Abstract
HOTAIR (homeobox transcript antisense RNA), one of the prototypical long non-coding RNAs, has been verified overexpressed in multiple carcinomas and has emerged as a promising novel anticancer target. Its well-established role is acting as a predictor of poor prognosis and promoting cancer cell metastasis. Recently, another important mission of HOTAIR was uncovered that targeting HOTAIR caused cancer cell apoptosis. Nevertheless, so far there is no published data elaborating the mechanism. Here, we report that microRNA miR-125a-5p decreases and releases caspase 2 to promote cancer cell apoptosis after HOTAIR knockdown. We applied siRNAs targeting HOTAIR to various cancer cells, and observed apoptosis in all of these cell lines. RNA sequencing detected that miR-125a-5p was decreased after HOTAIR knockdown and miR-125a-5p mimics could rescue the apoptosis induced by HOTAIR deficiency. Luciferase assays identified caspase 2, an initiator caspase, to be a new target of miR-125a-5p. Elevated expression and subsequent cleavage of caspase 2 was observed after HOTAIR knockdown or inhibition of miR-125a-5p. RNAi of caspase 2 could attenuate the apoptosis induced by HOTAIR knockdown. In 80 clinical colon cancer tissues, HOTAIR and miR-125a-5p levels were higher than adjacent tissues, whereas caspase 2 was lower. MiR-125a-5p expression level was significantly correlated with colon tumor size, lymph node metastasis and clinical stage. These findings indicate that miR-125a-5p decreases after HOTAIR knockdown to promote cancer cell apoptosis by releasing caspase 2. Our work reveals a previously unidentified apoptotic mechanism, which might be exploitable in anticancer drug development.
HOTAIR (homeobox transcript antisense RNA), the first discovered trans-acting long non-coding RNA (lncRNA), has been validated having an unignorable role in oncogenic progression. It was discovered aberrantly overexpressed in many kinds of cancer, including acute myeloid leukemia,1 cervical cancer,2 liver cancer,3 urothelial cancer,4 oral squamous cell carcinoma,5 breast cancer,6 glioma cancer,7, 8 ovarian cancer9 and gastric cancer,10, 11 suggesting a promising therapeutic target role in broad-spectrum cancer treatment. The high expression level of HOTAIR in tumors is a powerful predictor of eventual metastasis and bad prognosis.10, 11 The possible metastatic mechanism is that HOTAIR interacts with polycomb repressive complex 2 (PRC2) and lysine-specific demethylase 1A (LSD1) to epigenetically alter the expression of HOXD and some other select genes.12, 13 Although this metastasis-promoting theory is innovative and convincing, it might not fully illustrate HOTAIR's significant role in carcinogenesis.
Recently, a notable mission of HOTAIR that it is essential for cancer cell survival has been identified. It was reported that knockdown of HOTAIR sensitized HepG2 cells to TNF-α, doxorubicin and cisplatin, and the level of HOTAIR declined markedly in HeLa and MCF-7 cells when apoptosis induced by bleomycin happened.12, 13 Moreover, HOTAIR knockdown caused apoptosis in multiple cancer cell lines, and ectopic expression of HOTAIR reduced that.9, 14, 15, 16 Tumor shrinking effect of targeting HOTAIR has been validated in a mouse xenograft model.16 However, there is not a study reporting why HOTAIR knockdown leads to cancer cell apoptosis. As inducing apoptosis is an universal cancer therapeutic regimen, elucidating how apoptosis is triggered would be illuminating for cancer treatment. In this study, we discovered that microRNA miR-125a-5p decreased after HOTAIR knockdown, and its decline derepressed translation of its target, caspase 2(Casp2), and thus caused self-cleavage of CASP2 and activation of the mitochondrial apoptosis pathway. This newly uncovered apoptotic mechanism might be valuable in cancer therapy.
Results
HOTAIR knockdown leads to cancer cell apoptosis
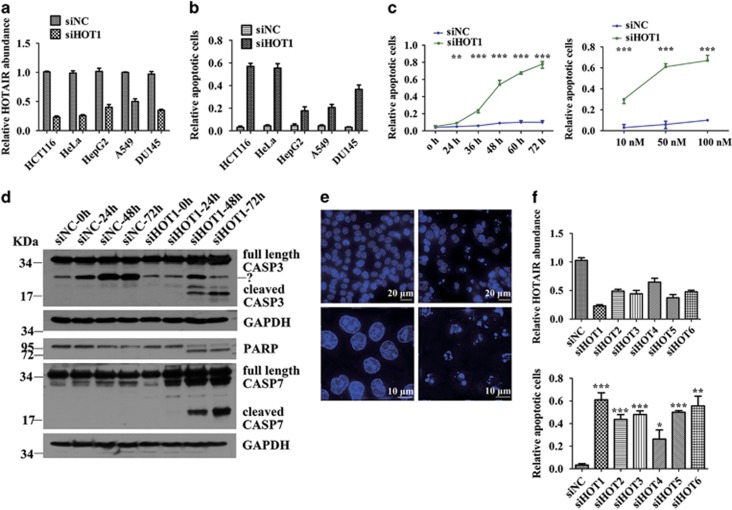
In order to find out whether HOTAIR is essential for cancer cell survival, we transfected a specific siRNA targeting HOTAIR (siHOT1) or scrambled siRNA (siNC) into HCT116, HeLa, HepG2, A549 and DU145 cells. Apoptotic assays were carried out after 48-h transfection. Obvious apoptosis was detected by flow cytometry (FCM) in all of these five cell lines. The amount of apoptotic cells was about 60% in HCT116 and HeLa cells, 20% in HepG2 and A549 cells, and 40% in DU145 cells (Figure 1b). Quantitative real-time PCR confirmed the effective knockdown of HOTAIR in all of these cell lines (Figure 1a). The amount of apoptotic cells significantly increased in a HOTAIR RNAi dose and time-dependent manner (Figure 1c, Supplementary Figures S1a and S1b). Caspase 3 (CASP3), Caspase 7 (CASP7) and poly ADP-ribose polymerase (PARP) cleavage was detected by western blot in HCT116 cells transfected with siHOT1 (Figure 1d). Morphology change in the compaction of nuclear chromatin is one of the characteristics of apoptotic execution. A remarkable degree of DNA compaction was observed in HCT116 cells transfected with siHOT1 (Figure 1e). To ensure these results were not off-target effects of RNA interference, we used six independent siRNA sequences targeting different sites of HOTAIR. Each siRNA depleted HOTAIR and led to concomitant apoptosis (Figure 1f, Supplementary Figure S1c). These data suggest that HOTAIR is vital for cancer cell survival and deficiency of HOTAIR leads to cancer cell apoptosis.
Figure 1.
HOTAIR knockdown leads to cancer cell apoptosis. (a) Quantitative real-time PCR measured the relative abundance of HOTAIR. (b) FCM detected the apoptotic situation of cells transfected with 50 nM siHOT1 or siNC for 48 h. (c) Apoptosis happened in a siHOT dose- and time-dependent manner in HCT116 cells. (d) Western blot detected the cleavage of CASP3, CASP7 and PARP. (e) Hoechst staining showed nuclear condensation of HCT116 cells. (f) Upper, quantitative real-time PCR measured the relative abundance of HOTAIR after 24-h transfection of 50 nM siHOT1-6 or siNC. Lower, FCM detected the apoptotic situation of cells transfected with 50 nM siHOT1-6 or siNC for 48 h. Bars represent mean±S.E.M. from three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 by Student's t-test. All experiments were performed in three biological repeats
MiR-125a-5p declines after HOTAIR knockdown and inhibits apoptosis
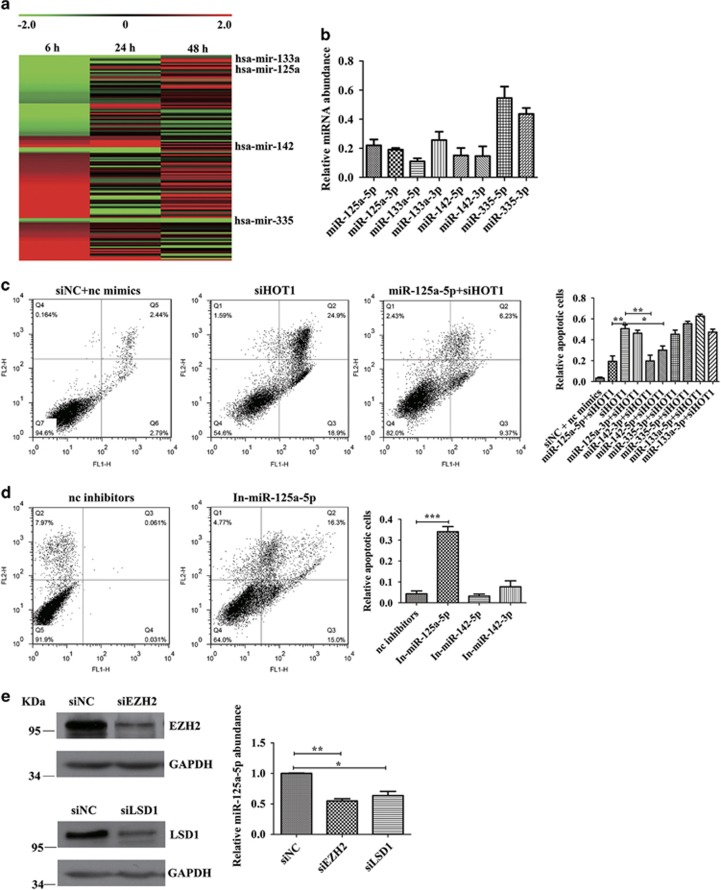
To figure out why HOTAIR is vital for cancer cell survival, we performed RNA deep sequencing to screen ncRNAs regulated by HOTAIR. Total RNA of cells transfected with siHOT1 for 6, 24 and 48 h was extracted and sequenced. Among the 8025 identified long ncRNAs, most are unannotated new lncRNAs and some are pri-miRNAs. Based on fold change >2 and P-value <0.05, 145 long ncRNAs were dysregulated at the sixth hour after siHOT1 transfection, among which the expression of pri-miRNAs hsa-mir-125a, hsa-mir-133a, hsa-mir-142 and hsa-mir-335 declined markedly (Figure 2a). The decline of these pri-miRNAs' processed mature miRNAs was validated with quantitative real-time PCR (Figure 2b). Considering that pri-miRNAs are processed to yield mature 22-nucleotide miRNAs to execute their function, we used miRNA mimics and inhibitors to explore the function of these miRNAs. To restore these miRNAs' expression level, we transfected miRNA mimics into HCT116 cells 6 h before the transfection of siHOT1. MiR-125a-3p, miR-133a-5p, miR-133a-3p, miR-335-5p and miR-335-3p did not rescue the apoptosis caused by HOTAIR knockdown, whereas miR-142-5p, miR-142-3p and miR-125a-5p markedly suppressed the apoptosis (Figure 2c). These data indicate that miR-142-5p, miR-142-3p and miR-125a-5p can inhibit apoptosis. We transfected inhibitors of these three miRNAs into HCT116 cells and observed that reduction of miR-125a-5p triggered apoptosis, whereas miR-142-5p and miR-142-3p did not (Figure 2d). Although miR-125a-5p and miR-125a-3p were both downregulated, only miR-125a-5p could reduce the apoptosis induced by HOTAIR deficiency. Inhibition of miR-125a-5p triggered apoptosis without any additional stimulation. These results suggest that miR-125a-5p, as a suppressor of apoptosis, has a significant role in HOTAIR knockdown induced apoptosis.
Figure 2.
MiR-125a-5p is reduced after HOTAIR knockdown. (a) Heat map of genes with significant induction (red) or repression (green) at sixth hour after 50 nM siHOT1 transfection into HCT116 cells (expressed as a ratio to HCT116 cells transfected with siNC, fold change >2, P-value <0.05, data are log2 transformed). (b) Quantitative real-time PCR validation of a representative panel of genes detected down-regulation by RNA-seq. Raw Ct values were normalized to U6 RNA. Bars represent mean±S.E.M. from three independent experiments. (c) FCM detected the rescue effect of miRNAs on apoptosis induced by HOTAIR knockdown. (d) Inhibition of miR-125a-5p caused apoptosis. (e) miR-125a-5p declined after RNAi of EZH2 and LSD1. Bars represent the mean±S.E.M. from three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 by Student's t-test. All experiments were performed in three biological repeats
To find out the mechanism that HOTAIR regulates the expression of miR-125a-5p, we silenced the co-factors of HOTAIR, enhancer of zeste homolog 2 (EZH2) and LSD1, using siRNAs, and detected the level of miR-125a-5p with quantitative real-time PCR. The level of miR-125a-5p decreased significantly with the suppression of EZH2 and LSD1 (Figure 2e), suggesting that the transcription of miR-125a-5p might be epigenetically regulated by HOTAIR and its co-factors EZH2 and LSD1.
CASP2 is a target of miR-125a-5p and an initiator in HOTAIR deficiency-induced apoptosis
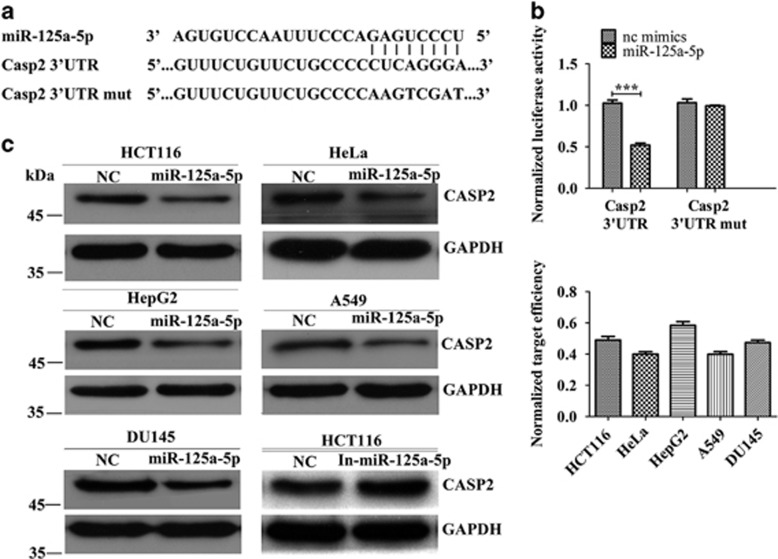
Given that miRNAs function by targeting mRNAs, we used in silico miRNA target prediction tools PicTar and TargetScan to predict potential target mRNAs of miR-125a-5p. Casp2 mRNA was predicted as high-confidence miR-125a-5p target. In all, 200-bp fragments containing wild-type or random mutated putative miR-125a-5p binding sites of Casp2 mRNA' 3′UTR were inserted into psiCHECK™-2 vector (Figure 3a). MiR-125a-5p reduced the luciferase activities of psiCHECK™-2-CASP2 by 50%, whereas the luciferase activities of psiCHECK™-2-CASP2-mut did not (Figure 3b). These data suggest that miR-125a-5p can bind to and repress the translation of Casp2 mRNA. To demonstrate that, we transfected 50 nM miR-125a-5p mimics or 100 nM miR-125a-5p inhibitors (In-miR-125a-5p) into HCT116, HeLa, HepG2, A549 and DU145 cells. Western blot detected about 50% decrease in CASP2 by miR-125a-5p mimics and onefold increase by miR-125a-5p inhibitors (Figure 3c). These results show that Casp2 mRNA is a target of miR-125a-5p.
Figure 3.
Casp2 is a target of miR-125a-5p. (a) Schematic outlines the predicted binding sites of miR-125a-5p on Casp2. (b) Luciferase reporter assay confirmed the target effect of miR-125a-5p on Casp2. (c) Western blot confirmed the target effect of miR-125a-5p on Casp2. Gray value of each CASP2 band was normalized to GAPDH to calculate target efficiency. Bars represent the mean±S.E.M. from three independent experiments. ***P<0.001 by Student's t -test. All experiments were performed in three biological repeats
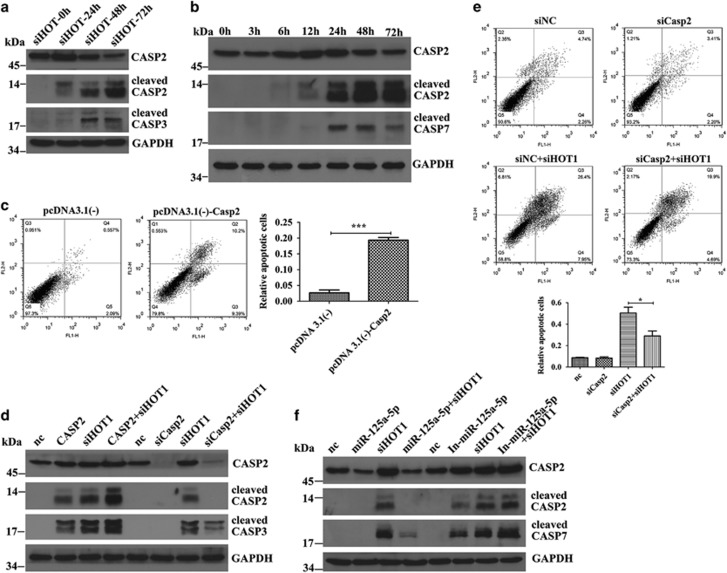
As miR-125a-5p declined after HOTAIR knockdown and miR-125a-5p targets Casp2, we checked the level of CASP2. As shown in Figure 4a, the level of full-length CASP2 markedly increased and then decreased after the silencing of HOTAIR. Cleavage of CASP2 and CASP3 was also detected. These data demonstrate that accumulation and activation of CASP2 happened after HOTAIR RNAi. It has been reported that CASP2 is an initiator protease of the mitochondrial apoptotic pathway and elevated CASP2 can cleave and activate itself to induce apoptosis. To confirm that, we transfected pcDNA3.1(-)-CASP2 into HCT116 cells. After 6 h, western blot detected a slight increase in full-length CASP2 protein but remarkable production of cleaved CASP2. Cleavage of CASP7 also occurred (Figure 4b). FCM detected about 20% early and advanced apoptotic cells about 36 h after pcDNA3.1(-)-CASP2 transfection (Figure 4c). These results prove that piling up of CASP2 contributes to self-activation, caspase cascade and apoptosis. We supposed that the accumulation of CASP2 protein after HOTAIR knockdown might be one of the incentives to apoptosis. To test that, we transfected siRNA targeting Casp2 into HCT116 cells. As shown in Figures 4d and e, the cleavage of CASP3 and the amount of apoptotic cells were diminished, suggesting that CASP2 contributed to apoptosis at upstream of the mitochondrial apoptotic pathway. When miR-125a-5p mimics were co-transfected with siHOT1, the increase in CASP2 was impaired and the cleavage of CASP2 and CASP3 was weakened. Consistently, inhibition of miR-125a-5p gave rise to increase in full-length CASP2 and aggravated cleavage of CASP2 and CASP7 (Figure 4f). These data indicate that CASP2 is modulated by miR-125a-5p to execute its apoptotic initiator role. This part of the results imply that when miR-125a-5p declines after HOTAIR knockdown, its target mRNA Casp2 is released, leading to accumulation of apoptosis initiator CASP2. The accumulated CASP2 cleaves itself and activates mitochondrial apoptosis pathway, finally resulting in apoptosis.
Figure 4.
CASP2 serves as an initiator in HOTAIR knockdown induced apoptosis. (a) Upregulation of full-length CASP2 and cleavage of CASP2 and CASP3 was detected after HOTAIR knockdown. (b) Overexpressed CASP2 cleaved itself and activated CASP7. (c) Overexpression of CASP2 induced apoptosis in HCT116 cells. (d) pcDNA3.1(-)-CASP2 aggravated activation of CASP3 and siCasp2 reduced that. (e) SiCasp2 rescued the apoptosis caused by siHOT1. (f) MiR-125a-5p diminished cleavage of CASP2 and CASP7 and inhibition of miR-125a-5p aggravated that. Bars represent the mean±S.E.M. from three independent experiments. *P<0.05, ***P<0.001 by Student's t-test. All experiments were performed in three biological repeats
Besides, we detected the level of P53 and B-cell lymphoma 2 (BCL-2) after silencing HOTAIR and found that the level of BCL-2 did not change while P53 was upregulated, implying a potential role of P53 in the apoptosis caused by HOTAIR deficiency (Supplementary Figure S2a). In addition, we confirmed that P53 was a target of miR-125a-5p (Supplementary Figures S2b–d). Taken together, after HOTAIR was knocked down, because of the declined expression of miR-125a-5p, its apoptotic targets CASP2 and P53 were accumulated and finally lead to apoptosis.
MiR-125a-5p is an independent marker of colon cancer progression
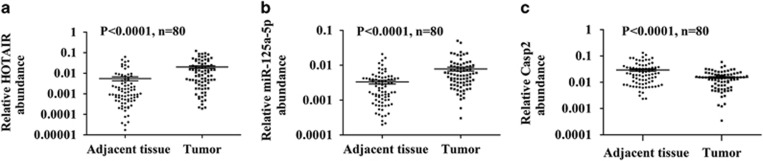
To evaluate the expression level of HOTAIR, miR-125a-5p and Casp2 in colon cancer cells, we performed quantitative real-time PCR in 80 paired cancerous and adjacent noncancerous tissues of colon cancer patients (Table 1). As shown in Figure 5, 63 cases (79%) exhibited higher levels of HOTAIR in tumors than in adjacent nontumorous tissues (mean ratio of 3.68-fold, P<0.0001). MiR-125a-5p level was significantly elevated in 64 cancerous tissues (80%) (mean ratio of 2.36-fold, P<0.0001), whereas Casp2 was decreased in 59 tumorous tissues (74%) (mean ratio of 0.52-fold, P<0.0001). Higher HOTAIR level has been proved to be associated with larger tumor size, advanced pathological stage, extensive metastasis and poorer survival. Based on the Mann–Whitney test, the high miR-125a-5p group showed greater incidence of bigger tumor size (P=0.023), lymph node metastasis (P=0.037) and clinical stage (P=0.025). No significant correlation between miR-125a-5p level and patient age (P=0.981) or gender (P=0.992) was found (Table 1). These results suggest that miR-125a-5p is an independent marker of colorectal cancer progression.
Table 1. Relationship of miR-125a-5p expression with clinicopathologic factors in colon cancer.
| Clinicopathologic factors | N (%), N=80 | Relative miR-125a-5p expression | P-value |
|---|---|---|---|
| Gender | P=0.992 | ||
| Male | 51 (64) | 3.49 (0.37–13.91) | |
| Female | 29 (36) | 5.06 (0.14–29.43) | |
| Age | |||
| ≤66 | 40 (50) | 4.27 (0.14–27.16) | P=0.981 |
| >66 | 40 (50) | 3.86 (0.53–29.43) | |
| Tumor size | P=0.023 | ||
| ≤5 cm | 51 (64) | 2.88 (0.14–14.16) | |
| >5 cm | 29 (36) | 6.14 (0.65–29.43) | |
| Lymph node metastasis | P=0.037 | ||
| Negative | 46 (58) | 2.90 (0.14–14.16) | |
| Positive | 34 (42) | 5.63 (0.37–29.43) | |
| Clinical stage | P=0.025 | ||
| I–II | 46 (58) | 2.87 (0.14–14.16) | |
| III –IV | 34 (42) | 5.67 (0.37–29.43) |
Figure 5.
HOTAIR, miR-125a-5p and Casp2 expression in 80 pairs human colon cancer tissues and adjacent noncancerous tissues. (a) The dot represents relative HOTAIR abundance of each sample. The expression level of HOTAIR was calculated by the 2ΔCt method, ΔCt=Ct(GAPDH)—Ct (HOTAIR). The Y axis was log10 transformed. (b) The expression level of miR-125a-5p was normalized to U6 RNA. (c) The expression level of Casp2 was normalized to GAPDH. The bars illustrated S.E.M. and the significant differences between samples were analyzed using Student's t-test
Discussion
HOTAIR, a typical trans-acting lncRNA, is highly expressed in a variety of cancers and thus have emerged as a potential anticancer target. Its well-established role is that the increased HOTAIR expression is a biomarker of poor prognosis, and it acts as a modular scaffold of histone modification complex PRC2 and LSD1 to regulate the expression of select genes and promote cancer cell metastasis.12, 17, 18 Recently, another significant role of HOTAIR was reported and that targeting HOTAIR led to cancer cell apoptosis. However, the mechanism remained unknown. In this study, we unraveled that miR-125a-5p decreased after HOTAIR knockdown, which brought about cleavage of proapoptosis protein CASP2, and therefore activated the mitochondrial apoptosis pathway.
Yang Z et al.12 reported that RNAi of HOTAIR sensitized HepG2 cells to TNF-α, doxorubicin and cisplatin. The expression of HOTAIR decreased in cells treated with calycosin, genistein or bleomycin.13, 17 HOTAIR knockdown induced apoptosis in multiple kinds of cancer cells, and overexpression of HOTAIR inhibited it.8, 9, 14, 15, 16 In our study, we observed apoptosis in all the cell lines transfected with siHOT and found a dose- and time-dependent correlation between cell viability and siHOT. Remarkable DNA compaction and pronounced CASP3, CASP7 and PARP cleavage took place after HOTAIR knockdown. All of these results confirmed the previous notion that HOTAIR deficiency led to cancer cell apoptosis. However, we did not detect any remedial effect of HOTAIR on apoptosis induced by cisplatin or TNF-α (data not shown), which was inconsistent with previous research.15 This inconsistency may be due to different apoptosis inducers. In summary, except for the role of promoting metastasis and predicting bad prognosis, HOTAIR is vital for cancer cell survival and its knockdown causes cancer cell apoptosis; therefore targeting HOTAIR might be promising in cancer therapy.
MiR-125 family is composed of three homologs, miR-125a, miR-125b-1 and miR-125-2. MiR-125a gene is located at 19q13 and processing of hsa-mir-125a generates miR-125a-5p and miR-125a-3p. They target different mRNAs and own controversial properties in different cellular context. MiR-125a-3p was reported suppressing proliferation and migration and inducing apoptosis in multiple cancer cells.18, 19 Unlike miR-125a-3p, miR-125a-5p acts as either oncogene or suppressor gene depending on the specific cell context. MiR-125a-5p is downregulated in breast cancer,20 ovarian cancer21 and lung cancer.22 It inhibits glioblastoma cell proliferation by targeting tafazzin.23 On the other hand, higher expression level of miR-125a-5p is observed in nasal pharyngeal cancer cells, multiple myeloma cells, human prostate cancer cells and the serum of non-small-cell lung cancer patients.24, 25, 26, 27 By targeting P53 directly, it promotes proliferation, migration and invasion of HONE1 cells.24 Inhibition of miR-125a-5p in multiple myeloma cells reduced cell growth, increased apoptosis and dampened cell migration.25 In our study, we observed that the level of pri-miRNA hsa-mir-125a and its mature forms miR-125a-5p and miR-125a-3p declined after inhibition of HOTAIR, and miR-125a-5p declined after inhibition of EZH2 and LSD1, which imply that the transcription of hsa-mir-125a might be managed by HOTAIR and its epigenetic co-factors EZH2 and LSD1. Although PRC2 complex was usually considered to suppress gene expression, research has proven that it can also promote gene expression.28, 29 Unlike miR-125a-3p, miR-125a-5p markedly reduced the apoptosis caused by HOTAIR knockdown and inhibition of miR-125a-5p led to apoptosis in HCT116 cells. We also observed that miR-125a-5p functions as an oncogene in colon cancer cells via targeting Casp2 and P53. Furthermore, just like HOTAIR, the amount of miR-125a-5p in colon cancerous tissues was more than that in adjacent healthy tissues, and it was positively correlated with tumor size, lymph node metastasis and clinical stage. These results reveal that miR-125a-5p is an oncogene in colon cancer cells and it is an independent marker for colon cancer. Since miR-125a-5p efficiently rescued the apoptosis triggered by HOTAIR knockdown, it's possible that miR-125a-5p acts at upstream of the apoptosis signaling pathway.
As CASP2 displays the properties of both initiator and effector caspase, it is hard to define CASP2 as a canonical initiator or effector. Like the initiator caspase 9 (CASP9), CASP2 contains a N-terminal caspase recruitment domain (CARD), followed by a large subunit containing the active site (p19) and a small subunit (p12).30 Its activation occurs by proximity-induced dimerization and autoproteolysis.31 Ectopic overexpression of CASP2 is sufficient for its activation.32, 33, 34 Activated CASP2 arouses caspase cascade and apoptosis via cleaving BH3 interacting domain death agonist (BID) and releasing cytochrome c into cytoplasm.35 In our study, CASP2 was upregulated and then cleaved after HOTAIR knockdown, and siCasp2 rescued the apoptosis caused by HOTAIR knockdown, suggesting that CASP2 served as an initiator in this process. Casp2 mRNA has been validated to be the target of miR-31,36 miR-34a,37 miR-17,34 and miR-96.38 In our study, we found Casp2 was targeted by miR-125a-5p in HCT116, HeLa, HepG2, A549 and DU145 cells. MiR-125a-5p diminished the cleavage of CASP2 and inhibition of miR-125a-5p aggravated that, implying that HOTAIR indirectly modulates the translation and activation of Casp2 via miR-125a-5p. In addition, we found that P53 was upregulated after HOTAIR knockdown, and confirmed that P53 was a target of miR-125a-5p in colon cancer cells, which implied that P53 also had a role in the apoptotic pathway triggered by HOTAIR inhibition. In summary, when HOTAIR was knocked down, the level of miR-125a-5p therewith descended, and thus Casp2 and P53 mRNA was released. The increased CASP2 and P53 caused apoptosis.
Reduced expression of Casp2 was reported in blood cancer and several solid tumors.39, 40, 41 High Casp2 level is associated with poor survival in human neuroblastoma patients.42 Even so, mutations of CASP2 are rare in various human cancers.43, 44 Therefore, direct mutational inactivation of CASP2 might not fully explain its decline or loss-of-function in human tumors. In our study, we observed for the first time that Casp2 was downregulated in clinical colon cancer tissues. The elevated expression of miR-125a-5p and its targeting effect on Casp2 might indirectly explain CASP2' dereliction of duty in cancer.
In conclusion, we find targeting HOTAIR can induce cancer cell apoptosis, in a manner depending on reducing the level of miR-125a-5p and activating of apoptotic initiator CASP2 and P53. Also, we find miR-125a-5p is an oncogene in colon cancer, and identified Casp2 is a new target of miR-125a-5p. For the first time, we explain the molecular mechanism of the apoptosis induced by HOTAIR knockdown and this multi-layer modulation might be valuable in cancer treatment strategy exploration.
Materials and Methods
Cell culture
HCT116, HeLa, A549, DU145 and HepG2 were purchased from ATCC (Manassas, VA, USA). These cancer cell lines were cultured in Dulbecco's modified Eagle's high glucose medium supplemented with 10% fetal bovine serum, 50 IU/ml penicillin, and 50 mg/ml streptomycin (Invitrogen, Carlsbad, CA, USA) and were maintained at 37 °C in a humidified incubator in the presence of 5% CO2.
Transfection
SiCasp2, siEZH2, siLSD1, six different siRNAs targeting HOTAIR (siHOT1–6) and scrambled negative control siRNA (siNC) were synthesized in GenePharma (Shanghai, China). Their sequences are listed in Supplementary Table S1. MicroRNA angomirs and antagomirs were purchased from RiboBio (Guangzhou, China). RNAs were transfected into cancer cells using Lipofectamine iMAX (Invitrogen).
RNA extraction and quantitative real-time PCR
Total RNA was extracted from tissues or cultured cells using TRIzol reagent (Invitrogen). Reverse transcription reactions were performed using the reverse transcription system kit (Aidlab, Beijing, China). Quantitative real-time PCR analysis was performed in triplicate with a CFX96 real-time system (Bio-Rad, Hercules, CA, USA) using GoTaq qPCR Master Mix (Promega, Madison, WI, USA). Relative mRNA and miRNA expression levels were normalized to GAPDH or U6 snoRNA and relative expression fold change was calculated with the 2−ΔΔCT method. Bulge-Loop™ miRNA qRT-PCR Primer Sets were purchased from RiboBio. Other genes' primer sequences are provided in Supplementary Table S2.
Western blot analysis
Antibodies against CASP2, CASP3, CASP7, PARP, EZH2 and P53 were purchased from Cell Signaling Technology (Danvers, BSN, USA). Antibody against GAPDH was purchased from Santa Cruz (Dallas, TX, USA). Antibody against BCL-2 was purchased from Proteintech (Chicago, IL, USA). Antibody against LSD1 was purchased from Abcam (Cambridge, UK).
Flow cytometric analysis of apoptosis
Cells were harvested at the indicated time points. After double staining with FITC-Annexin V and propidium iodide (Beyotime, Jiangsu, China), cells were analyzed with a flow cytometry (FCM, FACS Calibur, BD Biosciences, Franklin Lakes, NJ, USA) equipped with CellQuest software (BD Biosciences). Measurements were repeated independently three times.
RNA sequencing and expression analysis
Total RNA from cells transfected with siHOT1 for 6, 24 and 48 h was extracted using RNeasy mini kit (Qiagen, Venlo, The Netherlands). The quality and integrity of the RNA samples were examined using the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). After poly(A) selection, RNA was fragmented and then converted into cDNA sequencing library by using a TrueSeq DNA library preparation kit (Illumina, San Diego, CA, USA). The cDNA was end-repaired, adaptor ligated, PCR amplified and then sequenced on the Illumina Hiseq2000 platform with 100-bp pair-end sequencing strategy. In all, 4 GB of raw data were obtained for each sample. The SOAP software (http://soap.genomics.org.cn/, BGI, Guangzhou, China) was used to align the filtered reads to gencode.v19.long_noncoding_RNAs.gtf downloaded from ENCODE. Gene expression was calculated using the RPKM method (reads per kilobase transcriptome per million reads).
Luciferase reporter assay
In all, 200 bp of the Casp2 and P53 3′ UTR sequence containing the putative miR-125a-5p binding sites or the mutant miR-125a-5p binding sites were cloned into the psiCHECK™-2 vector to generate psiCHECK™-2-CASP2, psiCHECK™-2-CASP2-mut, psiCHECK™-2-P53, psiCHECK™-2-P53-mut plasmids. Cells grown in 24-well plates were transfected with 50 nM negative control miRNA mimics (nc miRNA mimics) or 50 nM miR-125a-5p mimics. Six hours later, plasmids were transfected. Luciferase activity was assayed 24 h after transfection, using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. All transfection experiments were performed in triplicate.
Clinical samples analysis
Totally 80 pairs of colorectal tumor tissues and adjacent noncancerous tissues were obtained from patients who underwent surgery at the ChaoYang Hospital between 2013 and 2014. All specimens were immediately frozen in liquid nitrogen and stored at −80 °C until RNA extraction. Informed consent was obtained from all the patients and no patient received chemotherapy or radiotherapy before surgery. Clinicopathologic data are listed in Table 1.
Statistical analysis
The Mann–Whitney test was used to estimate the significance of differences between miR-125a-5p expression of two different clinical groups. Other data were analyzed using Student's t-test. Data are shown as mean values S.E.M., and P-value <0.05 was considered statistically significant. All statistical analyses were performed using GraphPad Prism Software (La Jolla, CA, USA).
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (nos. 31470807, 31270872 and 31200610), National Key Basic Research Program of China (nos. 2010CB912203 and 2011CB915504) and funds from State Key Laboratory of Protein and Plant Gene Research, College of Life Sciences, Peking University.
Glossary
- HOTAIR
homeobox transcript antisense RNA
- lncRNA
long non-coding RNA
- PRC2
polycomb repressive complex 2
- EZH2
enhancer of zeste homolog 2
- LSD1
lysine-specific demethylase 1A
- Casp2
caspase 2
- siHOT
siRNA targeting HOTAIR
- siNC
scrambled siRNA
- CASP3
caspase 3
- CASP7
caspase 7
- PARP
poly ADP-ribose polymerase
- In-miR-125a-5p
miR-125a-5p inhibitors
- BCL-2
B-cell lymphoma 2
- CARD
caspase recruitment domain
- CASP9
caspase 9
- BID
BH3 interacting domain death agonist
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by E Candi
Supplementary Material
References
- Hao S, Shao Z. HOTAIR is upregulated in acute myeloid leukemia and that indicates a poor prognosis. Int J Clin Exp Pathol 2015; 8: 7223–7228. [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Mandal P, Sadhukhan T, Roy Chowdhury R, Ranjan Mondal N, Chakravarty B et al. Bridging links between long noncoding RNA HOTAIR and HPV oncoprotein E7 in cervical cancer pathogenesis. Sci Rep 2015; 5: 11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang H et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J Hepatol 2015; 63: 886–895. [DOI] [PubMed] [Google Scholar]
- Heubach J, Monsior J, Deenen R, Niegisch G, Szarvas T, Niedworok C et al. The long noncoding RNA HOTAIR has tissue and cell type-dependent effects on HOX gene expression and phenotype of urothelial cancer cells. Mol Cancer 2015; 14: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Xie H. Expression of long noncoding RNA-HOX transcript antisense intergenic RNA in oral squamous cell carcinoma and effect on cell growth. Tumour Biol 2015; 36: 8573–8578. [DOI] [PubMed] [Google Scholar]
- Wang YL, Overstreet AM, Chen MS, Wang J, Zhao HJ, Ho PC et al. Combined inhibition of EGFR and c-ABL suppresses the growth of triple-negative breast cancer growth through inhibition of HOTAIR. Oncotarget 2015; 6: 11150–11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Ren Y, Zhang J, Zhang C, Zhang K, Han L et al. HOTAIR is a therapeutic target in glioblastoma. Oncotarget 2015; 6: 8353–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori C, Kapranov P, Penas C, Peschansky V, Volmar C-H, Sarkaria JN et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci USA 2015; 112: 8326–8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JJ, Wang Y, Ding JX, Jin HY, Yang G, Hua KQ. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res 2015; 333: 238–248. [DOI] [PubMed] [Google Scholar]
- Zhang ZZ, Shen ZY, Shen YY, Zhao EH, Wang M, Wang CJ et al. HOTAIR long noncoding RNA promotes gastric cancer metastasis through suppression of poly r(C)-binding protein (PCBP) 1. Mol Cancer Ther 2015; 14: 1162–1170. [DOI] [PubMed] [Google Scholar]
- Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis 2014; 35: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011; 18: 1243–1250. [DOI] [PubMed] [Google Scholar]
- Ozgur E, Mert U, Isin M, Okutan M, Dalay N, Gezer U. Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin Exp Med 2013; 13: 119–126. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren X et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol 2015; 46: 2586–2594. [DOI] [PubMed] [Google Scholar]
- Chiyomaru T, Fukuhara S, Saini S, Majid S, Deng G, Shahryari V et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. J Biol Chem 2014; 289: 12550–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014; 25: 666–681. [DOI] [PubMed] [Google Scholar]
- Chen J, Lin C, Yong W, Ye Y, Huang Z. Calycosin and genistein induce apoptosis by inactivation of HOTAIR/p-Akt signaling pathway in human breast cancer MCF-7 cells. Cell Physiol Biochem 2015; 35: 722–728. [DOI] [PubMed] [Google Scholar]
- Yin F, Zhang JN, Wang SW, Zhou CH, Zhao MM, Fan WH et al. MiR-125a-3p regulates glioma apoptosis and invasion by regulating Nrg1. PloS One 2015; 10: e0116759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio-Many L, Grossman H, Shomron N, Chuderland D, Shalgi R. microRNA-125a-3p reduces cell proliferation and migration by targeting Fyn. J Cell Sci 2013; 126(Pt 13): 2867–2876. [DOI] [PubMed] [Google Scholar]
- Hsieh TH, Hsu CY, Tsai CF, Long CY, Chai CY, Hou MF et al. miR-125a-5p is a prognostic biomarker that targets HDAC4 to suppress breast tumorigenesis. Oncotarget 2015; 6: 494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellapa A, Bitterman P, Sharma S, Guirguis AS, Bahr JM, Basu S et al. Interleukin 16 expression changes in association with ovarian malignant transformation. Am J Obstet Gynecol 2014; 210: 272 e271–210. [DOI] [PubMed] [Google Scholar]
- Zhu WY, Luo B, An JY, He JY, Chen DD, Xu LY et al. Differential expression of miR-125a-5p and let-7e predicts the progression and prognosis of non-small cell lung cancer. Cancer Invest 2014; 32: 394–401. [DOI] [PubMed] [Google Scholar]
- Yuan J, Xiao G, Peng G, Liu D, Wang Z, Liao Y et al. MiRNA-125a-5p inhibits glioblastoma cell proliferation and promotes cell differentiation by targeting TAZ. Biochem Biophys Res Commun 2015; 457: 171–176. [DOI] [PubMed] [Google Scholar]
- Gao W, Chan JY, Wong TS. Curcumin exerts inhibitory effects on undifferentiated nasopharyngeal carcinoma by inhibiting the expression of miR-125a-5p. Clin Sci (London, England: 1979) 2014; 127: 571–579. [DOI] [PubMed] [Google Scholar]
- Leotta M, Biamonte L, Raimondi L, Ronchetti D, Di Martino MT, Botta C et al. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells. J Cell Physiol 2014; 229: 2106–2116. [DOI] [PubMed] [Google Scholar]
- Fu Y, Cao F. MicroRNA-125a-5p regulates cancer cell proliferation and migration through NAIF1 in prostate carcinoma. OncoTargets Ther 2015; 8: 3827–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R-J, Zheng Y-H, Wang P, Zhang J-Z. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol 2015; 8: 765–771. [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Sun X, Zhou X, Han L, Chen L, Shi Z et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget 2015; 6: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene 2008; 27: 6194–6206. [DOI] [PubMed] [Google Scholar]
- Butt AJ, Harvey NL, Parasivam G, Kumar S. Dimerization and autoprocessing of the Nedd2 (caspase-2) precursor requires both the prodomain and the carboxyl-terminal regions. J Biol Chem 1998; 273: 6763–6768. [DOI] [PubMed] [Google Scholar]
- Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell 1994; 78: 739–750. [DOI] [PubMed] [Google Scholar]
- Busbee PB, Nagarkatti M, Nagarkatti PS. Natural indoles, indole-3-carbinol (I3C) and 3,3'-diindolylmethane (DIM), attenuate staphylococcal enterotoxin B-mediated liver injury by downregulating miR-31 expression and promoting caspase-2-mediated apoptosis. PloS One 2015; 10: e0118506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Glaser SS, Francis H, Yang F, Han Y, Stokes A et al. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am J Pathol 2012; 181: 804–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic caspase-2. Science (New York, NY) 2012; 338: 818–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li K. MicroRNA-96 regulates RGC-5 cell growth through caspase-dependent apoptosis. Int J Clin Exp Med 2014; 7: 3694–3702. [PMC free article] [PubMed] [Google Scholar]
- Holleman A, den Boer ML, Kazemier KM, Beverloo HB, von Bergh AR, Janka-Schaub GE et al. Decreased PARP and procaspase-2 protein levels are associated with cellular drug resistance in childhood acute lymphoblastic leukemia. Blood 2005; 106: 1817–1823. [DOI] [PubMed] [Google Scholar]
- Yoo NJ, Lee JW, Kim YJ, Soung YH, Kim SY, Nam SW et al. Loss of caspase-2, -6 and -7 expression in gastric cancers. APMIS 2004; 112: 330–335. [DOI] [PubMed] [Google Scholar]
- Zohrabian VM, Nandu H, Gulati N, Khitrov G, Zhao C, Mohan A et al. Gene expression profiling of metastatic brain cancer. Oncol Rep 2007; 18: 321–328. [PubMed] [Google Scholar]
- Dorstyn L, Puccini J, Nikolic A, Shalini S, Wilson CH, Norris MD et al. An unexpected role for caspase-2 in neuroblastoma. Cell Death Dis 2014; 5: e1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Chung NG, Yoo NJ, Lee SH. Somatic mutation of proapoptotic caspase-2 gene is rare in acute leukemias and common solid cancers. Eur J Haematol 2011; 86: 449–450. [DOI] [PubMed] [Google Scholar]
- Kim MS, Kim HS, Jeong EG, Soung YH, Yoo NJ, Lee SH. Somatic mutations of caspase-2 gene in gastric and colorectal cancers. Pathol Res Pract 2011; 207: 640–644. [DOI] [PubMed] [Google Scholar]
- Read SH, Baliga BC, Ekert PG, Vaux DL, Kumar S. A novel Apaf-1-independent putative caspase-2 activation complex. J Cell Biol 2002; 159: 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Srinivasula SM, Druilhe A, Fernandes-Alnemri T, Alnemri ES. Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J Biol Chem 2002; 277: 13430–13437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.