Abstract
p53MutaGene is the first online tool for statistical validation of hypotheses regarding the effect of p53 mutational status on gene regulation in cancer. This tool is based on several large-scale clinical gene expression data sets and currently covers breast, colon and lung cancers. The tool detects differential co-expression patterns in expression data between p53 mutated versus p53 normal samples for the user-specified genes. Statistically significant differential co-expression for a gene pair is indicative that regulation of two genes is sensitive to the presence of p53 mutations. p53MutaGene can be used in ‘single mode' where the user can test a specific pair of genes or in ‘discovery mode' designed for analysis of several genes. Using several examples, we demonstrate that p53MutaGene is a useful tool for fast statistical validation in clinical data of p53-dependent gene regulation patterns. The tool is freely available at http://www.bioprofiling.de/tp53
Although p53 is probably the most intensively studied gene in cancer, the molecular basis for p53-mediated gene regulation is still largely unclear, in particular, in cancers where p53 is mutated.1 p53 transcriptional programmes are activated in response to a variety of signals, allowing p53 to control transcription of many genes and to have important role in many biological processes including tumour suppression. To date, over hundred genes have been shown to be direct transcriptional targets of p53. Although initially described solely as a transcriptional activator, p53 is now known to also mediate transcriptional repression,2 the regulation of translation3 and even the induction of a transcription-independent apoptotic response.4 Mutations in p53 lead to the loss of normal p53 function and are present in roughly 50% of all human cancers. These mutations in the TP53 gene very often lead not only to loss of wild-type p53 (wt-p53) functions but also to acquisition of additional properties (gain-of function, GOF) by mutant p53 (mut-p53). As part of its GOF activity, mut-p53 is commonly identified in a molecular complex with other proteins, enhancing or inhibiting their activity. Alteration of the activity of transcriptional factors (TFs) is a frequent mechanism of the function of mut-p53.5 Thus, mut-p53 alters gene regulation, leading to an addiction of the cancer cells to the expression of mut-p53.6 However, the molecular mechanism of mut-p53 activity is still largely unknown.
Significant efforts are still underway to understand the complexity of gene regulation in cancer. In a majority of cases, initial evidence of novel gene regulatory patterns is commonly discovered in vitro or in animal models.7 However, all these experimental settings have significant limitations in the degree to which they reproduce human cancer pathology and still demand extensive validation in the clinical setting. These limitations are often a bottleneck to upgrading novel-identified gene regulatory patterns from basic research to a translational stage. Statistical validation of gene regulatory patterns identified in vitro and in animal models can be performed by mining clinical gene expression data sets,8, 9, 10 many of which are currently available.11, 12, 13
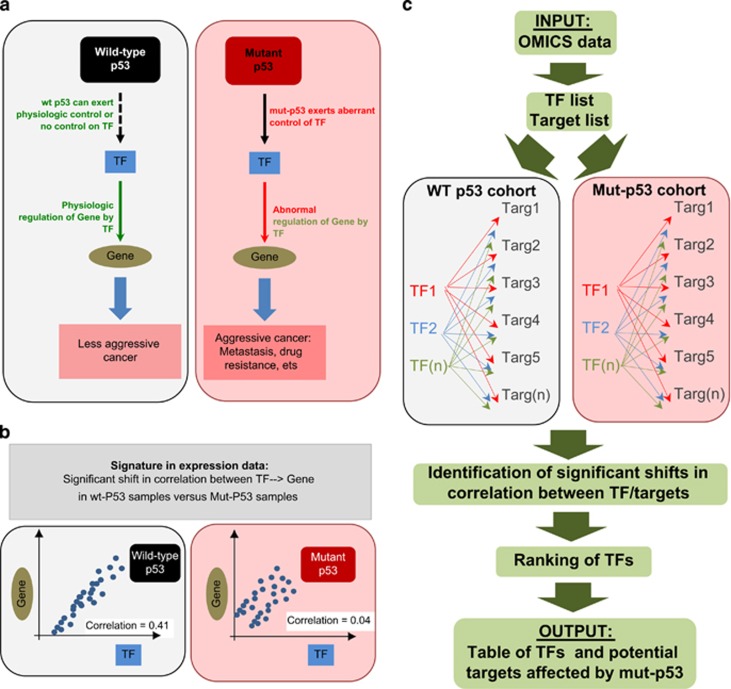
In many cases, data mining of gene expression data sets requires advanced bioinformatics skills. The demand for such services from the biological, clinical and pharmacological communities has stimulated development of various user-friendly online tools to exploit the plethora of publicly available clinical data.8, 9, 14, 15, 16, 17, 18 Despite this variety of available tools, at present, none are available to test the effects of p53 mutational status on gene regulation in cancer. The current in vitro animal-experimental pipeline is based on experimental validation of an abnormal control by mut-p53 of putative target proteins (such as TFs), supported by experimental evidence of abnormal regulation of downstream players (such as TF targets).1, 7 In clinical gene expression data, statistically significant differences between cohorts of mut-p53 and wt-p53 samples in the correlation of TF and TF target pairs would represent a signature of this regulatory model (Figures 1a and b).
Figure 1.
p53Mutagene allows statistical validation in clinical settings of mut-p53 effects on gene regulation. (a) Mut-p53 GOF has been associated with its ability to interact with proteins and altering their physiological activity. Therefore, in a tumour carrying a mutation in p53, the presence of mut-p53 proteins can affect the ability of transcriptional factors (TFs) to regulate their targets. According to this model, the correlation between a mut-p53-sensitive TF and its TF targets is different in a wt-p53 and mut-p53-expressing cohort of cancer patients. (b) In single mode, p53Mutagene computes TF/target correlation in the two cohorts of clinical samples (wt-p53 versus mut-p53) and estimates whether a statistically significant shift in correlation is observed. (c) p53Mutagene discovery mode: the list of putative TF is tested versus the list of putative TF targets (both lists are specified by the user). p53Mutagene computes correlations for all TF/target pairs in wt-p53 and mut-p53 cohorts. The final output will be the list of ranked TFs associated with putative targets, whose regulation (TF/target), is significantly different between wt-p53 and mut-p53 samples in gene expression data set
To address the wide interest of the scientific community on mut-p53-mediated gene regulation, we developed p53MutaGene, an online data mining tool for testing the effect of p53 mutational status on gene regulation in cancer. The tool is based on several clinical gene expression data sets annotated with p53 mutational status, and currently covers breast and colon cancers together with lung adenocarcinoma. p53MutaGene detects a shift in the correlation between mut-p53 and wt-p53-sample cohorts for user-specified genes. The tool can be used in ‘single mode' to test a specific pair of genes for sensitivity to p53 mutation (Figure 1b) or in ‘discovery mode' to screen genes to identify candidates whose regulation might be sensitive to p53 mutational status (Figure 1c). To demonstrate the potential utility of p53MutaGene, we have provided multiple examples of application of p53MutaGene (single mode) to validate well known p53-dependent gene regulatory models. We have also demonstrated the utility of p53MutaGene in ‘discovery mode' to identify potential candidates implicated in p53 regulatory programmes from the large list of candidate genes.
Results and Discussion
p53MutaGene: single mode
To test a single hypothesis, the user needs to specify two genes: the putative TF and the putative TF target (Figures 1a and b). As output, p53MutaGene provides two Pearson correlation coefficients between the input genes expression profiles in the mut-p53 versus the wt-p53 cohort. A statistically significant shift in correlation represents a signature in gene expression data that supports the hypothesis that regulatory relations between submitted genes are sensitive to mut-p53. Significance of differential co-expression (i.e., shift of correlation coefficients) is computed by random sampling procedure: the same size as the p53-mut subset is randomly selected (1000 times) from the whole set of samples (both wild type and p53-mut) that are annotated with p53 mutational status. Each time the correlation between TF/target is computed. The resulting distribution of correlation between two genes (of size 1000) is used to estimate significance of the correlation shift. In this case, the P-value is an observed probability to select randomly a subset of samples (of size equal to the number of p53-mut samples) with correlation between input genes high (negative shift p53-mut) or lower (positive shift in p53-mut) than correlation between input genes in the subset of p53-mut samples.
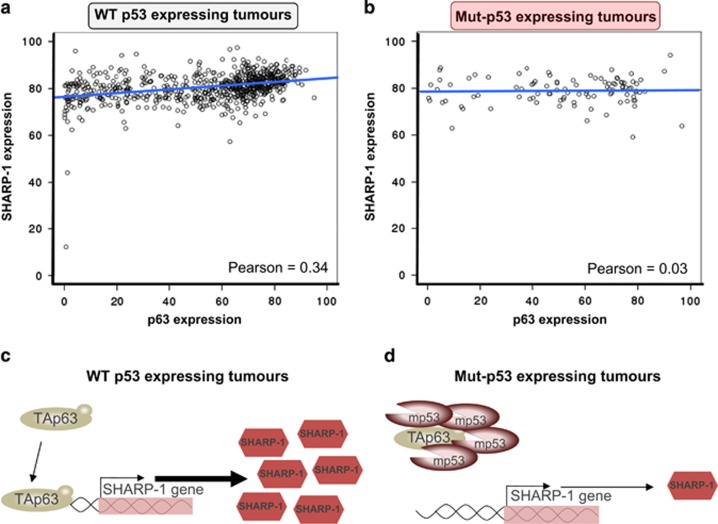
Recent literature has focused on the GOF of mut-p53 in the promotion of an invasive phenotype leading to metastasis. In this context, one of the well-established mechanisms by which mutant p53 exerts its GOF is the repression of the p53 family member p63.19 A direct physical interaction between these two proteins, consequent to the biochemical properties of mut-p53, leads to an inhibition of the transcriptional ability of the metastatic suppressor isoform TAp63.20, 21, 22 The basic helix-loop-helix family member e41 (BHLHE41), also known as Sharp-1/Dec-2, has also been implicated as a crucial regulator of the invasive and metastatic phenotype in breast cancer.7, 20 TAp63 promotes expression of Sharp-1 that in turn regulates the stability and pro-metastatic activity of hypoxia inducible factor 1α (HIF-1α). In vitro studies using cell lines, complemented by in vivo mouse models have been used to experimentally prove the ability of mut-p53 ability to alter the TAp63/Sharp-1 axis. Overexpression and silencing of mutant p53 modulates TAp63-dependent regulation of Sharp-1. We used p53MutaGene to validate this in vitro/in vivo experimental evidence in a clinical context. p63 and Sharp-1 were used as inputs for p53MutaGene (p63 as a TF and Sharp-1 as the TF target), and p53MutaGene reported statistically significant differential co-expression of p63/SHARP1 between mut-p53 and wt-p53 breast cancer patients (Figures 2a and b). A positive correlation between p63/SHARP1 is observed in the wt-p53 group (Pearson coefficient 0.34) while a lack of correlation (Pearson coefficient 0.03) is observed in p53-mut breast cancer samples. Therefore, the statistically significant negative shift in the p63/SHARP1 correlation provided by p53MutaGene is indicative of a signature in clinical gene expression data that supports mut-p53's role in altering the p63/Sharp-1 axis in breast cancer (Figures 2c and d). This example provides an overview of the potential applications of p53MutaGene ‘single mode', as a tool for validation of experimental data, as well as, a method to search for novel potential genes of interest in the investigation of mut-p53 roles in cancer biology.
Figure 2.
p53Mutagene predicts a significant effect of mut-p53 on the p63/Sharp-1 axis. (a and b) Correlation between p63 expression and Sharp-1 expression was computed in the two cohorts of wt-p53 and mut-p53-expressing breast cancers from the METABRIC data set. A statistically significant negative shift was observed in the Pearson correlation coefficient from 0.34 to 0.03 (P-value=0.003) suggesting a disruption by mut-p53 of the p63/Sharp-1 positive axis. (c and d) Schematic representation of the p63/Sharp-1 axis in the two possible scenarios of wt-p53 and mut-p53-expressing tumours. In wt-p53-expressing tumours, p63 is fully active and is therefore able to control expression (potentially via direct binding on the promoter) of Sharp-1 expression. In mut-p53-expressing tumours, p63 is sequestered and consequentially repressed by mut-p53. This results in disruption of the p63/Sharp-1 axis, which is observed in the correlation plot (b) as no increase in Sharp-1 expression parallels the p63-expression increase
p53MutaGene: discovery mode
Along with its ability to test single pairs (i.e., one TF and one TF target), p53MutaGene also has a discovery mode. In this case, the user inputs multiple genes that are putative TFs and multiple genes that are putative TF targets. For each pair (putative TF and putative TF target), the algorithm will perform the same analyses as in single mode. The output of a discovery mode is a ranked list of input TFs. The ranking is based on a number of significant models found for a submitted list of putative targets. Therefore, p53MutaGene in discovery mode ranks a list of putative TFs for a given list of TF targets based on available expression data (Figure 1c). Discovery mode can support the interpretation of large amounts of data, such as the results from omics studies of p53-dependent experimental settings (transcriptome analysis and/or chromatin-binding sequencing, etc.), where the output represents a list of genes (or several lists).
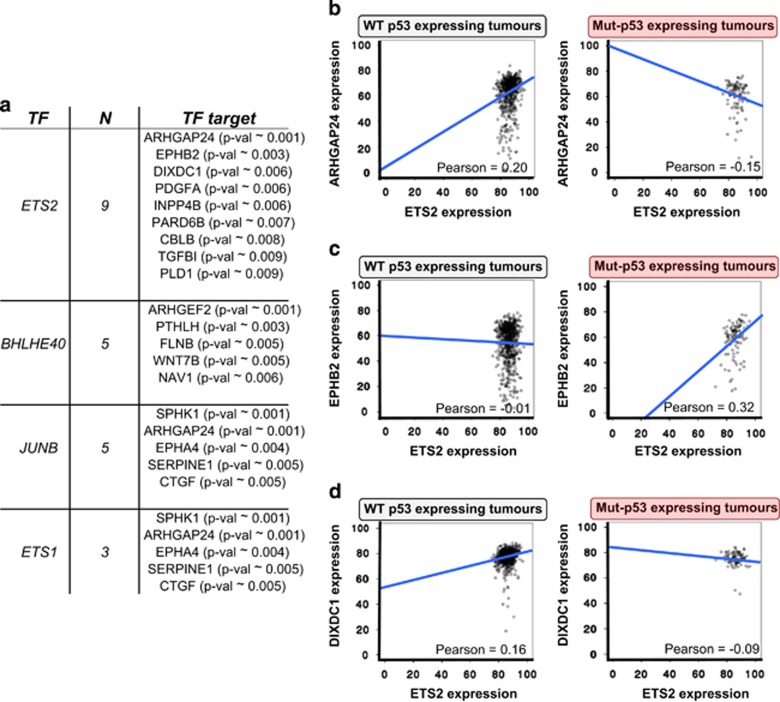
To demonstrate the potential of p53MutaGene discovery mode, we selected a previously published gene list generated by microarray analysis on MDA-MD-231 cells (expressing mut-p53 R280K) transfected with shp53 or a scrambled control construct and subsequently treated with TGFβ to activate the migration/invasion programme.20 The list consists of 72 genes differentially expressed after mut-p53 depletion. Four of these genes (ETS2, ETS1, BHLHE40, JUNB) are well known TFs. We included these genes in the TF list leaving all the remaining genes in a putative transcriptional target list. p53MutaGene discovery mode ranked ETS2 as the top TF for this list with nine putative targets having a significant shift of correlation. This indicates that ETS2 activity is sensitive to p53 mutational status (Figure 3a). ARHGAP24 (also known as FilGAP) was the highest scored target. ETS2/ARHGAP24 gene expression correlation has a strong negative shift in mut-p53 tumours (Figure 3b), indicating that mutated p53 can affect the ability of ETS2 to attenuate the expression of ARHGAP24 (Figure 3b). EPHB2 and DIXDC1 are the other top-ranked genes. The ETS2 correlation with EPHB2 and DIXDC1, respectively, showed a positive and a negative shift in p53 mutated samples (Figures 3c and d). All these top-ranked genes have been repeatedly associated with a metastatic phenotype. FilGAP, a Rho GTPase activating protein, has recently been identified as an actin-binding protein which has an important role in the central mechanotransduction element of the cytoskeleton23 and which has been implicated in invasion/metastasis. FilGAP expression and activity has been found to be altered in metastatic TNBC, highlighting a crucial role for this factor in metastatic suppression of breast cancer.24 DIXDC1 is a scaffold protein whose expression is often reduced in human metastatic cancer, leading to upregulation of Snail1 and consequentially increased cell invasion.25 Remarkably, wt-p53 inhibits EPHB2 expression to limit the pro-invasive properties of TGF-β3 in breast cancer.26 The correlation shift observed for these genes suggests that ETS2 exerts transcriptional activity on their promoters that can be affected by mut-p53. Strikingly, the shift of the correlation was always in agreement with the pro-metastatic role of mutated p53 in cancer. Therefore, considering the established role of FilGAP, DIXDC1 and EPHB2 in metastasis and the mut-p53-dependent sensitivity of ETS2/targets correlation detected by p53MutaGene, the analysis suggests that ETS2 is an effector of the mut-p53 pro-metastatic phenotype. Indeed, recent studies have revealed that ETS2 cooperates with mut-p53 to promote tumour progression. Mut-p53 increases the transcriptional expression of the ETS2 target genes, TDP and Pla2g16, in an ETS2-dependent manner.27, 28 Our analyses with p53MutaGene on transcriptomic experimental data led us to similar conclusions, potentially, extending our understanding of the mut-p53/ETS2 network. With a similar approach, p53MutaGene can assist in the interpretation of experimental data by shading light on completely novel networks affected by mut-p53.
Figure 3.
p53Mutagene discovery mode identified ETS2 as an effector of mut-p53 in alteration of gene networks. (a) In a gene list generated in a mut-p53 experimental setting (MDA-MB-231-shGFP or MDA-MB-231-shmut-p53), p53Mutagene discovery mode ranked ETS2 as the most affected TF and indicated nine potential targets. All of these targets show a significant shift in correlation with ETS2 in mut-p53-expressing breast cancers compared with wt-p53 (METABRIC data set). TF, transcriptional factor; N, number of targets. Specific P-values are indicated between brackets. (b–d) Plots depict a significant shift in correlation between ETS and ARHGAP24 (0.2 to −0.15) or EPHB2 (−0.01 to 0.32) or DIXDC1 (0.16 to −0.09) in mut-p53-expressing tumours compared with those expressing wt-p53
Materials and Methods
Gene expression data sets
Computation of p53MutaGene is based on several clinical gene expression cancer data sets. Samples in the data sets are additionally annotated with p53 mutational status. Statistics on the number of samples for each data set is presented in Table 1. For each data set, the same procedure was repeated. Each sample from the data set probes were ordered by expression value, and for each probe, expression rank was computed (i.e., rank 100 means the top expressed probe in the sample and rank 55 means that 55 per cent of probes have lower expression value in the sample). Expression rank reflects relative expression level and is more consistent, as it requires no normalisation, and thus introduces no normalisation bias.13, 18, 29 Only rank information is used for the purpose of this analysis.
Table 1. Statistics of the data sets used by p53MutaGene.
Single hypothesis mode
Pearson correlation is used as a measure of correlation between mRNA profiles (expression rank) of two genes. Correlation between two input genes (i.e., putative TF and putative TF target) is computed in a subset of p53 mutated samples. The expected correlation between the genes is computed based on a random permutation procedure: the same size as the p53-mut subset is randomly selected (1000 times) from the whole set of samples (both wild type and p53-mut) that are annotated with p53 mutational status. The resulting distribution of correlation between two genes (of size 1000) is used to estimate significance of the correlation shift. In this case, the P-value is an observed probability to select randomly a subset of samples (of size equal to the number of p53-mut samples) with correlation between input genes high (negative shift in p53-mut) or lower (positive shift in p53-mut) than correlation between input genes in the subset of p53-mut samples. In the case of multiple testing (the input gene can be mapped to multiple probes), a False Discovery Rate procedure is used to adjust computed P-values.30
Discovery mode
In discovery mode, the list of putative TF is tested versus the list of putative TF targets. For each pair (TF versus target), the same analysis is computed as described in the previous section. For each TF, the number of significant models (P-value<0.01) is counted. First, TFs are ranked based on the number of significant models among submitted targets. Second, for each TF genes from the list of putative targets are also ranked based on the P-value of differential correlation.
Acknowledgments
This work was supported by the UK Medical Research Council (MRC) and fundamental research program of the Russian State Academies of Sciences.
The authors declare no conflict of interest.
Footnotes
Edited by G Raschellà
References
- Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol 2013; 15: 2–8. [DOI] [PubMed] [Google Scholar]
- Ho J, Benchimol S. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ 2003; 10: 404–408. [DOI] [PubMed] [Google Scholar]
- Ewen ME, Miller SJ. p53 and translational control. Biochim Biophys Acta 1996; 1242: 181–184. [DOI] [PubMed] [Google Scholar]
- Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta 2009; 1787: 414–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev 2012; 26: 1268–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova EM, Yallowitz AR, Li D, Xu S, Schulz R, Proia DA et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 2015; 523: 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagner M, Enzo E, Forcato M, Zanconato F, Parenti A, Rampazzo E et al. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature 2012; 487: 380–384. [DOI] [PubMed] [Google Scholar]
- Antonov AV, Knight RA, Melino G, Barlev NA, Tsvetkov PO. MIRUMIR: an online tool to test microRNAs as biomarkers to predict survival in cancer using multiple clinical data sets. Cell Death Differ 2013; 20: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov AV, Krestyaninova M, Knight RA, Rodchenkov I, Melino G, Barlev NA. PPISURV: a novel bioinformatics tool for uncovering the hidden role of specific genes in cancer survival outcome. Oncogene 2014; 33: 1621–1628. [DOI] [PubMed] [Google Scholar]
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010; 123: 725–731. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012; 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Gostev M, Knight RA, Willis AE, Melino G, Antonov AV. DRUGSURV: a resource for repositioning of approved and experimental drugs in oncology based on patient survival information. Cell Death Dis 2014; 5: e1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov AV. BioProfiling.de: analytical web portal for high-throughput cell biology. Nucleic Acids Res 2011; 39: W323–W327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmann S, Lee W, Wong P, Rodchenkov I, Antonov AV. CCancer: a bird's eye view on gene lists reported in cancer-related studies. Nucleic Acids Res 2010; 38: W118–W123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundem G, Perez-Llamas C, Jene-Sanz A, Kedzierska A, Islam A, Deu-Pons J et al. IntOGen: integration and data mining of multidimensional oncogenomic data. Nat Methods 2010; 7: 92–93. [DOI] [PubMed] [Google Scholar]
- Oh K, Park HB, Byoun OJ, Shin DM, Jeong EM, Kim YW et al. Epithelial transglutaminase 2 is needed for T cell interleukin-17 production and subsequent pulmonary inflammation and fibrosis in bleomycin-treated mice. J Exp Med 2011; 208: 1707–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B et al. A mutant-p53/smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell 2009; 137: 87–98. [DOI] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 2009; 139: 1327–1341. [DOI] [PubMed] [Google Scholar]
- Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol 2011; 7: 285–295. [DOI] [PubMed] [Google Scholar]
- Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 2011; 478: 260–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M, Bao Y, Li Z, Li J, Gong M, Lam S et al. RASAL2 activates RAC1 to promote triple-negative breast cancer progression. J Clin Invest 2014; 124: 5291–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin JM, Svensson RU, Lou HJ, Winslow MM, Turk BE, Shaw RJ. An AMPK-independent signaling pathway downstream of the LKB1 tumor suppressor controls Snail1 and metastatic potential. Mol Cell 2014; 55: 436–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S, Wiercinska E, Teunisse AF, Lodder K, ten Dijke P, Jochemsen AG. Wild-type p53 inhibits pro-invasive properties of TGF-beta3 in breast cancer, in part through regulation of EPHB2, a new TGF-beta target gene. Breast Cancer Res Treat 2014; 148: 7–18. [DOI] [PubMed] [Google Scholar]
- Do PM, Varanasi L, Fan S, Li C, Kubacka I, Newman V et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev 2012; 26: 830–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S, Tu H, Kollareddy M, Pant V, Li Q, Zhang Y et al. Pla2g16 phospholipase mediates gain-of-function activities of mutant p53. Proc Natl Acad Sci USA 2014; 111: 11145–11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 2006; 10: 191–202. [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med 1990; 9: 811–818. [DOI] [PubMed] [Google Scholar]