Abstract
Genome size (or C-value) can present a wide range of values among eukaryotes. This variation has been attributed to differences in the amplification and deletion of different noncoding repetitive sequences, particularly transposable elements (TEs). TEs can be activated under different stress conditions such as interspecific hybridization events, as described for several species of animals and plants. These massive transposition episodes can lead to considerable genome expansions that could ultimately be involved in hybrid speciation processes. Here, we describe the effects of hybridization and introgression on genome size of Drosophila hybrids. We measured the genome size of two close Drosophila species, Drosophila buzzatii and Drosophila koepferae, their F1 offspring and the offspring from three generations of backcrossed hybrids; where mobilization of up to 28 different TEs was previously detected. We show that hybrid females indeed present a genome expansion, especially in the first backcross, which could likely be explained by transposition events. Hybrid males, which exhibit more variable C-values among individuals of the same generation, do not present an increased genome size. Thus, we demonstrate that the impact of hybridization on genome size can be detected through flow cytometry and is sex-dependent.
Keywords: genome size, flow cytometry, hybrids, Drosophila, transposable elements, AFLP markers
Introduction
Genome size, also known as C-value, is the measure of DNA mass per haploid nucleus (Gregory 2005b) and represents a crucial feature for the understanding of genome evolution and speciation (Kraaijeveld 2010). Although this value is constant within individuals, eukaryotic species present a wide variation in genome size, reaching differences higher than 600,000-fold (Gregory 2005a). The lack of correlation between organisms' genome size and their number of genes or their complexity was called the “C-value paradox,” an issue that was cleared up by the finding that genes are not the only (nor the major) components of genomes. It is now known that a large fraction of the genome of most eukaryotic organisms is noncoding repetitive DNA, including transposable elements (TEs), pseudogenes, introns, and satellites (Gregory 2005a). Together with polyploidization, transposition is considered to be one of the major forces of eukaryotic genome expansion (Kidwell 2002): for instance, the maize genome doubled its size during the last few million years after a series of transposition bursts (SanMiguel et al. 1996). In the Drosophila genus, some studies have demonstrated that TE amount can account for genome size variation between species (Boulesteix et al. 2006), as well as between populations of the same species (Vieira et al. 2002).
Although TE mobilization rates are usually low, spontaneous transposition bursts have been reported, often linked to different stressful conditions (reviewed in García Guerreiro 2012). Interestingly, some of these bursts seem to share timing with species radiation episodes (Rebollo et al. 2010). The merge of two different genomes during interspecific hybridization events can be considered a genomic stress condition, which has been shown to lead to transposition bursts in several species. For example, different macropodid hybrids present amplified centromeres due to the presence of TE-related sequences (O’Neill et al. 1998; Metcalfe et al. 2007), and retrotransposon proliferation has also been described in three sunflower species of hybrid origin (Ungerer et al. 2006).
In Drosophila, the first evidence of hybrid TE mobilization was the detection of a new insertion of the pDv111 element in Drosophila virilis–Drosophila littoralis hybrids by in situ hybridization (Evgen’ev et al. 1982). In the same way, an increase of transposition of the retrotransposon Osvaldo was reported in hybrids between Drosophila buzzatii and Drosophila koepferae (Labrador et al. 1999). More recently, a genome-wide study using AFLP markers in these same hybrids demonstrated that not only Osvaldo but at least 28 different TEs were mobilized (Vela et al. 2014), suggesting that transposition in D. buzzatii–D. koepferae hybrids is a widespread phenomenon. Other studies at a transcription level support the hypothesis of a TE derepression in hybrids between Drosophila species (Kelleher et al. 2012; Carnelossi et al. 2014; García Guerreiro 2015), as well as in hybrid lake whitefishes (Dion-Côté et al. 2014) and sunflowers (Renaut et al. 2014).
Massive bursts of transposition can cause drastic changes in genome size and composition. For instance, three hybrid-derived sunflower species present genome sizes 50% larger than parental species (Baack et al. 2005). This study shows that interspecific hybridization is a source of evolutionary novelties that may be at the origin of new species by the means of TE activation (reviewed in Fontdevila 2005; Rebollo et al. 2010). However, synthetic F1 and F6 hybrids between the same sunflower parental species do not present a genome increase, and neither do plants from hybrid-zone populations (Baack et al. 2005; Kawakami et al. 2011). These last results show that genome expansion is not a shared feature of all interspecific hybrids, which concurs with studies in other plants, such as oil palm, sea buckthorns, and grasses, where hybrids presented intermediate genome sizes between parental species (Mahelka et al. 2005; Zhou et al. 2010; Camillo et al. 2014).
In animals, despite the few studies describing TE activation in hybrids (O’Neill et al. 1998; Labrador et al. 1999; Metcalfe et al. 2007; Vela et al. 2014), information about the effect of hybridization on hybrid genome size is scarce. D. buzzatii and D. koepferae are two cactophilic species that only produce hybrid offspring when crossing D. buzzatii males with D. koepferae females—the reciprocal cross does not produce adult offspring (Marin et al. 1993). As previously mentioned, mobilization of different TEs in hybrids between these species has been reported by in situ hybridization, AFLPs and transposon display techniques (Labrador et al. 1999; Vela et al. 2011, 2014). We have estimated the genome size of these two parental species and their F1 hybrids, as well as three subsequent generations of backcrossed hybrids (fig. 1). Thus, the present work aims to analyze the impact of interspecific hybridization, at different stages of genomic introgression, on genome size of male and female Drosophila hybrids.
Fig. 1.—
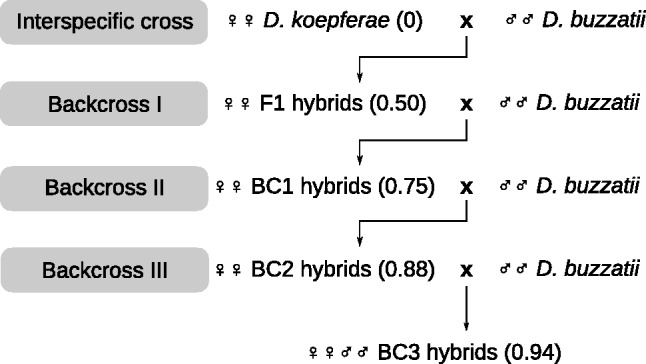
Diagram of crosses. A first interspecific massal cross of ten D. koepferae females with ten D. buzzatii males was followed by three subsequent backcrosses of ten hybrid females with ten D. buzzatii males. The D. buzzatii expected mean genome fraction of each generation is presented in parentheses.
Materials and Methods
Drosophila Stocks and Crosses
Six interspecific crosses were performed between ten D. buzzatii males (Bu28 strain) and ten D. koepferae females (Ko2 strain). Both strains are inbred lines originated by natural populations collected, respectively, in Bolivia and Argentina (Morán and Fontdevila 2014). Each cross was followed by three generations of backcrossing of ten hybrid females with ten D. buzzatii males. All stocks and crosses were reared at 25 °C in a standard Drosophila medium.
Genome Size Estimation
Genome size of D. buzzatii, D. koepferae, and their hybrids was estimated for males and females separately using flow cytometry technique. Nuclei were extracted from three heads of exactly 4 days-old flies, using D. virilis as internal control standard. Heads were homogenized in Galbraith buffer (30 mM trisodium citrate, 10−4 triton X-100, 2 μg/ml RNAse A, 20 mM MOPS, 21.3 mM MgCl2) with 0.1 mg/ml propidium iodide (pH 7.2). After two filtering steps through 140 and 30-micron nylon meshes, samples were analyzed on a FACSCanto II flow cytometer fitted with an argon laser (488 nm wavelength). The relative fluorescence intensity between our flies and D. virilis, whose genome size estimate is 0.34 pg (Gregory and Johnston 2008), was determined. We performed 5–6 biological replicates for parental samples and 8–10 for hybrids (supplementary table S3, Supplementary Material online).
Statistical Analyses
Comparisons between parental species genome sizes were performed using the nonparametric Wilcoxon rank sum test (Mann and Whitney 1947), while hybrid genome size estimates were compared to a single theoretical mean (specific to each generation) with the Wilcoxon signed-rank test (Wilcoxon 1945). The single theoretical value specific to each generation was calculated for males and females separately, as follows:
where is the D. buzzatii () or D. koepferae () mean genome fraction of each generation (for example, for BC1, and ) and is the mean genome size of D. buzzatii () or D. koepferae ().
AFLP Genotyping
AFLP technique was suitable for our study because it did not require prior information on our species sequences (D. koepferae available sequences are scarce) and had previously been used in our species and their hybrids (Morán and Fontdevila 2014; Vela et al. 2014). Markers were obtained following the protocol described in Vela et al. 2011, from six hybrid crosses used in a former study (Vela et al. 2014). Contrary to the previous study, where instability markers were checked, we here identified D. koepferae-specific markers for ten primer combinations (supplementary table S2, Supplementary Material online). The presence of these markers was then assessed in F1 and BC1 hybrids, as detailed in supplementary fig. S1, Supplementary Material online. Finally, we determined the mean number of markers found per individual per family as explained in supplementary table S2, Supplementary Material online.
Results and discussion
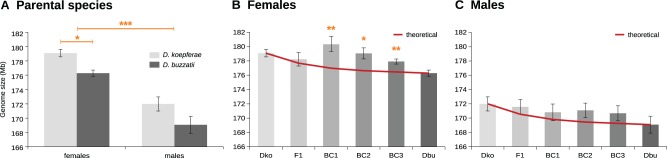
As a first goal, we have determined both D. buzzatii and D. koepferae parental genome sizes to assess differences between them, and also between males and females. Our results show that D. buzzatii presents a mean C-value of 176.28 Mb for females and 169.07 Mb for males (fig. 2A). These values are significantly higher (P = 0.022, supplementary table S1A, Supplementary Material online) than the 146–153 Mb previously reported by other authors (Guillén et al. 2015). It is known that differences in the estimated values can depend on the technique used (flow cytometry vs. densitometry), the analyzed tissues (heads vs. testes) or even on fly rearing conditions (Nardon et al. 2003). Furthermore, it is important to note that the genome size reference used in the former study (Guillén et al. 2015) was the Drosophila mojavensis genome assembly size, which could likely suppose an a priori underestimation due to assembling issues. Indeed, most of the repeated sequences are not assembled and we know they can contribute to genome size variation. On the other hand, intraspecific variation in Drosophila genome size among different strains or populations has been reported in several studies (Vieira et al. 2002; Bosco et al. 2007; Gregory and Johnston 2008; Ellis et al. 2014). These differences have been attributed to changes in TE (Vieira et al. 2002) and satellite DNA amounts (Bosco et al. 2007), and seem to be correlated with several life history traits and metabolism genes expression (Ellis et al. 2014).
Fig. 2.—
(A) Parental species mean genome size. *: P value < 0.05; **: P value < 0.01; ***: P value < 0.001 (Wilcoxon rank sum test W significant differences between species and sexes). (B and C) Mean genome size for parental species and all hybrid generations (gray bars) compared with theoretical mean values (red line) for female (B) and male (C) samples. Dbu: D. buzzatii; Dko: D. koepferae. Error bars represent standard error. *: P value < 0.05; **: P value < 0.01 Not useful (Wilcoxon signed-rank test V significant differences comparing experimental measures with the theoretical value).
In this study, we globally observe that parental females have significantly larger genomes than males (P = 1.48E-06, supplementary table S1A Supplementary Material online), with differences of approximately 7 Mb for both species (fig. 2A). Similar results have been described in Drosophila mauritania or Drosophila hydei (Girard and Hannon 2008), but Drosophila melanogaster presents equivalent genome sizes for both sexes (Vieira et al. 2002) and Drosophila simulans males exhibit larger genomes than females (Vieira et al. 2002). However, different results have been found in other strains of the latter two species (Gregory and Johnston 2008), indicating that genome size differences between males and females are strain-specific and likely depend on specific increases of repetitive DNA in the Y chromosome heterochromatin (Vieira et al. 2002). In our species, we expect females to have a higher genome size than males because X chromosome is known to be longer than Y (Wasserman 1962; Fontdevila et al. 1988). Interestingly, the standard errors observed within replicates are ≈2-fold higher in males than in females, showing that males present greater genome size variability (supplementary table S1, Supplementary Material online). The dynamic gene content of the Y chromosome, which also contains a high amount of repetitive sequences, might account for this diversity (Bernardo Carvalho et al. 2009).
Differences between species are significant for females (P = 0.015, supplementary table S1A, Supplementary Material online), with D. koepferae genome about 3 Mb larger than D. buzzatii (fig. 2A). No significant difference was observed in males (P = 0.126, supplementary table S1A, Supplementary Material online), which is probably due to the lower genome size and the higher variability found in male samples.
According to our null hypothesis, the genome size of hybrids (F1 and backcrosses) would present intermediate values between parental species and would be proportional to the D. buzzatii/D. koepferae genome fractions at each generation (fig. 1). Thus, we have compared the C-values of each hybrid generation to a theoretical weighted mean, reflecting the expected mean D. buzzatii introgression percentage in the hybrid genomes, assuming independent assortment of chromosomes during meiosis (see Materials and Methods). The accuracy of this assumption has been tested through AFLP genotyping of hybrids and parents: we have used 70 AFLP markers specific to D. koepferae and assessed which proportion of these markers is transmitted to hybrid progeny (see below).
In females, we show that the mean genome size of the four hybrid generations is higher than the theoretical value (fig. 2B), with statistically significant differences for the three backcrosses (supplementary table S1B, Supplementary Material online). The most striking results occur in the first backcross (BC1): the mean C-value (180.37 Mb) increases compared with the F1 generation (178.23 Mb), and is also higher than in both parental species (176.28 and 179.08 Mb). These results are concordant with the transposition-related instability observed previously in our hybrids, where new insertions of 28 different TEs, including retrotransposons and DNA transposons, were detected in the three backcrosses (Vela et al. 2014). In the case of F1, the vast majority of the detected instability markers were not transmitted to BC1, showing that the putative transposition events of F1 take place after meiosis (Vela 2012), which is also coherent with our results: somatic transpositions are not expected to cause a genome size increase. TE activation in hybrids seems to be caused by the failure of epigenetic repression mechanisms (Michalak 2009), such as histone methylation or small RNA biogenesis. In Drosophila ovaries, TEs are mainly regulated by piRNAs, a kind of small RNAs associated to Piwi proteins. Differences in piRNA pools between parental species, or incompatibilities between their piRNA pathway effector proteins, might lead to a TE silencing failure in hybrids. If a TE derepression took place in F1 ovaries at a transcriptional level, as shown for D. simulans–D. melanogaster hybrids (Kelleher et al. 2012), we would expect to detect new insertions in the following generations. Thus, new TE insertions could likely be responsible for the genome size increase observed after F1.
It is worth noting that other phenomena could also account for the observed genome expansion, such as the amplification of satellites or other noncoding repetitive sequences (Bosco et al. 2007), which are responsible for the large Drosophila orena genome (Boulesteix et al. 2006). On the contrary, polyploidization can be discarded, since early studies of D. buzzatii–D. koepferae hybrids, based in in situ hybridization (Labrador et al. 1999), never reported a case of hybrid abnormal karyotype due to genome duplication.
Finally, a transmission bias favoring the larger parental genome, D. koepferae, could also be consistent with our results (e.g., due to reduced recombination or differential gamete viability). In order to test this hypothesis, we have determined the inheritance of 70 D. koepferae-specific AFLP markers in F1 and BC1 hybrids from six different crosses. Our results, summarized in supplementary table S2, Supplementary Material online, show that almost 100% (92.9–97.1%) of the studied D. koepferae-specific markers are found in F1, as expected: all F1 individuals have an entire haploid copy of the D. koepferae genome. In the BC1, between 11.8% and 72.9% of the markers are found per individual (supplementary table S2, Supplementary Material online). This variability was also predictable, because inheritance of D. koepferae markers depends on the chromosomal assortment and recombination events occurring in each F1 gamete. Thus, it is not surprising that BC1 and BC2 hybrids present higher standard errors on genome size measurements than parental species (supplementary table S1B, Supplementary Material online and fig 2B). The average proportion of D. koepferae markers found in BC1, 32.4% (95% confidence interval: 11.6–53.2%), is lower than the expected mean of 50%, which suggests that either the transmission of the smallest parental genome (D. buzzatii) is favored in BC1 hybrids, or there is not any transmission bias. It is also worth noting than even considering the most extremely biased case (P = (½)5 = 0.03), in which 1) there is no recombination between D. buzzatii and D. koepferae chromosomes and 2) all individuals inherit all five D. koepferae chromosomes from their hybrid mothers; the D. koepferae genome fraction in backcrossed hybrids would be of 50% (as in F1). Assuming these improbable particulars, genome size estimates remain significantly higher than the expected for BC1 (Wilcoxon signed-rank test V = 49, P = 0.027).
Despite the genome size is higher than expected in all backcrosses (fig. 2B), its value actually decreases through generations after BC1 (fig. 2B). In rice (Oryza sativa), an important increase of Tos17 and RCS1 retrotransposons copy number was observed after introgression with Zizania latifolia, but no additional insertions were detected after a few generations (Liu and Wendel 2000), meaning that TE mobilization was by then controlled. Thereby, we can suppose that after a few generations of introgression, the preponderance of one of the parental genomes mitigates incompatibilities and palliates the hybridization effects. In this way, we can hypothesize that a greater transposition control in our hybrids would take place after BC1, which according to previous studies is true for the transposon Galileo, but not for Helena (high transposition rates observed also in BC2) and Osvaldo (higher transposition rates in BC3) (Vela et al. 2014). However, these elements represent only a small subset of these species’ TEs and may not be representative of the whole set behavior.
The simple backcrossing with D. buzzatii (species with smallest genome) could by itself lead to the observed genome size decrease after BC1, but active mechanisms involving genome reduction might also be involved, especially those implicated in TE control. For instance, it is known that internal and complete deletions of TE copies can act as a prevention mechanism against genome invasions (Petrov and Hartl 1998; Liu and Wendel 2000; Senerchia et al. 2015), the latter being guided by the presence of multiple TE copies through recombination events.
The observed genome increase in hybrid females could also be a technical artefact due to changes in chromatin topology. In hybrids, the failure to maintain chromatin integrity could improve the accessibility of DNA to fluorochromes (Nardon et al. 2003), resulting in an increase of genome size estimates. However, this hypothesis can be discarded because the lowest levels of chromatin compaction are expected in F1, whereas the highest genome size measures belong to BC1.
Regarding our hybrid males, it is worth mentioning that they are all sterile until BC3, when fertility is recovered for some individuals (Morán and Fontdevila 2014). Here, we show that all hybrid generations present intermediate genome size values between D. buzzatii and D. koepferae (fig. 2C and supplementary table S1B, Supplementary Material online). Although the mean C-value of each generation is higher than the theoretical, differences are not significant (supplementary table S1B, Supplementary Material online), meaning that the impact of hybridization and introgression on genome size is negligible in males. This seems contradictory with the fact that new TE insertions in our hybrids were also detected in males (Vela et al. 2014), where Osvaldo transcription rates were higher than in parents (García Guerreiro 2015). However, these male transposition events were thought to be partly somatic (Vela et al. 2014), and thus would not necessarily lead to a genome expansion. Furthermore, other transposons, such as Helena, seem to be repressed in hybrid males (Romero-Soriano and García Guerreiro 2016). This shows that TE regulation patterns differ between sexes and depend on the studied TEs, as proposed in a recent study (Senti et al. 2015). Indeed, the biogenesis of piRNAs has been shown to differ between males and females (Nagao et al. 2010; Siomi et al. 2010). Although we cannot rule out the involvement of particular TEs in the hybrid male sterile phenotype, our results suggest that, unlike hybrid females, males do not present a massive TE amplification.
Conclusions
We have shown that the increased transpositional activity previously reported in D. buzzatii–D. koepferae hybrids has an impact on hybrid female genome size. For the first time, an actual genome size increase due to interspecific hybridization has been described in animals. This allows us to validate flow cytometry as a technique to detect changes in C-value of Drosophila hybrids, probably due to transposition events. In males, the effects of hybridization are not significant, but we must note that changes in their genome size would lack direct evolutionary consequences, since they are all sterile until some individuals recover their fertility in BC3.
Supplementary Material
Supplementary figure S1 and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors want to thank M. Santos and two anonymous reviewers for helpful discussions. This work was supported by research grant CGL2013-42432-P from the Ministerio de Economía y Competitividad (Spain) and grant 2014 SGR 1346 from Generalitat de Catalunya to the Grup de Genòmica, Bioinformàtica i Biologia Evolutiva (GGBE). V.R.-S. was supported by a PIF PhD fellowship from the Universitat Autònoma de Barcelona (Spain).
Literature Cited
- Baack EJ, Whitney KD, Rieseberg LH. 2005. Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytol. 167:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo Carvalho A, Koerich LB, Clark AG. 2009. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 25:270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G, Campbell P, Leiva-Neto JT, Markow TA. 2007. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics 177:1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulesteix M, Weiss M, Biémont C. 2006. Differences in genome size between closely related species: the Drosophila melanogaster species subgroup. Mol Biol Evol. 23:162–167. [DOI] [PubMed] [Google Scholar]
- Camillo J, et al. 2014. Reassessment of the genome size in Elaeis guineensis and Elaeis oleifera, and its interspecific hybrid. Genomics Insights 7:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnelossi EAG, et al. 2014. Specific activation of an I-like element in Drosophila interspecific hybrids. Genome Biol Evol. 6:1806–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion-Côté AM, Renaut S, Normandeau E, Bernatchez L. 2014. RNA-seq reveals transcriptomic shock involving transposable elements reactivation in hybrids of young lake whitefish species. Mol Biol Evol. 31:1188–1199. [DOI] [PubMed] [Google Scholar]
- Ellis LL, et al. 2014. Intrapopulation genome size variation in D. melanogaster reflects life history variation and plasticity. PLoS Genet. 10:e1004522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgen’ev MB, Yenikolopov GN, Peunova NI, Ilyin YV. 1982. Transposition of mobile genetic elements in interspecific hybrids of Drosophila. Chromosoma 85:375–386. [DOI] [PubMed] [Google Scholar]
- Fontdevila A. 2005. Hybrid genome evolution by transposition. Cytogenet Genome Res. 110:49–55. [DOI] [PubMed] [Google Scholar]
- Fontdevila A, et al. 1988. Drosophila koepferae: a new member of the Drosophila serido (Diptera: Drosophilidae) superspecies taxon. Ann Entomol Soc Am. 81:380–385. [Google Scholar]
- García Guerreiro M. 2012. What makes transposable elements move in the Drosophila genome? Heredity 108:461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Guerreiro MP. 2015. Changes of Osvaldo expression patterns in germline of male hybrids between the species Drosophila buzzatii and Drosophila koepferae. Mol Genet Genomics. 290:1471–1483. [DOI] [PubMed] [Google Scholar]
- Girard A, Hannon GJ. 2008. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 18:136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. 2005a. Genome size evolution in animals. In: The Evolution of the Genome, edited by Gregory T.R. San Diego: Elsevier. pp. 3–87. [Google Scholar]
- Gregory TR. 2005b. The C-value enigma in plants and animals: a review of parallels and an appeal for partnership. Ann Bot. 95:133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR, Johnston JS. 2008. Genome size diversity in the family Drosophilidae. Heredity 101:228–238. [DOI] [PubMed] [Google Scholar]
- Guillén Y, et al. 2015. Genomics of ecological adaptation in cactophilic Drosophila. Genome Biol Evol. 7:349–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Dhakal P, Katterhenry AN, Heatherington CA, Ungerer MC. 2011. Transposable element proliferation and genome expansion are rare in contemporary sunflower hybrid populations despite widespread transcriptional activity of LTR retrotransposons. Genome Biol Evol.. 3:156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Edelman NB, Barbash DA. 2012. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 10:e1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115:49–63. [DOI] [PubMed] [Google Scholar]
- Kraaijeveld K. 2010. Genome size and species diversification. Evol Biol. 37:227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador M, Farré M, Utzet F, Fontdevila A. 1999. Interspecific hybridization increases transposition rates of Osvaldo. Mol Biol Evol. 16:931–937. [DOI] [PubMed] [Google Scholar]
- Liu B, Wendel JF. 2000. Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 43:874–880. [PubMed] [Google Scholar]
- Mahelka V, Suda J, Jarolímová V, Trávnicek P, Krahulec F. 2005. Genome size discriminates between closely related taxa Elytrigia repens and E. intermedia (Poaceae: Triticeae) and their hybrid. Folia Geobot. 40:367–384. [Google Scholar]
- Mann HB, Whitney DR. 1947. On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat. 18:50–60. [Google Scholar]
- Marin I, Ruiz A, Pla C, Fontdevila A. 1993. Reproductive relationships among ten species of the Drosophila repleta group from South America and the West Indies. Evolution 47:1616–1624. [DOI] [PubMed] [Google Scholar]
- Metcalfe CJ, et al. 2007. Genomic instability within centromeres of interspecific marsupial hybrids. Genetics 177:2507–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P. 2009. Epigenetic, transposon and small RNA determinants of hybrid dysfunctions. Heredity 102:45–50. [DOI] [PubMed] [Google Scholar]
- Morán T, Fontdevila A. 2014. Genome-wide dissection of hybrid sterility in Drosophila confirms a polygenic threshold architecture. J Hered. 105:381–396. [DOI] [PubMed] [Google Scholar]
- Nagao A, Mituyama T, Huang H, Chen D, Siomi MC. 2010. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 16:2503–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardon C, Weiss M, Vieira C, Biémont C. 2003. Variation of the genome size estimate with environmental conditions in Drosophila melanogaster. Cytometry A 55A:43–49. [DOI] [PubMed] [Google Scholar]
- O’Neill RJW, O’Neill MJ, Marshall Graves JA. 1998. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393:68–73. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Hartl DL. 1998. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol Biol Evol. 15:293–302. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Horard B, Hubert B, Vieira C. 2010. Jumping genes and epigenetics: towards new species. Gene 454:1–7. [DOI] [PubMed] [Google Scholar]
- Renaut S, Rowe HC, Ungerer MC, Rieseberg LH. 2014. Genomics of homoploid hybrid speciation: diversity and transcriptional activity of long terminal repeat retrotransposons in hybrid sunflowers. Philos Trans R Soc Lond B Biol Sci. 369:20130345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Soriano V, García Guerreiro MP. 2016. Expression of the retrotransposon Helena reveals a complex pattern of TE Deregulation in Drosophila Hybrids. PLoS One. doi: 10.1371/journal.pone.0147903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, et al. 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274:765–768. [DOI] [PubMed] [Google Scholar]
- Senerchia N, Parisod C, Parisod C. 2015. Genome reorganization in F1 hybrids uncovers the role of retrotransposons in reproductive isolation. Proc R Soc B Biol Sci. 282:20142874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti K-A, Jurczak D, Sachidanandam R, Brennecke J. 2015. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 29:1747–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Miyoshi T, Siomi H. 2010. piRNA-mediated silencing in Drosophila germlines. Semin Cell Dev Biol. 21:754–759. [DOI] [PubMed] [Google Scholar]
- Ungerer MC, Strakosh SC, Zhen Y. 2006. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr Biol. 16:R872–R873. [DOI] [PubMed] [Google Scholar]
- Vela Peralta DJ. 2012. Estudio de la inestabilidad genómica inducida por transposición en los híbridos interespecíficos de Drosophila buzzatii y Drosophila koepferae. Universitat Autònoma de Barcelona. [Google Scholar]
- Vela D, Fontdevila A, Vieira C, García Guerreiro MP. 2014. A genome-wide survey of genetic instability by transposition in Drosophila hybrids. PLoS One 9:e88992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela D, García Guerreiro MP, Fontdevila A. 2011. Adaptation of the AFLP technique as a new tool to detect genetic instability and transposition in interspecific hybrids. Biotechniques 50:247–250. [DOI] [PubMed] [Google Scholar]
- Vieira C, Nardon C, Arpin C, Lepetit D, Biémont C. 2002. Evolution of genome size in Drosophila. is the invader’s genome being invaded by transposable elements? Mol Biol Evol. 19:1154–1161. [DOI] [PubMed] [Google Scholar]
- Wasserman M. 1962. Cytological studies of the repleta group of the genus Drosophila. V. The mulleri subgroup. Univ Texas Publ. 6205:85–118. [Google Scholar]
- Wilcoxon F. 1945. Individual comparisons of grouped data by ranking methods. Biometrics Bull. 1:80–83. [DOI] [PubMed] [Google Scholar]
- Zhou X, et al. 2010. Genome size of the diploid hybrid species Hippophae goniocarpa and its parental species, H. rhamnoides ssp. sinensis and H. neurocarpa ssp. neurocarpa (Elaeagnaceae). Acta Biol Cracoviensia Ser Bot. 52:12–16 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.