Abstract
Tibetan chicken, unlike their lowland counterparts, exhibit specific adaptations to high-altitude conditions. The genetic mechanisms of such adaptations in highland chickens were determined by resequencing the genomes of four highland (Tibetan and Lhasa White) and four lowland (White Leghorn, Lindian, and Chahua) chicken populations. Our results showed an evident genetic admixture in Tibetan chickens, suggesting a history of introgression from lowland gene pools. Genes showing positive selection in highland populations were related to cardiovascular and respiratory system development, DNA repair, response to radiation, inflammation, and immune responses, indicating a strong adaptation to oxygen scarcity and high-intensity solar radiation. The distribution of allele frequencies of nonsynonymous single nucleotide polymorphisms between highland and lowland populations was analyzed using chi-square test, which showed that several differentially distributed genes with missense mutations were enriched in several functional categories, especially in blood vessel development and adaptations to hypoxia and intense radiation. RNA sequencing revealed that several differentially expressed genes were enriched in gene ontology terms related to blood vessel and respiratory system development. Several candidate genes involved in the development of cardiorespiratory system (FGFR1, CTGF, ADAM9, JPH2, SATB1, BMP4, LOX, LPR, ANGPTL4, and HYAL1), inflammation and immune responses (AIRE, MYO1F, ZAP70, DDX60, CCL19, CD47, JSC, and FAS), DNA repair, and responses to radiation (VCP, ASH2L, and FANCG) were identified to play key roles in the adaptation to high-altitude conditions. Our data provide new insights into the unique adaptations of highland animals to extreme environments.
Keywords: highland chicken, adaptive selection, gene ontology terms, admixture, extreme environment
Introduction
The Tibetan Plateau (also known as the Qinghai-Tibet Plateau) is located in western China and has an average elevation of more than 4,000 m above sea level. This elevated area, which is usually termed “the Roof of the World” or “the Third Pole,” is characterized by extreme environmental conditions, including low oxygen concentration, high intensity of ultraviolet (UV) radiation, and great fluctuations in temperature on a daily basis; these conditions impose severe physiological challenges to endothermic animals (Scheinfeldt and Tishkoff 2010; Cheviron and Brumfield 2012). A previous study indicated that humans migrating to high altitudes might suffer from acute mountain sickness, high-altitude pulmonary edema, and high-altitude cerebral edema (Srivastava et al. 2012). However, the Tibetan chicken is highly adapted to the extreme high-altitude environment and exhibits good performance in terms of survival and reproduction (Li and Zhao 2009).
Although the domestication of chickens dates back over 10,000 years (Xiang et al. 2014), the number of studies investigating the history of domesticated chickens on the Tibetan Plateau is limited. Tibetan chickens have been thought to have inhabited this region for more than 1,000 years, representing the most ancient chicken population living under high-altitude conditions (Zhang et al. 2007). Tibetan chickens are mainly distributed at altitudes of 2,200–4,100 m, and their appearance and behavior greatly resemble those of Red Jungle fowl (Gallus gallus) (Zhang et al. 2007; Wang et al. 2015). This native highland population is characterized by its small body size, outstanding foraging ability in the wild, and high hatchability under hypoxic conditions (Du 2004; Li and Zhao 2009; Xiang et al. 2014). Unlike their lowland counterparts, Tibetan chickens have several physiological parameters that efficiently promote the blood oxygen-carrying capacity, such as larger organs (heart, liver, and lungs), lower arterial oxygen partial pressure, lower venous blood pH, higher venous CO2 partial pressure, and higher hemoglobin concentration (Gou et al. 2007; Wei et al. 2007; Zhang et al. 2007; Zhang, et al. 2008). These unique characteristics that allow adaptation to high-altitude environments might have evolved under natural and human selection in Tibetan chickens.
High-throughput sequencing technologies allow the decoding of adaptation to extreme conditions in humans, plants, and animals (Mardis 2008; Turner et al. 2010; Treangen and Salzberg 2012; Liu et al. 2014; Zhou et al. 2014). Numerous studies have provided insights into the origin and genomic characters of high-altitude natives (Bigham et al. 2009, 2010; Xu et al. 2011; Scheinfeldt et al. 2012; Cai et al. 2013; Sun et al. 2015). Jeong et al. (2014) reported that the adaptation of Tibetans to high altitude could be ascribed to the genetic admixture of Sherpa and Han Chinese, and two positively selected genes (PSGs—EGLN1 and EPAS1) were identified to be responsible for this adaptation. Qu et al. (2013) reported that the expansion in genes involved in energy metabolism and hypoxia response might be associated with the adaptation of ground tit to the extreme conditions of the Tibetan Plateau. Li et al. (2013) found putative selection signatures in Tibetan wild boars, which might be related to genetic adaptation to hypoxia in high-altitude environments. Wang et al. (2015) showed that genes involved in the Ca2+ signaling pathway might be related to the adaptation of Tibetan chickens to hypoxic conditions at high altitudes. However, further studies are required to better understand the molecular mechanisms and physiological functions related to the adaptation of chicken to high-altitude environments, in order to improve the development of husbandry industry in these areas.
This study aimed to 1) investigate the genetic distinctions between highland and lowland chicken populations, 2) identify several differentially expressed genes (DEGs) in chicken embryos incubated under hypoxic conditions, and 3) determine the genetic relationships among different chicken populations and their corresponding genomic structure.
Materials and Methods
Sample Information
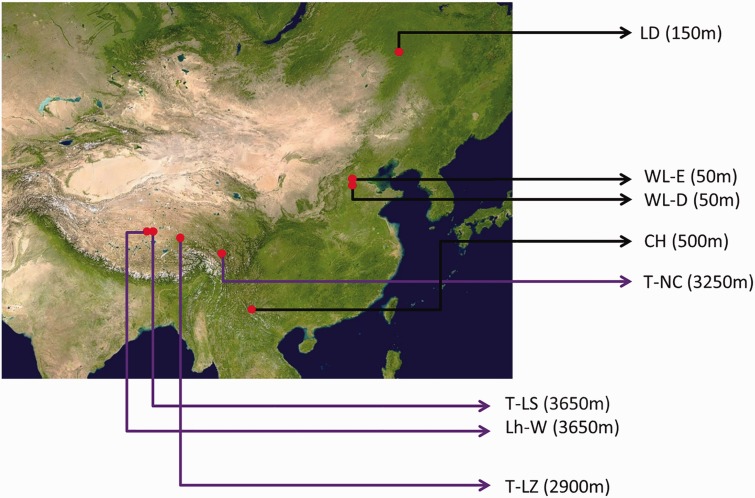
We used eight chicken populations from different locations (fig. 1), including four highland chicken populations (three Tibetan from Linzhi, Tibet [2,900 m altitude; T-LZ], Lahsa, Tibet [3,650 m altitude; T-LS], and Nixi, Yunnan Province [3,250 m altitude; T-NC], and one Lhasa White from Lhasa, Tibet [3,650 m altitude; Lh-W]) and four lowland chicken populations (two White Leghorn, line E reared at China Agricultural University, Beijing [50 m altitude; WL-E] and line D reared at Yukou Poultry, Beijing [50 m altitude; WL-D], one Chahua from Xishuangbanna, Yunnan Province [500 m altitude; CH], and one Lindian from Lindian, Heilong Jiang Province [150 m altitude; LD]). Peripheral blood samples were collected and stored at −20 °C for DNA resequencing. Eggs from T-LZ, Lh-W, CH, and WL-D were collected and incubated under hypoxic conditions (13% O2). Embryo heart tissues were collected at 16 days after incubation and stored at −80 °C for RNA sequencing. Required permission was obtained for sample collection; no endangered or protected species were included in this study. The experiments and animal care protocol were approved by the Animal Welfare Committee of the State Key Laboratory for Agro-Biotechnology of the China Agricultural University (approval number XK257).
Fig. 1.—
Geographic distribution of highland and lowland chicken populations used for sequencing analysis. T-LZ, 2,900 m altitude; T-LS, 3,650 m altitude; T-NC, 3,250 m altitude; Lh-W, 3,650 m altitude; WL-E, 50 m altitude; WL-D, 50 m altitude; CH, 500 m altitude; LD, 150 m altitude.
Sequencing and Assembly
Genomic DNA was isolated from blood samples by using phenol–chloroform extraction, and one sequencing library with an insert size of 200 bp was constructed using pooled DNA from ten individuals from each population. Eight libraries were constructed and then sequenced using Illumina Hiseq 2000 (Illumina, San Diego, CA) with 100-bp paired-end reads in two flow cells, according to the manufacturer’s instructions. Adapter sequences from raw reads were trimmed. The paired reads with low-quality bases (including single reads containing more than 3% of undetermined bases (N) or more than 15% of low-quality bases [Qphred ≤ 20]) were excluded, and the remaining short reads (clean reads) were aligned against the chicken reference genome (version 4.83) by using Burrows–Wheeler Aligner, allowing up to three mismatches (Li and Durbin 2009). Reads that were uniquely aligned in the genome were used for single nucleotide polymorphism (SNP) calling.
Total RNA was isolated from heart tissues of T-LZ, Lh-W, CH, and WL-D by using TRIzol (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. RNA sequencing libraries were prepared using the Illumina mRNA (Messenger RNA)-Seq Prep Kit (Illumina) and sequenced using Illumina HiSeq 2000 with 100-bp paired-end reads. In total, eight cDNA libraries were constructed, including two biological replicates for each population. After the adapter sequences were trimmed and reads containing more than 5% of undetermined bases (N) or more than 15% of low-quality bases (Qphred ≤ 20) were removed, the remaining reads were aligned against the chicken reference genome by using TopHat 2.0.1 (https://ccb.jhu.edu/software/tophat/index.shtml, last accessed February 26, 2016), allowing up to two mismatches. Reads that were uniquely mapped in the genome were used for DEG analysis. The DNA-seq data were deposited in the Sequence Read Archive (SRA) metadata with the accession no. SRP068802 and the RNA-seq profile was deposited in the Gene Expression Omnibus with the accession no. GSE77166.
SNP Detection and Analysis
SNP calling was performed using SAMtools with default settings (Li et al. 2009). SNPs were retrieved from highland and lowland chicken populations, forming two subsets. Unreliable SNPs that exhibited the following features were filtered out: 1) the quality of a corresponding base in reads was below 20; 2) the mapping quality was below 20; 3) the quality of an SNP calling was below 20; or 4) the depth of alternate bases was below four.
Phylogenetic Tree Construction
A phylogenetic tree was constructed using all identified SNPs by using genome resequencing. All variant call format (VCF) files generated from SNP calling were merged using VCFtools (Danecek et al. 2011). Homologous regions among different chicken populations were identified and extracted using SNPhylo (Lee et al. 2014). The corresponding SNPs in homologous regions were used to construct a neighbor-joining tree by using MEGA4 (http://www.megasoftware.net/mega4/mega.html, last accessed February 26, 2016) modeled on the p-distance between different chicken populations (Hall 2013).
Principle Component Analysis
Principle component analysis (PCA) was performed using the SNPRelate package. A pruned set of SNPs that were in approximate linkage equilibrium with each other was used in this package for PCA to avoid the strong influence of SNP clusters (Zheng et al. 2012). A covariance matrix of SNP data was calculated, and all possible genotypes of individual i at SNP j were assumed as homozygous for the reference alleles (scored as “0”) and heterozygous or homozygous for the nonreference alleles (scored as “1” and “2,” respectively). Covariance generated eigenvectors and their significance was calculated using Tracy–Widom test. Population means of PC1 and PC2 were plotted using R.
Structure Analysis
After the SNP data from VCF files were transformed to PLINK files by using Plink, the genetic structure of chicken populations was investigated using ADMIXTURE (https://www.genetics.ucla.edu/software/admixture/, last accessed February 26, 2016) based on the maximum-likelihood method (Alexander et al. 2009). Presumed ancestral populations (K) between 2 and 6 were run with 10,000 iterations.
Selective-Sweep Analysis
Selection signatures were identified by calculating the number of alleles at each SNP locus with sliding windows of 40 kb and a step size of 20 kb. The numbers of reads corresponding to the most and least abundant alleles (nMAJ and nMIN, respectively) were counted, and pooled heterozygosity (Hp) was calculated as follows:
Both population pools (highland and lowland) were analyzed in the same framework by transforming Hp to ZHp as follows:
where μHp is the mean of Hp values, and σHp is the standard deviation of Hp values in each window. In addition, a threshold of ZHp ≤ −6 was considered as the putative selective sweep, because it was the extreme lower end of the distribution (P < 0.002; supplementary fig. S1, Supplementary Material online) (Rubin et al. 2010).
Differences in SNP Distribution
Chi-square test was used to analyze the differences in SNP distribution, and significant P values were corrected with false discovery rate by multiplying with the number of reads that supported a single SNP locus (N). SNPs with corrected P value of <0.01 were considered as putative variants responsible for the unique adaptation to high-altitude environment.
DEG Analysis
The number of reads per kilobase of transcript per million mapped reads (RPKM) was used to calculate the levels of gene expression. Pearson’s correlation coefficients of the mean expression values between the highland populations Lh-W and T-LZ and the lowland populations CH and WL-D were tested using the cor.test function in R (https://www.r-project.org, last accessed February 26, 2016). DEGs were analyzed using DESeq (https://bioconductor.org, last accessed February 26, 2016). After the corresponding P values were determined, fold changes (log2Ratio) were estimated according to the normalized gene expression level in each sample. Identified genes with a fold change ≥1.5 and P ≤ 0.05 between highland and lowland populations were considered as DEGs.
Results
Genome Resequencing Data and SNP Calling
Approximately 153.2 Gb of raw data were obtained, and about 90% (89.2–97.91%) of raw reads had Qphred quality scores of >20 (supplementary table S1, Supplementary Material online). The mapping rates of eight chicken populations exceeded 95% (95.59–97.23%; supplementary table S2, Supplementary Material online). The reads without unique read alignments were discarded, whereas the remaining reads were used to analyze common genetic polymorphisms. About 11 M SNPs were called from each highland population, of which about 5 M was overlapped in the four populations, whereas about 10 M SNPs were called from each lowland population, of which 4 M was overlapped in the four populations (supplementary table S3 and fig. S2, Supplementary Material online). In all, 22,638,868 SNPs were identified from all populations, of which 19,661,985 was from highland populations and 18,437,763 from lowland populations (supplementary table S3, Supplementary Material online).
Phylogenetic Tree and Population Structure
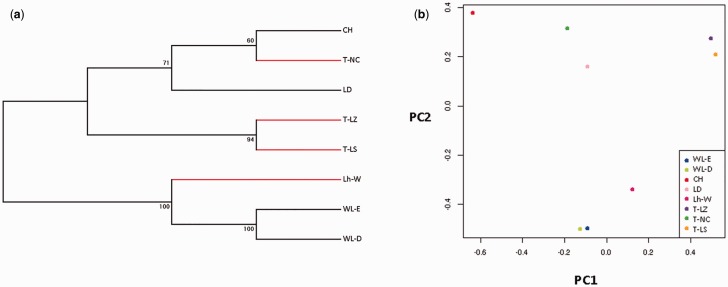
Chicken populations were grouped into two major clades: Tibetan chickens (T-NC, T-LZ, and T-LS) along with LD and CH lowland populations, and Lh-W, WL-E, and WL-D (fig. 2a). The two subclades of Tibetan chickens followed their geographical distributions rather than their breed ascription. PCA showed that Lhasa White formed a separate subclade between Tibetan chickens and the two lines of White Leghorn, confirming the breeding history of this population and supporting the effectiveness of our analysis (fig. 2b).
Fig. 2.—
Population variation analysis. (a) Phylogenetic tree and (b) principal component analysis of highland and lowland chicken populations. T-LZ, 2,900 m altitude; T-LS, 3,650 m altitude; T-NC, 3,250 m altitude; Lh-W, 3,650 m altitude; WL-E, 50 m altitude; WL-D, 50 m altitude; CH, 500 m altitude; LD, 50 m altitude.
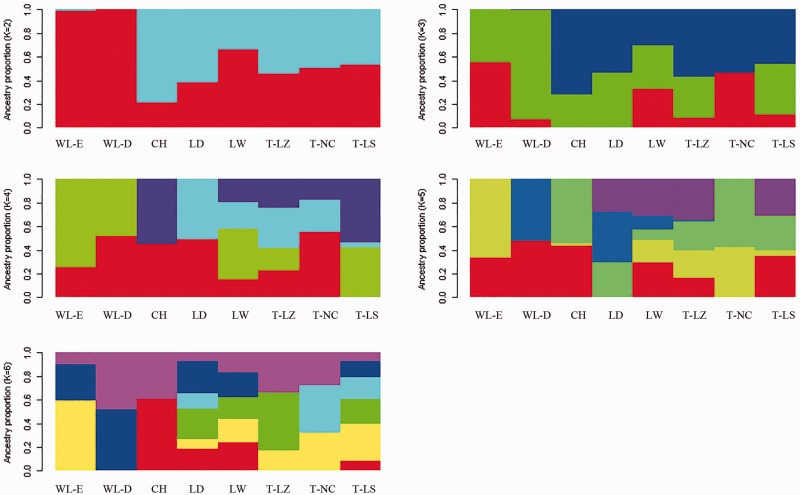
The number of presumed ancestral populations (K) varied from 2 to 6. A cluster with K = 5 was regarded as the most appropriate model, because the average likelihood value was the highest. The results revealed an evident admixture of Tibetan chickens with lowland populations (fig. 3).
Fig. 3.—
Population structure analysis of highland and lowland chicken populations. T-LZ, 2,900 m altitude; T-LS, 3,650 m altitude; T-NC, 3,250 m altitude; Lh-W, 3,650 m altitude; WL-E, 50 m altitude; WL-D, 50 m altitude; CH, 500 m altitude; LD, 150 m altitude.
Selective Sweeps and Positive Selected Genes
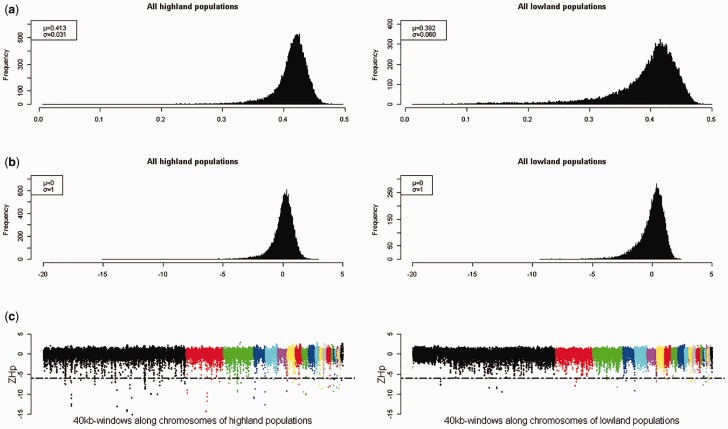
Natural or artificial selection can lead to an increased genetic homogeneity in certain selective sweep regions that exhibit obvious reduction in nucleotide diversity (Akey 2009; Wang et al. 2014). The genetic heterozygosity medians of highland and lowland populations were 0.413 and 0.392, respectively, indicating the different levels of selection sweep. The empirical distribution of Hp values was close to normal (fig. 4a); Hp values were transformed to ZHp values in order to simultaneously analyze both the populations (fig. 4b). The window numbers involved in the genomic analysis of highland and lowland populations were 44,384 and 44,382, respectively (supplementary table S4, Supplementary Material online). Unlike most of the loci that are subjected to genome-wide forces resulting from population demographic history such as genetic drift, only a subset of them can be influenced by selection. Loci under strong selective pressure were located at the extreme tails of the empirical distribution and recognized as outliers (Akey 2009). However, because of the complexity of populations and limited understanding of demographic history, the criteria for the definition of outliers are arbitrary. Rubin et al. (2010) suggested that the best way to identify loci under putative selective sweeps from those being affected by genome-wide forces was to apply various genomic data from different chicken populations to cross-reference and thus verify the criteria. In this study, considering that our analysis method was very similar to that of Rubin et al. (2010), who utilized a ZHp score of −6 or less as the criteria for putative sweeps, and loci with a ZHp score of −6 or less were located in the extreme lower end of the distribution, we believed that these regions were under positive selective sweeps (fig. 4c and supplementary fig. S1, Supplementary Material online). Only about 0.29% of the windows (140 out of 47,821) met this criteria in all chicken populations, and the corresponding fractions for the highland and lowland populations were 0.27% (n = 120) and 0.08% (n = 35), respectively (supplementary table S4 and fig. S3, Supplementary Material online). A total of 229 PSGs were identified in highland populations, of which 173 were highland-specific genes, whereas 56 were identified in the lowland populations (supplementary fig. S4, Supplementary Material online). Gene functional enrichment analysis of the 173 highland-specific PSGs was then performed, and the results showed that the top-ranked gene ontology (GO) terms were related to vasculature development, including angiogenesis, blood vessel development, and blood vessel morphogenesis (supplementary fig. S5, Supplementary Material online). Comparisons against the Kyoto Encyclopedia of Genes and Genomes (KEGG) indicated that several PSGs were clustered into several biological pathways, including melanogenesis, cytokine–cytokine receptor interaction, and the mitogen-activated protein kinase (MAPK) signaling pathway (supplementary fig. S6, Supplementary Material online).
Fig. 4.—
Analysis of positive gene selection in the chicken genome. (a) Distribution of heterozygosity (Hp) values in highland and lowland chicken populations for 40-kb windows. (b) Distribution of corresponding Z-transformed Hp values (ZHp) for 40-kb windows. (c) The lowest end of ZHp distribution in chromosomes 1–28. Horizontal dashed lines indicate the threshold at ZHp = −6.
Among the 173 highland-specific PSGs, GAS2L3 and ANO4, located in the same selected region, showed the lowest ZHp value of −13.07. GAS2L3 is a key regulator of cell division. The depletion of GAS2L3 in vivo results in defected cytokinesis, genomic instability, abnormal brain development, and tumorigenesis (Wolter et al. 2012; Sharaby et al. 2014). ANO4 (also known as TMEM16D) is involved in several biological processes such as ion transport, phospholipid scrambling, and membrane protein regulation; however, its exact role in cellular processes is not yet known. ANO1 and ANO2 have been reported to function as Ca2+-activated Cl− channels, which can regulate transepithelial ion transport, muscle contraction, and phototransduction (Pedemonte and Galietta 2014; Picollo et al. 2015). Seven PSGs (NFATC3, ADA, FGFR1, MEOX2, CTGF, DRD3, and ADAM9) were associated with cardiorespiratory ontogeny, angiogenesis, and response to hypoxia; FGFR1 is considered to be responsible for the neuronal recovery in mice after postnatal hypoxia (Fagel et al. 2009). CTGF is associated with angiogenesis and promoted endothelial cell growth via the activation of vascular endothelial growth factor (VEGF; Brigstock 2002). ADAM family members have been identified as PSGs in Tibetan yak and ground tit (Qiu et al. 2012; Cai et al. 2013). A previous study showed that the mRNA and protein expression levels of ADAM9 in prostate cancer cells are elevated by the regulation of reactive oxygen species under hypoxic stress conditions (Shen et al. 2013). In addition, the expression of ADAM9 in gastric cancer cells is upregulated by hypoxia. Three PSGs (VCP, ASH2L, and FANCG) were associated with DNA repair, cellular response to DNA damage stimulus, and response to radiation; seven PSGs (AIRE, MYO1F, ZAP70, DDX60, CCL19, SATB1, and CD47) with immune and inflammatory responses; and four PSGs (PLA2G15, HNF4ALPHA, STAR, and APOV1) with lipid transport and fatty acid metabolic process (table 1).
Table 1.
GO Annotations of PSGs in Highland Chicken Populations
| Gene ID | Genes | GO Accession | GO Term Name | ZHp |
|---|---|---|---|---|
| ENSGALG00000028373 | GAS2L3 | GO:0007050 | Cell cycle arrest | −13.07 |
| GO:0015629 | Actin cytoskeleton | |||
| GO:0005515 | Protein binding | |||
| GO:0003779 | Actin binding | |||
| ENSGALG00000011614 | ANO4 | GO:0016020 | Membrane | −13.07 |
| GO:0016021 | Integral to membrane | |||
| ENSGALG00000000771 | PLA2G15 | GO:0006629 | Lipid metabolic process | -9.48 |
| ENSGALG00000004285 | HNF4ALPHA | GO:0006629 | Lipid metabolic process | −6.16 |
| ENSGALG00000015134 | APOV1 | GO:0006629 | Lipid metabolic process | −7.67 |
| ENSGALG00000003242 | STAR | GO:0006869 | Lipid transport | −6.27 |
| ENSGALG00000003396 | NFATC3 | GO:0001569 | Patterning of blood vessels | −9.83 |
| GO:0007507 | Heart development | |||
| ENSGALG00000004170 | ADA | GO:0001666 | Response to hypoxia | −6.16 |
| GO:0030324 | Lung development | |||
| GO:0050728 | Negative regulation of inflammatory response | |||
| ENSGALG00000003311 | FGFR1 | GO:0007420 | brain development | −6.27 |
| GO:0001525 | Angiogenesis | |||
| GO:0030324 | lung development | |||
| GO:0048514 | Blood vessel morphogenesis | |||
| ENSGALG00000010794 | MEOX2 | GO:0001525 | Angiogenesis | −6.97 |
| ENSGALG00000002909 | CTGF | GO:0001525 | Angiogenesis | −9.63 |
| GO:0030324 | Lung development | |||
| ENSGALG00000015117 | DRD3 | GO:0002016 | Regulation of blood volume by renin-angiotensin | −8.51 |
| GO:0019216 | regulation of lipid metabolic process | |||
| GO:0045776 | Negative regulation of blood pressure | |||
| ENSGALG00000003398 | ADAM9 | GO:0000186 | Activation of MAPKK activity | −6.32 |
| GO:0030216 | Keratinocyte differentiation | |||
| GO:0042542 | Response to hydrogen peroxide | |||
| GO:0051384 | Response to glucocorticoid | |||
| GO:0051592 | Response to calcium ion | |||
| ENSGALG00000001327 | AIRE | GO:0006959 | Humoral immune response | −7.05 |
| GO:0042393 | Histone binding | |||
| ENSGALG00000001571 | MYO1F | GO:0045088 | Regulation of innate immune response | −6.5 |
| ENSGALG00000001486 | ZAP70 | GO:0006955 | Immune response | −6.5 |
| ENSGALG00000009639 | DDX60 | GO:0009615 | Response to virus | −7.22 |
| GO:0051607 | Defense response to virus | |||
| ENSGALG00000028256 | CCL19 | GO:0006955 | Immune response | −6.58 |
| GO:2000107 | Negative regulation of leukocyte apoptotic process | |||
| GO:0001768 | establishment of T cell polarity | |||
| ENSGALG00000011254 | SATB1 | GO:0016571 | Histone methylation | −6.63 |
| GO:0042110 | T cell activation | |||
| ENSGALG00000015355 | CD47 | GO:0050729 | Positive regulation of inflammatory response | −10.92 |
| GO:0050870 | positive regulation of T cell activation | |||
| ENSGALG00000001986 | VCP | GO:0006281 | DNA repair | −6.58 |
| ENSGALG00000003233 | ASH2L | GO:0006974 | Cellular response to DNA damage stimulus | −6.27 |
| ENSGALG00000002009 | FANCG | GO:0009314 | response to radiation | −6.58 |
Note.—ZHp, z-value of hetorozygosity.
Genes with Missense Mutations
The results of chi-square test indicated that 3,988 of 11,434 nonsynonymous SNPs were highly differentiated (corrected P < 0.01) between the two populations (supplementary fig. S7, Supplementary Material online) and were annotated to 2,490 genes. Gene enrichment analysis showed that several highly differentially distributed genes with nonsynonymous mutations were enriched in GO terms related to cardiorespiratory ontogeny, including cardiac muscle cell differentiation, cardiac cell differentiation, blood vessel development, blood circulation, systemic arterial blood pressure regulation, angiogenesis, heart and lung development, respiratory electron transport chain, respiratory tube development (supplementary fig. S8, Supplementary Material online), immune responses, DNA repair, apoptosis induction, response to UV radiation, lipid transport, and cytokine binding. Moreover, KEGG enrichment analysis indicated that the VEGF signaling pathway was probably responsible for hypoxia-initiated angiogenesis (supplementary fig. S9, Supplementary Material online).
PSGs with Nonsynonymous SNPs
In total, 44 nonsynonymous SNPs within the positive selected regions (ZHp ≤ −6; corrected P < 0.01) were identified; they were distributed among 36 PSGs, of which 1 (JPH2) was associated with cardiac muscle development. JPH2 can directly interact with Ca2+ to modulate Ca2+ handling, affecting excitation–contraction (EC) coupling and cardiac myocyte function. In rats, the downregulation of JPH2 impairs EC coupling and leads to cardiac hypertrophy (Garbino and Wehrens 2010). Four nonsynonymous SNPs (SATB1, SCLY, HP1BP3, and FGFR1) were associated with hypoxia responses. As one of the targets of miR-21 that is involved in the remodeling of hypoxia-induced pulmonary vascular remodeling, the DNA binding protein SATB1 is downregulated in the lungs of mice under low oxygen concentration (Yang et al. 2012). SCLY has been shown to be significantly lower in human hepatoma cells cultivated under hypoxia conditions and to hamper the biosynthesis of selenoprotein under oxidative stress (Becker et al. 2014). A previous study identified HP1BP3 as a key factor associated with hypoxia-induced oncogenesis through increased tumor cell viability, UV radiation resistance, and self-renewal (Dutta et al. 2014). FGFR1 is widely associated with various biological processes, including angiogenesis, positive regulation of the MAPK cascade, phosphorylation, blood vessel morphogenesis, brain and lung development, and ATP binding, which play significant roles in the adaptation to oxygen scarcity (Kohler et al. 2012; Gronych et al. 2013; Kuo et al. 2015). The average ZHp scores for total PSGs and PSGs with nonsynonymous SNPs were also calculated (supplementary fig. S10, Supplementary Material online). These results indicated that the average ZHp score of PSGs with nonsynonymous SNPs (−7.89) was lower than that of total PSGs (−7.46), suggesting possible stronger selective sweeps occurring on these genes.
Transcriptome Profiles of Embryonic Heart Tissue under Hypoxic Incubation
RNA-Seq generated 470,978,042 raw reads, of which about 96.10% (95.14–97.07%) had Qphred quality scores of >20 (supplementary table S5, Supplementary Material online). After low-quality reads and adaptors were trimmed off, 44.42 Gb of clean data was aligned against the chicken genome, and 84.09% (83.23–85.35%) of clean reads was mapped to the reference genome (supplementary table S6, Supplementary Material online). Of the mapped reads, 79.7% (75.4–82.0%) was located in exons, 5.5% (4.7–7.5%) in introns, and the remaining 14.8% (13.4–18.2%) in the intergenic regions (supplementary table S7, Supplementary Material online). RNA-seq experiments were biological duplicates with an R2 of 0.928–0.979 for the RPKM values of all positive expressive genes (supplementary fig. S11, Supplementary Material online), which suggested the high reliability of gene expression analysis.
Differentially Expressed Genes
DEG analysis revealed 132 genes in highland populations, including 64 downregulated and 68 upregulated DEGs. The top-ranked enriched GO terms were related to blood vessel development, angiogenesis, lung development, and respiratory system development (supplementary fig. S12, Supplementary Material online). Other enriched GO categories included response to endogenous stimulus, metal ion transport, muscle system process, and cell death regulation. In addition, KEGG analysis showed that several DEGs were clustered in the MAPK signaling pathway and cytokine–cytokine receptor interaction (supplementary fig. S13, Supplementary Material online). Further analysis of these genes indicated that five of the upregulated DEGs (BMP4, LOX, LPR, CTGF, and ANGPTL4) were associated with blood vessel development, angiogenesis, and lung development; three of them (BMP3, BMP4, and DUSP3) with MAPK cascade regulation; two (COLGALT2 and PRKAR2B) with fatty acid metabolic process; two (STC2 and ETV5) with oxidative stress response; and two (JSC and FAS) with immune responses. In addition, one downregulated DEG identified in highland populations (HYAL1) was associated with blood vessel development, angiogenesis, lung development, cellular response to UV radiation, and inflammation. DEGs along with PSGs and nonsynonymous SNPs highlighted the crucial role of cardiorespiratory function (especially vessel development) in the adaptation of highland chicken populations to the Tibetan Plateau.
Selective Signaling of DEGs and Differential Expression of PSGs
Further analysis was conducted to determine whether DEGs identified in RNA-Seq also underwent positive selection during the adaptive evolution to high altitude. The average ZHp score of DEGs (−0.26) was considerably smaller than that of the total expressed genes from RNA-Seq analysis (0.04) in embryonic heart tissue (supplementary fig. S14, Supplementary Material online), indicating the relatively low degree of heterozygosity and high fixation of these genes across the genome. Cross-reference of DEGs with PSGs showed only one overlapped gene (CTGF). The biological function of CTGF was associated with angiogenesis as mentioned above (Brigstock 2002). Of the 132 DEGs, 25 (17 upregulated and 8 downregulated) had nonsynonymous SNPs with differential allele distribution (corrected P ≤ 0.001). GO analysis showed that two (HYAL1 and BMP4) of them was related to angiogenesis regulation, lung and blood vessel development, and MAPK activity induction (Rothhammer et al. 2007; Tan et al. 2011; Liu et al. 2012); three (SGPL1, LPR, and MGAT4C) was involved in metabolic processes and protein binding (Rothhammer et al. 2007; Tan et al. 2011; Liu et al. 2012); one (HCTA1) was associated with ATP binding (Abdulhussein and Wallace 2014); and one (CTNNA3) played a role in cardiac muscle regulation (van Hengel et al. 2013). Moreover, the average fold change of the DEGs with nonsynonymous mutations (2.20) was higher than that of the total DEGs (2.04; supplementary fig. S15, Supplementary Material online).
Discussion
Diverse Origin of Tibetan Chickens
Our study suggested that Tibetan chickens have a diverse origin. This could possibly be because domesticated chickens had originated from multiple regions globally (Tixier-Boichard et al. 2011; Miao et al. 2013), and except for G. gallus, Gallus sonneratii also contributed to the genetic constitution of the original populations (Tixier-Boichard et al. 2011). Another explanation could be convergent evolution. The domestication of wild animals allowed humans to live in farming societies. With the expanding human population, domesticated chickens were transported to various regions of the world for trade or civilization (Tixier-Boichard et al. 2011). Ancient domesticated chickens were possibly reared at different altitudes on the Tibetan Plateau and gradually developed adaptive strategies to high-altitude conditions under the same selection pressure. Our study also revealed an evident admixture of Tibetan chickens and lowland populations, suggesting a complicated history of gene flow from lowland gene pools to that of the Tibetan chicken, owing to unintentional crossing or selective breeding for improving the performance of highland populations.
Genes Underlying Cardiorespiratory Development Might Be Involved in the Adaptation to Hypoxia
Hypoxia is the major type of environmental stress under high-altitude conditions, and it directly leads to systemic hypoxemia, which is the imbalance between metabolic demands and oxygen convective transport. When elevated metabolic requirements exceed the supply of oxygen due to the reduced partial pressure with increasing altitude, the capacity of cardiovascular and respiratory systems is suppressed (Tekin et al. 2010). Hypoxic conditions have been previously reported to result in hypotension in Red Jungle fowl (Lindgren and Altimiras 2011). The inadequate supply of oxygen also affects physiological performance and inhibits growth capacity (Semenza 2012). Hypoxia initiates the expression of hypoxia-induced factor-1a (HIF-1a) and further induces the expression of target genes such as VEGF and MAPK, which promote various systemic physiological changes such as angiogenesis, vascular development, respiratory system development, and cardiac cell differentiation (Giatromanolaki et al. 2010; Befani et al. 2012). HIF-dependent metabolic adaptations assist living cells to preserve their integrity, maintain energy supply, and retain their functionality under hypoxic conditions (Majmundar et al. 2010).
Our study identified several genes (FGFR1, CTGF, ADAM9, JPH2, SATB1, BMP4, LOX, LPR, ANGPTL4, and HYAL1) and biological pathways (VEGF signaling and MAPK signaling pathways) associated with cardiorespiratory development, indicating the crucial role of blood vessel development in the adaptation of Tibetan chickens to hypoxic conditions. The cardiorespiratory function affects the response of organisms to hypoxia (Giordano 2005), because genes and signaling pathways involved in this process can compensate for the scarcity of oxygen diffusion, improve the effectiveness of gas exchange, and maintain oxygen homeostasis. In addition, hypoxia promotes angiogenesis by the upregulation of HIF-1a and VEGF. Angiogenesis plays a crucial role in homeostatic mechanisms associated with vascular oxygen supply in hypoxia, and this process is regulated by the VEGF signaling pathway through the increased expression of HIF-1 (Semenza 2000). Moreover, VEGF-dependent angiogenesis is regulated by the activation of MAPK pathway. The MAPK signaling pathway affects the expression of VEGF and is associated with the binding of VEGF-1 to VEGF-2. The interaction between MAPK and VEGF can effectively increase angiogenesis (Olsson et al. 2006). A study on the adaptation of ground tits to the Tibetan Plateau also identified the enrichment of the MAPK signaling pathway by several PSGs (Qu et al. 2013).
Genes Associated with Inflammation Response Might Reflect the Effects of Long-Term Hypoxia and UV Radiation
Inflammation is an adverse effect of low-oxygen concentration under high-altitude conditions resulting from ischemia in the body organs (Bartels et al. 2013). In mice, short-term hypoxia can increase the circulation of proinflammatory cytokines and cause vascular leakage, resulting in the accumulation of inflammatory cells (Fujisaka et al. 2013). Inflamed tissues often become more hypoxic because of the imbalance between the increased cell metabolism and decreased metabolic output caused by oxidative injury (Eltzschig and Carmeliet 2011). Moreover, hypoxia-induced inflammatory response can stimulate the differentiation of T cells via the activation of hypoxia signaling pathways (Clambey et al. 2012).
Solar radiation can also activate inflammatory responses, including erythema, edema, and hyperplasia of the skin (Stoecklein et al. 2015). In mammals, high-dose UV radiation can cause DNA damage, cell apoptosis, tissue injury, inflammation, and cancer (Svobodova et al. 2012; Sklar et al. 2013), whereas dead or damaged cells release molecules that act as endogenous signals for the activation of inflammasome pathways and affect immune responses (Rock and Kono 2008). The immune system provides defense against inflammation and tissue injuries resulting from high-altitude conditions by enhancing multiple cytoprotective responses that promotes systematic adaptation to hypoxia and high UV radiation. In addition, hypoxia-induced innate and adaptive immunity responses are activated toward pathogens that cause inflammation (Schroder and Tschopp 2010).
In this study, several genes (AIRE, MYO1F, ZAP70, DDX60, CCL19, CD47, JSC, and FAS) and biological pathways (such as cytokine–cytokine receptor interaction) were found to be associated with inflammation and immune responses, which might reflect the adaptation process of highland chickens to hypoxia and UV radiation on the Tibetan Plateau. Cytokines are extracellular proteins that serve as crucial intercellular regulators of angiogenesis, repair process, homeostasis restoration, and inflammatory response (Mann 2015). Other genes (i.e., VCP, ASH2L, and FANCG) and biological pathways (melanogenesis signaling pathway) were also found to be associated with DNA repair process and responses to DNA damage stimulus and radiation, which might also indicate the adaptation of Tibetan chickens to UV radiation. Melanin is known to play an important role in the protection against high-dose solar radiation (Jablonski and Chaplin 2010); however, further studies are required to elucidate and better understand the underlying mechanisms.
Conclusion
This study provided new insights into the unique adaptation of highland animals to extreme environments. Genomic selective signature and transcriptome profile analyses were used in this study to identify candidate genes involved in cardiorespiratory system development (FGFR1, CTGF, ADAM9, JPH2, SATB1, BMP4, LOX, LPR, ANGPTL4, and HYAL1), inflammation and immune responses (AIRE, MYO1F, ZAP70, DDX60, CCL19, CD47, JSC, and FAS), DNA repair, and responses to radiation (VCP, ASH2L, and FANCG) with key roles in the adaptation of Tibetan chickens to highland environments. In addition, analysis of the candidate genes and SNPs might provide new insights into the unique adaptation of highland animals to extreme environments in avian species.
Supplementary Material
Supplementary tables S1–S7 and figures S1–S15 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (91331118 and 31272401) and Program for Changjiang Scholar and Innovation Research Team in University (IRT1191).
Literature Cited
- Abdulhussein AA, Wallace HM. 2014. Polyamines and membrane transporters. Amino Acids 46(3):655–660. [DOI] [PubMed] [Google Scholar]
- Akey JM. 2009. Constructing genomic maps of positive selection in humans: where do we go from here?. Genome Res. 19(5):711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels K, Grenz A, Eltzschig HK. 2013. Hypoxia and inflammation are two sides of the same coin. Proc Natl Acad Sci U S A. 110(46):18351–18352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker NP, et al. 2014. Hypoxia reduces and redirects selenoprotein biosynthesis. Metallomics 6(5):1079–1086. [DOI] [PubMed] [Google Scholar]
- Befani CD, et al. 2012. Bortezomib represses HIF-1alpha protein expression and nuclear accumulation by inhibiting both PI3K/Akt/TOR and MAPK pathways in prostate cancer cells. J Mol Med (Berl). 90(1):45–54. [DOI] [PubMed] [Google Scholar]
- Bigham A, et al. 2010. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 6(9):e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW, et al. 2009. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum Genomics. 4(2):79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigstock DR. 2002. Regulation of angiogenesis and endothelial cell function by connective tissue growth factor (CTGF) and cysteine-rich 61 (CYR61). Angiogenesis 5(3):153–165. [DOI] [PubMed] [Google Scholar]
- Cai Q, et al. 2013. Genome sequence of ground tit Pseudopodoces humilis and its adaptation to high altitude. Genome Biol. 14(3):R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, Brumfield RT. 2012. Genomic insights into adaptation to high-altitude environments. Heredity (Edinb) 108(4):354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clambey ET, et al. 2012. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc Natl Acad Sci U S A. 109(41):E2784–E2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZQ, et al. 2004. [Genetic diversity in Tibetan chicken]. Yi Chuan 26(2):167–171. [PubMed] [Google Scholar]
- Dutta B, Yan R, Lim SK, Tam JP, Sze SK. 2014. Quantitative profiling of chromatome dynamics reveals a novel role for HP1BP3 in hypoxia-induced oncogenesis. Mol Cell Proteomics. 13(12):3236–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig HK, Carmeliet P. 2011. Hypoxia and Inflammation. N Engl J Med. 364(7):656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagel DM, et al. 2009. Fgfr1 is required for cortical regeneration and repair after perinatal hypoxia. J Neurosci. 29(4):1202–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaka S, et al. 2013. Adipose tissue hypoxia induces inflammatory M1 polarity of macrophages in an HIF-1alpha-dependent and HIF-1alpha-independent manner in obese mice. Diabetologia 56(6):1403–1412. [DOI] [PubMed] [Google Scholar]
- Garbino A, Wehrens XH. 2010. Emerging role of junctophilin-2 as a regulator of calcium handling in the heart. Acta Pharmacol Sin. 31(9):1019–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giatromanolaki A, et al. 2010. Hypoxia and activated VEGF/receptor pathway in multiple myeloma. Anticancer Res. 30(7):2831–2836. [PubMed] [Google Scholar]
- Giordano FJ. 2005. Oxygen, oxidative stress, hypoxia, and heart failure. J Clin Invest. 115(3):500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, et al. 2007. Hypoxic adaptations of hemoglobin in Tibetan chick embryo: high oxygen-affinity mutation and selective expression. Comp Biochem Physiol B Biochem Mol Biol. 147(2):147–155. [DOI] [PubMed] [Google Scholar]
- Gronych J, Jones DTW, et al. 2013. In vivo and functional analysis of novel MAPK pathway activating mutations in childhood astrocytoma. Neuro-Oncology 151:28–29. [Google Scholar]
- Hall BG. 2013. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 30(5):1229–1235. [DOI] [PubMed] [Google Scholar]
- Jablonski NG, Chaplin G. 2010. Colloquium paper: human skin pigmentation as an adaptation to UV radiation. Proc Natl Acad Sci U S A. 107(Suppl. 2):8962–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, et al. 2014. Admixture facilitates genetic adaptations to high altitude in Tibet. Nat Commun. 5:3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler LH, et al. 2012. FGFR1 expression and gene copy numbers in human lung cancer. Virchows Arch. 461(3):353–354. [DOI] [PubMed] [Google Scholar]
- Kuo CH, et al. 2015. FGFR1 mediates recombinant thrombomodulin domain-induced angiogenesis. Cardiovasc Res. 105(1):107–117. [DOI] [PubMed] [Google Scholar]
- Lee TH, Guo H, Wang X, Kim C, Paterson AH. 2014. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhao C. 2009. Study on Tibetan Chicken embryonic adaptability to chronic hypoxia by revealing differential gene expression in heart tissue. Sci China C Life Sci. 52(3):284–295. [DOI] [PubMed] [Google Scholar]
- Li M, et al. 2013. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 45(12):1431–1438. [DOI] [PubMed] [Google Scholar]
- Lindgren I, Altimiras J. 2011. Sensitivity of organ growth to chronically low oxygen levels during incubation in Red Junglefowl and domesticated chicken breeds. Poult Sci. 90(1):126–135. [DOI] [PubMed] [Google Scholar]
- Liu C, et al. 2012. TAK1 promotes BMP4/Smad1 signaling via inhibition of erk MAPK: a new link in the FGF/BMP regulatory network. Differentiation 83(4):210–219. [DOI] [PubMed] [Google Scholar]
- Liu S, et al. 2014. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157(4):785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. 2010. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 40(2):294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DL. 2015. Innate immunity and the failing heart: the cytokine hypothesis revisited. Circ Res. 116(7):1254–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. 2008. The impact of next-generation sequencing technology on genetics. Trends Genet. 24(3):133–141. [DOI] [PubMed] [Google Scholar]
- Miao Y, et al. 2013. Chicken domestication: an updated perspective based on mitochondrial genomes. Heredity 110(3):277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. 2006. VEGF receptor signalling—in control of vascular function. Nat Rev Mol Cell Biol. 7(5):359–371. [DOI] [PubMed] [Google Scholar]
- Pedemonte N, Galietta LJV. 2014. Structure and function of tmem16 proteins (Anoctamin). Physiol Rev. 94(2):419–459. [DOI] [PubMed] [Google Scholar]
- Picollo A, Malvezzi M, Accardi A. 2015. TMEM16 proteins: unknown structure and confusing functions. J Mol Biol. 427(1):94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q, et al. 2012. The yak genome and adaptation to life at high altitude. Nat Genet. 44(8):946–949. [DOI] [PubMed] [Google Scholar]
- Qu Y, et al. 2013. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun. 4:2071. [DOI] [PubMed] [Google Scholar]
- Rock KL, Kono H. 2008. The inflammatory response to cell death. Annu Rev Pathol. 2008:99–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer T, Bataille F, Spruss T, Eissner G, Bosserhoff AK. 2007. Functional implication of BMP4 expression on angiogenesis in malignant melanoma. Oncogene 26(28):4158–4170. [DOI] [PubMed] [Google Scholar]
- Rubin CJ, et al. 2010. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464(7288):587–591. [DOI] [PubMed] [Google Scholar]
- Scheinfeldt LB, Tishkoff SA. 2010. Living the high life: high-altitude adaptation. Genome Biol. 11(9):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt LB, et al. 2012. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 13(1):R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. 2010. The inflammasomes. Cell 140(6):821–832. [DOI] [PubMed] [Google Scholar]
- Semenza GL. 2000. HIF-1: using two hands to flip the angiogenic switch. Cancer Metastasis Rev. 19(1-2):59–65. [DOI] [PubMed] [Google Scholar]
- Semenza GL. 2012. Hypoxia-inducible factors in physiology and medicine. Cell 148(3):399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharaby Y, et al. 2014. Gas213 is essential for brain morphogenesis and development. Dev Biol. 394(2):305–313. [DOI] [PubMed] [Google Scholar]
- Shen Z, et al. 2013. Both macrophages and hypoxia play critical role in regulating invasion of gastric cancer in vitro. Acta Oncol. 52(4):852–860. [DOI] [PubMed] [Google Scholar]
- Sklar LR, Almutawa F, Lim HW, Hamzavi I. 2013. Effects of ultraviolet radiation, visible light, and infrared radiation on erythema and pigmentation: a review. Photochem Photobiol Sci. 12(1):54–64. [DOI] [PubMed] [Google Scholar]
- Srivastava S, et al. 2012. Association of polymorphisms in angiotensin and aldosterone synthase genes of the renin-angiotensin-aldosterone system with high-altitude pulmonary edema. J Renin Angiotensin Aldosterone Syst. 13(1):155–160. [DOI] [PubMed] [Google Scholar]
- Stoecklein VM, et al. 2015. Radiation exposure induces inflammasome pathway activation in immune cells. J Immunol. 194(3):1178–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YB, et al. 2015. Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proc Natl Acad Sci U S A. 112(11):E1257–E1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svobodova AR, et al. 2012. DNA damage after acute exposure of mice skin to physiological doses of UVB and UVA light. Arch Dermatol Res. 304(5):407–412. [DOI] [PubMed] [Google Scholar]
- Tan JX, et al. 2011. Upregulation of HYAL1 expression in breast cancer promoted tumor cell proliferation, migration, invasion and angiogenesis. PLoS One 6(7):e22836. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tekin D, Dursun AD, Xi L. 2010. Hypoxia inducible factor 1 (HIF-1) and cardioprotection. Acta Pharmacol Sin. 31(9):1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tixier-Boichard M, Bed'Hom B, Rognon X. 2011. Chicken domestication: from archeology to genomics. C R Biol. 334(3):197–204. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Salzberg SL. 2012. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 13(1):36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, von Wettberg EJ, Hu TT, Nuzhdin SV. 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat Genet. 42(3):260–263. [DOI] [PubMed] [Google Scholar]
- van Hengel J, et al. 2013. Mutations in the area composita protein alphaT-catenin are associated with arrhythmogenic right ventricular cardiomyopathy. Eur Heart J. 34(3):201–210. [DOI] [PubMed] [Google Scholar]
- Wang G, Xie H, Peng MS, Irwin D, Zhang YP. 2014. Domestication genomics: evidence from animals. Ann Rev Anim Biosci. 2:65–84. [DOI] [PubMed] [Google Scholar]
- Wang MS, et al. 2015. Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol Biol Evol. 32(7):1880–1889. [DOI] [PubMed] [Google Scholar]
- Wei ZH, et al. 2007. Blood gas, hemoglobin, and growth of Tibetan chicken embryos incubated at high altitude. Poult Sci. 86(5):904–908. [DOI] [PubMed] [Google Scholar]
- Wolter P, et al. 2012. GAS2L3, a target gene of the DREAM complex, is required for proper cytokinesis and genomic stability. J Cell Sci. 125 (Pt 10):2393–2406. [DOI] [PubMed] [Google Scholar]
- Xiang H, et al. 2014. Early Holocene chicken domestication in northern China. Proc Natl Acad Sci U S A. 111(49):17564–17569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, et al. 2011. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol. 28(2):1003–1011. [DOI] [PubMed] [Google Scholar]
- Yang S, et al. 2012. miR-21 regulates chronic hypoxia-induced pulmonary vascular remodeling.”. Am J Physiol Lung Cell Mol Physiol. 302(6):L521–L529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wu CX, Chamba Y, Ling Y. 2007. Blood characteristics for high altitude adaptation in Tibetan chickens. Poult Sci. 86(7):1384–1389. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang XT, Chamba Y, Ling Y, Wu CX. 2008. Influences of hypoxia on hatching performance in chickens with different genetic adaptation to high altitude. Poult Sci. 87(10):2112–2116. [DOI] [PubMed] [Google Scholar]
- Zheng X, et al. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28(24):3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. 2014. Whole-genome sequencing of the snub-nosed monkey provides insights into folivory and evolutionary history. Nat Genet. 46(12):1303–1310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.