Abstract
Aspergillus ochraceoroseus and Aspergillus rambellii were isolated from soil detritus in Taï National Park, Ivory Coast, Africa. The Type strain for each species happens to be the only representative ever sampled. Both species secrete copious amounts of aflatoxin B1 and sterigmatocystin, because each of their genomes contains clustered genes for biosynthesis of these mycotoxins. We sequenced their genomes using a personal genome machine and found them to be smaller in size (A. ochraceoroseus = 23.9 Mb and A. rambellii = 26.1 Mb), as well as in numbers of predicted genes (7,837 and 7,807, respectively), compared to other sequenced Aspergilli. Our findings also showed that the A. ochraceoroseus Type strain contains a single MAT1-1 gene, while the Type strain of A. rambellii contains a single MAT1-2 gene, indicating that these species are heterothallic (self-infertile). These draft genomes will be useful for understanding the genes and pathways necessary for the cosynthesis of these two toxic secondary metabolites as well as the evolution of these pathways in aflatoxigenic fungi.
Keywords: Aspergillus ochraceoroseus, Aspergillus rambellii; genome sequence; phylogenomics; mating-type locus
Introduction
Aspergillus ochraceoroseus (ATCC 38873) and Aspergillus rambellii (ATCC 42001) are two fungal organisms that were first characterized in 1978 (Bartoli and Maggi 1978). Both were named A. ochraceoroseus until a more thorough comparison, decades later, revealed differences which resulted in renaming one of the isolates A. rambellii (Frisvad et al. 2005). These two isolates are the sole representatives of their respective species ever sampled, because constant civil unrest in the Ivory Coast makes further sampling a dangerous task, and also because no isolates of either species have been found anywhere else (Klich et al. 2003). These two species have not been found in agricultural or clinical environments, but they both secrete two important secondary metabolites (SMs): aflatoxin B1 (AFB1) and sterigmatocystin (ST). AFB1 is considered the most serious of the mycotoxins produced by any organism (Samuel et al. 2013). Frisvad et al. (2005) reported that A. rambellii secreted a greater concentration of AFB1 than any other aflatoxigenic species. Large quantities of ST are also secreted by both species. In many of the Aspergillus species from subgenus Circumdati, this less potent SM is most often an intermediate compound in the aflatoxin (AF) pathway, serving as a precursor in the synthesis of B1 and G1 AFs (Yu et al. 2004). ST has its own pathway of clustered genes in Aspergilli, mostly from subgenus Nidulantes, many of which are homologous to AF cluster genes but organized differently (Ehrlich et al. 2005). Whether or not these organisms are the ancestors to modern-day AF- and ST-producers, or the result of hybridization or horizontal gene transfer (HGT) events between AF and ST species, has yet to be determined. The genomes described here will allow for further studies that result in greater understanding of the evolution of aflatoxigenic Aspergillus species.
Comparative Analysis against Sequenced and Annotated Aspergillus Genomes
The draft genome assembly for A. ochraceoroseus was 23.9 Mb, composed of 2,644 contigs with an N50 of 330 and N50 length of 21,980 bp, and 48.48% G+C content. The A. rambellii genome was larger (26.1 Mb), composed of 3,102 contigs with an N50 of 206 and N50 length of 38,345 bp, and 46.16% G+C content. More genomic information for these Type strains is shown in table 1.
Table 1.
Genome Characteristics for the Aspergillus ochraceoroseus and Aspergillus rambellii Type Strains
| Genome Characteristic | Value |
|---|---|
| A. ochraceoroseus | |
| General | |
| Assembly size (bp) | 23,940,954 |
| CEGMA % completeness | 92.74 |
| Average depth | 36 |
| G+C (%) | 48.9 |
| Protein-coding genes | 7,837 |
| Protein-coding genes >100 amino acids | 7,631 |
| Predicted protein-coding sequences >100 amino acids | |
| Gene density (1 gene every n bp) | 3,054 |
| Median gene length (bp) | 1,799.9 |
| Mean gene length (bp) | 1,502 |
| Average number of exons per gene | 3.34 |
| A. rambellii | |
| General | |
| Assembly size (bp) | 26,069,208 |
| CEGMA % completeness | 95.97 |
| Average depth | 79 |
| G+C (%) | 46.2 |
| Protein-coding genes | 7,807 |
| Protein-coding genes >100 amino acids | 7,566 |
| Predicted protein-coding sequences >100 amino acids | |
| Gene density (1 gene every n bp) | 3,339 |
| Median gene length (bp) | 1,826.6 |
| Mean gene length (bp) | 1,517.5 |
| Average number of exons per gene | 3.37 |
The genomes of A. ochraceoroseus and A. rambellii were approximately 10 Mb smaller, and appeared to encompass 3,000–5,000 fewer predicted genes, than those for many other sequenced Aspergillus species (table 2). One species with the closest genome size and predicted gene counts to A. ochraceoroseus and A. rambellii was Aspergillus fumigatus (29 Mb and 9,296, respectively). One reason for A. ochraceoroseus and A. rambellii exhibiting smaller genome sizes likely related to the number of SM clusters found within these fungi. For example, A. fumigatus contained 18 putative SM clusters and Aspergillus nidulans contained 35 putative SM clusters, while species such as Aspergillus flavus (L-type) and Aspergillus nomius contained 45 and 53 SM clusters, respectively (Moore et al. 2015). The A. ochraceoroseus genome contained 18 putative SM clusters and the A. rambellii genome contained 20 putative SM clusters. It has been postulated that high genetic drift, coupled with deletions, in eukaryotes refines their genomes by removing regions that are not integral to the organism’s survival (Kelkar and Ochman 2012). From an evolutionary standpoint this could indicate that A. ochraceoroseus and A. rambellii represent a more streamlined genomic condition for aflatoxigenic Aspergilli. Unfortunately, no other isolates have been sampled for either species to aid with population-scale inferences, so their geographic isolation coupled with their lack of ubiquity adds to their value.
Table 2.
Genomic Comparison of Six Aspergillus Species
| Species Name | Genome Size (Mb) | Predicted Genes | References |
|---|---|---|---|
| A. flavus | 37 | 13,485 | Nierman et al. 2015 |
| A. fumigatus | 29 | 9,926 | Nierman et al. 2005 |
| A. nidulans | 30 | 10,605 | Galagan et al. 2005; Wortman et al. 2009 |
| A. nomius | 36 | 11,918 | Moore et al. 2015 |
| A. ochraceoroseus | 24 | 7,837 | |
| A. rambellii | 26 | 7,807 |
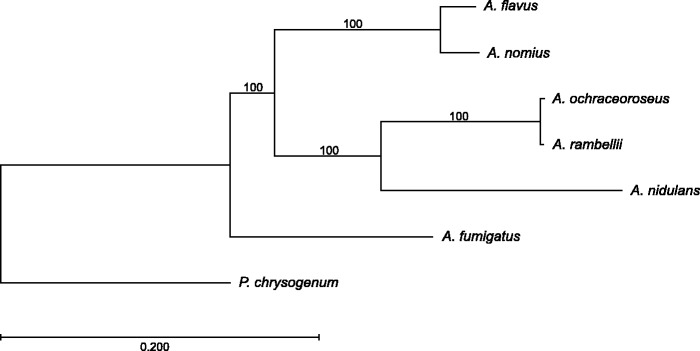
The availability of whole-genome data could help us to understand the origins, or evolution, of AF- and ST-producing (AF+ST) fungi. Phylogenomic comparison of A. ochraceoroseus and A. rambellii, with other Aspergillus species and the outgroup taxa Penicillium chrysogenum, indicated that these two species share a most recent common ancestor with A. nidulans (fig. 1). Even farther back in evolutionary time was their divergence from the AF-producing species representing section Flavi, preceded by divergence from a shared common ancestor with A. fumigatus (a nonproducer of AF or ST). Sequencing the genomes of more aflatoxigenic species will improve our understanding of how the AF or ST chemotype has evolved.
Fig. 1.—
Phylogenomic comparison of sequenced Aspergillus species reveals patterns of ancestry. This tree was inferred using multiple AF and/or ST producing Aspergillus species (A. flavus L, A. nidulans, A. nomius, A. ochraceoroseus, and A. rambellii) and one AF and ST nonproducing species (A. fumigatus), with Penicillium chrysogenum as the outgroup taxa.
Potential recombination or HGT among fungi may cloud a species’ true lineage; therefore, whole-genome comparisons offer a more holistic approach to lineage-based inferences. Previous single-locus phylogenies have shown the cladal relatedness of these ST-producing fungi (Klich et al. 2003; Frisvad et al. 2005), but the coproduction of AFB1 by A. ochraceoroseus and A. rambellii could indicate that the distant common ancestor shared with the strictly aflatoxigenic species may have been able to secrete both SMs, and that this ability was lost in A. nidulans at the time of divergence. Coalescence analysis of species containing the ST pathway may help to determine whether or not the ancestor of modern-day ST producers could also produce AFB1. An alternative scenario could be HGT of one or more genes, that allow for further synthesis of the ST intermediate to AFB1, from an aflatoxigenic species into these AF+ST producing species. Further research is required to confirm or refute the HGT scenario.
With regard to the AF+ST biosynthesis pathway, clustered genes in A. ochraceoroseus and A. rambellii share 99.4% identity and the same orientation. This confirmed a previous report by Cary et al. in which they characterized the A. ochraceoroseus AF+ST gene cluster. They found it to contain the same genes as the ST cluster of A. nidulans, but with a portion of the cluster (aflN to aflW) having reversed orientation (Cary et al. 2009). The basis for AFB1 synthesis in A. ochraceoroseus was unknown because two of the genes required for conversion of ST to AFB1 (aflP and aflQ) were not found within its AF+ST gene cluster. Recently, however, an ortholog of the aflQ gene was found to exist within the A. ochraceoroseus genome (Cary et al. 2012). BLAST query of the A. ochraceoroseus aflQ ortholog to the A. rambellii genome sequence revealed a putative aflQ ortholog sharing 99.95% identity. Phylogenetic analysis involving the aflQ gene from multiple AF and ST producing fungi revealed the ortholog in A. ochraceoroseus to be most similar to the most recent common ancestor than those of AF- or ST-producing species (Cary et al. 2012), which supports the existence of an ancestor that was able to produce and secrete both toxic SMs.
Previous research reported a possible heterothallic existence for A. ochraceoroseus and A. rambellii, with each species containing a single mating-type (MAT) idiomorph (Cary et al. 2012). Our findings from sequencing the genomes of these closely related species confirmed that A. ochraceoroseus contains a single (MAT1-1) gene and A. rambellii contains a single (MAT1-2) gene. The ability of these species to outcross with one another has not been reported. The other heterothallic Aspergillus species such as A. flavus and A. parasiticus have a MAT gene flanked by two conserved genes in close proximity: one for DNA lyase (APN) and one for cytoskeleton assembly control (SLA). The relative distances of these genes to the MAT1-1 gene in A. flavus and A. parasiticus are approximately 2,500 bp and 2,000 bp, respectively (Ramirez-Prado et al. 2008). For the MAT1-2 gene in these same two species, the distances are approximately 1,000 and 3,500 bp, respectively. For the A. ochraceoroseus MAT1-1 gene, there are over 51,000 bp of nucleotide sequence between it and the APN gene, and 2,300 bp of nucleotide sequence separating it from the SLA gene. The distances between each of the APN and SLA genes and the MAT1-2 gene in A. rambellii measured over 33,000 bp and 3,600 bp, respectively. The chromosomal location of the MAT locus in A. flavus and A. parasiticus is reported to be Chromosome VI (Ramirez-Prado et al. 2008), but this has not yet been determined for A. ochraceoroseus and A. rambellii. If the pathway cluster genes within these two AF+ST producing species are reflections of an ancestral organization that predates the divergence of AF and ST producers, then perhaps their MAT loci are also examples of a more relaxed [ancestral] organization and could impact their ability to outcross. There are conflicting theories about which form of sex (heterothallism vs. homothallism) is ancestral in fungi (Ni et al. 2011). If A. ochraceoroseus and A. rambellii are indeed reflective of an ancestor that produced AFB1 and ST, and that contained a more disorganized heterothallic mating type locus, then perhaps these species support the argument that heterothallism is ancestral to homothallism.
Materials and Methods
Genome Sequence and Annotation
We sequenced the genomes of the A. ochraceoroseus and A. rambellii Type strains species using a personal genome machine (PGM) from Life Technologies (Grand Island, New York). Template preparation and sequencing was conducted according to previously reported protocols (Moore et al. 2015). Totals of 2.92 M and 7.43 M reads were obtained for A. ochraceoroseus and A. rambellii, respectively. Their respective genome assemblies and annotations were performed as reported in Moore et al. (2015) with slight modifications. For example, we used CEGMA (Parra et al. 2007) to identify conserved genes which were used to train Augustus. Maker was then used to integrate ab initio gene predictions with protein homology evidence from the UniRef50 protein database (ftp://ftp.ncbi.nlm.nih.gov/refseq/release/fungi/, last accessed July 1, 2014) for A. rambellii, and the NCBI fungal RefSeq database (ftp://ftp.uniprot.org/pub/databases/uniprot/uniref/uniref50/uniref50.fasta.gz, last accessed July 1, 2014) for A. ochraceoroseus. The annotations were converted to NCBI submission format using Genome Annotation Generator [https://github.com/genomeannotation/GAG, last accessed October 14, 2015] and deposited at NCBI under BioSample project numbers PRJNA275128 (A. ochraceoroseus) and PRJNA275129 (A. rambellii).
Genomic Comparisons for Various Aspergillus Species
The Antibiotics-Secondary Metabolite Analysis Shell (anti-SMASH) program was used to predict SM clusters in A. ochraceoroseus and A. rambellii (Medema et al. 2011). This program is modeled to look for many types of backbone genes, but our estimates accounted for the presence of only three types: polyketide synthase (PKS), nonribosomal peptide synthetase (NRPS), and PKS-NRPS hybrid. Phylogenomic analysis involved protein orthologs, from multiple Aspergillus species that were identified by Proteinortho, and was performed using similar techniques as discussed in Moore et al. (2015). Ortholog groups that contained paralogs were not included. Penicillium chrysogenum was used as the outgroup taxa and we inferred the evolutionary relationships of A. ochraceoroseus and A. rambellii with two aflatoxigenic species from section Flavi (A. flavus L and A. nomius), an ST producer from section Nidulantes (A. nidulans) and A. fumigatus. The protein sequences for A. nidulans, A. flavus, and A. fumigatus were retrieved from FungiDB (accessed August 28, 2014). Penicillium chrysogenum was obtained from JGI (last accessed August 28, 2014). The A. nomius protein sequences were obtained from NCBI (last accessed August 28, 2014). The MAT locus comparisons were performed by BLAST query of A. flavus MAT, APN, and SLA genes to the A. ochraceoroseus and A. rambellii genomes. Distance mapping between the MAT genes and each flanking gene were performed using Sequencher software (Gene Codes Corporation, Ann Arbor, MI).
Acknowledgments
This work was supported by research funding from the Agricultural Research Service, an agency within the United States Department of Agriculture.
Literature Cited
- Bartoli A, Maggi O. 1978. Four new species of Aspergillus from Ivory Coast soil. Trans Br Mycol Soc. 71:383–394. [Google Scholar]
- Cary JW, Ehrlich KC, Beltz SB, Harris-Coward P, Klich MA. 2009. Characterization of the Aspergillus ochraceoroseus aflatoxin/sterigmatocystin biosynthetic gene cluster. Mycologia 101:352–362. [DOI] [PubMed] [Google Scholar]
- Cary JW, et al. 2012. Functional and phylogenetic analysis of the Aspergillus ochraceoroseus aflQ (ordA) gene ortholog. Mycologia. 104:857–864. [DOI] [PubMed] [Google Scholar]
- Ehrlich KC, Yu J, Cotty PJ. 2005. Aflatoxin biosynthesis gene clusters and flanking regions. J Appl Microbiol. 99:518–527. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Skouboe P, Samson RA. 2005. Taxonomic comparison of three different groups of aflatoxin producers and a new efficient producer of aflatoxin B1, sterigmatocystin and 3-O-methylsterigmatocystin, Aspergillus rambellii sp. nov. Sys Appl Microbiol. 28:442–453. [DOI] [PubMed] [Google Scholar]
- Galagan JE, et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 438:1105–1115. [DOI] [PubMed] [Google Scholar]
- Kelkar YD, Ochman H. 2012. Causes and consequences of genome expansion in fungi. Genome Biol Evol. 4:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klich MA, Cary JW, Beltz SB, Bennett CA. 2003. Phylogenetic and morphological analysis of Aspergillus ochraceoroseus. Mycologia 95:1252–1260. [DOI] [PubMed] [Google Scholar]
- Medema MH, et al. 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39:W339–W346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore GG, Mack BM, Beltz SB. 2015. Genomic sequence of the aflatoxigenic filamentous fungus Aspergillus nomius. BMC Genomics 16:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Sun S, Wang X, Heitman J. 2011. Sex in fungi. Annu Rev Genet. 45:405–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156. [DOI] [PubMed] [Google Scholar]
- Nierman WC, et al. 2015. Genome sequence of Aspergillus flavus NRRL 3357, a strain that causes aflatoxin contamination of food and feed. ASM Genome Announc. 3:e00168-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. [DOI] [PubMed] [Google Scholar]
- Ramirez-Prado JH, Moore GG, Horn BW, Carbone I. 2008. Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet Biol. 45:1292–1299. [DOI] [PubMed] [Google Scholar]
- Samuel SM, Aiko V, Panda P, Mehta A. 2013. Aflatoxin B1 occurrence, biosynthesis and its degradation. J Pure Appl Microbiol. 7:965–971. [Google Scholar]
- Wortman JR, et al. 2009. The 2008 update of the Aspergillus nidulans genome annotation: a community effort. Fungal Genet Biol. 46:S2–S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, et al. 2004. Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol. 70:1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]