Abstract
Our previous studies have demonstrated that inhibition of glycogen synthase kinase 3β (GSK3β) activity protects mice from acute liver failure (ALF), whereas its protective and regulatory mechanism remains elusive. Autophagy is a recently recognized rudimentary cellular response to inflammation and injury. The aim of the present study was to test the hypothesis that inhibition of GSK3β mediates autophagy to inhibit liver inflammation and protect against ALF. In ALF mice model induced by d-galactosamine (d-GalN) and lipopolysaccharide (LPS), autophagy was repressed compared with normal control, and d-GalN/LPS can directly induce autophagic flux in the progression of ALF mice. Autophagy activation by rapamycin protected against liver injury and its inhibition by 3-methyladenine (3-MA) or autophagy gene 7 (Atg7) small interfering RNA (siRNA) exacerbated liver injury. The protective effect of GSK3β inhibition on ALF mice model depending on the induction of autophagy, because that inhibition of GSK3β promoted autophagy in vitro and in vivo, and inhibition of autophagy reversed liver protection and inflammation of GSK3β inhibition. Furthermore, inhibition of GSK3β increased the expression of peroxisome proliferator-activated receptor α (PPARα), and the downregulated PPARα by siRNA decreased autophagy induced by GSK3β inhibition. More importantly, the expressions of autophagy-related gene and PPARα are significantly downregulated and the activity of GSK3β is significantly upregulated in liver of ALF patients with hepatitis B virus. Thus, we have demonstrated the new pathological mechanism of ALF that the increased GSK3β activity suppresses autophagy to promote the occurrence and development of ALF by inhibiting PPARα pathway.
Acute liver failure (ALF), an inflammation-mediated hepatocellular injury process, is a clinical syndrome that results from hepatocellular apoptosis and hemorrhagic necrosis.1 ALF frequently results from viral hepatitis, ingestion of drugs or toxic substances, or hepatic ischemia-reperfusion injury, among others. The prognosis for ALF is extremely poor, and there is currently no effective therapy for the end stage of the disease other than liver transplantation.2 Although the nature of ALF has been extensively studied, the mechanisms by which organ damage occurs are not completely understood.
Glycogen synthase kinases 3 are a group of ubiquitously expressed serine/threonine kinases that are initially found to regulate glycogen synthesis. There are two highly homologous isoforms, designated as glycogen synthase kinase 3 (GSK3)α and GSK3β, respectively. Constitutively active in resting cells, GSK3β has a broad range of substrates, and regulates cell activation, differentiation and survival.3, 4 Among the diverse functions that are regulated by GSK3β, inflammation has recently emerged as one of the major interesting focuses. Studies showed that GSK3β is an important positive regulator in inflammatory process.5, 6, 7, 8 GSK3β deletion results in embryonic lethality caused by severe liver degeneration during development.9 Particularly, GSK3β-deficient cells become more sensitive to tumor necrosis factor α (TNF-α)-induced apoptosis.10 Our studies have shown that the activity of GSK3β is promoted in the progression of ALF and inhibition of GSK3β mitigates liver inflammation to ameliorate ALF model of mice,11, 12 but its protective mechanisms are not well defined.
Macroautophagy (referred to hereafter as autophagy) is a highly evolutionarily conserved process found in virtually all types of eukaryotic cells. Autophagy involves the sequestration of regions of cytosol within double-membrane-bound compartments followed by lysosome-based degradation of the contents. Previous studies have suggested that autophagy represents an adaptive strategy by which cells can remove damaged organelles and enhance survival following bioenergetics-induced stress, and have multiple roles of autophagy in the regulation of cell death, differentiation and the anti-microbial response in mammals.13, 14, 15 In recent years, emerging evidence has indicated that the autophagy process may have an essential role for the host during bacterial clearance and may also interact with inflammatory processes, which consequently may impact the outcomes of disease progression.16, 17 There is a complex reciprocal relationship between autophagy pathway/proteins and inflammation.18, 19 Recent observations have revealed a relationship between autophagy and inflammasome-associated pro-inflammatory cytokine maturation in macrophages.20, 21
Given the above information, we speculated that autophagy activation may serve a protective function to restrain liver inflammation in cases of ALF. The study has also showed that inhibition of hepatocyte autophagy increases TNFα-dependent liver injury by promoting caspase-8 activation.22 So, we hypothesized that inhibition of GSK3β may promote autophagy to protect mice from ALF. To test these hypotheses, we used the ALF model induced by the co-injection of d-galactosamine (d-GalN) and lipopolysaccharide (LPS), which has been widely used to examine the underlying mechanisms of ALF,23, 24 to explore the protective mechanisms of GSK3β inhibition and its regulatory pathway in the context of ALF, and further measure the expression of autophagic gene in human liver samples from patients with ALF. Our findings demonstrate that inhibition of GSK3β increase autophagy to alleviate liver inflammation and protect mice from ALF mediated by peroxisome proliferator-activated receptor α (PPARα).
Results
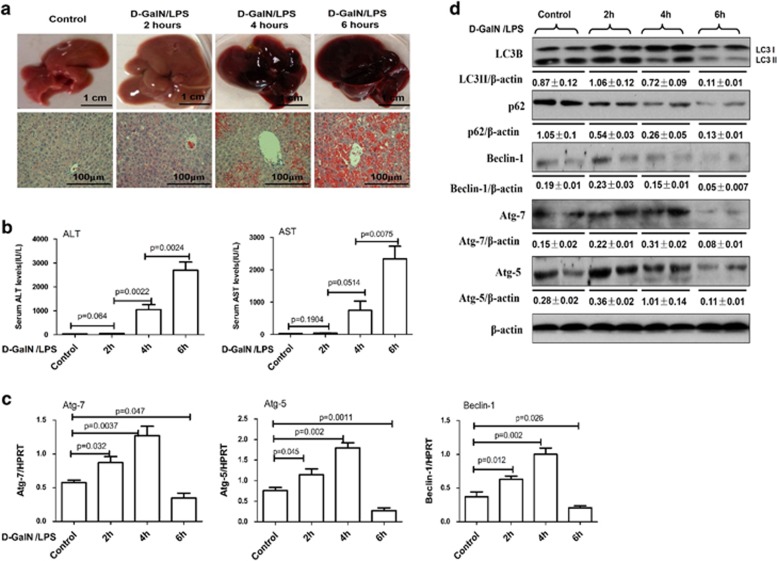
Dynamic profile of autophagy in the progression of d-GalN/LPS-induced ALF
We first investigated a possible association between autophagy and ALF induced by d-GalN/LPS treatment. We found that massive hepatic injury was apparent after 4–6 h as revealed by gross morphology of the liver; and the liver tissue of mice appears spotty hemorrhage at 2 h; the increased inflammation, hepatic lobules disorder, a large number of inflammatory cell infiltration and visible hepatocyte apoptosis or confluent necrosis are shown at 4 h; there are a lot of visible large necrosis areas in the liver tissue, the entire liver congestion at 6 h (Figure 1a). The results of gross morphology of the liver and hematoxylin and eosin (H&E) staining were in agreement with the increased serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) enzyme levels (Figure 1b). Therefore, a mouse model of ALF was successfully constructed at 6 h time point after the d-GalN/LPS treatment, which was employed for all subsequent experiments. Accompanying the liver injury, compared with normal mice, the mRNA or protein levels of autophagic gene including Atg5, Atg7, Beclin-1 and LC3II conversion were gradually upregulated after 2 h, but declined after 6 h (Figures 1c and d; Supplementary Figure 1). The p62 protein, also called sequestosome 1 (SQSTM1), binds directly to LC3 and GABARAP family proteins via a specific sequence motif. The protein is itself degraded by autophagy and may serve to link ubiquitinated proteins to the autophagic machinery to enable their degradation in the lysosome; as p62 accumulates when autophagy is inhibited, and decreased levels can be observed when autophagy is induced, p62 may be used as a marker to study autophagic flux.25 As showed in Figure 1d, p62 protein degradation is constantly increased in progression of d-GalN/LPS-induced liver injury. Taken together, these results indicated that autophagy is repressed in the mice ALF model.
Figure 1.
The regulation of autophagy during d-GalN/LPS induced mice ALF progression. Mice were intraperitoneally injected with d-GalN (700 mg/kg) and LPS (10 μg/kg) at 2, 4 and 6 h (12 mice per group). The mice in the control group (n=8) were injected with PBS only. (a). Representative livers and H&E staining of livers. (b) Serum AST and ALT enzyme levels. (c) Gene expressions of Beclin-1, Atg5 and Atg7 were measured by qRT-PCR in the livers. (d) Protein expression levels of LC3B, p62, Beclin-1, Atg5 and Atg7 were measured by western blot assays in the livers. A representative blot from two samples of every group is shown
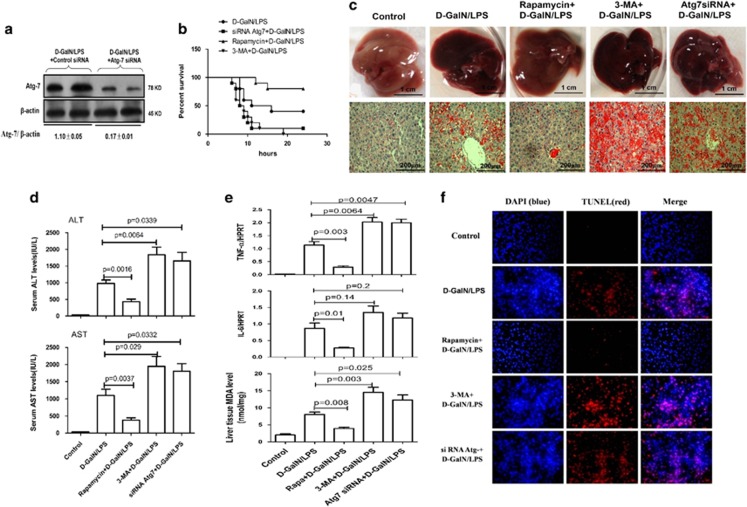
Enhanced autophagy ameliorates hepatotoxicity induced by d-GalN/LPS
We then evaluated the physiological roles of autophagy in d-GalN/LPS-induced ALF. Pretreatment with rapamycin for 2 h before d-GalN/LPS injection protected mice against ALF; and pretreatment with 3-methyladenine (3-MA) for 2 h or Atg7 siRNA for 24 h before d-GalN/LPS injection further aggravated the liver injury of ALF. The inhibition of liver Atg7-specific siRNA in vivo was confirmed by the reduced Atg7 protein levels in the mice (Figure 2a). In the survival analysis, the mice in the d-GalN/LPS control group began to perish 7 h after d-GalN/LPS injection, and the survival rate of these mice was 40% (4 of 10 mice) at the 24-h time point. By contrast, the survival rate after rapamycin treatment was 80% (8 of 10 mice); the survival rate after 3-MA or Atg7 siRNA treatment was 0% (0 of 10 mice) and 10% (1 of 10 mice), respectively (Figure 2b). The gross morphology of the liver after rapamycin treatment appeared substantially normal, and the liver architecture was well preserved, such as decreased inflammatory cell infiltration and less hepatocyte apoptosis or confluent necrosis; but pretreatment of d-GalN/LPS-induced mice with 3-MA or Atg7 siRNA results in more massive hepatic toxicity as revealed by gross morphology of the liver and the liver architecture (Figure 2c). Compared with the ALF group, the mice subjected to rapamycin treatment showed significantly lower sALT and sAST levels, the genes expression level of TNF-α and interleukin (IL)-6, and the level of malondialdehyde (MDA) in liver; but 3-MA or Atg7 siRNA pretreatment further aggravated liver functions, and significantly increased the level of TNF-α and MDA (Figures 2d and e); Moreover, by TUNEL (terminal deoxynucleotidyl transferasemediated dUTP nick-end labeling) assay, pretreatment with rapamycin significantly decreased hepatocyte apoptosis and 3-MA or Atg7 siRNA increased the hepatocyte apoptosis (Figure 2f). These results demonstrated that autophagy is critical for d-GalN/LPS-induced ALF and that its induction protects mice from liver injury.
Figure 2.
Autophagy protects against d-GalN/LPS-induced liver injury rapamycin+d-GalN/LPS-treated mice were administered rapamycin (2 mg/kg, i.p.) 2 h before d-GalN/LPS exposure (n=12); 3-MA+d-GalN/LPS-treated mice were administered 3-MA (10 mg/kg, i.p.) 2 h before d-GalN/LPS exposure (n=12); siRNA Atg7+d-GalN/LPS-treated mice were pretreated with siRNA Atg7 (3 mg/kg, i.v.) for 48 h before d-GalN/LPS exposure (n=12). Control mice were pretreated with vehicle (DMSO) 2 h before PBS injection (n=8). (a) Protein expression levels of Atg7 and β-actin were measured by western blotting. A representative blot for two samples from every group is shown. (b) The survival rate of mice was measured in the Atg7 siRNA +d-GalN/LPS-, rapamycin+d-GalN/LPS- and 3-MA+d-GalN/LPS-treated group and the d-GalN/LPS-treated group (10 mice per group). (c) Representative livers and H&E staining of livers from different groups. (d) Serum AST and ALT enzyme levels from different groups. (e) Gene expression levels of TNF-α and IL-6 were measured by qRT-PCR and the level of MDA was measured in livers from different groups. (f) TUNEL staining (red) liver tissue at 6 h after d-GalN/LPS administration. Representative of one experiment is shown. Original magnification × 200
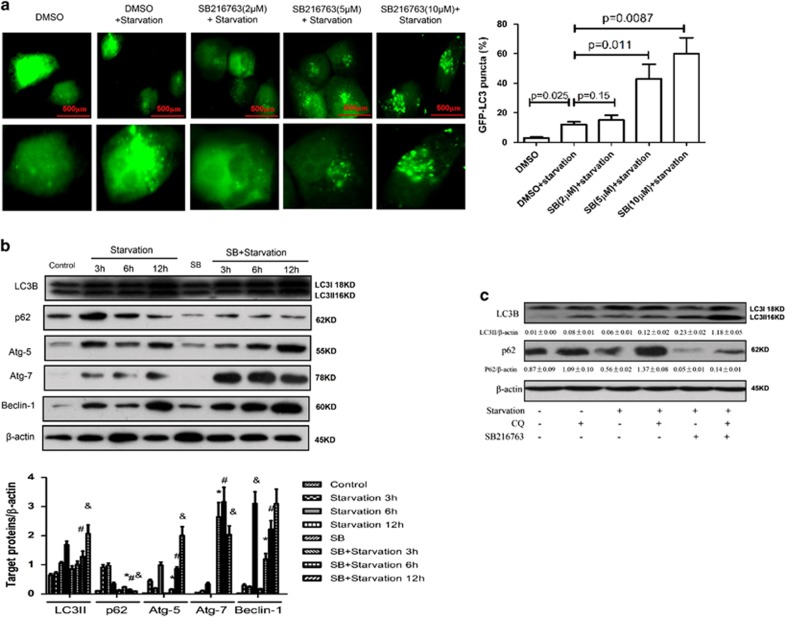
Inhibition of GSK3β promotes autophagy induced by starvation in vitro
Next, we investigated the regulation of autophagy by the effects of GSK3β inhibition on primary hepatocyte in response to starvation. We transfected the GFP-LC3 plasmid into hepatocyte to observe the formation of autophagosomes. As shown in Figure 3a, the GFP-LC3 signal was weak in the control cells, but was bright and punctate after SB216763 treatment, which is a specific inhibitor of GSK3β activity, in a dose-dependent manner. Western blot results showed that SB216763 treatment promoted the expression level of LC3II, Atg5, Beclin-1 and Atg7 proteins; meanwhile, SB216763 treatment also increased the degradation of p62 (Figure 3b). Furthermore, autophagic flux in hepatocyte was monitored after treatment with SB216763 in the presence or absence of chloroquine (CQ). The addition of CQ further increased LC3II expression, and decreased the degradation of p62 compared with cells treated with SB216763 and starvation (Figure 3c). Taken together, the results showed that inhibition of GSK3β promotes autophagic flux in hepatocyte treated by starvation in vitro.
Figure 3.
Inhibition of GSK3β further promotes autophagy induced by starvation in vitro. (a). Transfected GFP-LC3 plasmid for 12 h, the primary hepatocytes were pre-incubated with SB216763 (2, 5 or 10 μM) for 12 h to observe the formation of autophagosomes. (b) The primary hepatocytes were stimulated with or without SB216763 for 30 min, followed for 3, 6 or 12 h. The cell lysates were analyzed by western blot in different time. The levels of LC3B, p62, Atg7, Atg5, Beclin-1and β-actin were measured by western blotting. The graph of densitometry analysis should be shown (compared with starvation 3 h group: *P<0.05; compared with starvation 6 h group: #P<0.05; compared with starvation 12 h group: &P<0.05). (c) Inhibition of GSK3β induced autophagic flux in primary hepatocytes under starvation. Western blot analysis of expression of the LC3B and p62. A representative results from three independent experiments is shown. Densitometry analysis of the proteins was performed for each sample
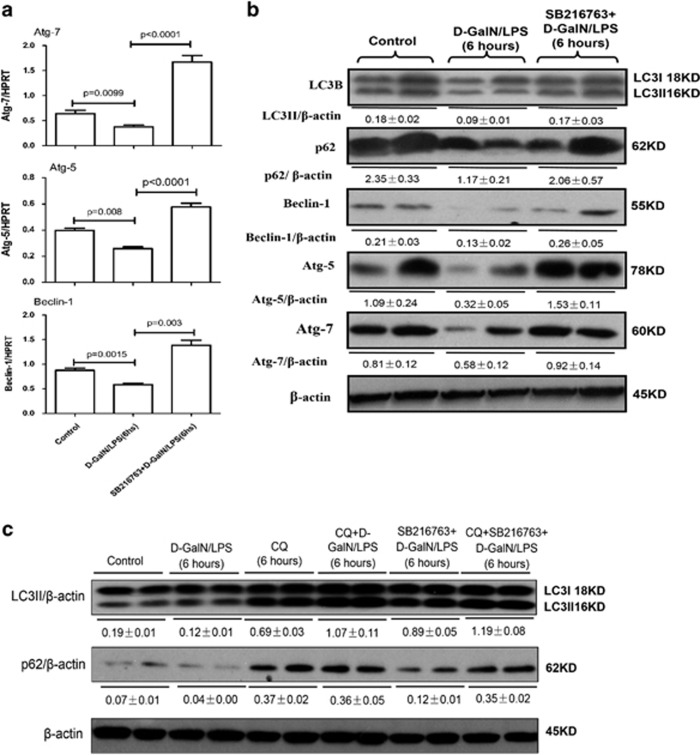
Inhibition of GSK3β promotes autophagy in liver of ALF mice induced by d-GalN/LPS
To further confirm our in vitro experimental findings, we investigated the regulation of autophagy by the effects of GSK3β inhibition on ALF mice induced by d-GalN/LPS. qRT-PCR (quantitative reverse transcription PCR) results showed that, compared with d-GalN/LPS-treated mice, GSK3β inhibition by SB216763 pretreatment in ALF mice promoted the expression of Atg7, Atg5 and Beclin-1 genes, these alterations were confirmed by western blot analyses; interestingly, GSK3β inhibition in ALF mice also promoted LC3II conversion and decreased p62 protein degradation in d-GalN/LPS-induced mice (Figures 4a and b). The CQ pretreatment further increased LC3II conversion and decreased p62 degradation compared with mice treated with SB216763 and d-GalN/LPS. Taken together, inhibition of GSK3β promotes autophagosomes and blocks autophagolysosome formation in d-GalN/LPS-induced ALF mice.
Figure 4.
Autophagy is promoted by GSK3β signaling molecule in vivo. The mice were pretreated with or without SB216763 (25 mg/kg, i.p.) for 2 h, followed by d-GalN/LPS for 6 h stimulation (n=10). CQ-treated mice were pretreated with CQ (60 mg/kg, i.p.) for 8 h (n=10); CQ+ d-GalN/LPS-treated mice were pretreated with CQ (60 mg/kg, i.p.) for 2 h before d-GalN/LPS exposure (n=10); CQ+SB216763+d-GalN/LPS-treated mice were pretreated with CQ (60 mg/kg, i.p.) and SB216763 for 2 h before d-GalN/LPS exposure (n=10). (a) Gene expression levels of autophagy-related proteins, including Atg7, Atg5 and Beclin-1 were measured by qRT-PCR in livers of control mice group, d-GalN/LPS-induced ALF mice group and SB216763 pretreament ALF group induced by d-GalN/LPS. (b) Protein expression levels of autophagy-related proteins, including LC3B, p62, Atg7, Atg5 and Beclin-1, were measured by western blotting in livers. Two representative blot for liver samples from each group were shown. (c) Protein expression levels of LC3B and p62 were measured by western blotting in livers. A representative blot for three samples from each group is shown
Inhibition of GSK3β protects mice from ALF through autophagy-mediated inflammatory response
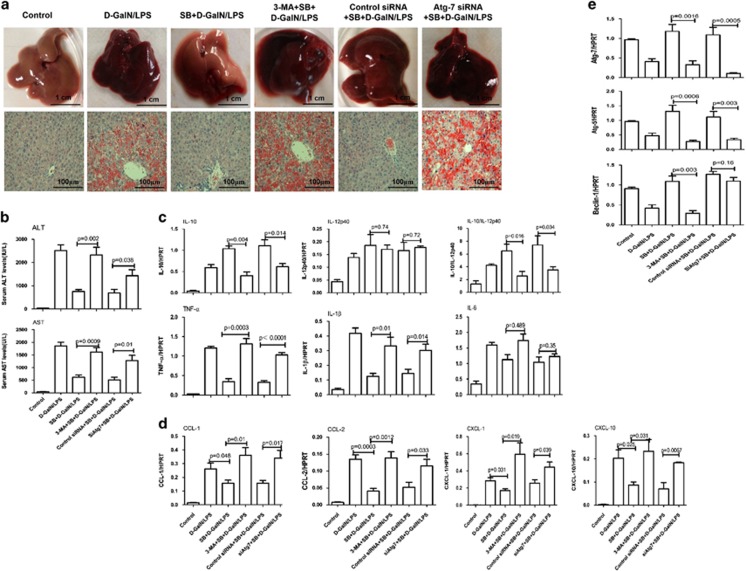
Next, we sought to explore whether GSK3β inhibition leads to the induction of autophagy to protect the liver from injury. The results showed that hepatic protection by SB216763 in ALF was partially negated by 3-MA or siRNA Atg7, which was evident by the relatively worse gross morphology, and the relatively less preserved liver architecture by histology (Figure 5a), and the significantly higher sALT and sAST levels (Figure 5b). Thus, these results demonstrated that hepatoprotective mechanisms of GSK3β inhibition depend on autophagy pathways.
Figure 5.
The inhibition of GSK3β activity protects mice from ALF through autophagy mechanisms. The mice were pretreated with or without SB216763 (25 mg/kg, i.p.) for 2 h, followed by d-GalN/LPS for 6 h stimulation (n=10). SB216763+3-MA+ d-GalN/LPS-treated mice were co-administered 3-MA (10 mg/kg) and SB216763 at 2 h before d-GalN/LPS exposure (n=10); SB216763+siAtg7+ d-GalN/LPS-treated mice were pretreated with Atg7 siRNA (3 mg/kg) for 48 h via tail vein injection and then administered SB216763 2 h before d-GalN/LPS exposure (n=11). (a) Representative livers and H&E staining of livers (200 ×) from different groups. (b) Serum AST and ALT enzyme levels from different groups. (c) Gene expression of cytokines including TNF-α, IL-1β and IL-6 at 6 h, IL-12p40 and IL-10 at 2 h after d-GalN/LPS injection. (d) Gene expression of chemokines including CCL-1, CCL-2, CXCL-1 and CXCL-10 at 6 h after d-GalN/LPS injection. (e) Gene expression of Atg7, Atg5 and Beclin-1 at 6 h after d-GalN/LPS injection
Given the ability of autophagy to elaborately regulate inflammation and GSK3β inhibition is also able to control liver inflammation in d-GalN/LPS-induced ALF, we sought to determine whether an autophagy pathway is required for GSK3β inhibition-mediated suppression of the inflammatory response. In the context of GSK3β inhibition in the ALF mice, 3-MA or siRNA Atg7 restored the gene expression of TNF-α, IL-1β, IL-6 and IL-12p40 (Figure 5c). Furthermore, autophagy inhibition upregulated the gene expression of chemokines again (Figure 5d). Meanwhile, compared with SB216763-pretreated ALF mice, 3-MA further reversed the genes expression of Atg7, Atg5 and Beclin-1, although Atg7 siRNA only reversed the genes expression of Atg5 (Figure 5e). These results demonstrated that the autophagy pathways induced by GSK3β inhibition may contribute to the suppression of liver inflammation in ALF.
Inhibition of GSK3β mediated promoting PPARα induces autophagy pathway
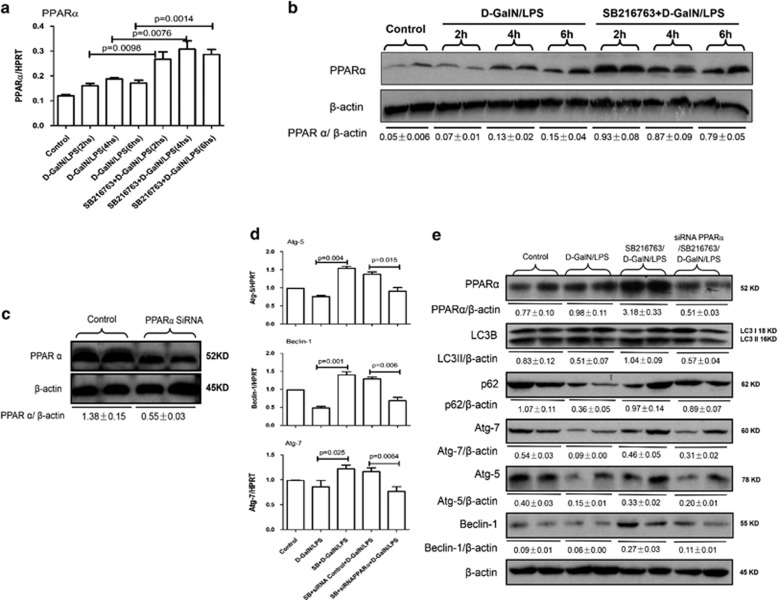
Because our previous study has shown that PPARα activation promotes autophagy to suppress the inflammatory response in ALF,26 we evaluated whether GSK3β inhibition promotes autophagy via a PPARα-dependent pathway in the context of ALF. qRT-PCR and western blotting results showed that, compared with d-GalN/LPS-treated mice, GSK3β inhibition by SB216763 pretreatment in ALF mice promoted the expression of PPARα gene and protein (Figures 6a and b). Next, we sought to confirm that GSK3β inhibition leads to the induction of autophagy by PPARα activation. We applied siRNA to knockdown PPARα and the inhibition of liver PPARα-specific siRNA in vivo was confirmed by the reduced PPARα levels in the mice (Figure 6c). Compared with SB216763-pretreated ALF mice induced by d-GalN/LPS, the PPARα siRNA pretreatment reversed the gene and protein expression of Atg7, Atg5 and Beclin-1 induced by SB216763; importantly, the PPARα siRNA also decreased the lipidation of LC3I to LC3II and mildly increased the degradation of p62 (Figures 6d and e). Moreover, we further explored whether GSK3β inhibition protects liver from injury through the induction of PPARα in ALF mice. The results showed that hepatic protection by SB216763 in ALF was partially reversed by PPARα siRNA, which was evident by the relatively worse gross morphology, and the relatively less preserved liver architecture by histology (Supplementary Figure 2A), and the significantly higher sALT and sAST levels compared with SB216763-pretreated ALF mice group (Supplementary Figure 2B). Furthermore, in the context of GSK3β inhibition in the ALF mice, PPARα siRNA restored the gene expression of pro-inflammatory cytokines, such as TNF-α, IL-1β, IL-12p40 and chemokines including CCL-1 (chemokine (C-C motif) ligand), CCL-2, CXCL-1 (chemokine (C-X-C motif) ligand) and CXCL-10 (Supplementary Figure 2C). Thus, these results demonstrated that inhibition of GSK3β induces autophagy pathway in a PPARα-dependent manner in ALF.
Figure 6.
Autophagy is regulated by GSK3β-PPARα signaling pathway in ALF. (a and b) The mice were pretreated with or without SB216763 (25 mg/kg, i.p.) for 2 h, followed by d-GalN/LPS for 2, 4 or 6 h stimulation(n=10 mice per every group). Gene and protein expression levels of PPARα at 2, 4 and 6 h after d-GalN/LPS injection in livers from every groups. A representative blot for two samples from each group is shown. (c) SiRNA control- or siRNA PPARα-treated mice were injected with SiRNA control or PPARα siRNA (3 mg/kg) for 48 h via tail vein injection (n=10); Mice were sacrificed 48 h after siRNA PPARα treatment. Protein expression level of PPARα was measured by western blotting in livers. A representative blot for two samples from each group is shown. (d and e) SB+siRNA PPARα+d-GalN/LPS-treated mice were pretreated with PPARα siRNA (3 mg/kg) for 48 h via tail vein injection and then administered SB216763 2 h before d-GalN/LPS exposure (n=12); SB+siRNA control+d-GalN/LPS-treated mice were pretreated with control siRNA (3 mg/kg) for 48 h via tail vein injection, then administered SB216763 2 h before d-GalN/LPS exposure (n=10). Gene expression levels of autophagy-related proteins were measured by qRT-PCR in livers. Protein expression levels of PPARα and autophagy-related proteins were measured by western blotting in livers. Two representative blot for liver samples from each group were shown
Decreased expression of autophagy in the liver of ALF patients with HBV infection
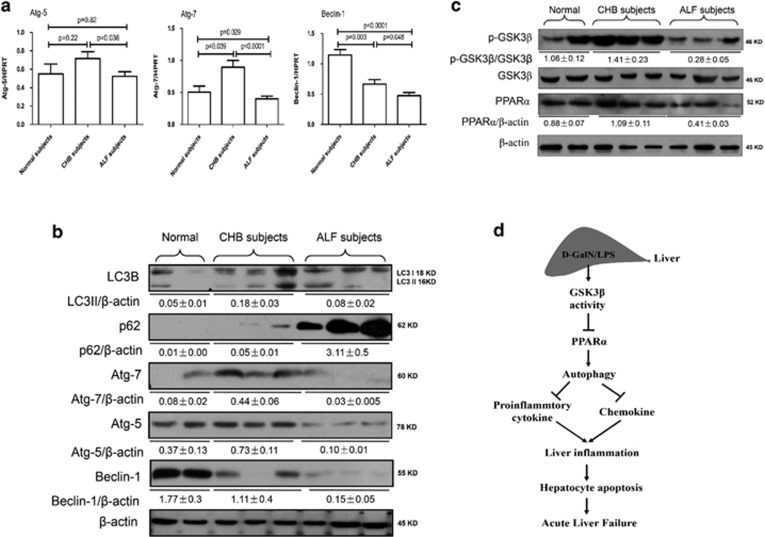
To determine whether autophagy participates in the progression of ALF in patients with hepatitis B virus (HBV) infection, we utilized liver tissues to measure the changes of autophagy among normal controls, chronic hepatitis B (CHB) patients and ALF patients with HBV infection. Relative to the normal controls, the qRT-PCR results showed that Atg7 and Atg5 gene expression levels were significantly increased in the cases of CHB, but were significantly reduced in the cases of ALF, but Beclin-1 gene expression was gradual decreased from the cases of CHB to ALF (Figure 7a); similar results were obtained by western blot analyses in liver tissue (Figure 7b); furthermore, the expression of LC3II was increased in CHB subjects and decreased in ALF subjects, and the degradation of p62 was significant decreased in the cases of ALF (Figure 7b). Moreover, relative to the normal controls, the western blotting results showed that the activity of GSK3β was downregulated in cases of CHB and upregulated in cases of ALF, and PPARα expression levels were not different in the cases of CHB, but were significantly reduced in the cases of ALF (Figure 7c). These results indicated that autophagy is depressed in ALF subjects induced by HBV infection, and it is likely to be regulated by GSK3β-PPARα pathway.
Figure 7.
Suppression of autophagy in the liver of ALF patients with HBV infection. (a) Gene expression levels of Atg7, Atg5 and Beclin-1 were measured by qRT-PCR in the livers of normal subjects (n=9), CHB patients (n=14) and ALF patients (n=19). The average target gene/HPRT ratios for each experimental group were plotted. (b) Protein expression levels of LC3B, p62, Atg7, Atg5 and Beclin-1 were measured by western blotting in the livers of normal subjects (n=9), CHB patients (n=14) and ALF patients (n=19). A representative blot from three samples of every group is shown. (c) Protein expression levels of p-GSK3β, GSK3β and PPARα were measured by western blotting in the livers of normal subjects (n=9), CHB patients (n=14) and ALF patients (n=19). A representative blot from three samples of every group is shown. (d) In the d-GalN/LPS-induced ALF mice, d-GalN/LPS treatment induces increase of GSK3β activity, and then suppresses the expression level of PPARα, which promotes down-regulation of autophagy and increases the expression of pro-inflammatory cytokines and chemokines. These events lead to incremental liver inflammation, and promote the hepatocyte apoptosis and ultimately induce the development of ALF
Discussion
In the present study, we demonstrated that autophagy is suppressed in d-GalN/LPS-induced ALF mice and ALF patients induced by HBV infection. Moreover, GSK3β inhibition resulted in elevated autophagy in vitro and in vivo, and autophagy inhibition reversed the hepatoprotective effect of GSK3β inhibition and restored the inflammatory response in ALF mice. Additionally, GSK3β inhibition promoted PPARα expression to increase autophagy in ALF mice. Hence, GSK3β-PPARα-autophagy pathway is essential to the liver inflammation mechanism in the ALF immune response cascade (as depicted in Figure 7d).
Although several previous investigations have suggested that autophagy serves as an adaptive response to cellular stress that avoids cell death,27 whether autophagy activation exerts protective properties in ALF mice models induced by d-GalN and LPS appears contradictory conclusions22, 28, 29 and the role of autophagy in the context of ALF patients induced by HBV infection are still scarce. Wang et al.28 and Amir et al.22 showed that autophagy is hepatoprotective in d-GalN/LPS-induced ALF mice model; but Li et al.29 showed that pretreatment with wortmannin alleviates d-GalN/LPS-induced acute liver injury by inhibit autophagy. Our results further confirmed that autophagy is promoted in the early phase and inhibited in the latter phase of liver injury (ALF phase) induced by d-GalN/LPS, and autophagy have a protective role in d-GalN/LPS-induced ALF mice.
What is the underlying molecular mechanism mediating the effect of autophagy in d-GalN/LPS-induced mice ALF models? The autophagy regulated by GSK3β-PPARα signaling pathway is a novel finding. Our lasted research shows that GSK3β has a key role in the pathogenesis of ALF, and the inhibition of GSK3β activity protect liver from acute injury by reducing liver inflammation.21 The studies have shown that GSK3β can regulate autophagy, but how to regulate autophagy, the different studies have given a different conclusion. The previous study suggested that lithium (an inhibitor of GSK3β activity) can induce autophagy by inhibiting inositol monophosphatase,30 however, the other study showed that lithium can reduce autophagy and apoptosis after neonatal hypoxia-ischemia.31 Inhibition of GSK3β activity using SB216763 or down-regulation of gene expression of GSK3β using interfering RNA, respectively, can promote autophagy to reduce cadmium-induced apoptosis;32 and the differential roles of GSK3β during myocardial ischemia and ischemia/reperfusion by regulating autophagy.33 In this paper, we definitely proved that GSK3β inhibition not only promotes autophagy but also has the differential influence on autophagolysosome formation in vivo and in vitro. In d-GalN/LPS-induced mice ALF, inhibition of GSK3β activity inhibits autophagolysosome formation and blocks the autophagic flux; but in starvation-treated hepatocyte with SB216763, inhibition of GSK3β promotes autophagic flux. The reason why such a difference may be due to the differential pathological mechanisms in vivo and in vitro experiments: for d-GalN/LPS-induced ALF mice, LPS-induced inflammation storm to cause a large number of liver cells apoptosis and necrosis; and, in vitro, the state of self-protection of hepatocyte is induced under starvation stress conditions. Whether or not, GSK3β is one of the important regulatory molecules of cell autophagy in the pathogenesis of ALF.
PPARs are members of the nuclear hormone receptor superfamily of ligand-activated transcription factors, with its subfamily consisting of three members: PPARα, PPARβ and PPARγ.34 PPARα has been reported to be involved in a number of cellular processes, including lipid and lipoprotein metabolism, apoptosis and inflammatory responses. PPARα-null mice exhibit an aggravated reaction to various inflammatory stimuli in the skin, blood vessels, intestine and lung,35, 36, 37, 38 and PPARα exhibits potent anti-inflammatory activity through suppressing nuclear factor-κB(NF-κB)and/or modulating the activation (phosphorylation) of signal transducer and activator of transcription1 (STAT1)-related inflammatory signaling in cultured neuronal cells.39, 40 Recently, in a model of lipopolysaccharide (LPS)-induced hepatic inflammatory injury, reduced PPARα expression was shown to be associated with increased tissue bacterial load in sepsis.41Additionally, a lack of PPARα exacerbates LPS-induced liver toxicity through STAT1 inflammatory signaling and increases oxidative/nitrosative stress.42 Our recent study found that, in both the mouse model and human ALF subjects, PPARα is significantly downregulated in the injured liver, and PPARα activation attenuates the inflammatory response to protect the liver from acute failure by promoting the autophagy pathway.26 In this paper, we further found that the inhibition of GSK3β activity increases PPARα expression and inhibition of PPARα restored the down-regulation of the autophagy induced by GSK3β inhibition in the liver. We demonstrated a new pathogenic mechanism of ALF: that the increased GSK3β activity-mediated PPARα suppress autophagy to promote the inflammatory response which induces hepatocyte apoptosis, as a result, which contributes to the development and progression of ALF.
In summary, this study adds to the general understanding of mechanisms of ALF and provides new insight into the importance of autophagy in regulating liver injury, partially regulated by a GSK3β-PPARα pathway. Therefore, autophagy activation represents a potent strategy to ameliorate ALF pathology. Further preclinical studies on autophagy agonists are warranted for the development of a clinically applicable therapeutic strategy against ALF.
Materials and Methods
Animal experiments
Male wild-type (WT, C57BL/6) mice (8–12 weeks of age) were purchased from Capital Medical University (Beijing, China) and housed in the Capital Medical University animal facility under specific pathogen-free condition and received humane care according to Capital Medical University Animal Care Committee guidelines.
The mice were intraperitoneally injected with d-GalN (700 mg/kg; Sigma, St. Louis, MO, USA) and LPS (10 μg/kg; InvivoGen, San Diego, CA, USA) to induce ALF or with saline in the control animals. Control mice received the same volume of saline. The specific inhibitor of GSK3β, SB216763 (25 mg/kg; Sigma), was given i.p. to mice 2 h before the administration of d-GalN/LPS. Autophagy is negatively regulated by mTOR, whose activity can be inhibited by rapamycin, a lipophilic macrolide antibiotic that is a well-established inducer of autophagy; 3-MA can block autophagy through its effect on class III phosphatidylinositol 3-kinase and CQ is common chemical medicine to inhibit the fuse of lysosomes and autophagosomes, which contributes to observe the autophagic flux. Therefore, for autophagy inhibition or induction, 3-MA (10 mg/kg; Sigma), CQ (60 mg/kg; Sigma) or rapamycin (2 mg/kg; Sigma) were given i.p. to mice 2 h before the administration of d-GalN/LPS. Furthermore, suppression of autophagy was also achieved by tail vein injection of siRNA for autophagy gene 7 (Atg7, 3 mg /kg). Suppression of PPARα was achieved by tail vein injection of siRNA for PPARα (3 mg/kg); the mice were sacrificed at various time points after d-GalN/LPS treatment, and liver and serum samples were collected for future analysis.
Serum aminotransferase activities
Plasma samples were taken from the mice at 6 h after d-GalN/LPS injection. Serum levels of ALT, AST as markers of hepatic damage were measured by using a multiparameteric analyzer (AU 5400, Olympus, Japan), according to an automated procedure.
Histopathological analysis
Liver tissues were fixed in formalin and embedded in paraffin wax, and sections in 5 μm were stained with H&E using a standard protocol, and then analyzed by light microscopy.
Quantitative real-time PCR analysis
Total RNA was isolated from hepatic samples using Trizol reagent according to the manufacturer's protocol. A total of 2.5 μg of RNA was reverse-transcribed into cDNA using SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). Quantitative-PCR was performed using the DNA Engine with Chromo 4 Detector (MJ Research, Waltham, MA, USA). The following were added to a final reaction volume of 20 μl: 1x SuperMix (Platinum SYBR Green qPCR Kit; Invitrogen); cDNA (2 μl); and 0.5 μM of each primer. The amplification conditions were as follows: 50 °C (2 min); 95 °C (5 min); followed by 50 cycles of 95 °C (15 s) and 60 °C (30 s).
Assay of MDA in liver
A part of liver tissue was prepared for homogenization with a buffer containing 0.15 M KCl to obtain 1:10 (w/v) homogenates. The homogenates were then centrifuged at 12 000 × g (4 °C) for 20 min to collect the supernatants. The concentrations of MDA activities were measured, as described previously43.
Western blot analyses
Protein was extracted from liver tissue in RIPA buffer together with phosphatase and protease inhibitors. Proteins in SDS-loading buffer were subjected to SDS-12% polyacrylamide gel electrophoresis (PAGE) and transferred to a PVDF membrane (Bio-Rad, Hercules, CA, USA). Antibodies against LC3B, Atg7, Atg5, Beclin-1, p62, β-actin (Cell Signaling Technology Inc., Santa Cruz, CA, USA) and PPARα (Abcam, Cambridge, MA, USA), were used for western blot analysis. The membranes were probed with primary antibodies (1:500–1000) in 10 ml of blocking buffer overnight at 4 °C. After washing, the membranes were further probed with the appropriate horseradish peroxidase-conjugated secondary antibody (1:2000) in 10 ml of blocking buffer for 1 h at room temperature. The Supersignal West Pico chemiluminescent substrates (Thermo Fisher Scientific, Rockford, IL, USA) were used for chemiluminescence development. Finally, the quantitative results of western blots were be assessed by ImageJ to make comparisons between different groups.
Atg7 or PPARα small interference RNA treatment in vivo
Autophagy was inhibited through the siRNA against Atg7 (3 mg/kg; Jima, Suzhou); its sequence is 5′-GCAUCAUCUUCGAAGUGAATT-3′. PPARα was inhibited through the siRNA against PPARα (3 mg/kg; Jima, Suzhou); its sequence is 5′-GAGAUCGGCCUGGCCUUCUAAACAU-3′. Atg7 or PPARα knockdown was achieved by siRNA using an Entranster in vivo transfection reagent (Engreen Biosystem Co, Beijing) via hydrodynamic tail vein injection in mice. Scrambled siRNA (3 mg/kg) was used as a control. The processes were performed following the manufacturer's instructions.
Isolation of primary mouse hepatocytes
The mouse livers were perfused with collagenase-containing Hanks' solution at 7 weeks of age, and viable hepatocytes were isolated by Percoll isodensity centrifugation as described.44
Starvation-induced autophagy in vitro
Starvation is the most extensively studied condition that induces autophagy. Transfected GFP-LC3 plasmid for 12 h, the primary hepatocytes were subjected to amino acid starvation by incubating for different hours with Earle's balanced salt solution. The percentage of cells with GFP-LC3 puncta were counted in the different groups receiving different treatment. GFP-positive cells were defined as cells that display bright, punctate staining. Approximately 50 cells were counted, and the experiment was repeated at least three times.
TUNEL assay
Apoptosis in liver sections was detected by TUNEL (red fluorescence) using the In Situ Cell Death Detection Kit (Roche, Indianapolis, IN, USA). Negative control was prepared through omission of terminal transferase. Positive controls were generated by treatment with DNase. Nuclei were stained with 49,6-diamino-2-phenylindole (DAPI, 1 μg/ml) for 10 min. Images were performed on a Nikon Eclipse E800 fluorescent microscope (Nikon Corp., Tokyo, Japan).
Human specimens
Normal liver tissues (n=10) were collected from four subjects undergoing hepatic resection for hepatic cyst, and three subjects undergoing hepatic resection for colorectal metastasis, three subjects undergoing hepatic resection for benign tumor, such as angeioma. CHB samples were obtained from the livers of 14 patients undergoing liver puncture biopsy. ALF samples were obtained from the livers of 19 patients with HBV infection undergoing liver transplantation. This study meets the ethical guidelines of the 1975 Declaration of Helsinki, and the study protocol was approved by the Medical Ethics Committee of Beijing YouAn Hospital. Informed consent was obtained from all patients. The clinical characteristics and details of the patients included in the study are shown in Table 1.
Table 1. General clinical characteristics of the different study groups.
| Normal subjects (n=10) | Chronic hepatitis B patients (n=14) | Acute liver failure patients (n=19) | P-value | |
|---|---|---|---|---|
| Age (years) | 39.4±3.6 | 32.4±4.1 | 41.6±5.3 | 0.11 |
| Gender (male/female) | 6/2 | 7/5 | 8/4 | 0.437 |
| Alanine aminotransferase (U/l) | 35.1±6.1 | 93.3±18.3 | 210.4±76.3 | 0.026 |
| Aspartate aminotransferase (U/l) | 30.6±3.9 | 62.6±15.4 | 305.8±44.6 | 0.038 |
| Serum bilirubin (μmol/l) | 8.8±2.9 | 20.9±6.3 | 196.1±50.6 | 0.01 |
| Prothrombin time (s) | 9±2.4 | 15±5.2 | 34.4±7.2 | 0.022 |
| Albumin (g/l) | 46.2±6.9 | 32.6±10.8 | 26.7±5.7 | 0.039 |
| Creatinin (μmol/l) | 73.6±21.5 | 80.1±29.0 | 95.4±32.8 | 0.042 |
| Hepatic encephalopathy score | — | — | 1.7±0.3 | — |
| Child-Pugh score | — | 6±0.6 | 13.7±2.4 | 0.031 |
| Model for end-stage liver disease score | — | — | 28.2±4.2 | — |
| HBsAg test | — | Positive | Positive | — |
Statistical analyses
The results are shown as the mean±S.E.M. The statistical analyses were performed using an unpaired Student's t-test, and P<0.05 (two tailed) was considered significant.
Acknowledgments
We thank Professor Yuanping Han (Si Chuan University) for improving the language and presentation of this manuscript. This study was supported by China National Key Project of the Twelfth Five-year Plan (2012ZX10002004-006, 2012ZX10004904-003-001, 2013ZX10002002-006-001), the National Natural Science Foundation of China (81270532, 81372094, 81300349), Wang Boen Liver Fibrosis Research Foundation of CFHPC (CFHPC20131031), Applied Research for the Clinical Characteristics of Capital (Z121107001012167), The Project of Construction of Innovative Teams and Teacher Career Development for Universities and Colleges Under Beijing Municipality (IDHT20150502) and High-level Technical Personnel Training Plan of Beijing Health System (2013-3-075).
Author contributions
FR, LZ, XZ, HS and TW performed the experiments; LB, SZ and YC analyzed clinical data and discussed the results; FR and LL drafted the manuscript; ZD and DC designed experiments and revised the manuscript. All authors contributed to manuscript editing and approval.
Glossary
- ALF
acute liver failure
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- Atg
autophagy gene
- CCL
chemokine (C-C motif) ligand
- CXCL
chemokine (C-X-C motif) ligand
- CHB
chronic hepatitis B
- CQ
chloroquine
- d-GalN
d-galactosamine
- HBV
hepatitis B virus
- IL-1β
interleukin-1β
- IL-6
interleukin-6
- GSK3β
glycogen synthase kinase 3β
- LPS
lipopolysaccharide
- PPARα
peroxisome proliferator-activated receptor α
- qRT-PCR
quantitative reverse transcription PCR
- siRNA
small interfering RNA
- 3-MA
3-methyladenine
- TNF-α
tumor necrosis factor-α
- TUNEL
terminal deoxynucleotidyl transferasemediated dUTP nick-end labeling
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by GM Fimia
Supplementary Material
References
- Rolando N, Wade J, Davalos M, Wendon J, Philpott-Howard J, Williams R. The systemic inflammatory response syndrome in acute liver failure. Hepatology 2000; 32: 734–739. [DOI] [PubMed] [Google Scholar]
- Hoofnagle JH, Carithers RJ, Shapiro C, Ascher N. Fulminant hepatic failure: summary of a workshop. Hepatology 1995; 21: 240–252. [PubMed] [Google Scholar]
- Cohen P, Goedert M. GSK3 inhibitors: development and therapeutic potential. Nat Rev Drug Discov 2004; 3: 479–487. [DOI] [PubMed] [Google Scholar]
- Rayasam GV, Tulasi VK, Sodhi R, Davis JA, Ray A. Glycogen synthase kinase 3: more than a namesake. Br J Pharmacol 2009; 156: 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Rehani K, Jope RS, Michalek SM. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat Immunol 2005; 6: 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurel E, Michalek SM, Jope RS. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3). Trends Immunol 2010; 31: 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Garcia CA, Rehani K, Cekic C, Alard P, Kinane DF et al. IFN-beta production by TLR4-stimulated innate immune cells is negatively regulated by GSK3-beta. J Immunol 2008; 181: 6797–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eléonore Beurel, Richard S. Jope. The paradoxical pro- and anti-apoptotic actions of GSK-3 in the intrinsic and extrinsic apoptosis signaling pathways. Prog Neurobiol 2006; 79: 173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature 2000; 406: 86–90. [DOI] [PubMed] [Google Scholar]
- Schwabe RF, Brenner DA. Role of glycogen synthase kinase-3 in TNF-alpha-induced NF-kappaB activation and apoptosis in hepatocytes. Am J Physiol Gastrointest Liver Physiol 2002; 283: G204–G211. [DOI] [PubMed] [Google Scholar]
- Linlin W, Feng R, Xiangying Z, Wen T, Shi H, Zheng S et al. Oxidative stress promotes d-GalN/LPS-induced acute hepatotoxicity by increasing glycogen synthase kinase 3β activity. Inflamm Res 2014; 63: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyan C, Feng R, Haiyan Z, Wen T, Piao Z, Zhou L et al. Inhibition of GSK3β ameliorates d-GalN/LPS-induced liver injury by reducing ERS-triggered apoptosis. Plos One 2012; 7: e45202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2001; 2: 211–216. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol 2007; 7: 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beth L, Noboru M, Herbert WV. Autophagy in immunity and inflammation. Nature 2011; 469: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander JSC, Stefan WR. Autophagy in inflammatory diseases. Int J Cell Biol 2011; doi: 10.1155/2011/732798. [DOI] [PMC free article] [PubMed]
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J 2008; 27: 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 2007; 27: 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 2008; 456: 264–268. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 2011; 12: 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir M, Zhao E, Fontana L, Rosenberg H, Tanaka K, Gao G et al. Inhibition of hepatocyte autophagy increases tumor necrosis factor-dependent liver injury by promoting caspase-8 activation. Cell Death Differ 2013; 20: 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignon A, Rouquet N, Fabre M, Martin S, Pagès JC, Dhainaut JF et al. LPS challenge in D-galactosamine-sensitized mice accounts for caspase-dependent fulminant hepatitis, not for septic shock. Am J Respir Crit Care Med 1999; 159: 1308–1315. [DOI] [PubMed] [Google Scholar]
- Nakama T, Hirono S, Moriuchi A, Hasuike S, Nagata K, Hori T et al. Etoposide prevents apoptosis inmouse liver with d-GalN/LPS-induced fulminant hepatic failure resulting inreduction of lethality. Hepatology 2001; 33: 1441–1450. [DOI] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Pankiv S, Øvervatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 2009; 452: 181–197. [DOI] [PubMed] [Google Scholar]
- Mingjin J, Feng R, Li Z, Xiangying Z, Li Z, Tao W et al. Peroxisome proliferator-activated receptor a activation attenuates the inflammatory response to protect the liver from acute failure by promoting the autophagy pathway. Cell Death Dis 2014; 5: e1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8: 741–752. [DOI] [PubMed] [Google Scholar]
- Wang K, Damjanov I, Wan YJ. The protective role of pregnane X receptor in lipopolysaccharide/D-galactosamine-induced acute liver injury. Lab Invest 2010; 90: 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang X, Wei Z, Mao H, Gao M, Liu Y et al. Pretreatment with wortmannin alleviates lipopolysaccharide/D-galactosamine-induced acute liver injury. Biochem Biophys Res Commun 2014; 455: 234–240. [DOI] [PubMed] [Google Scholar]
- Sovan S, Floto RA, Zdenek B, Imarisio S, Cordenier A, Pasco M. et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol 2005; 170: 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li H, Roughton K, Kroemer G, Blomgren K, Zhu C. Lithium reduces apoptosis and autophagy after neonatal hypoxia-ischemia. Cell Death Dis 2010; 1: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Shih YL, Kuo T, Ko WC, Shih CM. Cadmium toxicity toward autophagy through ROS-activated GSK3β in mesangial cells. Toxicol Sci 2009; 108: 124–131. [DOI] [PubMed] [Google Scholar]
- Peiyong Z, Sebastiano S, Jonathan G, Volpe M, Sadoshima J. Differential roles of GSK3β during myocardial Ischemia and ischemia/reperfusion. Circ Res 2011; 109: 502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota BP, Huang TH, Roufogalis BD. An overview on biological mechanisms of PPARs. Pharmacol Res 2005; 51: 85–94. [DOI] [PubMed] [Google Scholar]
- Delayre OC, Becker J, Guenon I, Lagente V, Auwerx J, Frossard N et al. PPAR alpha downregulates airway inflammation induced by lipopolysaccharide in the mouse. Respir Res 2005; 6: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di PR, Esposito E, Mazzon E, Genovese T, Muia C, Crisafulli C et al. Absence of peroxisome proliferators-activated receptors (PPAR) alpha enhanced the multiple organ failure induced by zymosan. Shock 2006; 26: 477–484. [DOI] [PubMed] [Google Scholar]
- Sheu MY, Fowler AJ, Kao J, Schmuth M, Schoonjans K, Auwerx J et al. Topical peroxisome proliferator activated receptor-alpha activators reduce inflammation in irritant and allergic contact dermatitis models. J Invest Dermatol 2002; 118: 94–101. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Abdelmegeed MA, Song BJ. Activation of PPARa by Wy-14,643 ameliorates systemic lipopolysaccharide-induced acute lung injury. Biochem Biophys Res Commun 2013; 436: 366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delerive P, De BK, Besnard S, Vanden BW, Peters JM, Gonzalez FJ et al. PPARα negatively regulates the vascular inflammatory gene response by negative cross talk with transcription factors NF-κB and AP-1. J Biol Chem 1999; 274: 32048–32054. [DOI] [PubMed] [Google Scholar]
- Lee JH, Joe EH, Jou I. PPAR-alpha activators suppress STAT1 inflammatory signaling in lipopolysaccharide-activated rat glia. Neuroreport 2005; 16: 829–833. [DOI] [PubMed] [Google Scholar]
- Stephen WS, Charles CC, Basilia Z, Hector RW. Reduced peroxisome proliferator-activated receptor α expression is associated with decreased survival and increased tissue bacterial load in sepsis. Shock 2012; 37: 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong HY, Ogyi P, Lauren EH, Mohamed AA, Kwan HM, Byoung JS. Lack of PPARα exacerbates lipopolysaccharide-induced liver toxicity through STAT1 inflammatory signaling and increased oxidative/nitrosative stress. Toxicol Lett 2011; 202: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZM, Wen T, Tan YF, Liu Y, Ren F, Wu H. Effects of salvianolic acid A on oxidative stress and liver injury induced by carbon tetrachloride in rats. Basic Clin Pharmacol Toxicol 2007; 100: 115–120. [DOI] [PubMed] [Google Scholar]
- Klaunig JE, Goldblatt PJ, Hinton DE, Lipsky MM, Chacko J, Trump BF. Mouse liver cell culture. I. Hepatocyte isolation. In Vitro 1981; 17: 913–925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.