Abstract
Numerous methods have been developed for immunogold labeling of thick, cryo-preserved biological specimens. However, most of the methods are permutations of chemical fixation and sample sectioning, which select and isolate the immunolabeled region of interest. We describe a method for combining immunogold labeling with cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) of the surface proteins of intact mammalian cells or the surface glycoproteins of assembling and budding viruses in the context of virus-infected mammalian cells cultured on EM grids. In this method, the cells were maintained in culture media at physiologically relevant temperatures while sequentially incubated with the primary and secondary antibodies. Subsequently, the immunogold-labeled specimens were vitrified and observed under cryo-conditions in the transmission electron microscope. Cryo-EM and cryo-ET examination of the immunogold-labeled cells revealed the association of immunogold particles with the target antigens. Additionally, the cellular structure was unaltered by pre-immunolabeling chemical fixation and retained well-preserved plasma membranes, cytoskeletal elements, and macromolecular complexes. We think this technique will be of interest to cell biologists for cryo-EM and conventional studies of native cells and pathogen-infected cells.
Keywords: cryo-electron microscopy (cryo-EM), cryo-electron tomography (cryo-ET), immuno-electron microscopy (immuno-EM), transmission electron microscopy (TEM)
Introduction
Cryo-electron microscopy (cryo-EM) technologies and techniques were established in order to retain specimen hydration during direct-image and diffraction data acquisition with the transmission electron microscope (TEM) (Taylor and Glaeser 1974). Much of the early work in the late 1970s and early 1980s focused on improving cryo-fixation approaches (Dubochet et al. 1988; Lepault et al. 1983), cryo-holders for specimen introduction into the TEM (Heide and Grund 1974; Jeng and Chiu 1987; Taylor et al. 1984), and low-dose data acquisition schemes for viruses, macromolecules, and small cells (Adrian et al. 1984; Frank et al. 1986; McDowall et al. 1986; Milne and Subramaniam 2009). Later, as microscopes became more automated and intermediate voltage microscopes were more accessible, cryo-electron tomography (cryo-ET) emerged as the leading approach for determining the three-dimensional (3D) structures of pleomorphic objects, such as asymmetric viruses and bacteria (Dierksen et al. 1995; Guerrero-Ferreira and Wright 2013; Koning and Koster 2009; Koster et al. 1992; Subramaniam et al. 2007). Cryo-EM imaging, combined with single particle analysis and cryo-ET united with sub-volume averaging, is a major branch of the structural biology continuum, and over the past forty years have matured to the level where many biological structures are now determined at sub-nanometer to atomic-level resolutions (Briggs 2013; Campbell et al. 2015; Jiang et al. 2015; Schur et al. 2015).
Of great interest to those who employ cryo-EM methods to investigate cellular ultrastructure and the localization patterns of complexes within and surrounding cells, as is the development and application of target-specific labeling strategies (Lucic et al. 2013). Already, the feasibility of antibody labeling for purified macromolecules, viruses, and isolated organelles has been demonstrated (Beck et al. 2007; Roos et al. 1996). These studies have enabled investigators to determine the 3D structures of low molecular weight proteins by single particle cryo-EM (Wu et al. 2012), where proteins localize on specific regions of a virus (Roos et al. 1996), and the structural rearrangements that occur when a neutralizing antibody binds with the target antigen (Bartesaghi et al. 2013; Dutta et al. 2014; Harris et al. 2013; Lin et al. 2013; Tran et al. 2012). However, similar procedures have not been widely applied to studies of whole, intact mammalian cells because of the concerns associated with retaining cell viability during immunolabeling; the thicknesses of cells and the impact this has on cryo-preservation and cryo-EM imaging; and early reports of antibody-induced membrane protein ‘capping’ on live cells (Ash et al. 1977; Ferrante and Thong 1979). Therefore, techniques were developed that combined the benefits of initial cryo-preservation of cells by high-pressure freezing with chemical fixation through freeze substitution, embedding in hydrophilic resins, sectioning, and immunolabeling. Although these more traditional approaches have been used for countless immunolocalization studies for conventional electron microscopy (McDonald 1999; Sawaguchi et al. 2004), there are a number of intrinsic issues associated with methods that include chemical fixation (Schnell et al. 2012). Most notably, fixation is required to prevent specimen degradation during subsequent steps of the procedure. However, fixation, dehydration, and embedding not only alter the antigen conformation, and thus hinder antibody binding, but also severely obscure or even remove ultrastructural detail. In addition, residual aldehydes from the fixative can potentially cross-link immuno-reagents to the cell or tissue and therefore lead to potential negative background immunolabeling. These additive negative effects limit the ability to resolve localization patterns at the molecular level via both conventional immuno-EM and hybrid cryo-immuno EM approaches.
In this study, we describe the procedures used for the immunogold labeling of live mammalian cells cultured on carbon-coated gold EM-support grids. Here, we performed native-state immunogold labeling of a cell-derived protein and of a viral glycoprotein. The first target protein labeled was tetherin (BST-2), a cellular restriction factor that inhibits the release of enveloped viruses from the plasma membrane of infected cells (Neil et al. 2008; Van Damme et al. 2008). We demonstrated site-specific labeling of tetherin and tetherin microdomains during the restriction of HIV-1 virus and virus-like particle (VLP) release from the plasma membrane of HT1080 cells. The second protein labeled was the fusion (F) glycoprotein of human respiratory syncytial virus (hRSV) in the context of an active infection of HEp-2 and HeLa cells. In this case, the hRSV F glycoprotein was labeled with a monoclonal antibody, palivizumab (IMpact-RSV Study Group 1998; Johnson et al. 1997), at sites of virus assembly, on assembling and budding virus particles, and on released virus particles. We also describe some of the challenges associated with the immunolabeling method and provide detailed approaches for optimum labeling and specimen preservation for both cryo-EM and conventional EM applications.
Materials & Methods
Antibodies and Reagents
Rabbit anti-tetherin antisera (Hammonds et al. 2010) was provided by Dr. Paul W. Spearman (Department of Pediatrics, Emory University, Atlanta, GA). Dr. Philip J. Santangelo (Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA) supplied the palivizumab, a humanized monoclonal antibody that targets the antigenic site II of the hRSV fusion (F) glycoprotein (IMpact-RSV Study Group 1998; Johnson et al. 1997). MedImmune, LLC, commercially manufactures palivizumab under the brand name Synagis for the prevention of hRSV infections in high-risk infants. The secondary antibodies, which included the 6-nm gold-conjugated goat anti-human IgG, F(ab’)2 fragment of goat anti-rabbit IgG, and protein G, were purchased from Electron Microscopy Sciences (Hatfield, PA).
Cell culture, Transfection, and Immunolabeling for Tetherin
HT1080 cells, obtained from the American Type Culture Collection (Manassas, VA), were maintained in high-glucose pyruvate DMEM (GIBCO; Grand Island, NY) supplemented with 10% fetal bovine serum (FBS), 5 mM penicillin-streptomycin, and 5 mM l-Glutamate. Cells were grown in an incubator at 37°C with 5% CO2. HT1080 cells (20,000-30,000) for tetherin immunogold labeling were seeded on pre-sterilized, 0.2% collagen coated Aclar disks or gold R2/1 Quantifoil TEM grids (Quantifoil, Germany) 24 hr before transfection.
HT1080 cells were co-transfected with either mCherry Gag (1:3 ratio codon optimized Gag) or with an HIV-1 Vpu- and Env-deficient provirus cDNA (Khan et al. 2001) (kindly provided by Klaus Strebel, NIAID, Bethesda, MA) and eGFP-tetherin, using jetPRIME (Polyplus-Transfection Inc.; New York, NY), according to the manufacturer’s instructions.
Immunogold labeling of tetherin on HIV-1 transfected HT1080 cells was carried out 16 hr post-transfection. Transfected cells on Aclar disks or gold Quantifoil TEM grids were maintained in growth medium in a 37°C incubator before immunogold labeling. Rabbit anti-tetherin antiserum (Hammonds et al. 2010) was added directly into the medium at a 1:200 dilution of the original stock solution (endpoint titer of 1×108/ml). Primary antibody incubation proceeded for 1.5 hr at 37°C, during which time the dishes were gently shaken periodically to facilitate antibody binding. After primary antibody incubation, cells were gently washed with medium four times for 30 sec each. The secondary antibodies, either the 6-nm gold-conjugated F(ab’)2 fragment of goat anti-rabbit IgG or protein G, were then added to the medium directly at a 1:20 dilution of the original stock solutions (10–20 μg/ml) and allowed to incubate for 1.5 hr at 37°C. The cells were washed after secondary antibody incubation as described above. Cells cultured on the Aclar disks were washed twice with phosphate-buffered saline (PBS) and then fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for conventional electron microscopy examination. Cells grown on the gold Quantifoil TEM grids were plunge-frozen immediately after the final wash with culture medium.
hRSV Purification
HEp-2 cells cultured in T75 flasks at 60% to 80% confluence were inoculated with RSV at a multiplicity of infection (MOI) of 0.01 in 3 ml serum-free medium for 1 hr, after which 12 ml of complete growth medium was added. The infection continued for approximately four days (>80% cytopathic effect, CPE), with 5 ml of complete media added each day. The flasks were then frozen at -80°C before purification. After thawing in a 37°C water bath, the contents of each flask were clarified at 1,902 × g for 7 min at 4°C. The supernatant was centrifuged using a 20% sucrose cushion at 90,353 × g (SW41 Ti, Beckman) for 3 hr at 4°C. The virus-containing pellet was resuspended in 100 μl Hanks Balanced Salt Solution (HBSS) after a brief wash and stored at -80°C. The final titer of the virus was ~2–5×108 plaque forming units per ml (PFU/ml). The purified virus was then used to infect the HeLa or HEp-2 cells (Alonas et al. 2014).
Cell Culture, RSV Infection, and Immunolabeling for hRSV F Glycoprotein
HeLa and HEp-2 cells (ATCC CCL-2 and CCL-23, respectively) were maintained in DMEM growth medium with 4.5 g/L d-glucose (Lonza), 4 mM l-glutamine (BioWhittaker), 10% fetal bovine serum (Sigma-Aldrich), and 1% penicillin-streptomycin (Sigma-Aldrich). Cells were grown in an incubator at 37°C with 5% CO2. For hRSV F glycoprotein immunogold labeling, 100,000–150,000 HEp-2 cells or HeLa cells were seeded on pre-sterilized, pre-incubated (with complete medium) Aclar disks or gold R2/1 Quantifoil TEM grids in MatTek dishes (MatTek Corp., MA) 4–8 hr prior to infection.
Cells for immunogold labeling were inoculated with an MOI of 10 with hRSV in serum-free media. Complete growth media was added after 1 hr. Infected cells were incubated at 37°C for 24 hr. Palivizumab was then added directly into the medium at 5 µg/ml and the cells were allowed to incubate for 1.5 hr at 37°C, during which time the culture plates and MatTek dishes were shaken gently to facilitate antibody binding. After primary antibody incubation, the cells were gently washed with complete medium 4 times for 30 sec each time. The secondary antibodies, 6-nm gold-conjugated goat anti-human IgG or protein G, were then added to the medium directly at 1:20 dilution of the original stock solutions (10–20 μg/ml) and allowed to incubate for 1.5 hr. Washes were repeated as described above after secondary antibody incubation. Cells on Aclar disks were washed twice with PBS and then fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for conventional transmission electron microscopy examination. Cells on the gold Quantifoil TEM grids were plunge-frozen immediately after the final wash with culture medium.
Immunogold Labeling Controls
Primary Antibody Specificity Control
HT1080 cells transfected with only HIV-1 Gag were incubated with anti-tetherin antiserum and 6-nm gold particles conjugated with secondary antibodies, as described above. Following immunogold labeling, cells were fixed for conventional electron microscopy examination. The HT1080 cell line does not constitutively express tetherin (Neil et al. 2008); therefore, one would not expect to see immunogold labeling in the absence of exogenous tetherin.
Capping and Patching Control
To test whether capping and patching occurred during the labeling process, we performed immunogold labeling of tetherin on transfected, chemically fixed HT1080 cells. The labeling was done following standard pre-embedding immunogold protocols (Hammonds et al. 2012).
Conventional TEM Sample Preparation and Imaging
Fixed cells on Aclar disks were washed twice with 0.1 M phosphate buffer (pH 7.4) after overnight fixation with 2.5% glutaraldehyde in the same buffer. Cells were then post-fixed with 1% osmium tetroxide in 0.1 M phosphate buffer (pH 7.4) for 1 hr. Following graded ethanol dehydration, cells were infiltrated, embedded, and then polymerized in Eponate 12 resin (Ted Pella Inc., Redding, CA).
Ultrathin sections were cut using a Leica Ultracut S ultramicrotome at a thickness of 70 nm. Sections were then stained with 5% uranyl acetate and 2% lead citrate and imaged on a Hitachi H-7500 transmission electron microscope (TEM, Hitachi High-Technologies, Japan) equipped with a SIA L12C 16-megapixel CCD camera (SIA, Duluth, GA) or a JEOL JEM-1400 TEM (JEOL Ltd., Japan) equipped with a Gatan US1000 2k×2k CCD camera (Gatan, Pleasanton, CA).
Cryo-TEM Sample Preparation, Imaging, and Image Processing
Prior to sample vitrification, a 4-μl aliquot of 10- or 20-nm gold nanoparticles (Sigma-Aldrich) was applied to the surface of the TEM grid. These gold nanoparticles were used for image alignment in the 3D tomographic reconstruction process (Kremer et al. 1996; Mastronarde 1997). Cells cultured on the gold Quantifoil TEM grids were vitrified by rapid immersion in liquid ethane using a Gatan CryoPlunger3 (Cp3) apparatus. Cryo-grids were transferred to a Gatan 914 high-tilt holder maintained at -178°C. Cryo-specimens were imaged with JEOL JEM-2200FS 200-kV field emission gun transmission electron microscope equipped with an in-column Omega energy filter (slit width 20 eV) and a Gatan US4000 4k×4k CCD camera. Polygon montages and tilt series were acquired using SerialEM software (Mastronarde 2005). A single-axis tilt series was collected over an angular range of -62° to 62°, with a 2° tilt increment. The total electron dose applied to the specimens did not exceed 120–150 e-/Å2. Tilt series images were acquired at 30,000× nominal magnification (calibrated pixel size of 0.737 or 0.764 nm) with -4 to -8-μm defocus applied. Tomographic reconstructions were generated with IMOD using the r-weighted back-projection algorithm (Kremer et al. 1996; Mastronarde 1997).
Results and Discussion
The rationale for this study was to develop a strategy for the improved identification of the 3D localization patterns of membrane-bound proteins associated with enveloped virus-infected cells while retaining structural information at a macromolecular resolution (2–10 nm). First, we examined tetherin placement at sites of HIV-1 VLP and HIV-1 virus assembly and release using HT1080 cells. Second, we studied hRSV F glycoprotein location and order using hRSV-infected HEp-2 and HeLa cells. In order to achieve our aims, we used a native immunolabeling process followed by rapid sample vitrification methods or chemical fixation and conventional TEM specimen processing that allowed for the proteins, viral particles, and cells to remain whole and well preserved.
In the protocol, each protein-specific primary antibody was applied to the cultured cells and allowed to incubate for 1.5 hr while the cells were maintained at 37°C with 5% CO2. The specimens were gently washed and then incubated with a 6-nm gold-conjugated secondary antibody for 1.5 hr at 37°C with 5% CO2. As with the studies of vaccinia virus assembly by Roos et al. (1996), it was necessary to use gold-conjugated secondary antibodies as an electron-dense marker in order to resolve the locations of the primary antibodies and target proteins along the cell and viral surfaces.
Targeted Labeling of Cellular and Viral Proteins
We sought to determine whether a native-immunolabeling approach would be specific to the protein targets of interest. We therefore examined the method as applied to the immunolabeling of tetherin on HIV-1 and tetherin co-transfected HT1080 cells. It has been well established that tetherin, an interferon-induced protein, when expressed, functions to inhibit the release of HIV-1 and other enveloped viruses (Jouvenet et al. 2009; Le Tortorec and Neil 2009; Mansouri et al. 2009; Radoshitzky et al. 2010; Sakuma et al. 2009). In addition, multiple immunofluorescence microscopy studies as well as conventional and immuno-TEM experiments have shown that tetherin and budding HIV-1 VLPs and HIV-1 viruses localize in specific domains along the plasma membrane (Billcliff et al. 2013; Fitzpatrick et al. 2010; Hammonds et al. 2012; Hammonds et al. 2010; Lehmann et al. 2011). However, many of the previous imaging experiments incorporated chemical fixation, which subsequently may have altered or compromised the ultrastructure of the cells and viruses, as well as the structure of the labeled target protein complexes. Cryo-EM imaging of the unfixed tetherin immunolabeled HT1080 cells highlighted the presence of 6-nm gold clusters at HIV-1 VLP and HIV-1 virus assembly and budding sites along the cellular plasma membrane (Figs. 1, 2). A similar distribution profile of immunolabeled tetherin was present on HT1080 cells that were native-immunolabeled and subsequently chemically fixed and processed for conventional TEM (Fig. 3). The black arrowheads in Figs. 1–3 and Supplementary Fig. 1A and 1B indicate a subset of the 6-nm gold clusters in the image field that are associated with immunolabeled tetherin. All of the 6-nm gold clusters are linked with tetherin on the cell plasma membrane, between the cell plasma membrane and HIV-1 VLPs and virions, or between extended networks of HIV-1 VLPs and virions.
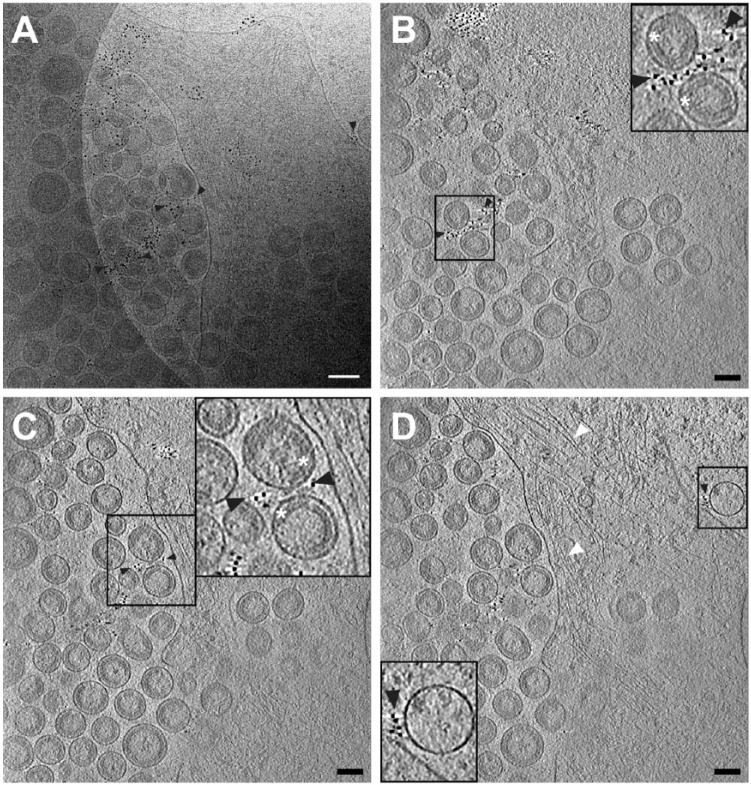
Figure 1.
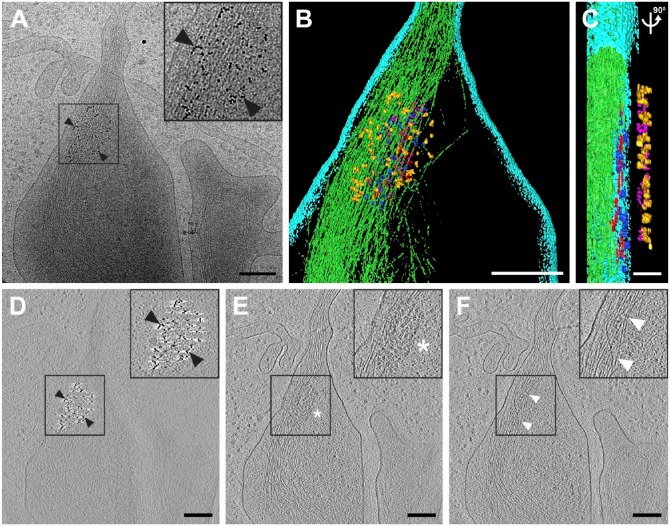
Cryo-electron tomography of immunolabeled-tetherin and HIV-1 virus-like particles (VLPs) attached to an HT1080 cell. (A) The central projection image from a tilt series of HIV-1 VLPs tethered to the edge of an HT1080 cell. Image was low-pass filtered using ImageJ (Gaussian blur, Schneider et al., 2012). (B–D) Slices (7.64 nm) near the bottom (B), through the middle (C), and at the top (D) of the 3D reconstruction, showing a visible, ordered Gag lattice in the HIV-1 VLPs and immunogold-labeled tetherin located on the plasma membrane (D) and on HIV-1 VLPs (B–D), as indicated by black arrowheads. Cytoskeletal elements are visible, as indicated by white arrowheads in a 3D tomographic slice (D). Insets in (B–D) (black boxes) are 2× magnification. White asterisks indicate the immature HIV-1 Gag lattice in insets for panels (B) and (C). Gold fiducial markers, 20 nm in diameter, were added to the sample and used as image alignment aids during the 3D tomographic reconstruction process. Scale, 50 nm.
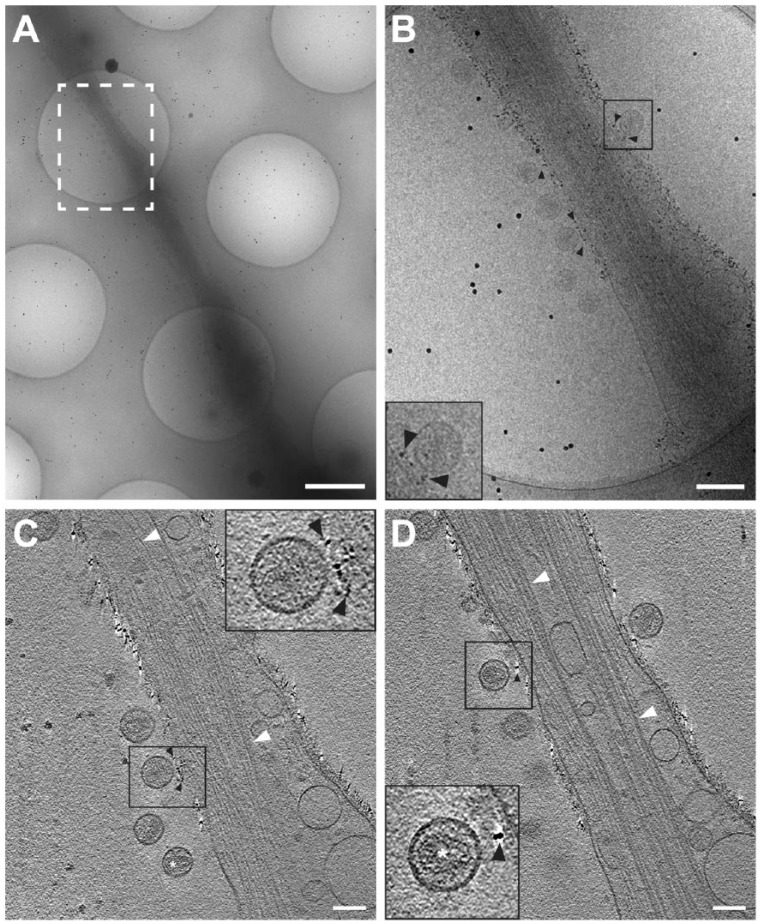
Figure 2.
Cryo-electron tomography (Cryo-ET) of immunolabeled tetherin and HIV-1 virions attached to an HT1080 cell microspike. (A) Cryo-TEM 2D image montage of HIV-1 tethered to a thin HT1080 cell extension. The dashed white box highlights the area of enlargement in (B). (C and D) Slices (8.84 nm) through the 3D reconstruction; black arrowheads indicate immunogold-labeled tetherin located between HIV-1 virions and the plasma membrane. Preserved cytoskeletal elements are visible in the 2D images and the 3D tomographic slices, as indicated by white arrowheads. White asterisk was placed over typical mature HIV-1 conical cores in panel (C) and the inset for panel (D). Insets in (B–D) (black boxes) are 2× magnification. Gold fiducial markers, 20-nm in diameter, were added to the sample and used as image alignment aids during the 3D tomographic reconstruction process. Scale (A) 500 nm; (A) and (B–D) 100 nm.
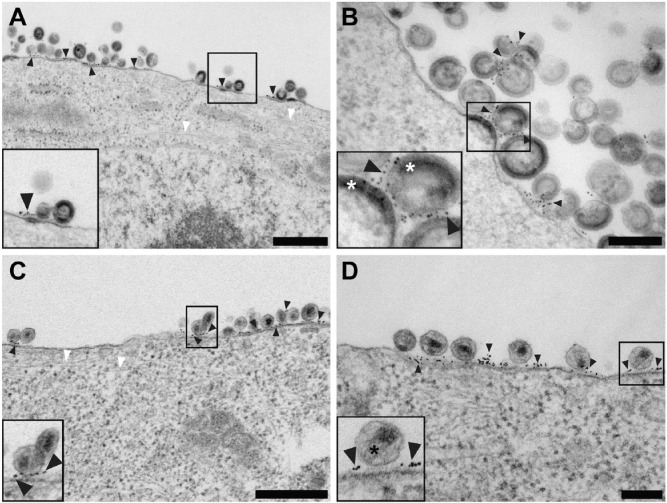
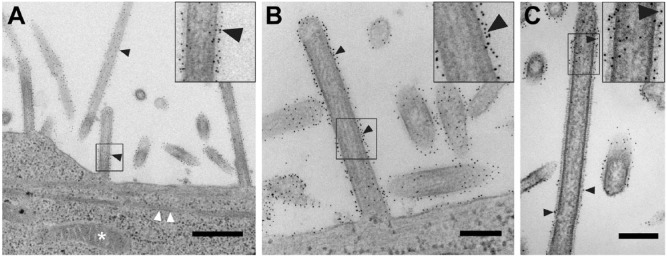
Figure 3.
Conventional transmission electron microscopy of native immunolabeled tetherin on HIV-1 VLPs and HIV-1 virions attached to HT1080 cells. (A and B) Low and high magnification images, respectively, of native immunolabeled tetherin on the plasma membrane of an HT1080 cell and HIV-1 VLPs. Black arrowheads point to immunogold labeling. (A and C) Actin filaments extend through the cell, as indicated by white arrowheads. (C and D) Low and intermediate magnification images, respectively, of native immunolabeled tetherin on the plasma membrane of an HT1080 cell and HIV-1 virions. Black arrowheads point to immunogold labeling. White asterisks are placed over the immature HIV-1 Gag lattice in the inset of panel (B). Black asterisks cover the mature HIV-1 conical core in the inset of panel (D). Insets in (A–D) (black boxes) are 2× magnification. Scale (A, C) 500 nm; (B, D) 200 nm.
Capping and patching was reported in late 1970s for some membrane proteins when bivalent binding molecules, such as immunoglobulin, were applied to the live cells (Ash et al. 1977; Ferrante and Thong 1979). It was speculated that the cross-linking effect of immunoglobulin caused the proteins to redistribute within the plasma membrane and form either multiple patches or a single cap. This rearrangement of the proteins could potentially inhibit the determination of native membrane protein distribution patterns. We therefore examined whether aggregates of antibodies noted by unusual gold particle localization patterns, i.e., patches or caps, were present on the cell surfaces of the native-immunolabeled specimens, as compared with conventionally processed samples. If this phenomenon were present, it would obscure the identification of tetherin-rich regions along the plasma membrane and at sites of virus assembly and budding. We determined that antibody-induced capping and patching did not occur, because, in images of both the native-immunolabeled and conventionally processed specimens, the distribution of antibody immunolabeling was equivalent (Supplementary Fig. 1A, 1B). We also assessed the level of immunolabeling specificity; here, we did not transfect the cells with tetherin but followed with the native-immunolabeling procedure and standard processing steps. The absence of tetherin in the cells resulted in the complete loss of tetherin labeling (Supplementary Fig. 1C).
We then examined another distinct biological system to validate the performance of the native-immunolabeling protocols. Here, we wished to resolve the presence and arrangement of the hRSV F glycoprotein on virus particle assembly and budding from HeLa and HEp-2 cells. HRSV presents three surface glycoproteins: the fusion (F) and the attachment (G) glycoproteins, and the small hydrophobic (SH) protein, which is significantly smaller in size (McLellan et al. 2013). The two major glycoproteins (F and G) work in concert to establish and maintain an active infection in host cells; explicitly, the epithelial cells of the lower respiratory tract. HRSV is a major viral pathogen that causes acute lower respiratory tract infections mainly in infants, young children, and immunosuppressed adults (Nair et al. 2010). To combat the virus, a series of neutralizing antibodies have been, and continue to be, developed against both glycoproteins. Currently, only one that targets the F glycoprotein is used in the prevention of hRSV infections in high-risk cases, palivizumab (IMpact-RSV Study Group 1998; Johnson et al. 1997); it is this antibody that we used for our analyses. Cryo-EM and cryo-ET imaging of hRSV particles revealed both unlabeled and immunolabeled glycoproteins along the entire length of viral filaments and at sites of virus assembly at the cell plasma membrane (Figs. 4, 5; Supplementary Fig. 2; Supplementary Movie 2). The immunolabeling of the hRSV F glycoprotein was consistent with that which has been previously reported. The hRSV F glycoprotein, when labeled for direct stochastic optical reconstruction microscopy (dSTORM) imaging, was present along the length of viral filaments (Alonas et al. 2014), which is consistent with our results. However, at EM-level resolution, where we are able to resolve the individual glycoproteins (Kiss et al. 2014; Liljeroos et al. 2013), we expected to find the distribution of the immunolabeled F glycoproteins to be impacted by: (1) the ratio and organization of the F and G glycoproteins on the virus, and (2) the steric hindrance/accessibility associated with the primary and 6-nm gold-conjugated secondary antibody with the F glycoprotein antigenic site II (McLellan et al. 2011). The native immunolabeling of the hRSV F glycoprotein underscores this level of glycoprotein organization (Figs. 4, 5; Supplementary Fig. 2). We examined whether employing different secondary antibodies would alter immunolabeling efficiency and the distribution of the gold particles. No variability in label distribution was noted when either the 6-nm gold-conjugated goat anti-human IgG or protein G was applied (Supplementary Fig. 2). Conventional TEM processing and imaging of samples post-immunolabeling disclosed comparable labeling efficiencies and arrangements of the 6-nm gold clusters along hRSV viral filaments and at the hRSV assembly sites along the plasma membrane (Fig. 6).
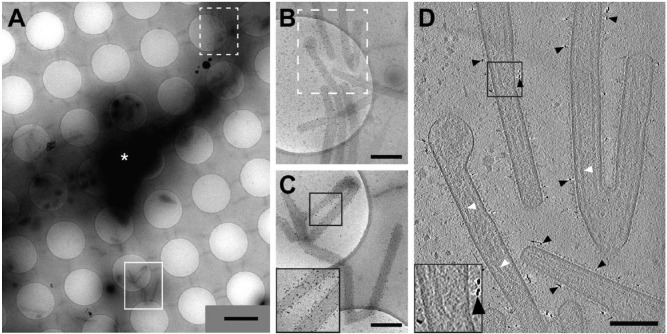
Figure 4.
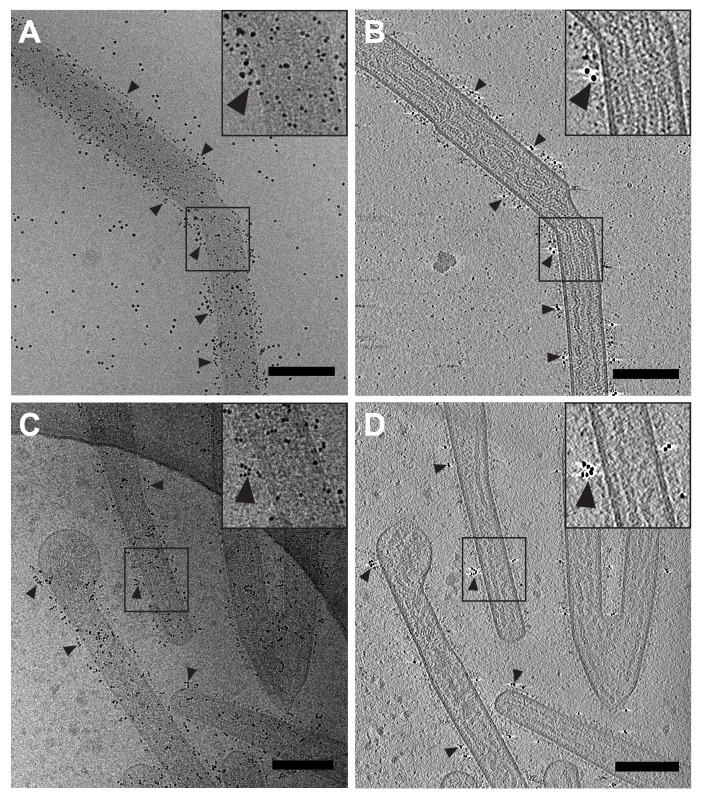
Cryo-transmission electron microscopy and cryo-electron tomography of hRSV-infected HeLa cells with the hRSV F glycoprotein immunolabeled. (A) Low magnification montage of released, filamentous hRSV particles and a HeLa cell extension (white asterisk). (B and C) Higher magnification views of the montage in (A) highlighting immunolabeling of hRSV F along the viral filaments. (B–C) Dashed box (B) and solid box (C) are indicated in (A). (D) Slice (7.64 nm) from the 3D reconstruction of the area in (B) illustrating hRSV filaments with surface glycoproteins and immunogold (black arrowheads). Note the presence of ribonucleoprotein (RNP) in some of the viral filaments (white arrowheads). Insets in (C) and (D) (black boxes) are 2× magnification. Gold fiducial markers, 20 nm in diameter, were added to the sample and used as image alignment aids during the 3D tomographic reconstruction process. Scale (A) 2 μm; (B, C) 500 nm; (D) 200 nm.
Figure 5.
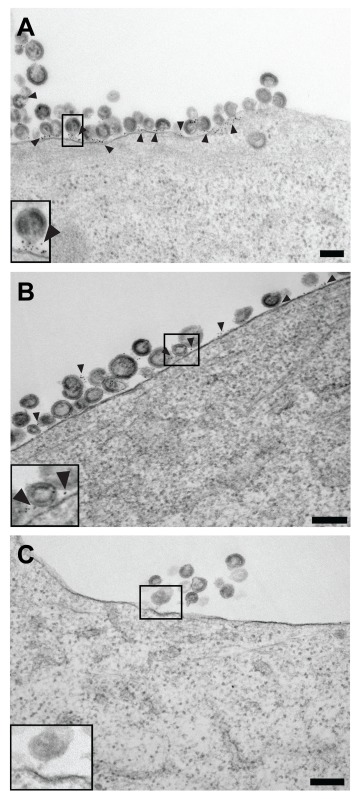
Early hRSV assembly site detected by native immunolabeling. (A) 2D cryo-electron microscopy image of an hRSV assembly site; the 6-nm gold particles indicate the location of the hRSV F glycoprotein (black arrowheads) at the upper surface of the plasma membrane. (B and C) Segmented 3D volume of the hRSV and cellular macromolecules. (B) Top view. (C) Cut-away and side view of (B) with 90° rotation applied. The cell membrane is presented in cyan. The filamentous actin network is noted in green. The matrix protein is depicted in blue. The glycoprotein densities are highlighted in magenta. Red tubular densities correspond to ribonucleoprotein (RNP). Gold densities are the 6-nm gold particles conjugated to the secondary antibody. (D–F) Slices (7.64 nm) through the reconstructed 3D volume showing the gold on the top (D, black arrowheads), a quarter-plane slice noting the hRSV viral proteins (E, white asterisk), and a bottom slice highlighting the actin filaments (F, white arrowheads). Inset in A is 2× magnification, insets in D-F are 1.5× magnification. Gold fiducial markers, 20 nm in diameter, were added to the sample and used as image alignment aids during the 3D tomographic reconstruction process. Scale (A, B, D–F) 200 nm; (C) 50 nm.
Figure 6.
Conventional transmission electron microscopy (TEM) of hRSV with the native F glycoprotein immunolabeled. (A–C) Black arrowheads point to immunogold labeling along viral filaments both at assembly sites and of released hRSV filaments. (A) White arrowheads note actin filaments and the white asterisk points to a mitochondrion. (B) Assembling viral filament extended from the plasma membrane. (C) Intermediate magnification image of filamentous hRSV virion with immunogold labeling of the F glycoproteins. Inset in (A) is 3× magnification, insets in (B) and (C) are 2× magnification. Scale (A) 500 nm and (B and C) 200 nm.
Preservation of Cellular Ultrastructure
One significant compromise researchers make when immunolabeling a specimen for EM is with respect to ultrastructure preservation. Most protocols require chemical fixation using either paraformaldehyde or a combination of paraformaldehyde and glutaraldehyde and, in some cases, post-fixation with osmium tetroxide, in order to stop biological activity and stabilize cell architecture. However, chemical fixation might disrupt antigenicity of the target macromolecules and may alter the cellular ultrastructure. Therefore, we examined the 2D projection images and 3D reconstructions of the cells and viruses to address whether the native immunolabeling technique would inadvertently deteriorate the stability of known cellular and viral structures. In all cryo-preserved samples of the HIV-1 cDNA and tetherin co-transfected HT1080 cells, cellular structures, such as membranes, the cytoskeleton, intracellular vesicles, and macromolecular complexes were intact and well preserved (Figs. 1, 2; Supplementary Movie 1). The morphology and architecture of the native-immunolabeled cryo-preserved cells was comparable to unlabeled cells also preserved by vitrification [data not shown and Strauss et al. (2014)]. In particular, long tracts and extended networks of actin filaments were well resolved in the areas associated with HIV-1 virus assembly, budding, and restriction (Fig. 2; Supplementary Movie 1). We also studied the images and 3D volumes of the cryo-preserved hRSV-infected HeLa cells and determined that cellular structures were intact and consistent with those of unlabeled cells (Fig. 5; Supplementary Movie 2). As with the HT1080 cells, HeLa cells also contained extensive actin networks and distinct cell membranes, a hallmark of well-preserved cellular structure.
Three-Dimensional Structure and Localization Information
The main rationales behind the development and application of this native-immunolabeling approach for cells and virus-infected cells were to: 1) rapidly identify regions of interest on a cell, 2) determine the 3D spatial relationships between the immunolabeled entity and other macromolecular complexes, and 3) retain cellular and viral ultrastructural integrity. We used 2D cryo-EM imaging to quickly pinpoint regions of importance and cryo-ET to generate 3D volumes of released viruses and the virus-transfected or virus-infected cells. The gold labels were critical for locating sections of the grids with labeled virus, in the case of hRSV (Fig. 4), or regions of the cells where viral particles were assembling (Figs. 4, 5), or for HIV-1 association with tetherin (Figs. 1, 2). The 6-nm gold particle conjugates used for immunolabeling the target proteins could be clearly differentiated from the 10-nm or 20-nm gold particles used for cryo-ET image alignment (Figs. 2, 5; Supplementary Fig. 2; Supplementary Movies 1, 2).
In our analysis of the 3D volumes of the tetherin-labeled HIV-1 cDNA and tetherin co-transfected HT1080 cells, we were able to identify and assess the nature of tetherin along the plasma membrane. As has been reported previously, we show that tetherin-enriched regions resided as discrete areas, or microdomains, on the plasma membrane (Billcliff et al. 2013; Hammonds et al. 2012). The tetherin-containing regions were also closely associated with regions of HIV-1 VLP and virus assembly and budding. Immunolabeled tetherin macromolecules were resolved as small fibrous extensions on the cell membrane, between the cell membrane and viral particles, and between viral particles (Fig. 2; Supplementary Movie 1). Characteristic HIV-1 VLP and virus particle structures were preserved in all samples examined (Benjamin et al. 2005; Briggs et al. 2006; Briggs et al. 2009; Carlson et al. 2008; Wright et al. 2007). HIV-1 VLPs ranged from 78 nm to 182 nm in diameter (mean ± SD, 115 nm ± 20; n=178) and contained highly ordered regions of the Gag lattice (Fig. 1). Both immature Gag-containing and mature, cored HIV-1 virus particles were observed bound to the plasma membrane of the HT1080 cells by filamentous extensions (Fig. 2; Supplementary Movie 1).
We examined the 3D volumes of the hRSV viral filaments and hRSV-infected cells immunolabeled for the F glycoprotein to determine: 1) the location and organization of the F glycoprotein on the viral filaments, 2) the relationship between the F glycoprotein and other hRSV structural proteins located in the interior of the viral filaments, and 3) discrete sites of virus assembly on the cell surface. The placement of the immunolabeled hRSV F glycoproteins was easily determined both along extending viral filaments (Fig. 4) and at sites of assembly (Fig. 5; Supplementary Movie 2). The arrangement of the hRSV F glycoprotein along viral filaments was consistent with that which has been reported previously (Kiss et al. 2014; Liljeroos et al. 2013). The matrix protein and ribonucleoprotein (RNP) complex were also resolved along the viral membrane in close proximity to the glycoproteins (Fig. 4; Supplementary Fig. 2) (Kiss et al. 2014; Liljeroos et al. 2013). Native immunolabeling of the hRSV F glycoprotein also proved useful for identifying the location of an early stage of virus assembly (Fig. 5; Supplementary Movie 2). Studies of this early event will play a significant role in our understanding of the coordination of hRSV viral proteins during assembly and subsequent budding at the plasma membrane. 2D cryo-EM imaging allowed us to locate a small patch of 6-nm gold particles on the plasma membrane of an hRSV-infected HeLa cell (Fig. 5A). The 3D tomographic reconstruction revealed the hRSV glycoproteins on the membrane surface and the matrix protein and the RNP complex coalescing under the membrane. Actin filaments were also visualized in the cell cytoplasm beneath areas containing the viral proteins. Results from the hRSV experiments highlight that native immunolabeling of surface proteins provides guidance for cryo-ET data collection; in this case, assistance for locating the early stages of hRSV assembly.
In summary, we have demonstrated the practicality of a native-immunolabeling approach for cells and virus-infected or virus-transfected cells that can be used for both cryo-EM and conventional EM strategies. This methodology provides a rapid means for immunolabeling proteins and protein complexes present on the membranes of cells and viruses. The ultrastructure of the immunolabeled cells was retained to high fidelity, contrary to conventional immunolabeling strategies. When the cryo-preserved labeled specimens were imaged by cryo-EM, the regions of interest were readily identified and the 3D spatial relationships between specific complexes were determined at macromolecular resolution. Immunolabeled specimens that were processed for conventional TEM also retained plasma membrane integrity, cytoplasm and cytoskeleton organization, and virus architecture.
In this study, we used ‘indirect’ immunolabeling, whereby a gold particle-conjugated secondary antibody was applied. To further expand the utility of native immunolabeling for correlative imaging from light microscopy to electron microscopy, fluorescently labeled secondary antibodies conjugated with gold particles could be employed. This strategy would facilitate the light microscopic identification of specific cells grown on a substrate prior to sample preservation for cryo-TEM or scanning electron microscopy (SEM) imaging. In addition, future investigations to improve the method will include employing gold-conjugated primary antibodies to ‘directly’ immunolabel the target and the use of smaller gold particles to minimize size-associated steric hindrances.
The focus of this study was to investigate the localization patterns of proteins and complexes present on the membrane surfaces of cells and virus-infected or virus-transfected cells. Further improvements and advancements are required for the targeted native-state, electron-dense labeling of macromolecules within the interior of cells and viruses. Current conventional, pre-embedding immunolabeling methods of whole cells generally necessitate the permeabilization of the plasma membrane for the introduction of antibodies and marker-conjugated antibodies into the cell cytoplasm (Hayat 1995). However, the process of permeabilizing the plasma membrane may alter membrane integrity or cell ultrastructure, neither of which is ideal for cryo-EM structural studies. Consequently, several approaches are under development and incorporate the use of ferritin (Wang et al. 2011), quantum dots (Gold et al. 2014), or metallothionein (Diestra et al. 2009; Fernandez de Castro et al. 2014; Mercogliano and DeRosier 2007) fusion constructs for the targeted labeling of macromolecules. However, even though each technique has shown promise in specific biological instances, none have been widely used in cryo-EM analyses due to incompatibilities with cell types, inability of the target protein to assemble properly, or cytotoxicity associated with the reagents. Concurrent with the exploration of electron-dense probes for cryo-EM, high-precision correlative light and electron microscopy (CLEM) approaches have been realized (Briegel et al. 2010; Jun et al. 2011; Schorb and Briggs 2014). These advances may have eliminated some of the need for using an electron-dense marker to determine the placement of complexes within a cell. However, immunolabeling with electron-dense markers will remain an essential tool for all areas of electron microscopy because it permits the assessment of interactions between complexes at resolutions not readily achievable by fluorescence microscopy.
Finally, we did not computationally ‘remove’ gold particles from the 2D images or 3D reconstructions (Kremer et al. 1996). This approach, combined with the use of smaller gold particles may be advantageous for improved 3D structure interpretation. Due to the utility and easy implementation of the method we have described for labeling thicker biological specimens, such as whole mammalian cells, for cryo-EM investigations, we believe it will be an advantageous technique for ultrastructural analyses of many cellular systems.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgments
We thank Ms. Jeannette Taylor of the Emory University Robert P. Apkarian Integrated Electron Microscopy Core for assistance.
Footnotes
Competing Interests: The authors declared no potential competing interests with respect to the research, authorship, and/or publication of this article.
Author Contributions: HY and ERW conceived and designed the experiments; HY, JDS, ZK, EA, RSD, CMH, KML, and JEH performed the experiments; PJS and PWS provided reagents and materials; HY, JDS, ZK, EA, CMH, and ERW wrote the paper.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by Emory University, Children’s Healthcare of Atlanta, the Center for AIDS Research at Emory University (P30 AI050409), and the Georgia Research Alliance to E.R.W.; NSF grant 0923395 to E.R.W; the James B. Pendleton Charitable Trust to P.W.S. and E.R.W.; and public health service grants F32GM112517 to J.D.S, AI101775 to E.R.W., and AI058828 and S10 RR025679 to P.W.S.
References
- Adrian M, Dubochet J, Lepault J, Mcdowall AW. (1984). Cryo-electron microscopy of viruses. Nature 308: 32-6. [DOI] [PubMed] [Google Scholar]
- Alonas E, Lifland AW, Gudheti M, Vanover D, Jung J, Zurla C, Kirschman J, Fiore VF, Douglas A, Barker TH, Yi H, Wright ER, Crowe JE, Jr., Santangelo PJ. (2014). Combining single RNA sensitive probes with subdiffraction-limited and live-cell imaging enables the characterization of virus dynamics in cells. ACS Nano 8: 302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash JF, Louvard D, Singer SJ. (1977). Antibody-induced linkages of plasma membrane proteins to intracellular actomyosin-containing filaments in cultured fibroblasts. Proc Natl Acad Sci U S A 74: 5584-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. (2013). Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol 20: 1352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Lucic V, Forster F, Baumeister W, Medalia O. (2007). Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature 449: 611-5. [DOI] [PubMed] [Google Scholar]
- Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. (2005). Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J Mol Bio 346: 577-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billcliff PG, Rollason R, Prior I, Owen DM, Gaus K, Banting G. (2013). CD317/tetherin is an organiser of membrane microdomains. J Cell Sci 126: 1553-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Chen S, Koster AJ, Plitzko JM, Schwartz CL, Jensen GJ. (2010). Correlated light and electron cryo-microscopy. Methods Enzymol 481: 317-41. [DOI] [PubMed] [Google Scholar]
- Briggs JA. (2013). Structural biology in situ–the potential of subtomogram averaging. Curr Opin Struct Biol 23: 261-7. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Grunewald K, Glass B, Forster F, Krausslich HG, Fuller SD. (2006). The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14: 15-20. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. (2009). Structure and assembly of immature HIV. Proc Natl Acad Sci U S A 106: 11090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MG, Veesler D, Cheng A, Potter CS, Carragher B. (2015). 2.8 A resolution reconstruction of the 20 S proteasome using cryo-electron microscopy. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA, Briggs JA, Glass B, Riches JD, Simon MN, Johnson MC, Muller B, Grunewald K, Krausslich HG. (2008). Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe 4: 592-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierksen K, Typke D, Hegerl R, Walz J, Sackmann E, Baumeister W. (1995). Three-dimensional structure of lipid vesicles embedded in vitreous ice and investigated by automated electron tomography. Biophys J 68: 1416-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diestra E, Fontana J, Guichard P, Marco S, Risco C. (2009). Visualization of proteins in intact cells with a clonable tag for electron microscopy. J Struct Biol 165: 157-68. [DOI] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, Mcdowall AW, Schultz P. (1988). Cryo-electron microscopy of vitrified specimens. Q Rev Biophys 21: 129-228. [DOI] [PubMed] [Google Scholar]
- Dutta M, Liu J, Roux KH, Taylor KA. (2014). Visualization of retroviral envelope spikes in complex with the V3 loop antibody 447-52D on intact viruses by cryo-electron tomography. J Virol 88: 12265-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez De Castro I, Sanz-Sanchez L, Risco C. (2014). Metallothioneins for correlative light and electron microscopy. Methods Cell Biol 124: 55-70. [DOI] [PubMed] [Google Scholar]
- Ferrante A, Thong YH. (1979). Antibody induced capping and endocytosis of surface antigens in Naegleria fowleri. Int J Parasitol 9: 599-601. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick K, Skasko M, Deerinck TJ, Crum J, Ellisman MH, Guatelli J. (2010). Direct restriction of virus release and incorporation of the interferon-induced protein BST-2 into HIV-1 particles. PLoS Pathog 6: e1000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Wagenknecht T, Verschoor A. (1986). A new method for three-dimensional reconstruction of single macromolecules using low-dose electron micrographs. Ann N Y Acad Sci 483: 77-87. [DOI] [PubMed] [Google Scholar]
- Gold VA, Ieva R, Walter A, Pfanner N, Van Der Laan M, Kuhlbrandt W. (2014). Visualizing active membrane protein complexes by electron cryotomography. Nat Commun 5: 4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Ferreira RC, Wright ER. (2013). Cryo-electron tomography of bacterial viruses. Virology 435: 179-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammonds J, Ding L, Chu H, Geller K, Robbins A, Wang JJ, Yi H, Spearman P. (2012). The tetherin/BST-2 coiled-coil ectodomain mediates plasma membrane microdomain localization and restriction of particle release. J Virol 86: 2259-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammonds J, Wang JJ, Yi H, Spearman P. (2010). Immunoelectron microscopic evidence for Tetherin/BST2 as the physical bridge between HIV-1 virions and the plasma membrane. PLoS Pathog 6: e1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AK, Bartesaghi A, Milne JL, Subramaniam S. (2013). HIV-1 envelope glycoprotein trimers display open quaternary conformation when bound to the gp41 membrane-proximal external-region-directed broadly neutralizing antibody Z13e1. J Virol 87: 7191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayat MA. (1995). Immunogold-Silver Staining – Principles, Methods, and Applications, New York, NY, CRC Press, Inc. [Google Scholar]
- Heide HG, Grund S. (1974). [Deep-freeze link for transport of water-containing biological objects to the electron microscope]. J Ultrastruct Res 48: 259-68. [DOI] [PubMed] [Google Scholar]
- Jeng TW, Chiu W. (1987). High resolution cryo system designed for JEM 100CX electron microscope. Ultramicroscopy 23: 61-6. [DOI] [PubMed] [Google Scholar]
- Jiang J, Pentelute BL, Collier RJ, Zhou ZH. (2015). Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature doi: 10.1038/nature14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S, Oliver C, Prince GA, Hemming VG, Pfarr DS, Wang SC, Dormitzer M, O’grady J, Koenig S, Tamura JK, Woods R, Bansal G, Couchenour D, Tsao E, Hall WC, Young JF. (1997). Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J Infect Dis 176: 1215-24. [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, Mcnatt M, Hatziioannou T, Bieniasz PD. (2009). Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol 83: 1837-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Ke D, Debiec K, Zhao G, Meng X, Ambrose Z, Gibson GA, Watkins SC, Zhang P. (2011). Direct visualization of HIV-1 with correlative live-cell microscopy and cryo-electron tomography. Structure 19: 1573-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Aberham C, Kao S, Akari H, Gorelick R, Bour S, Strebel K. (2001). Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J Virol 75: 7252-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss G, Holl JM, Williams GM, Alonas E, Vanover D, Lifland AW, Gudheti M, Guerrero-Ferreira RC, Nair V, Yi H, Graham BS, Santangelo PJ, Wright ER. (2014). Structural Analysis of Respiratory Syncytial Virus Reveals the Position of M2-1 Between the Matrix Protein and the Ribonucleoprotein Complex. J Virol 88: 7602-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning RI, Koster AJ. (2009). Cryo-electron tomography in biology and medicine. Ann Anat 191: 427-45. [DOI] [PubMed] [Google Scholar]
- Koster AJ, Chen H, Sedat JW, Agard DA. (1992). Automated microscopy for electron tomography. Ultramicroscopy 46: 207-27. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, Mcintosh JR. (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71-6. [DOI] [PubMed] [Google Scholar]
- Le Tortorec A, Neil SJ. (2009). Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol 83: 11966-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Rocha S, Mangeat B, Blanchet F, Uji IH, Hofkens J, Piguet V. (2011). Quantitative multicolor super-resolution microscopy reveals tetherin HIV-1 interaction. PLoS Pathog 7: e1002456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepault J, Booy FP, Dubochet J. (1983). Electron microscopy of frozen biological suspensions. J Microsc 129: 89-102. [DOI] [PubMed] [Google Scholar]
- Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. (2013). Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci U S A 110: 11133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Cheng N, Hogle JM, Steven AC, Belnap DM. (2013). Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy. J Immunol 191: 884-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Rigort A, Baumeister W. (2013). Cryo-electron tomography: the challenge of doing structural biology in situ. J Cell Biol 202: 407-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, Moses AV, Fruh K. (2009). Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi’s sarcoma-associated herpesvirus. J Virol 83: 9672-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol 120: 343-52. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152: 36-51. [DOI] [PubMed] [Google Scholar]
- Mcdonald K. (1999). High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol Biol 117: 77-97. [DOI] [PubMed] [Google Scholar]
- Mcdowall AW, Smith JM, Dubochet J. (1986). Cryo-electron microscopy of vitrified chromosomes in situ. EMBO J 5: 1395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclellan JS, Ray WC, Peeples ME. (2013). Structure and function of respiratory syncytial virus surface glycoproteins. Curr Top Microbiol Immunol 372: 83-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclellan JS, Yang Y, Graham BS, Kwong PD. (2011). Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol 85: 7788-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercogliano CP, Derosier DJ. (2007). Concatenated metallothionein as a clonable gold label for electron microscopy. J Struct Biol 160: 70-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JL, Subramaniam S. (2009). Cryo-electron tomography of bacteria: progress, challenges and future prospects. Nat Rev Microbiol 7: 666-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O’brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. (2010). Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375: 1545-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451: 425-30. [DOI] [PubMed] [Google Scholar]
- Radoshitzky SR, Dong L, Chi X, Clester JC, Retterer C, Spurgers K, Kuhn JH, Sandwick S, Ruthel G, Kota K, Boltz D, Warren T, Kranzusch PJ, Whelan SP, Bavari S. (2010). Infectious Lassa virus, but not filoviruses, is restricted by BST-2/tetherin. J Virol 84: 10569-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos N, Cyrklaff M, Cudmore S, Blasco R, Krijnse-Locker J, Griffiths G. (1996). A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J 15: 2343-55. [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Noda T, Urata S, Kawaoka Y, Yasuda J. (2009). Inhibition of Lassa and Marburg virus production by tetherin. J Virol 83: 2382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaguchi A, Mcdonald KL, Forte JG. (2004). High-pressure freezing of isolated gastric glands provides new insight into the fine structure and subcellular localization of H+/K+-ATPase in gastric parietal cells. J Histochem Cytochem 52: 77-86. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell U, Dijk F, Sjollema KA, Giepmans BN. (2012). Immunolabeling artifacts and the need for live-cell imaging. Nat Methods 9: 152-8. [DOI] [PubMed] [Google Scholar]
- Schorb M, Briggs JA. (2014). Correlated cryo-fluorescence and cryo-electron microscopy with high spatial precision and improved sensitivity. Ultramicroscopy 143: 24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schur FK, Hagen WJ, Rumlova M, Ruml T, Muller B, Krausslich HG, Briggs JA. (2015). Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature 517: 505-8. [DOI] [PubMed] [Google Scholar]
- Strauss JD, Hammonds JE, Spearman PW, Wright ER. (2014). Structural Characterization of Tethered HIV-1 VLPs by Light Microscopy and Cryo-Electron Tomography. Microscopy and Microanalysis 20: 1256-1257. [Google Scholar]
- Subramaniam S, Bartesaghi A, Liu J, Bennett AE, Sougrat R. (2007). Electron tomography of viruses. Curr Opin Struct Biol 17: 596-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM. (1974). Electron diffraction of frozen, hydrated protein crystals. Science 186: 1036-7. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Milligan RA, Raeburn C, Unwin PN. (1984). A cold stage for the Philips EM300 electron microscope. Ultramicroscopy 13: 185-90. [DOI] [PubMed] [Google Scholar]
- The IMpact-RSV Study Group. (1998). Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 102: 531-7. [PubMed] [Google Scholar]
- Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. (2012). Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog 8: e1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. (2008). The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3: 245-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Mercogliano CP, Lowe J. (2011). A ferritin-based label for cellular electron cryotomography. Structure 19: 147-54. [DOI] [PubMed] [Google Scholar]
- Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. (2007). Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J 26: 2218-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Avila-Sakar A, Kim J, Booth DS, Greenberg CH, Rossi A, Liao M, Li X, Alian A, Griner SL, Juge N, Yu Y, Mergel CM, Chaparro-Riggers J, Strop P, Tampe R, Edwards RH, Stroud RM, Craik CS, Cheng Y. (2012). Fabs enable single particle cryoEM studies of small proteins. Structure 20: 582-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.