S1097-2765(00)00119-2. [DOI] [PubMed] [Google Scholar]
- Hirst RA, Gosai B, Rutman A, Guerin CJ, Nicotera P, Andrew PW, O’Callaghan C. Streptococcus pneumoniae deficient in pneumolysin or autolysin has reduced virulence in meningitis. J Infect Dis. 2008;197:744–751. doi: 10.1086/527322. [DOI] [PubMed] [Google Scholar]
- Hotze EM, Wilson-Kubalek EM, Rossjohn J, Parker MW, Johnson AE, Tweten RK. Arresting pore formation of a cholesterol-dependent cytolysin by disulfide trapping synchronizes the insertion of the transmembrane beta-sheet from a prepore intermediate. J Biol Chem. 2001;276:8261–8268. doi: 10.1074/jbc.M009865200. [DOI] [PubMed] [Google Scholar]
- Hotze EM, Heuck AP, Czajkowsky DM, Shao Z, Johnson AE, Tweten RK. Monomer-monomer interactions drive the prepore to pore conversion of a beta-barrel-forming cholesterol-dependent cytolysin. J Biol Chem. 2002;277:11597–11605. doi: 10.1074/jbc.M111039200. [DOI] [PubMed] [Google Scholar]
- Korchev YE, Bashford CL, Pederzolli C, Pasternak CA, Morgan PJ, Andrew PW, Mitchell TJ. A conserved tryptophan in pneumolysin is a determinant of the characteristics of channels formed by pneumolysin in cells and planar lipid bilayers. Biochem J. 1998;329:571–577. doi: 10.1042/bj3290571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C, Dudkina NV, Lukoyanova N, Hodel AW, Farabella I, Pandurangan AP, Jahan N, Pires Damaso M, Osmanovic D, Reboul CF, et al. Stepwise visualization of membrane pore formation by suilysin, a bacterial cholesterol-dependent cytolysin. eLife. 2014;3:e04247. doi: 10.7554/eLife.04247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JA, Susanto O, Jenkins MR, Lukoyanova N, Sutton VR, Law RH, Johnston A, Bird CH, Bird PI, Whisstock JC, et al. Perforin forms transient pores on the target cell plasma membrane to facilitate rapid access of granzymes during killer cell attack. Blood. 2013;121:2659–2668. doi: 10.1182/blood-2012-07-446146. [DOI] [PubMed] [Google Scholar]
- Lukoyanova N, Saibil HR. Friend or foe: the same fold for attack and defense. Trends Immunol. 2008;29:51–53. doi: 10.1016/j.it.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Lukoyanova N, Kondos SC, Farabella I, Law RH, Reboul CF, Caradoc-Davies TT, Spicer BA, Kleifeld O, Traore DA, Ekkel SM, et al. Conformational changes during pore formation by the perforin-related protein pleurotolysin. PLoS Biol. 2015;13:e1002049. doi: 10.1371/journal.pbio.1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchioretto M, Podobnik M, Dalla Serra M, Anderluh G. What planar lipid membranes tell us about the pore-forming activity of cholesterol-dependent cytolysins. Biophys Chem. 2013;182:64–70. doi: 10.1016/j.bpc.2013.06.015. [DOI] [PubMed] [Google Scholar]
- Menestrina G, Bashford CL, Pasternak CA. Pore-forming toxins: experiments with S. aureus alpha-toxin, C. perfringens theta-toxin and E. coli haemolysin in lipid bilayers, liposomes and intact cells. Toxicon. 1990;28:477–491. doi: 10.1016/0041-0101(90)90292-F. [DOI] [PubMed] [Google Scholar]
- Metkar SS, Wang B, Catalan E, Anderluh G, Gilbert RJ, Pardo J, Froelich CJ. Perforin rapidly induces plasma membrane phospholipid flip-flop. PLoS ONE. 2011;6:e24286. doi: 10.1371/journal.pone.0024286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metkar S, Marchioretto M, Antonini V, Lunelli L, Wang B, Gilbert RJC, Anderluh G, Roth R, Pooga M, Pardo J, et al. Perforin oligomers form arcs in cellular membranes: a locus for intracellular delivery of granzymes. Cell Death Dis. 2015;22:78–85. doi: 10.1038/cdd.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PJ, Hyman SC, Byron O, Andrew PW, Mitchell TJ, Rowe AJ. Modeling the bacterial protein toxin, pneumolysin, in its monomeric and oligomeric form. J Biol Chem. 1994;269:25315–25320. [PubMed] [Google Scholar]
- Mulvihill E, van Pee K, Mari SA, Muller DJ, Yildiz O. Directly observing the lipid-dependent self-assembly and pore-forming mechanism of the cytolytic toxin listeriolysin O. Nano Lett. 2015;15:6965–6973. doi: 10.1021/acs.nanolett.5b02963. [DOI] [PubMed] [Google Scholar]
- Nollmann M, Gilbert R, Mitchell T, Sferrazza M, Byron O. The role of cholesterol in the activity of pneumolysin, a bacterial protein toxin. Biophys J. 2004;86:3141–3151. doi: 10.1016/S0006-3495(04)74362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberley TD, Duncan JL. Characteristics of streptolysin O action. Infect Immun. 1971;4:683–687. doi: 10.1128/iai.4.6.683-687.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno-Iwashita Y, Iwamoto M, Mitsui K, Ando S, Nagai Y. Protease-nicked theta-toxin of Clostridium perfringens, a new membrane probe with no cytolytic effect, reveals two classes of cholesterol as toxin-binding sites on sheep erythrocytes. Eur J Biochem. 1988;176:95–101. doi: 10.1111/j.1432-1033.1988.tb14255.x. [DOI] [PubMed] [Google Scholar]
- Ota K, Leonardi A, Mikelj M, Skocaj M, Wohlschlager T, Kunzler M, Aebi M, Narat M, Krizaj I, Anderluh G, et al. Membrane cholesterol and sphingomyelin, and ostreolysin A are obligatory for pore-formation by a MACPF/CDC-like pore-forming protein, pleurotolysin B. Biochimie. 2013;95:1855–1864. doi: 10.1016/j.biochi.2013.06.012. [DOI] [PubMed] [Google Scholar]
- Palmer M, Valeva A, Kehoe M, Bhakdi S. Kinetics of streptolysin O self-assembly. Eur J Biochem/FEBS. 1995;231:388–395. doi: 10.1111/j.1432-1033.1995.tb20711.x. [DOI] [PubMed] [Google Scholar]
- Palmer M, Harris R, Freytag C, Kehoe M, Tranum-Jensen J, Bhakdi S. Assembly mechanism of the oligomeric streptolysin O pore: the early membrane lesion is lined by a free edge of the lipid membrane and is extended gradually during oligomerization. EMBO J. 1998;17:1598–1605. doi: 10.1093/emboj/17.6.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podack ER, Dennert G. Assembly of two types of tubules with putative cytolytic function by cloned natural killer cells. Nature. 1983;302:442–445. doi: 10.1038/302442a0. [DOI] [PubMed] [Google Scholar]
- Podobnik M, Marchioretto M, Zanetti M, Bavdek A, Kisovec M, Cajnko MM, Lunelli L, Dalla Serra M, Anderluh G. Plasticity of listeriolysin O pores and its regulation by pH and a unique histidine. Sci Rep. 2015;5:9623. doi: 10.1038/srep09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praper T, Sonnen AF, Viero G, Kladnik A, Froelich CJ, Anderluh G, Dalla Serra M, Gilbert RJ. Human perforin employs different avenues to damage membranes. J Biol Chem. 2011;286:2946–2955. doi: 10.1074/jbc.M110.169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul CF, Whisstock JC, Dunstone MA. A new model for pore formation by cholesterol-dependent cytolysins. PLoS Comput Biol. 2014;10:e1003791. doi: 10.1371/journal.pcbi.1003791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul CF, Whisstock J, Dunstone MA. Giant MACPF/CDC pore forming toxins: a class of their own. Biochim Biophys Acta. 2016;1858:475–486. doi: 10.1016/j.bbamem.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Rosado CJ, Buckle AM, Law RH, Butcher RE, Kan WT, Bird CH, Ung K, Browne KA, Baran K, Bashtannyk-Puhalovich TA, et al. A common fold mediates vertebrate defense and bacterial attack. Science. 2007;317:1548–1551. doi: 10.1126/science.1144706. [DOI] [PubMed] [Google Scholar]
- Shatursky O, Heuck AP, Shepard LA, Rossjohn J, Parker MW, Johnson AE, Tweten RK. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: a novel paradigm for pore-forming toxins. Cell. 1999;99:293–299. doi: 10.1016/S0092-8674(00)81660-8. [DOI] [PubMed] [Google Scholar]
- Shepard LA, Heuck AP, Hamman BD, Rossjohn J, Parker MW, Ryan KR, Johnson AE, Tweten RK. Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: an alpha-helical to beta-sheet transition identified by fluorescence spectroscopy. Biochemistry. 1998;37:14563–14574. doi: 10.1021/bi981452f. [DOI] [PubMed] [Google Scholar]
- Solovyova AS, Nollmann M, Mitchell TJ, Byron O. The solution structure and oligomerization behavior of two bacterial toxins: pneumolysin and perfringolysin O. Biophys J. 2004;87:540–552. doi: 10.1529/biophysj.104.039974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen AF, Henneke P. Structural biology of the membrane attack complex. Sub Cell Biochem. 2014;80:83–116. doi: 10.1007/978-94-017-8881-6_6. [DOI] [PubMed] [Google Scholar]
- Sonnen AF, Plitzko J, Gilbert RJC. Incomplete pneumolysin oligomers form membrane pores. R Soc Open Biol. 2014;4:140044. doi: 10.1098/rsob.140044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SE, D’Angelo ME, Paintavigna S, Tabor RF, Martin LL, Bird PI. Assembly of streptolysin O pores assessed by quartz crystal microbalance and atomic force microscopy provides evidence for the formation of anchored but incomplete oligomers. Biochim Biophys Acta. 2015;1848:115–126. doi: 10.1016/j.bbamem.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Tilley SJ, Orlova EV, Gilbert RJ, Andrew PW, Saibil HR. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell. 2005;121:247–256. doi: 10.1016/j.cell.2005.02.033. [DOI] [PubMed] [Google Scholar]
- Tschopp J. Ultrastructure of the membrane attack complex of complement. Heterogeneity of the complex caused by different degree of C9 polymerization. J Biol Chem. 1984;259:7857–7863. [PubMed] [Google Scholar]
- Tweten RK. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect Immun. 2005;73:6199–6209. doi: 10.1128/IAI.73.10.6199-6209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweten RK, Hotze EM, Wade KR. The unique molecular choreography of giant pore formation by the cholesterol-dependent cytolysins of gram-positive bacteria. Annu Rev Microbiol. 2015;69:323–340. doi: 10.1146/annurev-micro-091014-104233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskoboinik I, Smyth MJ, Trapani JA. Perforin-mediated target-cell death and immune homeostasis. Nat Rev Immunol. 2006;6:940–952. doi: 10.1038/nri1983. [DOI] [PubMed] [Google Scholar]
- Weaver JC. Molecular basis for cell membrane electroporation. Ann N Y Acad Sci. 1994;720:141–152. doi: 10.1111/j.1749-6632.1994.tb30442.x. [DOI] [PubMed] [Google Scholar]
- Young JD, Hengartner H, Podack ER, Cohn ZA. Purification and characterization of a cytolytic pore-forming protein from granules of cloned lymphocytes with natural killer activity. Cell. 1986;44:849–859. doi: 10.1016/0092-8674(86)90007-3. [DOI] [PubMed] [Google Scholar]
- Young JD, Nathan CF, Podack ER, Palladino MA, Cohn ZA. Functional channel formation associated with cytotoxic T-cell granules. Proc Nat’l Acad Sci USA. 1986;83:150–154. doi: 10.1073/pnas.83.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Podack ER, Cohn ZA. Properties of a purified pore-forming protein (perforin 1) isolated from H-2-restricted cytotoxic T cell granules. J Exp Med. 1986;164:144–155. doi: 10.1084/jem.164.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LH, Joag SV, Zheng LM, Lee CP, Lee YS, Young JD. Perforin-mediated myocardial damage in acute myocarditis. Lancet. 1990;336:1019–1021. doi: 10.1016/0140-6736(90)92486-2. [DOI] [PubMed] [Google Scholar]