Abstract
New multivalent RGD-containing macromolecules were designed by exploiting two orthogonal chemoselective ligations. They were next applied to a competitive cell adhesion assay and used for the non invasive optical imaging of tumour in small animals.
Keywords: Amino Acid Sequence; Animals; Oligopeptides; Biological Assay; Cell Adhesion; Cell Line; Chemistry, Organic; Chromatography, High Pressure Liquid; Humans; Mice; Molecular Sequence Data
Introduction
The design and the synthesis of targeting molecules for diagnostic and therapeutic applications represent a major goal in cancer medicine. To this end, peptide ligands for various targets have been identified using combinatorial libraries1 or phage display method.2 To attain improved activity and receptor selectivity, it is often essential to restrict the conformational space of peptides by using them in a cyclic form.3 In this context, cyclic peptides encompassing RGD (Arg-Gly-Asp) sequence have served as the basis for the development of potent peptide ligands used to selectively target the αV β3 integrin.4 The latter represents an attractive target for cancer therapeutic purposes.5 Furthermore, it is well known that the multivalent display of a ligand enhances the binding strength of the ligand to its receptor and can promote receptor-mediated internalisation of the bound entity.6 Today, the principle of multivalency has then been recognized as an important strategy for the design of synthetic ligands.7 The effect of multivalency in ligand binding was particularly demonstrated for glycoconjugates,8 and for peptide ligands.9 Enhancements of biological activity were especially obtained from multivalent RGD (Arg-Gly-Asp) peptide ligand used to target cell surface receptors such as αV β3 integrin.10
Recently, we have shown that tetrameric RGD-containing scaffolds exhibit desirable biological properties for tumour imaging11 and for targeted drug delivery.12 These compounds contain a cluster of four copies of a cyclo[-RGDfK-] monomer grafted onto a cyclic decapeptide scaffold (Fig. 1). Preliminary work aimed at studying the effect of the multivalency parameter in terms of interaction between the ligand and the target receptor and examining the contribution of each c[-RGDfK-] motif. For this purpose, we designed an array of peptide derivatives containing from one to four copies of the c[-RGDfK-] monomer (Fig. 1).13 In order to obtain ligands with similar shape, similar steric hindrance and close molecular weights, which is essential for their comparison in vitro, we opted to substitute c[-RGDfK-] for non sense c[-RβADfK-] motifs in the ligands whose valency was lower than four. We used a combinatory assembling strategy to explore all possible positions of the RGD motifs on the cyclodecapeptide scaffold. Consequently, we were unable to isolate the different isomers that differ in the position of cyclic RGD pentapeptides onto the cyclodecapeptide scaffold.
Fig. 1.

Structure of clustered RGD-containing compounds.
To overcome this problem, the formation of two successive chemoselective linkages (“click–click” chemistry) enables a direct access to complex structures.14 Such reactions, in particular the azide-alkyne cycloaddition, can be achieved under conditions which are fully compatible with biological environments.15 We recently reported an orthogonal chemoselective ligation strategy that allows access to well defined biomolecular assemblies by exploiting the Huisgen dipolar cycloaddition and the oxime bond formation.16 Following this strategy, herein we describe the synthesis of new multivalent RGD compounds such as the fluorescent glycoconjugate 1 (Scheme 1). The incorporation of the carbohydrate moiety may provide an enhanced solubility and clearance. With the molecules in hand, we then concentrated our work on assessing biological activities to determine the potency of the different RGD-containing compounds.
Scheme 1.
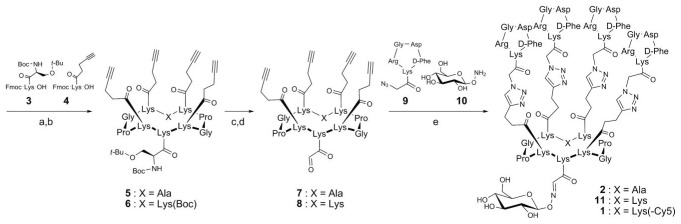
Synthesis of compounds 1–2. a) Standard Fmoc/t-Bu Solid-Phase Peptide Synthesis; b) PyBOP (1 equiv.), DIPEA (4 equiv.), 1 h; c) TFA/H2 O (95 : 5), 2 h; d) NaIO4 (10 equiv.), 30 min; e) 10 (3 equiv.), t-BuOH/H2 O–AcOH (50 : 45 : 5), 2 h then 9 (6 equiv.), Cu(0) microsize powder (0.5 mg), pH 7.0, 18 h; For X = Lys, then Cy5–OSu (1 equiv.), DMF, DIPEA (pH 8), 3 h. Compounds 3 and 4 were prepared as previously described.16,19
Results and discussion
Chemical assemblies
Scheme 1 illustrates the approach used for the synthesis of compounds 1–2. The biomolecular assembling process implies two chemoselective ligations (click–click chemistry): the oxime ligation17 and the Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC).18 To introduce suitable functions within peptide moities, we synthesized building blocks such as compounds 3 and 4 which contain protected serine (masked aldehyde) and alkyne groups, respectively (Scheme 1). The use of building blocks during the solid-phase peptide synthesis (SPPS) reduces the number of steps involved for the construction of such conjugates.19
In this context, linear peptides encompassing chemoselective ligations were prepared following rigorously the standard Fmoc/t-Bu SPPS procedure using PyBOP as coupling reagent. The head-to-tail cyclizations provided the desired cyclodecapeptide scaffolds 5 and 6. Deprotection of serine residue using a concentrated TFA solution followed by a subsequent oxidation with periodate20 afforded key intermediates 7 and 8, isolated in sufficient purity to carry out subsequent chemoselective assemblies. In parallel, RGD-containing cyclopentapeptide 9 bearing the prerequisite azide function and aminooxy-carbohydrate 10 were prepared as described.16,21 Very recently, we have shown that cyclopeptide assemblies are possible by means of orthogonal oxime and copper-mediated click reactions in a stepwise or in a one-pot approach.16 The latter method is much desired, as it avoids lengthy separation process and purification of intermediates while increasing overall chemical yield. Biomolecular ligations of azidopeptide 9 and aminooxy-carbohydrate 10 were then performed on either molecular scaffold 7 or 8. Peptides 7 and carbohydrate 10 (3 equiv.) were applied under mild acidic conditions using a solution containing dilute acetic acid. Rapid oxime ligation of 10 was observed (Fig. 2). Neutralizing the pH and addition of 9 (6 equiv.) and copper microsize powder resulted in complete disappearance of the intermediate and the exclusive formation of the expected compound 2 in a 64% yield.
Fig. 2.
One-pot chemoselective assembly of peptides 7, 9, and carbohydrate 10. HPLC traces are shown at 2 h and 18 h. Int = intermediate.
To evaluate the RGD-containing compounds for further in vivo studies, we decided to introduce a fluorescent reporter such as Cyanine 5 (Cy5) because its near-infra red (NIR) band (λem = 670 nm) can penetrate tissue up to 6 cm allowing non invasive optical imaging in small animals. Furthermore, this NIR band is free from interfering biofluorescence. For this purpose, the intermediate 11 was synthesized according to the procedure described above. Cy5 dye was then introduced at the lysine side-chain of 11 under neutral conditions (pH 8.0) affording the fluorescent conjugate 1 in 72% yield after RP-HPLC purification. Compounds 1 and 2 were characterized by ES-MS and the observed molecular weights were found to be in excellent agreement with the calculated values.
To study the contribution of each c[-RGDfK-] motif, an array of molecules 12–19 was designed and prepared according to the method previously described (Fig. 3).12a,16 Briefly, RGD ligands and nonsense RβAD peptides were introduced onto the scaffold by using respectively the Huisgen dipolar cycloaddition and orthogonal oxime bond formation, the latter providing shorter linker.
Fig. 3.

Structure of compounds 12–19.
Biological assays
The adhesion potency of the different multivalent RGD-containing peptides was first determined using a traditional ELISA-type inhibition assay. In this experiment, we measured the efficiency of peptides to compete with vitronectin, the natural substrate of the αV β3 integrin, when binding to HEK-β3 cells that overexpress αV β3 receptors. HEK-β3 cells were therefore incubated with soluble compounds 2, 12–18 at 37 °C onto vitronectin-coated assay plates. The IC50 values, or concentration of compounds required to inhibit 50% of the cells from attaching to vitronectin, are reported in Table 1. As expected the negative control peptide 13 did not inhibit cell adhesion to vitronectin as reported for similar compounds.10d,13 Increasing the number of RGD motifs from 1 to 3 gradually improved the potency of the ligand to compete with vitronectin. We reasoned that the observed multivalent effect arises from a statistical rebinding of the RGD-containing compound due to the high local concentration of RGD moieties. This phenomenon was observed for dendrimer scaffolds.8f It is worth noting that compounds 16 and 17 that differ in the position of the RGD units onto the cyclodecapeptide scaffold show similar IC50 (respectively, 7.1 and 8.2 μM). The position of the RGD peptides onto the cyclodecapeptide scaffold does not improve the affinity of the molecule. Compounds that display four RGD units (i.e. molecules 2, 12 and 14) showed potent inhibitory effect. Nevertheless, IC50 values for compounds 2 and 14 (respectively, 3.8 and 4.1 μM) are slightly lower than the value obtained for compound 12 (4.9 μM) encompassing shorter oxime linkers. Surprisingly, the molecule 15 encompassing three RGD units displayed the best IC50 (2.8 μM). We previously showed that compounds including three or four RGD ligands exhibit close IC50.13 We argue that the shorter oxime linker used to graft non-sense RβAD peptide within molecule 15 generates less steric hindrance than unbound RGD moieties within molecules 2, 12 or 13 while the RGD-containing compound binds to αV β3 receptor.
Table 1.
Competitive cell adhesion assay
| Compounds
|
IC50/μMa | Standard deviation/μMa | |
|---|---|---|---|
| # | RGD unit/molecule | ||
| 12 | 4 | 4.85 | 0.19 |
| 13 | 0 | NIb | — |
| 14 | 4 | 4.10 | 0.08 |
| 15 | 3 | 2.80 | 0.15 |
| 16 | 2 | 7.13 | 0.36 |
| 17 | 2 | 8.22 | 0.16 |
| 18 | 1 | 48.81 | 0.24 |
| 2 | 4 | 3.77 | 0.11 |
Values were determined from three separate experiments;
NI, no inhibition was observed.
Then, fluorescent derivatives 1 and 19 have been comparatively evaluated in vivo in order to pinpoint the influence of the carbohydrate moiety. We measured the capacity of the molecules to target tumour in mice. Fig. 4 shows typical FRI images of nude mice bearing an subcutaneous human TS/A-pc tumour at different time points after intravenous (iv) injection of 10 nmol fluorescent molecules (i.e. 1 or 19) (see also the ESI†). A strong signal is observed in the kidneys reflecting the prominent and fast renal excretion of RGD-containing molecules as previously shown.11 One hour postinjection, fluorescent molecules accumulate in the tumour but the whole body is also fluorescent due to the presence of unbound circulating molecules. The average values for the tumour/skin ratios were found to be similar for mice treated with 1 and for mice treated with 19 (respectively 1.39 ± 0.37 and 1.32 ± 0.15) (see Table S1 in the ESI†). The contrast (tumour/skin ratio) was found to be statistically better with 1 (Fig. 4D) 3 h after iv injection while the ratio was lower at late time. In comparison, tumour/skin ratio for mice treated with 19 reaches its maximum at 6 h, and then slowly decreases (see the ESI†). These experimental results are in good agreement with a better clearance of the carbohydrate-containing compound 1.
Fig. 4.

Representative images of optical imaging of subcutaneous tumour-bearing mice observed at (A–C) 1 h and (B–D) 3 h after iv injection of (A–B) 10 nmol 19 and (C–D) 10 nmol 1.
Conclusions
We have expanded the scope of click–click chemistry by gaining access to new RGD-containing macromolecules. For instance, we have shown that biomolecular assembly combining carbohydrate and peptides is possible by means of orthogonal oxime and copper-mediated click reactions in a one-pot synthesis. This approach is part of the general trend of organic chemistry taking control of macromolecule synthesis to produce well-defined constructs that could likely become the rule in drug applications. The ensuing RGD compounds were then evaluated through competitive cell adhesion assays and in vivo experiments. The results obtained highlight the utility of a clustered ligand, and as expected the grafting of an additional carbohydrate enhances clearance of the RGD-containing compound. It is worth noting that our approach is not limited to integrin ligands, it may be conceptually exploited to synthesize other sophisticated macromolecular conjugates.
Experimental
Cyclodecapeptide scaffolds 5
Linear decapeptides were assembled on 2-chlorotritylchloride® resin (150 mg, loading of 0.8 mmol g−1) using the general procedure (see the ESI†) by using building blocks 3 and 4. The cyclization reaction were carried out in DMF using linear peptide (172 mg, 100 μmol, 0.5 mM) and PyBOP (1 equiv.) for 1 h at room temperature. After completion of the reaction, the solvent was evaporated and the cyclic peptide 5 was obtained as a white solid powder after ether precipitation (161 mg, 100 μmol, quantitative yield). Mass spectrum (ES-MS, positive mode) calc for C79 H122 N16 O18 : 1583.95, found m/z: 1584.0.
Cyclodecapeptide scaffolds 6
Following the procedure previously described and starting with linear peptide (171 mg, 92 μmol), cyclic peptide 5 was obtained as a white solid powder (167 mg, 96 μmol, 96% yield). Mass spectrum (ES-MS, positive mode) calc for C87 H137 N17 O20 1741.16, found m/z: 1740.9.
Cyclodecapeptide scaffolds 7
Full deprotection of peptide 5 (161 mg, 100 μmol) was carried out in a solution containing 10 mL of TFA/H2 O (95 : 5) for 2 h at room temperature. The product was isolated after removal of solvents under reduced pressure and precipitation from Et2 O. A serine oxidation by an aqueous solution containing NaIO4 (10 equiv.) afforded the peptide 7. The crude product was directly purified by using RP-HPLC affording the compound 7 as a white powder. (72 mg, 48 μmol, 48% yield). Mass spectrum (ES-MS, positive mode) calc for C69 H101 N15 O16 : 1396.67, found m/z : 1396.7.
Cyclodecapeptide scaffolds 8
Following the procedure previously described and starting with cyclic peptide 6 (167 mg, 96 μmol), peptide 8 was obtained as a white powder. (60 mg, 41 μmol, 43% yield). Mass spectrum (ES-MS, positive mode) calc for C72 H108 N16 O16 1453.76, found m/z : 1453.8.
Carbohydrate 10
Compound 10 was prepared as previously described.21
Peptide 2
To a solution containing the cyclodecapeptide 7 (5 mg, 3.5 μmol) in 500 μL tBuOH/H2 O–AcOH (50 : 45 : 5) were added the carbohydrate 10 (3 equiv.). The reaction mixture was stirred for 2 h at room temperature. Then, the pH was adjusted to 8 by addition of a NaHCO3 solution (10%) and the compound 9 c[-RGDfK(COCH2 N3)-] (6 equiv.) and Cu(0) microsize powder (5 equiv.) were added. The reaction mixture was stirred overnight at room temperature and centrifuged for 5 min. The solution was then purified by RP-HPLC to give the desired compound 2 (9.8 mg, 2.3 μmol, yield 64%). Mass spectrum (ES-MS, positive mode) calc for C191 H280 N64 O53 4320.76, found m/z 4320.5.
Peptide 11
To a solution containing the cyclodecapeptide 8 (5 mg, 3.4 μmol) in 500 μL tBuOH/H2 O–AcOH (50 : 45 : 5) were added the carbohydrate 10 (3 equiv.). The reaction mixture was stirred for 2 h at room temperature. Then, the pH was adjusted to 8 by addition of a NaHCO3 solution (10%) and the compound 9 c[-RGDfK(COCH2 N3)-] (6 equiv.) and Cu(0) microsize powder (5 equiv.) were added. The reaction mixture was stirred overnight at room temperature and centrifuged for 5 min. The solution was then purified by RP-HPLC to give the desired compound 11 (8.7 mg, 2 μmol, yield 58%). Mass spectrum (ES-MS, positive mode) calc for C194 H287 N65 O53 4377.85, found m/z 4377.7.
Peptide 1
The peptide 11 (7.0 mg, 1.59 μmol) was dissolved in 1 mL of anhydrous DMF and the pH adjusted with DIPEA to pH 9. The solution was added to Cy™ 5 Mono NHS Ester (1.2 mg, 1.59 μmol) and stirred for 3 h at room temperature. The product was then purified by RP-HPLC affording the fluorescent peptide 11 as a deep blue solid powder (5.77 mg, 1.14 mmol, yield 72%). Mass spectrum (ES-MS, positive mode) calc for C227 H324 N67 O60 S2 5015.65, found 5016.7
Peptide 19
The peptide 14 (3.0 mg, 0.73 μmol) was dissolved in 1 mL of anhydrous DMF and the pH adjusted with DIPEA to pH 9. The solution was added to CyTM 5 Mono NHS Ester (0.54 mg, 0.73 μmol) and stirred for 3 h at room temperature. The product was then purified by RP-HPLC affording the fluorescent peptide 14 as a deep blue solid powder (2.5 mg, 0.53 mmol, yield 73%). Mass spectrum (ES-MS, positive mode) calc for C216 H308 N65 O53 S2 4727.39, found 4727.4.
Peptides 12–18
Peptides 12–16 were prepared as previously described.16
Competitive cell adhesion assays
Competitive assay was carried out as described.12 Briefly, 96-well assay plates were coated for 1 h at room temperature with 5 μg mL−1 vitronectin in PBS and blocked for 30 min with 3% bovine serum albumin (BSA). Varying amounts of peptides were added simultaneously with 105 trypsinated HEK-β3 cells to the wells and the plate was incubated for 30 min at 37 °C. Wells were rinsed three times with cold PBS to remove vitronectin-unbound cells. Attached cells were then fixed with methanol, stained with methylene blue and quantified. The activity of peptides is expressed as IC50 values (concentration of peptide necessary to inhibit 50% of cell attachment to the vitronectin substrate) and determinates from triplicates in three separate experiments.
Fluorescence Reflectance Imaging (2D-FRI)
Female NMRI nude mice (8–10 weeks old, n = 6) were injected subcutaneously with human TS/A-pc cells (1 × 106 cells per mouse). After tumor growth (~10 days), anaesthetized mice were injected intravenously with 10 nmol of Cy5-containing peptide. Mice were illuminated by 633 nm light-emitting diodes equipped with interference filters. Fluorescence images were acquired during 100 ms.
Acknowledgments
This work was supported by the Université Joseph Fourier, the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Institut National du Cancer (INCA), the Nanoscience Foundation and NanoBio (Grenoble).
Footnotes
Electronic supplementary information (ESI) available: General procedures for peptide synthesis; Synthesis of 9; RP-HPLC profiles and ESI-MS analysis of 5, 6, 7, 8, 11, 1, 2 and 19; General biological procedures. See DOI: 10.1039/c0ob00070a
Notes and references
- 1.Thompson LA, Ellman JA. Chem Rev. 1996;96:555. doi: 10.1021/cr9402081. [DOI] [PubMed] [Google Scholar]
- 2.Ladner RC, Sato AK, Gorzelany J, de Souza M. Drug Discovery Today. 2004;9:525. doi: 10.1016/S1359-6446(04)03104-6. [DOI] [PubMed] [Google Scholar]
- 3.Kessler H. Angew Chem, Int Ed Engl. 1982;21:512. [Google Scholar]
- 4.(a) Aumailley M, Gurrath M, Müller G, Calvete J, Timpl R, Kessler H. FEBS Lett. 1991;291:50. doi: 10.1016/0014-5793(91)81101-d. [DOI] [PubMed] [Google Scholar]; (b) Ellerby HM, Arap W, Ellerby LM, Kain R, Andrusiak R, Del Rio G, Krajewski S, Lombardo CR, Rao R, Ruoslahti E, Bredesen DE, Pasqualini R. Nat Med. 1999;5:1032. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 5.(a) Hynes RO. Nat Med. 2002;8:918. doi: 10.1038/nm0902-918. [DOI] [PubMed] [Google Scholar]; (b) Temming K, Schiffelers RM, Molema G, Kok RJ. Drug Resist Updates. 2005;8:381. doi: 10.1016/j.drup.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Mammen M, Choi SK, Whitesides GM. Angew Chem, Int Ed. 1998;37:2754. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 7.Kiessling LL, Gestwicki JE, Strong LE. Curr Opin Chem Biol. 2000;4:696. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 8.(a) Pagé D, Zanini D, Roy R. Bioorg Med Chem. 1996;4:1949. doi: 10.1016/s0968-0896(96)00177-0. [DOI] [PubMed] [Google Scholar]; (b) Sprengard U, Schudok M, Kretzschmar G, Kunz H. Angew Chem, Int Ed Engl. 1996;35:321. [Google Scholar]; (c) Dimick SM, Powell SC, McMahon A, Moothoo N, Naismith JH, Toone EJ. J Am Chem Soc. 1999;121:10286. [Google Scholar]; (d) Kitov PI, Sadowska JM, Mulvey G, Armstrong GD, Ling H, Pannu NS, Read RJ, Bundle DR. Nature. 403:669. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]; (e) Fulton DA, Stoddart JF. Bioconjugate Chem. 2001;12:655. doi: 10.1021/bc0100410. [DOI] [PubMed] [Google Scholar]; (f) Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. J Am Chem Soc. 2002;124:14922. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]; (g) Renaudet O, Křenek K, Bossu I, Dumy P, Kádek A, Adámek D, Vaněk O, Kavan D, Gažák R, Šulc M, Bezouška K, Křen V. J Am Chem Soc. 2010;132:6800. doi: 10.1021/ja101296t. [DOI] [PubMed] [Google Scholar]; (h) Arosio D, Fontanella M, Baldini L, Mauri L, Bernardi A, Casnati A, Sansone F, Ungaro R. J Am Chem Soc. 2005;127:3660. doi: 10.1021/ja0444029. [DOI] [PubMed] [Google Scholar]
- 9.(a) Yim CB, Boerman OC, de Visser M, de Jong M, Dechesne AC, Rijkers DTS, Liskamp RMJ. Bioconjugate Chem. 2009;20:1323. doi: 10.1021/bc900052n. [DOI] [PubMed] [Google Scholar]; (b) Trouche N, Wieckowski S, Sun W, Chaloin O, Hoebeke J, Fournel S, Guichard G. J Am Chem Soc. 2007;129:13480. doi: 10.1021/ja073169m. [DOI] [PubMed] [Google Scholar]
- 10.(a) Maynard HD, Okada SY, Grubbs RH. J Am Chem Soc. 2001;123:1275. doi: 10.1021/ja003305m. [DOI] [PubMed] [Google Scholar]; (b) Kok RJ, Schraa AJ, Bos EJ, Moorlag HE, Ásgeirsdóttir SA, Everts M, Meijer DKF, Molema G. Bioconjugate Chem. 2002;13:128. doi: 10.1021/bc015561+. [DOI] [PubMed] [Google Scholar]; (c) Thumshirn G, Hersel U, Goodman SL, Kessler H. Chem-Eur J. 2003;9:2717. doi: 10.1002/chem.200204304. [DOI] [PubMed] [Google Scholar]; (d) Boturyn D, Coll JL, Garanger E, Favrot MC, Dumy P. J Am Chem Soc. 2004;126:5730. doi: 10.1021/ja049926n. [DOI] [PubMed] [Google Scholar]; (e) Montet X, Funovics M, Montet-Abou K, Weissleder R, Josephson L. J Med Chem. 2006;49:6087. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]
- 11.(a) Jin Z, Josserand V, Razkin J, Garanger E, Boturyn D, Favrot MC, Dumy P, Coll JL. Mol Imaging. 2006;5:188. [PubMed] [Google Scholar]; (b) Razkin J, Josserand V, Boturyn D, Jin Z, Dumy P, Favrot M, Coll JL, Texier I. ChemMedChem. 2006;1:1069. doi: 10.1002/cmdc.200600118. [DOI] [PubMed] [Google Scholar]; (c) Jin Z, Razkin J, Josserand V, Boturyn D, Grichine A, Texier I, Favrot MC, Dumy P, Coll JL. Mol Imaging. 2007;6:43. [PubMed] [Google Scholar]; (d) Sancey L, Ardisson V, Riou LM, Ahmadi M, Marti-Batlle D, Boturyn D, Dumy P, Fagret D, Ghezzi C, Vuillez JP. Eur J Nucl Med Mol Imaging. 2007;34:2037. doi: 10.1007/s00259-007-0497-z. [DOI] [PubMed] [Google Scholar]; (e) Sancey L, Dufort S, Josserand V, Keramidas M, Rome C, Faure AC, Foillard S, Roux S, Boturyn D, Tillement O, Koenig A, Boutet J, Rizo P, Dumy P, Coll JL. Int J Pharm. 2009;379:309. doi: 10.1016/j.ijpharm.2009.05.034. [DOI] [PubMed] [Google Scholar]; (f) Dimastromatteo J, Riou LM, Ahmadi M, Pons G, Pellegrini E, Broisat A, Sancey L, Gavrilina T, Boturyn D, Dumy P, Fagret D, Ghezzi C. J Nucl Cardiol. 2010;17:435. doi: 10.1007/s12350-010-9191-9. [DOI] [PubMed] [Google Scholar]
- 12.(a) Foillard S, Jin Z, Garanger E, Boturyn D, Favrot M, Coll JL, Dumy P. ChemBioChem. 2008;9:2326. doi: 10.1002/cbic.200800327. [DOI] [PubMed] [Google Scholar]; (b) Foillard S, Sancey L, Coll JL, Boturyn D, Dumy P. Org Biomol Chem. 2009;7:221. doi: 10.1039/b817251j. [DOI] [PubMed] [Google Scholar]
- 13.Garanger E, Boturyn D, Coll JL, Favrot MC, Dumy P. Org Biomol Chem. 2006;4:1958. doi: 10.1039/b517706e. [DOI] [PubMed] [Google Scholar]
- 14.(a) Grandjean C, Rommens C, Gras-Masse H, Melnyk O. Angew Chem, Int Ed. 2000;39:1068. [PubMed] [Google Scholar]; (b) Aucagne, Leigh DA. Org Lett. 2006;8:4505. doi: 10.1021/ol061657d. [DOI] [PubMed] [Google Scholar]; (c) Kele P, Mez G, Achatz D, Wolfbeis OS. Angew Chem, Int Ed. 2009;48:344. doi: 10.1002/anie.200804514. [DOI] [PubMed] [Google Scholar]; (d) Xiao J, Tolbert TJ. Org Lett. 2009;11:4134. doi: 10.1021/ol9016468. [DOI] [PubMed] [Google Scholar]; (e) Lee DJ, Mandal K, Harris PWR, Brimble MA, Kent SBH. Org Lett. 2009;11:5270. doi: 10.1021/ol902131n. [DOI] [PubMed] [Google Scholar]; (f) Vieyres A, Lam T, Gillet R, Franc G, Castonguay A, Kakkar A. Chem Commun. 2010;46:1875. doi: 10.1039/b924888a. [DOI] [PubMed] [Google Scholar]; (g) Meyer A, Spinelli N, Dumy P, Vasseur JJ, Morvan F, Defrancq E. J Org Chem. 2010;75:3927. doi: 10.1021/jo100599m. [DOI] [PubMed] [Google Scholar]
- 15.(a) Lim RKV, Lin Q. Chem Commun. 2010;46:1589. doi: 10.1039/b925931g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lutz JF. Angew Chem, Int Ed. 2008;47:2182. doi: 10.1002/anie.200705365. [DOI] [PubMed] [Google Scholar]
- 16.Galibert M, Dumy P, Boturyn D. Angew Chem, Int Ed. 2009;48:2576. doi: 10.1002/anie.200806223. [DOI] [PubMed] [Google Scholar]
- 17.(a) Rose K. J Am Chem Soc. 1994;116:30. [Google Scholar]; (b) Shao J, Tam JP. J Am Chem Soc. 1995;117:3893. [Google Scholar]
- 18.(a) Tornøe CW, Meldal M. In: Peptides: The Wave of the Future. Lebl H, Houghten RA, editors. American Peptide Society; San Diego: 2001. p. 263. [Google Scholar]; (b) Tørnoe CW, Christensen C, Meldal M. J Org Chem. 2002;67:3057. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]; (c) Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem, Int Ed. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Foillard S, Ohsten Rasmussen M, Razkin J, Boturyn D, Dumy P. J Org Chem. 2008;73:983. doi: 10.1021/jo701628k. [DOI] [PubMed] [Google Scholar]
- 20.Nicolet BH, Shinn LA. J Am Chem Soc. 1939;61:1615. [Google Scholar]
- 21.Renaudet O, Dumy P. Tetrahedron Lett. 2001;42:7575. [Google Scholar]