Abstract
Aberrant activation of the EGF receptor (EGFR) contributes to many human cancers by activating the Ras-MAPK and other pathways. EGFR signaling is augmented by Src-family kinases, but the mechanism is poorly understood. Here, we show that human EGFR preferentially phosphorylates peptide substrates that are primed by a prior phosphorylation. Utilizing peptides based on the sequence of the adaptor protein Shc1, we show that Src mediates the priming phosphorylation, promoting subsequent phosphorylation by EGFR. Importantly, the doubly phosphorylated Shc1 peptide binds more tightly to the Ras activator Grb2, a key step in activating the Ras-MAPK pathway, than singly phosphorylated peptides. Finally, a crystal structure of EGFR in complex with a primed Shc1 peptide reveals the structural basis for EGFR substrate specificity. These results provide a molecular explanation for the integration of Src and EGFR signaling with downstream effectors such as Ras.
The epidermal growth factor receptor (EGFR) is a transmembrane receptor tyrosine kinase that plays a critical role in regulating cell proliferation, differentiation, migration, and apoptosis1. EGFR is comprised of an extracellular ligand-binding domain, an intracellular tyrosine kinase domain, and a C-terminal tail that contains several autophosphorylation sites. Ligand binding induces EGFR dimerization, which activates the kinase domain by an allosteric mechanism, and leads to signaling through the Ras-MAPK, PI3K-AKT, and other pathways2.
Aberrant EGFR signaling, through overexpression and/or mutation, contributes to many solid tumors, including non-small cell lung cancer (NSCLC), head and neck cancer, and cancers of the breast, ovary, prostate, pancreas and colon. As a result, EGFR is a key drug target and therapeutics targeting mutant EGFR have proven successful for the treatment of several cancers3. However, despite the clinical success of EGFR inhibitors, most patients that initially respond eventually become resistant4.
An important element of EGFR signaling is its cooperativity with the cytoplasmic tyrosine kinase Src5. Src enhances EGF-induced Ras-MAPK signaling and also mediates transactivation of EGFR by GPCRs, cytokine receptors, and integrins6–9. This functional interaction between Src and EGFR may also play a role in tumorigenesis. For example, concurrent overexpression of Src and EGFR is often observed in human cancers and has been shown to produce a synergistic increase in proliferation, transformation, and tumorigenesis in model systems10–12. Further, mutant EGFRs found in NSCLC require cooperativity with Src for transformation13,14. Finally, the expression and activation of Src has recently emerged as a potential mediator of resistance to EGFR inhibitors15–17.
The mechanism underlying the functional interaction between EGFR and Src is poorly understood; however, it has been proposed to be dependent, in part, on Src phosphorylation of Tyr845 of EGFR6,7. Tyr845 is in the activation loop of EGFR and phosphorylation at homologous residues in other tyrosine kinases is generally required for activation18. However, phosphorylation of Tyr845 is not required for EGFR catalytic activity or EGF-induced activation of the Ras-MAPK pathway 6,19. In addition, Src-mediated activation affects only a subset of EGFR functions and correlates with a different pattern of downstream protein phosphorylations as compared to EGF-mediated activation8,20. These results suggest that additional mechanisms underlie the ability of Src to augment selective aspects of EGFR function.
To further investigate the functional interaction between EGFR and Src, we used a peptide library approach to examine their substrate specificities. Surprisingly, we found that EGFR preferentially phosphorylates substrates with a phosphotyrosine at the +1 position relative to the phosphorylation site. We identified several candidate EGFR phosphorylation sites, including Tyr239 of the adaptor protein Shc1, that have a phosphotyrosine at the +1 position in vivo. Using synthetic peptides based on the sequence surrounding Tyr239 of Shc1, we found that Src preferentially phosphorylates Tyr240, promoting subsequent phosphorylation of Tyr239 by EGFR. We also found that, compared to the singly phosphorylated peptides, the doubly phosphorylated Shc1 peptide binds more tightly to Grb2, a key step in activating the Ras-MAPK pathway. Finally, we determined the crystal structure of the EGFR kinase domain in complex with a Shc1 peptide ‘primed’ at Tyr240 and this structure reveals the structural basis for EGFR substrate specificity. Together, these results reveal a previously unknown characteristic of EGFR signaling that contributes to the functional synergy between EGFR and Src.
RESULTS
Determination of the EGFR optimal substrate motif
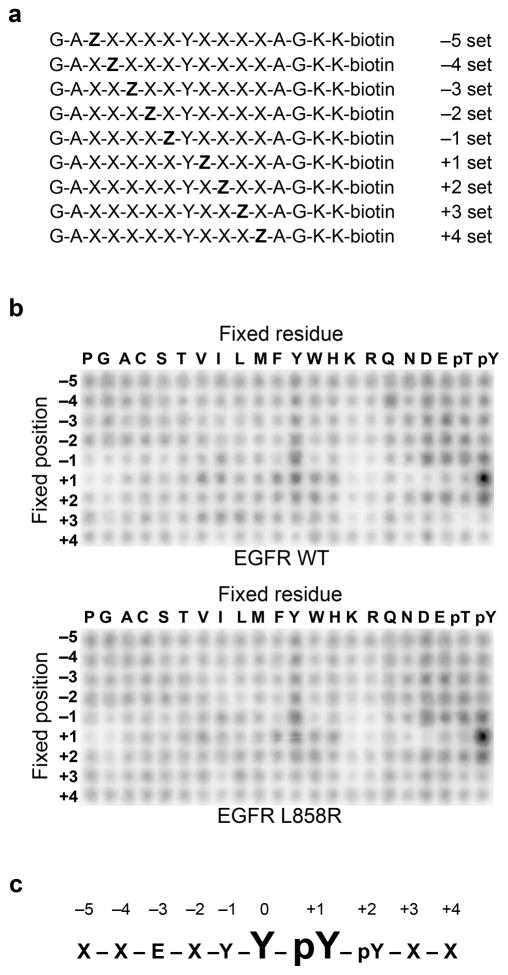
To determine the substrate specificity of EGFR, we used a positional scanning peptide library (PSPL) approach21. The PSPL approach utilizes a set of 198 biotinylated peptide libraries, each containing a tyrosine fixed at the central position and one additional amino acid fixed at flanking positions (Fig. 1a). An advantage of the PSPL approach, as compared to other methods used to profile kinase substrate specificity, is that phosphorylated amino acids are also fixed at the flanking positions enabling the identification of kinases whose specificity determinants include a priming phosphorylation. The other positions in each library contain a degenerate mixture of all amino acids, excluding cysteine and tyrosine.
Figure 1.
Determination of EGFR optimal substrate motif. (a) Schematic representation of the peptide libraries used in the positional scanning peptide library assay (Z, fixed amino acid; X, degenerate mixture of amino acids). (b) Peptide library results for wild-type and L858R EGFR kinase domains. Representative images from 3 independent experiments are shown. Quantification is provided in Supplementary Table 1. (c) Primary and secondary selections determined from the EGFR peptide library results. An “X’ denotes no selectivity.
The peptide libraries were simultaneously phosphorylated in solution using recombinant EGFR kinase domain and γ-32P-ATP. Aliquots of each reaction were then spotted in parallel on a streptavidin-coated membrane and the relative preference for each amino acid at each position was determined by the level of radiolabel incorporation into the corresponding peptides. Surprisingly, we found that EGFR has a strong preference for phosphotyrosine at the +1 position relative to the phosphorylation site (Fig. 1b). The remaining positions are less important for EGFR substrate recognition, with a minor preference for acidic amino acids at several positions N-terminal to the central tyrosine, in agreement with previous studies22. We also determined the optimal substrate motif for the L858R mutant of EGFR, which is found frequently in non-small cell lung cancer23. Because this mutation lies in the activation loop of the kinase domain, a region critical for binding of substrates, it could potentially alter EGFR substrate specificity. However, we found that the L858R mutant has a motif that is essentially identical to wild-type EGFR (Fig. 1b). Phosphorylation of the peptide library with phosphotyrosine at the +1 position was blocked by the EGFR-specific inhibitor gefitinib, confirming that the results were not due to a contaminating kinase (Supplementary Fig. 1). Data from three independent peptide library assays were averaged to generate a consensus EGFR phosphorylation motif (Fig. 1c and Supplementary Table 1).
Identification of candidate EGFR phosphorylation sites
The PSPL results suggest that cellular substrates of EGFR are likely to have a phosphotyrosine one position C-terminal to the phosphorylation site. The primary cellular substrate of EGFR, like other receptor tyrosine kinases, is the receptor itself. Ligand binding induces EGFR dimerization and autophosphorylation of tyrosine residues in the intracellular region of the receptor and these phosphorylated tyrosines serve as docking sites for effector molecules. The docked effector molecules, and other cytoplasmic proteins, can also be phosphorylated by EGFR, linking the activated receptor to downstream signaling pathways. The intracellular region of EGFR does not contain any Y-Y sequences; therefore, the optimal phosphorylation motif determined in the peptide library is likely to be a determinant of EGFR specificity for downstream substrates rather than autophosphorylation.
To identify potential downstream substrates that have a phosphotyrosine one position C-terminal to the predicted phosphorylation site, we used PhosphoSitePlus (http://www.phosphosite.org). PhosphoSitePlus is a database of experimentally observed post-translational modifications from Cell Signaling Technologies24. We searched the database to find tyrosine phosphorylation sites that are sensitive to treatment with EGFR-specific inhibitors and have a phosphotyrosine at the +1 position. Interestingly, many of the predicted phosphorylation sites occur in proteins with known roles in EGFR signaling (Table 1). However, of these predicted substrates, only Shc1 has been reported to be directly phosphorylated by EGFR25. Shc1 is an adaptor protein that integrates the activity of various signaling proteins, including RTKs, cytoplasmic kinases, integrins, and GPCRs, with the Ras-MAPK pathway26. Phosphorylation of Shc1 at Tyr239 creates a pTyr-X-Asn-X-motif, a binding site for the SH2 domain of Grb2, leading to activation of the Ras-MAPK pathway27,28.
Table 1.
Candidate EGFR phosphorylation sites identified in PhosphoSitePlus
| Symbol | Name | Site | Sequence |
|---|---|---|---|
| MLLT4 | Adhesion protein afadin | 1656 | RQEEGYySRLE |
| C9orf150 | Chromosome 9 open reading frame 150 | 227 | KLDSEYyCFG_ |
| CD34 | Adhesion protein CD34 | 329 | LGEDPYyTENG |
| EFNB1 | Membrane protein of the Ephrin family | 343 | SPANIYyKV__ |
| EIF2A | Eukaryotic translation initiation factor 2A | 250 | KTGASYyGEQT |
| EMD | Nuclear membrane protein emerin | 94 | GYNDDYyEESY |
| PTK2 | Focal adhesion kinase FAK | 576 | MEDSTYyKASK |
| ERBB2 | Receptor tyrosine kinase HER2 | 1221 | AFDNLYyWDQD |
| JAK1 | Janus kinase 1 | 1034 | ETDKEYyTVKD |
| LSR | Lipolysis stimulated lipoprotein LISCH | 406 | SMRVLYyMEKE |
| MET | HGF receptor tyrosine kinase | 1234 | MYDKEYySVHN |
| ERRFI1 | EGFR Negative regulator MIG-6 | 394 | VSSTHYyLLPE |
| PKP4 | Adhesion protein plakophilin 4 | 420 | YEGRTYySPVY |
| PVRL3 | Nectin family adhesion protein PVRL3 | 510 | ERPMDYyEDLK |
| SHC1 | Adaptor protein Shc1 | 239 | PPDHQYyNDFP |
| INPPL1 | Inositol phosphatase SHIP-2 | 986 | FNNPAYyVLEG |
| SKT | Skeletal development protein SKT | 244 | ESRNVYyELND |
| TANC2 | Ankyrin-repeat containing protein TANC2 | 1919 | LLEDDYySPHG |
The PhosphoSitePlus database was searched for sites that conform to the EGFR YpY motif and whose phosphorylation correlates with EGFR activity. Putative phosphorylation sites are shown in bold. Phosphorylated tyrosines are shown in lower case.
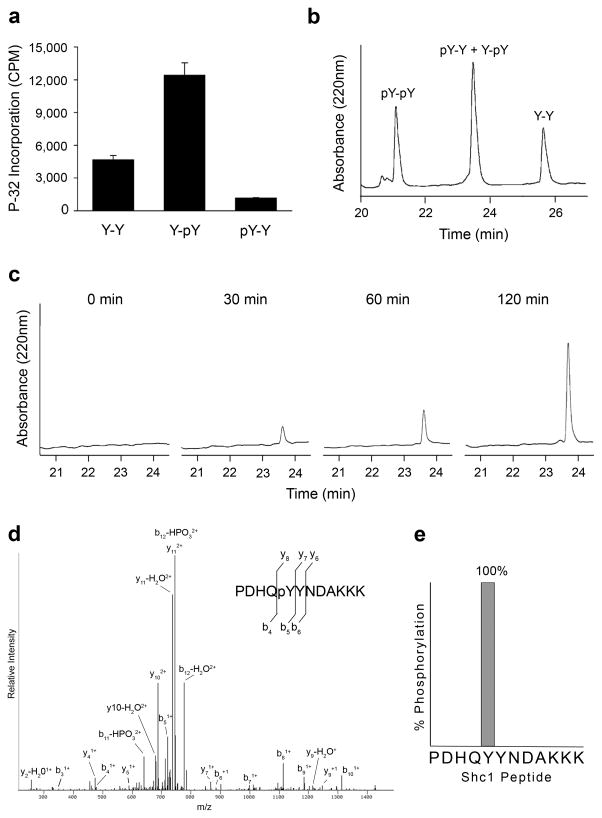
To test whether prior phosphorylation also enhances EGFR activity toward the identified candidate substrates, we generated synthetic peptides corresponding to the sequences surrounding the predicted Shc1 and MET phosphorylation sites (Table 1). Consistent with the PSPL results, both peptides were substantially better substrates for EGFR when the tyrosine at the +1 position was phosphorylated (Fig. 2a and Supplementary Fig. 2).
Figure 2.
Shc1 is phosphorylated by EGFR at Tyr239. (a) EGFR kinase assays with synthetic peptides corresponding to the sequence surrounding Tyr239 of Shc1 (PDHQYYNDAKKK = Y-Y; PDHQYpYNDAKKK = Y-pY; PDHQpYYNDAKKK = pY-Y). Error bars, s.d. (n=3 technical replicates) (b) Reversed-phase HPLC separation of Shc1 peptide standards (PDHQpYpYNDAKKK = pY-pY). The retention times of each peptide were established by individual injections (data not shown). (c) HPLC analysis of an EGFR kinase assay with the Shc1 Y-Y peptide at multiple time points. (d) Mass spectrometry based analysis of the 120 min peak in panel C. The LC-MS/MS spectrum for the singly phosphorylated peptide PDHQpYYNDAKKK is shown. (e) Percentage of peptide spectra detected by mass spectrometry phosphorylated at Tyr239 or Tyr240.
EGFR phosphorylation of Shc1 is enhanced by a priming kinase
While it has been reported that EGFR regulates the phosphorylation of Tyr239 and Tyr240 of Shc1, whether one or both sites are directly phosphorylated by EGFR has not been examined25. The above results show that EGFR phosphorylation of the Shc1 peptide at Tyr239 is enhanced by phosphorylation at Tyr240. This coordinated regulation could result if EGFR sequentially phosphorylates both sites. In this scenario, EGFR would phosphorylate the unmodified substrate at Tyr240 with low efficiency. Once the initial phosphorylation occurs, the substrate would be activated for subsequent phosphorylation at Tyr239. A similar mechanism has been proposed for SR protein kinase 1 (SRPK1), which ‘primes’ and then sequentially phosphorylates arginine-serine dipeptide repeats in RS proteins29. Consistent with this possible mechanism, EGFR has weak but detectable activity toward the unmodified Shc1 peptide (Fig. 2a). Alternatively, a different kinase may prime Shc1 at Tyr240, enhancing EGFR phosphorylation of Tyr239.
To determine which mechanism most likely explains EGFR phosphorylation of Shc1, we analyzed the product of EGFR phosphorylation of the unmodified Shc1 peptide by HPLC and tandem mass spectrometry (LC-MS/MS). If EGFR sequentially phosphorylates both sites, it would be expected that the initial product generated would be peptides phosphorylated at Tyr240. Over time, the doubly phosphorylated product would then accumulate. If a different kinase primes Shc1, the product generated would be peptides phosphorylated at Tyr239 and the doubly phosphorylated product should not be detected.
To analyze the reaction between EGFR and the unmodified Shc1 peptide, we first synthesized the three possible product peptides: the product of phosphorylation at Tyr239 (pY-Y), the product of phosphorylation at Tyr240 (Y-pY), and the product of phosphorylation at both sites (pY-pY). We then determined retention times for each product peptide and the substrate peptide (Y-Y) by reverse-phase HPLC. While complete separation was achieved between the substrate, mono-, and doubly phosphorylated peptides, both mono-phosphorylated peptides (pY-Y, Y-pY) had the same retention time (Fig. 2b). Aliquots of an in vitro kinase reaction containing EGFR and the unphosphorylated Shc1 peptide were then monitored at different time points under the same HPLC conditions. Even after 2 hours, mono-phosphorylated peptides were the only product detected (Fig. 2c). To determine whether the mono-phosphorylated product generated was the peptide phosphorylated at Tyr239, Tyr240, or a mixture, the mono-phosphorylated peak was collected and analyzed by LC-MS/MS. Of all the phosphopeptide spectra identified by LC-MS/MS in the mono-phosphorylated peak (>50 spectra), 100% were phosphorylated at Tyr239 (Fig. 2d and 2e). No peptides phosphorylated at Tyr240 were detected after rigorous inspection of the fragment ions. Moreover, the peptide phosphorylated at Tyr239 (pY-Y) was a very unfavorable substrate for phosphorylation by EGFR (Fig. 2a). Together, these results are consistent with a mechanism wherein EGFR specifically phosphorylates Tyr239 of Shc1 and phosphorylation of Tyr240 by a different kinase enhances this activity.
Because the peptide phosphorylated at Tyr239 is not a substrate for further phosphorylation by EGFR (no pY-pY peptides were detected in Fig. 2c), the reaction with the Shc1 peptide can be assumed to follow a standard Michaelis-Menten mechanism. Therefore, we compared the Km and kcat values for EGFR phosphorylation of the unmodified peptide with the peptide primed at Tyr240. The rate constant (kcat) was similar for both peptides (Table 2). However, the priming phosphorylation decreased the Km for the peptide by approximately 4-fold.
Table 2.
EGFR steady-state kinetic parameters for Shc1 peptides
| Peptide | Sequence | Km (μM) | kcat (s−1) | kcat/Km (μM−1s−1) |
|---|---|---|---|---|
| Y-Y | PDHQYYNDAKKK | 1829 ± 172 | 0.060 ± 0.006 | 3.34E-5 |
| Y-pY | PDHQYpYNDAKKK | 488 ± 34 | 0.063 ± 0.008 | 1.30E-4 |
Kinetic parameters were determined by unweighted, nonlinear least squares analysis. Values represent means from three separate experiments ± s.e.m. The phosphorylated tyrosine is underlined.
Src primes Shc1 for EGFR phosphorylation
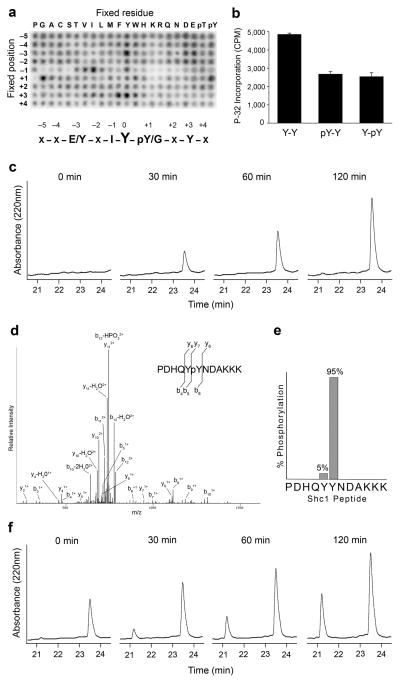
The results presented above suggest that prior phosphorylation by a priming kinase enhances EGFR activity towards Shc1, and potentially other substrates. We were therefore interested in identifying the kinase, or kinases, responsible for the priming phosphorylation. In the case of Shc1, Src has been previously reported to coordinately phosphorylate both Tyr239 and Tyr24025,27. We were therefore interested in further examining if Src sequentially phosphorylates both sites or if Src preferentially phosphorylates Ty239 or Tyr240. First, we examined the substrate specificity of Src using the PSPL assay. Src was considerably less specific than EGFR and displayed activity toward many of the peptides comprising the PSPL assay (Fig. 3a). Src had strong preferences for acidic residues in the −3 position, hydrophobics, especially Isoleucine, at the −1 position, and aromatics at the +3 position (Supplementary Table 2). Interestingly, tyrosine was a relatively strong selection at several positions relative to the phosphorylation site (particularly +3 and −3), consistent with a potential role for Src in priming substrates (Fig. 3a). Src treated with the Src-family inhibitor dasatinib did not exhibit selectivity at these positions, confirming that the results were not due to a contaminating kinase (Supplementary Fig. 1).
Figure 3.
Src primes Shc1 for EGFR phosphorylation by phosphorylating Tyr240. (a) Peptide library results for Src. Representative image from 3 independent experiments is shown. Quantification is provided in Supplementary Table 2. (b) Src kinase assays with synthetic peptides corresponding to the sequence surrounding Tyr239 of Shc1 (PDHQYYNDAKKK = Y-Y; PDHQYpYNDAKKK = Y-pY; PDHQpYYNDAKKK = pY-Y). Error bars, s.d. (n=3 technical replicates) (c) HPLC analysis at of a Src kinase assay with the Shc1 Y-Y peptide at multiple time points. (d) Mass spectrometry based analysis of the 120 min peak in panel C. The LC-MS/MS spectrum for the singly phosphorylated peptide PDHQYpYNDAKKK is shown. (e) Percentage of peptide spectra detected by LC-MS/MS phosphorylated at Tyr239 or Tyr240. (f) HPLC analysis of an in vitro kinase assay containing Src, EGFR and the Y-Y Shc1 peptide at multiple time points.
We then tested Src activity toward the unmodified and phosphorylated peptides corresponding to the sequence surrounding Tyr239 of Shc1. Src phosphorylated each peptide. However, in contrast to EGFR, the unphosphorylated Shc1 peptide was a better substrate for Src than peptides with either Tyr239 or Tyr240 already phosphorylated (Fig. 3b). To determine which site(s) on the unmodified Shc1 peptide were phosphorylated by Src, aliquots of the reaction were monitored at different time points using the same HPLC conditions as were used in the analysis of EGFR specificity. Similar to the results seen with EGFR, mono-phosphorylated peptides were the predominant product of the reaction with Src (Fig. 3c). Mass spectrometric analysis of the mono-phosphorylated peak indicated that 95% of all phosphopeptide spectra detected were phosphorylated at Tyr240 (Fig. 3d and 3e). Only if both Src and EGFR were included in the reaction, was the peptide phosphorylated at both Tyr239 and Tyr240 produced (Fig. 3f). Together, these results suggest that Src phosphorylates Tyr240 of Shc1, priming it for phosphorylation at Tyr239 by EGFR.
Phosphorylation of Tyr239 enhances Shc1 binding to Grb2
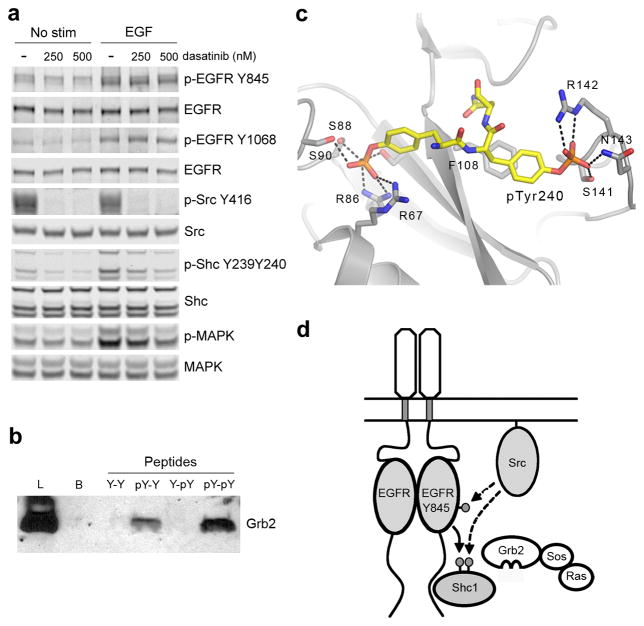
To determine if Src and EGFR coordinately phosphorylate Tyr240 and Tyr239 of Shc1 in cells, we utilized a phospho-specific Shc antibody that only detects Shc1 when it is phosphorylated at both sites (Supplementary Fig. 3.). When MCF10A cells, which have high Src activity, were stimulated with EGF, significant phosphorylation of both Tyr239 and Tyr240 was detected (Fig. 4a). However, when Src activity was blocked with the Src inhibitor dasatinib, the dual phosphorylation induced by EGF was almost completely abolished (Fig. 4a). These results are consistent with our in vitro data and indicate that the combined activities of Src and EGFR generate doubly phosphorylated Shc1 in cells.
Figure 4.
Dual phosphorylation of Tyr239 and Tyr240 is controlled by EGFR and Src in cells and enhances Shc1 binding to Grb2. (a) Immunoblot analysis of cell lysates prepared from MCF10A cells pretreated with dasatinib for 1 hour then stimulated with 1ng/mL EGF for 3 min. (b) Pulldown assay and anti-Grb1 immunoblot. Samples are lysates from A431 cells incubated with the indicated biotinylated phosphopeptides. (Lysate = L; Control Beads = B; DHQYYNDFPGKE = Y-Y; DHQYpYNDFPGKE = Y-pY; DHQpYYNDFPGKE = pY-Y; DHQpYpYNDFPGKE = pY-pY). (c) Structure of a pseudo-peptide based on the sequence of Shc1 phosphorylated at Tyr239 and Tyr240 (mAZ-pY-(αME)pY-N-NH2) bound to the SH2 domain of Grb2 (PDB ID:1JYQ)31. Hydrogen bonds are indicated with dashed lines. (d) Model for the integration of EGFR and Src signaling by coordinated phosphorylation of Shc1. EGFR phosphorylation of Shc1 at Tyr239 creates a binding site for Grb2, leading to activation of the Ras-MAPK pathway. Active Src enhances activation by phosphorylating Tyr845 of the EGFR as well as Tyr240 of Shc1. Phosphorylation of Tyr240 of Shc1 potentiates phosphorylation at Tyr239 by the EGFR and enhances the binding of Grb2. Full-size images are shown in Supplementary Data Set 1.
As discussed above, it is well established that Src activity augments MAPK signaling initiated by EGFR5. For example, cells in which both Src and EGFR are overexpressed or coactivated show a synergistic increase in total Shc phosphorylation, Shc1-Grb2 binding, and MAPK activation10,11,30. Our results suggest that one mechanism by which Src can enhance EGF-induced MAPK signaling is by phosphorylating Tyr240 of Shc1 to promote EGFR phosphorylation of Tyr239. A potential second mechanism is by enhancing the phosphorylation-induced interaction between Shc1 and Grb2. To test this possibility, we generated synthetic peptides corresponding to phosphorylated Tyr239 and Tyr240 of Shc1. The peptides were immobilized, incubated with cell lysates, and Grb2 binding to each peptide was detected by immunoblotting (Fig. 4b). As expected, the peptide phosphorylated at Tyr239 (pY-Y) bound Grb2 whereas the peptide phosphorylated at Tyr240 (Y-pY, which lacks the pTyr-X-Asn-X Grb2 binding motif) did not bind to Grb2. Importantly, the peptide phosphorylated at both Tyr239 and Tyr240 (pY-pY) bound significantly more Grb2 than the peptide phosphorylated at Tyr239 alone (Fig. 4b). Consistent with this observed enhancement in the interaction between Shc1 and Grb2, generation of doubly phosphorylated Shc1 correlates with enhanced MAPK activation in cells (Fig. 4a).
A crystal structure of Grb2 in complex with a pseudo-peptide based on the sequence of Shc1 doubly phosphorylated at Tyr239 and Tyr240 has been reported31. In the structure, the phosphate of phosphorylated Tyr240 makes several interactions with the Grb2 SH2 domain (Fig 4c). These interactions confer a 3-fold increase in the affinity of doubly phosphorylated Shc1 for Grb2 as compared to phosphorylation at Tyr239 alone31. Taken together, these results indicate that Src and EGFR coordinately regulate the phosphorylation of Shc1 Tyr240 and Tyr239 in cells and the dual phosphorylation enhances the ability of Shc1 to bind Grb2 (Fig. 4d).
Structural basis for EGFR substrate specificity
In order to identify the molecular basis for EGFR substrate specificity, we determined the crystal structure of the EGFR L858R kinase domain in complex with the Shc1 peptide primed at Tyr240 (PDHQYpYNDF; crystallographic and refinement statistics are provided in Table 3). Like other protein kinases, the EGFR kinase domain has an N-terminal lobe, comprised of an antiparallel β-sheet and single α-helix (C-helix), and a predominantly α-helical C-terminal lobe (Fig. 5a). The activation loop (A-loop) lies near the interface of the two lobes and provides part of the platform for peptide binding (discussed below). In the structure of the EGFR-Shc1 peptide complex, several Shc1 peptide residues (proline at P-4, aspartic acid at P-3, and phenylalanine at P+4) did not have well defined electron density and are not included in the model (Supplementary Fig. 4).
Table 3.
Data collection and refinement statistics
| EGFR L858R + Shc1 peptide | EGFR L858R + Optimal peptide | |
|---|---|---|
| Data collection | ||
| Space group | I23 | I23 |
| Cell dimensions | ||
| a, b, c (Å) | 143.9, 143.9, 143.9 | 144.6, 144.6, 144.6 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0–2.6 (2.69–2.6)a | 50.0–2.8 (2.9–2.8)a |
| Rmerge | 0.068 (0.477) | 0.062 (0.473) |
| I / σI | 13.8 (2.3) | 16.6 (2.2) |
| Completeness (%) | 99.0 (99.5) | 98.8 (99.3) |
| Redundancy | 3.4 (3.1) | 3.4 (3.1) |
| Refinement | ||
| Resolution (Å) | 38.5–2.6 | 34.1–2.8 |
| No. reflections | 15207 | 12361 |
| Rwork / Rfree | 0.182/0.266 | 0.192/0.222 |
| No. atoms | ||
| Protein | 2501 | 2495 |
| Water | 129 | 63 |
| B factors | ||
| Protein | 59.5 | 69.7 |
| Water | 61.3 | 65.9 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.010 | 0.010 |
| Bond angles (°) | 1.180 | 1.146 |
Values in parentheses are for highest-resolution shell.
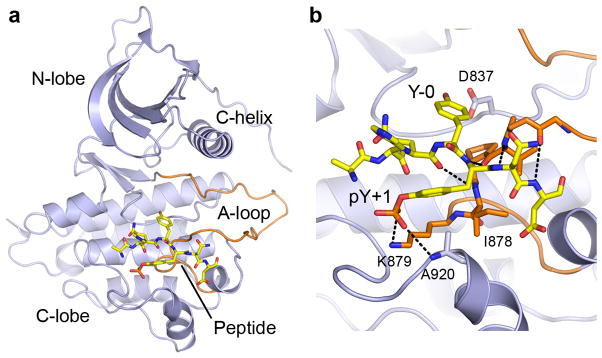
Figure 5.
Shc1 phosphopeptide binding to the EGFR kinase domain. (a) Overall structure of the EGFR L858R kinase domain bound to a Shc1 peptide phosphorylated at Tyr240 (PDHQYpYNDF). The peptide is shown in stick form. (b) Detailed view of interactions between EGFR and the primed Shc1 substrate peptide. Hydrogen bonds are indicated with dashed lines.
The Shc1 peptide binds in an extended conformation to the C-lobe and A-loop of EGFR, similar to the mode of substrate binding in other protein kinase-peptide substrate complexes (Fig. 5a). A short, antiparallel β-strand interaction is formed between the peptide and the C-terminal segment of the A-loop. Four hydrogen bonds are formed between the Shc1 peptide backbone at positions P-1 to P+3 and the mainchain of the EGFR kinase domain from residues Gly874 to Ile878. The hydroxyl oxygen of Tyr239 (Y-0) is hydrogen bonded to Asp837, the catalytic base, and is in position for phosphoryl transfer. Surprisingly, the only other sequence-specific interaction observed between EGFR and the Shc1 peptide occur with phosphoTyr240 (pY+1). The phosphate group binds the ε-amino nitrogen of Lys879 and the main chain nitrogen of Ala920 (Fig. 5b). The binding site formed by Lys879 and Ala920 in the EGFR is only weakly basic, consistent with the observation that phosphorylation of Tyr240 enhances, but is not required for, EGFR activity.
The position equivalent to EGFR Lys879 is conserved in the other active members of the ERBB family of receptor tyrosine kinases, ERBB2 and ERBB4. We therefore tested whether ERBB2 and ERBB4, like EGFR, have specificity for primed substrates. Using the PSPL assay, we found that both ERBB2 and ERBB4 also had a strong preference for phosphotyrosine at the +1 position relative to the phosphorylation site (Supplementary Fig. 5).
We also determined the crystal structure of the EGFR kinase domain in complex with a substrate peptide having the optimal sequence as determined in the PSPL assay. In addition to the strong selection for phosphotyrosine at the +1 position, EGFR has weak preferences for acidic residues, particularly N-terminal to the phosphorylation site, and hydrophobic residues, particularly at the +3 position (Fig. 1 and Supplementary Table 1). The optimal substrate peptide (DEEDYpYEIP) binds to the EGFR kinase domain in a mode that is nearly identical to that observed with the Shc1 peptide, with the phosphotyrosine at the +1 position forming interactions with the ε-amino nitrogen of Lys879 and the main chain nitrogen of Ala920 (Supplementary Fig. 4). Surprisingly, despite having an optimized sequence, only one additional substrate side chain interaction is observed between the aspartic acid at P-1 and Arg841 of EGFR. In previously reported structures of tyrosine kinase domains in complex with substrate peptides (for example IRK, IGF1R, and ABL), hydrophobic contacts between amino acids C-terminal to the phosphorylation site and hydrophobic pockets on the surfaces of the enzymes are the predominant interactions observed32–34. While the surface of EGFR has a structurally equivalent hydrophobic pocket (formed from the side chains of Val876, Ile878 and Ile886), the Isoleucine at the +3 position of the optimal substrate peptide but did not have well defined electron density. These structural observations further suggest that, other than phosphotyrosine at +1, the primary sequence surrounding the phosphorylation site may have little influence on EGFR specificity.
DISCUSSION
Cooperativity with Src is a key element of EGFR signaling and plays an important functional role in EGFR-driven cancers; however, the underlying mechanism is poorly understood. Here, we provide a molecular explanation for how Src activity is integrated with EGFR signaling. We found that EGFR preferentially phosphorylates substrates that are primed by a prior phosphorylation and, in the case of the adaptor protein Shc1, we showed that Src mediates the priming phosphorylation. Importantly, the dual phosphorylation of Shc1 by Src and EGFR significantly enhanced Shc1 binding to Grb2, a key step in activating the Ras-MAPK pathway, beyond EGFR phosphorylation alone (Fig. 4d).
Evidence that EGFR selectivity for phosphotyrosine primed substrates is relevant in cells comes from the observation that many tyrosine phosphorylation sites in cells driven by EGFR mutations have a phosphotyrosine at the +1 position35. We used the PhosphositePlus database to identify a subset of these sites that have also been shown to be sensitive to treatment with EGFR-specific inhibitors (Table 1). In the case of Shc1 Tyr239, we showed that Src can function as the priming kinase and catalyze the priming phosphorylation at Tyr240. However, it is likely that Src primes multiple substrates for EGFR phosphorylation. For example, a recent study showed that EGFR phosphorylation of Mig6, a putative tumor suppressor and feedback inhibitor of EGFR, is enhanced by prior phosphorylation of an adjacent tyrosine by Src36. If Src primes multiple EGFR substrates, it will be important to determine if this is a mechanism by which Src promotes signaling through a subset of pathways downstream of EGFR or if Src induces the phosphorylation of a different set of substrates.
The crystal structure of the EGFR kinase domain in complex with a Shc1 peptide phosphorylated at Tyr240 provides a molecular explanation for EGFR specificity for primed substrates. Interestingly, in both this structure and a structure of EGFR in complex with an optimized substrate peptide, the major sequence-specific interaction was between EGFR and the phosphate of the phosphotyrosine at P+1 (Fig. 5 and Supplementary Fig. 4). This observation suggests that prior phosphorylation is the dominant sequence feature recognized by EGFR, not the sequence of the surrounding amino acids. A lack of stringent primary sequence requirements suggests that EGFR has the potential to phosphorylate substrates primed by many different tyrosine kinases and this could serve as a mechanism of signal integration with multiple signaling pathways.
In the case of Shc1, we showed that Src preferentially phosphorylates Tyr240 to promote EGFR phosphorylation of Tyr239. However, our PSPL results indicate that Src has relatively strong selectivity for tyrosine at several positions surrounding the phosphorylation site (Fig. 3a). These results suggest that Src could potentially catalyze priming phosphorylations at multiple positions flanking tyrosine phosphorylation sites. Substrate priming may therefore be a general characteristic of Src signaling and may be the mechanism by which Src participates in signaling downstream of several receptor tyrosine kinases5.
Our current work indicates that the SH2 domain of Grb2 binds doubly phosphorylated Shc1 with greater affinity than Shc1 phosphorylated at Tyr239 alone (Fig. 4b). We previously investigated the optimal binding motif for the Grb2 SH2 domain28. We found that Asn at the +2 position relative to the phosphotyrosine dominated the binding selectivity (Asn is found at the +2 position relative to pY239 of Shc1) and subsequent work elucidated the structural basis for this specificity37. However, our original study only investigated selectivity for the 20 natural amino acids; selectivity for phosphorylated amino acids within the optimal binding motif was not addressed. Our current results, together with several recent studies showing increased affinity of SH2 domains for doubly phosphorylated peptides, raise the possibility that the presence of multiple phosphorylations within an SH2 domain recognition motif may be a common mechanism that enables complex regulation of SH2 domain-target protein interactions38–41.
Finally, it is well established that protein kinase substrate selectivity is strongly influenced by the sequence immediately surrounding the phosphorylation site. Until recently, selectivity for phosphorylated amino acids was thought to be unique to the serine/threonine protein kinases glycogen synthase kinase-3 and casein kinase 142,43. However, recent high-resolution phosphoproteomics analyses have made it clear that the sequences surrounding phosphorylation sites often contain additional sites of phosphorylation43. For example, a survey of multiply phosphorylated proteins showed that more than half of all phosphorylation sites are within 4 amino acids of another site44. Based on these observations, proximal phosphorylations would be expected to be an important selectivity determinant for far more protein kinases. One explanation for why so few kinases with this selectivity have been identified is that phosphorylated amino acids had to be excluded from previous methods used to determine protein kinase selectivity in vitro. Consistent with this, recent studies utilizing the PSPL approach have identified several additional kinases with selectivity for phosphorylated substrates46–49. These observations, together with results described here, suggest that coordinated substrate phosphorylation by two or more kinases is likely to be a common mechanism of signal integration that contributes to both normal and oncogenic protein kinase signaling.
ONLINE METHODS
Positional Scanning Peptide Library (PSPL) assay
Recombinant, wild-type and L858R EGFR kinase domains (residues 696–1022) and recombinant Src were generated as previously described50,51. Recombinant ERRB2 was from Life Technologies and recombinant ERBB4 was from SignalChem. The sequence of each of the 198 peptide libraries is G-A-X-X-X-Z-X-Y-X-X-X-X-A-G-K-K-biotin (Z=fixed amino acid, X=equimolar mixture of amino acids excluding Tyr and Cys). Aqueous stocks of each library were prepared in 50mM Hepes, pH 7.4 and arrayed in 384-well plates containing 50mM Tris pH 7.5, 10mM MgCl2, 10mM MnCl2, 2mM DTT. Each row in the array contained libraries with a different fixed position relative to the central tyrosine and in each column that position is fixed to a different amino acid. EGFR or Src and γ-32P ATP were added to the wells and the entire set of peptide libraries was phosphorylated in parallel at 30° for 1–2 hours. The final concentrations of the reaction components were 50μM peptide library, 200nM EGFR or 70nM Src, and 100μM ATP at a specific activity of 1mCi γ-32P-ATP/μmol in a total volume of 10μL. Following the reaction, 2μL aliquots from each well were simultaneously transferred to an avidin-coated membrane (Promega SAM2 biotin capture membrane) using a 384-slot pin replicator (VP Scientific). Membranes were washed sequentially with 0.1% SDS in TBS, 2M NaCl + 1%H3PO4, and 2M NaCl, then dried. Phosphate incorporation into each library was quantified using a phosphorimager and the ImageQuant 5.2 software from Molecular Dynamics. The amount of phosphate incorporated into each well was divided by the average of all the wells in a row to give the selectivity value for a given amino acid relative to the other residues at the same position. Therefore, values greater than 1 represent positive selections and values less than 1 represent negative selections.
In vitro kinase assays
Peptides based on the sequences surrounding MET Tyr1234 (MYDKEYYSVHN) and Shc1 Tyr239 (PPDHQYYNDFP) were synthesized, HPLC purified, and verified by mass spectrometry. Lysine residues were added to the C-terminus of the peptides to ensure quantitative binding in phosphocellulose assays. The relative activity of EGFR or Src for each peptide was determined in reactions containing 100nM EGFR or 100nM Src, kinase buffer (50mM Tris pH 7.5, 10mM MgCl2, 10mM MnCl2, 2mM DTT), 200μM ATP at a specific activity of 1mCi γ-32P-ATP/μmol, and 400μM peptide. After incubation at 30° for 30min, aliquots of each reaction were spotted onto P81 phosphocellulose papers and washed extensively with 75mM H3P04. Filters were dried and radioactivity determined by scintillation counting. Values from control reactions in which the substrate peptide was omitted were subtracted from experimental values. All reactions were done in triplicate.
For the HPLC analysis of the reaction products, the concentrations of EGFR and Src were adjusted to compensate for different specific activities. The final concentrations of the reaction components were 1μM EGFR or 5μM Src, 500μM Shc1 peptide, and 1mM ATP in a 100μL reaction volume. For the reaction containing both Src and EGFR, the reaction was initiated by the addition of Src and after a 60min preincubation, EGFR was added. At various times, 25μL aliquots of the reactions were removed and quenched with EDTA. Quenched aliquots were analyzed by HPLC on a 4.6 × 150mM Agilent Eclipse Plus C18 column. Peptides were eluted with a 30 min linear gradient of 0–30% solvent B with a flow rate of 1.0 ml/min where solvent A contained 0.1% trifluoroacetic acid and solvent B contained 0.1% trifluoroacetic acid in 100% acetonitrile. The elution profile was monitored by absorbance at 220nm. The retention times of each peptide was established by individual injections under the same HPLC conditions. To identify the sites on the Shc1 peptide phosphorylated by EGFR or Src, the reaction products were analyzed by reversed-phase microcapillary/tandem mass spectrometry (LC/MS/MS).
Kinetic parameters for EGFR phosphorylation of the Shc1 peptides were determined using the phosphocellulose assay. Increasing concentrations of peptide (0 – 12mM) were incubated with 100nM EGFR in kinase buffer supplemented with 2μCi γ-32P-ATP/reaction for 30 min at 30°. Initial rates were determined from the linear portion of the reaction (<5% substrate consumption). Kinetic parameters were determined by fitting data to the Michaelis Menten equation by use of nonlinear least squares analysis of initial rates. Data shown are the average of three separate experiments, each done in triplicate.
Tandem mass spectrometry (LC/MS/MS)
LC/MS/MS was performed using an EASY-nLC nanoflow HPLC (Thermo Fisher Scientific) with a self-packed 75 μm id × 15 cm C18 column coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) in the data-dependent acquisition and positive ion mode at 300 nL/min. Peptide ions from the predicted Shc1 peptide phosphorylation sites were also targeted in MS/MS mode for quantitative analyses. MS/MS spectra collected via collision induced dissociation (CID) were searched against the concatenated target and decoy (reversed) single entry Shc1 sequence and full Swiss-Prot protein databases using Sequest (Proteomics Browser Software, Thermo Fisher Scientific) with differential modifications for Ser/Thr/Tyr phosphorylation (+79.97) and the sample processing artifacts Met oxidation (+15.99), and deamidation of Asn and Gln (+0.984). False discovery rates (FDR) of peptide hits were estimated below 1.5% based on reversed (background) database hits. Passing MS/MS spectra were manually inspected to be sure that all b- and y- fragment ions aligned with the assigned sequence and the pTyr modification sites were correctly assigned. Quantification of phospho- peptides was achieved by assessing the number of spectral counts (identified peptides) for each assigned pTyr site.
Cell culture and immunoblotting
MCF10A cells were purchased from ATCC and maintained in HuMEC Medium (Life Technologies). Cells were authenticated by DNA fingerprinting analysis (STR analysis) and confirmed to be free of mycoplasma contamination by QF-PCR. Cells were incubated 16 hours in growth-factor-free medium before pretreatment with dasatinib (250 or 500nM) for 1 hour and stimulation with 1ng/mL EGF for 3 minutes. Cells were then lysed in RIPA buffer supplemented with protease inhibitors (Roche) and phosphatase inhibitors (Calbiochem). Proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and subject to immunoblotting. Antibodies against phospho-EGFR Y845 (cat. no. 6963), phospho-EGFR Y1068 (cat. no. 2236), phospho-Src (cat. no. 6943), Src (cat. no. 2123), phospho-Shc (cat. no. 2434), and phospho-p44/42 MAPK (cat. no. 4370) were purchased from Cell Signaling Technology. Antibodies against EGFR (cat. no. 610016), Shc (cat. no. 610878), and p44/42 MAPK (cat. no. 610123) were purchased from BD Biosciences. The anti-Grb2 antibody (cat. no. sc-255) was from Santa Cruz Biotechnology. Validation for each antibody is provided on the manufacturers’ websites. Original images of blots used in this study can be found in Supplementary Data Set 1.
Shc1 phosphopeptide binding assay
Biotinylated phosphopeptides corresponding to the sequence surrounding Shc1 Tyr239 (Biotin- AHA-DHQYYNDFPGKE) were immobilized on streptavidin-sepharose beads and incubated with A431 cell lysate. Beads were collected by centrifugation and washed. Bound proteins were eluted with SDS sample buffer and resolved by SDS-PAGE. Proteins were transferred to nitrocellulose and probed by western blotting with an anti-GRB2 antibody.
Structure determination
The EGFR L858R kinase domain was crystallized as previously described50. Phosphopeptides based on the Shc1 Tyr239 phosphorylation site (PDHQYpYNDF) and the optimal EGFR substrate motif (DEEDYpYEIP), were synthesized, HPLC purified, and verified by mass spectrometry. Peptides were introduced into the crystals by soaking the crystals for 4 hours in reservoir solution supplemented with 10mM peptide. This solution was also used as the cryoprotectant. Diffraction data were collected at Argonne National Laboratory ID24 beamline at 100K. Data were processed and merged with HKL200052. The structure was determined by the Difference Fourier method using the EGFR L858R kinase structure (PDB 2ITV)50 as the starting model. Simulated-annealing in CNS was then used to obtain a less biased model and 2Fo-Fc and Fo-Fc maps for manual model inspection and adjustment53. Repeated rounds of manual refitting and crystallographic refinement were performed using COOT54,55. The peptides were modeled into the closely fitting positive Fo-Fc electron density and then included in following refinement cycles. Detailed statistics are in Table 3.
Supplementary Material
Supplementary Figure 1. EGFR and Src peptide library assays using specific inhibitors. (a) Phosphorylation of the pY+1 peptide library by EGFR WT or EGFR L858R in the presence of increasing concentrations of gefitinib. Activity is graphed as percent of control (no gefitinib). (b) Phosphorylation of the I–1, G+1, pY+1 and Y+3 peptide libraries by Src in the presence of increasing concentrations of dasatinib. Activity is graphed as percent of control (no dasatinib).
Supplementary Figure 2. EGFR activity towards peptides based on the sequence surrounding MET Tyr1234. Peptides were phosphorylated in vitro with recombinant EGFR (MYDKEYYSVHNKK = Y–Y; MYDKEYpYSVHNKK = Y-pY). Error bars, s.d. (n=3 technical replicates).
Supplementary Figure 3. Specificity of phospho-Shc pY239/240 antibody. Serial dilutions of peptides corresponding to the sequence surrounding Tyr239 of Shc1 were spotted on nitrocellulose and probed with a phospho-Shc pY239/240 antibody (Cell Signaling Technology Cat #2434; PDHQYYNDAKKK = Y–Y; PDHQYpYNDAKKK = Y-pY; PDHQpYYNDAKKK = pY-Y; PDHQpYpYNDAKKK = pY-pY). 40nmol corresponds to 2μL of a 20mM stock solution of each peptide (20mM is the solubility limit of the peptides in Tris buffer).
Supplementary Figure 4. Crystal structures of EGFR in complex with peptides. (a) (b) Electron density of peptides in complex with EGFR L858R kinase domain. Fo-Fc difference omit maps (in which the peptides were omitted) for the optimal peptide (a) or the primed Shc1 peptide (b) contoured at 3σ. (c) Interactions between the EGFR L858R kinase domain and the optimal substrate peptide (DEEDYpYEIP). Hydrogen bonds are indicated with dashed lines.
Supplementary Figure 5. Peptide library results for ERBB2 and ERBB4.
Supplementary Table 1. EGFR selectivity values. Following the positional scanning peptide library assay, phosphate incorporation into each library was quantified using a phosphorimager and the ImageQuant 5.2 software from Molecular Dynamics. Each value was then divided by the average phosphate incorporation for all amino acids at that position to yield a selectivity value for each amino acid at that position relative to the phosphoacceptor site. The values shown are the averages of three independent experiments. Values greater than 1.0 represent positive selections and values less than 1.0 represent negative selections. Strong selections are shown in bold. Note that selections for tyrosine could arise from phosphorylation at the fixed tyrosine rather than (or in addition to) the central tyrosine.
Supplementary Table 2. Src selectivity values. Src selectivity values were determined as described in Supplementary Table 1. Strong selections are shown in bold.
Acknowledgments
We thank S. Breitkopf and M. Yuan for help with mass spectrometry experiments and Y. Zheng for help with HPLC experiments. We thank B. Murray for critical reading of the manuscript. This work was partially supported by US National Institutes of Health grants 2P01CA120964 (J.M.A.) and S10OD010612 (J.M.A.).
Footnotes
Accession codes
Coordinates for the crystal structures reported here have been deposited in the Protein Data Bank under accession codes PDB 5CZI (Shc1 peptide) and 5CZH (optimal peptide).
AUTHOR CONTRIBUTIONS
M.J.B. designed and conducted all biochemical, enzymatic, and cell-based experiments. C. Y. determined all crystal structures. C.A.G and J.L.J., together with M.J.B., performed peptide library experiments. J. M. A. executed mass spectrometry-based experiments. M.J.E conceived and supervised structural experiments. I.A. and A.J.C. conceived and supervised cell-based experiments. L.C.C, together with M.J.B, conceived the study and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Yarden T, Sliwkowski MX. Untangling the ErbB signaling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 4.Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors – impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bromann PA, Korkaya H, Courtneidge SA. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene. 2004;23:7957–7968. doi: 10.1038/sj.onc.1208079. [DOI] [PubMed] [Google Scholar]
- 6.Tice DA, Biscardi JS, Nickles AL, Parsons SJ. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc Natl Acad Sci USA. 1999;96:1415–1420. doi: 10.1073/pnas.96.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biscardi JS, et al. c-Src mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem. 1999;274:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 8.Moro L, et al. Integrin-induced epidermal growth factor (EGF) receptor activation requires c-Src and p130Cas and leads to phosphorylation of specific EGF receptor tyrosines. J Biol Chem. 2002;277:9405–9414. doi: 10.1074/jbc.M109101200. [DOI] [PubMed] [Google Scholar]
- 9.Boerner JL, Biscardi JS, Silva CM, Parsons SJ. Transactivating agonists of the EGF receptor require Tyr845 phosphorylation for induction of DNA synthesis. Mol Carcinog. 2005;44:262–273. doi: 10.1002/mc.20138. [DOI] [PubMed] [Google Scholar]
- 10.Biscardi JS, Belsches AP, Parsons SJ. Characterization of human epidermal growth factor receptor and c-Src in human breast tumor cells. Mol Carcinogen. 1998;21:261–272. doi: 10.1002/(sici)1098-2744(199804)21:4<261::aid-mc5>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 11.Maa MC, Leu TH, McCarley DJ, Schatzman RC, Parsons SJ. Potentiation of EGF receptor-mediated oncogenesis by c-SRC: implications for the etiology of multiple human cancers. Proc Natl Acad Sci USA. 1995;92:6981–6985. doi: 10.1073/pnas.92.15.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimri M, et al. Modeling breast cancer-associated c-SRC and EGFR overexpression in human MECs: c-Src and EGFR cooperatively promote aberrant three-dimensional acinar structure and invasive behavior. Cancer Res. 2007;67:4164–4172. doi: 10.1158/0008-5472.CAN-06-2580. [DOI] [PubMed] [Google Scholar]
- 13.Fu Y-N, et al. EGFR mutants found in non-small cell lung cancer show different levels of sensitivity to suppression of Src: implications in targeting therapy. Oncogene. 2008;27:957–965. doi: 10.1038/sj.onc.1210684. [DOI] [PubMed] [Google Scholar]
- 14.Chung BM, et al. The role of cooperativity with Src in oncogenic transformation mediated by non-small cell lung cancer-associated EGF receptor mutants. Oncogene. 2009;28:1821–1832. doi: 10.1038/onc.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller KL, Hunter LA, Ethier SP, Boerner JL. Met and c-Src cooperate to compensate for loss of epidermal growth factor receptor kinase activity in breast cancer cells. Cancer Res. 2008;68:3314–3322. doi: 10.1158/0008-5472.CAN-08-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheeler DL, et al. Epidermal growth factor receptor cooperates with Src family kinases in acquired resistance to cetuximab. Cancer Biol Ther. 2009;8:696–703. doi: 10.4161/cbt.8.8.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filosto S, Baston DS, Chung S, Becker CR, Goldkorn T. Src mediates cigarette smoke-induced resistance to tyrosine kinase inhibitors in NSCLC cells. Mol Cancer Ther. 2013;12:1579–1590. doi: 10.1158/1535-7163.MCT-12-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases: controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 19.Gotoh N, Tojo A, Hino M, Yazaki Y, Shibuya M. A highly conserved tyrosine residue at codon 845 within the kinase domain is not required for the transforming activity of human epidermal growth factor receptor. Biochem Biophys Res Commun. 1992;186:768–774. doi: 10.1016/0006-291x(92)90812-y. [DOI] [PubMed] [Google Scholar]
- 20.Mueller KL, Powell K, Madden JM, Eblen ST, Boerner JL. EGFR tyrosine 845 phosphorylation-dependent proliferation and transformation of breast cancer cells require activation of p38 MAPK. Transl Oncol. 2012;5:327–334. doi: 10.1593/tlo.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutti JE, et al. A rapid method for determining protein kinase phosphorylation specificity. Nat Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 22.Songyang Z, et al. Catalytic specificity of protein tyrosine kinases is critical for selective signaling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 23.Chan SK, Gullick WJ, Hill ME. Mutations of the epidermal growth factor receptor in non-small cell lung cancer - Search and destroy. Eur J Cancer. 2006;42:17–23. doi: 10.1016/j.ejca.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 24.Hornbeck PV, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in human and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato K, et al. Tyrosine residues 239 and 240 of Shc are phosphatidylinositol 4,5-bisphosphate-dependent phosphorylation sites by c-Src. Biochem Biophys Re Commun. 1997;240:399–404. doi: 10.1006/bbrc.1997.7667. [DOI] [PubMed] [Google Scholar]
- 26.Wills MKB, Jones N. Teaching an old dogma new tricks: twenty years of Shc adaptor signaling. Biochem J. 2012;447:1–16. doi: 10.1042/BJ20120769. [DOI] [PubMed] [Google Scholar]
- 27.Van der Geer P, Wiley S, Gish GD, Pawson T. The Shc adaptor protein is highly phosphorylated at conserved, twin tyrosine residues (Y239/240) that mediate protein-protein interactions. Curr Biol. 1996;6:1435–1444. doi: 10.1016/s0960-9822(96)00748-8. [DOI] [PubMed] [Google Scholar]
- 28.Songyang Z, et al. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngo JCK, et al. A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol Cell. 2008;29:563–576. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luttrell LM, et al. Role of c-Src tyrosine kinase in G protein-coupled receptor and Gβγ subunit-mediated activation of mitogen-activated protein kinases. J Biol Chem. 1996;271:19443–19450. doi: 10.1074/jbc.271.32.19443. [DOI] [PubMed] [Google Scholar]
- 31.Nioche P, et al. Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor. J Mol Biol. 2002;315:1167–1177. doi: 10.1006/jmbi.2001.5299. [DOI] [PubMed] [Google Scholar]
- 32.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5573–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Favelyukis S, Till JH, Hubbard SR, Miller WT. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat Struct Biol. 2001;8:1058–1063. doi: 10.1038/nsb721. [DOI] [PubMed] [Google Scholar]
- 34.Levinson NM, et al. A Src-like inactive conformation in the abl tyrosine kinase domain. PLos Biol. 2006;4:753–767. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo A, et al. Signaling networks assembled by oncogenic EGFR and c-MET. Proc Natl Acad Sci USA. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park E, et al. Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6. Nat Struct Mol Biol. 2015;22:703–711. doi: 10.1038/nsmb.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogura K, Tsuchiya S, Terasawa H, Yuzawa S, Hatanaka H, Mandiyan V, Schlessinger J, Inagaki F. Solution structure of the SH2 domain of Grb2 complexed with the Shc-derived phosphotyrosine-containing peptide. J Mol Biol. 1999;289:439–445. doi: 10.1006/jmbi.1999.2792. [DOI] [PubMed] [Google Scholar]
- 38.Lubman OY, Waksman G. Structural and thermodynamic basis for the interaction of the Src SH2 domain with the activated form of the PDGF beta-receptor. J Mol Biol. 2003;328:655–668. doi: 10.1016/s0022-2836(03)00344-9. [DOI] [PubMed] [Google Scholar]
- 39.Chen CH, Martin VA, Gorenstein NM, Geahlen RL, Post CB. Two closely spaced tyrosines regulate NFAT signaling in B cells via Syk association with Vav. Mol Cell Biol. 2011;31:2984–2996. doi: 10.1128/MCB.05043-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groesch TD, Zhou F, Mattila S, Geahlen RL, Post CB. Structural basis for the requirement of two phosphotyrosine residues in signaling mediated by Syk tyrosine kinase. J Mol Biol. 2006;356:1222–1236. doi: 10.1016/j.jmb.2005.11.095. [DOI] [PubMed] [Google Scholar]
- 41.Weber T, Schaffhausen B, Liu Y, Gunther UL. NMR structure of the N-SH2 of the p85 subunit of phosphoinositide 3-kinase complexed to a doubly phosphorylated peptide reveals a second phosphotyrosine binding site. Biochemistry. 2000;39:15869–15869. doi: 10.1021/bi001474d. [DOI] [PubMed] [Google Scholar]
- 42.Fiol CJ, Mahrenholz AM, Wang Y, Roeske RW, Roach PJ. Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase 2 and glycogen synthase kinase 3. J Biol Chem. 1987;262:14042–14048. [PubMed] [Google Scholar]
- 43.Flotow H, et al. Phosphate groups as substrate determinants for casein kinase 1 action. J Biol Chem. 1990;265:14264–14269. [PubMed] [Google Scholar]
- 44.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 45.Schweiger R, Linial M. Cooperativity within proximal phosphorylation sites is revealed from large-scale proteomics data. Biol Direct. 2010;5:6. doi: 10.1186/1745-6150-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hutti JE, et al. IkappaB kinase beta phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF-kappaB pathway. Mol Cell Biol. 2007;27:7451–7461. doi: 10.1128/MCB.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis TL, et al. Structural recognition of an optimized substrate for the ephrin family of receptor tyrosine kinases. FEBS J. 2009;276:4395–4404. doi: 10.1111/j.1742-4658.2009.07147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouskila M, et al. TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, activity, localization, and development. Biochem J. 2011;437:157–167. doi: 10.1042/BJ20110276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen S, et al. Tyrosine kinase BMX phosphorylates phosphotyrosine-primed motif mediating the activation of multiple receptor tyrosine kinases. Sci Signal. 2013;6:ra40. doi: 10.1126/scisignal.2003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yun C-H, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-SRC. Nature. 1997;385:595–601. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 52.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology, Volume 276: Macromolecular Crystallography, Part A. Academic Press; New York, NY: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 53.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. EGFR and Src peptide library assays using specific inhibitors. (a) Phosphorylation of the pY+1 peptide library by EGFR WT or EGFR L858R in the presence of increasing concentrations of gefitinib. Activity is graphed as percent of control (no gefitinib). (b) Phosphorylation of the I–1, G+1, pY+1 and Y+3 peptide libraries by Src in the presence of increasing concentrations of dasatinib. Activity is graphed as percent of control (no dasatinib).
Supplementary Figure 2. EGFR activity towards peptides based on the sequence surrounding MET Tyr1234. Peptides were phosphorylated in vitro with recombinant EGFR (MYDKEYYSVHNKK = Y–Y; MYDKEYpYSVHNKK = Y-pY). Error bars, s.d. (n=3 technical replicates).
Supplementary Figure 3. Specificity of phospho-Shc pY239/240 antibody. Serial dilutions of peptides corresponding to the sequence surrounding Tyr239 of Shc1 were spotted on nitrocellulose and probed with a phospho-Shc pY239/240 antibody (Cell Signaling Technology Cat #2434; PDHQYYNDAKKK = Y–Y; PDHQYpYNDAKKK = Y-pY; PDHQpYYNDAKKK = pY-Y; PDHQpYpYNDAKKK = pY-pY). 40nmol corresponds to 2μL of a 20mM stock solution of each peptide (20mM is the solubility limit of the peptides in Tris buffer).
Supplementary Figure 4. Crystal structures of EGFR in complex with peptides. (a) (b) Electron density of peptides in complex with EGFR L858R kinase domain. Fo-Fc difference omit maps (in which the peptides were omitted) for the optimal peptide (a) or the primed Shc1 peptide (b) contoured at 3σ. (c) Interactions between the EGFR L858R kinase domain and the optimal substrate peptide (DEEDYpYEIP). Hydrogen bonds are indicated with dashed lines.
Supplementary Figure 5. Peptide library results for ERBB2 and ERBB4.
Supplementary Table 1. EGFR selectivity values. Following the positional scanning peptide library assay, phosphate incorporation into each library was quantified using a phosphorimager and the ImageQuant 5.2 software from Molecular Dynamics. Each value was then divided by the average phosphate incorporation for all amino acids at that position to yield a selectivity value for each amino acid at that position relative to the phosphoacceptor site. The values shown are the averages of three independent experiments. Values greater than 1.0 represent positive selections and values less than 1.0 represent negative selections. Strong selections are shown in bold. Note that selections for tyrosine could arise from phosphorylation at the fixed tyrosine rather than (or in addition to) the central tyrosine.
Supplementary Table 2. Src selectivity values. Src selectivity values were determined as described in Supplementary Table 1. Strong selections are shown in bold.