Abstract
The predominantly aquatic order Alismatales, which includes approximately 4,500 species within Araceae, Tofieldiaceae, and the core alismatid families, is a key group in investigating the origin and early diversification of monocots. Despite their importance, phylogenetic ambiguity regarding the root of the Alismatales tree precludes answering questions about the early evolution of the order. Here, we sequenced the first complete plastid genomes from three key families in this order: Potamogeton perfoliatus (Potamogetonaceae), Sagittaria lichuanensis (Alismataceae), and Tofieldia thibetica (Tofieldiaceae). Each family possesses the typical quadripartite structure, with plastid genome sizes of 156,226, 179,007, and 155,512 bp, respectively. Among them, the plastid genome of S. lichuanensis is the largest in monocots and the second largest in angiosperms. Like other sequenced Alismatales plastid genomes, all three families generally encode the same 113 genes with similar structure and arrangement. However, we detected 2.4 and 6 kb inversions in the plastid genomes of Sagittaria and Potamogeton, respectively. Further, we assembled a 79 plastid protein-coding gene sequence data matrix of 22 taxa that included the three newly generated plastid genomes plus 19 previously reported ones, which together represent all primary lineages of monocots and outgroups. In plastid phylogenomic analyses using maximum likelihood and Bayesian inference, we show both strong support for Acorales as sister to the remaining monocots and monophyly of Alismatales. More importantly, Tofieldiaceae was resolved as the most basal lineage within Alismatales. These results provide new insights into the evolution of Alismatales as well as the early-diverging monocots as a whole.
Keywords: complete plastid genome, genome evolution, phylogenomics, Alismatales, basal monocots
Introduction
As currently defined, the monocot order Alismatales is a cosmopolitan and highly diverse group comprising 14 families with approximately 166 genera and approximately 4,500 species. Alismatales is often divided into three clades: Araceae, Tofieldiaceae, and 12 families composing the superorder Alismatiflorae (Dahlgren et al. 1985), which are also generally known as Helobiae (Tomlinson 1982) or core alismatids (Stevens 2001 onwards; Angiosperm Phylogeny Group 2009; Iles et al. 2013; Les and Tippery 2013). The core alismatids can be further subdivided into two informal groups based on floral characteristics: a “petaloid” and a “tepaloid” clade (Posluszny and Charlton 1993; Posluszny et al. 2000). Petaloid alismatids are composed of three families, Alismataceae (including Limnocharitaceae), Butomaceae, and Hydrocharitaceae, whereas tepaloid alismatids are composed of nine families: Aponogetonaceae, Cymodoceaceae, Juncaginaceae, Ma-undiaceae, Posidoniaceae, Potamogetonaceae, Ruppiaceae, Scheuchzeriaceae, and Zosteraceae (Posluszny and Charlton 1993; Posluszny et al. 2000).
Alismatales is one of the most important orders widely associated with the aquatic habitat within the angiosperms (Barrett et al. 1993; Ackerman 1995; Les and Schneider 1995). They have undergone extensive diversification, displaying all major aquatic life-forms, including emergent, floating-leaved, free-floating, submersed, and fully aquatic, and exceptionally high diversity of flower morphology and development as well (Posluszny and Charlton 1993; Les et al. 1997; Posluszny et al. 2000). The core alismatids are mostly fully aquatic and represent the greatest adaptive radiation of freshwater plants. This group also includes all marine angiosperms (seagrasses; den Hartog 1970), which are unique as they possess highly reduced flowers, underwater pollination and specialized pollen (Ducker et al. 1978; Furness and Banks 2010).Furthermore, these marine lineages within the core alismatids are believed to have independently evolved at least three to four times during their recolonization of the sea (Papenbrock 2012).On the other hand, water-pollinated angiosperms (hydrophily) a real most entirely restricted to the core alismatids with only a few species found in Ceratophyllaceae and Callitrichaceae (Les 1988; Philbrick 1993; Les et al. 1997). The shift of pollination system from aerial to water and colonization of marine habitats within Alismatales are thus regarded as considerable evolutionary events for flowering plants.
Within the monocots, there are three informal groupings: basal monocots (Acorales and Alismatales), lilioid monocots (Asparagales, Dioscoreales, Liliales, Pandanales, and Petrosaviales), and commelinoid monocots (Arecales, Commelinales, Dasypogonales, Poales, and Zingiberlaes). The near-basal phylogenetic position of the Alismatales order in monocots makes it potentially important in inferring the evolution of early diverging monocots (Chase et al. 2006; Graham et al. 2006; Angiosperm Phylogeny Group 2009; Iles et al. 2013). Based on morphological evidence, the most archaic monocots have been considered to be aquatic alismatids (Stebbins 1974; Thorne 1976; Cronquist 1981; Takhtajan 1991), net-veined groups (e.g., Dioscoreaceae, Dahlgren et al. 1985; Melanthiales, Thorne 1992), or Tofieldiaceae (Walker 1986; Tamura 1998). Based on early molecular studies of monocots, however, Acoraceae was inferred as the first branch and Alismatales the second branch to diverge within monocots (Chase et al. 1993; Duvall, Clegg, et al. 1993; Duvall, Learn, et al.1993), a finding that has received substantial support from the majority of subsequent molecular phylogenetic analyses (Chase et al. 2006; Duvall et al. 2006; Givnish et al. 2006; Graham et al. 2006; Li and Zhou 2007). In contrast, a few phylogenetic studies employing mitochondrial DNA supported Alismatales as the most basal group of monocots, with Acorus nested within it as a sister group to the alismatid families (Qiu et al. 2000; Davis et al. 2004; Petersen et al. 2006, 2016). It has been suggested that the controversial placement of Acorus within Alismatales is mainly caused by the nature of different data sets used for phylogenetic analyses, that is, the relative proportion of mitochondrial, plastid, and nuclear DNA utilized (Petersen et al. 2016).
Although the first comprehensive phylogeny of Alismatales with comprehensive taxon sampling was proposed by Les et al. (1997) based on the plastid rbcL gene, the internal structure of the phylogeny has long remained contentious. The major dispute centers on the basal group of this order, involving the relative phylogenetic arrangement of Araceae, Tofieldiaceae, and the core alismatid families. Some studies have suggested Tofieldiaceae to be the basal group of Alismatales (Graham et al. 2006; Iles et al. 2013), although bootstrap support (BS) for this relationship is low. In one extreme case, this family was placed outside the Alismatales order based on analysis of nuclear PHYC sequences (Duvall and Ervin 2004). In contrast, other molecular phylogenetic studies resolved Araceae as the basal group of Alismatales, with Tofieldiaceae and the core alismatid families as the sister group (Chase et al. 2006; Givnish et al. 2006; Soltis et al. 2011; Nauheimer et al. 2012). Furthermore, in an additional study on combined analyses of plastid DNA and 18S rDNA sequences, the group comprising Araceae and Tofieldiaceae was revealed as sister to the core alismatids (Li and Zhou 2007).
Compared with previous smaller data sets consisting of either a single or a few genes, genome-scale data sets, which provide increased character information, have been heralded as having the potential to significantly advance our ability to resolve historically difficult phylogenies (Rokas et al. 2003; Lemmon EM and Lemmon AR 2013; Soltis et al. 2013). Recently, complete plastid genomes have been rapidly sequenced from across the green plant tree of life due to the availability of next-generation sequencing platforms. Consequently, plastid genome-scale data have been used successfully to address various phylogenetic problems from deep to shallow hierarchical levels of plants (Parks et al. 2009; Barrett and Davis 2012; Ma et al. 2014; Ruhfel et al. 2014). In addition, the evolution of plastid genomes has also been deduced from plastid phylogenomic analyses (Kumar et al. 2011; Barrett et al. 2012; Salichos and Rokas 2013).
Because they are rare events, changes in the plastid genome can be extremely useful as phylogenetic markers for resolving phylogenies. In most land plants, the plastid genome is a single circular molecule of 120–160 kb consisting of one large single copy (LSC), one small single-copy (SSC), and two inverted repeat (IR) regions(Wicke et al. 2011). Gene content and order and genome organization are relatively conserved in the plastid genome, and the evolutionary rate of plastid DNA sequence is usually slow (Wicke et al. 2011). Nevertheless, major genomic structural changes, such as gene losses, large inversions, and contraction or expansion of IR regions, were observed in the evolution of plastid genomes from certain angiosperm lineages. For example, the plastid genes rpl22, infA, and accD were lost in the legumes (Fabaceae), Lemnoideae (Araceae), and Acoraceae, respectively (Doyle et al. 1995; Goremykin et al. 2005; Leebens-Mack et al. 2005; Mardanov et al. 2008; Wang and Messing 2011), whereas three genes (accD, ycf1, and ycf2) were all lost in Poaceae (Guisinger et al. 2010). Further, two and three inversions restricted to the plastid genomes of Asteraceae and Poaceae have been identified, respectively (Jansen and Palmer 1987; Kim et al. 2005; Guisinger et al. 2010), and a 50 kb inversion occurred in the plastid genome of all papilionoid legumes, supporting monophyly of this clade (Doyle et al. 1995; Wojciechowski et al. 2004; Jansen et al. 2008). Finally, the organization of genes flanking IR-LSC junctions can be used to distinguish monocots from other angiosperms, as the former have a trnH-rps19 gene cluster nearby those junctions (Wang et al. 2008).
In Alismatales, only seven plastid genomes belonging to two families, including five genera in Araceae (Colocasia, Lemna, Spirodela, Wolffia, and Wolffiella) and two in Hydrocharitaceae (Elodea and Najas), have been previously sequenced. All these genomes exhibit moderate genomic variations. The infA gene was lost from the plastid genomes in subfamily Lemnoideae (Araceae) (Mardanov et al. 2008; Wang and Messing 2011). One to several ndh genes were inferred to be independently lost in multiple lineages of the alismatid families, and this loss may be correlated with the altered physiological constraints required for life in aquatic environments (Martín and Sabater 2010; Iles et al. 2013; Peredo et al. 2013). Similarly, 11 genes were lost in the plastid genome of Najas flexilis, a freshwater plant with underwater pollination (Peredo et al. 2013).
Here, we report the complete plastid genome sequences of three species belonging to three families within Alismatales: Potamogeton perfoliatus (Potamogetonaceae), Sagittaria lichuanensis (Alismataceae), and Tofieldia thibetica (Tofieldiaceae). These represent the first plastid genomes sequenced from these families. Using these sequenced genomes, we examine plastid genomic features of the Alismatales and investigate the evolution of plastid genomes in the early diverging monocots. Additionally, protein-coding sequences from common genes shared among the plastid genomes of 22 select taxa are used to estimate phylogenetic relationships within Alismatales by different tree-building methods, with a particular emphasis placed on determining the basal group of this order.
Materials and Methods
Plant Material and Plastid Genome Sequencing
Based on previous phylogenies of Alismatales (Les et al. 1997; Iles et al. 2013; Petersen et al. 2016) and available plastid genomes from this order (Mardanov et al. 2008; Wang and Messing 2011; Ahmed et al. 2012; Huotari and Korpelainen 2012; Peredo et al. 2013), we chose to sample the following taxa from additional major lineages of Alismatales: P. perfoliatus (Potamogetonaceae; collected from Chengjiang county, Yunnan), S. lichuanensis (Alismataceae; from Wuhan Botanical Garden), and T. thibetica (Tofieldiaceae; from Shangri-La county, Yunnan). Fresh young leaves of these taxa were collected in the field for DNA isolation. Voucher specimens (14CS9221 for P. perfoliatus; Yi14056 for S. lichuanensis; and 11CS3064 for T. thibetica) were deposited at the herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN).
Total DNA was extracted from about 100 mg of leaf material according to a modified CTAB method (Doyle 1987; Yang et al. 2014), and quality was assessed by agarose gel electrophoresis. Following Yang et al. (2014), we amplified the entire plastid genome with long-range polymerase chain reaction (PCR). Briefly, amplification was performed using Takara PrimeSTAR GXL DNA polymerase (TAKARA BIO INC.) in 25-µl reaction mixtures containing 30–100 ng of DNA template and 0.5 µM of each of nine primer pairs (as described in Yang et al. [2014]). Subsequently, the nine long-range PCR products were pooled together in roughly equal mass mixtures for Illumina sequencing. These mixtures were fragmented and then used to construct short-insert libraries (500 bp) according to the manufacturer’s protocol (Illumina). DNA libraries from different species were indexed by tags and pooled together in one lane of Illumina’s Miseq for sequencing (paired-end, 250 bp) at the Germplasm Bank of Wild Species (KUN).
We used a combination of de novo and reference-guided strategies to assemble plastid genomes. First, Illumina short reads were assembled into contigs using SOAPdenovo with a k-mer length of 63–75 (Li et al. 2010). Next, using BLAST (http://blast.ncbi.nlm.nih.gov/) with default parameters, output scaffolds/contigs larger than 1,000 bp were mapped to the four plastid reference genomes of Alismatales: Colocasia esculenta (NC_016753), Elodea canadensis (NC_018541), Lemna minor (NC_010109), and Spirodela polyrhiza (NC_015891). Finally, the order of aligned scaffolds/contigs was determined according to these reference genomes, and any gaps that were present were resolved by mapping the raw reads to the assembly.
Genome Annotation and Comparison
The three complete plastid genomes were annotated using the plastid genome annotation package DOGMA (Wyman et al. 2004). Intron positions, as well as start and stop codons of protein-coding genes, were manually adjusted, if necessary, based on the four reference genomes (see above). The identified tRNA genes were further confirmed using tRNA scan-SE version 1.23 (Lowe and Eddy 1997). The annotated GenBank files of the three plastid genomes were used to draw gene maps using the Organellar Genome DRAW tool (OGDRAW) (Lohse et al. 2007). The final annotated plastid genomes were deposited into GenBank (Accession numbers: KT899950–KT899952).
We compared these three newly sequenced plastid genomes with those of three representative taxa from Alismatales: Colocasia esculents (NC_016753), E. canadensis (NC_018541), and L. minor (NC_010109). Acorus calamus (NC_007407) and Amborella trichopoda (NC_005086) were also included in our comparative analyses, as these taxa are representative of the early diverging lineages of monocots and angiosperms, respectively. Any large genomic structural events, such as gain or loss of genes, gene order rearrangement, and IR region expansion or contraction events, were recorded.
Phylogenetic Analyses
To estimate phylogenetic relationships within Alismatales, 22 taxa with available complete plastid genomes were compared, including ten taxa from Alismatales (three sequenced here) and, as outgroups, five taxa from other major monocot clades, six basal angiosperms, and one eudicot (supplementary table S1, Supplementary Material online). Complete plastid genome sequences with annotation information were downloaded from NCBI for each taxa. Ten taxa of Alismatales were sampled from five families and represented all major clades of the Alismatales phylogeny, taking the probable basal group of Alismatales into consideration.
The nucleotide sequences of 79 common protein-coding genes in the plastid genomes of 22 sampled taxa were extracted from each genome, of which 17 were pseudogenes in certain lineages and were thus treated as missing data in these lineages (supplementary table S2, Supplementary Material online). The nucleotide sequences of each gene were aligned by MUSCLE with default settings (Edgar 2004) and were manually adjusted according to amino acid codes if necessary. Subsequently, these genes were concatenated and the alignment was 75,597 bp for the 79-gene data set. To assess the possible effect of missing data on phylogeny estimation, we built a 62-gene data set with the 17 pseudogenes excluded (supplementary table S2, Supplementary Material online), and the alignment for this data set was 52,098 bp. In addition to these two main data sets, we also performed phylogenetic analyses on the 79-gene alignment subsets containing either first and second codon positions or third codon positions only to explore the effect of potential sequence saturation at third codon.
Bayesian and maximum likelihood (ML) analyses were first performed on the four data matrices described above in an unpartitioned way. In addition to unpartitioned analyses, we also conducted partitioned analyses for the 79 and 62-gene data sets. For the 79-gene data set, we used three different partitioning strategies: partitioned by codon, partitioned by gene, and a partition scheme determined by the software PartitionFinder v.1.1.1 (Lanfear et al. 2012). For the 62-gene data set, we only employed the software PartitionFinder to select the optimum partitioning scheme. We run PartitionFinder to find the optimum scheme among 79 or 62 genes and three codon positions for the 79 and 62-gene data sets, respectively, using the Bayesian information criterion and the “rcluster” algorithm. As a result, the 79 and 62 gene data sets were divided into 33 and 27 partitions, respectively.
Best-fitting substitution models for phylogenetic analyses were primarily determined using Akaike Information Criterion (Posada and Buckley 2004) in the program jModeltest (Darriba et al. 2012). The general time reversible (GTR) + I + Gmodel was selected for all the four data sets of 79-gene, 62-gene, combined first and second positions, and third positions. For the partitioned analyses under the scheme determined by PartitionFinder, the best models (GTR + G or GTR + I + G) identified simultaneously by it were used. The GTR + G model was used for all ML analyses implemented in RAxML version 8.0.20, as suggested in the manual (Stamatakis 2014). Nonparametric bootstrapping implemented in the “fast bootstrap” algorithm of RAxML used 200 replicates. For Bayesian analyses, a more parameterized model was used when the selected model (supplementary table S3, Supplementary Material online) was not available in MrBayes version 3.2.5 (Ronquist and Huelsenbeck 2003). In partitioned analyses, each subset was given its own model and all parameters were unlinked except those for branch lengths and topology. The Markov chain Monte Carlo (MCMC) algorithm was run with two independent chains with a random starting tree and default priors for 2,000,000 generations with trees sampled every 1,000 generations. The MCMC convergence was assumed when the average standard deviation of split frequencies reached 0.01 or less. The first 25% of trees from all runs were discarded as burn-in, and the remaining trees were used to construct majority-rule consensus trees.
Results
Overall Structure and Gene Pool
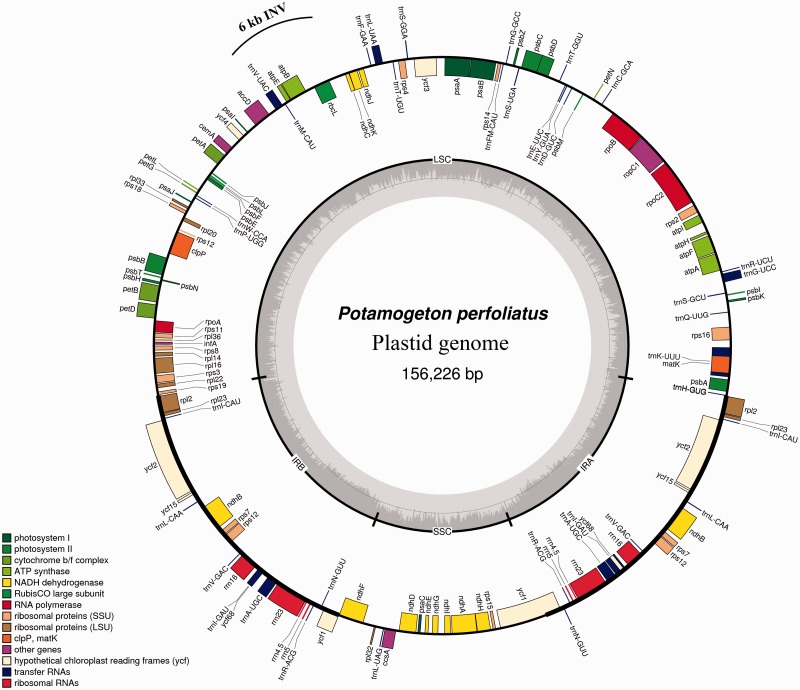
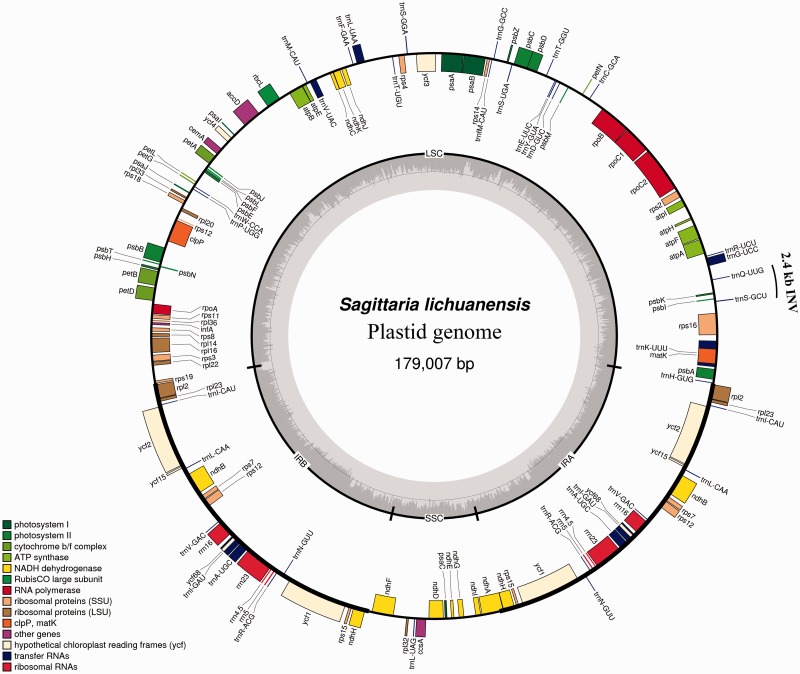
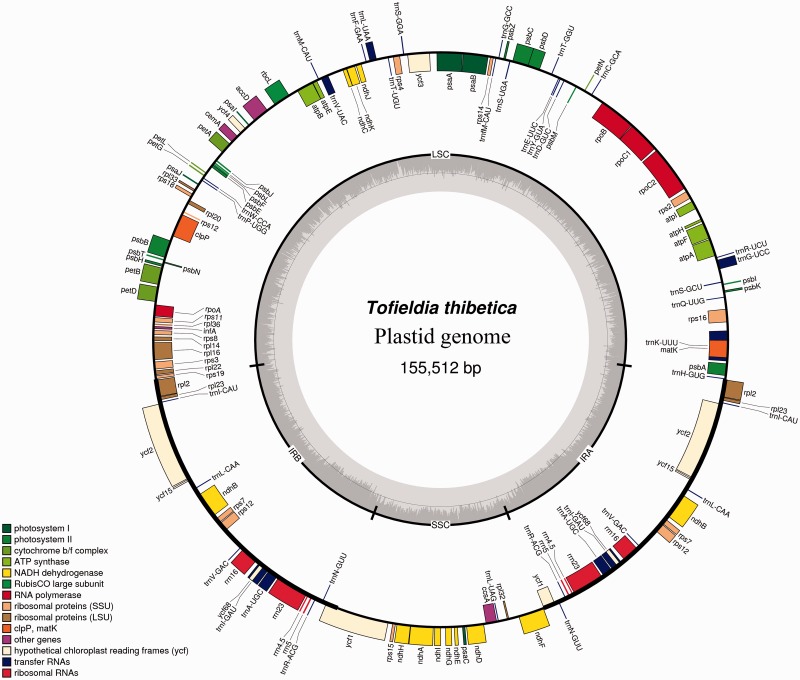
We selected Potamogeton, Sagittaria, and Tofieldia as representatives of the three families in the order Alismatales for plastid genome sequencing. Illumina sequencing generated 1,676,798 paired-end raw reads for Potamogeton, 1,104,080 for Sagittaria, and 1,425,322 for Tofieldia, with an average sequencing depth greater than 1,100×. The combination of de novo and reference-guided assembly successfully produced the complete nucleotide sequences for all three species. The complete plastid genomes of Potamogeton, Sagittaria, and Tofieldia are 156,226, 179,007, and 155,512 bp in length, respectively (see table 1 and figs. 1–3). All these plastid genomes possess a typical quadripartite structure found in the vast majority of angiosperms, including a pair of IRs of 25,612, 33,302, and 26,389 bp for Potamogeton, Sagittaria, and Tofieldia in length, respectively (table 1). The sizes of the LSC and SSC regions range from 84,584 to 99,125 bp and from 13,288 to 18,238 bp in these three genomes, respectively (table 1). Overall GC content of the plastid genomes is similar across the three species (Potamogeton, 36.5%; Sagittaria, 37.4%; Tofieldia, 36.8%). And IR regions have a higher GC content (41.2–42.8%) compared to that of the LSC and SSC regions (34.2–35.3% and 29.6–31.6%, respectively; table 1).
Table 1.
Comparison of Major Features of Am. trichopoda, A. calamus, and Six Representative Alismatales Taxa
| Feature | Am. trichopoda | A. calamus | T. thibetica | L. minor | C. esculenta | E. canadensis | S. lichuanensis | P. perfoliatus |
|---|---|---|---|---|---|---|---|---|
| Entire plastid size (bp) | 162,686 | 153,821 | 155,512 | 165,955 | 162,424 | 156,700 | 179,007 | 156,226 |
| LSC length (bp) | 90,970 | 84,149 | 84,584 | 89,906 | 89,670 | 86,194 | 99,125 | 86,764 |
| IR length (bp) | 26,651 | 25,697 | 26,389 | 31,223 | 25,273 | 26,348 | 33,302 | 25,612 |
| SSC length (bp) | 18,414 | 18,278 | 18,150 | 13,603 | 22,208 | 17,810 | 13,288 | 18,238 |
| G–C (%) | ||||||||
| Total genome | 38.3 | 38.6 | 37.4 | 35.7 | 36.1 | 37 | 36.8 | 36.5 |
| IR | 43.1 | 42.5 | 42.8 | 40.1 | 42.4 | 43 | 41.2 | 42.7 |
| LSC | 36.6 | 37.2 | 35.3 | 33.5 | 34.4 | 34.8 | 34.7 | 34.2 |
| SSC | 33.3 | 33.4 | 31.6 | 30.1 | 29 | 30.5 | 30.6 | 29.6 |
| Number of genes (different/total) | 113/129 | 112/132 | 113/130 | 112/130 | 112/130 | 113/129 | 113/133 | 113/130 |
| Number of genes duplicated in IR | 16 | 20 | 17 | 18 | 18 | 16 | 20 | 17 |
| Number of genes with introns (with 2 introns) | 18 (2) | 18 (2) | 18 (2) | 18 (2) | 18 (2) | 18 (2) | 18 (2) | 18 (2) |
| Number of protein-coding genes (total/in IR) | 79 (6) | 78 (8) | 79 (6) | 78 (7) | 78 (7) | 79 (6) | 79 (9) | 79 (6) |
| Number of rRNA genes | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Number of tRNAgenes (total/in IR) | 36 (6) | 38 (8) | 37 (7) | 37 (7) | 37 (7) | 36 (6) | 37 (7) | 37 (7) |
Fig. 1.—
Map of the complete plastid genome of P. perfoliatus. INV, inversion.
Fig. 2.—
Map of the complete plastid genome of S. lichuanensis. INV, inversion.
Fig. 3.—
Map of the complete plastid genome of T. thibetica.
The SSC region in the plastid genome of Tofieldia is in reverse orientation relative to other Alismatales species and the majority of other angiosperms (fig. 1), however, this does not indicate any changes in gene order as the SSC can exist in two orientations in the plastid genome (Palmer 1983). Although the genomic reconfiguration of the three newly sequenced plastid genomes is generally similar to those of published Alismatales plastid genomes, certain genomic rearrangements were observed in these genomes (figs. 1–3). For example, an approximately 2.4 kb inversion comprised the trnS-GCU, psbI, psbK, and trnQ-UUG genes in the LSC region occurs in the plastid genome of Sagittaria relative to other Alismatales species. Similarly, an approximately 6 kb inversion comprised the rbcL, atpB, atpE, trnM-CAU, and trnV-UAC genes in the LSC region is found in the plastid genome of Potamogeton but is absent in other Alismatales species. Interestingly, while such large inversions in the plastid genome are generally associated with the presence of short direct or IRs located at the ends of insertions (Downie and Palmer 1992; Rogalski et al. 2006), no repeats were identified surrounding the ends of the two inversions described above.
While the plastid genomes of Potamogeton, Sagittaria, and Tofieldia contain the same number of 113 unique genes, 17 (for Potamogeton), 20 (for Sagittaria), and 18 (for Tofieldia) of them are duplicated in IR regions, thus giving a total of 130–133 genes per plastid genome (table 1). Among these 113 unique genes, 79 were protein-coding genes, 30 were transfer RNA genes, and 4 were ribosomal RNA genes. As in most angiosperms, 18 genes contained introns, 15 of which contained a single intron whereas three (clpP, rps12, and ycf3) had two introns. There were six hypothetical coding regions (ycf1, ycf2, ycf3, ycf4, ycf15, and ycf68) identified in the three plastid genomes, but two (ycf15 and ycf68) showed notably long insertions and deletions disrupting the reading frame, implying that these two genes are very likely pseudogenes. As is typical for plastid genomes, the codon ATG was most often used as the start codon of protein-coding genes. In addition, the genes ndhD and rpl2 contained an ACG start codon in Tofieldia; rpl2 contained an ATA and ACG start codon in Sagittaria and Tofieldia, respectively; and rps19 contained a GTG start codon in Tofieldia, ndhB contained an ACG start codon in Potamogeton, and cemA contained a GTG start codon in all three genera. However, it should be noted that the ACG start codon from these genes has been suggested to undergo C-to-U RNA editing in the transcripts (Tsudzuki et al. 2001).
Genome Comparisons
Table 1 summarizes the general features of six plastid genomes representing different families within Alismatales, one related monocot (A. calamus), and the probable earliest diverging lineage within angiosperms, Am. trichopoda (Mathews and Donoghue 1999; Leebens-Mack et al. 2005; Jansen et al. 2007; Soltis et al. 2011). Overall, the plastid genomes of Alismatales are larger than Acorus and more similar to Amborella. In addition, these genomes showed moderate variability in size, ranging from 155 kb in Tofieldia to 179 kb in Sagittaria. However, size changes varied across the LSC, SSC, and IR regions of the Alismatales plastid genomes, with overall size variability largely attributable to IR regions. The IR region was expanded to 31 kb in Lemna and 33 kb in Sagittaria while had a constant size of 25–26 kb in all other members. Likewise, the LSC region was expanded to 99 kb in Sagittaria, thereby making it the largest genome within Alismatales, despite having the smallest SSC region. The plastid genome of Sagittaria was roughly 25 and 16 kb larger than Acorus and Amborella, respectively. On the other hand, the plastid genome of Tofieldia was the smallest among the Alismatales species, with the decrease in size mainly occurring in the LSC region (table 1). Despite such genome size variation, Alismatales plastid genomes each contained the same 113 unique genes, aside from Lemna for which the protein-coding gene infA was lost. Four rRNA genes, 30 unique tRNA genes, and intron content in the protein-coding genes were conserved in all these genomes. Mean GC content was 36.6% in Alismatales, which was lower relative to both Acorus (38.6%) and Amborella (38.3%) because of the greater GC content in the LSC and SSC regions for these two genera.
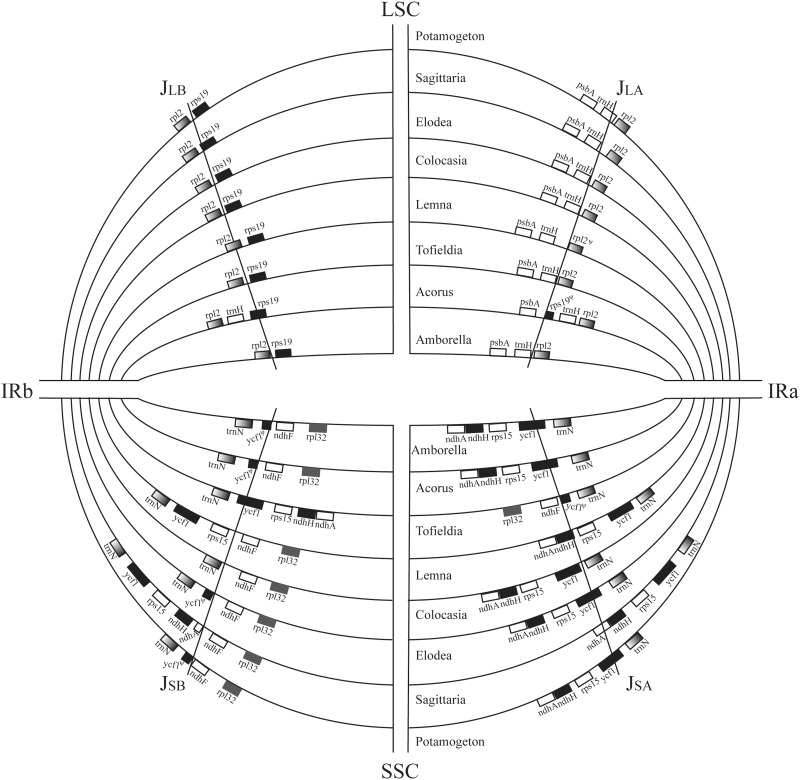
Variation in gene composition at the junctions between IR and LSC/SSC regions of Tofieldia, Sagittaria, and Potamogeton was observed relative to those of the other three Alismatales taxa, to Acrorus, and to Amborella (fig. 4). The trnH gene spanned the junction between inverted repeat A (IRa) and LSC regions in Potamogeton, whereas it was otherwise completely confined either within LSC regions (other Alismatales taxa and Amborella) or within IR regions (Acrorus).The junction between inverted repeat B (IRb) and LSC regions was located between the rps19 and rpl2 genes in all Alismatales, with the exception of Lemna in which the junction was located in the rpl2 gene. In contrast, the junctions between IR and SSC regions in the Alismatales plastid genomes experienced greater variation. Similar to what was found in Acrorus and Amborella, the ycf1 gene spanned the junction between IRa and SSC regions, with part of this gene having also been duplicated in the IRb region, in the plastid genomes of Elodea, Potamogeton, and Tofieldia. In Colocasia, however, the ycf1 gene was wholly located in the SSC region. In addition to ycf1, the rps15 and rps15-ndhH genes were completely duplicated in the IR regions of Lemna and Sagittaria, respectively.
Fig. 4.—
Comparison of junction positions (JLB, JLA, JSB, and JSA) between IR and single-copy regions. Included are six representative Alismatales taxa and two outgroups (Amborella and Acrorus).
Phylogenetic Analysis
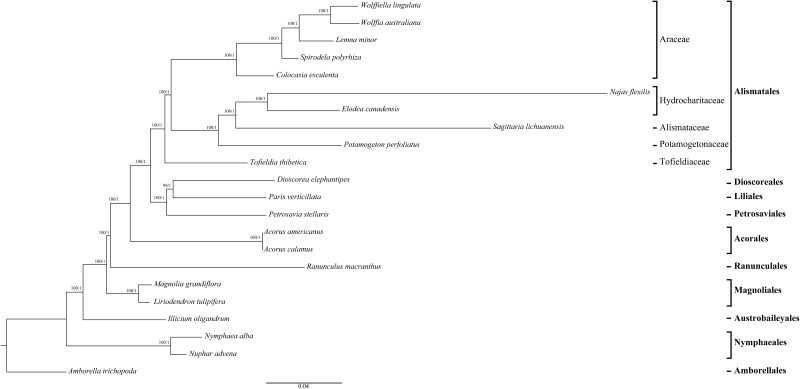
To explore phylogenetic relationships between major lineages in Alismatales and Acrorus, we generated trees using ten taxa of Alismatales, two Acrorus taxa, three related monocots, and seven taxa representing other major angiosperms lineages (supplementary table S1, Supplementary Material online).We assembled a data matrix consisting of 79 plastid protein-coding genes with approximately 3.5% missing data at the nucleotide level due to 17 of which were pseudogenes in certain lineages. Unpartitioned ML and Bayesian analyses of this data set produced identical tree topologies, with 100% BS form ML and 1.0 Bayesian posterior probabilities (PPs) at nearly every node (fig. 5). The results from phylogenetic analyses of the complete 62-gene data set with the 17 pseudogenes excluded were quite similar (supplementary fig. S1, Supplementary Material online).In addition, the trees produced by separate analyses of 79-gene data set partitioned by codon position, gene locus, and partitioning scheme identified by PartitionFinder had identical topologies with similarly high support values across both ML and Bayesian methods, as did the partitioned analyses of the 62-gene data set (supplementary fig. S1, Supplementary Material online; table 2).Finally, we also obtained the identical topologies in analyses of different codons (first and second positions combined and third positions) derived from the 79-gene data set (supplementary figs. S1 and S2, Supplementary Material online).
Fig. 5.—
Phylogenetic tree of 22 taxa based on 79 plastid protein-coding genes using unpartitioned ML and Bayesian inference (BI). Numbers at each node are bootstrap support values/Bayesian posterior probabilities. Ordinal and higher level group names follow APG III.
Table 2.
Bayesian Inference (BI) Posterior Probability and ML Bootstrap Support Values for Selected Nodes of the Phylogeny from Analyses of 79 and 62 Plastid Protein-Coding Genes Data Sets
| Lineage |
79 ptDNA, 75,597 Characters |
62 ptDNA, 52,098 Characters |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ML |
BI |
ML |
BI |
|||||||||
| Unpartitioned | p1 | p2 | p3 | Unpartitioned | p1 | p2 | p3 | Unpartitioned | p3 | Unpartitioned | p3 | |
| Monocots | 100 | 100 | 100 | 100 | 1 | 1 | 0.99 | 1 | 100 | 100 | 1 | 1 |
| Monocots with Acoraceae excluded | 100 | 100 | 100 | 100 | 1 | 1 | 1 | 1 | 100 | 100 | 1 | 1 |
| Alismatales | 100 | 100 | 100 | 100 | 1 | 1 | 1 | 1 | 100 | 100 | 1 | 1 |
| Araceae + core alismatids | 99 | 96 | 100 | 100 | 1 | 1 | 1 | 1 | 99 | 100 | 1 | 1 |
| Core alismatids | 100 | 100 | 100 | 100 | 1 | 1 | 1 | 1 | 100 | 100 | 1 | 1 |
Note.—p1, partitioned by all three codon positions; p2, partitioned by the 79 genes; p3, partitioned by the modeling strategy using PartitionFinder.
As results were very similar among all tree topologies, we only present the results of the tree topology from the analysis of the unpartitioned 79 protein-coding genes (fig. 5). Acorus was not placed within the Alismatales, but it was instead resolved as sister to the other monocots with maximum support across all analyses (supplementary figs. S1 and S2, Supplementary Material online; table 2). Monophyly of Alismatales was also 100% BS and 1.0 PP supported. Within Alismatales, the three major clades (Araceae, Tofieldiaceae, and the core alismatid families) were recovered with strong support for relationships among them (fig. 5).The Tofieldia was determined as the earliest diverging lineage within it, receiving strong support (BS > 90%, PP = 1.0) in almost all analyses (supplementary figs. S1 and S2, Supplementary Material online; table 2). Only in the analysis of the combined first and second codon positions from the 79-gene data set, the phylogenetic placement of Tofieldia received moderate support (BS = 80%, PP = 1.0; supplementary fig. S2, Supplementary Material online).Araceae was found to be sister to the core alismatids.
Discussion
Evolution of Alismatales Plastid Genome
In this study, we compiled the complete plastid genome sequences of Potamogeton (Potamogetonaceae), Sagittaria (Alismataceae), and Tofieldia (Tofieldiaceae) using Illumina sequencing. To date, there are now five families whose plastid genomes have been sequenced within Alismatales (the three reported for the first time here plus Araceae and Hydrocharitaceae (Mardanov et al. 2008; Wang and Messing 2011; Ahmed et al. 2012; Huotari and Korpelainen 2012; Peredo et al. 2013). The addition of these three genomes provides new insights into the evolution of the Alismatales plastid genome.
With a size of 179,007 bp, the plastid genome of Sagittaria is the largest plastid genome among monocots and the second largest among angiosperms (∼39 kb smaller than Pelargonium x hortorum (Geraniaceae) at 217,942 bp (Chumley et al. 2006) sequenced to date. On the other hand, the Tofieldia plastid genome is the smallest one (155,512 bp) within Alismatales. Although sequences of the IR region in the plastid genome are generally highly conserved, the expansion or contraction of the IR region is largely responsible for the overall size variation among plastid genomes (Chang et al. 2006; Raubeson et al. 2007; Ahmed et al. 2012). We find that, in Alismatales, length variation in both IR and LSC regions contribute to overall plastid genome size variation.
Many early-diverging monocot taxa display gene loss and order rearrangement events in their plastid genomes, such as loss of accD in Acorus and loss of rps16 in Dioscorea (Goremykin et al. 2005; Leebens-Mack et al. 2005; Hansen et al. 2007). In many Alismatales plastid genomes, including those of L. minor, Sp. polyrhiza, Wolffia australiana, and Wolffiella lingulata, loss of the gene infA has been frequently observed (Mardanov et al. 2008; Wang and Messing 2011). Furthermore, the loss of 11 ndh genes has occurred in the N. flexilis plastid genome, and the loss of many photosynthesis-related genes and a high number of rearrangements has occurred in the Petrosavia stellaris plastid genome (Peredo et al. 2013; Logacheva et al. 2014). In contrast to those monocots, the three plastid genomes reported here have not undergone any gene losses. Although loss of ndh genes has occurred in some taxa within the core alismatid clade (e.g., Najas, Posidonia, and Thalassia; Iles et al. 2013), these genes are present in the plastid genomes of two alismatid taxa: Potamogeton and Sagittaria. In addition, gene order is generally collinear among the Alismatales plastid genomes without major rearrangement. Only 2.4 and 6 kb inversions involving four and five genes occurred in the plastid genome of Sagittaria and Potamogeton, respectively. These inversions may have resulted from recombination between repeats (Downie and Palmer 1992; Rogalski et al. 2006); however, as we could not identify any significant repeats associated with these inversions, future study is required to tease this apart.
Junctions between IR and LSC/SSC regions are highly variable among angiosperms. Plastid genomes of the majority of taxa in the monocot order contain a trnH-rps19 gene cluster near the IRa-LSC junction (Wang et al. 2008),with complete trnH and partial rps19 sequences duplicated in the IR region (e.g., in Acorus and Dioscorea). Nevertheless, except for Potamogeton, in which portions of the trnH gene are indeed duplicated in the IR region, all Alismatales plastid genomes lack the trnH-rps19 gene cluster, with these two genes being instead wholly confined in the LSC region (fig. 4).This junction type of Alismatales is more similar to basal angiosperms and most eudicots than to other monocots, and it could therefore be deemed a specific feature distinguishing Alismatales from the remaining orders within monocots. Furthermore, the trnH gene spanning the IRa-LSC junction has only been reported in a few early-diverging angiosperms (e.g., Chloranthus oldhamii, C. spicatus, Sarcandra glabra, and Canella winterana) and basal eudicots (Ranunculus japonica and R. macranthus) (Wang et al. 2008), and Potamogeton is the first genus within monocots to have this type described. With respect to the IR-SSC junction, the IR region has been largely expanded in the plastid genomes of Lemna and Sagittaria to include the additional genes rps15 and ycf1 and further to ndhH and part of ndhA locus in the latter. Such a large expansion has only been previously observed in Pelargonium × hortorum, which has the largest plastid genome in angiosperms(Chumley et al. 2006).
Resolution of the Root of Alismatales
Monophyly of each major group within order Alismatales (Araceae, Tofieldiaceae, and the alismatids) is an important prerequisite for understanding the basal-most lineage of Alismatales, yet a certain degree of uncertainty has existed in the literature regarding monophyly of one specific group: Tofieldiaceae. Tofieldiaceae is a small family with about 20 species, and monophyly of this group has been confirmed using plastid DNA (matK and the noncoding trnL–trnL–trnF region) from 17 species (Azuma and Tobe 2011). However, using the same rbcL sequences, both Tamura et al. (2004) and Chen et al. (2013) suggested the positioning of Isidrogalvia schomburgkiana, as embedded within the family Petrosaviaceae or as sister to Narthecium within Nartheciaceae. This unlikely result has since been called into question as the I. schomburgkiana sample used in Tamura et al. (2004) was suspected as being either contaminated DNA of Nartheciaceae or misidentified (Azuma and Tobe 2011). Moreover, all pertinent molecular phylogenetic studies to date have consistently resolved the monophyly of the aquatic alismatids and Araceae (Les et al. 1997; Chase et al. 2006; Cabrera et al. 2008; Iles et al. 2013; Petersen et al. 2016). Monophyly of Tofieldiaceae and the core alismatids has been further supported by specific pollen characters, that is, diaperturate and inaperturate pollen grains, respectively (Luo et al.,forthcoming).
Given this majority of support for monophyly within Alismatales, we can gain more understanding regarding the root of Alismatales. Based on DNA analyses, Acorus has often been recovered as the first lineage to diverge within extant monocots (Chase et al. 2006; Givnish et al. 2006; Soltis et al. 2011). In contrast, inclusion of Acorus within Alismatales has been proposed based on mitochondrial loci (Petersen et al. 2006, 2016). Results using low or single copy nuclear genes, however, appear more congruent with plastid than mitochondrial data (Morton 2011; Zhang et al. 2012). Furthermore, based on our plastid genomic structural evidence, the difference observed between the IR-LSC junction type of Acorus and that of Alismatales does not support inclusion of Acorus within Alismatales (fig. 4). Instead, our plastid phylogenomic analyses strongly support Acorus as sister to the remaining monocots (BS = 100%, PP = 1.0; table 2).As placement of Acorus may be highly dependent on the balance between plastid and mitochondrial data, with Acorus being more likely placed as sister to other monocots when plastid data is dominant (Petersen et al. 2016), inclusion of more nuclear genes in our phylogenomic analyses should be employed in the future to resolve the potential discord between plastid and mitochondrial data.
After placing Acorus as sister to the monocots, the three complete plastid genomes sequenced here, which represent three key groups of Alismatales (the possible earliest diverging family of Alismatales (Tofieldiaceae), a petaloid alismatid family (Alismataceae), and a tepaloid alismatid family (Potamogetonaceae), help us to further resolve the root within this group. According to recent phylogenetic studies, including one analysis using 17 plastid regions of 31 taxa from 13 families (Iles et al. 2013) and another combining five mitochondrial genes, two plastid genes, and morphology of 58 taxa from 13 families (Petersen et al. 2016), Tofieldiaceae represents the earliest diverging lineage of Alismatales. However, both of these studies found negligible or weak support for this position of Tofieldiaceae within Alismatales. In contrast, utilizing whole plastid genome sequence data, we recovered strong support for both monophyly of Alismatales and Tofieldiaceae as the earliest-diverging lineage within it (fig. 5 and table 2).
Although poorly studied with respect to its relationship to the core alismatids within Alismatales, for example, having been previously assigned to various orders such as Liliales (Melchior 1964), Melanthiales (Dahlgren et al. 1985; Takhtajan 1997), and Nartheciales (Thorne 2000), Tofieldiaceae has morphological features that indicate a high degree of similarity to Acorus in comparison to core Alismatales. For example, Delpino (1903) placed Acorus not in Acoraceae but with Tofieldia, because both have unifacial, equitant, and distichous leaves. In addition, a secretory tapetum is recorded in both Acorus and Tofieldia, while all other Alismatales are plasmodial (Furness and Rudall 2001). However, it should be pointed out here that these shared morphological characters between Acorus and Tofieldia might be symplesiomorphies for early diverging monocots. On the other hand, Remizowa et al. (2010) proposed that the septal nectaries in early monocots was instructive that Tofieldiaceae rather than Araceae could represent the most basal lineage of Alismatales. Taken together, we could consider that such morphological evidence lends further support to Tofieldiaceae as one of the most archaic monocots (Walker 1986; Tamura 1998), and it consequently helps to improve our understanding of the evolution of the earliest lineage of Alismatales and this order as a whole.
Supplementary Material
Supplementary figures S1 and S2 and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Ting-Shuang Yi, Jie Cai, and Ting Zhang for assistance in obtaining material and Steven Callen for critical edits of the manuscript. This work was supported by the National Key Basic Research Program of China (No. 2014 CB954100), a Major International Joint Research Project of National Natural Science Foundation of China (NSFC) (No. 31320103919), and partially supported by an NSFC project (No. 31270272), and Youth Innovation Promotion Association of Chinese Academy of Sciences (2015321).
Literature Cited
- Ackerman JD. 1995. Convergence of filiform pollen morphologies in seagrasses: functional mechanisms. Evol Ecol. 9:139–153. [Google Scholar]
- Ahmed I, et al. 2012. Mutational dynamics of aroid chloroplast genomes. Genome Biol Evol. 4:1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 161:105–121. [Google Scholar]
- Azuma H, Tobe H. 2011. Molecular phylogenetic analyses of Tofieldiaceae (Alismatales): family circumscription and intergeneric relationships. J Plant Res. 124:349–357. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Davis JI. 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 99:1513–1523. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Davis JI, Leebens-Mack J, Conran JG, Stevenson DW. 2012. Plastid genomes and deep relationships among the commelinid monocot angiosperms. Cladistics 29:65–87. [DOI] [PubMed] [Google Scholar]
- Barrett SC, Eckert CG, Husband BC. 1993. Evolutionary processes in aquatic plant populations. Aquat Bot. 44:105–145. [Google Scholar]
- Cabrera LI, et al. 2008. Phylogenetic relationships of aroids and duckweeds (Araceae) inferred from coding and noncoding plastid DNA. Am J Bot. 95:1153–1165. [DOI] [PubMed] [Google Scholar]
- Chang C-C, et al. 2006. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 23:279–291. [DOI] [PubMed] [Google Scholar]
- Chase MW, et al. 1993. Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann Mo Bot Gard. 80:528–580. [Google Scholar]
- Chase MW, et al. 2006. Multigene analyses of monocot relationships: a summary. Aliso 22:63–75. [Google Scholar]
- Chen L-Y, Chen J-M, Gituru RW, Wang Q-F. 2013. Eurasian origin of Alismatidae inferred from statistical dispersal–vicariance analysis. Mol Phylogenet Evol. 67:38–42. [DOI] [PubMed] [Google Scholar]
- Chumley TW, et al. 2006. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 23:2175–2190. [DOI] [PubMed] [Google Scholar]
- Cronquist A. 1981. An integrated system of classification of flowering plants. New York: Columbia University Press. [Google Scholar]
- Dahlgren RM, Clifford HT, Yeo PF. 1985. The families of the monocotyledons: structure, evolution, and taxonomy. Berlin: Springer-Verlag. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JI, et al. 2004. A phylogeny of the monocots, as inferred from rbcL and atpA sequence variation, and a comparison of methods for calculating jackknife and bootstrap values. Syst Bot. 29:467–510. [Google Scholar]
- Delpino F. 1903. Aggiunte alla teoria della classificazione delle Monocotiledoni. Mem. Acad. Bologna Ser. V. 10:569–584. [Google Scholar]
- den Hartog C. 1970. The seagrasses of the world. North-Holland Publ. Co.: Amsterdam. [Google Scholar]
- Downie SR, Palmer JD. 1992. Use of chloroplast DNA rearrangements in reconstructing plant phylogeny In: Soltis PS, Soltis DE, Doyle JJ, editors. Molecular systematics of plants. New York: Chapman & Hall; p. 14–35. [Google Scholar]
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Doyle JJ, Doyle JL, Palmer JD. 1995. Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst Bot. 20:272–294. [Google Scholar]
- Ducker SC, Pettitt J, Knox R. 1978. Biology of Australian seagrasses: pollen development and submarine pollination in Amphibolis antarctica and Thalassodendron ciliatum (Cymodoceaceae). Aust J Bot. 26:265–285. [Google Scholar]
- Duvall MR, Clegg MT, et al. 1993. Phylogenetic hypotheses for the monocotyledons constructed from rbcL sequence data. Ann Mo Bot Gard. 80:607–619. [Google Scholar]
- Duvall MR, Ervin AB. 2004. 18S gene trees are positively misleading for monocot/dicot phylogenetics. Mol Phylogenet Evol. 30:97–106. [DOI] [PubMed] [Google Scholar]
- Duvall MR, Learn GH, Eguiarte LE, Clegg MT. 1993. Phylogenetic analysis of rbcL sequences identifies Acorus calamus as the primal extant monocotyledon. Proc Natl Acad Sci U S A. 90:4641–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall MR, Mathews S, Mohammad N, Russell T. 2006. Placing the monocots: conflicting signal from trigenomic analyses. Aliso 22:79–90. [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness CA, Banks H. 2010. Pollen evolution in the early-divergent monocot order Alismatales. Int J Plant Sci. 171:713–739. [Google Scholar]
- Furness CA, Rudall PJ. 2001. Pollen and anther characters in monocot systematics. Grana 40:17–25. [Google Scholar]
- Givnish TJ, et al. 2006. Phylogenetic relationships of monocots based on the highly informative plastid gene ndhF: evidence for widespread concerted convergence. Aliso 22:28–51. [Google Scholar]
- Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH. 2005. Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol. 22:1813–1822. [DOI] [PubMed] [Google Scholar]
- Graham SW, et al. 2006. Robust inference of monocot deep phylogeny using an expanded multigene plastid data set. Aliso 22:3–20. [Google Scholar]
- Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK. 2010. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol. 70:149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DR, et al. 2007. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol Phylogenet Evol. 45:547–563. [DOI] [PubMed] [Google Scholar]
- Huotari T, Korpelainen H. 2012. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene 508:96–105. [DOI] [PubMed] [Google Scholar]
- Iles WJ, Smith SY, Graham SW. 2013. A well-supported phylogenetic framework for the monocot order Alismatales reveals multiple losses of the plastid NADH dehydrogenase complex and a strong long-branch effect In: Wilkin P, Mayo SJ, editors. Early events in monocot evolution. Cambridge: Cambridge University Press; p. 1–28. [Google Scholar]
- Jansen RK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 104:19369–19374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Palmer JD. 1987. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae). Proc Natl Acad Sci U S A. 84:5818–5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Wojciechowski MF, Sanniyasi E, Lee S-B, Daniell H. 2008. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol Phylogenet Evol. 48:1204–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-J, Choi K-S, Jansen RK. 2005. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol Biol Evol. 22:1783–1792. [DOI] [PubMed] [Google Scholar]
- Kumar S, Filipski AJ, Battistuzzi FU, Pond SLK, Tamura K. 2011. Statistics and truth in phylogenomics. Mol Biol Evol. 29:457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701. [DOI] [PubMed] [Google Scholar]
- Leebens-Mack J, et al. 2005. Identifying the basal angiosperm node in chloroplast genome phylogenies: sampling one's way out of the Felsenstein zone. Mol Biol Evol. 22:1948–1963. [DOI] [PubMed] [Google Scholar]
- Lemmon EM, Lemmon AR. 2013. High-throughput genomic data in systematics and phylogenetics. Annu Rev Ecol Evol Syst. 44:99–121. [Google Scholar]
- Les DH. 1988. Breeding systems, population structure, and evolution in hydrophilous angiosperms. Ann Mo Bot Gard. 75:819–835. [Google Scholar]
- Les DH, Cleland MA, Waycott M. 1997. Phylogenetic studies in Alismatidae, II: evolution of marine angiosperms (seagrasses) and hydrophily. Syst Bot. 22:443–463. [Google Scholar]
- Les DH, Schneider EL. 1995. The Nymphaeales, Alismatidae, and the theory of an aquatic monocotyledon origin In: Rudall PJ, Cribb PJ, Cutler DF, Humphries CJ, editors. Monocotyledons: systematics and evolution. Kew, England: Royal Botanic Gardens; p. 23–42. [Google Scholar]
- Les DH, Tippery NP. 2013. In time and with water… the systematics of alismatid monocotyledons In: Wilkin P, Mayo SJ, editors. Early events in monocot evolution. Cambridge: Cambridge University Press; p. 1–28. [Google Scholar]
- Li R, et al. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20:265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-X, Zhou Z-K. 2007. The higher-level phylogeny of monocots based on matK, rbcL and 18S rDNA sequences. J Syst Evol. 45: 113–133. [Google Scholar]
- Logacheva MD, Schelkunov MI, Nuraliev MS, Samigullin TH, Penin AA. 2014. The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biol Evol. 6:238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, et al. 2015. Evolution of angiosperm pollen. 3. Monocots. Ann Mo Bot Gard. 101:406–455. [Google Scholar]
- Ma P-F, Zhang Y-X, Zeng C-X, Guo Z-H, Li D-Z. 2014. Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Syst Biol. 64:933–950. [DOI] [PubMed] [Google Scholar]
- Mardanov AV, et al. 2008. Complete sequence of the duckweed (Lemna minor) chloroplast genome: structural organization and phylogenetic relationships to other angiosperms. J Mol Evol. 66:555–564. [DOI] [PubMed] [Google Scholar]
- Martín M, Sabater B. 2010. Plastid ndh genes in plant evolution. Plant Physiol Biochem. 48:636–645. [DOI] [PubMed] [Google Scholar]
- Mathews S, Donoghue MJ. 1999. The root of Angiosperm phylogeny inferred from duplicate phytochrome genes. Science 286:947–950. [DOI] [PubMed] [Google Scholar]
- Melchior H. 1964. Liliflorae In: Melchior H, editor. A. Engler's Syllabus der Pflanzenfamilien. Vol 2. Berlin: Gebrüder Borntraeger; Berlin. p. 513–543. [Google Scholar]
- Morton CM. 2011. Newly sequenced nuclear gene (Xdh) for inferring angiosperm phylogeny 1. Ann Mo Bot Gard. 98:63–89. [Google Scholar]
- Nauheimer L, Metzler D, Renner SS. 2012. Global history of the ancient monocot family Araceae inferred with models accounting for past continental positions and previous ranges based on fossils. New Phytol. 195:938–950. [DOI] [PubMed] [Google Scholar]
- Palmer JD. 1983. Chloroplast DNA exists in two orientations. Nature 301:92–93. [Google Scholar]
- Papenbrock J. 2012. Highlights in seagrasses' phylogeny, physiology, and metabolism: what makes them special? ISRN Bot. ID103892. [Google Scholar]
- Parks M, Cronn R, Liston A. 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 7:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peredo EL, King UM, Les DH. 2013. The plastid genome of Najas flexilis: adaptation to submersed environments is accompanied by the complete loss of the NDH complex in an aquatic angiosperm. PLoS One 8:e68591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G, et al. 2006. Mitochondrial data in monocot phylogenetics. Aliso 22:52–62. [Google Scholar]
- Petersen G, et al. 2016. Phylogeny of the Alismatales (monocotyledons) and the relationship of Acorus (Acorales?). Cladistics: 32:141–159. [DOI] [PubMed] [Google Scholar]
- Philbrick CT. 1993. Underwater cross-pollination in Callitriche hermaphroditica (Callitrichaceae): evidence from random amplified polymorphic DNA markers. Am J Bot. 80:391–394. [Google Scholar]
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike Information Criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53:793–808. [DOI] [PubMed] [Google Scholar]
- Posluszny U, Charlton W. 1993. Evolution of the helobial flower. Aquat Bot. 44:303–324. [Google Scholar]
- Posluszny U, Charlton WA, Les DH. 2000. Modularity in helobial flowers In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melbourne (Australia: ): CSIRO; p. 63–74. [Google Scholar]
- Qiu YL, et al. 2000. Phylogeny of basal angiosperms: analyses of five genes from three genomes1. Int J Plant Sci. 161:S3–S27. [Google Scholar]
- Raubeson LA, et al. 2007. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics 8:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remizowa MV, Sokoloff DD, Rudall P. 2010. Evolutionary history of the nonocot flower 1. Ann Mo Bot Gard. 97:617–645. [Google Scholar]
- Rogalski M, Ruf S, Bock R. 2006. Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 34:4537–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, Carroll SB. 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425:798–804. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. 2014. From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 14:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichos L, Rokas A. 2013. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497:327–331. [DOI] [PubMed] [Google Scholar]
- Soltis DE, et al. 2011. Angiosperm phylogeny: 17 genes, 640 taxa. Am J Bot. 98:704–730. [DOI] [PubMed]
- Soltis DE, et al. 2013. The potential of genomics in plant systematics. Taxon 62:886–898. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. 1974. Flowering plants: evolution above the species level. Cambridge: Belknap Press. [Google Scholar]
- Stevens PF. 2001. onwards. Angiosperm phylogeny website. Version 12, November 23, 2015. St. Louis: Missouri Botanical Garden. Available from: http://www.mobot.org/MOBOT/research/APweb/.
- Takhtajan AL. 1991. Evolutionary trends in flowering plants. New York: Columbia University Press. [Google Scholar]
- Takhtajan AL. 1997. Diversity and classification of flowering plants. New York: Columbia University Press. [Google Scholar]
- Tamura M, Fuse S, Azuma H, Hasebe M. 2004. Biosystematic studies on the family Tofieldiaceae I. Phylogeny and circumscription of the family inferred from DNA sequences of matK and rbcL. Plant Biol. 6:562–567. [DOI] [PubMed] [Google Scholar]
- Tamura MN. 1998. Nartheciaceae In: Kubitzki K, editor. The families and genera of vascular plants, III, flowering plants, monocotyledons: Liliaceae (except Orchidaceae). Berlin: Springer; p. 381–392. [Google Scholar]
- Thorne RF. 1976. A phylogenetic classification of the Angiospermae. Evol Biol. 9: 35–106. [Google Scholar]
- Thorne RF. 1992. An updated phylogenetic classification of the flowering plants. Aliso 13:365–389. [Google Scholar]
- Thorne RF. 2000. The classification and geography of the monocotyledon subclasses Alismatidae, Liliidae and Commelinidae In: Nordenstam B, El-Ghazaly G, Kassas M, editors. Plant systematics for the 21st century. London: Portland Press; p. 75–124. [Google Scholar]
- Tomlinson P. 1982. Anatomy of the monocotyledons VII. Helobiae (Alismatidae) (including the seagrasses). Oxford: Clarendon Press. [Google Scholar]
- Tsudzuki T, Wakasugi T, Sugiura M., 2001. Comparative analysis of RNA editing sites in higher plant chloroplasts. J Mol Evol. 53:327–332. [DOI] [PubMed] [Google Scholar]
- Walker J. 1986. Classification and evolution of the monocotyledons. Am J Bot. 73:746–746. [Google Scholar]
- Wang R-J, et al. 2008. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 8:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Messing J. 2011. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS One 6:e24670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, Müller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76:273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ. 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot. 91:1846–1862. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031. [DOI] [PubMed] [Google Scholar]
- Zhang N, Zeng L, Shan H, Ma H. 2012. Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytol. 195:923–937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.