Inhibitory neurons regulate the adaptation of neural circuits to sensory experience1, but the molecular mechanisms by which experience controls the connectivity between different types of inhibitory neurons2,3 to regulate cortical plasticity are largely unknown. We found that exposure of dark-housed mice to light induces a gene program in cortical VIP neurons that is strikingly distinct from that induced in excitatory neurons and other subtypes of inhibitory neurons. We identify IGF-1 as one of several activity-regulated genes that are specific to VIP neurons, and demonstrate that IGF-1 functions cell-autonomously in VIP neurons to increase inhibitory synaptic input onto these neurons. Our findings further suggest that in cortical VIP neurons, experience-dependent gene transcription regulates visual acuity by activating the expression of IGF-1, thus promoting the inhibition of disinhibitory neurons3–5 and affecting inhibition onto cortical pyramidal neurons.
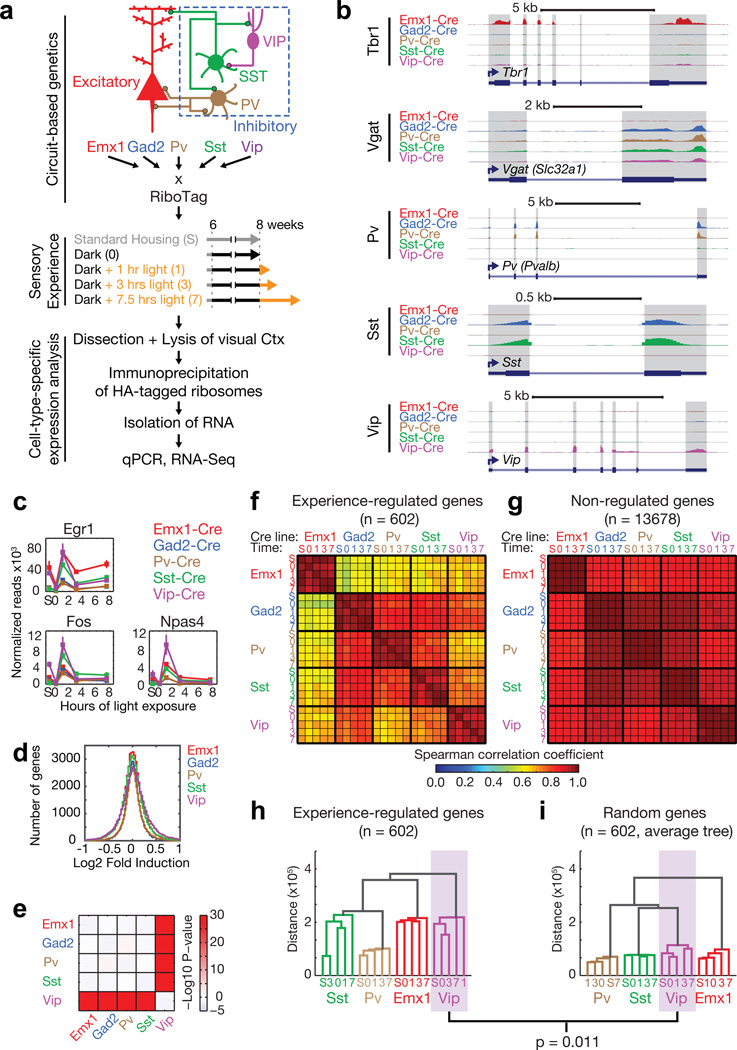
To explore how sensory experience affects gene expression in VIP neurons, we examined this process in the visual cortex of adult mice that were housed in standard conditions, in complete darkness (i.e. dark-housed), or dark-housed and then exposed to light for increasing amounts of time6,7 (Fig. 1a). Light deprivation for as little as 12 hours drives robust gene expression after light exposure, and increasing durations of dark-housing accentuate the gene induction response (Extended Data Fig. 1a) irrespective of the phase of the circadian rhythm (Extended Data Fig. 1b). To purify RNA selectively from VIP-expressing and other inhibitory neuron subtypes, we generated mice that were heterozygous for alleles of either Vip-Cre, Sst-Cre or Pv-Cre, and were also heterozygous for the Rpl22-HA (RiboTag) allele8, which expresses an HA-tagged ribosomal protein specifically in Cre-expressing neurons (Fig. 1a). For purposes of comparison, we also purified ribosome-bound RNA from excitatory and inhibitory neurons, labeled by Emx1-Cre or Gad2-Cre.
Figure 1. VIP neurons mount a unique transcriptional response to sensory experience.
a) Approach for purifying ribosome-bound RNA from Cre-expressing neurons in visual cortex following manipulation of visual experience.
b) Representative RNA-Seq tracks of cell-type-specific marker genes (exons shaded).
c) Line plots showing the expression levels of selected early-induced genes in each Cre-line at each time point (n=3, bars = SEM).
d) Histograms showing the distribution of all observed fold-changes for each Cre-line (mean of three replicates).
e) Color-coded matrix showing the −Log10 P-value of pairwise corrected two-tailed t-tests of the fold-change distributions between each Cre-line.
f, g) Matrices of Spearman correlation coefficients computed from the expression levels of experience-regulated genes (f) or genes not regulated by experience (g) (mean of three replicates).
h, i) Cladograms of (h) all experience-regulated genes created by using the mean values of each gene in each sample or (i) average cladogram created from 1000 random sets of 602 random genes using the mean expression values of each gene in each sample (p=0.011, Monte Carlo test).
By quantitative real-time PCR (qPCR), we find that mRNAs for cell-type-specific marker genes are highly enriched in the appropriate samples (Extended Data Fig. 1c) and that light exposure induces the expression of early-response genes in each Cre line (Extended Data Fig. 1d). To quantify experience-induced gene expression at a genome wide level, we performed RNA-Seq on RNA isolated from the dark-housed/light-exposed RiboTag-mice (Supplemental Table 1) (Fig 1b,c, Extended Data Fig. 2a,b). This analysis identified the genes whose levels change reproducibly in response to visual stimulation in at least one Cre line (n = 602; see Supplemental Table 2 and Methods) and thus allowed us to ask how levels of these experience-regulated genes are correlated across the different neuronal subtypes compared to non-regulated genes (n = 13,678) (Fig. 1d–i). We found that the expression of experience-regulated genes is strikingly dissimilar across different neuronal subtypes when compared to genes that are not regulated by sensory experience (irrespective of differences in the number or expression levels of experience-regulated genes; Fig. 1f–i, Extended Data Fig. 3a–d). While unique subsets of experience-responsive genes were identified in each neuronal subtype (Fig. 2a,b, Extended Data Fig. 3e–f), VIP neurons are the most responsive to sensory stimulation (Fig. 1d,e) and possess an experience-induced gene expression program that is strikingly distinct from the other neuronal subtypes analyzed (Fig. 1h,i). This suggests that in VIP neurons the experience-dependent gene program may have a unique function in adapting the cortex’s neural circuits to sensory experience.
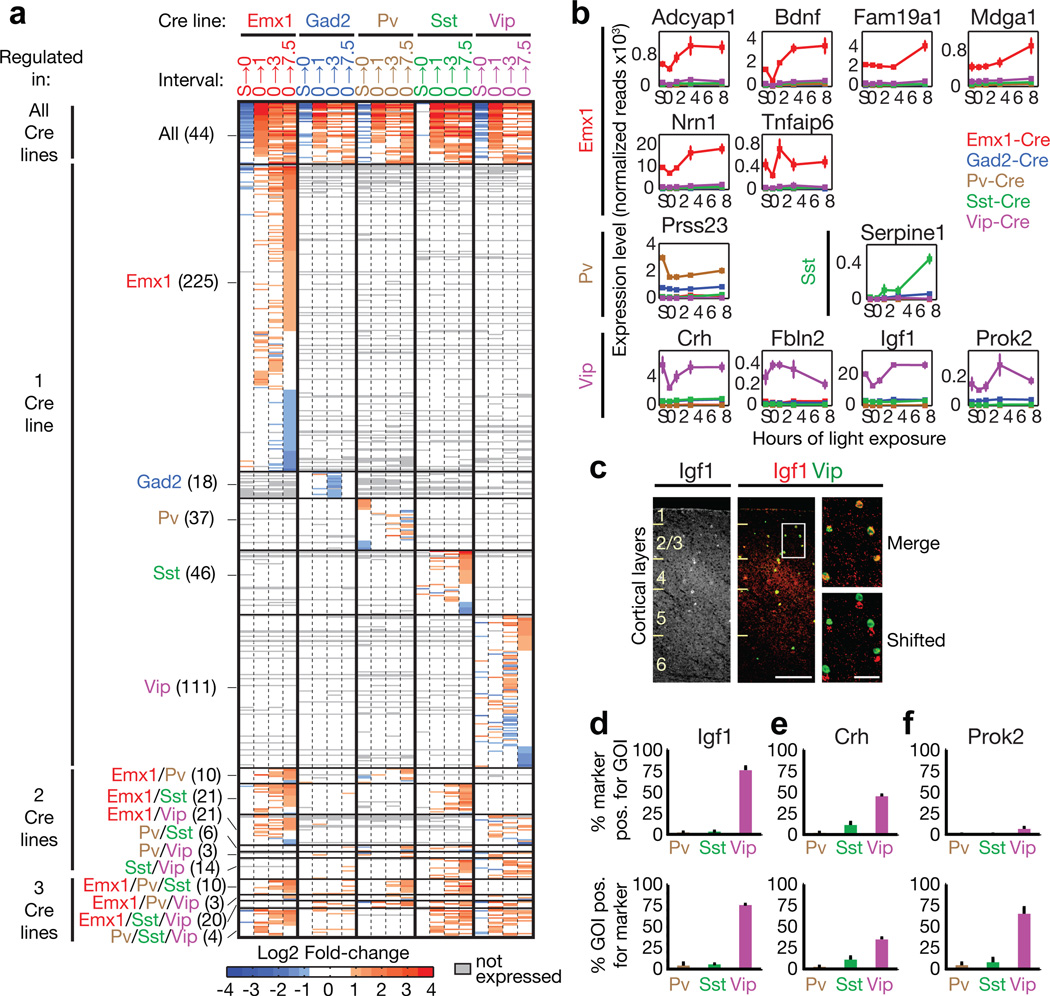
Figure 2. IGF-1 is an experience-induced cell-type-specific secreted factor in VIP neurons.
a) Heatmap showing the fold-change of all experience-regulated genes across all stimulus intervals.
b) Line plots of secreted factors that are experience-regulated and expressed in a cell-type-specific manner (n=3, bars = SEM).
c) Fluorescent In situ hybridization for Igf1 and Vip in mouse visual cortex after dark housing and light exposure for 7.5 hours (white box = magnified area; scale bar = 200 µm in main image, 50 µm in magnification).
d–f) Quantification of fluorescent in situ hybridization for Igf1, Crh, Prok2, and inhibitory markers in visual cortices of dark-housed/light-exposed mice (n=3, bars represent SEM).
We hypothesized that experience-regulated genes that are specifically expressed and selectively regulated in VIP neurons are likely to have important functions in regulating the synaptic connectivity onto VIP neurons. Thus, we first identified the mRNAs that are specifically enriched in each subtype (Extended Data Fig. 4a; Methods) and cross-referenced these genes with the list of experience-regulated genes (Extended Data Fig. 4b). This analysis identified 31 genes that are both cell-type-specific and experience-regulated, 11 of which are specific to VIP neurons (Supplemental Table 4). Strikingly, secreted molecules are significantly over-represented in this gene set (GO-term ‘Secreted’ p = 0.002) and each type of neuron has its own set of cell-type-specific experience-regulated secreted factors, including four experience-induced secreted molecules that are specific to VIP neurons (Igf1, Crh, Prok2, Fbln2; Fig. 2b, Supplemental Table 4).
We next performed fluorescent in situ hybridization (FISH) on sections of visual cortices of dark-housed/light-exposed mice to quantify the percentage of cells that co-express an inhibitory subtype marker and the respective secreted factor (Fig. 2c–f). Of the four secreted factors, Igf1 is the one factor that is expressed in the vast majority of VIP neurons, and whose expression is highly enriched in these neurons (Fig. 2d). We were unable to reliably identify Fbln2-expressing cells, and Prok2 was expressed nearly exclusively in a sparse subpopulation of VIP neurons (Fig. 2f), consistent with the low expression level of these genes in the RiboTag-Seq experiments (Extended Data Fig. 4c). While the FISH analysis revealed that in the cortex Crh is highly enriched in VIP neurons compared to PV and SST neurons (Fig. 2e), this gene is also expressed in Pv-/Sst-/Vip-negative cortical interneurons9. Since IGF-1 is the sole experience-induced secreted factor that is selectively expressed in most VIP neurons, we focused our subsequent analysis on IGF-1.
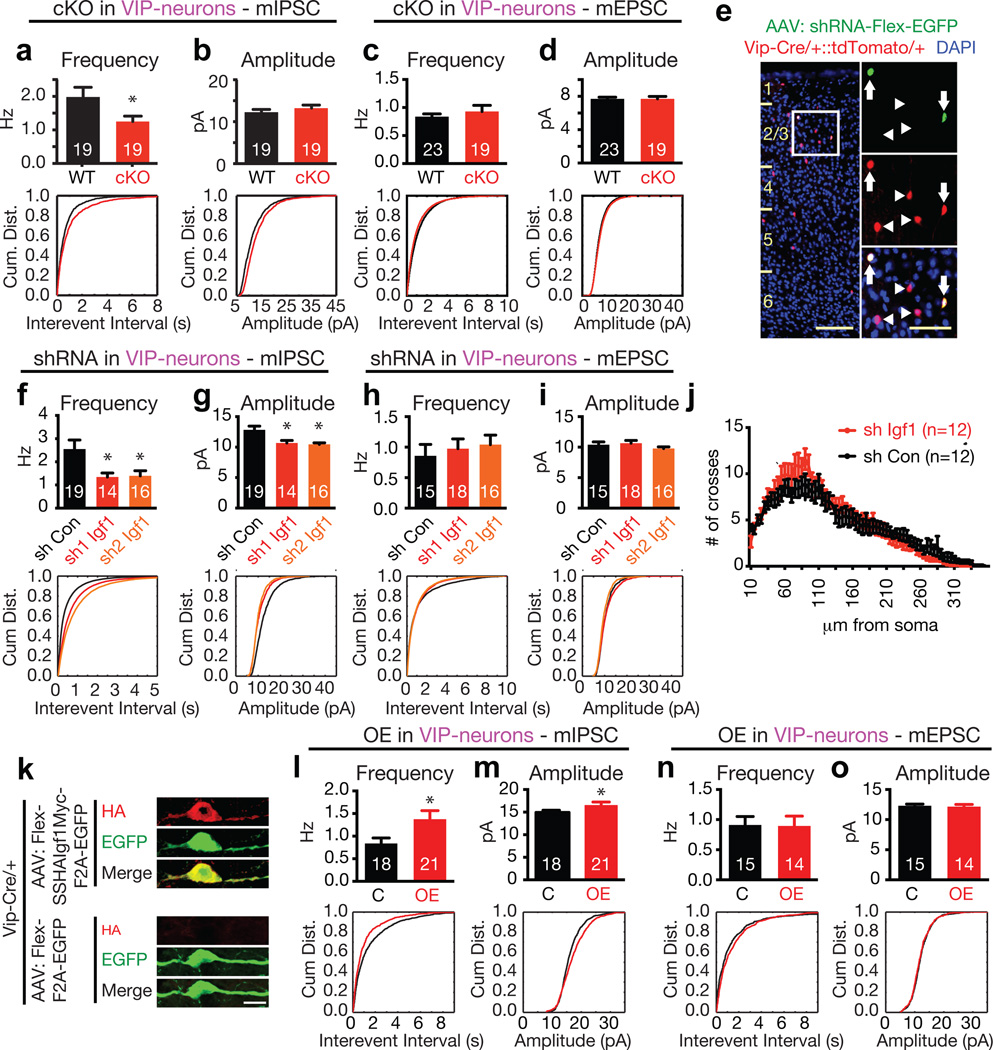
Previous reports have suggested that IGF-1 is synthesized in the cortex10, but the function of cortical-derived IGF-1 was unknown. Global disruption of the Igf1 gene results in abnormally small animals with smaller brains that contain smaller neurons with dendrites that are less branched and contain fewer synapses11–13, and the effects of IGF-1 on brain development and function are due at least in part to IGF-1 that is produced by non-neural tissues and then enters the brain14. To investigate specifically VIP neuron-derived IGF-1, we crossed Vip-Cre mice to both IGF-1 conditional knockout mice15 and Cre-reporter mice. Disruption of Igf1 specifically in VIP neurons had no effect on the thickness of the cortical layers, on the number and layer distribution of VIP neurons, or on the size of VIP neuron cell bodies at postnatal day 21 (i.e. P21) (Extended Data Fig. 5a–d). To test whether VIP neuron-derived IGF-1 affects excitatory and/or inhibitory inputs to VIP neurons, we recorded miniature inhibitory or excitatory postsynaptic currents (mIPSCs or mEPSCs) in VIP neurons in acute visual cortex slices; we found that conditional deletion of Igf1 in VIP neurons leads to a significant reduction in mIPSC frequency (Fig. 3a) but not amplitude (Fig. 3b). Since conditional deletion of Igf1 had no effect on the frequency or amplitude of excitatory mEPSCs on VIP neurons (Fig. 3c,d), these findings suggest that VIP neuron-derived IGF-1 specifically enhances inhibitory synaptic input onto VIP neurons.
Figure 3. IGF-1 promotes inhibitory inputs to VIP neurons in a cell autonomous manner.
a–d) Bar graph and cumulative distribution of the frequency and interevent intervals of mIPSCs or mEPSCs recorded from Igf1 WT or cKO VIP neurons (mIPSC frequency: p = 0.046; amplitude p = 0.3. mEPSC frequency: p = 0.44; amplitude p = 0.9, Mann-Whitney U-Test).
e) Example image of sparsely infected VIP neurons upon injection of AAV-shRNA-hUbc-Flex-EGFP into mice expressing tdTomato in all VIP neurons (white box = magnified area; arrows = infected VIP neurons; arrowheads = non-infected VIP neurons; scale bars, 100 µm in main image, 50 µm in magnification)
f–i) Bar graph and cumulative distribution of mIPSC/mEPSC frequency, interevent interval and amplitude recorded from VIP neurons sparsely infected with control or Igf1 shRNAs (mIPSC Frequency: shRNA1 p = 0.05, shRNA2 p = 0.042. mIPSC Amplitude: shRNA1 p = 0.004, shRNA2 p = 0.001. mEPSC Frequency: shRNA1 p = 0.13, shRNA2 p = 0.07. mEPSC Amplitude: p = 0.77, shRNA2 p = 0.44. Mann-Whitney U-Test).
j) Sholl analyses of VIP neurons infected with control or Igf1 shRNA (p = 0.76, two-way repeated-measures ANOVA).
k) Expression of epitope-tagged Igf1.4 in VIP-neurons. Cortices of P3 Vip-Cre/+ mice were injected with AAVs driving Cre-dependent expression of SSHAIgf1.4Myc-F2A-EGFP (top) or F2A-EGFP (bottom) and stained at P20 for HA (red) and EGFP (green) (Scale-bar = 10 µm).
l–o) Bar graphs and cumulative distribution plots showing mIPSC/mEPSC amplitude and frequency / interevent interval in VIP neurons infected with control or IGF1-OE AAVs. (mIPSC: Amplitude, p = 0.05; Frequency, p = 0.02. mEPSC: Amplitude, p = 0.55; Frequency, p = 0.86. Mann Whitney U-Test)
a–d, f–j, l–o) Numbers inside bars indicate the number of cells recorded
To test whether IGF-1 functions cell-autonomously to regulate inhibitory input onto the cell from which it is expressed, we used a virus-based approach to acutely knock down Igf1 expression in only a few VIP neurons. We generated shRNA-constructs against Igf1 (Extended Data Fig. 6a,b), injected low titer AAVs expressing the shRNA and Cre-dependent EGFP into the visual cortex of P14-15 Vip-Cre mice, and recorded mIPSCs and mEPSCs one week later (P20–P22) in EGFP-positive VIP neurons that are surrounded by non-infected VIP neurons (Fig. 3e). This sparse and acute knockdown of Igf1 in VIP neurons using either of two distinct shRNAs against Igf1 resulted in a marked reduction in mIPSC frequency and amplitude as compared to VIP neurons infected with a control shRNA (Fig. 3f,g), but had no significant effect on mEPSCs (Fig. 3h,i). These effects are not due to altered VIP neuron morphology (Fig. 3j, Extended Data Fig. 6c), indicating that VIP neuron-derived IGF-1 acutely promotes inhibition onto VIP neurons in a cell-autonomous manner.
To determine if VIP neuron-derived IGF-1 regulates inhibitory inputs onto other types of cortical neurons, we adopted a protocol that leads to widespread infection of neurons in the cortex (see Methods). Injecting AAVs into Vip-, Pv- or Sst-Cre mice to label these cells with EGFP while knocking down Igf1 in VIP neurons, we recorded mIPSCs from each cell type and found that early knockdown of Igf1 in the cortex decreases mIPSCs frequency in VIP neurons, but does not affect mIPSCs onto PV, SST, or pyramidal neurons (Extended Data Fig. 6e–l). Furthermore, ELISA-based analysis of IGF-1 levels in the blood of mice whose cortices were injected with these viruses demonstrated that removing IGF-1 from VIP neurons did not alter the level of serum-derived IGF-1 (Extended Data Fig. 6d). While we formally cannot exclude that serum-derived IGF-1 contributes to inhibition onto VIP neurons, this finding indicates that the decrease in mIPSCs in VIP neurons expressing Igf1 shRNAs is due at least in part to reduced Igf1 expression in VIP neurons. Thus, VIP neuron-derived IGF-1 regulates the inhibitory inputs onto the VIP neuron in which it is produced, probably via local release from VIP neurons. Consistent with this idea, we find that the Igf1 splice-variant expressed by VIP neurons encodes an isoform of IGF-1 containing a Heparin binding domain (Igf1.4; Extended Data Fig. 7a) that may limit the diffusion of IGF-1 and facilitate its local action16.
We next overexpressed IGF-1 in VIP neurons by injecting Vip-Cre mice with an AAV construct that drives expression of an epitope-tagged version of IGF-1 together with EGFP in a Cre-dependent manner (Fig. 3k; Extended Data Fig. 7b,c) and assessed the effect on mIPSCs and mEPSCs (Fig. 3l–o). We find that when overexpressed in VIP neurons, IGF-1 selectively promotes inhibition onto VIP neurons, as it has no effect on mEPSCs in these cells. Likewise, ectopic expression of IGF-1 in SST and excitatory neurons (by intracortical injections into Sst- or Emx1-Cre mice, respectively; Extended Data Fig. 7d–g) leads to a similar increase in mIPSC frequency in these cells. These findings raise the possibility that the selective expression of Igf1 in VIP neurons is required for the proper organization and function of cortical circuits, as aberrant IGF-1 expression could enhance inhibition indiscriminately within cortical circuits by signaling through IGF-1 receptors that are ubiquitously expressed in these neurons (Extended Data Fig. 7h–j).
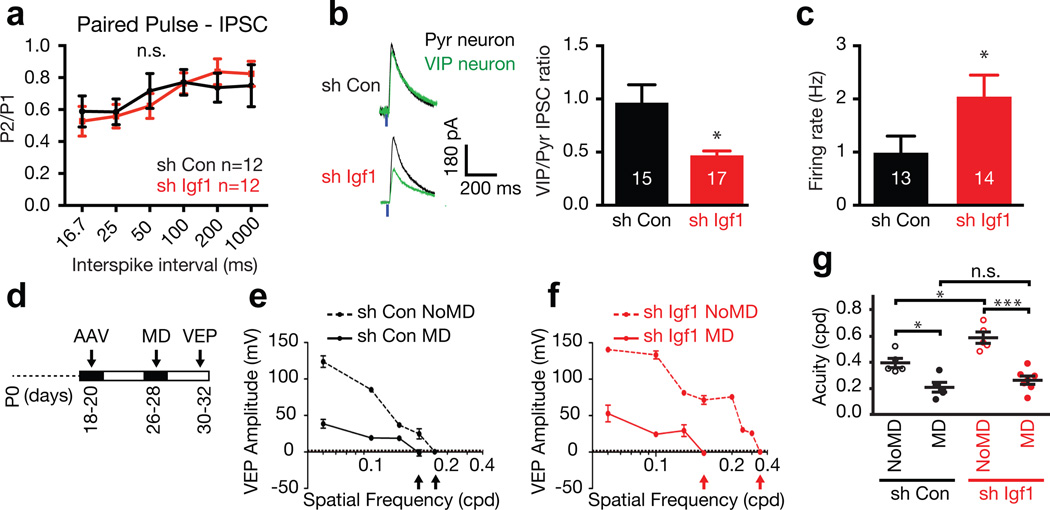
The change in mIPSC frequency upon Igf1 knockdown in VIP neurons could reflect a change in the presynaptic probability of release and/or a reduction in inhibitory synapse number and/or strength. By paired-pulse stimulation we find that Igf1 knockdown in VIP neurons does not significantly alter the probability of release of vesicles from either inhibitory (Fig. 4a) or excitatory (Extended Data Fig. 7k) terminals that synapse onto VIP neurons. To test evoked inhibition, we co-injected Vip-Cre mice with either Igf1 or control shRNA AAVs and an AAV encoding the excitatory light-activated ion channel ReachR17. Performing paired voltage clamp recordings from EGFP-positive VIP neurons and neighboring pyramidal cells to control for variation in stimulation intensity, we found that the strength of light-evoked inhibition onto VIP neurons is decreased when Igf1 expression is knocked down in VIP neurons (Fig. 4b). These experiments suggest that the primary site of IGF-1 action is post-synaptic and indicate that experience-dependent activation of Igf1 expression increases the number and/or strength of functional inhibitory synapses that form on VIP neurons. To test whether this IGF-1 dependent increase in inhibition alters the frequency of action potentials in these neurons, we performed cell-attached recordings from EGFP-labeled VIP neurons expressing control or Igf1 shRNAs. We find that VIP neurons lacking IGF-1 fire action potentials at a significantly higher rate than controls (Fig 4c). Given that VIP neurons disinhibit cortical circuits, it seems likely that this decreased firing of VIP neurons might alter how the cortex responds to sensory experience. To begin to investigate this possibility, we next assessed the effect of knocking down Igf1 expression on visual cortex plasticity.
Figure 4. VIP neuron-derived IGF-1 regulates VIP neuron function and regulates visual acuity in an experience-dependent manner.
a) Paired-pulse recordings from VIP neurons infected with control or Igf1 shRNA (p = 0.96, two-way ANOVA).
b) Left: average traces of light-evoked IPSCs (eIPSC) from paired recordings of VIP neurons infected with control or Igf1 shRNA (green traces) and neighboring pyramidal neurons (black traces). Right: quantification of eIPSC amplitude of the VIP neuron after infection with AAVs expressing control or Igf1 shRNA normalized, to the eIPSC amplitude of the paired pyramidal neuron (p = 0.01, Mann-Whitney U-Test).
c) Average firing rate of VIP neurons infected with Igf1 or control shRNA (p = 0.04, Mann-Whitney U-Test).
b, c) Numbers inside bars indicate the number of cells recorded
d) Schematic of the schedule for monocular deprivation (MD) experiments. e–f) Representative traces of visually evoked potential (VEP) amplitude as a function of spatial frequency (cpd) in the contralateral visual cortex of mice that received bilateral injections of AAVs expressing Igf1 or control shRNA into their visual cortices and were subjected to MD or not.
g) VIP neuron-derived IGF-1 restricts visual acuity in an experience-dependent manner. Visual acuity in mice injected with AAVs expressing Igf1 or control shRNA with no MD or MD (P24 – P28; control shRNA NoMD, n = 5; control shRNA MD, n = 5; Igf1 shRNA NoMD, n = 5; Igf1 shRNA MD, n = 7; p* < 0.05, p*** < 0.0001, n.s. – not significant, One way ANOVA-Tukey post test).
Cortical inhibition regulates ocular dominance (OD) plasticity1,18 and visual acuity19, and hyper-activation of VIP neurons drives a form of adult cortical plasticity20. To determine whether knocking down Igf1 expression in VIP neurons affects visual cortex function, we injected control or Igf1 shRNA AAVs into the binocular zone of visual cortices of P18 Vip-Cre mice and recorded visual-evoked potentials (VEPs) between P28 and P32 (Fig 4d, Extended Data Fig. 8a). Stimulation of the contralateral or ipsilateral eye with gratings at low spatial frequency elicited robust VEPs both under control and Igf1 knockdown conditions (Extended Data Fig. 8b–d). Furthermore, the ratio between the contralateral and ipsilateral eye’s response (C/I ratio) was similar in the presence or absence of Igf1 (Extended Data Fig. 8e), indicating that basic visual cortex function is not obviously disrupted upon Igf1 knockdown. Remarkably, when we assessed visual acuity of the contralateral eye by increasing the spatial frequency of the gratings presented, the mice injected with AAVs expressing Igf1 shRNA exhibited significantly increased visual acuity as compared to mice injected with control AAVs (Fig. 4–g).
To test whether the effect of Igf1 knockdown is experience-dependent, we next monocularly deprived (i.e. MD) mice for a brief period of time, beginning at the peak time of ocular dominance plasticity (i.e. at P26-28, Fig. 4d). After four days of MD, we recorded VEPs from the visual cortex contralateral to the deprived eye and quantified the C/I ratio upon stimulation at low spatial frequency as well as the visual acuity upon stimulation of the deprived eye. Brief MD led to a reduction in the C/I-ratio in mice injected with AAVs expressing either control or Igf1 shRNA; this is a consequence of the reduction in the contralateral response (Extended Data Fig. 8b–e)18. Notably, when we tested visual acuity after brief MD, both Igf1 and control shRNA injected mice exhibited similar levels of amblyopia (i.e. loss of visual acuity) in the deprived eye (Fig. 4e–g), despite the higher visual acuity in the Igf1 shRNA injected mice that were not monocularly deprived (Fig. 4e–g). These findings indicate that VIP-derived IGF-1 regulates visual acuity in an experience-dependent manner and may function as a sensory-dependent brake on cortical plasticity. The observation that in response to sensory experience IGF-1 in VIP neurons controls inhibition, taken together with the previous finding that experience induces BDNF in excitatory neurons to regulate excitatory-inhibitory balance21,22, suggests a model in which each type of neuron within a cortical circuit expresses a unique set of experience-induced secreted factors that control specific synaptic inputs onto the neuron and plasticity within a neural circuit6,23.
Methods (online only)
Visual Stimulation
For calibrating the duration of the dark housing period prior to light exposure, C57Bl6 wild-type mice were housed in a standard light cycle until they were placed in constant darkness for varying amounts of time prior to analysis at postnatal day 56. At P56, all mice were either sacrificed in the dark (dark-housed condition) or light-exposed for 1, 3, or 7.5 hours before being sacrificed. The eyes of all animals were enucleated (for the dark-housed condition, enucleation was performed in the dark) before dissection of the visual cortex in the light.
For RiboTag-experiments, mice were reared in a standard light cycle and then housed in constant darkness for two weeks starting from P42; at P56, all mice were either sacrificed in the dark (dark-housed condition) or light-exposed for 1, 3, or 7.5 hours before being sacrificed. Additional cohorts of mice for the “Standard” condition were housed in a standard light cycle until P56 when they were sacrificed. The eyes of all animals were enucleated (for the dark-housed condition, enucleation was performed in the dark) before dissection of the visual cortex in the light.
RNA isolation, Reverse Transcription, qPCR-analysis
Total RNA was extracted with Trizol reagent (Sigma) according to manufacturer’s instructions, and RNA quality was assessed on a 2100 BioAnalyzer (Agilent); all RNAs were treated with DNAseI (Invitrogen) prior to reverse transcription. For the cloning of riboprobes, total RNA was extracted from whole adult C57Bl6 wild-type mouse brains and cDNA was prepared using SuperScript® II kit (Life Technologies). For real-time quantitative PCR experiments aimed at calibrating the duration of the dark housing period, total RNA was extracted for each sample from the visual cortices of one animal. For real-time quantitative PCR experiments aimed at testing the efficacy of shRNA constructs directed against Igf1, total RNA was isolated from two pooled 24-wells of cultured cortical neurons for each condition. For qPCR-experiments, RNA was reverse transcribed with the High Capacity cDNA Reverse Transcription kit (Life Technologies). Real-time quantitative PCR reactions were performed on the LightCycler 480 system (Roche) with LightCycler® 480 SYBR Green I Master. Reactions were run in duplicates, triplicates or quadruplicates, and beta-Actin (ActB) or Tubulin beta 3 (Tubb3) levels were used as an endogenous control for normalization using the ΔΔCt method24. Real-time PCR primers were designed using the Universal ProbeLibrary (Roche) as exon-spanning whenever possible and answered the following criteria: linear amplification over 3 orders of magnitude of target concentration, no amplification product in control samples that were not reverse-transcribed (i.e. control for contamination with genomic DNA), no amplification product in control samples where no template was added (i.e. control for primer dimers), amplification of one singular product as determined by melt-curve analysis and analysis of the product in agarose gel electrophoresis and sequencing of the PCR product. The qPCR primers used in this study are listed in Supplementary Table 6.
For analysis of light-induced gene expression in wild-type mice, the gene expression levels were analyzed in four mice (two males and two females) at each time point. The data were calculated as fold-changes relative to the average of the overnight dark-housed condition and normalized to the average of the maximally induced time point. Data in figures represent the mean and SEM of four mice.
For assessing Igf1-levels in cortical cultures infected with shRNA-expressing lentiviral constructs, qPCRs were performed in quadruplicates for each condition and fold-changes were calculated relative to the non-infected non-stimulated cultures. Data were normalized to the maximally induced condition in each biological replicate, and data in figures represent the mean and SEM of three biological replicates.
RiboTag-purifications, RiboTag-qPCR and RiboTag-Seq
Immunoprecipitation and purification of ribosome associated RNA was performed essentially as described6,8, with minor modifications: lysis of the samples was performed in the presence 10 mM Ribonucleoside Vanadyl Complex (NEB, Ipswich, MA), and immunoprecipitation was performed with a different anti-HA antibody (HA-7, 12 µg/IP, Sigma). Briefly, the visual cortices were dissected, flash frozen in liquid nitrogen and then kept at −80°C until further processing. Visual cortices from three individual animals (each sample contained both male and female animals) were pooled for each biological replicate, and three biological replicates were performed. After lysis of the tissues and prior to immunopurification, a small fraction of lysate of each sample (i.e. “input”) was set aside and total RNA was extracted with Trizol reagent followed by the RNEasy Micro Kit’s procedure (Qiagen, Valencia, CA). After immunopurification of the ribosome-associated RNAs, RNA quality was assessed on a 2100 BioAnalyzer (Agilent, Palo Alto, CA) and RNA amounts were quantified using the Qubit 2.0 Fluorometer (Life Technologies). Only samples with RIN numbers above 8.0 were considered for analysis by qPCR and RNA-Seq. For all RNA samples of sufficient integrity, 5–10 ng of RNA were SPIA-amplified with the Ovation RNA Amplification System V2 (NuGEN, San Carlos, CA), yielding typically 5–8 µg of cDNA per sample.
Quantitative RT-PCR was performed as described above and relative expression levels were determined in every experiment by normalizing the Ct-values to those of beta-Actin (ActB) from the 0 hour input using the ΔΔCt method24. To determine the fold-enrichment (IP/Input), the actin-normalized expression levels for every time point of every biological replicate were averaged, and the grand averages from the IP and Input were divided to find the IP/Input ratio. To calculate fold-induction for each biological replicate, each time point was divided by the maximal value occurring in that biological replicate, such that the maximal value was set to 1 in each biological replicate. The mean and standard error were calculated at each time point from these normalized values. All samples were analyzed by qPCR for purity and light-induced gene expression prior to analysis by high throughput sequencing.
RNA-sequencing and analysis
SPIA-amplified samples from RiboTag-immunoprecipitated fractions for each of the 5 stimulus conditions and each of the 5 Cre lines were prepared as described above and processed in triplicate (75 samples total). For preparing Seq-libraries, 2 µg of each amplified cDNA were fragmented to a length of 200–400 bp using a Covaris S2 sonicator (Acoustic wave instruments) using the following parameters: duty cycle: 10%, intensity: 5, cycles per burst: 200, time: 60 seconds, total time: 5 minutes. After validating the fragment length of the sonicated cDNA using a 2100 BioAnalyzer (Agilent, Palo Alto, CA), 2 µg of the fragmented cDNA were used for Seq library preparation using the PrepX DNA kit on an Apollo 324 robot (IntegenX). The quality of completed Seq-libraries was assessed using a 2100 BioAnalyzer (Agilent, Palo Alto, CA) and the completed libraries were sequenced on an Illumina HiSeq 2000 instrument, following the manufacturer's standard protocols for single-end 50 bp sequencing with single index reads. Sequencing typically yielded 30 – 80M usable non-strand-specific reads per IP sample. Reads were mapped to the mm9 genome using TopHat (v2.0.13) and Bowtie (2.1.0.0)25. On average, ~70% of mapped IP reads were uniquely mapped to the mm9 genome allowing for 0 mismatches and were therefore assignable to genic features (one RiboTag-Seq library (Sst-Cre, Standard-housing, Biological replicate 2) was excluded from analysis due to low mappability). Values from all IP libraries were normalized using Cufform (v2.2.1), and values from the Cuffnorm output file ‘genes-Count_Table’ (normalized reads) were taken as a proxy for gene expression. P-values were generated for each Cre-line for each Dark-Light conditions using Cuffdiff (v2.2.1) using the timeseries (-T) flag based on 3 biological replicates.
Identification and Classification of Experience-Regulated Transcripts
To identify transcripts regulated by visual experience, for each biological replicate of each Cre line, the fold-change in normalized reads was calculated for each gene at every time point (dark-housed/standard-housed, 1 hour light/dark-housed, 3 hours light/dark-housed; 7.5 hours light/dark-housed). Genes were flagged as experience-regulated in a given Cre-line if they met the following conditions in at least one sample: 1) p-value < 0.005, 2) mean fold-change of 2-fold or greater, 3) fold-changes of 2 or higher in 2 of 3 biological replicates, 4) the mean expression value in at least one sample must be above absolute expression threshold (set at the 40th percentile of all observed values).
To determine in which Cre lines genes were regulated by experience, genes were simply classified according to the above criteria. However, for this analysis we excluded the Gad2-Cre line, since Pv-, Sst-, and Vip-Cre all label subsets of the neurons labeled by Gad2-Cre. However, we did detect genes regulated solely in Gad2-Cre, but no other Cre lines; we reasoned that these genes are likely regulated by experience in a population of 5HT3aR+/VIP− neurons that are contained in Gad2-Cre but none of the other Cre lines.
We classified the set of experience-regulated genes into categories ‘early’, ‘late’, and ‘long-term’ based on the fastest kinetics observed. When genes were found to be elevated and/or suppressed at multiple time points, we assigned them to the categories based on the most rapid observed change. For example, while Fos levels are elevated over dark housing at 1, 3, and 7.5 hours of light exposure and suppressed after two-weeks of dark housing, Fos is classified as ‘early-up’ because it is elevated at 1 hour after light exposure.
Linkage Analysis
All linkage analysis was performed using the ‘single’ method and ‘Cityblock’ metric using Matlab’s linkage function. To determine the branch-order significance of the cladogram resulting from clustering of the 602 experience-regulated genes, we generated 1000 cladograms from 602 sets of random expressed genes (including experience-regulated genes, with replacement) and asked how often we generated a cladogram with an identical branch order at the level of the Cre lines. Only 11 sets of 1000 random genes sets generated an identical tree. For the purposes of this analysis, we only compared the branches above the level of the individual Cre line.
Identification of Cell-Type-Specific Transcripts
To identify cell-type-enriched transcripts, an enrichment score was calculated for every transcript in every Cre line for each biological replicate. This enrichment score was calculated by dividing the maximum expression value observed in a given Cre line by the maximum expression value observed across all conditions for all other Cre lines (GABAergic subtypes were not required to be enriched above Gad2-Cre). The enrichment scores for a set of known cell-type-specific genes were evaluated (Vglut1, Tbr1, Pvalb, Sst, Vip), and our threshold was set at the enrichment score of the-cell-type specific gene with the lowest score (Slc17a7/Vglut1, at 5.5-fold-enriched in Emx1-Cre). Transcripts were considered to be expressed in a cell-type-specific manner (or “highly enriched”) in a given Cre line if their mean enrichment score was above this threshold and if the enrichment score exceeded this threshold in 2 out of 3 biological replicates.
Cloning of riboprobes, knockdown and expression constructs
Cloning of all constructs was done using standard cloning techniques, and the integrity of all cloned constructs was validated by DNA sequencing. Templates for the riboprobes for Igf1, Gad1, Pvalb, Sst and Vip were prepared by PCR-amplification of cDNA fragments generated from total RNA isolated from adult C57Bl6 mouse brains (see Supplementary Table 7 for primer sequences) and cloning of the respective PCR fragments into the pBlueScript II vector (Agilent Technologies).
Lentiviral shRNA constructs were generated by cloning shRNA stem loop sequences against Igf1 (Igf1 shRNA #1: GGTGGATGCTCTTCAGTTC; Igf1 shRNA #2: TGAGGAGACTGGAGATGTA) and Luciferase (Luc, control: ACTTACGCTGAGTACTTCG) into a modified version of pLentiLox3.726 in which the CMV promoter driving the expression of EGFP was replaced with an hUbc promoter and in which the LoxP sites surrounding the hUbc-EGFP cassette were removed. The loop sequence used in these shRNA-constructs is based on miR-25 (cctctcaacactgg)27. ShRNA-expressing AAV-constructs (pAAV-U6-shRNA-hUbc-Flex-EGFP) were made by first replacing the Flex-GFP-Gephyrin cassette in pAAV-Flex-GFP-Gephyrin22 with a Flex-EGFP cassette (resulting in pAAV-hUbc-Flex-EGFP) and then transferring the U6-shRNA cassettes from the pLentiLox constructs to pAAV-hUbc-Flex-EGFP.
AAV constructs for the Cre-conditional co-expression of epitope-tagged IGF1.4 and EGFP or of EGFP alone were cloned by synthesizing the gBlocks (Integrated DNA Technologies) and using the gBlocks as templates for PCR amplification; the respective PCR products were then cloned into the pAAV-hUbc-Flex-EGFP (see above) by replacing the EGFP with the respective insert. This strategy yielded plasmids termed pAAV-hUbc-Flex-SSHAIgf1.4Myc-F2A-EGFP and pAAV-hUbc-Flex-F2A-EGFP, whereby the Cre-dependent inserts were driven by a human ubiquitin promoter (hUbc). The sequence for Igf1.4 was based on NM_001111275 (bp 277–752) and was modified in the following way: an HA epitope (TATCCtTATGATGTTCCAGATTATGCT) was inserted in frame between the Igf1.4 signal sequence and the beginning of the coding sequencing (cds) of Igf1.4, Igf1.4 was rendered resistant to the shRNA against Igf1 by introducing silent mutations into the target sequences specified above (sh1: TGTTGACGCGCTCCAATTT; sh2: TACGCCGGTTAGAAATGTA) and the followings tags were inserted in frame 3’ to the Igf1.4 cds: Myc epitope (GAACAAAAACTCATCTCAGAAGAGGATCTG), Furin cleavage site (CGGGCCAAGCGG) and a 2A peptide (GGCAGTGGAGAGGGCAGAGGAAGTCTTCTAACATGCGGTGACGTGGAGGAGAATCCCGGCCCT). The sequence for EGFP was based of the published sequence of EGFP. For pAAV-hUbc-Flex-F2A-EGFP a gBlock was synthesized containing the Furin cleavage site followed by the 2A site and EGFP. Detailed sequences are available upon request.
Double-fluorescent ISH
For double-fluorescent in situ hybridization (FISH), wild-type C57Bl6 mice were dark-housed and light-exposed for 7.5 hours as described above. After light exposure, the brains were dissected and fresh frozen in Tissue-Tek Cryo-OCT compound (Fisher Scientific) on dry ice and stored at −80°C until use.
FISH for Igf1 was essentially done as described28,29: riboprobes were prepared by in vitro transcription of linearized plasmids containing the template of the respective probe. Riboprobes for Igf1 were labeled with UTP-11-Digoxigenin, while the riboprobes for the subtype markers (Gad1, Pvalb, Sst, Vip) were labeled with UTP-12-Fluorescein (Roche); all riboprobes were hydrolyzed to lengths of 200–400 bp after synthesis and validated for labeling with Dioxigenin or Fluorescein. For in situ hybridization, coronal sections (20 µm thick) of the visual cortex were cut on a cryostat and fixed in 4% paraformaldehyde for 10 minutes. Endogenous peroxidases were inactivated by treating the sections for 15 minutes in 0.3% H2O2 in PBS, and acetylation was performed as described. Prehybridization was done overnight at room temperature, and hybridization was performed under stringent conditions at 71.5°C. Following hybridization, stringency washes in SSC were performed as described at 65°C. For immunological detection of the first probe (Igf1), the tissue was first treated with a blocking step for 1 hour in blocking buffer (B2) at room temperature before the anti-Digoxigenin-POD antibody (Roche) was applied at a concentration of 1:1000 in blocking buffer for 1 hour at room temperature. Following three washes in buffer B1 and an additional wash in buffer TNT (0.1 M Tris-HCl pH 7.5, 0.15 M NaCl, 0.05% Tween20), the Igf1 probe was detected by exposing the sections at room temperature in the dark for 20 minutes to TSA™ Plus Cy3 reagent (Perkin Elmer) diluted 1:100 in TSA working solution, after which the sections were washed three times in TNT buffer. Before the immunological detection of the second probe, the peroxidases for detecting the first probe were inactivated by treating the sections for 30 minutes with 3% H2O2, followed by three washes in PBS. After an additional blocking step in blocking buffer for 1 hour at room temperature, the anti-Fluorescein-POD antibody (Roche) was applied at a concentration of 1:1000 in blocking buffer overnight at 4°C. Following three washes in buffer B1 and an additional wash in buffer TNT, the probes of the subtype markers were detected by exposing the sections at room temperature in the dark for 15 minutes to TSA™ Plus Cy5 reagent (Perkin Elmer) diluted 1:100 in TSA working solution, after which the sections were washed three times in TNT buffer. Finally, the sections were counterstained with DAPI (4',6-Diamidino-2-Phenylindole, Molecular Probes) and mounted using Fluoromount-G (Southern Biotech). In each experiment, controls for hybridization specificity were included (sense probe for Igf1) as well as controls for ensuring the specificity of the immunological detection of the Digoxigenin- and Fluorescein-labeled riboprobes.
FISH for Crh, Prok2 and Fbln2 was done using the RNAscope system (Advanced Cell Diagnostic); this was necessary since no reliable signal could be detected with the method described above for Igf1 FISH using DIG-labeled riboprobes. RNAscope probes for all genes were synthesized by ACD and all experiments were done according to the ACD’s protocol for fresh frozen brain sections.
For quantifying of the expression pattern of all genes of interest (GOI, i.e. Igf1, Crh and Prok2; Fbln2 could not be detected reliably), the visual cortices in each section were imaged on a Zeiss Axio Imager microscope with a 10× objective and 3 × 5 fields-of-view were “stitched” into one compound image; in all cases, image exposures were kept constant throughout a given experiment for each channel. Compound images of each visual cortex were then imported to Photoshop, and additional layers were created for each probe (i.e. one layer for the GOI and one layer for the subtype marker in each compound image). The cells positive for each probe were then marked with a dot in the new respective layer by two independent investigators in a blinded manner (one investigator marking GOI-positive cells and the other investigator marking subtype marker-positive cells). Finally, the layers containing the dots of the identified positive cells were compiled into a separate image file together with the DAPI-layer and imported into ImageJ. In ImageJ, the images were analyzed in a blinded manner by defining the visual cortex and its layers as regions-of-interest (ROI) based on the DAPI staining and quantifying the number of cells positive for either one or both markers per ROI. For each combination of probes (GOI together with each of the subtype markers), two visual cortices from four animals were analyzed (a total of 8 visual cortices for each combination).
Virus production and neuronal cultures
Concentrated lentiviral stocks were prepared and titrated essentially as described30. AAV stocks were prepared at the University of North Carolina (UNC) Vector Core and at the Children’s Hospital Boston Vector Core; see also Supplementary Table 8 for further details on AAV stocks.
Primary cultures of cortical neurons were prepared from E16.5 mouse embryos as described6. Briefly, 3×105 neurons per well were plated in 24-well dishes coated with Poly-D-Lysine (20 µg/mL) and Laminin (3.4 µg/mL). Cultures were maintained in Neurobasal medium supplemented with B27 (Invitrogen), 1 mM L-glutamine, and 100 U/mL penicillin/streptomycin, and one third of the media in each well was replaced every other day. For testing of viral shRNA constructs, the cultures were infected at DIV 3 with concentrated viral stocks for 5 hours at an MOI of 6. After infection, the cultures were washed twice in plain Neurobasal medium after which the conditioned medium was returned to the dish and the cultures were continued to be maintained as described. At DIV 7, neuronal cultures were treated overnight with 1 µM TTX and 100 µM AP-5 to silence spontaneous activity before the cultures were depolarized at DIV 8 with 55 mM extracellular KCl as described6 and lysed in Trizol after 6 hours of stimulation.
Western Blot for testing of IGF-1 constructs and ELISA for determining serum IGF-1 levels
HEK293T cells were used for testing the expression and the biological activity of the epitope tagged Igf1.4 constructs. HEK293T cells were cultured in DMEM (Life Sciences) containing 10% FCS and Penicilin/Streptomycin. Cells were transfected using Lipofectamine (Life Technologies) and 18 hours post transfection, the medium was replaced with DMEM containing 0.1% FCS; 42 hours post transfection, the conditioned media were collected, spun down to remove cell debris and used immediately for stimulating non-transfected HEK293T that were serum starved for 3 days in DMEM containing 0.1 % FCS. The conditioned media were applied to the serum starved cells for 15 minutes at 37°C after which the cells were lysed in boiling SDS sample buffer and subjected to Western Blot analysis essentially as described6,31. For detecting the (phosphorylated) Igf1-receptor, the following antibodies were used: anti IGF-I Receptor β (D23H3) XP® Rabbit mAb (#9750, Cell Signaling, 1:1000) and anti Phospho-IGF-I Receptor β (Tyr1135/1136)/Insulin Receptor β (Tyr1150/1151) (19H7) Rabbit mAb (#3024, Cell Signaling, 1:1000). For determining serum IGF-1 levels, we used the IGF-1 Quantikine ELISA kit (R&D Systems), following the manufacturer’s instructions (P3 VIP-Cre heterozygous pups were injected intracortically with the respective AAV and bled at P20).
Perfusions, immunohistochemistry and morphological analysis of Igf1-cKO visual cortices
Mice were anesthetized with 10% ketamine and 1% xylazine in PBS by intraperitoneal injection. When fully anaesthetized, the animals were transcardially perfused with ice cold PBS for five minutes followed by fifteen minutes of cold 4% PFA in PBS. Brains were dissected and post-fixed for one hour at 4°C in 4% PFA, followed by three washes (each for 30 minutes) in cold PBS, and cryoprotection overnight in 20% sucrose in PBS at 4°C. The following day, brains were placed in Tissue-Tek Cryo-OCT compound (Fisher Scientific), frozen on dry ice and stored at −80°C. Coronal sections (20 µm thick) of the visual cortices were subsequently cut using a Leica CM1950 cryostat and used for subsequent experiments.
For immuno-labeling, the slides were blocked for 1 hour with PBS containing 5% normal goat serum and 0.1% Triton X-100 (blocking solution). The samples were incubated overnight with different primary antibodies diluted in blocking solution, washed three times with PBS and then incubated for 45 min at room-temperature with secondary antibodies and/or Hoechst stain (ThermoFisher Scientific). Slides were mounted in FluoromountG (Southern Biotech) and imaged on a Zeiss Axio Imager microscope. The following antibodies were used: Mouse-anti-HA (HA-7, Sigma; 1:1000), Chicken-anti-GFP (GFP-1020, Aves labs ; 1:1500), Goat Anti-Mouse IgG (H+L) Alexa Fluor® 488 (Highly Cross-Adsorbed, Life Technologies; 1:1000), Goat anti-Chicken IgY (H+L) Alexa Fluor® 488 (Life Technologies; 1:1000).
For analyzing the brains of Igf1 VIP-Cre WT and cKO mice, brains of three-week-old WT and cKO littermates were placed on the same slide to minimize variation. After cryosectioning, the slides were either counterstained immediately or stored at −20°C before they were counterstained and imaged. Counterstaining was done with DAPI (4',6-Diamidino-2-Phenylindole, Molecular Probes) in PBS for 15–30 minutes at room temperature, after which the sections were washed once in PBS and mounted in Fluoromount-G (SouthernBiotech). For cell counting experiments, coronal visual cortex sections were imaged using a Zeiss Axio Imager microscope with a 10× objective and typically, 3 × 5 fields-of-view were “stitched” into one compound image. In all cases, image exposures were kept constant throughout a given experiment for each channel. Custom ImageJ and MATLAB macros were used to quantify the area of each cortical layer, the number tdTomato-positive cells per layer, and the size of tdTomato-postive cells. Briefly, regions of interest (ROI) encompassing the visual cortex and its layers were defined based on the DAPI counterstaining. While the width of these ROIs was kept constant throughout the analysis of all sections, the height of the ROIs was adjusted in each image according to the DAPI counterstaining in each section and the areas of each layer in each section were recorded. For analyzing the number and soma size of tdTomato-postive cells in each layer, a threshold for each channel was determined based on multiple user-defined negative regions. Channels were thresholded and binarized, and a mask of each channel was created. The number of tdTomato-cells positive was determined by taking the logical AND of the DAPI and tdTomato channel masks and counting the number of components greater than 4 pixels in size in the double overlap of the masks of the two channels in each layer ROI. The soma size was calculated as the area of these double-overlapping components. Three animals per genotype and 4–6 visual cortex sections per animal were analyzed, and these data were used to determine the mean and SEM of the values reported for each genotype.
Stereotaxically guided surgery and intra-cortical injections of AAV constructs
All surgeries were performed according to protocols approved by the Harvard University Standing Committee on Animal Care and were in accordance with federal guidelines. Surgeries were performed on mice between P14 and P15. Animals were deeply anaesthetized by inhalation of isoflurane (initially 3–5% in O2, maintained with 1–2%) and secured in the stereotaxic apparatus (Kopf). Animal temperature was maintained at 37°C. The fur was shaved and scalp cleaned with betadine and 100% ethanol three times before an incision was made to expose the skull. Injections into the visual cortex were made by drilling a ~0.5 mm burrhole (approximately 2.7 mm lateral, 0.5 mm anterior to lambda) through the skull, inserting a glass pipette to a depth of 200–400 µm and injecting 250 nL of the respective AAV construct at a rate of 100 nL min−1. Five minutes post-injection, the glass pipette was retracted, the scalp sutured and the mouse returned to its home cage. All animals were monitored for at least one hour post-surgery and at 12 hours intervals for the next 5 days. Post-operatively, analgesic (flunixin, 2.5 mg per kg) was administered at 12 hour intervals for 72 hours.
For neonatal injections, pups postnatal day 3–5 were anaesthetized on ice for 2–3 minutes, and secured to a stage where their head was supported using a clay mold using standard lab tape. A beveled glass pipette was lowered into visual cortex (approximately 2 mm lateral, 0.2 mm anterior to lambda), and 50 nL of the respective AAV virus was injected at a rate of 23 nL/sec. Injections were made into eight sites (4 on each hemisphere), and the mouse was then allowed to recover on a 37° warm plate before being returned to the home cage.
For bilateral stereotaxic intra-cortical injections of AAV constructs for visual plasticity experiments, surgeries were performed on mice between P18 and P20. Animals were anesthetized with isofluorane gas (1– 2% in O2), and body temperature was maintained at around 37°C with a heating pad during surgery. The head was held in place by standard mouse stereotaxic frame. The fur was shaved and scalp cleaned with betadine and 100% ethanol three times before an incision was made to expose the skull. Burr holes were drilled into the skull at the point of injection guided by sterotaxic coordinates and blood vessel patterns (approximately 2 mm and 2.7 mm lateral, 0.5 mm anterior to lamba) on both hemispheres. A 28-gauge Hamilton® syringe (701RN) was inserted to a depth of 200–300 µm and 250 nL of the respective AAV construct was injected at the rate of 50 mL/min. Five minutes post-injection, Hamilton syringe was retracted, the scalp sutured and the mouse returned to its home cage. All animals were monitored for at least one hour post-surgery. Post-operatively, analgesic (meloxicam, 5–10 mg/kg) was administered every 24 hours for 2 days.
Electrophysiology
Coronal sections (300 µm thick) containing the primary visual cortex were cut from P19–P21 mice using a Leica VT1000S vibratome in ice-cold choline dissection media (25 mM NaHCO3, 1.25 mM NaH2PO4, 2.5 mM KCl, 7 mM MgCl2, 25 mM glucose, 0.5 mM CaCl2, 110 mM choline chloride, 11.6 mM ascorbic acid, 3.1 mM pyruvic acid). Slices were incubated in artificial cerebral spinal fluid (ACSF, contains 127 mM NaCl, 25 mM NaHCO3, 1.25 mM NaH2PO4, 2.5 mM KCl, 2.5 mM CaCl2, 1 mM MgCl2, 25 mM glucose) at 32°C for 30 minutes immediately after cutting, and subsequently at room temperature. All solutions were saturated with 95% O2/5% CO2, and slices were used within 6 hours of preparation. Whole-cell voltage-clamp recordings were performed in ACSF at room temperature from neurons in primary visual cortex that were identified under fluorescent and DIC optics. Recording pipettes were pulled from borosilicate glass capillary tubing with filaments using a P-1000 micropipette puller (Sutter Instruments) and yielded tips of 2–5.5 MΩ resistance. All experiments were recorded with pipettes filled with 135 mM Cesium Methanesulfonate, 15 mM HEPES, 0.5 mM EGTA, 5 mM TEA-Cl, 1 mM MgCl2, 0.16 mM CaCl2, 2 mM Mg-ATP, 0.3 mM Na-GTP, 10 mM Phosphocreatine (Tris), and 2 mM QX-314-Cl. Osmolarity and pH were adjusted to 310 mOsm and 7.3 with Millipore water and CsOH, respectively. Recordings were sampled at 20 kHz and filtered at 2 kHz. mEPSCs were isolated by holding neurons at −70 mV and exposing them to 0.5 µM tetrodotoxin, 50 µM picrotoxin and 25 µM cyclothiazide and were blocked by application of 25 µM NBQX and 50 µM CPP. mIPSCs were isolated by holding neurons at 0 mV and exposing them to 0.5 µM tetrodotoxin, 25 µM NBQX, and 50 µM CPP and were blocked by 50 µM picrotoxin. Data were acquired using either Clampex10 or custom Matlab software, using either an Axopatch 200B or Multiclamp 700B amplifier, and digitized with a DigiData 1440 data acquisition board (Axon Instruments) or a PCIe-6323 (National Instruments). For measuring miniature postsynaptic currents (minis), cells were allowed to stabilize for at least two minutes.
For paired pulse experiments, no drugs were used in the ACSF. A stimulating electrode (ISO-Flex, A.M.P.I.) was positioned approximately 100 µm below the cell, and 0.1 ms electrical pulses were given while adjusting the stimulus intensity and electrode position until the first pulse was between 100 and 500 pA. Inter-stimulus interval was varied and 10 seconds elapsed between each sweep. Pulse amplitudes were obtained from average sweeps of at least 10 trials. Cells were held at 0 mV to record IPSCs and −70 mV to record EPSCs.
For evoked IPSCs, no drugs were used in the ACSF. Simultaneous paired whole-cell recordings were obtained from a EGFP-expressing VIP neuron and a morphologically identified pyramidal neuron located not more than 5 cell bodies away from the VIP neuron. Both cells were held at 0 mV, and a 5 ms light pulse from a blue LED (Thorlabs) was used to evoke IPSCs. Light intensity and the objective position were varied until the VIP neuron IPSC amplitude was between 200 and 500 pA. Average light power at 470 nm varied from between 0.3 and 0.7 mW over the course of the experiment. Reported ratios were obtained by dividing IPSC amplitudes obtained from an average trace of at least 10 trials.
For electrophysiology experiments, N was set to min n=10 to detect 20% effect size with power 0.95. For experiments to determine average firing rate of VIP neurons, a modified ACSF that promotes increased action potential firing was used containing, 3.5 mM KCl and 0.8 mM CaCl2. Cell-attached patch recordings were obtained from EGFP positive cells. Cells that did not fire an action potential in the first 30 seconds of recording were discarded, and recordings were maintained for at least 30 ten-second sweeps. Average firing rate was determined from the first sweep to the last recorded sweep in which an action potential occurred.
Miniature IPSC and EPSC data were analyzed using Axograph X. Events were identified using a variable amplitude template-based strategy. Templates for each event type were defined as follows: mEPSC: 0.25 ms rise time, 3 ms decay τ, amplitude threshold of −3xSD local noise; mIPSC: 1 ms rise time, 50 ms decay τ, amplitude threshold of 2.5xSD local noise. Local noise was determined by calculating the standard deviation of the current in a 5 ms window before event rise onset. Templates lengths extended 25 ms after rise onset in the case of mEPSCs and 50 ms after rise onset in the case of mIPSCs. Events were discarded if they had a rise time outside the range of 0–3 ms. Statistical significance for all recorded parameters between genotypes was evaluated using a Mann-Whitney U-test on the mean values from individual neurons in a given experiment.
Minis were additionally evaluated for significance using both a Kolmogorov-Smirnov test (KS-test) and Monte Carlo test. For these tests, 50 random minis were sampled from each neuron in each condition to obtain a continuous distribution for each condition that equally weighted each cell in that condition: these distributions are the data shown in the cumulative distribution graphs. One hundred random events were randomly sampled from these distributions for KS-test; and for Monte Carlo tests, 100 random events were randomly sampled from each distribution 1000 times (with replacement), and the means were compared. All significant differences in mini amplitude and frequency were found to be significant by Monte Carlo Test, KS-Test, and Mann-Whitney U-Test of cell means. Since the Mann-Whitney test was found to be the most stringent test, the P-values from Mann-Whitney tests are reported. All data was analyzed blind to genotype or experimental condition. In all conditions, series resistance, holding potential, cell capacitance, and input resistance were recorded and were not found to be significantly different except where noted. Statistical tests were performed using Graphpad Prism and Matlab.
Sholl Analysis
VIP neurons were filled with a patch pipette containing 1% Alexa 647 Hydrazide and the internal solution was allowed to dialyze for at least 30 minutes before slices were fixed in 4% paraformaldehyde for 1 hour at room temperature. Slices were then washed three times for 30 minutes in PBS before slices were mounted in Fluormount-G (Southern Biotech). Images were acquired using a Zeiss Axio Imager microscope with a 20× objective with the use of an Apotome (Zeiss). Neurons were reconstructed using NeuronJ (ImageJ), and Sholl analysis was performed using a custom script in Matlab.
Monocular Deprivation (MD) procedure
Eyelids were trimmed and sutured under isoflurane anesthesia (1– 2% in O2) as previously described31. The integrity of the suture was checked daily and mice were used only if the eyelids remained closed throughout the duration of the deprivation period. One eye was closed for 4 days starting between P26 to P28. The eyelids were reopened immediately prior to recording, and the pupil was checked for clarity.
Mouse visual evoked potential (VEP)
VEPs were recorded from anesthetized mice (50 mg/kg Nembutal and 0.12 mg Chlorprothixene) using standard techniques described previously32. The contra- and the ipsi-lateral eye of the mouse were presented with horizontal black and white sinusoidal bars that alternated contrast (100%) at 2 Hz. A tungsten electrode was inserted into the binocular visual cortex at 2.8 mm from the midline where the visual receptive field was approximately 20° from the vertical meridian. VEPs were recorded by filtering the signal from 0.1–100 Hz and amplifying 10,000 times. VEPs were measured at the cortical depth where the largest amplitude signal was obtained in response to a 0.05 cpd stimulus (400–600 µm); 3–4 repetitions of 20 trials each were averaged in synchrony with the abrupt contrast reversal. The signal was baseline corrected to the mean voltage of the first 50 ms post stimulus onset. VEP amplitude was calculated by finding the minimum voltage (negative peak) within a 50–150 ms post stimulus onset time window. Acuity was calculated only from the deprived eye. For each different SF (spatial frequency), 3–4 repetitions of 20 trials each were averaged in synchrony with the abrupt contrast reversal. VEP amplitude was plotted against the log of the different SF, and the threshold of visual acuity was determined by linear extrapolation to 0 uV.
Animal Husbandry and Colony Management
Igf1 conditional knockout mice15, Ai9 tdTomato reporter mice33, Emx1-Cre34, Pv-Cre35, Gad2-Cre, Sst-Cre, Vip-Cre36 and Ribotag mice8 are available from The Jackson Laboratory.
For routine experimentation, animals were genotyped using a PCR-based strategy; PCR primer sequences are available at the The Jackson Laboratory’s website. For RiboTag experiments, mice homozygous for the RiboTag allele were crossed to mice homozygous for the Cre allele and all experiments were performed with mice double heterozygous for both the RiboTag and the Cre alleles. For Igf1 cKO experiments, mice heterozygous for the Igf1 conditional allele (Igf1flx/wt) and homozygous for the Vip-Cre allele were crossed to mice heterozygous for the Igf1 conditional allele and homozygous for the tdTomato reporter allele. Resulting littermates all had one copy of the Vip-Cre transgene and the tdTomato Cre-reporter and yielded Igf1wt/wt and Igf1flx/flx littermates for experimentation. For injections of AAV constructs in the visual cortices of Cre mice (Vip-, Pv-, Sst-, or Emx1-Cre), mice homozygous for the Cre allele were crossed to wild-type C57Bl6 mice and offspring heterozygous for the Cre allele were used for experiments. The use of animals was approved by the Animal Care and Use Committee of Harvard Medical School and/or the University of California Berkeley.
Extended Data
Extended Data Figure 1. Validation of the sensory stimulation protocol and the RiboTag-based cell-type-specific purification of mRNA.
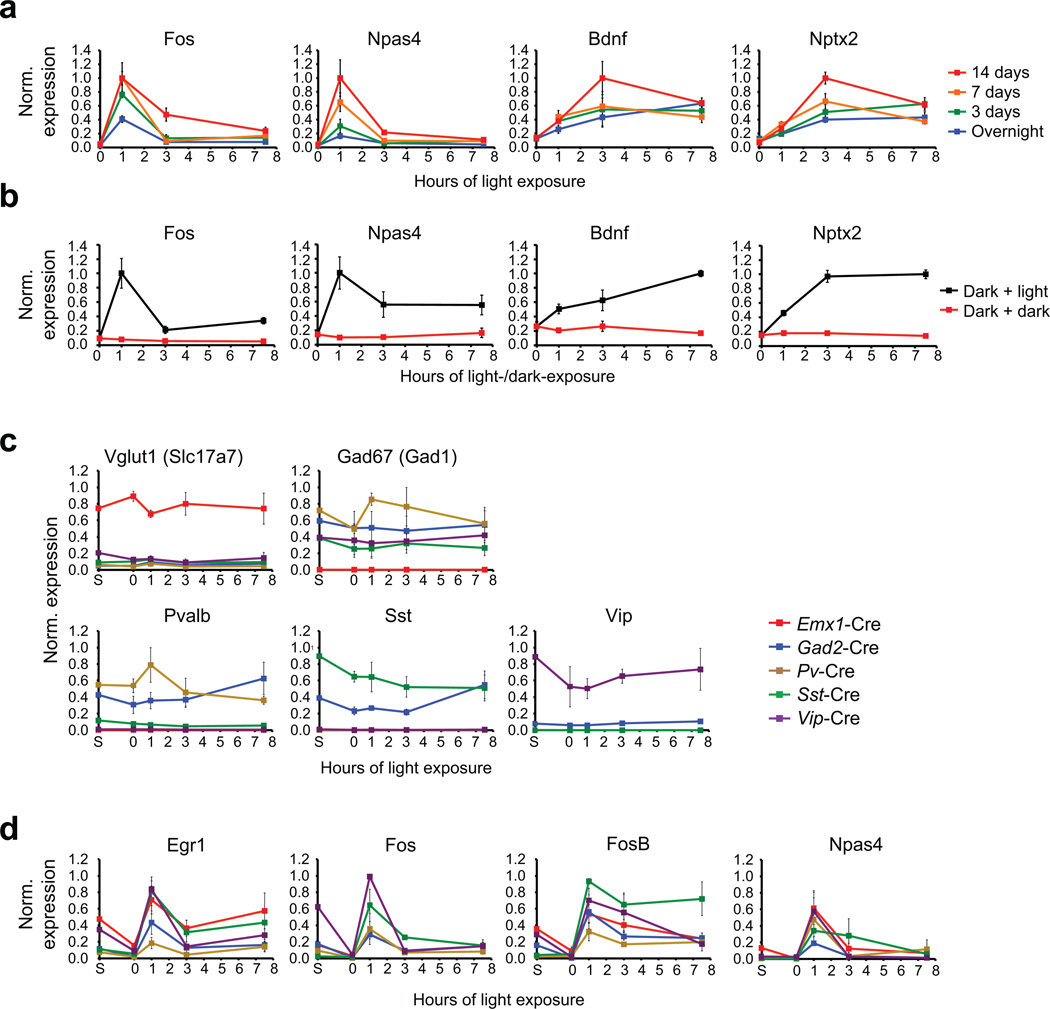
a) Quantitative real-time PCR (qPCR) for known experience-regulated genes on RNA isolated from the visual cortex of mice that were dark-housed for varying durations (overnight, 3 days, 7 days or 14 days) and then either sacrificed in the dark or exposed to light for 1, 3, or 7.5 hours, and then sacrificed. Data are normalized to the maximal value in each dataset and represent the mean and standard error of four biological replicates.
b) qPCR for known experience-regulated genes on RNA isolated from the visual cortex of mice that were dark-housed for 14 days and then either exposed to light for 1, 3, or 7.5 hours (dark + light, black) or kept in the dark during these hours (dark + dark, red). All mice of a given time point were dissected in very close temporal proximity. Data are normalized to the maximal value in each dataset and represent the mean and standard error of four biological replicates.
c) qPCR for known cell-type-specific marker genes on RNA isolated from RiboTag mice expressing Cre in distinct neuronal subtypes. Data are normalized to the maximal value in each dataset and represent the mean and standard error of three biological replicates.
d) qPCR for known early-induced transcription factors on RNA isolated from RiboTag mice expressing Cre in distinct neuronal subtypes. Data are normalized to the maximal value in each dataset and represent the mean and standard error of three biological replicates.
Extended Data Figure 2. Validation of the RiboTag-Seq approach.
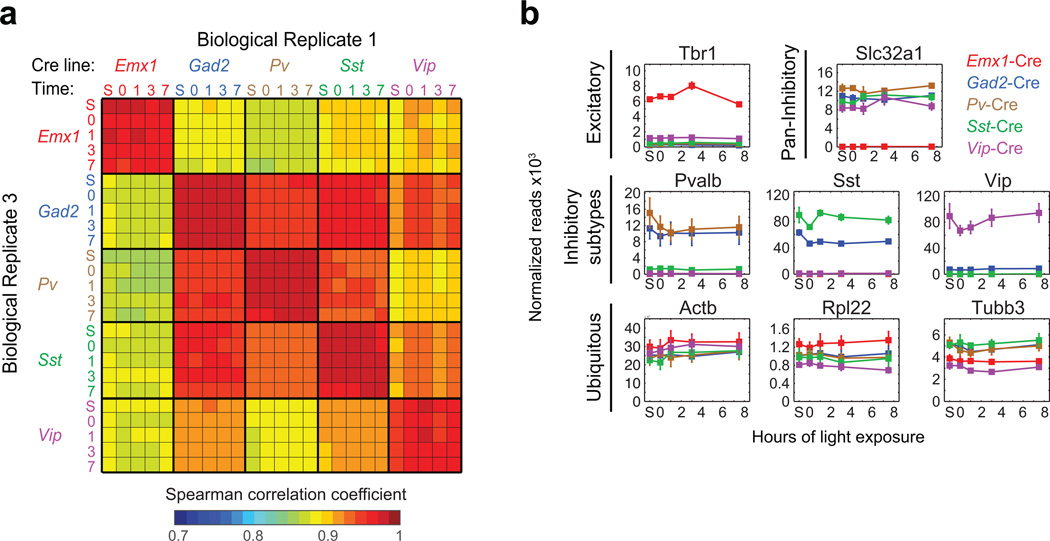
a) Matrix of Spearman correlation coefficients between biological replicates across all samples (Scale of correlation coefficients extends from 0.7 to 1, see color bar) (S = standard housing, 0 = dark-housed only, 1/3/7.5 = 1/3/7.5 hours of light exposure after dark housing).
b) Line plots of RNA-Seq data showing the expression values (normalized reads across all exons of a gene) for cell-type-specific marker genes and ubiquitously expressed house-keeping genes in different Cre lines (Emx1-Cre: red; Gad2-Cre: blue, Pv-Cre: brown, Sst-Cre: green, and Vip-Cre: purple) across all time points of the experiment. Data represent the mean and standard error of three biological replicates.
Extended Data Figure 3. Characterization of the experience-induced gene programs in subtypes of cortical neurons.
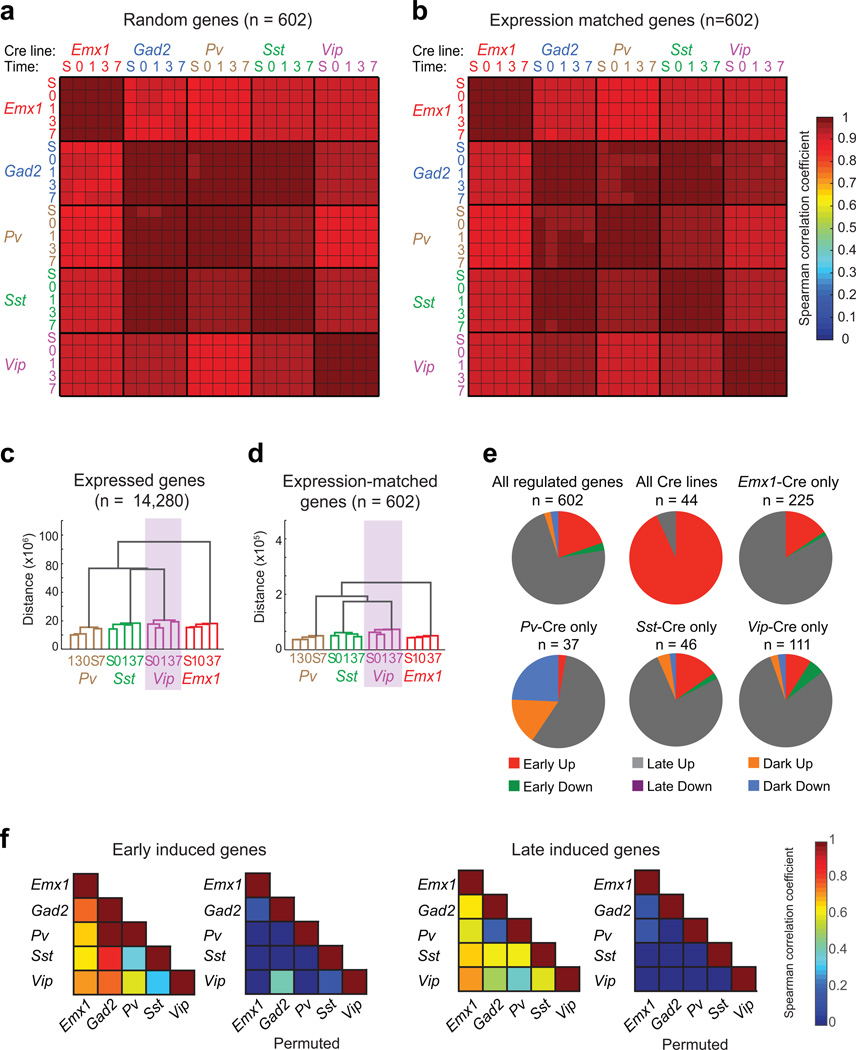
a) Average matrix of Spearman correlation coefficients computed from the expression values of 1000 random sets of 602 genes (including experience-regulated genes, with replacement).
b) Matrix of the Spearman correlation coefficient computed from the expression levels of control transcripts that match the expression distribution of experience-regulated genes (n = 602).
c) Cladogram resulting from hierarchical clustering of all samples (except samples from Gad2-Cre). Cladograms were computed using the mean expression values (i.e. normalized reads across all exons of a gene) for all expressed transcripts (n = 14,280).
d) Cladogram resulting from hierarchical clustering of the mean expression values of a set of control transcripts that match the expression distribution of experience-regulated genes (n = 602).
e) Pie charts showing the subdivision of experience-regulated genes on the basis of kinetics in each set of Cre lines (red: rapidly induced; gray: induced with delayed kinetics; orange: induced only after two weeks of dark housing; green: rapidly suppressed; magenta: suppressed with delayed kinetics; blue: suppressed only after two weeks of dark housing).
f) Left, matrix of Spearman correlation coefficients between Cre lines computed using the mean expression values (normalized reads across all exons of a gene) of early-induced genes one hour after light exposure. Right, matrix of Spearman correlation coefficients between Cre lines computed using the mean expression values of late-induced genes 7.5 hours after light exposure. For each matrix, the correlations upon permuting the expression values are also shown. (Colorbar at right; scale begins at zero.)
Extended Data Figure 4. Characterization of cell-type-specific and experience-induced gene in subtypes of cortical neurons.
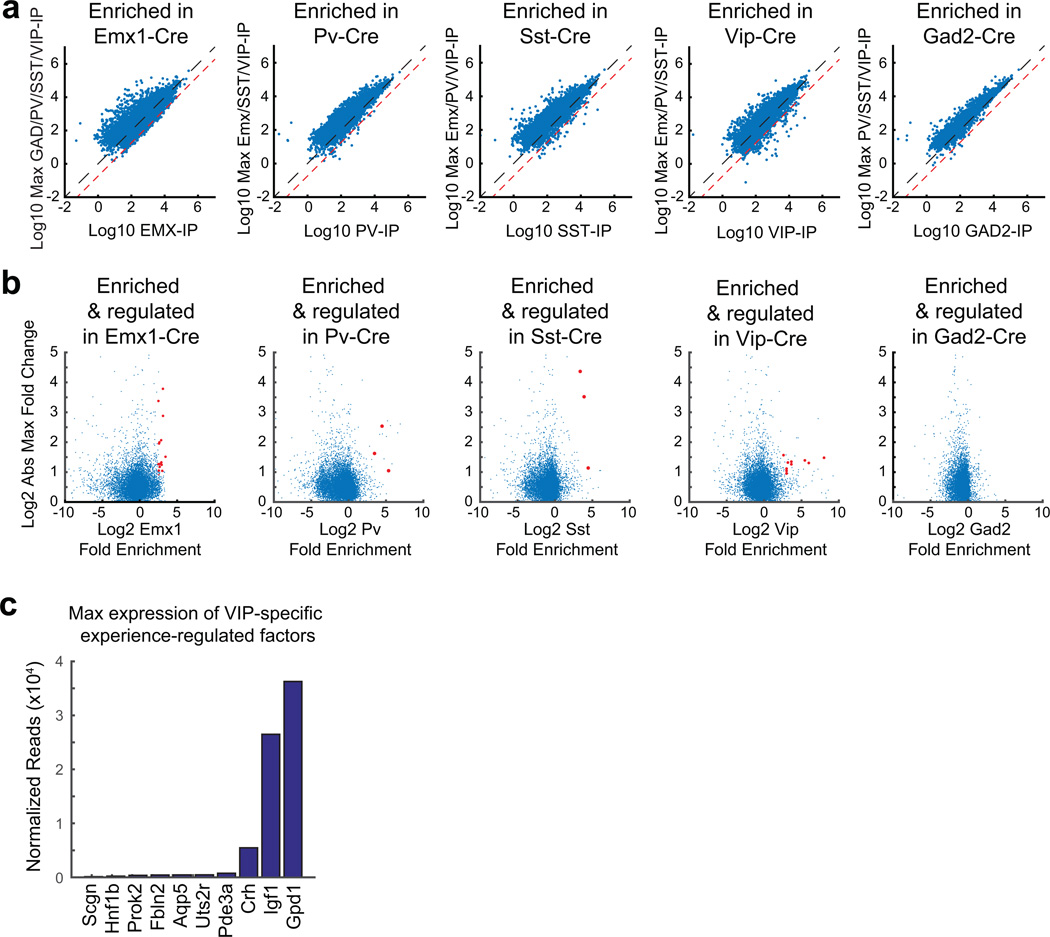
a) Scatter plots showing the Log10 expression values for each expressed gene in a given Cre line (x-axis) plotted against the maximum Log10 expression values for that gene found in all other Cre lines (y-axis). Black line denotes unity, and the red line is the 5.5-fold enrichment threshold set to include Vglut1 as a cell-type-specific gene in Emx1-Cre. Data represent the mean values of three biological replicates.
b) Scatter plots of all expressed genes, for each Cre line plotting the mean Log2 fold-enrichment in that Cre line (x-axis) against the mean Log2 of the absolute value of the maximum fold-change observed in that Cre line. Data represent the mean values of three biological replicates. Genes that pass both enrichment and induction thresholds in 3 of 3 biological replicates are shown in red.
c) Bar graph showing the maximum expression value (in normalized reads) for VIP-neuron-specific experience-regulated genes.
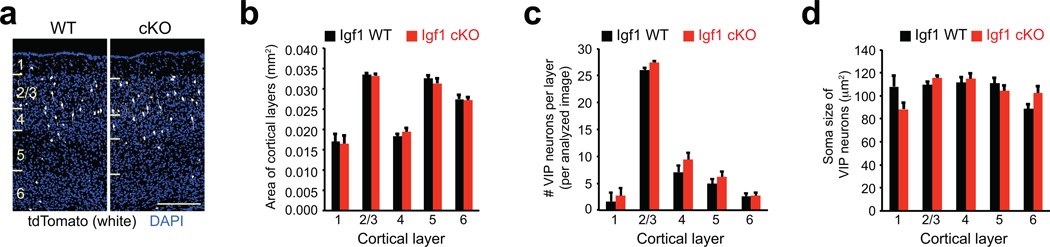
Extended Data Figure 5. Conditional knockout of Igf1 in VIP neurons does not affect cortical morphology or gross morphology of VIP neurons.
a) Example image of cortices from Igf1 WT (Vip-Cre/+, LSL-tdTomato/+, Igf1 wt/wt) or cKO (Vip-Cre/+, LSL-tdTomato/+, Igf1 flx/flx) mice. VIP neurons are labeled in white, with DAPI shown in blue (cortical layers are indicated on the left, scale bar = 200 µm).
b–d) Bar graphs showing the area of each cortical layer (b), number of VIP neurons per image per layer (c), or soma size of VIP neurons (d) in Igf1 WT (black) or cKO (red) mice. Data represent the mean and standard error of three biological replicates.
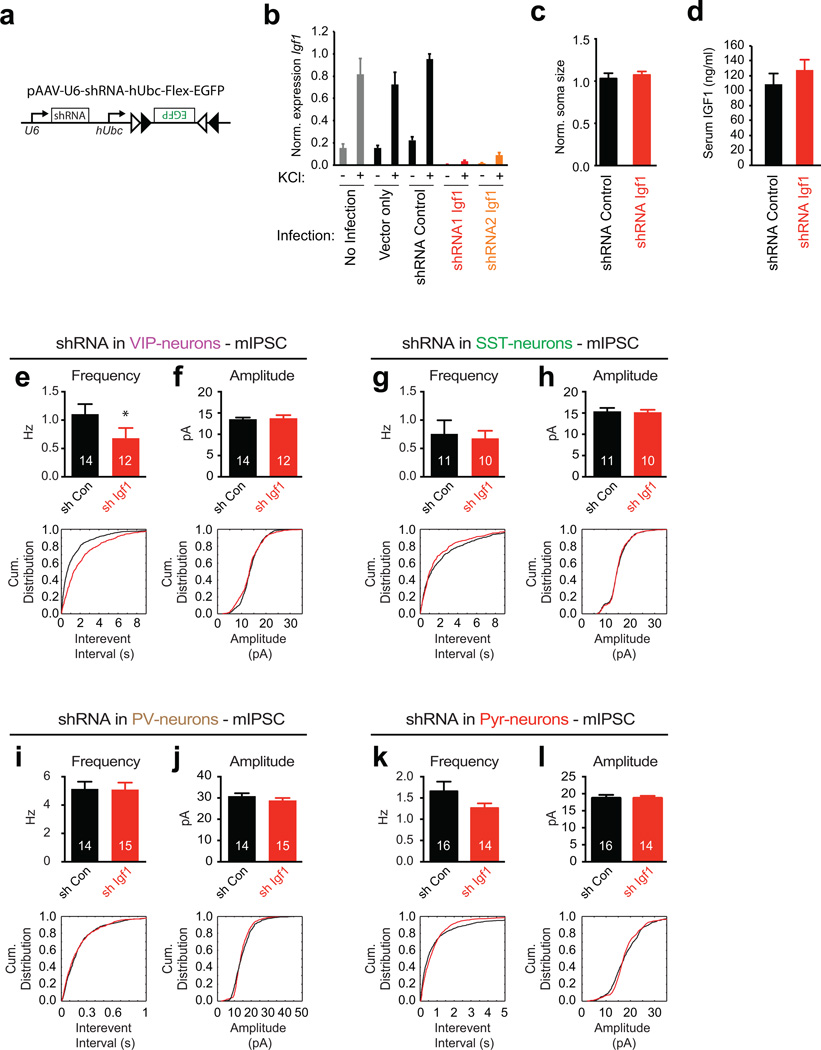
Extended Data Figure 6. Igf1 knockdown in VIP neurons affects inhibitory inputs onto VIP neurons but not onto neighboring neurons.
a) AAV shRNA constructs. shRNA cassettes against Igf1 or a control gene (Luc) were cloned downstream of the U6 promoter into an AAV vector that drives Cre-dependent expression of EGFP.
b) qPCR validation of the efficacy of Igf1 shRNA constructs. Cultured cortical neurons were infected with lentiviral constructs either expressing no shRNA (Vector only), a control shRNA (against Luc) or shRNAs against Igf1. Four days post infection the cultures were quieted overnight with TTX and AP-5 and then harvested either prior to or after being depolarized with 55 mM KCl for 6 hours; RNA was then isolated and qPCR was performed. Data are normalized to the maximal value in each replicate and represent the mean and standard error of three biological replicates.
c) Bar graph showing normalized soma size of P21 visual cortex VIP neurons infected with control shRNA or shRNA targeting Igf1 (shRNA Control: n = 103, shRNA Igf1: n = 174, p =0.41, Mann Whitney U-Test).
d) Bar graphs showing the levels of IGF-1 in the serum of P20 mice that were injected intracortically with AAVs driving the expression of control shRNA (black) or Igf1 shRNA. Data represent the mean and SEM of the serum IGF-1 levels of 4 mice per group.
e–l) Bar graphs and cumulative distribution plots showing mIPSC amplitudes and frequency/interevent interval upon early widespread knockdown of Igf1 in VIP (e,f), SST (g,h), PV (i,j) and excitatory (k,l) neurons after injection of AAVs into P3 cortices of the respective Cre-mice. VIP neurons (identified by EGFP-positive cells in Vip-Cre mice): control shRNA and Igf1 shRNA (Amplitude p = 0.96, Frequency p = 0.04). SST neurons (identified by EGFP-positive cells in Sst-Cre mice): control and Igf1 shRNA (Amplitude p = 0.89, Frequency p = 0.55). PV neurons (identified by EGFP-positive cells in Pv-Cre mice): control and Igf1 shRNA (Amplitude p = 0.084, Frequency p = 0.93). Pyramidal neurons (identified by morphology): control and Igf1 shRNA (Amplitude p = 0.84, Frequency p = 0.15). (All p values are derived from Mann Whitney U-Tests; numbers inside bars indicate the number of cells recorded)
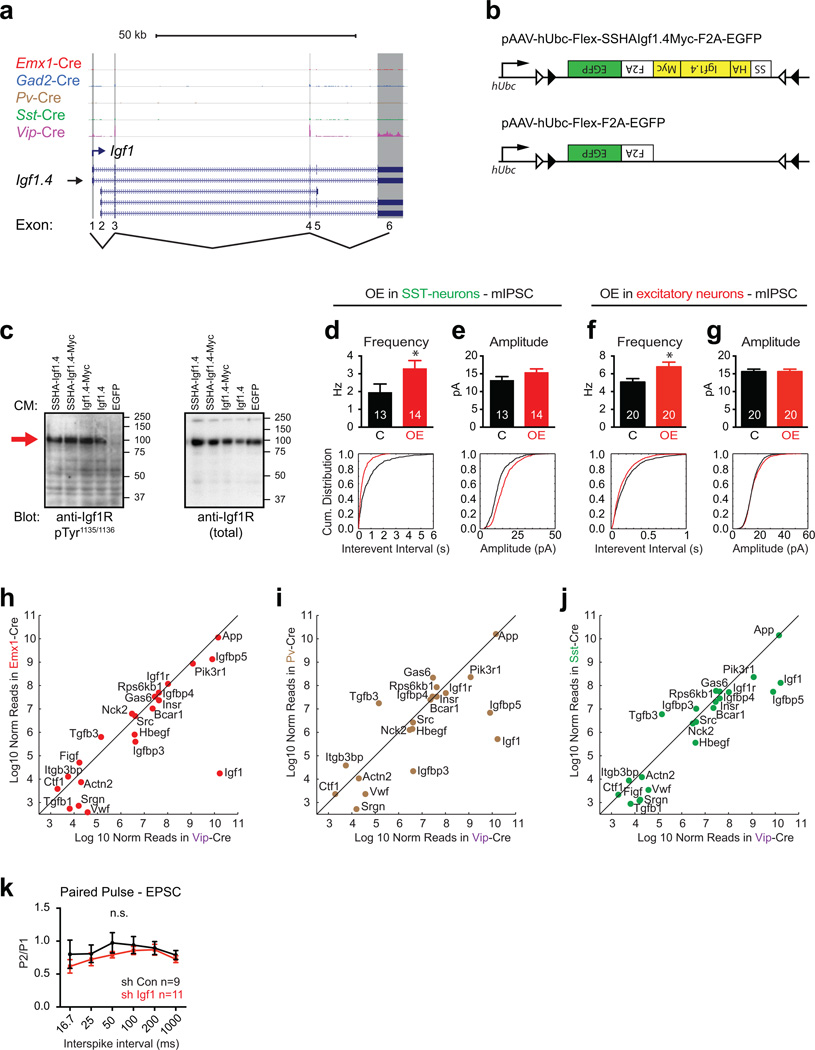
Extended Data Figure 7. Effects of IGF-1 overexpression in excitatory and Sst-positive neurons.
a) RiboTag-Seq identifies Igf1.4 as the major Igf1-isoform expressed in VIP neurons. Representative tracks of histograms of the RNA-Seq reads in each Cre line across the Igf1 genomic locus. Data is from the 7.5 hour light exposure RiboTag-Seq datasets.
b) AAV constructs for the Cre-dependent expression of HA-/Myc-tagged IGF-1 (Igf1.4) and EGFP (i.e. IGF1-OE, top) or of EGFP alone (i.e. control, bottom). F2A = Furin cleavage site followed by the 2A peptide; black and white triangles represent a Cre-dependent Flex-switch.
c) Western blot analysis of IGF1-receptor activation status in lysates of serum starved HEK293T cells that were stimulated with conditioned media (CM) containing epitope-tagged isoforms of IGF-1. CM was produced by transfecting HEK293T cells with the respective construct and collecting the culture media. IGF1-receptor is detected with antibodies against either activated IGF1-receptor (anti Igf1R pTyr1136/1138) or total IGF-1 receptor (anti Igf1R total). (Molecular weight markers are on the right and the arrow indicates the band of the IGF1-receptor).
d,e) Bar graphs and cumulative distribution plots showing mIPSC frequency / interevent interval (d) and amplitude (e) of mIPSCs recorded from EGFP positive neurons in P20 Sst-Cre mice that were intracortically injected with AAVs driving the expression of control (C) or IGF1-OE (OE) constructs (Amplitude, p = 0.16; Frequency, p = 0.01; Mann Whitney U-Test; numbers inside bars indicate the number of cells recorded).
f,g) Bar graphs and cumulative distribution plots showing mIPSC frequency / interevent interval (f) and amplitude (g) of mIPSCs recorded from EGFP positive neurons in P20 Emx1-Cre mice that were intracortically injected with AAVs driving the expression of control (black, n = 20) or IGF1-OE AAVs (red, n = 20). (Amplitude, p = 0.99; Frequency, p = 0.01, Mann Whitney U-Test)
h–j) Scatter plots of IGF-1-interacting proteins showing the Log10 normalized mean expression values in Vip-Cre neurons versus each of the other Cre lines (h - Emx1, i - Pv, j - Sst).
k) Quantification of EPSC paired-pulse recordings from VIP neurons infected with control shRNA (black n = 9) or Igf1 shRNA (red n = 11) expressing AAVs. The ratio of the second EPSC amplitude divided by the first EPSC amplitude is plotted against inter-stimulus interval (p = 0.1, two-way ANOVA).
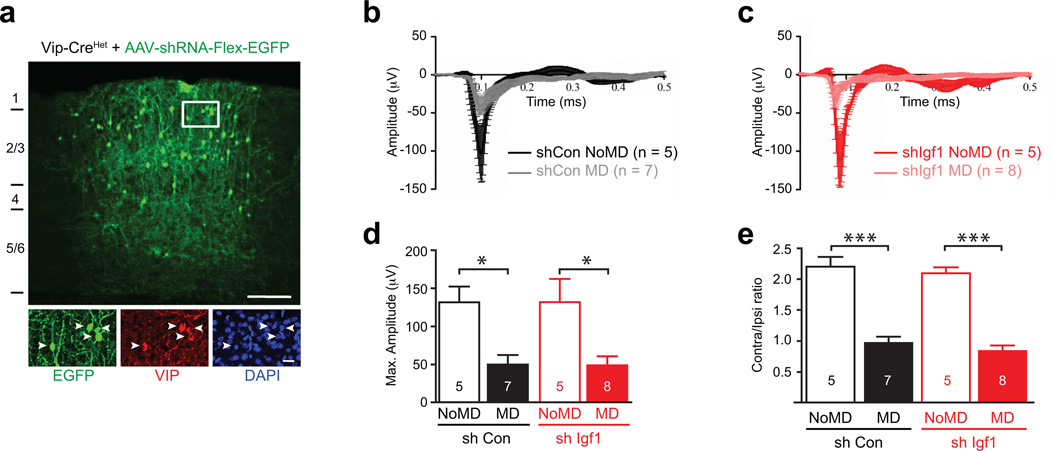
Extended Data Figure 8. VIP neuron-derived IGF-1 does not disrupt ocular dominance plasticity.
a) Widespread infection of VIP neurons by AAV-shRNA-hUbc-Flex-EGFP. High-titer injection of AAVs into the visual cortex of P18-20 Vip-Cre/+ mice leads to infection of the majority VIP neurons (green = EGFP, red = anti-VIP, blue = DAPI, arrowheads = infected VIP neurons; Scale bars 150 µm, 20 µm in the inlet).
b–c) Average of VEP traces recorded in the visual cortices of mice that were injected with AAVs expressing control shRNA (black/grey) or shRNA against Igf1 (red/pink) shRNA and that were (grey, pink) or were not (black, red) subjected to monocular deprivation in the eye contralateral to the recording site (MD vs NoMD, respectively).
d) Monocular deprivation induces a significant reduction in the VEP amplitude in response to low spatial frequency stimulation in mice that had AAVs expressing control shRNA and Igf1 shRNA injected into their visual cortices (control shRNA NoMD, n = 5 mice; control shRNA MD, n = 7 mice; Igf1 shRNA NoMD, n = 5; Igf1 shRNA MD, n = 8. p* < 0.05, Mann Whitney U-Test).
e) Mice that had AAVs expressing control shRNA (black) and Igf1 shRNA (red) injected into their visual cortices display normal ocular dominance plasticity as monocular deprivation (MD) induces a shift to the ispilateral eye in both groups (control shRNA NoMD, n = 5 mice; control shRNA MD, n = 7; Igf1 shRNA NoMD, n = 5 ; Igf1 shRNA MD, n = 8 mice. p** < 0.0001, One-way ANOVA-Tukey post test).
Supplementary Material
Acknowledgments
We thank C. Chen for help with electrophysiology experiments, E. Griffith and T. Cherry for critical reading of the manuscript, P. Zhang for managing the mouse colony and the HMS Biopolymers Facility Next-Gen Sequencing Core for their expertise in constructing Seq libraries and sequencing of the library samples. The ReachR-tdTomato construct was a kind gift from J. Lin, and we thank M. Li for production of the ReachR-virus. This work was funded by fellowships by the Human Frontiers Science Program and the Swiss National Science Foundation (I.S) and the National Institute of Health grant NS028829 (M.E.G.).
Footnotes
Author Contributions
Experiments were designed by A.R.M., I.S., and M.E.G. Experiments were conducted and analyzed by A.R.M., I.S., E.C., A.P., C.P.T., J.E.B., C.M.B., and D.A.H. Experiments were supervised by H.A., M.F., and M.E.G. The manuscript was prepared by A.R.M., I.S., and M.E.G.
Raw data and processed values from RiboTag-Seq have been submitted to the NCBI Gene Expression Omnibus under the accession number GSE77243.
The authors have no competing financial interests.
References
- 1.Hensch TK. Critical period plasticity in local cortical circuits. Nat Rev Neurosci. 2005;6:877–888. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]
- 2.Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Devel Neurobio. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfeffer CK, Xue M, He M, Huang ZJ, Scanziani M. Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nat. Neurosci. 2013;16:1068–1076. doi: 10.1038/nn.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S, Kruglikov I, Huang ZJ, Fishell G, Rudy B. A disinhibitory circuit mediates motor integration in the somatosensory cortex. Nat. Neurosci. 2013;16:1662–1670. doi: 10.1038/nn.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pi H-J, et al. Cortical interneurons that specialize in disinhibitory control. Nature. 2013;503:521–524. doi: 10.1038/nature12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spiegel I, et al. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majdan M, Shatz CJ. Effects of visual experience on activity-dependent gene regulation in cortex. Nat. Neurosci. 2006;9:650–659. doi: 10.1038/nn1674. [DOI] [PubMed] [Google Scholar]
- 8.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubota Y, et al. Selective coexpression of multiple chemical markers defines discrete populations of neocortical GABAergic neurons. Cerebral Cortex. 2011;21:1803–1817. doi: 10.1093/cercor/bhq252. [DOI] [PubMed] [Google Scholar]
- 10.Bondy CA. Transient IGF-I gene expression during the maturation of functionally related central projection neurons. J. Neurosci. 1991;11:3442–3455. doi: 10.1523/JNEUROSCI.11-11-03442.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 12.Cheng CM, et al. Insulin-like growth factor 1 is essential for normal dendritic growth. J. Neurosci. Res. 2003;73:1–9. doi: 10.1002/jnr.10634. [DOI] [PubMed] [Google Scholar]
- 13.Cao P, Maximov A, Südhof TC. Activity-Dependent IGF-1 Exocytosis Is Controlled by the Ca2+-Sensor Synaptotagmin-10. Cell. 2011;145:300–311. doi: 10.1016/j.cell.2011.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishijima T, et al. Neuronal activity drives localized blood-brain-barrier transport of serum insulin-like growth factor-I into the CNS. Neuron. 2010;67:834–846. doi: 10.1016/j.neuron.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Liu JL, et al. Insulin-like growth factor-I affects perinatal lethality and postnatal development in a gene dosage-dependent manner: manipulation using the Cre/loxP system in transgenic mice. Mol. Endocrinol. 1998;12:1452–1462. doi: 10.1210/mend.12.9.0162. [DOI] [PubMed] [Google Scholar]
- 16.Hede MS, et al. E-peptides control bioavailability of IGF-1. PLoS ONE. 2012;7:e51152. doi: 10.1371/journal.pone.0051152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat. Neurosci. 2013;16:1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fagiolini M, Hensch TK. Inhibitory threshold for critical-period activation in primary visual cortex. Nature. 2000;404:183–186. doi: 10.1038/35004582. [DOI] [PubMed] [Google Scholar]
- 19.Davis MF, et al. Inhibitory Neuron Transplantation into Adult Visual Cortex Creates a New Critical Period that Rescues Impaired Vision. Neuron. 2015;86:1055–1066. doi: 10.1016/j.neuron.2015.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu Y, Kaneko M, Tang Y, Alvarez-Buylla A, Stryker MP. A cortical disinhibitory circuit for enhancing adult plasticity. eLife. 2015;4:e05558. doi: 10.7554/eLife.05558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong EJ, McCord AE, Greenberg ME. A Biological Function for the Neuronal Activity-Dependent Component of Bdnf Transcription in the Development of Cortical Inhibition. Neuron. 2008;60:610–624. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503:121–125. doi: 10.1038/nature12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turrigiano G. Too many cooks? Intrinsic and synaptic homeostatic mechanisms in cortical circuit refinement. Annu. Rev. Neurosci. 2011;34:89–103. doi: 10.1146/annurev-neuro-060909-153238. [DOI] [PubMed] [Google Scholar]
References
- 24.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubinson DA, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 27.Schopman NCT, Liu YP, Konstantinova P, Brake, ter O, Berkhout B. Optimization of shRNA inhibitors by variation of the terminal loop sequence. Antiviral Res. 2010;86:204–211. doi: 10.1016/j.antiviral.2010.02.320. [DOI] [PubMed] [Google Scholar]
- 28.Spiegel I, et al. A central role for Necl4 (SynCAM4) in Schwann cell-axon interaction and myelination. Nat. Neurosci. 2007;10:861–869. doi: 10.1038/nn1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 30.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 31.Gordon JA, Stryker MP. Experience-dependent plasticity of binocular responses in the primary visual cortex of the mouse. J. Neurosci. 1996;16:3274–3286. doi: 10.1523/JNEUROSCI.16-10-03274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durand S, et al. NMDA receptor regulation prevents regression of visual cortical function in the absence of Mecp2. Neuron. 2012;76:1078–1090. doi: 10.1016/j.neuron.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorski JA, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. Journal of Neuroscience. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hippenmeyer S, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. Plos Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.