Abstract
Background:
The fetal inflammatory response (FIR) in placental membranes to an intrauterine infection often precedes premature birth raising neonatal mortality and morbidity. However, the precise molecular events behind FIR still remain largely unknown, and little has been investigated at gene expression level.
Methods:
We collected publicly available microarray expression data profiling umbilical cord (UC) tissue derived from the cohort of extremely low gestational age newborns (ELGANs) and interrogate them for differentially expressed (DE) genes between FIR and non–FIR-affected ELGANs.
Results:
We found a broad and complex FIR UC gene expression signature, changing up to 19% (3,896/20,155) of all human genes at 1% false discovery rate. Significant changes of a minimum 50% magnitude (1,097/3,896) affect the upregulation of many inflammatory pathways and molecules, such as cytokines, toll-like receptors, and calgranulins. Remarkably, they also include the downregulation of neurodevelopmental pathways and genes, such as Fragile-X mental retardation 1 (FMR1), contactin 1 (CNTN1), and adenomatous polyposis coli (APC).
Conclusion:
The FIR expression signature in UC tissue contains molecular clues about signaling pathways that trigger FIR, and it is consistent with an acute inflammatory response by fetal innate and adaptive immune systems, which participate in the pathogenesis of neonatal brain damage.
Fetal inflammatory response (FIR) in histological chorioamnionitis (HCA) is defined by the detection of polymorphonuclear cells within the wall of vessels in umbilical cord (UC) (funisitis) and chorionic plate. HCA is a surrogate of intra-amniotic infection and intrauterine inflammation, and its frequency increases with lower GA at birth (1). FIR is considered an advanced stage of HCA and in preterm infants increases the risk for perinatal mortality and morbidity, particularly of neonatal brain damage (2,3). The latter is one of the most severe short-and long-term complications of preterm birth (PB), particularly in extremely low gestational age newborns (ELGANs)—preterm infants born before 28 wk of gestation (4).
Here, we describe the results of a gene expression analysis of FIR in UC tissue using data from a previously published study by Cohen et al. (5). In that study, the authors profiled gene expression using microarrays in UC tissue at birth from a cohort of n = 54 ELGANs. Their goal was to search for gene expression changes in bronchopulmonary dysplasia and, in fact, did not find any with false discovery rate (FDR) < 5%, corresponding to an uncorrected P < 10−6. However, exploring the extensive clinicopathological data from the ELGAN cohort (4) (Supplementary Tables S1–S3 online) and the molecular metadata from the microarray chips (Supplementary Table S4 online), we have been able to obtain an appropriate experimental design that has allowed us to interrogate these data for differentially expressed (DE) genes between FIR and non–FIR-affected ELGANs.
Results
UC tissue carries a FIR gene expression signature in ELGANs
Using state-of-the-art statistical techniques and software for processing and analyzing microarray gene expression data, we discarded six samples, from the initial n = 54, that did not meet specific quality control criteria (Supplementary Figures S1–S6 and Supplementary Table S5 online). Because FIR status was also missing in some of the samples, the final data set analyzed in this paper consisted of n = 43 samples in which 18 were derived from FIR-affected infants and 25 from unaffected ones. After background correction and normalization of the microarray samples, a differential expression analysis with linear models (6), in which batch and other sources of FIR-unrelated variation were adjusted (7), showed that up to 19% of genes (3,896 out of 20,155) changed their expression at 1% FDR.
The magnitude of change among these 3,896 genes ranged from 1.12 to 32.7-fold difference between FIR and non–FIR-affected ELGANs. Using a minimum 1.5-fold change cutoff, we called 1,097 DE genes. From these, 592 were upregulated and 505 were downregulated (Figure 1 and Supplementary Tables S6 and S7 online). A functional enrichment analysis of these genes with the gene ontology (GO) database (8) yielded 542 and 35 significant GO terms (FDR < 10% and odds ratio (OR) > 1.5) by up and downregulated genes, respectively (Figure 2 and Supplementary Tables S8 and S9 online).
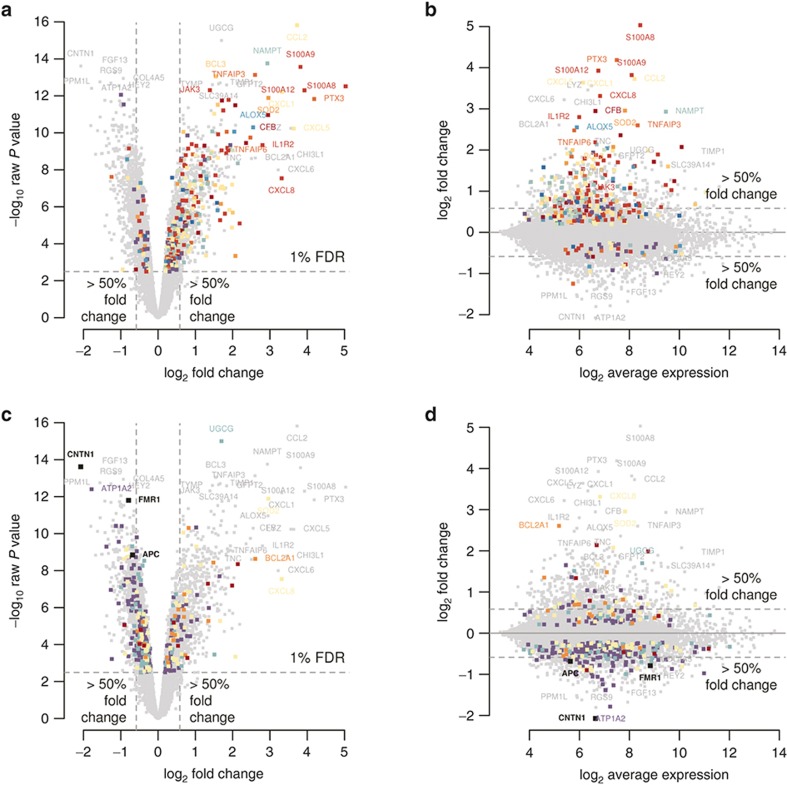
Figure 1.
Differential gene expression caused by FIR. (a,c) Volcano plot of the significance level of gene expression changes between FIR and non-FIR infants in −log10 scale (y-axis), as function of their fold-change (x-axis) in log2 scale. The horizontal dashed line indicates the threshold for a multiple-test correction above which 3,896 genes change significantly their expression at FDR < 1%. Vertical dashed lines indicate a minimum magnitude of 1.5-fold change in expression, met by 1,097 genes with FDR < 1%. Genes highlighted in colors enrich significantly the following gene sets: (a) Complement cascase (carmine), Cytokine signaling (dark red), TNF superfamily signaling (red), reactive oxygen species/glutathione/cytotoxic granules (salmon), Th17 cell response (orange), Adhesion/extravasation/migration (yellow), NF-kB signaling (yellow–green), innate pathogen detection (light green), leukocyte signaling (green), eicosanoid signaling (turquoise), phagocytosis/Ag presentation (blue), MAPK signaling (violet); (c) MSC genes (carmine), apoptosis genes (salmon), senescence genes (yellow), APC targets (green), FMRP targets (violet). (b,d) MA-plot of the magnitude of gene expression changes (y-axis) as function of the average expression (x-axis), both in log2 scale. Horizontal dashed lines indicate a minimum magnitude of 1.5-fold change in expression. Colors highlight the same gene sets as in a and c.
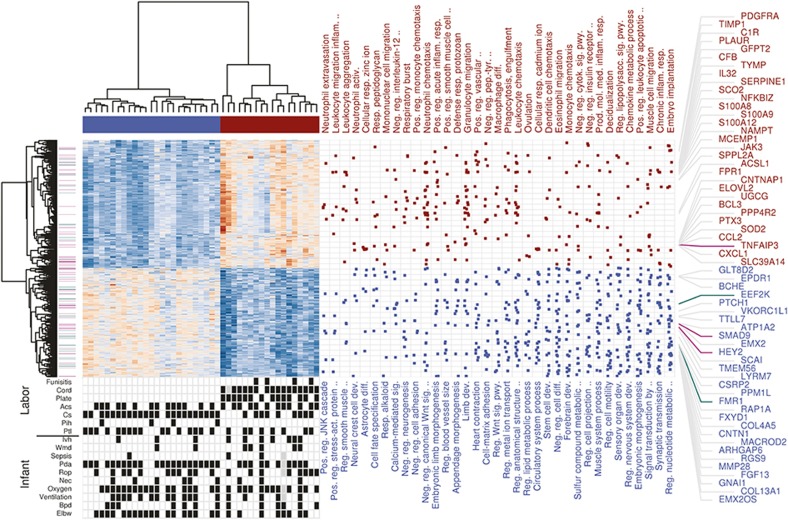
Figure 2.
Functional enrichment analysis of the FIR expression signature. Heatmap of expression values for 1,097 DE genes with FDR < 1% and minimum 1.5-fold change between FIR and non-FIR infants, obtained after removing FIR-unrelated variability. Dendrograms on the x and y-axes represent the hierarchical clustering of samples and genes, respectively. Leaves on the gene dendrogram are color coded according to whether genes encode for transcription factors (violet) or RNA-binding proteins (green), while those on the sample dendrogram are color coded according to FIR status where red indicates samples derived from FIR-affected infants and blue unaffected ones. The right dot-matrix represents DE genes (y-axis) belonging to GO terms (x-axis) significantly enriched (FDR < 10% and OR > 1.5) by upregulated (top) and downregulated (bottom) DE genes. Only the top-35 GO terms with highest OR from left to right are reported. The top 60 DE genes with largest fold change are on the right-hand side. At the bottom left, a dot-matrix representation of infant phenotypes is provided including Ivh, Wmd, Pda, Rop, Nec, Oxygen (>27 d of oxygen), ventilation (>7 d of ventilation), Bpd, Elbw (weight < 1,000 g), and labor conditions including cord (neutrophils in UC), plate (neutrophils in chorionic plate), Acs, Cs, Pih, and Ptl.
Upregulation of innate and adaptive immune pathways in FIR
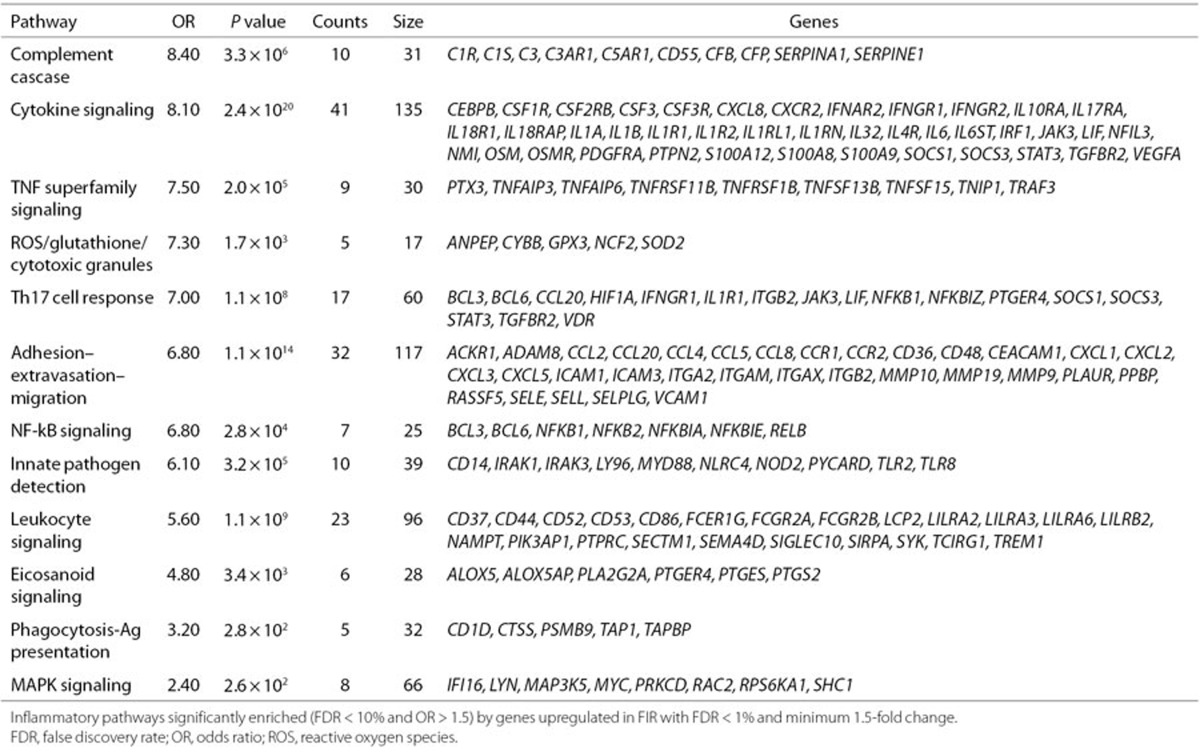
We found that genes involved in the innate immune response were among the top 10 upregulated genes with largest fold change (Supplementary Table S6 online), such as calgranulins (S100A8, S100A9, S100A12), the lyzome (LYS), chemokines (CXCL8, CXCL1, CXCL5, CCL2), and the pentraxin 3 (PTX3). The GO enrichment analysis of upregulated genes also showed multiple significant GO terms (FDR < 10%, OR > 1.5) of the innate immune system. Among the top 10 enriched GO terms with largest OR (Figure 2 and Supplementary Table S8 online), we found terms related to the activation of neutrophils (e.g., neutrophil extravasation), monocytes (e.g., mononuclear cell migration), and oxidative stress (OS) (respiratory burst). We also tested the enrichment of a targeted collection of 17 immune pathways (16 innate (9) and one T helper type 17 (TH 17) response (10)) by the subset of significant 592 upregulated genes at 1% FDR with a minimum 1.5-fold change. Using one-tailed Fisher's exact tests, we found 12 of these 17 immune pathways to be significantly enriched at 10% FDR with OR > 1.5. More concretely, we found enrichment of DE genes in the TH 17 response pathway and in 11 innate response pathways (Table 1). Next to the enrichment of DE genes in the TH 17 response pathway, we found additional evidence of an adaptive immune response in FIR by enriched GO terms such as positive regulation of T cell migration (OR = 8.18) and positive regulation of B cell activation (OR = 3.58).
Table 1. Significantly upregulated genes classified into inflammatory pathways.
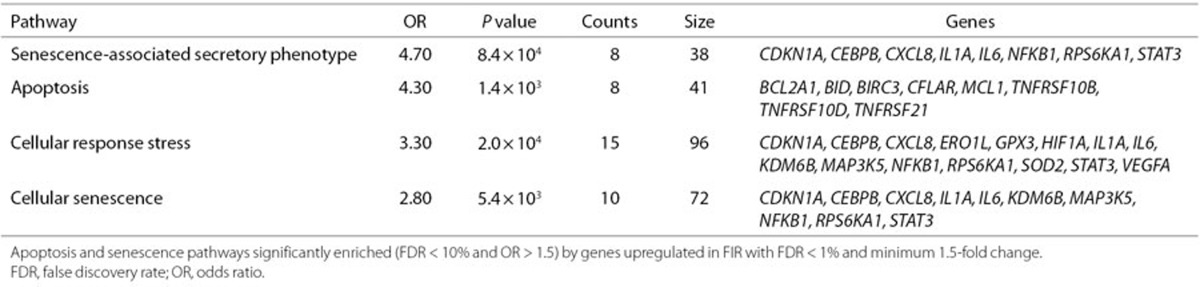
As a result of the activation of the innate and adaptive immune systems, the inflamed tissue may show cellular adaptations such as apoptosis and cellular senescence. We found enrichment of upregulated DE genes in apotosis (9) and senescence (11) gene sets (FDR < 10% and OR > 1.5); see Table 2. Finally, we also interrogated the Human Phenotype Ontology (HPO) database (12) and found a significant enrichment of upregulated genes in HPO terms (FDR < 10% and OR > 1.5) related to skin, dental and respiratory infections (Supplementary Table S10 online). This suggests that some of the upregulated gene expression changes associated with FIR are similar to those caused by infection of surface tissues.
Table 2. Significantly upregulated genes classified into apoptosis and senescence pathways.
Downregulation of neurodevelopmental pathways in FIR
We found enrichment of downregulated DE genes in GO terms associated with neurodevelopment (see Figure 2 and Supplementary Table S9 online), such as neural crest cell development (OR = 6.25), astrocyte differentiation (OR = 5.51), forebrain development (OR = 2.33), and synaptic transmission (OR = 1.90). Among the most significantly downregulated genes (Figure 1 and Supplementary Table S7 online), we found CNTN1, FMR1, and APC, in which CNTN1 is a cell adhesion molecule that plays a role in the orderly progression of cortical development (13), FMR1 encodes the Fragile-X mental retardation RNA-binding protein whose loss of function causes Fragile-X syndrome and which is essential for cognitive development by regulating translation in neurons (14), and APC encodes an RNA-binding protein implicated in RNA localization and required for human neuron and axon migration (15). We mapped the FMRP and APC target genes found by HITS-CLIP (14,15) to our data set and observed a significant enrichment of downregulated genes at FDR < 1% in these two gene sets (one-tailed Fisher's exact P = 1.08 × 10−6, OR = 1.49 and P = 0.01, OR = 1.44, respectively; Figure 1).
These findings were unexpected because there is no neuronal tissue in UC. Interestingly, we observed enrichment of the GO terms stem cell development (OR = 1.89) and stem cell differentiation (OR = 1.75), and in a gene signature of 19 selective markers for UC mesenchymal stem cells (MSCs) that mapped to our data set (16), we also detected a significant enrichment by upregulated genes (one-tailed Fisher's exact P = 0.001, OR = 11.64, Figure 1).
A molecular signature of severe FIR
Advanced histological patterns of FIR are associated with perinatal brain damage (17); therefore, gene expression profiles of UC tissue from infants with “severe FIR” can potentially shed light on molecular mechanisms leading to adverse neurological outcome. Despite the limited sample size of n = 43, we observed in our data an increasing fraction of intraventricular hemorrhage (IVH) and white matter disease (WMD) cases among FIR-affected over unaffected infants (Supplementary Table S2 online), which grows markedly when considering only the n = 17 infants who were not administered antenatal glucocorticoids (ACS), and even significantly in the case of IVH. (two-tailed Fisher's exact P = 0.005 and P = 0.1 for WMD; see Supplementary Table S3 online).
Using the joint IVH and WMD condition as surrogate for severe FIR, we compared the expression profiles of infants affected by all three conditions FIR, IVH, and WMD (n = 4) with those not affected by any of them (n = 22) and found 183 DE genes with FDR < 1%, 151 of them with a minimum 1.5-fold change (Supplementary Table S11 online). These figures increased more than threefold up to 570 and 566 genes, respectively, when restricting the comparison to infants who were not administered with ACS (n = 3 FIR/IVH/WMD vs. n = 10 non-FIR/non-IVH/non-WMD); see Supplementary Table S12 online. In this comparison, we observed that calgranulin genes remain among the top 10 upregulated genes. However, many of the expression changes occurred in genes that either did not change significantly their expression or did it below a 50% magnitude, in the comparison between FIR-affected and non–FIR-affected infants from the whole data set of n = 43 samples. For instance, the nerve injury-induced protein 2 gene (NINJ2), which is involved in nerve regeneration after nerve injury, is significant at FDR < 1% in both cases but its significance level and fold change increase, respectively, from P = 2.45 × 10−3 to P = 9.79 × 10−5 and from 1.3-fold to 3.7-fold.
A GO functional analysis among the 310/566 upregulated DE genes with FDR < 1% and minimum 1.5-fold change yielded 70 significantly enriched GO terms (FDR < 10% and OR > 1.5), 4 of which were exclusively enriched by upregulated genes called DE only in the joint FIR/IVH/WMD comparison: oxygen transport, hydrogen peroxide catabolic process, tetrapyrrole metabolic process, and homeostasis of number of cells (Supplementary Table S13 online). Downregulated genes (256/566) enriched only two GO terms (FDR < 20% and OR > 1.5) associated with neurogenesis (negative regulation of neurogenesis) and morphogenetic processes in central nervous system development (negative regulation of cytoskeleton organization; see Supplementary Table S14 online). These results show that expression changes in UC tissue, occurring under a joint FIR/IVH/WMD condition, convey a molecular signature of severe FIR concealing additional clues on molecular mechanisms associated with neonatal brain damage.
Discussion
We have shown that FIR is characterized by a broad and complex acute inflammatory molecular expression signature in UC tissue of ELGANs. Previously, Madsen-Bouterse et al. (18) explored gene expression changes in UC blood leukocytes of preterm infants with fetal inflammatory response syndrome and funisitis (FIR) (n = 10) vs. a control group (n = 10), and they reported a total of 541 genes changing significantly at 5% FDR.
Among the 20,155 genes analyzed in our study, 483 from the 541 genes reported by Madsen-Bouterse et al. (18) mapped to our data, overlapping significantly with our FIR signature (one-tailed Fisher's exact P < 2.2 × 10−16 with OR = 2.67 for DE genes with FDR < 1% and P < 2.2 × 10−16 with OR = 3.58 for DE genes with FDR < 1% and minimum 1.5-fold change). The comparison of the log2-fold changes (Supplementary Figure S7 online) of these overlapping genes shows that even though there are important differences in the RNA source and microarray technology employed in both studies, the magnitude of expression change shows a positive and significant correlation in every subset of genes derived at different significance levels (0.39 < ρ < 0.51 with P < 3.16 × 10−4).
There is, however, an important discrepancy in the extent of the expression changes reported by Madsen-Bouterse et al. (18) and us. We found a total of 3,896 DE genes with FDR < 1% between the UC tissue of FIR and non-FIR infants, a figure that increases up to 5,461 with FDR < 5%, one order of magnitude above the 541 DE genes found by the study of Madsen-Bouterse et al. (18) at the same significance level. To rule out the possibility that this discrepancy is due to our larger sample size (n = 43 vs. n = 20), we carried out a simulation study matching the fetal inflammatory response syndrome group sample composition, showing that on average one expects 4,532 DE genes with FDR < 5% (Supplementary Figure S8 online). Therefore, we can conclude that the main difference between the Madsen-Bouterse et al.'s (18) study and ours has a biological explanation, and more concretely, on the origin of biological samples that points to the participation of UC cells, apart from UC blood leukocytes, in the FIR molecular signature.
The immune cells that trigger the production of inflammatory mediators in UC blood in FIR are not yet fully characterized (19). Madsen-Bouterse et al. (18) did not find differential expression of the IL6 gene in UC blood leukocytes. Diverse studies (20,21) have failed to detect differences in the concentration of the mRNA encoding for interleukin (IL)-6 in UC blood leukocytes between infants with elevated concentration of IL-6 in UC blood vs. control, and the authors have proposed that other cells than UC blood leukocytes participate in the production of inflammatory mediators. We did find differential expression in the IL6 gene and in multiple other inflammatory genes that participate in the innate immune response (Supplementary Table S6 online). We found DE genes related to neutrophil activation and migration (e.g., CXCL8, CXCL1, CXCL2); activation of endothelium (e.g., ICAM1, ICAM3, SELE, ITGB2); production of reactive oxygen species and OS (e.g., SOD2, GPX3, MGST1); secretion of cytokines (e.g., IL1B, NAMPT, MMP9); synthesis of leukotrienes (e.g., ALOX5), and prostaglandins (e.g., PTGES). On the other hand, Hecht et al. (22) recently demonstrated that ELGANs with funisitis (FIR) had elevated blood concentrations of multiple inflammatory mediators during the first 3 days after birth. Thus, altogether, it suggests the hypothesis that UC cells participate in the onset of FIR, which can result in a systemic neonatal inflammation after birth.
The origin of HCA in PB has been traditionally attributed to an ascending intrauterine infection. However, recent evidence (23) suggests that one of the initial events of HCA could be a microbial invasion of the amniotic cavity. Gillaux et al. (24) have shown the capacity of amniotic epithelial cells to trigger an immune response through TLR recognition. We found upregulation of TLR2 and TLR8 genes, and genes encoding proteins that participate in the downstream signaling pathways associated with TLR activation (e.g., MYD88, CD14, IRAK3, NFKB1, and NFKB2). In addition, genes encoding for S100 calgranulin proteins (S100A8, S100A9, and S100A12), which can act as damage-associated molecular patterns (“alarmins”), are among the most upregulated genes in FIR and severe FIR (Supplementary Tables S6 and S12 online). Among the body of evidence consistent with these results, Buhimschi et al. (25) has shown that the calgranulin S100A12 is associated with clinically significant funisitis in amniotic fluid. On the other hand, Kim et al. (26) has demonstrated that skin samples from fetal autopsies, which had chorioamnionitis with fetal vasculitis (FIR) on placental examination, showed perivascular inflammatory cell infiltration by neutrophils, lymphocytes, and histiocytes in the superficial dermis, and TLR-2 immunoreactivity in the skin. Interestingly, we found a significant enrichment of upregulated genes in HPO terms related to skin infections (Supplementary Table S10 online). In summary, it is biologically plausible that microbial invasion of the amniotic cavity triggers the onset of downstream molecular pathways by UC epithelial cells and TLR recognition, which initiate the production of inflammatory mediators and the recruitment of blood leukocytes to UC tissue (Figure 3). Afterwards, the production of calgranulins and reactive oxygen species can act as alarmins, enhancing FIR. These results support the role of calgranulins as biomarkers of intra-amniotic inflammation in PB (25).
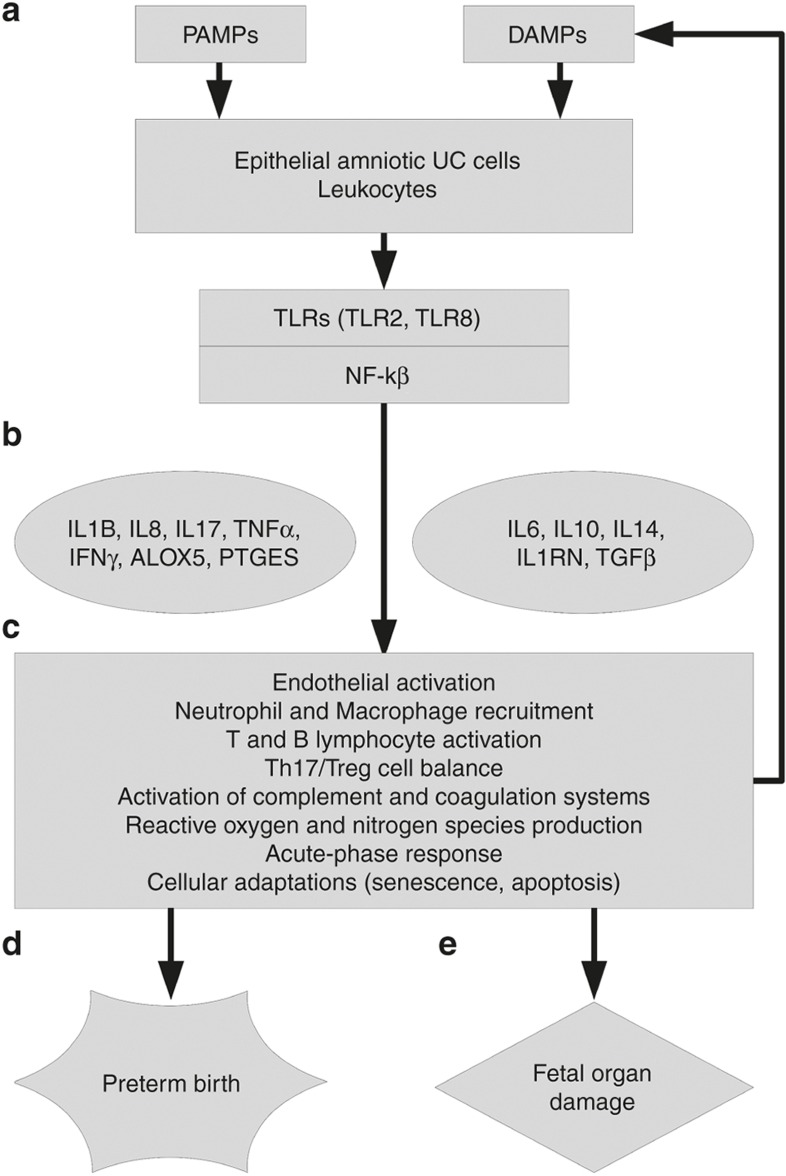
Figure 3.
Molecular architecture of FIR. (a) UC microbial invasion is detected by epithelial amniotic cells and leukocytes in UC tissue. TLRs identify PAMPs, and DAMPs, producing and enhancing FIR and participating in the activation of the NF-kB signaling pathway. (b) The transcriptional response of NF-kB activates proinflammatory mediators and anti-inflammatory ones. (c) The state of multiple cell types and tissues is affected by the acute inflammatory cascade. (d) The activation of proinflammatory pathways can trigger the production of labor intermediates and promote parturition onset. (e) The acute inflammatory response can damage fetal organs through its mediators and immune cells.
The role of the adaptive immune system has not been fully investigated in comparison to the innate system in FIR. In preterm infants, Duggan et al. (27) and Luciano et al. (28) have demonstrated that neonatal brain damage and PB were associated with T-cell activation. In premature infants, these cells have a bias to differentiate in TH 17 cells, which play a prominent role in the body defense against extracellular microbes at epithelial barriers. In our study, we found a significant enrichment of DE genes in the TH 17 response pathway (e.g., HIF1A) and in GO terms related to T- and B-cell activation (Table 1 and Supplementary Table S8 online). Interestingly, HIF1A encodes for a transcription factor protein that can induce the differentiation of TH 17 vs. T-regulatory cells. Thus, our results corroborate the activation of the fetal adaptive immune system in FIR, concretely of TH 17 cells, and support the evidence of an adaptive immune response in intrauterine inflammation during PB (Figure 3). This has been shown to occur through the induction of a TH 17 cell response by the activation of the IL-1/IL-17 axis (29).
The cellular adaptation of UC tissue to FIR is of pathogenic interest, yet it has been poorly investigated. Senescence is one such cellular adaptations in response to noxious stimuli such as OS, and it is defined by an irreversible cell-cycle growth arrest, and by the synthesis of multiple inflammatory mediators (senescence-associated secretory phenotype). We found endogenous enzymatic antioxidant encoding genes (SOD2, GPX3, MGST1) and matrix metalloproteinases (MMP9, MMP10, MMP19) upregulated in FIR (Supplementary Table S6 online) and a significant enrichment of multiple OS-related GO terms (e.g., superoxide anion generation; Supplementary Table S8 online), of senescence pathways, and of a UC MSCs expression signature (Table 2). OS damage and the activation of senescence pathways have been proposed as candidate pathogenic mechanisms in PB and neonatal diseases (30,31). Thus, the acute inflammatory cascade and OS generated during FIR may damage MSCs, which could enter into a senescence state with senescence-associated secretory phenotype that in turn may also participate and enhance FIR (Figure 3).
Accumulated evidence indicates that intrauterine inflammation participates in the genesis of encephalopathy of prematurity. While epidemiological studies have demonstrated that FIR increases the risk of brain damage, few investigations have explored pathogenic mechanisms behind this association. We found that downregulated genes enriched significantly GO terms related to fetal development, particularly of brain (Supplementary Table S9 online). We observed downregulation of neurodevelopmental genes such as FMR1, APC, and their targets, and CNTN1, which are essential for cognitive development (13,14,15). Despite its limited sample size (n = 43), the data set analyzed in this article recapitulates the significant association between FIR and IVH after adjusting by ACS (Supplementary Table S3 online), reported in larger studies (32). Using the perinatal neurological status (IVH and WMD), we found a molecular signature of severe FIR and evidence of the effect of the ACS therapy at molecular level. Overall, these findings are consistent with the hypothesis that expression changes in UC caused by strong intrauterine inflammation conceal clues on neurological damage.
One possible molecular mechanism for this hypothesis is the presence of MSCs in UC, which have the capacity to differentiate into neurons and could be damaged by FIR. Given that we observed the differential expression of key genes and pathways associated with MSCs in UC tissue, we postulate that UC MSCs could constitute a useful system for modeling perinatal brain damage in ELGANs. In fact, several studies (33) have already used human pluripotent stem cells, obtained from tissue of patients affected by diverse disorders, including neurological diseases, for investigating mechanisms of disease. In turn, hypotheses on precise pathogenic molecular mechanisms, such as the loss of function of neurodevelopmental genes (e.g., CNTN1, FMR1, and APC) provide an array of potential therapeutic targets (34) to fight against such an adverse neonatal outcome.
Methods
The bioinformatic and statistical analyses described in this section were performed using R version 3.1.3 and multiple add-on software packages deposited at the Comprehensive R Archive Network and at the Bioconductor project (35) release 3.0. Scripts reproducing the analyses described in this section are available in the Supplementary Data online.
Subjects and clinical data
The data analyzed in this article were generated by Cohen et al. (5) using UC samples derived from preterm newborns at 23 to 28 GA weeks enrolled in the ELGAN study (4), and therefore, informed consent was not required to conduct the bioinformatic and statistical analyses presented in this article. A full description of the ELGAN study, sample collection, and preparation is provided elsewhere (4,5). Phenotypic and clinical data were obtained through the ELGAN Publication and Data Analysis committee for the purpose of this research and are described in Supplementary Tables S1 to S3 online. The clinical record of each infant included the presence or absence of neutrophils in UC and chorionic plate. From these two factors, we built a binary variable indicating the activation of FIR when neutrophils were present in either the UC or the chorionic plate and, conversely, indicating the lack of FIR when neutrophils were absent from both UC and chorionic plate. For six infants, FIR status could not be determined due to missing values occurring in these two factors and that precluded using the previous rule.
Data import, quality assessment, and normalization
Raw data CEL files corresponding to the n = 54 samples of RNA in UC, hybridized on Affymetrix HG-U133 Plus 2.0 gene expression microarray chips and initially published by Cohen et al. (5) were downloaded from the Gene Expression Omnibus (National Center for Biotechnology Information, Bethesda, MD) using the accession GSE8586. Batch processing information was derived from the scanning timestamp stored in CEL files as previously described (36), creating a batch indicator variable that divided the samples into three different batches. The cross-classification of infants by FIR status and batch indicator showed no correlation between the primary outcome of our analysis, FIR status, and sample batch processing (Supplementary Table S4 online).
We applied seven quality assessment diagnostics on the whole set of n = 54 samples and flagged by visual inspection those showing potentially problematic features (Supplementary Figures S1 to S6 online). We ranked samples by the number of flagged diagnostics (Supplementary Table S5 online) and discarded six that failed in the majority of them. Raw data from the remaining n = 48 samples were normalized into Affymetrix probesets using fRMA (37) leading to an expression data set of P = 54,675 probesets by n = 48 samples. At this point, we also excluded from further analysis five samples for which the FIR status was missing, ending with a final set of n = 43 samples.
Nonspecific filtering and differential expression analysis
A first nonspecific filtering step was performed to remove probesets of little interest for the rest of the analysis. Concretely, we removed probesets without annotation to Entrez gene identifiers, probesets annotated to a common Entrez gene keeping the one with highest variability measured by the IQR, and Affymetrix control probesets. This resulted in an expression data set of P = 20,155 probesets in one-to-one correspondence with Entrez genes, by n = 43 samples.
Using the top 10% most variable (IQR) genes, we estimated covariates that capture variability unrelated to FIR status and batch, with surrogate variable analysis (7) that yielded seven such covariates. For each gene g, we defined the following linear model,
where the expression profile yg of gene g is a linear function of the FIR status variable xF, the batch indicator variable xB, and the seven covariates xS1, ..., xS7 estimated by surrogate variable analysis. Using limma (6), we fitted this linear model to the every gene expression profile and calculated moderated t-statistics for the coefficient estimating the effect between FIR and non–FIR-affected infants and their corresponding P values for the null hypothesis of no-differential expression.
We considered discarding nine fractions, from 10% to 90%, of genes with lower variability (IQR). In the remaining genes from each fraction, we adjusted the previously calculated P values by FDR and tallied the number of genes with FDR < 1%. In a second nonspecific filtering step, we removed from further analysis the fraction of genes with lower variability that maximized the amount of detected differential expression (38) at 1% FDR. Supplementary Figure S9 online shows that this fraction was 40% (8,062 genes from 20,155), which enabled adjusting by FDR on a reduced data set of 12,093 genes providing a set of 3,896 genes changing significantly at 1% FDR, 1,097 of them with a minimum 1.5-fold change.
When analyzing the subset of n = 17 infants who were not administered with ACS, we introduced two changes to this procedure. First, we used a factorial design without intercept term, estimating all the three-way effects of FIR, IVH, and WMD. Then, we calculated the moderated t-statistic based on the contrast that compared infants affected by all three FIR, IVH, and WMD effects against those not affected by any of them. Second, due to the small sample size, we removed from the model the xB term corresponding to the batch indicator variable and let surrogate variable analysis to estimate any effects unrelated to the contrast of interest.
Differential expression analysis with smaller sample size
From the entire data set of n = 43 infants, we generated 100 bootstrapped data sets with a sample-group composition identical to the study by Madsen-Bouterse et al. (18). Infants in each bootstrapped data set were randomly sampled with replacement ensuring that there were n = 10 FIR-affected infants, n = 10 non-FIR ones, and at least one sample per batch. In each bootstrapped data set, we carried out a differential expression analysis identical to the one performed on the whole data set. The number of genes, P values, and absolute log2-fold changes shown in Supplementary Figure S8 online correspond to those with FDR < 5% to match the significance level employed by Madsen-Bouterse et al. (18).
Functional enrichment analysis
Functional enrichment analyses with the GO and the HPO databases were performed using a conservative implementation of the conditional hypergeometric test (39) in the GOstats Bioconductor package (40), where a significant conditioning term was identified not only on the basis of a P value cutoff, but also using minimum OR and gene-set size thresholds. In all GO and HPO enrichment analyses, we used a P value cutoff of 0.01, a minimum OR of 1.5, and a minimum and maximum gene set sizes of 5 and 300 genes, respectively. Among the previously identified GO or HPO terms, we discarded those whose average IQR across their respective genes was below the 90% of such values in a simulated distribution with random gene sets of matching size. On the remaining GO or HPO terms passing these nonspecific filters, P values were adjusted by FDR, and we reported as significantly enriched those GO or HPO terms with FDR < 10%, OR > 1.5, and at least five genes enriching them. When testing functional enrichment separately by up and downregulated genes, the gene universe was restricted to the subsets of 10,868 genes with positive log2 fold change and 9,287 genes with negative log2 fold change, respectively. Sources for the targeted gene sets shown in Figure 1 and Tables 1 and 2 are described in the Supplementary Table S15 online.
Statement of Financial Support
This study was supported by a grant from the Spanish Ministry of Economy and Competitiveness (TIN2011-22826).
Disclosure
The authors declare no conflict of interest.
Acknowledgments
The authors thank the ELGAN PAD committee and Jennifer Cohen, Linda Van Marter, Yao Sun, Elizabeth Allred, Alan Leviton, and Isaac Kohane for granting access to the relevant ELGAN data, whose collection was supported by the National Institute of Neurological Disorders and Stroke (1 U01 NS 40069-01A2). We are grateful to E. Allred and A. Leviton for their feedback on earlier versions of this manuscript and to S.A. Madsen-Bouterse for sharing with us the complete list of 541 DE genes and fold changes reported in her previous study.
Supplementary Material
References
- Dammann O, Allred EN, Leviton A, et al. Fetal vasculitis in preterm newborns: interrelationships, modifiers, and antecedents. Placenta 2004;25:788–96. [DOI] [PubMed] [Google Scholar]
- Lau J, Magee F, Qiu Z, Houbé J, Von Dadelszen P, Lee SK. Chorioamnionitis with a fetal inflammatory response is associated with higher neonatal mortality, morbidity, and resource use than chorioamnionitis displaying a maternal inflammatory response only. Am J Obstet Gynecol 2005;193:708–13. [DOI] [PubMed] [Google Scholar]
- Leviton A, Paneth N, Reuss ML, et al. Maternal infection, fetal inflammatory response, and brain damage in very low birth weight infants. Developmental Epidemiology Network Investigators. Pediatr Res 1999;46:566–75. [DOI] [PubMed] [Google Scholar]
- O'Shea TM, Allred EN, Dammann O, et al.; ELGAN study Investigators. The ELGAN study of the brain and related disorders in extremely low gestational age newborns. Early Hum Dev 2009;85:719–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J, Van Marter LJ, Sun Y, Allred E, Leviton A, Kohane IS. Perturbation of gene expression of the chromatin remodeling pathway in premature newborns at risk for bronchopulmonary dysplasia. Genome Biol 2007;8:R210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007;3:1724–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza MJ, McCall CE, Li L, Isaacs WB, Xu J, Chang BL. Assembly of inflammation-related genes for pathway-focused genetic analysis. PLoS One 2007;2:e1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, et al. A validated regulatory network for Th17 cell specification. Cell 2012;151:289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinky F, Nativ N, Stelzer G, et al. PathCards: multi-source consolidation of human biological pathways. Database (Oxford) 2015;2015: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Doelken SC, Mungall CJ, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res 2014;42:D966–D974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzoca A, Corsi P, Polizzi A, et al. F3/Contactin acts as a modulator of neurogenesis during cerebral cortex development. Dev Biol 2012;365:133–51. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011;146:247–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Quan J, Nowakowski DW, et al. APC is an RNA-binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell 2014;158:368–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Mazor Y, Rinon A, et al. LifeMap Discovery™: the embryonic development, stem cells, and regenerative medicine research portal. PLoS One 2013;8:e66629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas AA, Faye-Petersen OM, Sims B, et al. Histological characteristics of the fetal inflammatory response associated with neurodevelopmental impairment and death in extremely preterm infants. J Pediatr 2013;163:652–7.e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen-Bouterse SA, Romero R, Tarca AL, et al. The transcriptome of the fetal inflammatory response syndrome. Am J Reprod Immunol 2010;63:73–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alquen D, Kramer BW, Seidenspinner S, et al. Activation of umbilical cord endothelial cells and fetal inflammatory response in preterm infants with chorioamnionitis and funisitis. Pediatr Res 2005;57:263–9. [DOI] [PubMed] [Google Scholar]
- Singh B, Merchant P, Walker CR, Kryworuchko M, Diaz-Mitoma F. Interleukin-6 expression in cord blood of patients with clinical chorioamnionitis. Pediatr Res 1996;39:976–9. [DOI] [PubMed] [Google Scholar]
- Berner R, Niemeyer CM, Leititis JU, et al. Plasma levels and gene expression of granulocyte colony-stimulating factor, tumor necrosis factor-alpha, interleukin (IL)-1beta, IL-6, IL-8, and soluble intercellular adhesion molecule-1 in neonatal early onset sepsis. Pediatr Res 1998;44:469–77. [DOI] [PubMed] [Google Scholar]
- Hecht JL, Fichorova RN, Tang VF, Allred EN, McElrath TF, Leviton A; Elgan Study Investigators. Relationship between neonatal blood protein concentrations and placenta histologic characteristics in extremely low GA newborns. Pediatr Res 2011;69:68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Romero R, Gervasi MT, et al. Widespread microbial invasion of the chorioamniotic membranes is a consequence and not a cause of intra-amniotic infection. Lab Invest 2009;89:924–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaux C, Méhats C, Vaiman D, Cabrol D, Breuiller-Fouché M. Functional screening of TLRs in human amniotic epithelial cells. J Immunol 2011;187:2766–74. [DOI] [PubMed] [Google Scholar]
- Buhimschi CS, Buhimschi IA, Abdel-Razeq S, et al. Proteomic biomarkers of intra-amniotic inflammation: relationship with funisitis and early-onset sepsis in the premature neonate. Pediatr Res 2007;61:318–24. [DOI] [PubMed] [Google Scholar]
- Kim YM, Romero R, Chaiworapongsa T, Espinoza J, Mor G, Kim CJ. Dermatitis as a component of the fetal inflammatory response syndrome is associated with activation of Toll-like receptors in epidermal keratinocytes. Histopathology 2006;49:506–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan PJ, Maalouf EF, Watts TL, et al. Intrauterine T-cell activation and increased proinflammatory cytokine concentrations in preterm infants with cerebral lesions. Lancet 2001;358:1699–700. [DOI] [PubMed] [Google Scholar]
- Luciano AA, Yu H, Jackson LW, Wolfe LA, Bernstein HB. Preterm labor and chorioamnionitis are associated with neonatal T cell activation. PLoS One 2011;6:e16698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallapur SG, Presicce P, Senthamaraikannan P, et al. Intra-amniotic IL-1β induces fetal inflammation in rhesus monkeys and alters the regulatory T cell/IL-17 balance. J Immunol 2013;191:1102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Auten RL. Maturation of the antioxidant system and the effects on preterm birth. Semin Fetal Neonatal Med 2010;15:191–5. [DOI] [PubMed] [Google Scholar]
- Menon R. Oxidative stress damage as a detrimental factor in preterm birth pathology. Front Immunol 2014;5:567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Furuhashi M, Ishikawa K, et al.; Neonatal Research Network Japan. The effects of antenatal corticosteroids therapy on very preterm infants after chorioamnionitis. Arch Gynecol Obstet 2014;289:1185–90. [DOI] [PubMed] [Google Scholar]
- Unternaehrer JJ, Daley GQ. Induced pluripotent stem cells for modelling human diseases. Philos Trans R Soc Lond B Biol Sci 2011;366:2274–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci 2013;16:1530–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 2010;11:733–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA). Biostatistics 2010;11:242–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci USA 2010;107:9546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006;22:1600–7. [DOI] [PubMed] [Google Scholar]
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics 2007;23:257–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.