Abstract
Unraveling the complex network of neural circuits that form the nervous system demands tools that can manipulate specific circuits. The recent evolution of genetic tools to target neural circuits allows an unprecedented precision in elucidating their function. Here we describe two general approaches for achieving circuit specificity. The first uses the genetic identity of a cell, such as a transcription factor unique to a circuit, to drive expression of a molecule that can manipulate cell function. The second uses the spatial connectivity of a circuit to achieve specificity: one genetic element is introduced at the origin of a circuit and the other at its termination. When the two genetic elements combine within a neuron, they can alter its function. These two general approaches can be combined to allow manipulation of neurons with a specific genetic identity by introducing a regulatory gene into the origin or termination of the circuit. We consider the advantages and disadvantages of both these general approaches with regard to specificity and efficacy of the manipulations. We also review the genetic techniques that allow gain- and loss-of-function within specific neural circuits. These approaches introduce light-sensitive channels (optogenetic) or drug sensitive channels (chemogenetic) into neurons that form specific circuits. We compare these tools with others developed for circuit-specific manipulation and describe the advantages of each. Finally, we discuss how these tools might be applied for identification of the neural circuits that mediate behavior and for repair of neural connections.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-016-0425-7) contains supplementary material, which is available to authorized users.
Keywords: Circuit manipulation, Circuit specificity, Genetic tools, Optogenetics, Chemogenetics
Introduction/Limits of Lesions
Cell biology has gained enormous insight into the function of individual molecules using loss-of-function with genetic deletion (e.g., knockout and silencing) and gain-of-function studies with amplification (e.g., overexpression). Systems neuroscience is now poised to make similar large-scale advances by understanding how alteration of specific neural circuits affects nervous system function. Systems neuroscience seeks to understand how groups of neurons are organized into circuits, how neural circuits mediate brain function, and how this understanding can be used to improve function. While many reviews have been written about the tools that have been created to manipulate function, such as optogenetics and chemogenetics, we approach this problem from the systems neuroscience perspective. That is, if one wants to test circuit function, which approach is best? In this review, we try to answer that question.
Lesions of the brain, both those that occur spontaneously in humans and those performed experimentally in animals, form the core of what we know about how neurological function is localized in the brain. For example, the neurologist Paul Broca described patients who sustained damage to the left inferior frontal gyrus and could not speak [1, 2]. These case studies taught us that this region (dubbed Broca’s area) controls production of speech. Focal brain lesions, especially those from ischemic stroke, have further localized function through its loss [3, 4]. The function of other areas of the brain could be revealed through gain of function, with electrical stimulation [5]. The brain mapping studies of Wilder Penfield, for example, revealed that motor cortex is arranged somatotopically, with different regions encoding movement of different body segments [6].
However, these classic loss- and gain-of-function techniques have limitations. Brain lesions are irreversible and can only be tested once. This can be overcome with a variety of reversible inactivation techniques—drugs, cooling, and brain stimulation that inactivate surrounding neural tissue. These techniques can produce “virtual lesions” that are reversible [7]. However, reversible techniques are also invasive, and surgery itself can alter neurological function. Finally, and most importantly, these lesion techniques lack specificity. Regions manipulated with lesions or electrical stimulation might consist of multiple cell types and circuits, even though the region is very small. This limitation has been overcome with genetic tools.
This review is divided into 2 sections. The first describes the general approaches to achieving circuit specificity, “genetic identity” and “spatial control”. In the second section, we compare the genetic tools that allow loss of function (silencing) and gain of function (increased neuronal activity) within specific circuits. By the end of the review, we intend that the reader understand from a systems neuroscience perspective how tools for manipulating neuron function might be applied to modify specific neural circuits.
Two Methods for Achieving Circuit Specificity
To achieve circuit specific manipulation, genetic tools have been applied using two general approaches. The first is to use the genetic identity of a neuron to drive expression of a gene that can alter its function. This identity can be a transcription factor unique to a circuit; often, transcription factors instruct circuit formation in the first place and some of them retain circuit-specificity in adulthood. Other defining characteristics of a neuron can also be used, such as the neurotransmitter or calcium-binding protein that it expresses. Circuit-specific manipulation is achieved by inserting a gene that can manipulate function into the region of these signature elements. In doing so, cells containing those signature elements are manipulated, while other cells in the region of the injection are not perturbed.
The second major strategy for achieving circuit-specificity is to use knowledge of the connectivity of a circuit to manipulate it. This system introduces two genes, one at the origin of a circuit and one at the termination. When the two genetic elements combine, they can alter the function only in the targeted circuit. Neurons near the origin that terminate elsewhere and neurons that terminate in the same location but originate elsewhere are not affected because they only contain 1 of the 2 necessary elements.
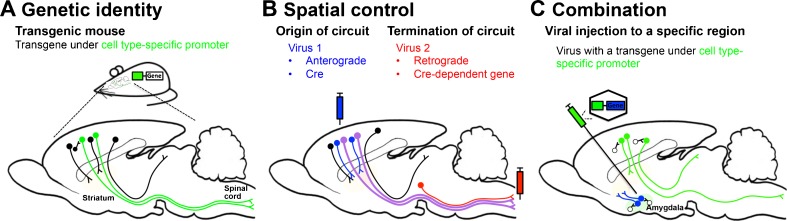
To make this distinction more tangible, we offer graphical examples of achieving circuit-specific manipulation using these two approaches in Fig. 1. Figure 1a demonstrates how neurons can be targeted according to genetic identity. The mammalian neocortex has a laminar organization, with each of the 6layers serving distinct functions. Our laboratory [8, 9] and others have a strong interest in neurons projecting from layer 5 of motor cortex to subcortical targets. Projection neurons of motor cortex are characterized largely by the set of transcription factors during development and cell type-specific genes in maturity [10–12]. This genetic identity can be used to target each projection neurons. Figure 1a shows that neurons expressing a gene under the control of green promoter project to the spinal cord (green neurons), whereas other neurons projecting to the striatum or superior colliculus (among many other targets) do not express the green gene. The gene under the green promoter is the cell type-specific gene of the neurons projecting to the spinal cord accordingly. When a transgene to label or manipulate a neuron is inserted under control of the green promoter, only the neurons projecting to the spinal cord express the transgene. These neurons can then be controlled by the transgene to perform gain- and loss-of-function studies selectively within the projection to the spinal cord.
Fig. 1.
Strategies for targeting specific neurons and a circuit. (a) Genetic identity. The connected boxes in the animal are transgenic constructs. The green box represents the promoter region of a cell type-specific gene (genetic identity), which is expressed by the green neurons in cortex and the connected box indicates a genetic tool. The transgenic animal expresses a genetic tool exclusively in the green neurons (circuit). (b) Spatial control. The known locations of the origin and termination of the circuit are used to target the circuit. The virus 1 (blue syringe) with Cre recombinase transduces neurons anterogradely, and virus 2 (red syringe) carrying Cre-dependent genetic tool transduces neurons retrogradely. The neurons (blue) having their cell body in the cortex are infected with virus 1 and the neurons having their axon terminal in the spinal cord are infected with virus 2. However, only the neurons that have their origin in the cortex and termination in the spinal cord are double-infected (purple) and express the genetic tool. (c) Combination. In order to manipulate excitatory pyramidal neurons selectively in amygdala, virus with a genetic tool (blue box) under the control of a excitatory pyramidal neuron-specific promoter region (green box) is injected to the amygdala. Although excitatory pyramid neurons are ubiquitous in the brain (green neurons), only amygdala excitatory pyramidal neurons (blue neurons with green outline) and not γ-aminobutyric acid-ergic neurons (white) are express genetic tools, owing to the spatial control of viral injection
Specific circuits can be targeted based on their connectivity rather than genetic identity [13, 14]. As shown in Fig. 1B, projection neurons in cortex project their axons to striatum and spinal cord, among many other targets. The projection neurons innervating the spinal cord can be targeted by injecting an anterograde virus (blue) into the motor cortex and a retrograde virus (red) into the spinal cord. Only the neurons co-infected with both viruses (purple) express a gene that can manipulate its function. We call this approach “spatial control” because it uses information about the origin and the termination of a circuit to achieve circuit specificity. In the following sections, we will discuss these 2 approaches to target circuits of interest and provide examples of each.
Genetic Identity
The development of brain connections is determined, in part, by a series of transcription factors and other genes that help to guide axons to their proper terminations. A similar transcriptional profile helps to determine function during maturity [10, 11, 13, 14]. Therefore, these cell type-specific genes are attractive targets to manipulate specific types of neurons. With transgenic mouse technology, the cell type-specific expression of transgenes has been accomplished by cloning them under the control of the promoter regions of cell type-specific genes [15–20]. For example, promoters of calcium/calmodulin kinase II alpha (CaMKIIα) and adenosine2A receptor drive transgene expression in forebrain neurons and striatopallidal medium spiny neurons, respectively [15, 21].
Neurons can be manipulated according to which neurotransmitter they use. Within motor cortex, 2 main neural subtypes are projection neurons, which employ glutamate, and interneurons, which express γ-aminobutyric acid (GABA) [13]. These differences have been used to modify selectively glutamatergic projection neurons. Selective expression of a genetic tool in the glutamatergic neurons in hindbrain and spinal cord has been achieved by inserting a transgene under the control of vesicular glutamate transporter 2 (Vglut2) promoter [18]. Vglut2 is the dominant glutamate transporter expressed by in these regions [22, 23]. In this transgenic mouse, the genetic tool was found only in Vglut2-positive neurons in the central nervous system and exclusively activated by photostimulation. Manipulation of the targeted neurons in the spinal cord and hindbrain showed that glutamatergic neurons in the spinal cord are critical for generating rhythm in the central pattern generators and the activation of glutamatergic reticulospinal neurons in hindbrain area is sufficient to activate directly the spinal locomotor network [18]. Likewise, the promoter regions of vesicular GABA transporter, choline acetyltransferase or tryptophan hydroxylase 2 have been used to target GABAergic, cholinergic, or serotonergic neurons, respectively [19, 24–27].
Genes that define subsets of projection neurons can be used to achieve tract specificity. In the motor system, the corticospinal system can be differentiated from other corticofugal pathways (e.g., innervating thalamus, basal ganglia, and brain stem) by several genes, including Fezf2, Diap3, and Crym [10, 11, 28]. A transgenic mice line with green fluorescent protein (GFP) under the control of the promoter region of Crym demonstrated strong and specific labeling of the corticospinal tract with GFP [12]. The specificity of this approach suggests that the corticospinal tract could be manipulated relatively selectively by inserting a transgene that alters function under control of the mu-Crystallin promoter. These reports implicate the potential use of theses corticospinal neuron-specific genes for targeting corticospinal tract. Moreover, a transgenic mouse that expresses Cre recombinase under the control of mu-Crystallin promoter is available from the Gene Expression and Nervous System Atlas (GENSAT). This makes manipulation of the corticospinal tract using this genetic identity possible in the near future.
There are 2 major advantages in use of transgenic mice expressing genetic tools under the control of cell type-specific promoters (Table 1). First, when transgenes are introduced into the germline, genetic manipulation is noninvasive, as opposed to spatial control, which involves injections of virus into the origin and termination of a tract. Second, the cell type-specific expression of the transgene covers all the targeted cells and the expression level is stable, homogenous, and reproducible [18, 29, 30]. This offers great advantages for comparing manipulations across experiments.
Table 1.
Comparison between strategies for targeting specific cell type and circuit
| Genetic identity | Spatial control | |
|---|---|---|
| Definition | Use of a promoter region of an endogenous gene that defines a neuronal subtype to drive an exogenous gene | Use of the location where a group of neurons and/or their axon terminal exist to deliver an exogenous gene or stimulus |
| Methods | Transgenic animals | Viral injection Local stimulation |
| Advantages | Strong, stable, homogeneous and reproducible expression Cell type-specific expression Noninvasive Germline |
Connectivity-based specificity Cell type-specific expression when combined with genetic identity Flexibility of experimental design Applicable to various species |
| Disadvantages | Lack of connection-based specificity Possibly multiple types of neurons can express the genetic tool The same type of neurons can be in multiple regions of brain Limited species available (mouse) Might affect expression of endogenous genes |
Multiple injection required for large region Invasive Skilled injection and surgery required |
Despite great advantages, this strategy has several limitations (Table 1). Neurons are not characterized by expression of a single gene. Instead, the combination of several transcription factors and their temporal expression help to determine neuron subtypes and their projection patterns. Therefore, multiple types of neurons could express the targeted genes. Also, the specific cell types defined with a single gene can be found in multiple regions of the nervous system [18, 23].
Thus, the most critical and important limitation of genetic control is that it lacks the ability to target a specific circuit. In many cases, the genetic basis for a difference in projections is not known. For example, a neuron in motor cortex may project to multiple subcortical targets, yet the genetic basis for these targets is unknown. In addition, a single cortical neuron can branch at multiple levels and innervate several subcortical targets [31]. This limits the specificity with which a genetic control can regulate specificity of a target. These limitations can be overcome with spatial control strategies.
In addition, transgenesis might affect the expression of endogenous genes. There are 2 different strategies to generate transgenic mice, termed random and positional transgenesis according to the site of transgene integration. The positional transgenesis approach, knock-in, inserts transgene into mouse genome at a specific location, such as Rosa26 and HPRT locus, while random transgenesis, which usually uses bacterial artificial chromosome (BAC), integrates the transgene at a random location. These days, the BAC transgenic approach has been widely used owing to its advantages, including ease of use and the availability of large library [32–37]. However, endogenous gene expression can be altered because the delivery of BAC may contain coding regions or promoters of other genes [20, 38]. Indeed, a BAC transgenic mouse line expressing enhanced GFP under the control of D2 receptor promoter had increased expression of the D2 receptor and behavioral hypersensitive to D2 receptor agonists [38].
Finally, the overwhelming majority of transgenic animals are mice, and transgenic animals of other species are limited. The number of transgenic rats and higher mammals, such as cats and monkeys, are highly restricted [39]. While technical advances have made the creation of transgenic rats and larger mammals easier [40], the high cost of creating and maintaining these animals and the large number of mouse transgenic lines makes the mouse the most practical choice for most studies. However, the neural systems in a mouse may not adequately model human neural systems. This presents a challenge both to delineating neural circuits that are pertinent to humans and also for the translational potential of these tools.
Spatial Control
Specific circuits can be targeted, based on their connectivity, using 2 viruses, the one injected at the origin and the other virus injected at the termination of the targeted circuit (Fig. 1B). Only the neurons co-infected with both viruses express the genetic tool and are manipulated. This dual viral infection system has been applied and controlled diverse circuits [41–44]. This strategy provides the most flexibility to target circuits; one needs only to know the origin and termination of a circuit. For example, a new connection induced by timely combined interventions after stroke was targeted with this strategy. The only information available to target this connection was the location of the origin and termination of the connection. One virus was injected to the ipsilateral hemisphere, the origin of the circuit, and the other virus was injected into spinal cord of impaired forelimb side, the termination of the circuit. Inactivation with genetic tools proved the necessity of the new circuit for the improvement of the functional recovery [41]. This strategy is made possible by 2 technological advances: binary systems and viruses that transduce neurons retrogradely.
As the name implies, a binary system consists of 2 genes—one gene drives the expression of the other gene when they are delivered together to the same neuron. Gene 1 usually encodes a transcription activator or a DNA recombinase that binds to a special element on DNA molecules and induces transcription or DNA recombination. Gene 2 is the gene whose expression is controlled through these special DNA elements. Thus, one can manipulate a specific circuit by delivering gene 1 and gene 2 to its termination and origin, respectively. For example, injection of a virus with Cre recombinase (gene 1) into the spinal cord and another virus with a Cre-dependent chemogenetic tool (gene 2) into motor cortex allowed manipulation of the corticospinal tract [41].
The most widely and frequently used binary system is the Cre/lox site-specific recombination system. Cre recombinase catalyzes DNA recombination between two loxP sites in a DNA molecule [45]. The 8 base pair-long asymmetric spacer in a loxP site is responsible for the directionality of the site. When 2 loxP sites in a DNA molecule are in the same orientation, Cre excises the intervening DNA segment. When 2 loxP sites have the opposite orientations, the intervening DNA segment is inverted [45]. These recombination reactions have been used for controlling gene expression. The floxed-stop system uses excision of a transcriptional stop sequence. A stop sequence, which contains several stop codons, is inserted between 2 loxP sites in the same orientation in front of the transgene to prevent its expression. In the presence of Cre, the stop codon is excised and the transgene is expressed [45, 46]. In the flip-excision system, the transgene is encoded reversed and double-floxed between loxP sites, rendering it unable to be expressed [47, 48]. However, in the presence of Cre, the transgene is flipped into the sense orientation and permanently expressed [48, 49].
In addition to the spatial control of gene expression, temporal control with Cre recombination has been achieved with ligand-activated Cre. Cre has been fused with a modified estrogen receptor (CreER), which is sensitive only to synthetic ligand tamoxifen [50–52]. The cytoplasmic CreER is translocated into the nucleus where the DNA recombination occurs only in the presence of tamoxifen. Thus, the expression of Cre-dependent gene, which is introduced with flox-stop or flip-excision system, can be activated temporally [52].
Another binary system used widely for control of a transgene is the tetracycline-regulated gene expression system. This system is based on transcription factors controlled by tetracycline. The first element of this system is tetracycline-controlled transactivator. The second element is a gene that can manipulate cellular activity under the control of a tetracycline-response element. When a cell is doubly infected, the gene used to manipulate a cell can be regulated by administration of tetracycline. This system adds a layer of control—the tetracycline response element can either repress or activate expression of a transgene in the presence of tetracycline. Tetracycline-controlled transactivator represses gene transcription [53, 54], while reverse tetracycline-controlled transactivator activates the expression of the transgene [54–56].
An essential element of any spatial control system is the need for 2 viruses, one that infects the cell body and the other that infects axons and transports the transgene retrogradely to the cell body. Retrograde viruses have been more difficult to create. The first solution to this problem was to use viruses known to cause disease through retrograde transport, such as rabies and herpes simplex virus [57–60]. Although these viruses can be modified to prevent transmission to the experimenter, they can be difficult to produce and can cause inflammation and injury to nervous tissue. Now, several serotypes of adeno-associated virus (AAV) such as AAV6, AAV8, and AAV9 are characterized to transduce neurons retrogradely [61–65], and lentivirus has been modified with its glycoprotein fused with rabies virus glycoprotein for retrograde transduction [41, 42, 66, 67]. These viruses are easy to produce, result in less inflammation than herpes simplex and rabies-derived vectors, and have long duration and high level of transgene expression [68–72].
There are several advantages of the spatial control strategy (Table 1). Most important is the degree of specificity achieved, including the ability to target selectively newly formed connections that cannot be defined with genetic identity [41]. Indeed, the dual virus infection strategy has been used to prove necessity of newly sprouted axons for functional improvement [41]. In addition, this strategy can be applied to circuits in any species, as the tools are inserted with viruses and not introduced through germline manipulation. Moreover, the manipulation of different circuits can be achieved easily by injecting viruses at different locations, providing tremendous experimental flexibility.
The greatest disadvantage of this approach is the need for invasive injections of viruses. The injections can cause damage by mechanical injury to the tissue, by the harmful effects of the viruses themselves, or by the immune reaction to them [73]. In addition, the resulting gene expression can be weak or uneven compared with the expression of transgenes. In addition, circuits that are widely dispersed, particularly in larger animals, require multiple injections of viruses [41, 42]. For diffusely projecting systems (ascending monoaminergic systems of the brain stem) or for systems that originate in large areas (corticospinal system), a dual virus approach may not be practical.
Combining Genetic Identity and Spatial Control
Combining the strategies of genetic identity and spatial control can maximize the advantages of each approach (Table 1). The simplest combination is injecting virus that encodes a genetic tool under the control of a cell type-specific promoter (Fig. 1C). The spatial control is achieved by targeted injection, and the genetic control is achieved using a cell-type specific promoter. As shown in Fig. 1C, the pyramidal neurons in the lateral nucleus of amygdala can be targeted using this approach. The viral injection is made into the amygdala, which contains both excitatory pyramidal neurons (green) and GABAergic neurons (white) [74]. Only the excitatory pyramidal neurons express the transgene (blue with green outline) because the transgene was inserted under the control of CaMKIIα promoter (green) [75]. This promoter is expressed in excitatory neurons, not GABAergic neurons, in the following brain regions: lateral nucleus of amygdala, cerebral cortex, hippocampus, thalamus, brain stem nuclei, and spinal cord [76, 77]. However, excitatory neurons in other regions, including cerebral cortex (green in Fig. 1C) were not infected because the injection was contained to the amygdala. Thus, in this example, viral injection produced the spatial specificity and CaMKIIα promoter produced the cell type specificity.
The targeting strategies using genetic identity and location of the circuit can be combined into a binary system. A virus encoding genetic tools under a constitutive promoter with a binary system is injected into a brain region of a cell type-specific driver mouse, which express drivers such as Cre and tetracycline-controlled transactivator in a specific type of neurons. That accomplishes cell type and region-specific targeting with strong expression of genetic tools. For example, AAV encoding a Cre-dependent fluorescent protein, neurotoxin, or chemogenetic tool was injected into the medullary reticular formation ventral part (MdV) of the Vglut2–Cre mouse to label, ablate, or manipulate the glutamatergic circuit from MdV in order to test the roles of MdV in the forelimb movement control [78]. Hundreds of cell type-specific driver mice lines are available [79–86]. Moreover, driver mice lines of Cre/lox system with the combination of other binary systems [87, 88], including the tetracycline-regulated gene expression system [89], enable powerful approaches for manipulation of a wide range of neurons in the nervous system with spatial and temporal specificity.
The most widely used approach of using a binary system with a combination strategy is targeting the origin of the circuit. Viruses carrying a driver-dependent genetic tool have been injected into the brain region of the cell type-specific driver mouse lines where the circuit originates [90–96]. In this approach, the driver transgene defines the cell types, and the injection of virus provides regional specificity. For example, in order to target cholinergic interneurons, viruses encoding Cre-dependent genetic tools were injected into various brain regions of a mouse line that encodes Cre under the control of choline acetyltransferase promoter [91, 97, 98]. Although strong Cre expression is found in multiple brain regions, including striatum, brain stem, and hippocampus [83, 84, 98–101], the expression of genetic tools was restricted only in the brain regions targeted with viral injection [91, 97, 98].
The combination strategy can also use a driver mouse line and a retrograde virus encoding a driver-dependent genetic tool. For example, serotonergic neurons in the raphe nuclei project their axons to multiple areas of the brain, including the basolateral amygdala [102]. This circuit was targeted specifically by injecting a retrograde virus carrying a Cre-dependent genetic tool into the basolateral amygdala of serotonin reuptake transporter–Cre mice [64]. The Cre mice defined the cell type and the retrograde AAV defined the specific circuit.
A limitation still remains, even though targeting strategies provide precious specificity. Axons branch and make connections with neurons not only in the main target region, but also in several regions of brain on the way to the target [31, 103, 104]. For instance, the cortico-subthalamic nucleus projection has collaterals that innervate multiple other brain regions, including striatum, superior colliculus, pontine nucleus, and spinal cord [104]. Therefore, activation and inhibition of a specific circuit with optogenetic or chemogenetic tools still affect other systems through these collaterals. Indeed, a challenge of any circuit dissection or manipulation is that the nervous system is a highly integrated network. Any perturbation of a circuit in that network is likely to have distant, though milder, effects.
Genetic Toolbox for Circuit Manipulation
A variety of genetic tools have been created to manipulate cellular activity. Optogenetics uses light-activated channels, while chemogenetics uses receptors responsive only to the designed ligand [105, 106]. Here, we discuss the variety of genetic tools available, and the advantages and disadvantages in using each; these are summarized in Table 2.
Table 2.
Comparison of tools for manipulating activity of neurons and circuits
| Tools | Action | Actuators | Inducer | Mechanism | Onset | Advantages | Disadvantages |
|---|---|---|---|---|---|---|---|
| Optogenetics | Inhibitory | Holorhodopsin Archaerhodopsin Cruxhalorhodopsin |
Yellow–red Green–yellow Red |
Chloride pump Proton pump Chloride pump |
Milliseconds | Diversity of tools High temporal specificity Spatial specificity from local stimulation |
Invasiveness of stimulation Specialized equipment for stimulation |
| Excitatory | Channelrhodopsin family | Blue–red | Cation channel | ||||
| Chemogenetics - DREADDs |
Inhibitory | Modified muscarinic receptors | Clozapine-N-oxide Salvinorin B |
Potassium channel | Minutes | Prolonged effect Noninvasive stimulation No specialized equipment needed |
Fewer tools available |
| Excitatory | Modified muscarinic receptors | Clozapine-N-oxide | PLC pathway cAMP pathway |
||||
| Chemogenetics - Ligand-gated ion channels | Inhibitory | PSAM/PSEM/Gly | PSEM | Chloride channel | Minutes | ||
| Excitatory | PSAM/PSEM/5-HT | PSEM | Cation channel | ||||
| Inducible tetanus neurotoxin | Inhibitory | Tetanus neurotoxin | Doxycycline | VAMP2 cleavage | Days | Long-lasting effect | Slow onset |
| Neurotoxins | Inhibitory | Diphtheria toxin | Doxycycline Diphtheria toxin |
Protein synthesis inhibition | Permanent | Strong inactivation with lesion | Irreversible, permanent lesion |
| Saporin | – |
DREADDs = designer receptor exclusively activated by designer drug; PSAM = pharmacologically selective activator molecule; PSEM = pharmacologically selective effector modules; Gly = glycine; 5-HT = 5-hydroxytryptamine; PLC = phospholipase C; cAMP = cyclic adenosine monophosphate; VAMP2 = vesicle associated membrane protein 2
Optogenetics
Optogenetics is a technique that uses optical stimulation of genetically encoded light-sensitive channels and pumps to achieve neuromodulation [105, 107, 108]. Optogenetics was born with the discovery of light-sensitive microbial membrane proteins [107, 109–111]. Channelrhodopsins, light-sensitive cation channels, rapidly open and depolarize neurons in response to blue light (460 nm) [110, 111]. In contrast, halorhodopsin and archaerhodopsin inhibit neuronal activity. Halorhodopsin is a light-sensitive inward chloride pump that silences neuronal firing in response to yellow light (580 nm) [112]; archaerhodopsin is a proton pump responsive to yellow–green light (550 nm) [113].
One of the main strengths of optogenetics is the diversity of tools available. Through molecular engineering, these tools have been optimized to allow for faster alteration of channels [114, 115], response to different wavelengths [116–120], more robust gene expression [114, 117, 121, 122], and channels with stepwise kinetics [117, 123, 124]. Having tools activated with different wavelengths makes it possible to manipulate the same neuron bidirectionally with both excitatory and inhibitory channels.
The other main advantage in using optogenetic tools is precise temporal control: opsins can manipulate neuronal activity within milliseconds (Table 2) [124]. With this advantage, it is possible to generate the typical firing rates or patterned firing of neurons, and to manipulate the activity of neurons to mimic the changes in neuronal activity time-locked to a specific environmental event or key moment during behavior by using optic stimulation [125, 126]. Optogenetic manipulation has been used, for example, to alter the firing rate of dopamine neurons in the ventral tegmental area, which is critical for coding reward [125]. The frequency of the light stimulation was critical to how the animal responds to a stimulus, indicating that the firing frequency of this reward circuit is critical to its function [39, 125]. This temporal precision allows manipulation of specific neurons or circuits in a time-locked behavior, such as unexpected reward [125, 126].
The other strength of optogenetics is spatial specificity from local stimulation. The targeting of light for optogenetics is similar to the spatial control provided viral injection. Whereas the light can vary by intensity and size of the optical fiber, viral injection can vary by the volume of the injection and the titer of the viral particle. While delivery of light can be achieved many ways, there are limitations to delivering either a highly targeted light or light to activate a broad region of the brain.
Optogenetic manipulation also has disadvantages to overcome (Table 2). The major limitation is in the invasive method of delivering light. For the delivery of light stimulation, surgery is required to implant an optical fiber or other indwelling light source into or near the brain region of interest. Although successfully applied to live and free-moving animals, this can limit the movement of animal and induce brain damage [127–129]. Recent efforts have been made to overcome this limitation with wireless head-mounted systems [130–134]. Although this system made it easy to apply optogenetics to free-moving animals, it is still invasive—wireless headstage-mounted systems still need to implant optical fiber into the brain. To reduce invasiveness channels that respond to longer wavelength light have been developed; longer wavelength light penetrates tissue more deeply than short [135–140]. Red light-activated opsins such as excitatory red light-activated channelrhodopsin and inhibitory red-shifted cruxhalorhodopsin (Jaws) can be applied to the surface of the brain or through a thinned skull [119, 120].
Chemogenetics
Chemogenetics is the technique of giving a drug to activate exogenously expressed receptors for neuromodulation. To achieve specificity, the drug ligand and the receptor are modified so that they only interact with each other and not endogenous receptors or ligands.
Designer Receptor Exclusively Activated by Designer Drug
This chemogenetic tool employs design-based drugs to activate G protein-coupled receptors that have been modified to respond only to the novel ligand [106, 141]. G protein-coupled receptors play crucial roles as modulator in many physiological processes [142–144]. In neurons, G protein-coupled receptors can activate, inhibit, or modulate neuronal firing depending on their specific downstream effectors [143]. Using this natural heterogeneity of function, chemogenetics has modified G protein-coupled proteins to modulate several cellular functions [141, 145–147].
Like optogenetics, designer receptor exclusively activated by designer drug (DREADDs) can either activate or silence neurons. The first DREADD system mutated the muscarinic acetylcholine receptor so that it is only activated by clozapine-N-oxide, an inactive metabolite of the central nervous system drug clozapine [146, 148, 149]. The muscarinic DREADD receptor is coupled to Gq, which is an excitatory channel that depolarizes neurons and enhances their excitability through phospholipase C [146, 150]. A chimeric muscarinic–adrenergic receptor DREADD known as GsD also activates neurons but via cyclic adenosine monophosphate-mediated signaling [149, 151]. In contrast, the human M4 muscarinic DREADD receptor is coupled to Gi and leads to neuronal silencing through opening inwardly rectifying potassium channels [146].
A previous limitation of DREADDs was their dependence on the same drug, clozapine-N-oxide, for activation. To overcome this limitation, a new set of receptor and ligand was recently developed [152, 153]. KORD, the new DREADD, is a kappa-opioid receptor coupled to inhibitory Gi protein and selectively responsive to an inert ligand, salvinorin B [153–155]. With 2 chemogenetic tools, the same circuit can be manipulated bidirectionally, activating or inactivating, with different ligands.
Engineered Ligand-gated Ion Channels
Another chemogenetic tool uses chemical activation of ion channels modified to respond selectively to synthetic ligands [141, 152]. The α7 nicotinic acetylcholine receptor (nAchR) consists of 2 domains that are extracellular ligand-binding domain (LBD) and ion pore domain (IPD) [156]. The α7 nAchR LBD was mutated to be sensitive only to specific ligands [141, 152]. The mutated LBDs are called pharmacologically selective actuator modules (PSAM) and the specific ligands are called pharmacologically selective effector modules (PSEM) [141, 152].
Various combinations of PSAM/PSEM/IPD enable both activation and inactivation of neurons. The α7 nAchR LBD behaves as an independent actuator module and can be transplanted to other ion channels with similar structures [157, 158]. Thus, PSAMs are spliced with IPDs of various other ion channels to generate different ionic conductance in response to the specific ligands [152]. For example, PSAM was fused with the IPD of a serotonin receptor that generates cation conductance in order to activate neurons [152, 157], and the IPD of the chloride-selective glycine receptor that generates chloride conductance to silence the neurons in response to PSEM [152, 158]. This PSAM/PSEM/glycine silencing system is successfully applied to inactivate MdV–spinal motorneuron circuit and the inactivation of this circuit showed its critical roles in skilled motor function of the forelimb [78].
Major differences between chemogenetic and optogenetic tools are the timing of channel opening and invasiveness (Table 2). In terms of time, light-activated channels open on the millisecond time scale, while chemogenetic tools take minutes to hours to alter function after administration of the drug ligand. In terms of choosing of tools for experiments, optogenetics allow precise temporal alterations of function. Chemogenetics, especially DREADDs are more appropriate than optogenetics for changes of behaviors, which requires prolonged testing as DREADD activity is sustained for several hours after 1 injection of the ligand [41, 150, 151, 153]. In addition, chemogenetics also can be done without a head-mounted light source and invasive fiber optics.
Inducible Tetanus Neurotoxin
Another genetic tool used to inhibit circuit activity is the inducible tetanus neurotoxin system. Tetanus neurotoxin light chain (TeNT) blocks synaptic transmission through proteolytic cleavage of a synaptic vesicle protein [159–162]. As the light chain of tetanus neurotoxin blocks synaptic transmission, it is reversible; the heavy chain of tetanus neurotoxin is the element that causes sustained depolarization and cell injury. The inducible TeNT system is composed of 3 elements. The first element is the reverse tetracycline transactivator; the second is TeNT under the control of tetracycline-responsive element; the third is administration of doxycycline, a tetracycline derivative. When the 2 genetic elements are delivered to a neuron and doxycycline is given, TeNT is expressed. The circuit is inactivated through blocking neurotransmission. This system has been used successfully to inactivate neurons and circuits selectively and reversibly [41, 42, 163, 164]. For instance, in order to test the roles of the circuit between propriospinal neurons and spinal motor neurons in dexterous hand movement of primates, AAV encoding reverse tetracycline transactivator was injected into the origin of the circuit where the propriospinal neurons are located, and retrograde gene transfer lentivirus carrying TeNT under the control of tetracycline responsive element was injected into the termination of the circuit where the spinal motor neurons reside. When doxycycline is administrated, only the propriospinal neurons expressed TeNT and the animals showed impairment of reach and grasp movements during doxycycline administration and recovery of the hand function during discontinuance of doxycycline administration [42]. Inactivation could be repeated many times.
The inducible TeNT system is also appropriate to test the roles of specific neurons and circuits in prolonged behavioral changes such as functional recovery after neurological injuries because manipulation with this system is sustained for days to weeks during doxycycline administration [41]. Doxycycline is given orally in drinking water, which is an advantage of this system. Like chemogenetics, the intervention is noninvasive after the initial viral injections, which enables animals to conduct complicated behavioral tasks freely.
Despite several strengths, the inducible TeNT system has weaknesses (Table 2). This system is based on protein expression after doxycycline treatment. Thus, it has a relatively long latency of several days to reach to the maximum inactivation level after stimulation. The latency can be various from 2 to 7 or more days according to the length of the targeted circuit because TeNT has to travel from the soma to the axon terminal [41–43]. This long time period allows other circuits to adapt to the loss of transmission within the targeted circuit [42, 43]. In addition, doxycycline is an antibiotic that may cause physiological changes that might affect animal’s behavior.
Cell-Specific Neurotoxins
Other neurotoxins can be used to alter circuits by killing neurons. Diphtheria toxin kills cells by inhibiting protein synthesis [165]. By using binary systems, spatiotemporal ablation of neurons has been accomplished in specific neurons. One gene is expressed in a genetically defined set of neurons. Another gene codes for the neurotoxin that kills the cells with a known genetic identity [166–168]. Saporin, a toxin protein from a plant Saponara officinalis, can also be used for cell type or circuit-specific ablation. Saporin kills cells by inactivating ribosomes thereby inhibiting translation [169]. Targeting specific cell types and circuits has been accomplished with local injection of modified saporins that is conjugated with various proteins including cell type-specific antibodies and the neurotransmitter substance P [170–177]. Some neurotoxins, such as tetanus toxin and botulinum toxin are retrogradely transported by presynaptic neurons. When they are injected into the termination of a circuit they can be used to deliver toxins retrogradely [178–180].
The main advantage of these toxins is that they can be combined with systems that control retrograde transport to selectively kill connected neurons. For example, mice do not normally express diphtheria toxin receptor [181, 182]. Conditional expression of the receptor allows only the genetically modified cells to take up the toxin. In this approach, when diphtheria toxin was administrated, it was transferred only into the targeted cells and induced cell death [78, 182, 183]. Cholera toxin B fragment and wheat germ agglutinin are highly sensitive retrograde tracers [184–186]. Neurotoxins, including diphtheria toxin, botulinum toxin, ricin, and saporin, have been conjugated with these retrograde tracers to kill connected neurons [187–190]. For example, respiratory motor neurons were with intrapleural injections of cholera toxin B-conjugated saporin [190].
The cell type-specific neurotoxins allow elimination of specific cell types with circuit specificity. However, because they result in permanent cell loss, the function of the targeted cells is lost once, with subsequent adaptation to the injury.
Conclusions
Selective manipulation of specific neurons and circuits has been a longstanding goal of neuroscience. Unlike classical systems neuroscience tools, the strategies and genetic tools discussed in this review have the capacity to unravel the roles of individual circuits in brain function. Now we can manipulate specific circuits with genetic tools in various time windows from milliseconds to weeks. We can target them based on their genetic identity, connectivity, or a combination thereof. Owing to the abundance of transgenic mouse strains, a myriad of manipulations is possible when using mice. However, when using other animals such as rat, cat, and monkey, viral transduction remains the most viable strategy.
In addition to elucidating function, these can be used to manipulate neural circuits for therapy, which is the subject of this issue. These tools have been applied to animal models of various neurological diseases. For Parkinson’s disease [191, 192], the goal has been to modulate the basal ganglia circuits in a way that is achieved with deep brain stimulation but with better spatial and temporal control. For stroke [193], excitatory optogenetics has been used to strengthen the function of intact circuits so that they can restore lost motor function. After spinal cord injury [194, 195], optical control of spinal neurons distal to the lesion could establish control of circuits that have been disconnected from brain control. In epilepsy [196–198], the goal has been to reduce cortical excitability, which can be accomplished with activation of inhibitory neurons or inactivation of excitatory ones.
For translation to humans, these tools have significant barriers to overcome. First, all of the tools involve gene therapy in neurons, which is both challenging to achieve and potentially dangerous. In addition to the invasiveness of injection, viruses can be toxic, both in the short- and long-term. Even if it can be done safely, achieving stable expression of genetic tools is a challenge. Finally, introducing light into the human brain for optogenetic control also involves risk and technical challenge.
One promising area for the use of circuit-specific tools is optogenetics for retinal disease [199, 200]. In retinitis pigmentosa, for example, photoreceptors are lost. However, as other neurons, including retinal ganglion cells, are spared, they could be used to transduce light into neural activity by inserting optogenetic channels. Retinal disease presents an attractive target because the retina is more accessible that the central nervous system and because optogenetic channels may be able to substitute for endogenous rhodopsin function (see the review by Sagdullaev in this issue [201]). Until the genetic tools overcome the various obstacles for use in people, electrical stimulation of neural circuits will likely remain the clinical tool for modulation of neural circuits.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Required Author Forms Disclosure forms provided by the authors are available with the online version of this article. (PDF 3166 kb)
Acknowledgments
We thank Edmund Hollis for critical reading of manuscript and advice. This work was supported by National Institute of Neurological Disorders and Stroke–National Institutes of Health (to J.B.C., K08 NS073796 and 1R01NS092875) and the New York State Spinal Cord Injury Research Program (DOH01-Fellow-2015-00012 to H.P.).
References
- 1.Broca P. Remarques Sur le Siége de la Faculté Du Langage Articulé, Suivies D’une Observation D’aphémie (Perte de la Parole) Bull Soc Anat. 1861;6:330–357. [Google Scholar]
- 2.Nishitani N, Schürmann M, Amunts K, et al. Broca’s region: from action to language. Physiology (Bethesda) 2005;20:60–69. doi: 10.1152/physiol.00043.2004. [DOI] [PubMed] [Google Scholar]
- 3.Ochfeld E, Newhart M, Molitoris J, et al. Ischemia in Broca area is associated with Broca aphasia more reliably in acute than in chronic stroke. Stroke. 2010;41:325–330. doi: 10.1161/STROKEAHA.109.570374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin JH. The corticospinal system: from development to motor control. Neuroscientist. 2005;11:161–173. doi: 10.1177/1073858404270843. [DOI] [PubMed] [Google Scholar]
- 5.Gross CG. The discovery of motor cortex and its background. J Hist Neurosci. 2007;16:320–331. doi: 10.1080/09647040600630160. [DOI] [PubMed] [Google Scholar]
- 6.Snyder PJ, Whitaker HA. Neurologic heuristics and artistic whimsy: the cerebral cartography of Wilder Penfield. J Hist Neurosci. 2013;22:277–291. doi: 10.1080/0964704X.2012.757965. [DOI] [PubMed] [Google Scholar]
- 7.Lomber SG. The advantages and limitations of permanent or reversible deactivation techniques in the assessment of neural function. J Neurosci Methods. 1999;86:109–117. doi: 10.1016/S0165-0270(98)00160-5. [DOI] [PubMed] [Google Scholar]
- 8.Carmel JB, Kimura H, Berrol LJ, et al. Motor cortex electrical stimulation promotes axon outgrowth to brain stem and spinal targets that control the forelimb impaired by unilateral corticospinal injury. Eur J Neurosci. 2013;37:1090–1102. doi: 10.1111/ejn.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmel JB, Kimura H, Martin JH. Electrical stimulation of motor cortex in the uninjured hemisphere after chronic unilateral injury promotes recovery of skilled locomotion through ipsilateral control. J Neurosci. 2014;34:462–466. doi: 10.1523/JNEUROSCI.3315-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arlotta P, Molyneaux BJ, Chen J, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 11.Molyneaux BJ, Arlotta P, Hirata T, et al. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 12.Fink KL, Strittmatter SM, Cafferty WBJ. Comprehensive corticospinal labeling with mu-crystallin transgene reveals axon regeneration after spinal cord trauma in ngr1– /– mice. J Neurosci. 2015;35:15403–15418. doi: 10.1523/JNEUROSCI.3165-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molyneaux BJ, Arlotta P, Menezes JRL, et al. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 14.Greig LC, Woodworth MB, Galazo MJ, et al. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayford M, Bach ME, Huang YY, et al. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- 16.Arenkiel BR, Peca J, Davison IG, et al. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Peca J, Matsuzaki M, et al. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proc Natl Acad Sci U S A. 2007;104:8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hägglund M, Borgius L, Dougherty KJ, et al. Activation of groups of excitatory neurons in the mammalian spinal cord or hindbrain evokes locomotion. Nat Neurosci. 2010;13:246–252. doi: 10.1038/nn.2482. [DOI] [PubMed] [Google Scholar]
- 19.Zhao S, Ting JT, Atallah HE, et al. Cell type–specific channelrhodopsin-2 transgenic mice for optogenetic dissection of neural circuitry function. Nat Methods. 2011;8:745–752. doi: 10.1038/nmeth.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolisnyk B, Guzman MS, Raulic S, et al. ChAT-ChR2-EYFP mice have enhanced motor endurance but show deficits in attention and several additional cognitive domains. J Neurosci. 2013;33:10427–10438. doi: 10.1523/JNEUROSCI.0395-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrell MS, Pei Y, Wan Y, et al. A Gαs DREADD mouse for selective modulation of cAMP production in striatopallidal neurons. Neuropsychopharmacology. 2013;38:854–862. doi: 10.1038/npp.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kullander K, Butt SJB, Lebret JM, et al. Role of EphA4 and EphrinB3 in local neuronal circuits that control walking. Science. 2003;299:1889–1892. doi: 10.1126/science.1079641. [DOI] [PubMed] [Google Scholar]
- 23.Fremeau RT, Voglmaier S, Seal RP, et al. VGLUTs define subsets of excitatory neurons and suggest novel roles for glutamate. Trends Neurosci. 2004;27:98–103. doi: 10.1016/j.tins.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Shen M, Yu Y, et al. Optogenetic activation of GABAergic neurons in the nucleus accumbens decreases the activity of the ventral pallidum and the expression of cocaine-context-associated memory. Int J Neuropsychopharmacol. 2014;17:753–763. doi: 10.1017/S1461145713001570. [DOI] [PubMed] [Google Scholar]
- 25.Ren J, Qin C, Hu F, et al. Habenula “cholinergic” neurons corelease glutamate and acetylcholine and activate postsynaptic neurons via distinct transmission modes. Neuron. 2011;69:445–452. doi: 10.1016/j.neuron.2010.12.038. [DOI] [PubMed] [Google Scholar]
- 26.Ma M, Luo M. Optogenetic activation of basal forebrain cholinergic neurons modulates neuronal excitability and sensory responses in the main olfactory bulb. J Neurosci. 2012;32:10105–10116. doi: 10.1523/JNEUROSCI.0058-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito H, Yanase M, Yamashita A, et al. Analysis of sleep disorders under pain using an optogenetic tool: possible involvement of the activation of dorsal raphe nucleus-serotonergic neurons. Mol Brain. 2013;6:59. doi: 10.1186/1756-6606-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue K, Terashima T, Nishikawa T, et al. Fez1 is layer-specifically expressed in the adult mouse neocortex. Eur J Neurosci. 2004;20:2909–2916. doi: 10.1111/j.1460-9568.2004.03763.x. [DOI] [PubMed] [Google Scholar]
- 29.Liësi P, Salonen EM, Dahl D, et al. Thy-1 is neuronal and glial surface antigen which interacts with matrix protein and plasminogen activator 9. Exp Brain Res. 1990;79:642–650. doi: 10.1007/BF00229332. [DOI] [PubMed] [Google Scholar]
- 30.Théveniau M, Durbec P, Gennarini G, et al. Expression and release of phosphatidylinositol anchored cell surface molecules by a cell line derived from sensory neurons. J Cell Biochem. 1992;48:61–72. doi: 10.1002/jcb.240480110. [DOI] [PubMed] [Google Scholar]
- 31.O’Leary DD, Terashima T. Cortical axons branch to multiple subcortical targets by interstitial axon budding: implications for target recognition and “waiting periods”. Neuron. 1988;1:901–910. doi: 10.1016/0896-6273(88)90147-X. [DOI] [PubMed] [Google Scholar]
- 32.Marra MA, Kucaba TA, Dietrich NL, et al. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandler KJ, Chandler RL, Broeckelmann EM, et al. Relevance of BAC transgene copy number in mice: Transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm Genome. 2007;18:693–708. doi: 10.1007/s00335-007-9056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong S, Yang XW, Li C, et al. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kγ origin of replication. Genome Res. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osoegawa K, Tateno M, Woon PY, et al. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10:116–128. [PMC free article] [PubMed] [Google Scholar]
- 36.McPherson JD, Marra M, Hillier L, et al. A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- 37.Gong S, Zheng C, Doughty ML, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 38.Kramer PF, Christensen CH, Hazelwood LA, et al. Dopamine D2 receptor overexpression alters behavior and physiology in Drd2-EGFP mice. J Neurosci. 2011;31:126–132. doi: 10.1523/JNEUROSCI.4287-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witten IB, Steinberg EE, Lee SY, et al. Recombinase-driver rat lines: tools, techniques, and optogenetic application to dopamine-mediated reinforcement. Neuron. 2011;72:721–733. doi: 10.1016/j.neuron.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 41.Wahl AS, Omlor W, Rubio JC, et al. Neuronal repair. Asynchronous therapy restores motor control by rewiring of the rat corticospinal tract after stroke. Science. 2014;344:1250–1255. doi: 10.1126/science.1253050. [DOI] [PubMed] [Google Scholar]
- 42.Kinoshita M, Matsui R, Kato S, et al. Genetic dissection of the circuit for hand dexterity in primates. Nature. 2012;487:235–238. doi: 10.1038/nature11206. [DOI] [PubMed] [Google Scholar]
- 43.Sooksawate T, Isa K, Matsui R, et al. Viral vector-mediated selective and reversible blockade of the pathway for visual orienting in mice. Front Neural Circuits. 2013;7:162. doi: 10.3389/fncir.2013.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oguchi M, Okajima M, Tanaka S, et al. Double virus vector infection to the prefrontal network of the macaque brain. PLoS One. 2015;10:e0132825. doi: 10.1371/journal.pone.0132825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 46.Bareyre FM, Kerschensteiner M, Misgeld T, et al. Transgenic labeling of the corticospinal tract for monitoring axonal responses to spinal cord injury. Nat Med. 2005;11:1355–1360. doi: 10.1038/nm1331. [DOI] [PubMed] [Google Scholar]
- 47.Schnütgen F, Doerflinger N, Calléja C, et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- 48.Sohal VS, Zhang F, Yizhar O, et al. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atasoy D, Aponte Y, Su HH, et al. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metzger D, Clifford J, Chiba H, et al. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci U S A. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feil R, Brocard J, Mascrez B, et al. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feil R, Wagner J, Metzger D, et al. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 53.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berens C, Hillen W. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur J Biochem. 2003;270:3109–3121. doi: 10.1046/j.1432-1033.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- 55.Hillen W, Berens C. Mechanisms underlying expression of TN10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 56.Gossen M, Freundlieb S, Bender G, et al. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 57.LaVail JH, Topp KS, Giblin PA, et al. Factors that contribute to the transneuronal spread of herpes simplex virus. J Neurosci Res. 1997;49:485–496. doi: 10.1002/(SICI)1097-4547(19970815)49:4<485::AID-JNR9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 58.Kelly RM, Strick PL. Rabies as a transneuronal tracer of circuits in the central nervous system. J Neurosci Methods. 2000;103:63–71. doi: 10.1016/S0165-0270(00)00296-X. [DOI] [PubMed] [Google Scholar]
- 59.Larsen DD. Retrograde tracing with recombinant rabies virus reveals correlations between projection targets and dendritic architecture in layer 5 of mouse barrel cortex. Front Neural Circuits. 2008;1:5. doi: 10.3389/neuro.04.005.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng H. Efficient retrograde transport of adeno-associated virus type 8 to spinal cord and dorsal root ganglion. Hum Gene Ther. 2010;21:87–97. doi: 10.1089/hum.2009.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aschauer DF, Kreuz S, Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS One. 2013;8:e76310. doi: 10.1371/journal.pone.0076310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Löw K, Aebischer P, Schneider BL. Direct and retrograde transduction of nigral neurons with AAV6, 8, and 9 and intraneuronal persistence of viral particles. Hum Gene Ther. 2013;24:613–629. doi: 10.1089/hum.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rothermel M, Brunert D, Zabawa C, et al. Transgene expression in target-defined neuron populations mediated by retrograde infection with adeno-associated viral vectors. J Neurosci. 2013;33:15195–15206. doi: 10.1523/JNEUROSCI.1618-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salegio EA, Samaranch L, Kells AP, et al. Axonal transport of adeno-associated viral vectors is serotype-dependent. Gene Ther. 2013;20:348–352. doi: 10.1038/gt.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kato S, Kobayashi K, Inoue K, et al. A lentiviral strategy for highly efficient retrograde gene transfer by pseudotyping with fusion envelope glycoprotein. Hum Gene Ther. 2011;22:197–206. doi: 10.1089/hum.2009.179. [DOI] [PubMed] [Google Scholar]
- 67.Kato S, Kuramochi M, Takasumi K, et al. Neuron-specific gene transfer through retrograde transport of lentiviral vector pseudotyped with a novel type of fusion envelope glycoprotein. Hum Gene Ther. 2011;22:1511–1523. doi: 10.1089/hum.2011.111. [DOI] [PubMed] [Google Scholar]
- 68.Peel AL, Klein RL. Adeno-associated virus vectors: activity and applications in the CNS. J Neurosci Methods. 2000;98:95–104. doi: 10.1016/S0165-0270(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 69.Jacobsson J, Lundberg C. Lentiviral vectors for use in the central nervous system. Mol Ther. 2006;13:484–493. doi: 10.1016/j.ymthe.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 70.Wickersham IR, Finke S, Conzelmann KK, et al. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–49. doi: 10.1038/nmeth999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Urban A, Rancillac A, Martinez L, et al. Deciphering the neuronal circuitry controlling local blood flow in the cerebral cortex with optogenetics in PV::Cre transgenic mice. Front Pharmacol. 2012;3:105. doi: 10.3389/fphar.2012.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kantor B, Bailey RM, Wimberly K, et al. Methods for gene transfer to the central nervous system. Adv Genet. 2014;87:125–197. doi: 10.1016/B978-0-12-800149-3.00003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ciesielska A, Hadaczek P, Mittermeyer G, et al. Cerebral infusion of AAV9 vector-encoding non-self proteins can elicit cell-mediated immune responses. Mol Ther. 2012;21:158–166. doi: 10.1038/mt.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McDonald AJ. Neuronal organization of the lateral and basolateral amygdaloid nuclei in the rat. J Comp Neurol. 1984;222:589–606. doi: 10.1002/cne.902220410. [DOI] [PubMed] [Google Scholar]
- 75.Johansen JP, Hamanaka H, Monfils MH, et al. Optical activation of lateral amygdala pyramidal cells instructs associative fear learning. Proc Natl Acad Sci U S A. 2010;107:12692–12697. doi: 10.1073/pnas.1002418107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Benson DL, Isackson PJ, Gall CM, et al. Contrasting patterns in the localization of glutamic acid decarboxylase and Ca2+/calmodulin protein kinase gene expression in the rat central nervous system. Neuroscience. 1992;46:825–849. doi: 10.1016/0306-4522(92)90188-8. [DOI] [PubMed] [Google Scholar]
- 77.Jones EG, Huntley GW, Benson DL. Alpha calcium/calmodulin-dependent protein kinase II selectively expressed in a subpopulation of excitatory neurons in monkey sensory-motor cortex: comparison with GAD-67 expression. J Neurosci. 1994;14:611–629. doi: 10.1523/JNEUROSCI.14-02-00611.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Esposito MS, Capelli P, Arber S. Brainstem nucleus MdV mediates skilled forelimb motor tasks. Nature. 2014;508:351–356. doi: 10.1038/nature13023. [DOI] [PubMed] [Google Scholar]
- 79.Gong S, Doughty M, Harbaugh CR, et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taniguchi H, He M, Wu P, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanaka KF, Matsui K, Sasaki T, et al. Expanding the repertoire of optogenetically targeted cells with an enhanced gene expression system. Cell Rep. 2012;2:397–406. doi: 10.1016/j.celrep.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 82.Harris JA, Hirokawa KE, Sorensen SA, et al. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits. 2014;8:76. doi: 10.3389/fncir.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Madisen L, Mao T, Koch H, et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci. 2012;15:793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hägglund M, Dougherty KJ, Borgius L, et al. Optogenetic dissection reveals multiple rhythmogenic modules underlying locomotion. Proc Natl Acad Sci U S A. 2013;110:11589–11594. doi: 10.1073/pnas.1304365110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gerfen CR, Paletzki R, Heintz N. GENSAT BAC Cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron 2013;80:1368–1383. [DOI] [PMC free article] [PubMed]
- 87.Sadowski PD. The Flp recombinase of the 2-microns plasmid of Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1995;51:53–91. doi: 10.1016/S0079-6603(08)60876-4. [DOI] [PubMed] [Google Scholar]
- 88.Anastassiadis K, Fu J, Patsch C, et al. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis Model Mech. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- 89.Madisen L, Garner AR, Shimaoka D, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuhlman SJ, Huang ZJ. High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS One. 2008;3:e2005. doi: 10.1371/journal.pone.0002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Witten IB, Lin SC, Brodsky M, et al. Cholinergic interneurons control local circuit activity and cocaine conditioning. Science. 2010;330:1677–1681. doi: 10.1126/science.1193771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krashes MJ, Koda S, Ye C, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice Michael. J Clin Invest. 2011;121:2–6. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krashes MJ, Shah BP, Madara JC, et al. An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature. 2014;507:238–242. doi: 10.1038/nature12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhan C, Zhou J, Feng Q, et al. Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. J Neurosci. 2013;33:3624–3632. doi: 10.1523/JNEUROSCI.2742-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Soumier A, Sibille E. Opposing effects of acute versus chronic blockade of frontal cortex somatostatin-positive inhibitory neurons on behavioral emotionality in mice. Neuropsychopharmacology. 2014;39:2252–2262. doi: 10.1038/npp.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Perova Z, Delevich K, Li B. Depression of Excitatory synapses onto parvalbumin interneurons in the medial prefrontal cortex in susceptibility to stress. J Neurosci. 2015;35:3201–3206. doi: 10.1523/JNEUROSCI.2670-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Threlfell S, Lalic T, Platt NJ, et al. Striatal dopamine release is triggered by synchronized activity in cholinergic interneurons. Neuron. 2012;75:58–64. doi: 10.1016/j.neuron.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 98.Yi F, Catudio-Garrett E, Gabriel R, et al. Hippocampal “cholinergic interneurons” visualized with the choline acetyltransferase promoter: anatomical distribution, intrinsic membrane properties, neurochemical characteristics, and capacity for cholinergic modulation. Front Synaptic Neurosci. 2015;7:4. doi: 10.3389/fnsyn.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Armstrong DM, Saper CB, Levey AI, et al. Distribution of cholinergic neurons in rat brain: demonstrated by the immunocytochemical localization of choline acetyltransferase. J Comp Neurol. 1983;216:53–68. doi: 10.1002/cne.902160106. [DOI] [PubMed] [Google Scholar]
- 100.Oda Y, Nakahishi I. The distribution of cholinergic neurons in the central nervous system. Histol Histopathol. 2000;15:825–834. doi: 10.14670/HH-15.825. [DOI] [PubMed] [Google Scholar]
- 101.Newman EL, Gupta K, Climer JR, et al. Cholinergic modulation of cognitive processing: insights drawn from computational models. Front Behav Neurosci. 2012;6:1–19. doi: 10.3389/fnbeh.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hornung JP. The human raphe nuclei and the serotonergic system. J Chem Neuroanat. 2003;26:331–343. doi: 10.1016/j.jchemneu.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 103.O’Leary DD, Koester SE. Development of projection neuron types, axon pathways, and patterned connections of the mammalian cortex. Neuron. 1993;10:991–1006. doi: 10.1016/0896-6273(93)90049-W. [DOI] [PubMed] [Google Scholar]
- 104.Kita T, Kita H. The subthalamic nucleus is one of multiple innervation sites for long-range corticofugal axons: a single-axon tracing study in the rat. J Neurosci. 2012;32:5990–5999. doi: 10.1523/JNEUROSCI.5717-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Deisseroth K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci. 2015;18:1213–1225. doi: 10.1038/nn.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Urban DJ, Roth BL. DREADDs (designer receptors exclusively activated by designer drugs): chemogenetic tools with therapeutic utility. Annu Rev Pharmacol Toxicol. 2015;55:399–417. doi: 10.1146/annurev-pharmtox-010814-124803. [DOI] [PubMed] [Google Scholar]
- 107.Deisseroth K, Feng G, Majewska AK, et al. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nagel G, Ollig D, Fuhrmann M, et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 110.Nagel G, Szellas T, Huhn W, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boyden ES, Zhang F, Bamberg E, et al. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 112.Zhang F, Wang LP, Brauner M, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 113.Chow BY, Han X, Dobry AS, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lin JY, Lin MZ, Steinbach P, et al. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gunaydin LA, Yizhar O, Berndt A, et al. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 116.Zhang F, Prigge M, Beyrière F, et al. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat Neurosci. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yizhar O, Fenno LE, Prigge M, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Prigge M, Schneider F, Tsunoda SP, et al. Color-tuned channelrhodopsins for multiwavelength optogenetics. J Biol Chem. 2012;287:31804–31812. doi: 10.1074/jbc.M112.391185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin JY, Knutsen PM, Muller A, et al. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci. 2013;16:1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chuong AS, Miri ML, Busskamp V, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat Neurosci. 2014;17:1123–1129. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang H, Sugiyama Y, Hikima T, et al. Molecular determinants differentiating photocurrent properties of two channelrhodopsins from Chlamydomonas. J Biol Chem. 2009;284:5685–5696. doi: 10.1074/jbc.M807632200. [DOI] [PubMed] [Google Scholar]
- 122.Berndt A, Schoenenberger P, Mattis J, et al. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc Natl Acad Sci U S A. 2011;108:7595–7600. doi: 10.1073/pnas.1017210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Berndt A, Yizhar O, Gunaydin LA, et al. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 124.Rein ML, Deussing JM. The optogenetic (r)evolution. Mol Genet Genomics. 2012;287:95–109. doi: 10.1007/s00438-011-0663-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim KM, Baratta MV, Yang A, et al. Optogenetic mimicry of the transient activation of dopamine neurons by natural reward is sufficient for operant reinforcement. PLoS One. 2012;7:e33612. doi: 10.1371/journal.pone.0033612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bass CE, Grinevich VP, Gioia D, et al. Optogenetic stimulation of VTA dopamine neurons reveals that tonic but not phasic patterns of dopamine transmission reduce ethanol self-administration. Front Behav Neurosci. 2013;7:1–10. doi: 10.3389/fnbeh.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 128.Polikov VS, Tresco PA, Reichert WM. Response of brain tissue to chronically implanted neural electrodes. J Neurosci Methods. 2005;148:1–18. doi: 10.1016/j.jneumeth.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 129.Xu HT, Pan F, Yang G, et al. Choice of cranial window type for in vivo imaging affects dendritic spine turnover in the cortex. Nat Neurosci. 2007;10:549–551. doi: 10.1038/nn1883. [DOI] [PubMed] [Google Scholar]
- 130.Wentz CT, Bernstein JG, Monahan P, et al. A wirelessly powered and controlled device for optical neural control of freely-behaving animals. J Neural Eng. 2011;8:046021. doi: 10.1088/1741-2560/8/4/046021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim TI, McCall JG, Jung YH, et al. Injectable, cellular-scale optoelectronics with applications for wireless optogenetics. Science. 2013;340:211–216. doi: 10.1126/science.1232437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kwon KY, Lee H-M, Ghovanloo M, et al. Design, fabrication, and packaging of an integrated, wirelessly-powered optrode array for optogenetics application. Front Syst Neurosci. 2015;9:69. doi: 10.3389/fnsys.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Montgomery KL, Yeh AJ, Ho JS, et al. Wirelessly powered, fully internal optogenetics for brain, spinal and peripheral circuits in mice. Nat Methods. 2015;12:969–974. doi: 10.1038/nmeth.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Park SI, Shin G, Banks A, et al. Ultraminiaturized photovoltaic and radio frequency powered optoelectronic systems for wireless optogenetics. J Neural Eng. 2015;12:056002. doi: 10.1088/1741-2560/12/5/056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li X, Gutierrez DV, Hanson MG, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci U S A. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huber D, Petreanu L, Ghitani N, et al. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hira R, Honkura N, Noguchi J, et al. Transcranial optogenetic stimulation for functional mapping of the motor cortex. J Neurosci Methods. 2009;179:258–263. doi: 10.1016/j.jneumeth.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 138.Drew PJ, Shih AY, Driscoll JD, et al. Chronic optical access through a polished and reinforced thinned skull. Nat Methods. 2010;7:981–984. doi: 10.1038/nmeth.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Scott NA, Murphy TH. Hemodynamic responses evoked by neuronal stimulation via channelrhodopsin-2 can be independent of intracortical glutamatergic synaptic transmission. PLoS One. 2012;7:e29859. doi: 10.1371/journal.pone.0029859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jacques SL. Optical properties of biological tissues: a review. Phys Med Biol. 2013;58:R37–R61. doi: 10.1088/0031-9155/58/11/R37. [DOI] [PubMed] [Google Scholar]
- 141.Sternson SM, Roth BL. Chemogenetic tools to interrogate brain functions. Annu Rev Neurosci. 2014;37:387–407. doi: 10.1146/annurev-neuro-071013-014048. [DOI] [PubMed] [Google Scholar]
- 142.Schöneberg T, Schulz A, Biebermann H, et al. Mutant G-protein-coupled receptors as a cause of human diseases. Pharmacol Ther. 2004;104:173–206. doi: 10.1016/j.pharmthera.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 143.Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85:1159–1204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- 144.Allen JA, Roth BL. Strategies to discover unexpected targets for drugs active at G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2011;51:117–144. doi: 10.1146/annurev-pharmtox-010510-100553. [DOI] [PubMed] [Google Scholar]
- 145.Coward P, Wada HG, Falk MS, et al. Controlling signaling with a specifically designed Gi-coupled receptor. Proc Natl Acad Sci U S A. 1998;95:352–357. doi: 10.1073/pnas.95.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Armbruster BN, Li X, Pausch MH, et al. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Conklin BR, Hsiao EC, Claeysen S, et al. Engineering GPCR signaling pathways with RASSLs. Nat Methods. 2008;5:673–678. doi: 10.1038/nmeth.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Weiner DM, Meltzer HY, Veinbergs I, et al. The role of M1 muscarinic receptor agonism of N-desmethylclozapine in the unique clinical effects of clozapine. Psychopharmacology (Berl) 2004;177:207–216. doi: 10.1007/s00213-004-1940-5. [DOI] [PubMed] [Google Scholar]
- 149.Guettier J-M, Gautam D, Scarselli M, et al. A chemical-genetic approach to study G protein regulation of beta cell function in vivo. Proc Natl Acad Sci U S A. 2009;106:19197–19202. doi: 10.1073/pnas.0906593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Alexander GM, Rogan SC, Abbas AI, et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Farrell MS, Roth BL. Pharmacosynthetics: reimagining the pharmacogenetic approach. Brain Res. 2013;1511:6–20. doi: 10.1016/j.brainres.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Magnus CJ, Lee PH, Atasoy D, et al. Chemical and genetic engineering of selective ion channel-ligand interactions. Science. 2011;333:1292–1296. doi: 10.1126/science.1206606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vardy E, Robinson JE, Li C, et al. A new DREADD facilitates the multiplexed chemogenetic interrogation of behavior. Neuron. 2015;86:936–946. doi: 10.1016/j.neuron.2015.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ansonoff MA, Zhang J, Czyzyk T, et al. Antinociceptive and hypothermic effects of Salvinorin A are abolished in a novel strain of kappa-opioid receptor-1 knockout mice. J Pharmacol Exp Ther. 2006;318:641–648. doi: 10.1124/jpet.106.101998. [DOI] [PubMed] [Google Scholar]
- 155.Hooker JM, Munro TA, Béguin C, et al. Salvinorin A and derivatives: protection from metabolism does not prolong short-term, whole-brain residence. Neuropharmacology. 2009;57:386–391. doi: 10.1016/j.neuropharm.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Corringer PJ, Novere NL, Changeux JP. Nicotinic receptors at the amino acid level. Annu Rev Pharmacol Toxicol. 2000;40:431–458. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- 157.Eisele JL, Bertrand S, Galzi JL, et al. Chimaeric nicotinic-serotonergic receptor combines distinct ligand binding and channel specificities. Nature. 1993;366:479–483. doi: 10.1038/366479a0. [DOI] [PubMed] [Google Scholar]
- 158.Grutter T, Carvalho LP, Dufresne V, et al. Molecular tuning of fast gating in pentameric ligand-gated ion channels. Proc Natl Acad Sci U S A. 2005;102:18207–18212. doi: 10.1073/pnas.0509024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Link E, Edelmann L, Chou JH, et al. Tetanus toxin action: inhibition of neurotransmitter release linked to synaptobrevin proteolysis. Biochem Biophys Res Commun. 1992;189:1017–1023. doi: 10.1016/0006-291X(92)92305-H. [DOI] [PubMed] [Google Scholar]
- 160.Schiavo G, Benfenati F, Poulain B, et al. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 161.Südhof TC. The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- 162.Schoch S, Deák F, Königstorfer A, et al. SNARE function analyzed in synaptobrevin/VAMP knockout mice. Science. 2001;294:1117–1122. doi: 10.1126/science.1064335. [DOI] [PubMed] [Google Scholar]
- 163.Yamamoto M, Wada N, Kitabatake Y, et al. Reversible suppression of glutamatergic neurotransmission of cerebellar granule cells in vivo by genetically manipulated expression of tetanus neurotoxin light chain. J Neurosci. 2003;23:6759–6767. doi: 10.1523/JNEUROSCI.23-17-06759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nakashiba T. Transgenic inhibition of synaptic transmission reveals role of CA3 output in hippocampal learning. Science. 2008;1260:1260–1264. doi: 10.1126/science.1151120. [DOI] [PubMed] [Google Scholar]
- 165.Collier RJ. Diphtheria toxin: mode of action and structure. Bacteriol Rev. 1975;39:54–85. doi: 10.1128/br.39.1.54-85.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gogos JA, Osborne J, Nemes A, et al. Genetic ablation and restoration of the olfactory topographic map. Cell. 2000;103:609–620. doi: 10.1016/S0092-8674(00)00164-1. [DOI] [PubMed] [Google Scholar]
- 167.Sakamoto M, Imayoshi I, Ohtsuka T, et al. Continuous neurogenesis in the adult forebrain is required for innate olfactory responses. Proc Natl Acad Sci U S A. 2011;108:8479–8484. doi: 10.1073/pnas.1018782108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Han L, Ma C, Liu Q, et al. A subpopulation of nociceptors specifically linked to itch. Nat Neurosci. 2013;16:174–182. doi: 10.1038/nn.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stirpe F, Gasperi-Campani A, Barbieri L, et al. Ribosome-inactivating proteins from the seeds of Saponaria officinalis L. (soapwort), of Agrostemma githago L. (corn cockle) and of Asparagus officinalis L. (asparagus), and from the latex of Hura crepitans L. (sandbox tree) Biochem J. 1983;216:617–625. doi: 10.1042/bj2160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wenk GL, Stoehr JD, Quintana G, et al. Behavioral, biochemical, histological, and electrophysiological effects of 192 IgG-saporin injections into the basal forebrain of rats. J Neurosci. 1994;14:5986–5995. doi: 10.1523/JNEUROSCI.14-10-05986.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wiley RG, Kline Iv RH. Neuronal lesioning with axonally transported toxins. J Neurosci Methods. 2000;103:73–82. doi: 10.1016/S0165-0270(00)00297-1. [DOI] [PubMed] [Google Scholar]
- 172.Yoder RM, Pang KCH. Involvement of GABAergic and cholinergic medial septal neurons in hippocampal theta rhythm. Hippocampus. 2005;15:381–392. doi: 10.1002/hipo.20062. [DOI] [PubMed] [Google Scholar]
- 173.Antonucci F, Alpar A, Kacza J, et al. Cracking down on inhibition: selective removal of GABAergic interneurons from hippocampal networks. J Neurosci. 2012;32:1989–2001. doi: 10.1523/JNEUROSCI.2720-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hamlin AS, Windels F, Boskovic Z, et al. Lesions of the basal forebrain cholinergic system in mice disrupt idiothetic navigation. PLoS One. 2013;8:e53472. doi: 10.1371/journal.pone.0053472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Suzuki R, Rahman W, Rygh LJ, et al. Spinal-supraspinal serotonergic circuits regulating neuropathic pain and its treatment with gabapentin. Pain. 2005;117:292–303. doi: 10.1016/j.pain.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 176.Potts JT, Fong AY, Anguelov PI, et al. Targeted deletion of neurokinin-1 receptor expressing nucleus tractus solitarii neurons precludes somatosensory depression of arterial baroreceptor-heart rate reflex. Neuroscience. 2007;145:1168–1181. doi: 10.1016/j.neuroscience.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Akiyama T, Nguyen T, Curtis E, et al. A central role for spinal dorsal horn neurons that express neurokinin-1 receptors in chronic itch. Pain. 2015;156:1240–1246. doi: 10.1097/j.pain.0000000000000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Antonucci F, Rossi C, Gianfranceschi L, et al. Long-distance retrograde effects of botulinum neurotoxin A. J Neurosci. 2008;28:3689–3696. doi: 10.1523/JNEUROSCI.0375-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Restani L, Giribaldi F, Manich M, et al. Botulinum neurotoxins A and E undergo retrograde axonal transport in primary motor neurons. PLoS Pathog. 2012;8:e1003087. doi: 10.1371/journal.ppat.1003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Papagiannopoulou D, Vardouli L, Dimitriadis F, et al. Retrograde transport of radiolabelled botulinum neurotoxin type A to the CNS after intradetrusor injection in rats. BJU Int 2015 Apr 24 [Epub ahead of print]. [DOI] [PubMed]
- 181.Naglich JG, Eidels L. Isolation of diphtheria toxin-sensitive mouse cells from a toxin-resistant population transfected with monkey DNA. Proc Natl Acad Sci U S A. 1990;87:7250–7254. doi: 10.1073/pnas.87.18.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Buch T, Heppner FL, Tertilt C, et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- 183.Han JH, Kushner SA, Yiu AP, et al. Selective erasure of fear memory. Science. 2009;323:1492–1496. doi: 10.1126/science.1164139. [DOI] [PubMed] [Google Scholar]
- 184.Gonatas NK, Harper C, Mizutani T, et al. Superior sensitivity agglutinin. J Histochem Cytochem. 1979;27:728–734. doi: 10.1177/27.3.90065. [DOI] [PubMed] [Google Scholar]
- 185.Masco D, Van de Walle M, Spiegel S. Interaction modulates of ganglioside GM1 with the B subunit of cholera toxin growth and differentiation of neuroblastoma. J Neurosci. 1991;11:2443–2452. doi: 10.1523/JNEUROSCI.11-08-02443.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lanciego JL, Wouterlood FG. A half century of experimental neuroanatomical tracing. J Chem Neuroanat. 2011;42:157–183. doi: 10.1016/j.jchemneu.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 187.Kaneda Y, Uchida T, Mekada E, et al. Entry of diphtheria toxin into cells: possible existence of cellular factor(s) for entry of diphtheria toxin into cells was studied in somatic cell hybrids and hybrid toxins. J Cell Biol. 1984;98:466–472. doi: 10.1083/jcb.98.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Oeltmann TN, Wiley RG. Wheat germ agglutinin-ricin A-chain conjugate is neuronotoxic after vagal injection. Brain Res. 1986;377:221–228. doi: 10.1016/0006-8993(86)90862-0. [DOI] [PubMed] [Google Scholar]
- 189.Chaddock J a, Purkiss JR, Friis LM, et al. Inhibition of vesicular secretion in both neuronal and nonneuronal cells by a retargeted endopeptidase derivative of Clostridium botulinum neurotoxin type A. Infect Immun 2000;68:2587–2593. [DOI] [PMC free article] [PubMed]
- 190.Nichols NL, Vinit S, Bauernschmidt L, et al. Respiratory function after selective respiratory motor neuron death from intrapleural CTB–saporin injections. Exp Neurol. 2015;267:18–29. doi: 10.1016/j.expneurol.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dell’Anno MT, Caiazzo M, Leo D, et al. Remote control of induced dopaminergic neurons in parkinsonian rats. J Clin Invest. 2014;124:3215–3229. doi: 10.1172/JCI74664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Seeger-Armbruster S, Bosch-Bouju C, Little STC, et al. Patterned, but not tonic, optogenetic stimulation in motor thalamus improves reaching in acute drug-induced Parkinsonian rats. J Neurosci. 2015;35:1211–1216. doi: 10.1523/JNEUROSCI.3277-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cheng MY, Wang EH, Woodson WJ, et al. Optogenetic neuronal stimulation promotes functional recovery after stroke. Proc Natl Acad Sci U S A. 2014;111:12913–12918. doi: 10.1073/pnas.1404109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Alilain WJ, Li X, Horn KP, et al. Light-induced rescue of breathing after spinal cord injury. J Neurosci. 2008;28:11862–11870. doi: 10.1523/JNEUROSCI.3378-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Towne C, Montgomery KL, Iyer SM, et al. Optogenetic control of targeted peripheral axons in freely moving animals. PLoS One. 2013;8:e72691. doi: 10.1371/journal.pone.0072691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kätzel D, Nicholson E, Schorge S, et al. Chemical-genetic attenuation of focal neocortical seizures. Nat Commun. 2014;5:3847. doi: 10.1038/ncomms4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Krook-Magnuson E, Soltesz I. Beyond the hammer and the scalpel: selective circuit control for the epilepsies. Nat Neurosci. 2015;18:331–338. doi: 10.1038/nn.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Krook-Magnuson E, Szabo GG, Armstrong C, et al. Cerebellar directed optogenetic intervention inhibits spontaneous hippocampal seizures in a mouse model of temporal lobe epilepsy. eNeuro. 2014;1:e.2014. doi: 10.1523/ENEURO.0005-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Marc R, Pfeiffer R, Jones B. Retinal prosthetics, optogenetics, and chemical photoswitches. ACS Chem Neurosci. 2014;5:895–901. doi: 10.1021/cn5001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Barrett JM, Degenaar P, Sernagor E. Blockade of pathological retinal ganglion cell hyperactivity improves optogenetically evoked light responses in rd1 mice. Front Cell Neurosci. 2015;9:1–14. doi: 10.3389/fncel.2015.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sagdullaev Neurotherapeutics 2016;13:2. in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Required Author Forms Disclosure forms provided by the authors are available with the online version of this article. (PDF 3166 kb)