Abstract
The relationship between the TP53 Arg72Pro polymorphism (rs1042522) and the risk of leukemia remains controversial. Consequently, we performed a meta-analysis to accurately evaluate the association between TP53 Arg72Pro polymorphism and leukemia risk. A comprehensive search was conducted to find all eligible studies of TP53 Arg72Pro polymorphism and leukemia risk. Fourteen case-control studies, with 2,506 cases and 4,386 controls, were selected for analysis. The overall data failed to indicate a significant association between TP53 Arg72Pro polymorphism and the risk of leukemia (C vs. G: OR = 1.09, 95% CI = 0.93–1.26; CC vs. GC + GG: OR = 1.23, 95% CI = 0.96–1.57). In a subgroup analysis of clinical types, an increased risk was observed in the acute lymphocytic leukemia (ALL) subgroup (CC vs. GC + GG: OR = 1.73; 95% CI = 1.07–2.81) but not in the acute myeloid leukemia (AML) subgroup. In the subgroup analysis, no significant associations with ethnicity and the source of the controls were observed. In conclusion, the results suggest that there is no association between TP53 Arg72Pro polymorphism and the risk of leukemia, but the CC genotype may increase the risk of ALL TP53 Arg72Pro polymorphism CC genotype may increase the risk of ALL but is not associated with AML. Further large-scale, well-designed studies are needed to confirm our results.
Leukemia is a malignant tumor of the hematopoietic systems. It has a poor prognosis due to a range of complicated features1. Based on the speed of onset and cytogenetic analysis, leukemia was divided into four main types2: acute lymphocytic leukemia (ALL), chronic lymphocytic leukemia (CLL), acute myeloid leukemia (AML), and chronic myeloid leukemia (CML). Although leukemogenesis studies have been conducted for many years, the mechanisms underlying the development of this hematologic malignancy remain unclear. Multiple risk factors are thought to be implicated in leukemia, and genetic factors, such as constitutional genetic variation in components of DNA damage response pathways, have become the focus of research3,4.
The TP53 tumor suppressor gene, which is located on chromosome 17p13, plays a pivotal role in the maintenance of genomic stability through encoding of the TP53 protein. The TP53 protein functions by regulating cell cycle arrest, DNA repair, apoptosis, and gene transcription to mediate cellular responses to DNA damage5. Frequent mutations and differential expression of TP53 in various cancers highlight the significance role of p53 in carcinogenesis and tumor progression6,7. A functional single-nucleotide polymorphism (SNP) at codon 72 of TP53 gene (rs1042522), encoding a transversion of G to C (Arg to Pro), has been demonstrated to be associated with interindividual differences of TP53 expression to malignant tumors, including leukemia8. In addition, it has been reported that the Arg72 variant induces apoptosis markedly better compared to the Pro72 variant9. Thus, this genetic polymorphism holds promise as a potential biomarker for leukemia.
To date, numerous studies have investigated the relationship between the TP53 Arg72Pro polymorphism and predisposition to leukemia, but the impact of TP53 Arg72Pro polymorphism on leukemia was still conflicting due to inconsistent findings in individual studies. It was necessary to quantitatively summarize the evidence using the gradually accumulated data. Therefore, the current meta-analysis of the 14 most recent and relevant case-control studies involving 2,506 cases and 4,386 controls was performed to provide a more precise estimate of the associations.
Results
Characteristics of studies
A total of 14 case-control studies that examined the association of TP53 Arg72Pro polymorphism with leukemia were included in the meta-analysis10,11,12,13,14,15,16,17,18,19,20,21,22. As shown in the flowchart of the selection process of the included studies (Fig. 1), one article was excluded because it was conducted on overlapping populations with another eligible study23; one article was excluded because the gene distribution of control group deviated from the HWE24; and one article involved two independent case-control studies that were considered separately, resulting in two studies14. Of the 14 studies, 8 were conducted in Asian populations, 5 in Caucasian populations, and one in a mixed population. There were 7 AML studies and 5 ALL studies, the remaining studies were CML, CLL, ATL and mixed types. There were 12 population-based studies and 2 hospital-based studies. The main characteristics of the selected studies and the genotype distribution of the TP53 Arg72Pro polymorphism are summarized in Table 1.
Figure 1. Flow chart of publication selection procedure.
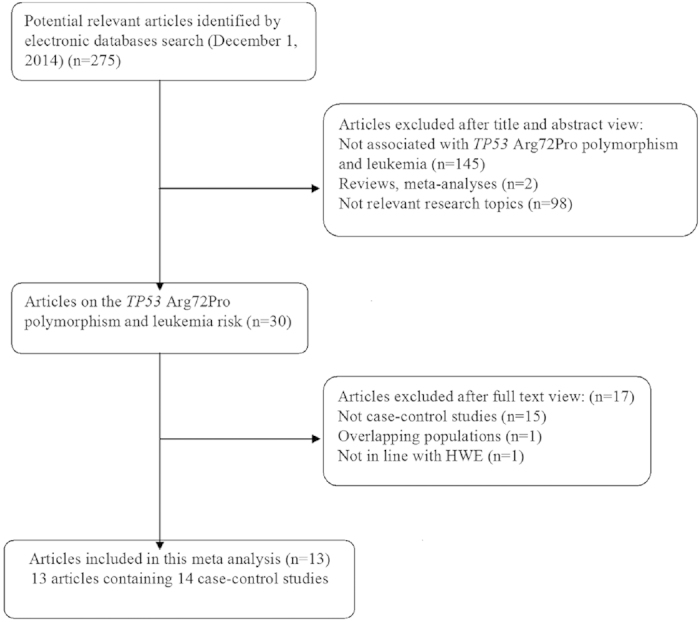
Table 1. The characteristics of studies included in the meta-analysis.
| First author | Year | Country | Ethnicity | Source of control | Clinical types | Sample size Case/Control | Cases | Controls | HWE | Methods of genotyping | Quality scores | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GC | CC | GG | GC | CC | ||||||||||
| Nakano | 2000 | Japan | Asian | Population based | AML | 200/188 | 82 | 93 | 25 | 59 | 95 | 34 | 0.69 | SSCP | 13 |
| Bergamaschi | 2004 | Italy | Caucasian | Population based | CML | 96/174 | 49 | 37 | 10 | 106 | 61 | 7 | 0.63 | PCR-RFLP | 11 |
| Takeuchi | 2005 | Japan | Asian | Hospital based | ATL | 87/89 | 33 | 38 | 16 | 32 | 37 | 20 | 0.15 | PCR-RFLP | 10 |
| Kochethu | 2006 | UK | Caucasian | Population based | CLL | 203/97 | 119 | 62 | 22 | 44 | 40 | 13 | 0.42 | PCR-RFLP | 11 |
| Ellis | 2008a | USA | Caucasian | Population based | AML | 80/2224 | 42 | 35 | 3 | 1255 | 838 | 131 | 0.57 | Taqman | 15 |
| Ellis | 2008b | UK | Caucasian | Population based | AML | 91/798 | 53 | 31 | 7 | 459 | 289 | 50 | 0.62 | PCR-RFLP | 14 |
| Phang | 2008 | Singapore | Asian | Population based | MIX | 44/160 | 13 | 25 | 6 | 56 | 72 | 32 | 0.32 | PCR-RFLP | 11 |
| Xiong | 2009 | China | Asian | Hospital based | AML | 231/128 | 52 | 127 | 52 | 39 | 64 | 25 | 0.89 | PCR-RFLP | 12 |
| Do | 2009 | US | Caucasian | Population based | ALL | 114/414 | 50 | 45 | 19 | 234 | 154 | 26 | 0.92 | Taqman | 13 |
| Shi | 2011 | China | Asian | Population based | AML | 180/555 | 55 | 84 | 41 | 171 | 289 | 95 | 0.15 | MALDI-TOF | 13 |
| Chauhan | 2012 | India | Asian | Population based | AML | 131/199 | 38 | 71 | 22 | 51 | 112 | 36 | 0.06 | PCR-RFLP | 12 |
| ALL | 99/199 | 28 | 43 | 28 | 51 | 112 | 36 | 0.06 | PCR-RFLP | 12 | |||||
| Dunna | 2012 | India | Asian | Population based | ALL | 147/245 | 59 | 67 | 21 | 79 | 123 | 43 | 0.68 | PCR-RFLP | 12 |
| AML | 141/245 | 64 | 44 | 33 | 79 | 123 | 43 | 0.68 | PCR-RFLP | 12 | |||||
| Chen | 2013 | China | Asian | Population based | ALL | 174/356 | 39 | 90 | 45 | 113 | 183 | 60 | 0.33 | PCR-RFLP | 13 |
| Perim | 2013 | Brazil | Mixed | Population based | ALL | 54/58 | 24 | 23 | 7 | 37 | 19 | 2 | 0.82 | PCR-RFLP | 10 |
HWE, Hardy-Weinberg equilibrium; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; SSCP, Single-Strand Conformation Polymorphism.
ALL, Acute lymphoblastic leukemia; AML, Acute myeloid leukemia; CLL, Chronic lymphocytic Leukemia; CML, Chronic myeloid leukemia; ATL, Adult T-cell leukemia.
Meta-analysis Results
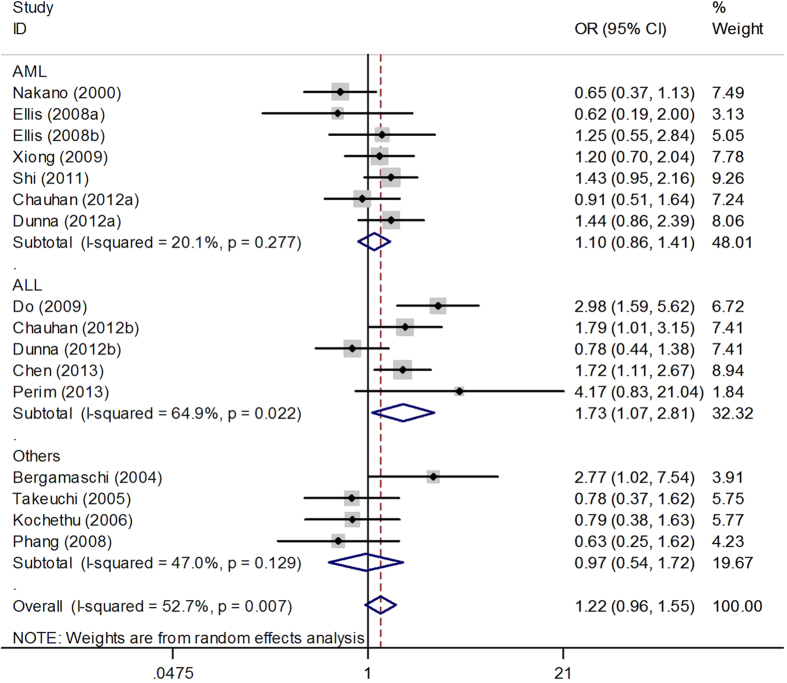
Table 2 lists the main results of this meta-analysis. The results of pooling all studies showed that there was no statistically significant association between TP53 Arg72Pro polymorphism and the risk of leukemia (C vs. G: OR = 1.09, 95% CI = 0.93–1.26; CC vs. GG: OR = 1.24, 95% CI = 0.91–1.69; CC + GC vs. GG: OR = 1.04, 95% CI = 0.85–1.28; CC vs. GC + GG: OR = 1.23, 95% CI = 0.96–1.57) (see Supplementary Fig. S1). Likewise, no significant association was found in the stratified analysis by ethnicity. In the subgroup analysis by type of leukemia, we found no association between TP53 Arg72Pro polymorphism and AML in all genetic models. However, we found a significant association between TP53 Arg72Pro polymorphism and ALL under the recessive model (OR = 1.73, 95% CI = 1.07–2.81, P = 0.025) (Fig. 2). When stratified restricting by control sources, negative results were observed in population-based controls and hospital-based controls.
Table 2. Meta-analysis of the association between TP53 Arg72Pro polymorphism and leukemia.
| Subgroup | NO. | C vs. G | CC vs. GG | CC + GC vs. GG | CC vs. GC + GG | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95% CI) | Ph | POR | OR (95% CI) | Ph | POR | OR(95% CI) | Ph | POR | OR(95% CI) | Ph | POR | ||
| Overall | 14 | 1.09(0.93–1.26) | 0.000 | 0.272 | 1.24(0.91–1.69) | 0.001 | 0.203 | 1.04(0.85–1.28) | 0.001 | 0.530 | 1.23(0.96–1.57) | 0.014 | 0.106 |
| Ethnicity | |||||||||||||
| Asian | 8 | 1.02(0.86–1.20) | 0.012 | 0.847 | 1.07(0.77–1.49) | 0.024 | 0.678 | 0.98(0.76–1.27) | 0.011 | 0.894 | 1.16(0.96–1.38) | 0.130* | 0.120* |
| Caucasian | 5 | 1.15(0.83–1.59) | 0.002 | 0.415 | 1.44(0.68–3.04) | 0.005 | 0.337 | 1.11(0.78–1.59) | 0.018 | 0.552 | 1.44(0.76–2.72) | 0.022 | 0.263 |
| Mixed | 1 | 2.11(1.15–3.86) | – | 0.016 | 5.40(1.03–28.19) | – | 0.046 | 2.20(1.03–4.70) | – | 0.041 | 4.17(0.83–21.04) | – | 0.084 |
| Source of control | |||||||||||||
| Population-based | 13 | 0.88(0.74–1.04) | 0.000 | 0.127 | 0.78(0.55–1.10) | 0.001 | 0.158 | 0.74(0.56–0.98) | 0.005 | 0.038 | 0.93(0.75–1.16) | 0.004 | 0.529 |
| Hospital-based | 2 | 0.90(0.71–1.16) | 0.197* | 0.423 | 0.83(0.51–1.37) | 0.185* | 0.469 | 0.97(0.63–1.49) | 0.353* | 0.885 | 0.81(0.55–1.18) | 0.214* | 0.267 |
| Clinical types | |||||||||||||
| AML | 7 | 0.98(0.88–1.10) | 0.179* | 0.730* | 1.00(0.79–1.27) | 0.208* | 0.986* | 0.91(0.72–1.16) | 0.048 | 0.448 | 1.11(0.89–1.37) | 0.277* | 0.351* |
| ALL | 5 | 1.33(0.97–1.82) | 0.002 | 0.078 | 1.83(0.95–3.50) | 0.003 | 0.069 | 1.26(0.84–1.90) | 0.008 | 0.266 | 1.73(1.07–2.81) | 0.022 | 0.025 |
| Others# | 4 | 0.97(0.68–1.39) | 0.035 | 0.882 | 0.95(0.61–1.48) | 0.087* | 0.827* | 0.97(0.73–1.28) | 0.056* | 0.823* | 0.93(0.62–1.40) | 0.129 | 0.722* |
OR odds ratio; 95% CI, 95% confidence interval; POR, pool P value; Ph, P value of heterogeneity test; NA, data not available.
*Estimates for fixed-effects model; otherwise, random-effects model was used.
#Others include chronic lymphocytic leukemia, chronic myeloid leukemia, adult T-cell leukemia, and mixed types.
Figure 2. Forest plot of OR with 95% CI of leukemia associated with the TP53 Arg72Pro polymorphism according to the clinical type group (recessive model).
Sensitivity analysis and Publication bias
A sensitivity analysis was performed by sequentially omitting individual studies of TP53 Arg72Pro polymorphism, and the corresponding pooled ORs were not materially changed (see Supplementary Fig. S2). Begg’s funnel plot and Egger’s test were performed to access the publication bias in this meta-analysis. Funnel plot shapes did not reveal any obvious evidence of asymmetry, and the P value of the Egger’s tests was greater than 0.05. The results suggested that publication bias was not evident in this meta-analysis (see Supplementary Fig. S3).
Discussion
The association between TP53 genetic mutation and the susceptibility to tumor risk has been confirmed in several functional studies with genetically modified mice, and the results have indicated that mice lacking the inactivating mutation in one TP53 allele developed fewer tumors than the mice harboring the mutation25. Moreover, the TP53 Arg72Pro polymorphism is the most studied SNP that may be associated with tumor risk. Therefore, it is biologically plausible that this polymorphism has significant association with leukemia progression.
However, previous a meta-analysis has reported that TP53 Arg72Pro polymorphism was not associated with leukemia risk, even in subgroup analyses by ethnicities and types of leukemia26. In the present meta-analysis, with 5 more studies and a larger number of subjects, we reached some meaningful conclusions. The overall summary results of our meta-analysis also suggested no statistically significant risk of TP53 Arg72Pro polymorphism to leukemia, and no significant association was found in a subgroup analysis by ethnicity. When stratified by types of leukemia, we observed a significant association between TP53 Arg72Pro polymorphism and ALL risk under the recessive model but not in the AML subjects. In the previous meta-analysis, which was limited by the number of studies, the authors only performed subgroup analyses on the association between TP53 Arg72Pro polymorphism, the risk of AML (n = 5), and the risk of other types of leukemia (n = 4), and the results were consistent with ours. Thus, our study confirms the association between TP53 Arg72Pro polymorphism and the risk of leukemia found in previous analysis. However, further studies with larger patient cohorts are needed to clarify this association.
ALL is more common in children, who comprise over 80% of all acute leukemia cases. Childhood ALL shows a consistent association with miscarriages26, and the TP53 Arg72Pro polymorphism has been implicated in implantation failure27 and in recurrent miscarriages28. However, the association is controversial. Fraga et al. reported no association was observed between TP53 Arg72Pro polymorphism and pregnancy loss risk, but the interaction of the TP53 Arg/Arg (rs1042522) and MDM2 TT (rs2279744) genotypes was shown to increase the risk to pregnancy loss29. This feature of childhood ALL makes it an interesting target for the study of TP53 pathway genes. TP53 Arg72Pro polymorphism may be a modifier of risk for childhood ALL susceptibility and might interact with MDM2. The Arg72 variant is more efficient than the Pro72 variant at inducing apoptosis. The greater apoptotic potential of the Arg72 protein is caused by greater interaction of this protein with MDM2, which facilitates the nuclear export, and probably also the subsequent mitochondrial localization31,32. Consistent with their biological relationship, our current analysis provides evidence that the TP53 Arg72Pro polymorphism is associated with an increased risk of developing ALL. To the best of our knowledge, this meta-analysis is the first to investigate the effect of TP53 Arg72Pro polymorphism on ALL. Furthermore, due to a limited number of available published studies, further studies with larger sample sizes are needed to reach a more convincing conclusion.
In interpreting our results of this meta-analysis, some limitations must be addressed. First, our meta-analysis was based on unadjusted estimates. However, a more precise analysis might be conducted if individual data were available, such an analysis would have allowed us to adjust for other covariates, such as age, family history and environmental factors. Second, the TP53 Arg72Pro polymorphism might influence susceptibility to leukemia with other factors, but we did not conduct relative research, such as the gene-gene and gene-environment interactions, because the data were insufficient. Third, we could not perform further subgroup stratification analysis by ages because of the limited number of published studies. Finally, publication bias is possible because no attempts were made to identify unpublished studies, although the results of our Begg’s funnel plot and Egger’s test revealed no evidence of publication bias in the allele model.
In conclusion, this meta-analysis did not find any evidence of an association between TP53 Arg72Pro polymorphism and the risk of leukemia in the overall studies, without the influence of ethnic diversity. However, a statistically significant association was found there was a stronger power trend towards a risk between the TP53 Arg72Pro polymorphism and ALL. Further studies with larger sample sizes and well-designed studies are required to investigate the associations between TP53 Arg72Pro polymorphism and leukemia.
Materials and Methods
Identification of Eligible Studies
A comprehensive literature search of studies published through December 2014 was performed using the PubMed, EMBASE, Web of Knowledge and the Chinese National Knowledge Infrastructure (CNKI) databases. Various combinations of the terms “TP53 or P53”, “polymorphism or variant”, and “leukemia or leucocythemia” were used to screen for potentially relevant studies. The search was conducted without language restrictions. Additional articles were located manually through the references of the related reports.
Inclusion and Exclusion Criteria
The following inclusion criteria were applied: a) studies evaluating the association between TP53 Arg72Pro polymorphism and the risk of leukemia, b) case-control or cohort studies, c) studies with a control genotype distribution in HWE, and d) studies with sufficient genotype data for calculating the odds ratio (OR) with 95% confidence intervals (95% CIs). If studies included overlapping subjects, only the study with a larger sample size was selected. Studies were excluded if one of the following existed: a) not a case-control study, b) no sufficient data were reported, and c) case reports or reviews.
Data Extraction
Two investigators (Dai and Sun) independently collected the data from each article using a standardized protocol. Disagreement was resolved by discussion with the other co-authors. The following information from the eligible studies was extracted: the first author’s surname, publication year, country of origin, ethnicity, source of control, sample sizes of cases and controls, number of genotypes, P-value for Hardy-Weinberg equilibrium (HWE), and genotyping methods. The study design was categorized as a hospital-based study (controls from the hospitalized patients) or a population-based study (controls from the healthy population).
Quality score assessment
The quality of the studies was independently assessed by the two reviewers (S.J., and J.S) according to the scale for quality assessment (see Supplementary Table S1). These scores were based on both traditional epidemiological considerations and cancer genetic issues. Any disagreement was resolved by discussion between the two reviewers. Total scores ranged from 0 (worst) to 15 (best).
Statistical analysis
All statistical analyses were performed using Stata software version 12.0 (Stata Corp., College Station, TX, USA), and all P values were two-sided. The Hardy-Weinberg equilibrium of controls for each study was tested using the χ2 test (significant at the 0.05 level). An OR with the corresponding 95% CI was used to assess the strength of the association between the TP53 Arg72Pro polymorphism and leukemia susceptibility according to allelic contrast (C vs. G), homozygote comparisons (CC vs. GG), dominant models (CC + GC vs. GG) and recessive models (CC vs. GC + GG). The significance of the summary OR was determined with a Z-test, and a p value of less than 0.05 was considered to be statistically significant. Between-study heterogeneity was assessed by calculating the Q-statistic and then quantified using the I2 value. When the effects were assumed to be heterogeneous (P < 0.05, I2 > 50%), the random-effects model was used; otherwise, the fixed-effects model was more appropriate33,34. Sensitivity analysis was used to test the stability of pooled studies through sequential omission of individual studies. Potential publication bias was estimated with a Begg’s funnel plot and corroborated with the Egger’s test (P < 0.05 was considered to indicate significant publication bias)35,36. Additionally, subgroup analyses of ethnicity, source of control and clinical types were performed.
Additional Information
How to cite this article: Tian, X. et al. Association between TP53 Arg72Pro polymorphism and leukemia risk: a meta-analysis of 14 case-control studies. Sci. Rep. 6, 24097; doi: 10.1038/srep24097 (2016).
Supplementary Material
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (No. 81573654).
Footnotes
The authors declare no competing financial interests.
Author Contributions Designed the study: J.Y. and T.X. Searched databases and collected full-text papers: D.S. and J.S. Extracted and analyzed the data: S.J. and J.S. Statistical analyses: J.Y. and D.S. Wrote the manuscript: T.X. All authors reviewed the manuscript.
References
- Estey E. H. Prognostic factors in acute myelogenous leukemia. Leukemia 15, 670–2 (2001). [DOI] [PubMed] [Google Scholar]
- Vardiman J. W. et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 114, 937–51 (2009). [DOI] [PubMed] [Google Scholar]
- Ma Z. et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet 28, 220–1 (2001). [DOI] [PubMed] [Google Scholar]
- Di Bernardo M. C. et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet 40, 1204–10 (2008). [DOI] [PubMed] [Google Scholar]
- Levine A. J. p53, the cellular gatekeeper for growth and division. Cell 88, 323–331 (1997). [DOI] [PubMed] [Google Scholar]
- Vousden K. H. & Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer 2, 594–604 (2002). [DOI] [PubMed] [Google Scholar]
- Nigro J. M. et al. Mutations in the p53 gene occur in diverse human tumour types. Nature 342, 705–8 (1989). [DOI] [PubMed] [Google Scholar]
- Pim D. & Banks L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer 108, 196–9 (2004). [DOI] [PubMed] [Google Scholar]
- Dumont P., Leu J. I., Della Pietra A. C. 3rd, George D. L. & Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33, 357–365 (2003). [DOI] [PubMed] [Google Scholar]
- Nakano Y. et al. Poor clinical significance of p53 gene polymorphism in acute myeloid leukemia. Leuk Res 24, 349–52 (2000). [DOI] [PubMed] [Google Scholar]
- Bergamaschi G. et al. TP53 codon 72 polymorphism in patients with chronic myeloid leukemia. Haematologica 89, 868–9 (2004). [PubMed] [Google Scholar]
- Takeuchi S. et al. P53 codon 72 polymorphism is associated with disease progression in adult T-cell leukaemia/lymphoma. Br J Haematol 131, 552–3 (2005). [DOI] [PubMed] [Google Scholar]
- Kochethu G. et al. Two germ line polymorphisms of the tumour suppressor gene p53 may influence the biology of chronic lymphocytic leukaemia. Leuk Res 30, 1113–8 (2006). [DOI] [PubMed] [Google Scholar]
- Ellis N. A. et al. MDM2 SNP309 and TP53 Arg72Pro interact to alter therapy-related acute myeloid leukemia susceptibility. Blood 112, 741–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang B. H., Linn Y. C., Li H. & Sabapathy K. MDM2 SNP309 G allele decreases risk but does not affect onset age or survival of Chinese leukaemia patients. Eur J Cancer 44, 760–6 (2008). [DOI] [PubMed] [Google Scholar]
- Xiong X. et al. Risk of MDM2 SNP309 alone or in combination with the p53 codon 72 polymorphism in acute myeloid leukemia. Leuk Res 33, 1454–8 (2009). [DOI] [PubMed] [Google Scholar]
- Do T. N., Ucisik-Akkaya E., Davis C. F., Morrison B. A. & Dorak M. T. TP53 R72P and MDM2 SNP309 polymorphisms in modification of childhood acute lymphoblastic leukemia susceptibility. Cancer Genet Cytogenet 195, 31–6 (2009). [DOI] [PubMed] [Google Scholar]
- Shi J. Y. et al. Genetic variations of DNA repair genes and their prognostic significance in patients with acute myeloid leukemia. Int J Cancer 128, 233–8 (2011). [DOI] [PubMed] [Google Scholar]
- Chauhan P. S. et al. High order interactions of xenobiotic metabolizing genes and P53 codon 72 polymorphisms in acute leukemia. Environ Mol Mutagen 53, 619–30 (2012). [DOI] [PubMed] [Google Scholar]
- Dunna N. R. et al. TP53 codon 72 polymorphism and risk of acute leukemia. Asian Pac J Cancer Prev 13, 347–50 (2012). [DOI] [PubMed] [Google Scholar]
- Chen J., Zhu B., Chen J. & Li Y. Genetic variations in MDM2 and P53 genes confer risk for adult acute lymphoblastic leukemia in a Chinese population. DNA Cell Biol 32, 414–9 (2013). [DOI] [PubMed] [Google Scholar]
- de Lourdes Perim A. et al. CXCL12 and TP53 genetic polymorphisms as markers of susceptibility in a Brazilian children population with acute lymphoblastic leukemia (ALL). Mol Biol Rep 40, 4591–6 (2013). [DOI] [PubMed] [Google Scholar]
- Chauhan P. S. et al. Association of glutathione S-transferase, EPHX, and p53 codon 72 gene polymorphisms with adult acute myeloid leukemia. DNA Cell Biol 30, 39–46 (2011). [DOI] [PubMed] [Google Scholar]
- El-Danasouri N. M., Ragab S. H., Rasheed M. A., Ali El Saadany Z. & Abd El-Fattah S. N. MDM2 SNP309 and p53 codon 72 genetic polymorphisms and risk of AML: an Egyptian study. Ann Clin Lab Sci 44, 449–54 (2014). [PubMed] [Google Scholar]
- Donehower L. A. et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–21 (1992). [DOI] [PubMed] [Google Scholar]
- Weng Y. et al. p53 codon 72 polymorphism and hematological cancer risk: an update meta-analysis. PLos One 7, e45820 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorak M. T., Pearce M. S., Hammal D. M., McNally R. J. & Parker L. Examination of gender effect in birth weight and miscarriage associations with childhood cancer (United Kingdom). Cancer Causes Control 18, 219–28 (2007). [DOI] [PubMed] [Google Scholar]
- Kay C., Jeyendran R. S. & Coulam C. B. p53 tumour suppressor gene polymorphism is associated with recurrent implantation failure. Reprod Biomed Online 13, 492–6 (2006). [DOI] [PubMed] [Google Scholar]
- Pietrowski D. et al. Recurrent pregnancy failure is associated with a polymorphism in the p53 tumour suppressor gene. Hum Reprod 20, 848–51 (2005). [DOI] [PubMed] [Google Scholar]
- Fraga L. R. et al. p53 signaling pathway polymorphisms associated to recurrent pregnancy loss. Mol Biol Rep 41, 1871–7 (2014). [DOI] [PubMed] [Google Scholar]
- Boyd S. D., Tsai K. Y. & Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol 2, 563–568 (2000). [DOI] [PubMed] [Google Scholar]
- Geyer R. K., Yu Z. K. & Maki C. G. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol 2, 569–573 (2000). [DOI] [PubMed] [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–48 (1959). [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–88 (1986). [DOI] [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–34 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuck A. E., Rubenstein L. Z. & Wieland D. Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. BMJ 316, 469–70 (1998). [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.