There was a time in medical genetics when intellectual disability (ID), autism, and other neurodevelopmental symptoms were predominantly viewed within the context of distinct clinical syndromes. By definition, a syndrome has a circumscribed pattern of phenotypic features that can be quantified into objective prevalence estimates, such as a 44% risk for a cardiac defect in a newborn with Down syndrome. From a practical standpoint, knowing the prevalence of associated clinical features in genetic syndromes allows targeted assessment and anticipatory medical management of these conditions. Binary categorizations (i.e., “present” versus “absent”) work well for symptoms such as congenital structural defects and most other physical aspects of a clinical phenotype. Traditionally, cognitive and behavioral features of genetic syndromes have been described in the same categorical way as physical findings, based on prevalence statistics that imply an all-or-none chance for symptoms such as ID and psychiatric disorders.
Recent trends toward more fine-grained research on cognitive and behavioral phenotypes, along with genomic evidence that blurs the lines among formerly distinct psychiatric diagnoses, have revealed the inadequacy of using a categorical model to describe neurodevelopmental outcomes in genetic disorders.1 For example, parents of an infant newly diagnosed with 22q11.2 deletion syndrome worry whether their child will be among the 30% with ID or the estimated 70% without this diagnosis, not appreciating that the syndrome's intellectual deficits occur along a continuum, where the threshold demarcating ID is set, by convention, at a particular point. In addition, the diagnosis of well-described, discrete genetic syndromes has been overtaken by newly identified copy-number variants (CNVs) and single-gene variants about which little is yet known, apart from a vague risk for a broad array of cognitive and behavioral symptoms. These are challenging times for geneticists and genetic counselors, but even more so for families who leave the medical genetics clinic with many worries and few focused answers about neurodevelopmental risk.
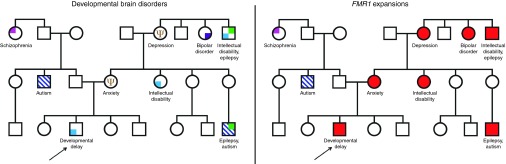
Emerging new perspectives in psychiatry,1 based partly on genomic findings, provide an opportunity to reevaluate our approach to the assessment and discussion of intellectual and behavioral prognoses in the medical genetics setting. Whole-genome copy-number and sequencing studies have revealed that identical genetic causes are common among apparently distinct developmental and psychiatric conditions.2,3,4 Shared etiological underpinnings now directly connect a host of seemingly unrelated disorders, including autism and schizophrenia. From a genomics perspective, the reconceptualization and merging of childhood developmental and adult-onset psychiatric disorders have profound implications for pedigree construction, risk modeling, and anticipatory guidance. The traditional pedigree that separates autism, ID, bipolar disorder, and schizophrenia as distinct and unrelated conditions in a family can no longer be considered valid (Figure 1). In many cases these multiple diagnostic subtypes of brain dysfunction represent variable expressivity of a single underlying genetic cause. Developmental brain disorders reflect varying degrees of dysfunction along a continuum of heritable human traits, including intelligence, social abilities, and motor skills.4 All humans fall somewhere along the functional continuum for these quantitative traits, with categorical diagnoses such as autism and ID defined by an artificial threshold at one end of a spectrum.
Figure 1.
At left, a traditional pedigree representing a large family with categorically defined developmental brain disorders. On the right, the same pedigree indicating inherited FMR1 expansions with variable expressivity.
Family background has long been known to play an important role in influencing patterns of phenotypic expression in genetic syndromes and common diseases. Parental height, for example, is a significant predictor of adult stature in girls with Turner syndrome,5 whereas family history modulates lifetime risk for cardiovascular disease. Phenotypic variability in women with BRCA1/2 mutations has been linked to genomic modifying factors that may ultimately be used to further refine their lifetime cancer risk.6 Relatively few studies have directly examined the relationship between parental functioning and neurodevelopmental outcomes in children with genetic syndromes. Significant intelligence quotient correlations between probands and their first-degree relatives have been documented in Down, Klinefelter, Prader-Willi, fragile X, and 22q11.2 deletion syndromes.7 Our recent family study of de novo 16p11.2 deletions8 demonstrated that neurodevelopmental outcomes in children with this CNV represent a predictable “shift” from expected functioning, based in part on parental background across multiple domains.
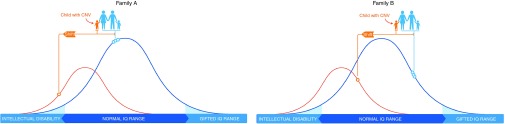
Such studies have important implications for genetic counseling and medical genomics practice. Depending on the parental starting point for quantitative traits such as intelligence and social abilities, the shift in functioning due to a specific CNV may or may not cause a child to reach the defined threshold for a diagnosable clinical disorder (Figure 2). For example, the empiric risk for autism in children with a de novo 16p11.2 deletion is about 15%.8 The deletion confers a 2.2 standard deviation deleterious effect on social behavior, a highly heritable and continuously distributed trait that can be reliably measured using the Social Responsiveness Scale.9 An infant with a de novo 16p11.2 deletion whose parents have higher than average social abilities, as measured on the Social Responsiveness Scale, actually has a lower risk for autism than one born to parents whose personalities are naturally skewed toward the less sociable end of the scale.8
Figure 2.
The blue curve represents the normal distribution of intelligence quotient (IQ) in the general population. The smaller, orange curve shows the IQ distribution for individuals with a particular copy-number variant (CNV). Circles on each curve indicate IQ scores for specific family members. In both family A and family B, this CNV confers the same magnitude of deleterious impact (“shift”) on a child's IQ. In family A, the CNV shifts the affected child's IQ into the range of intellectual disability. In family B, because the family IQ starting point is higher, the CNV shift does not reach the defined threshold for intellectual disability. Even in family B, however, the shift has an effect, lowering the child's IQ from where it would have been without the CNV.
Although research in this area is still evolving, it is likely that different CNVs and single-gene disorders have distinct profiles of deleterious impact on various functional domains. The well-established 25% risk for schizophrenia among individuals with a 22q11.2 deletion does not hold true for those with fragile X syndrome, for example, although the likelihood of autism in fragile X syndrome is much higher. In addition, family studies are unlikely to refine prognosis for some disorders, including those with severe cognitive effects and a narrow range of phenotypic variability. Parental studies may prove particularly helpful in focusing the prognosis for conditions with milder and more variable constellations of cognitive and behavioral features, including the growing number of pathogenic but poorly characterized CNVs involving psychiatric symptoms.
A potential future application of family studies might be the development of neurodevelopmental risk algorithms, similar to those in use for cancer and cardiovascular genetic counseling. Such algorithms could take into account variables that compound developmental risk, including the results of parental assessments on relevant functional domains, such as cognition, social behavior, and motor skills; the known deleterious effect on those domains for a specific genetic condition; other diagnosis-specific variables (e.g., gender, the presence of congenital anomalies); and environmental factors, such as prematurity. The result could be a customized profile that would ideally guide intervention by identifying a child's main area(s) of neurodevelopmental vulnerability. A child with a 15q11.2–13.1 duplication, for example, might have a particularly high risk for developing autism, warranting early and intensive behavioral therapy. Another child with the same genetic diagnosis might have a relatively low risk for autism but a heightened chance for significant ID. By better quantifying the type and magnitude of neurodevelopmental risks in children with various CNVs and sequence variations, clinicians could more effectively pinpoint areas for proactive intervention, as opposed to the current approach of “watchful waiting.” The ability to preemptively identify subgroups of children with distinct neurodevelopmental vulnerabilities will become increasingly important as pharmaceuticals that target dysfunctional molecular pathways in the brain enter clinical practice in future decades.
In light of genomic evidence and the rapidly evolving psychiatric landscape, traditional models of pedigree interpretation and categorical description are no longer adequate to explain the neurodevelopmental aspects of genetic disorders. Gone are the days when an empiric prevalence figure for ID or autism can simply be listed in the same breath as a cleft lip or a club foot. Counseling about neurodevelopmental prognosis for genetic disorders needs to more accurately convey the continuously distributed nature of intellectual and behavioral traits, as well as their cross-connections with clinical psychiatric diagnoses. Those discussions could potentially be enhanced by family studies that identify vulnerabilities and target interventions. Such an approach is wholly consistent with efforts to expand “precision medicine” beyond cancer treatment to other areas of medical practice.10
Could the future of medical genetics management include assessment of measures of social responsiveness, motor skills, language abilities, and cognitive functioning in a genetically diagnosed child, as well as his parents? This notion is intriguing, but one that must be carefully researched and approached with great caution. Genetics has a well-documented and checkered past with regard to family studies that resulted in errant social policies, including involuntary sterilization, based on naive notions about the heritability of intelligence, criminality, and antisocial behavior. The future goal of family studies in the context of neurodevelopmental disorders should be to enhance prognostic focus and maximize the appropriateness of interventions to improve outcomes. Regarding family studies for intellectual and behavioral traits, medical genetics now has a rare opportunity for a “do-over,” with the benefit of hindsight in a (hopefully) more socially enlightened era. Meticulous research on clinical applications of neurodevelopmental “shift,” in close collaboration with families and with careful consideration of social consequences, may allow us to get it right the second time around.
Disclosure
D.H.L. is a consultant to Natera, Inc. The other authors declared no conflict of interest.
Acknowledgments
This work was supported by the National Institute of Mental Health of the National Institutes of Health under award RO1MH074090. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Insel T, Cuthbert B, Garvey M, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry 2010;167:748–751. [DOI] [PubMed] [Google Scholar]
- Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet 2009;25:528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Brkanac Z, Coe BP, et al. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet 2011;7:e1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca A, Myers SM, Challman TD, Moreno-De-Luca D, Evans DW, Ledbetter DH. Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol 2013;12:406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A, Kauli R, Pertzelan A, et al. Final height of girls with Turner's syndrome: correlation with karyotype and parental height. Acta Paediatr 1995;84:550–554. [DOI] [PubMed] [Google Scholar]
- Gaudet MM, Kuchenbaecker KB, Vijai J, et al.; KConFab Investigators; Ontario Cancer Genetics Network; HEBON; EMBRACE; GEMO Study Collaborators; GENICA Network. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet 2013;9:e1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski P, Radoeva D, Fremont W, Kates WR, Antshel KM. Is child intelligence associated with parent and sibling intelligence in individuals with developmental disorders? An investigation in youth with 22q11.2 deletion (velo-cardio-facial) syndrome. Res Dev Dis 2014;35:3582–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca A, Evans DW, Ledbetter DH, et al. The role of parental cognitive, behavioral, and motor profiles in clinical variability in individuals with chromosome 16p11.2 deletions. JAMA Psychiatr 2015:72:119–126. [DOI] [PubMed] [Google Scholar]
- Constantino JN, Davis SA, Todd RD, et al. Validation of a brief quantitative measure of autistic traits: comparison of the social responsiveness scale with the autism diagnostic interview-revised. J Autism Dev Disord 2003;33:427–433. [DOI] [PubMed] [Google Scholar]
- Insel T, Cuthbert BN. Brain disorders? Precisely. Precision medicine comes to psychiatry. Science 2015;348:499–500. [DOI] [PubMed] [Google Scholar]