Abstract
Eustigmatophyceae (Ochrophyta, Stramenopiles) is a small algal group with species of the genus Nannochloropsis being its best studied representatives. Nuclear and organellar genomes have been recently sequenced for several Nannochloropsis spp., but phylogenetically wider genomic studies are missing for eustigmatophytes. We sequenced mitochondrial genomes (mitogenomes) of three species representing most major eustigmatophyte lineages, Monodopsis sp. MarTras21, Vischeria sp. CAUP Q 202 and Trachydiscus minutus, and carried out their comparative analysis in the context of available data from Nannochloropsis and other stramenopiles, revealing a number of noticeable findings. First, mitogenomes of most eustigmatophytes are highly collinear and similar in the gene content, but extensive rearrangements and loss of three otherwise ubiquitous genes happened in the Vischeria lineage; this correlates with an accelerated evolution of mitochondrial gene sequences in this lineage. Second, eustigmatophytes appear to be the only ochrophyte group with the Atp1 protein encoded by the mitogenome. Third, eustigmatophyte mitogenomes uniquely share a truncated nad11 gene encoding only the C-terminal part of the Nad11 protein, while the N-terminal part is encoded by a separate gene in the nuclear genome. Fourth, UGA as a termination codon and the cognate release factor mRF2 were lost from mitochondria independently by the Nannochloropsis and T. minutus lineages. Finally, the rps3 gene in the mitogenome of Vischeria sp. is interrupted by the UAG codon, but the genome includes a gene for an unusual tRNA with an extended anticodon loop that we speculate may serve as a suppressor tRNA to properly decode the rps3 gene.
Keywords: Eustigmatophyceae, evolution, phylogenomics, split genes, Stramenopiles, suppressor tRNA
Introduction
Mitochondria are organelles of the eukaryotic cell that evolved early in the eukaryote evolution from an endosymbiotic α-proteobacterium (Lang and Burger 2012). An extremely reduced endosymbiont genome is still maintained in most mitochondria, except some mitochondrion-related organelles of anaerobic protists that have dispensed with the genome completely (Makiuchi and Nozaki 2014). The least derived mitochondrial genomes (mitogenomes) known were found in an obscure protist group called jakobids and harbor up to 100 genes (including protein-coding genes and genes for noncoding RNA molecules) (Burger et al. 2013), whereas mitogenomes of Myzozoa (a subgroup of alveolates) code for only three proteins (Burger et al. 2012). The architecture of the mitogenome also varies considerably among eukaryotic lineages; whereas the most common structure is a contiguous circular-mapping DNA molecule, a contiguous linear genome or genomes segmented into multiple circular or linear molecules are known from some taxa (Gray et al. 2004; Burger et al. 2012). Although considerable progress has been made in exploring the diversity and evolution of mitogenomes across the whole span of the eukaryote phylogeny, representative genome sequences are still limited or altogether lacking for a large number of lineages, especial microbial eukaryotes (protists) (Smith 2016). Hence, further sampling is necessary to get a fuller picture of the evolutionary history of mitochondria and their genomes.
Ochrophyta (hereafter ochrophytes; often called heterokontophytes or stramenopile algae) are probably the largest and ecologically most significant algal phylum. They are defined by the presence of a secondary plastid of a rhodophyte origin combined with a suite of features that characterize an even more inclusive group of eukaryotes called stramenopiles or heterokonts (which additionally include such groups as oomycetes, labyrinthulids, opalinids, or bicosoecids) (Adl et al. 2012). Ochrophytes are currently classified into some 15 classes, although the independent status of some of these classes is debated and novel phylogenetic lineages have been discovered that cannot be accommodated into any of the established classes (Andersen 2004; Yang et al. 2012; Cavalier-Smith and Scoble 2013). The most specious ochrophyte class are diatoms (Bacillariophyceae sensu lato), whose biology has been extensively studied owing to their enormous importance in freshwater and marine ecosystems. Other familiar ochrophyte groups include the multicellular brown algae (Phaeophyceae), the golden algae (Chrysophyceae), or the class Raphidophyceae remarkable for comprising algae often forming toxic blooms. At least two mitogenome sequences have been determined for each of the above-mentioned ochrophyte classes (supplementary table S1, Supplementary Material online), providing an insight into the mitogenome diversity in ochrophytes. However, our knowledge about the mitogenomes of most ochrophyte classes remains limited or even completely lacking.
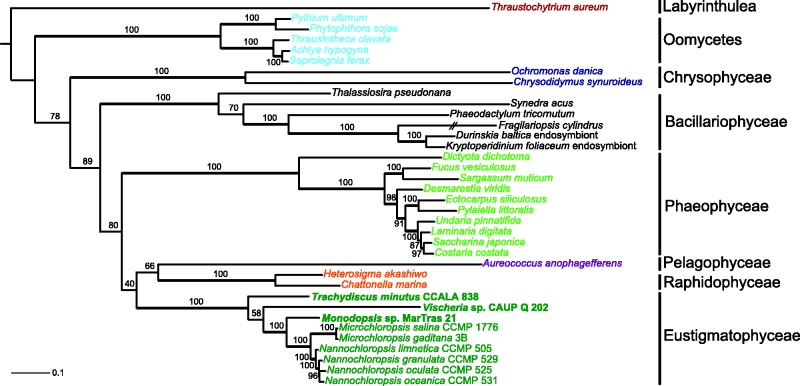
An ochrophyte lineage that had remained rather obscure for nearly 40 years since its description (Hibberd and Leedale 1970) only to rise to prominence in the past few years is the class Eustigmatophyceae. Most known eustigmatophytes were originally considered as members of the class of yellow-green algae (Xanthophyceae), but ultrastructural, biochemical, and finally molecular phylogenetic data pointed to their status as an independent class. According to the most recent multi-gene phylogenetic analyses, Eustigmatophyceae are most closely related to a clade comprising the classes Chrysophyceae (including Synurophyceae) and Synchromophyceae, whereas the “true” Xanthophyceae are related to Phaeophyceae and some additional less well-known classes (Yang et al. 2012; Ševčíková et al. 2015). The most familiar taxon of the class Eustigmatophyceae is the genus Nannochloropsis, recently split by segregating two species into a separate genus Microchloropsis (Fawley et al. 2015), comprising several predominantly marine species of microalgae that are considered to have a big potential for biotechnologies, especially for production of biofuels owing to their capability to accumulate large amounts of lipids (Wang et al. 2014).
The significance of Nannochloropsis/Microchloropsis has been recently attested by several genome-sequencing projects (Radakovits et al. 2012; Vieler et al. 2012; Corteggiani Carpinelli et al. 2014; Wang et al. 2014), making eustigmatophytes an ochrophyte class with one of the most extensive genomic resources (second only to diatoms). These initiatives delivered not only more or less complete nuclear genome sequences, but together with additional dedicated studies (Wei et al. 2013; Starkenburg et al. 2014) provided complete organellar (plastid and mitochondrial) genome sequences for virtually all Nannochloropsis/Microchloropsis species known, including multiple isolates of the same species (supplementary table S1, Supplementary Material online). Analyses of the mitogenomes provided a lot of interesting evolutionary and functional insights, but without data from a broader sample of eustigmatophytes, it is unsure to what extent the features of Nannochloropsis/Microchloropsis mitogenomes are representative for eustigmatophytes in general. Indeed, recent studies has revealed an unexpected phylogenetic diversity of eustigmatophytes, including the existence of two deeply separated principal lineages, Eustigmatales and Goniochloridales (Přibyl et al. 2012; Fawley et al. 2014). In frame of a broader project to map the diversity and evolutionary history of eustigmatophytes, we set out to sequence and characterize mitogenomes of eustigmatophyte species representing lineages with different phylogenetic distances from Nannochloropsis/Microchloropsis.
Here, we provide a detailed comparative analysis of three new eustigmatophyte mitogenome sequences. We targeted the following taxa: 1) a new isolate belonging to the genus Monodopsis, which represents a lineage (together with the genus Pseudotetraëdriella) sister to Nannochloropsis/Microchloropsis, together constituting the family Monodopsidaceae; 2) a new isolate of the genus Vischeria, which represents a more distantly related clade still within Eustigmatales; 3) Trachydiscus minutus, a representative of Goniochloridales (the phylogenetic placement of the studied taxa as inferred from the 18S rRNA gene is apparent from supplementary fig. S1, Supplementary Material online). This sampling strategy allowed us to obtain a relatively comprehensive overview of the evolution of eustigmatophyte mitogenomes across the phylogeny of the whole group.
Materials and Methods
Algal Cultures, DNA Isolation, Sequencing, and Assembly of Mitogenome Sequences
A culture of T. minutus strain CCALA 838, isolated and characterized by Přibyl et al. (2012), was obtained from the Culture Collection of Autotrophic Organisms (CCALA), Academy of Sciences of the Czech Republic, Třeboň (kindly sent by the curator Pavel Přibyl). The strains Vischeria sp. CAUP Q 202 and Monodopsis sp. MarTras21 were kindly donated by Pavel Škaloud and Martina Pichrtová, respectively (Charles University in Prague, Faculty of Science, Prague, Czech Republic). Vischeria sp. CAUP Q 202 (hereafter Vischeria) was isolated in 2003 from soil of a ventarole at the Boreč Hill (Czech Republic) and the strain is available from the Culture Collection of Algae of the Charles University in Prague (CAUP; http://botany.natur.cuni.cz/algo/caup.html, last accessed February 19, 2016). Monodopsis sp. MarTras21 (hereafter Monodopsis) was isolated in 2009 from a small pond in a wet hummock meadow, Petuniabukta bay, Svalbard (Norway). The cultures were maintained on solid Bold’s Basal Medium (Nichols 1973) with and without vitamins (B1, B12) at 17 °C with continual illumination, or in liquid Bold’s Basal Medium in a Multi-Cultivator MC1000-OD (Photon System Instruments, Drasov, Czech Republic) at room temperature with continual illumination (60 E) and aeration.
Total DNA from all species was isolated from the algal cultures by modified Dellaporta protocol (Dellaporta et al. 1983) and purified by phenol–chloroform extraction. Sequencing was performed by a combination of the 454 pyrosequencing and Illumina methods (T. minutus) or using Illumina alone (MiSeq 150 bp paired-end one lane for Monodopsis, HiSeq 100 bp paired-end one lane for Vischeria). Details on DNA extraction and purification, library preparation, and sequencing are provided in supplementary methods, Supplementary Material online. 454 reads were assembled using Newbler (v 2.8. Roche), Illumina reads were trimmed using Trimmomatic (Bolger et al. 2014) and assembled using SPAdes 2.5.1 assembler (Bankevich et al. 2012). The complexity was reduced using Flash (Magoč and Salzberg 2011). The assemblies were searched for contigs containing genes homologous to known reference mitochondrial genes, revealing a single circular-mapping counting for each assembly. Errors were identified in the 454-based assembly of the T. minutus mitogenome, which were corrected by mapping Illumina reads using Bowtie2 mapper (Langmead and Salzberg 2012) and by manual verification. The newly determined mitogenome sequences were deposited at GenBank with accession numbers KU501220–KU501222.
For comparative purposes, we improved the assembly of the mitogenome sequence of the pelagophyte Aureococcus anophagefferens. The mitogenome of this species was excluded from the whole-genome assembly published previously for this species (Gobler et al. 2011), but a scaffold representing a major portion of the mitogenome (scaffold_85) could be downloaded from the A. anophagefferens genome database at the Joint Genome Institute (http://genome.jgi-psf.org/Auran1/Auran1.home.html, last accessed February 19, 2016). We used the original Sanger WGS reads available from the Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/wgs/ [last accessed February 19, 2016], prefix ACJI01) and Illumina-sequenced transcriptomic data available in Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/SRX275708, last accessed February 19, 2016) to iteratively fill gaps in the scaffold and to extend its ends. By this procedure, which led us to recruit into the assembly a small scaffold previously released as a part of the nuclear genome assembly (scaffold_2027, GenBank accession number ACJI01005131.1), we managed to obtain a circular-mapping sequence and to close all but one gap. Some regions in the assembled A. anophagefferens mitogenome sequence (available as supplementary data set S1, Supplementary Material online) are based solely on the transcriptome data, so this hybrid sequence must be interpreted with caution, as the actual genome sequence may look different due to transcript splicing or editing.
Genome Annotation and Analyses
Initial annotation of the newly sequenced eustigmatophyte mitogenomes was obtained using MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl, last accessed February 19, 2016). Predicted genes were individually checked to confirm their identity and borders using BLAST. In cases where the predicted protein appeared to possibly lack the actual N-terminus (as indicated by comparison with homologous proteins), the upstream genomic sequence was checked for the presence of an alternative initiation codon. AUG or other triplets (UUG) known to be used as initiation codons (Peabody 1989) in mitochondria were considered, except the orfY gene of Monodopsis, which appears to use AUU as the initiation codon (based on conservation of the N-terminal sequence of the OrfY protein). Intergenic regions were screened for possible features (short genes etc.) missed by MFannot by BLAST. 5S rRNA genes were identified by searching the mitogenomes with the covariance models developed by Valach et al. (2014) using the program INFERNAL 1.1rc2 (version from December 2012). Best E-value scores were reached using the covariance model for A + T-rich 5S rRNAs (mtAT-5S). The output of the search was used as a basis for building secondary structure models for the respective 5S rRNA molecules, which were further manually refined based on similarity with previously published model for Microchloropsis (=Nannochloropsis) salina 5S rRNA (Valach et al. 2014). The predicted secondary structures were visualized using VARNA (Darty et al. 2009). Circular genome maps were generated by combining outputs of the programs GenomeVx (Conant and Wolfe 2008) (gene order) and OGDraw (Lohse et al. 2013) (GC content graphs).
Whole-genome alignment of eustigmatophyte mitogenome sequences was generated using progressiveMauve (Darling et al. 2010) using default settings. Sequences of nuclear genes used for targeted analyses were identified with BLAST used to search various public databases (see supplementary table S2, Supplementary Material online) and our unpublished genome and transcriptome assemblies for T. minutus, Vischeria, and Monodopsis (individual sequences extracted from these assemblies were deposited at GenBank with accession numbers KU501223–KU501236). Possible presence of mitochondrial targeting signal (transit peptides) in proteins was tested using TargetP 1.1 (Emanuelsson et al. 2000). Transmembrane regions in TatA and OrfZ proteins were predicted using TMHMM 2.0 with default settings (Krogh et al. 2001). Multiple alignments of all protein sequences were obtained using the program MAFFT version 7 (http://mafft.cbrc.jp/alignment/server/ [last accessed February 19, 2016], Katoh and Standley 2013) with either default settings or (in case of Nad11 and Rps3) the L-INS-i iterative refinement method. The presence of a termination codon interrupting the coding sequence of the Vischeria rps3 gene in the gene transcript was tested by reverse transcription polymerase chain reaction (RT-PCR). Briefly, RNA was isolated using the Trizol method and treated by DNase I (TURBO DNA-free Kit, ThermoFisher Scientific, Waltham), cDNA was synthesized using Tetro cDNA Synthesis Kit (Bioline Inc.), and PCR was carried out using MyRED Taq polymerase (Bioline Inc.) and gene-specific primers.
For comparative purposes, the updated assembly of the A. anophagefferens mitogenome (see above) was annotated using MFannot, but we did not attempt to systematically improve the automatic annotation or to check the identity of the predicted unidentified open reading frames (ORFs). The output of the MFannot run on the A. anophagefferens sequence is included in the Supplementary data set S2, Supplementary Material online. During the comparative and phylogenetic analyses of mitochondrial genes in ochrophytes, we encountered a number of errors in the delimitation of actual coding sequences or even omissions of some genes in the previously published annotations of ochrophyte mitogenome sequences. Updated lists of genes present in different ochrophyte mitogenomes can be found in supplementary table S3, Supplementary Material online, details on the corrections or revised annotations of the genes are provided in supplementary table S4, Supplementary Material online.
Phylogenetic Analyses
Selected previously sequenced mitogenomes from diverse stramenopiles (supplementary table S1, Supplementary Material online) were combined with the newly sequenced genomes to perform phylogenetic analyses of individual homologous genes and of a concatenated alignment of 24 most conserved proteins (4,370 amino acid positions). Sequences were aligned using MAFFT and unreliably aligned positions were removed using program GBLOCKS 0.91b program (Castresana 2000) on the Gblocks server (http://molevol.cmima.csic.es/castresana/Gblocks_server.html, last accessed February 19, 2016), and the alignments were concatenated using FASconCAT (Kück and Muesemann 2010). A maximum likelihood (ML) phylogenetic analysis applying mixed/partitioned model, the protein substitution matrix general time reversible (GTR) and empirical base frequencies was carried out using RAxML-HPC BlackBox (7.3.2) (Stamatakis 2006) at the CIPRES Portal (http://www.phylo.org/sub_sections/portal/ [last accessed February 19, 2016], Cyberinfrastructure for Phylogenetic Research, San Diego Supercomputing Center; Miller et al. 2010). Phylogenetic analyses for individual proteins (the 24 conserved proteins used for the multigene analysis, Nad11, and release factor proteins) were performed as described above, except using the LG substitution model. For the Atp1 protein two analyses were carried out: an ML analysis with the GTR model using RAxML-HPC as described above, and Baysian inference using MrBayes (3.1.2) (Huelsenbeck and Ronquist 2001) at the CIPRES Portal. The substitution model was GTR+Γ+I, two runs (each with two chains) were run for 5,000,000 generations, trees were sampled every 100 generations, the consensus tree was obtained by summarizing the sampled trees after excluding the first 25% trees as burn-in. All phylogenetic trees were displayed by iTOL (http://itol.embl.de/ [last accessed February 19, 2016]; Letunic and Bork 2011).
Results and Discussion
Eustigmatophyte Mitogenomes Exhibit a Standard and Well-Conserved Gene Content
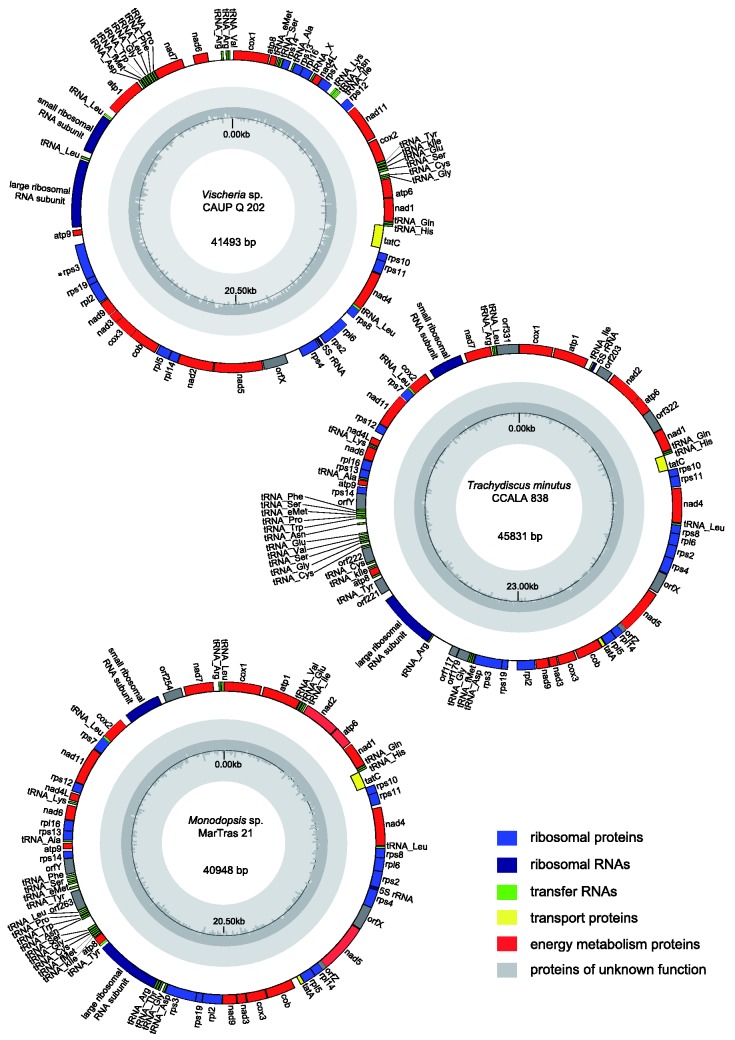
All three newly sequenced mitogenomes are circular-mapping. They are similar in size and gene content to previously sequenced mitogenomes of eustigmatophytes and other ochrophytes (fig. 1 and table 1; supplementary table S3, Supplementary Material online). The GC content fits into the range known from ochrophyte mitogenomes sequenced so far (24–38%), but within eustigmatophytes Vischeria sp. exhibits a somewhat lower GC content (27.3%) than the other species (31.4–33.7%). Three genes for ribosomal RNA, including 16S, 23S, and 5S RNA (see below), were found in all three genomes. The number of tRNA genes ranges from 27 to 29, the difference stemming mainly from the fact that some tRNA genes have duplicated in some species (for a more detailed discussion on tRNA genes see a separate section below). Mitochondrial genes for tmRNA (ssrA) were identified in jakobids (Keiler et al. 2000; Burger et al. 2013) and more recently in oomycetes (Hafez et al. 2013), a lineage relatively close to eustigmatophytes. However, no ssrA gene could be detected in any of the available eustigmatophyte mitogenomes, despite using a presumably highly sensitive covariance model built from the previously defined mitochondrial ssrA genes. We also did not find any group I or group II introns in the newly sequenced genomes, hence the group IIA intron previously found in the cox1 gene in Nannochloropsis oculata CCMP 525 (Starkenburg et al. 2014) remains the only mitochondrial intron reported from eustigmatophytes so far.
Fig. 1.—
Mitogenome maps of the eustigmatophytes Trachydiscus minutus, Vischeria sp. CAUP Q 202, and Monodopsis sp. MarTras21. Genes are shown as blocks facing outside if transcribed in the clockwise direction or facing inside if transcribed in a counter-clockwise direction. The assignment of the genes into different functional categories is indicated by their different colors. The plot in the inner circle shows the GC content, with the black line marking 50%.
Table 1.
Basic Features of Eustigmatophyte Mitochondrial Genomes
| T. minutus CCALA 838 | Vischeria sp. CAUP Q 202 | Monodopsis sp. MarTras21 | N. oceanica LAMB0001a | M. salina CCMP1776 a | |
|---|---|---|---|---|---|
| Length (bp) | 45,831 | 41,493 | 40,948 | 38,065 | 41,992 |
| GC (%) | 32.52 | 27.32 | 33.73 | 31.85 | 31.4 |
| tRNA genes | 27 | 28 | 29 | 26 | 26 |
| rRNA genes | 3 | 3 | 3 | 3 | 3 |
| Protein-coding genes | 36 | 36 | 36 | 36 | 37 |
| Conserved ORFs | 3 | 1 | 3 | 3 | 3 |
| Lineage-specific ORFs | 7 | 0 | 2 | 0 | 3 |
aGene numbers may differ from the original publication, as they reflect an updated annotation (see supplementary tables S3 and S4, Supplementary Material online).
The protein-coding gene set is highly conserved in eustigmatophyte mitogenomes (table 1; supplementary table S3, Supplementary Material online). Except the duplication of the cox1 gene in Microchloropsis species (then known as Nannochloropsis gaditana and Nannochloropsis salina) reported previously (Wei et al. 2013), the presence of some hypothetical short ORFs specific for individual species, and the loss of three genes in Vischeria sp. (see below), all eustigmatophytes have the same set of protein coding genes. All genes with an assigned functional annotation previously identified in other stramenopile mitogenomes are present, except rpl31 and rpl10 so far detected only in brown algae and diatoms, respectively (supplementary table S3, Supplementary Material online; Burger and Nedelcu 2012). One more such gene may be rps1, which we identified in the mitogenome of the pelagophyte A. anophagefferens and which lacks reliably identified orthologs in mitogenomes of other ochrophytes (table 1). The presence of a clear rps1 ortholog in the A. anophagefferens mitogenome is interesting, because previous surveys encountered the gene in mitogenomes of only very few eukaryotes, namely jakobids, malawimonads, and some land plants (Kannan et al. 2014).
Although the 5S rRNA gene was not mentioned in the original reports on the Nannochloropsis/Microchloropsis mitogenomes (Wei et al. 2013; Starkenburg et al. 2014), its presence was recently documented by Valach et al. (2014). Our analyses of newly sequenced mitogenomes extend the occurrence of the gene to mitogenomes of eustigmatophytes in general (fig. 1; supplementary fig. S2 and table S3, Supplementary Material online). In all three cases we obtained a typical three-domain consensus structure, but the helix I of the Trachydiscus 5S rRNA displays several unpaired bases, suggesting an alternative, permuted structure similar to that suggested for brown algal 5S rRNA (supplementary fig. S2, Supplementary Material online; Valach et al. 2014).
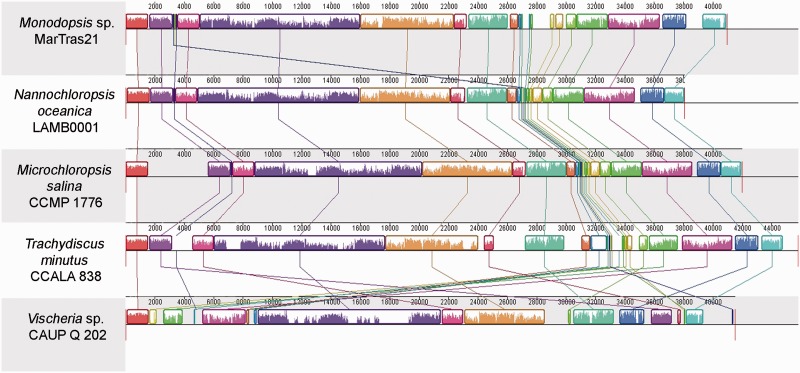
The Mitochondrial Gene Order Has Been Extensively Modified in the Vischeria Lineage
All eustigmatophyte mitogenomes share the same global architecture with all genes, except tatC, located on the same strand (fig. 1; see also Wei et al. 2013; Starkenburg et al. 2014). However, rearrangements of the mitogenomes via gene duplications and translocations have occurred along the eustigmatophyte phylogeny, and their distribution exhibits a striking pattern (fig. 2; supplementary fig. S3, Supplementary Material online). The highest similarity in the gene order across genera is exhibited by the pair T. minutus—Nannochloropsis/Microchloropsis. Given the fact that T. minutus and Nannochloropsis/Microchloropsis represent the two principal eustigmatophyte clades (Eustigmatales and Goniochloridales), their highly similar mitochondrial gene order probably reflects only a limited number of rearrangements in the lineages leading to these two taxa. As explained in detail in supplementary Results and Discussion, Supplementary Material online, the mitogenome of T. minutus appears to have retained the ancestral gene order (except duplication of the trnC gene and perhaps also emergence of nonconserved ORFs), whereas various rearrangements could be mapped to different branches of the Eustigmatales order. Specifically, all Eustigmatales share a translocation of the rn5 gene, whereas members of the Monodopsidaceae share a translocation of the trn(f)M gene. A few additional changes in the gene order in the Monodopsidacae are specific for the Monodopsis lineage (duplications and translocations of several tRNA genes) and for the genus Microchloropsis (duplication of the cox1 gene).
Fig. 2.—
Whole mitochondrial genome alignments of five phylogenetically diverse eustigmatophytes. Linearized newly obtained (Trachydiscus minutus, Vischeria sp. CAUP Q 202, Monodopsis sp. MarTras21) and previously published (Microchloropsis salina and Nannochloropsis oceanica) mitogenome sequences were aligned using progressiveMauve starting with the cox1 gene. Corresponding conserved synteny blocks are depicted in the same colors and the plot inside each block reflects the level of sequence similarity. The ruler above each genome represents nucleotide positions.
In contrast, a dramatic evolution of the mitochondrial gene order has occurred along the Vischeria lineage (fig. 2). Seven blocks of genes fully collinear between Vischeria and at least some other eustigmatophytes can be recognized (supplementary fig. S3, Supplementary Material online). Two larger blocks comprise 16 and 7 genes, while the remaining collinear blocks comprise only two or three genes. A relatively large region of the Vischeria genome (comprising three of the seven blocks mentioned above) lost complete collinearity with other eustigmatophyte mitogenomes due to readily traceable events: loss of the tatA and orfZ genes and replacement of the latter gene by the nad2 gene moved from elsewhere (supplementary fig. S3, Supplementary Material online). The rest of the genome is, however, nearly completely reshuffled with only a few three- or two-gene collinear blocks retained.
The Vischeria Lineage Has Experienced Accelerated Evolution of Mitochondrial Gene Sequences
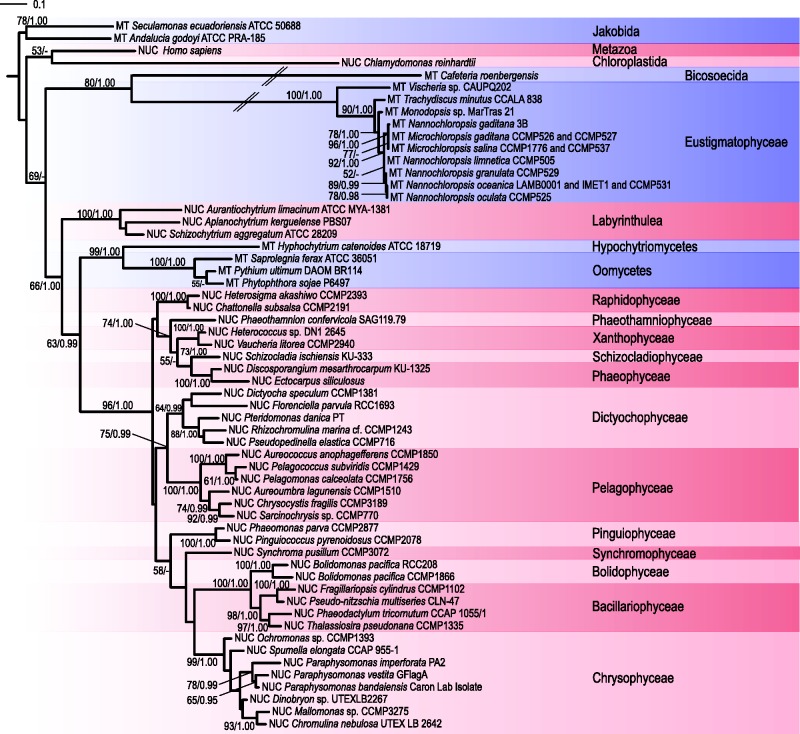
Mitogenome sequences are a valuable source of data for inferring phylogenetic relationships among species (see, e.g., Lang et al. 2002; Pombert et al. 2004; Bernt et al. 2013). The availability of mitogenome sequences for a diverse set of stramenopile lineages prompted us to conduct a phylogenomic analysis using 24 conserved proteins encoded by mitogenomes. The resulting tree (fig. 3) is consistent with the monophyletic origin of all ochrophyte classes represented in the data set by more than one mitogenome and with the established relationships within the classes. However, relationships among the ochrophyte classes suggested by the tree are not congruent with the current view of the ochrophyte phylogeny that have emerged from analyses of nuclear and plastid genes (Yang et al. 2012; Janouškovec et al. 2015; Ševčíková et al. 2015). For example, eustigmatophytes have been shown to robustly group with chrysophytes (Yang et al. 2012; Ševčíková et al. 2015), which is not recapitulated in the mitochondrial phylogenomic tree. The aberrant topology of the tree together with relatively low boostrap support values for the deep branches point to limitations of the method of phylogenetic inference employed and/or peculiar features of the mitochondrial gene sequence evolution.
Fig. 3.—
Phylogenomic analysis of mitogenomes in stramenopiles. The phylogenetic tree is based on an ML analysis of sequences of 24 proteins (4,370 amino acid positions) encoded by mitochondrial genes. The root is arbitrarily placed between the labyrinthulid Traustochytrium aureum and remaining stramenopiles (oomycetes and ochrophytes), reflecting the well-established basal position of labyrinthulids with respect to oomycetes and ochrophytes (Tsui et al. 2009). Bootstrap support values higher than 50% are shown at branches. Species are colored according to their affiliation to different stramenopile groups. The scale bar indicates the number of substitutions per site.
Indeed, the phylogenetic analysis of mitogenomes in ochrophytes is perhaps complicated due to the apparently very different degree of sequence divergence of mitochondrial genes in different ochrophytes groups, as evident from the varying branch lengths in the tree (fig. 3). Compare, for example, the branches of eustigmatophytes (except Vischeria, see below) and chrysophytes, which belong among the least and the most divergent, respectively. Interestingly, the pattern of branch lengths exhibited by individual ochrophyte taxa in the mitochondrial phylogenomic analysis is very different from that observed in the phylogenomic analysis of plastid protein sequences. For example, eustigmatophytes form markedly longer branches than most other ochrophytes in the plastid phylogenomic analysis (Ševčíková et al. 2015), while as noted above, their branches in the mitochondrial phylogenomic analyses are among the shortest. This suggests that the (average) substitution rate of different organellar genomes is not correlated in ochrophytes.
Notably, Vischeria has a markedly longer branch in the mitochondrial phylogenomic analysis than the other eustigmatophytes sampled (it is twice as along from the eustigmatophyte root as the second most divergent lineage, the genus Microchloropsis; see fig 3). We checked trees inferred individually for the 24 conserved proteins used in the phylogenomic analysis and 19 of them agree with the multigene tree in having Vischeria placed as the longest eustigmatophyte branch at the base of other Eustigmatales, whereas four trees exhibit an apparently artificial topology with Vischeria drawn to the base of all eustigmatophytes (data not shown). This indicates that the Vischeria lineage has experienced accelerated evolution of mitochondrial gene sequences compared to other eustigmatophyte, which perhaps also accounts for the relatively low bootstrap support value for the monophyly of Eustigmatales in the mitochondrial multigene tree (fig. 3).
Eustigmatophytes Are the Only Ochrophyte Group with an atp1 Gene in the Mitochondrial Rather than Nuclear Genome
Mitogenomes of Nannochloropsis/Microchloropsis spp. were previously found to harbor the atp1 gene, exceptionally among all ochrophyte mitogenomes sequenced by that time (Wei et al. 2013; Starkenburg et al. 2014). We can now extend its occurrence to eustigmatophytes in general, as it was found in all three new genomes. On the other hand, atp1 is missing from available mitogenome sequences from other ochrophytes (supplementary table S3, Supplementary Material online). We searched available genomic and transcriptomic data from ochrophytes and other stramenopiles to establish a detailed map of the cellular location of the atp1 gene in this group (details on this analysis are provided in the supplementary Results and Discussion, Supplementary Material online). These searches revealed two key points. First, a mitochondrial atp1 gene has been retained not only in eustigmatophytes, but also some nonalgal stramenopile lineages (including oomycetes, hyphochytriomycetes, and bicosoecids). Second, nuclear atp1 versions were identified in most stramenopile species, except those where evidence for the mitochondrial copy exists (see fig. 4; supplementary table S2, Supplementary Material online, and supplementary Results and Discussion, Supplementary Material online).
Fig. 4.—
ML phylogenetic tree of Atp1 sequences (459 aligned amino acid residues) in stramenopiles. The root of the tree is arbitrarily placed between sequences from jakobids and other eukaryotes. Bootstrap support values and posterior probabilities are indicated at branches when higher than 50% and 0.95, respectively. The scale bar indicates the number of substitutions per site. Long branches (crossed by a double line) were reduced to 50% of their original lengths to save space. Nuclear or mitochondrial location of the gene is indicated by different colors and abbreviations (NUC = red, MT = blue).
We conducted a phylogenetic analysis of Atp1 protein sequences to investigate the relationship between the mitochondrial and nuclear versions of the atp1 gene. The resulting phylogenetic tree (fig. 4) reveals three notable points. First, Atp1 sequences encoded by the mitochondrial genes in eustigmatophytes and the bicosoecid Cafeteria roenbergensis form extremely long branches in the tree, contrasting with the branches of other atp1 genes. The reasons for the elevated substitution rate of these genes cannot be ascribed simply to their cellular location, as the mitochondrial atp1 genes in oomycetes and Hyphochytrium catenoides seems to have evolved at a similar rate as nuclear atp1 genes. Second, the phylogenetic position of the Atp1 sequence from Vischeria with respect to sequences from other eustigmatophytes is inconsistent with the phylogenetic relationships within the group (see above and supplementary fig. S1, Supplementary Material online). Our preferred explanation is that the recovered topology is an artefact caused by a changed mode of evolution of the atp1 gene the Vischeria lineage, because taking the inferred topology at face value would necessitate a relatively complex evolutionary scenario involving incomplete lineage sorting or hidden paralogy. Third, the whole eustigmatophyte Atp1 cluster branches off outside a strongly supported monophyletic group of nucleus-encoded ochrophyte Atp1 sequences, which otherwise show relationships fairly consistent with the known phylogeny of ochrophyte species. To test for a possible different evolutionary origin (e.g., due to horizontal gene transfer) of the atp1 genes in eustigmatophytes and other ochrophytes, we carried out a phylogenetic analysis of a broader set of Atp1 sequences including a wider sampling of eukaryotic and bacterial homologs, but no evidence for such a possibility was found (data not shown).
We therefore consider two possible evolutionary scenarios for the atp1 gene in ochrophytes. The first assumes that the inheritance of the atp1 gene in ochrophytes is simply vertical, but the extreme divergence of eustigmatophyte sequences causes a phylogenetic artefact driving them outside the expected position among Atp1 sequences from other ochrophytes. If this is true, then at least four independent translocations of the atp1 gene to the nuclear genome need to be invoked (considering the phylogenetic position of eustigmatophytes as established by Yang et al. [2012] and Ševčíková et al. [2015]), specifically in the lineages leading to: 1) a common ancestor of diatoms, bolidophytes, pelagophytes, and dictyochophytes (collectively constituting Khakista sensu Riisberg et al. [2009]); 2) pinguiophytes; 3) a common ancestor of chrysophytes and synchromophytes; and 4) a common ancestor of raphidophytes and the PX clade (phaeophytes, xanthophytes and a few additional minor groups). The alternative scenario assumes that an ochrophyte ancestor harbored both the mitochondrial and the nuclear atp1 gene and one or the other copy was later differentially lost in different ochrophyte lineages. Direct evidence for the absence of a mitochondrial atp1 gene is lacking for many non-eustigmatophyte lineages of ochrophytes, so we cannot exclude the possibility that ochrophytes possessing both nuclear and mitochondrial atp1 versions are eventually found. This would support the latter explanation for the inferred phylogeny of Atp1 sequences. However, as a taxonomically broad analysis of mitochondrial genes suggested that the atp1 gene has frequently been moved from the mitochondrial to the nuclear genome (Kannan et al. 2014), we believe that the first scenario assuming independent mitochondrion-to-nucleus atp1 translocations in different ochrophyte lineages is more likely.
The Relocation of the 5′-Part of a Split nad11 Gene into the Nuclear Genome Is a Eustigmatophyte Synapomorphy
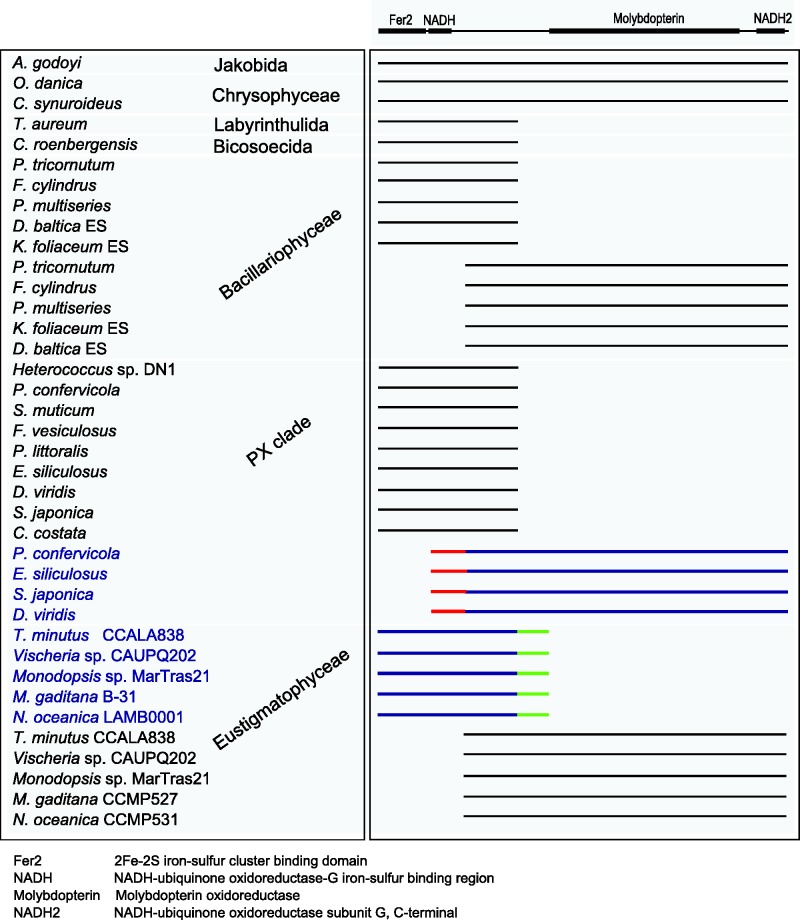
The largest (≈75 kDa) subunit of the mitochondrial NADH:ubiquinone oxidoreductase (called NDUFS1 in humans) is a protein comprising an N-terminal region with three binding sites for iron-sulfur (FeS) clusters and a C-terminal region homologous to the molybdopterin-binding enzymes (Sazanov 2007). It is encoded by a nuclear gene in many eukaryotes, but some protists have retained the respective gene, nad11, in their mitogenomes (Kannan et al. 2014). A previous analysis of the Nannochloropsis/Microchloropsis mitogenomes revealed that they harbor a truncated version of the nad11 gene encoding only the C-terminal part of the Nad11 protein, whereas the region corresponding to the N-terminal part is missing (Starkenburg et al. 2014). Here we show that such a truncated nad11 gene occurs in all eustigmatophyte mitogenomes sequenced (fig. 5), indicating that the split occurred already before the divergence of the two principal eustigmatophyte clades (Eustigmatales and Goniochloridales). The sister lineage of the Eustigmatophyceae includes the class Chrysophyceae (Yang et al. 2012; Ševčíková et al. 2015), for which two mitogenomes have been reported, both with an intact nad11 gene (fig. 5), hence we can infer that the loss of the 5′-part of the nad11 gene from the mitogenome is a synapomorphic feature for eustigmatophytes.
Fig. 5.—
Full and split Nad11 proteins in stramenopiles. The figure shows a schematic alignment of full-length proteins and those representing separate N- and C-terminal parts. Proteins shown in black are encoded by mitochondrial genes, proteins shown in blue are encoded by nuclear genes. Predicted mitochondrial targeting sequences are highlighted in red, unique C-terminal extensions of the nucleus-encoded parts of eustigmatophyte Nad11 proteins are highlighted in green. Functional conserved domains identified in the Nad11 protein by Pfam (Finn et al. 2014; http://pfam.xfam.org/) are shown above the alignment, the meaning of the domain labels is explained at the bottom of the scheme.
Starkenburg et al. (2014) also noticed the existence of a gene in a draft Microchloropsis (=Nannochloropsis) salina nuclear genome sequence that may encode the missing N-terminal domain of Nad11, but no detailed analysis was provided. We screened our preliminary data from nuclear genomes of the three eustigmatophyte species characterized here as well as available genome sequence for Nannochloropsis/Microchloropsis spp. and found a conserved gene encoding a protein corresponding to the missing N-terminal part including the FeS-binding domain followed by a novel C-terminal extension (fig. 5). It is therefore likely that a complete Nad11 protein is assembled in eustigmatophyte mitochondria from two separate polypeptides encoded by genes in different genomes. Interestingly, the nuclear genome-encoded parts of the Nad11 protein have no apparent N-terminal mitochondrial transit peptide in either eustigmatophyte species analyzed. This does not mean that such a protein cannot reach the mitochondrion, as nuclear genome-encoded mitochondrial proteins lacking obvious N-terminal transit peptides are known from different eukaryotes (Garg et al. 2015). It is also possible that the C-terminal extension of the nucleus-encoded parts of the Nad11 protein could serve as a signal for mitochondrial import, as has been previously described, for example, for a DNA helicase protein in the yeast Saccharomyces cerevisiae (Lee et al. 1999).
Interestingly, eustigmatophytes are not the only group with a split nad11 gene (fig. 5). Although some diatoms exhibit an intact nad11 gene in their mitogenome, two separate ORFs corresponding to the 5′- and 3′-part of the nad11 genes were found in the mitogenomes of different diatom species (Oudot-Le Secq and Green 2011; Imanian et al. 2012), reflecting at least one independent nad11 split event. The bicosoecid C. roenbergensis, the labyrinthulid Thraustochytrium aureum, and diverse brown algae (Phaeophyceae) were reported to harbor in their mitogenomes a shortened nad11 gene encoding only the N-terminal part of the Nad11 protein (Gray et al. 2004; Oudot-Le Secq et al. 2006), that is, the region that is nuclear genome-encoded in eustigmatophytes. In contrast, a gene corresponding to the 3′-end region of the nad11 gene missing from these mitogenomes was found to be located in the nuclear genome of at least one species, Ectocarpus siliculosus, and to encode a protein with a predicted mitochondrial transit peptide (Oudot-Le Secq and Green 2011).
We analyzed available ochrophyte genome and transcriptome sequence data to pinpoint the origin of the nad11 split in the phaeophyte lineage (fig. 5; supplementary table S2, Supplementary Material online). Transcripts encoding a separate C-terminal part of the Nad11 protein with a putative mitochondrial transit peptide were identified not only in various phaeophytes, but also in Phaeothamnion confervicola, a representative of the related class Phaeothamniophyceae. The transcriptome of the same species also includes an apparently mitochondrial polycistronic transcript encoding the N-terminal part of Nad11. The partial genome data released for a member of another phaeophyte relative, the xanthophyte Heterococcus sp. DN1 (Nelson et al. 2013), revealed the presence of a fragment of the mitogenome sequence including a separate gene encoding the N-terminal Nad11 part. A gene encoding the C-terminal part could not be identified in the Heterococcus sequence data, possibly because of their highly incomplete nature. Based on these observations, we posit that the nad11 gene was split in a common ancestor of Phaeophyceae, Xanthophyceae, and Phaeothamniophyceae, possibly in a common ancestor of the whole PX clade after it had diverged from the lineage leading to Raphidophyceae (a sister group of the PX clade exhibiting an intact mitochondrial nad11 gene).
Altogether, stramenopiles show an interesting tendency to split their nad11 at the equivalent position between the functionally separate N-terminal and C-terminal domains (fig. 5). At least four independent split events have been identified so far: in the eustigmatophyte lineage, within diatoms, in an ancestor of the PX clade, and once or twice in nonphotosynthetic stramenopiles (the number depends on whether the truncated nad11 genes in bicosoecids and labyrinthulids stem from the same evolutionary event, which is presently unknown because of poor sampling and unresolved stramenopile phylogeny). Furthermore, phylogenetic analyses of the N-terminal and C-terminal domains of Nad11 proteins are consistent with the idea that the nuclear genes coding for the N-terminal domain in eustigmatophytes and the C-terminal domain in phaeophytes result from the original intact mitochondrial genes moved to the nuclear genome rather than from an external source (supplementary fig. S4, Supplementary Material online). Such a tendency for splitting of mitochondrial genes and relocation of the separate parts to the nuclear genome is not without precedent. For example, the cox2 gene is known to have been split independently in at least two lineages, Myzozoa and Chlorophyceae; whereas both split parts are now located in the nuclear genome in Myzozoa and chlamydomonadalean green algae, one of the parts has been retained in the mitochondrial genome in remaining chlorophyceans (Waller and Keeling 2006; Rodríguez-Salinas et al. 2012).
Novel Features of the Mitochondrial Protein-Coding Gene Set: tatA and Three Conserved Unknown Proteins Specific for Eustigmatophytes
ORFs encoding hypothetical proteins lacking annotated homologs are commonly found in newly sequenced organellar genomes. A series of such ORFs was also identified in mitogenomes of Nannochloropsis/Microchloropsis species (Wei et al. 2013; Starkenburg et al. 2014). We used the expanded sampling of eustigmatophyte mitogenomes provided by our sequencing projects and probed into the origin and identity of these ORFs.
The first significant conclusion of our analyses is the identification of one of these ORFs as the gene tatA. This gene codes for a subunit of the twin-arginine translocase, which in prokaryotes and plastids mediates translocation of fully folded proteins across the plasma or thylakoid membrane (Sargent 2007). Two subunits of the translocase, TatA and TatC, appear to be generally conserved in diverse prokaryotes as well as in plastids (Berks 2015). The presence of TatC homologs encoded by some mitogenomes was for the first time noted by Bogsch et al. (1998) many years ago. Subsequently, the existence of mitochondrial tatA genes was recognized in the primitive mitogenomes of jakobids (Jacob et al. 2004). A recent review of algal mitogenomes (Burger and Nedelcu 2012) noted the presence of tatA in other eukaryotic groups, including diatoms, raphidophytes, and the chrysophyte Chrysodidymus synuroideus, but no details on the genes were provided. Here we show that tatA is indeed present in mitogenomes of diverse ochrophytes, including eustigmatophytes except Vischeria (supplementary table S2, Supplementary Material online). The encoded TatA proteins are short (52–55 amino acid residues) and poorly conserved, but they all share a predicted transmembrane region close to their N-terminus (fig. 6). A putative functional Tat translocase consisting of both TatA and TatC subunits thus seems to be common in ochrophyte mitochondria, but its actual function (i.e., specific substrates translocated across the inner mitochondrial membrane) remains to be determined.
Fig. 6.—
TatA proteins encoded by stramenopile mitogenomes. Alignment of TatA sequences from eustigmatophytes (green) and other groups: Chrysophyceae (blue), Raphidophyceae (orange), Pelagophyceae (violet), Bacillariophyceae (brown), and Jakobida (black). Conservation of aligned positions is indicated by similarity shading (>35%). Transmembrane domains predicted by the TMHMM program are highlighted in red. GenBank accession numbers of the protein sequences or genomic coordinates for genes not annotated in the current mitogenome annotation are given on the right.
In addition to tatA, three more mitochondrial ORFs, denoted by us orfX, orfY, and orfZ, are conserved across the whole phylogenetic breadth of eustigmatophytes (supplementary fig. S5, Supplementary Material online). One of them is present in every species, but the two remaining ORFs appear to be missing from Vischeria, apparently as a result of secondary loss, as no homologs could be found even in the nuclear genome and transcriptome sequences (data not shown). We attempted to identify possible homologs of these three genes outside eustigmatophytes, but no conclusive results could be obtained even using the highly sensitive tool HHpred server (Söding et al. 2005). A salient feature of the proteins encoded by orfZ is the presence of two predicted transmembrane regions (supplementary fig. S5, Supplementary Material online), suggesting that it is probably a protein of the inner mitochondrial membrane. At the moment is it impossible to determine whether orfX, orfY, and orfZ are de novo created genes that emerged as evolutionary innovations of the eustigmatophyte lineage, extremely divergent versions of genes gained by an eustigmatophyte ancestor by horizontal gene transfer from an unknown source, or drastically modified descendants of genes present in ancestral eukaryotic mitogenomes, such as some of the ribosomal protein genes exceptionally identified in mitogenomes of some ochrophyte lineages (rpl31 in phaeophytes and rps1 in A. anophagefferens, see above and supplementary table S3, Supplementary Material online).
Novel Features of the Eustigmatophyte Mitochondrial tRNA Gene Set: Possible Re-Emergence of trnT and a Putative Suppressor tRNA
Annotation and a comparative analysis of tRNA genes in eustigmatophyte mitogenomes revealed several aspects worth mentioning. First, virtually all eustigmatophyte mitogenomes share the same core set of 26 tRNA genes corresponding to an initiator tRNA (tRNA-fMet) plus tRNAs carrying 19 different amino acids, five of them, specifically glycine, serine, arginine, leucine, and isoleucine, represented by two (three in the case of leucine) isoacceptor tRNAs (supplementary table S3, Supplementary Material online). This tRNA set enables eustigmatophyte mitochondria to decode all codons (when super-wobble pairing of the third codon position is considered; see Lang et al. 2012), except a gene for tRNA-Thr. It was previously suggested that the trnT gene was lost already from the mitogenome of an ancestor of all stramenopiles and that tRNA-Thr is imported into the mitochondrion from the cytosol in this group (Gray et al. 2004; Burger and Nedelcu 2012).
Second, lineage-specific duplications of some tRNA genes are responsible for the varying numbers of tRNA genes in different eustigmatophyte genomes. While some of these duplications appear to have only trivial biological implications (e.g., the existence of more or less similar paralogs with no apparent functional difference), several cases stand out as particularly interesting. One such case concerns a mitochondrial tRNA gene in Monodopsis adjacent to the trnR(ucu) gene, and as suggested by a simple phylogenetic analysis of tRNA gene sequences (supplementary fig. S6, Supplementary Material online), perhaps evolutionarily derived from a duplicated trnR(ucu) copy. However, the anticodon of the respective tRNA molecule is UGU (fig. 7), which would suggest that the tRNA recognized codons for threonine. It is, therefore, possible, that a trnT gene, inferred to have been lost in a stramenopile ancestor (see above), has been recreated in the Monodopsis lineage by neofunctionalization of a paralog of a trnR gene. A similar case has been described from the yeast S. cerevisiae, where a novel trnT gene was apparently created from a paralog of trnH (Su et al. 2011). The re-emergence of a trnT gene in the Monodopsis mitogenome would eliminate the need of the mitochondrion to import the respective tRNA molecule from the cytosol, but whether this is indeed the case needs to be tested experimentally.
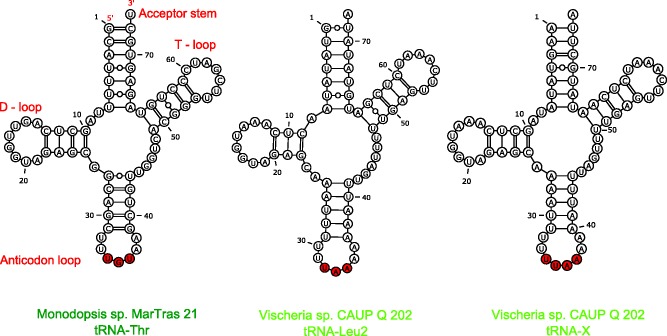
Fig. 7.—
Unusual mitochondrial tRNAs in Monodopsis and Vischeria. The figure shows predicted secondary structures of the Monodopsis tRNA-Thr molecule specified by the trnT(ugu) gene and the Vischeria tRNA-Leu2 and tRNA-X molecules specified by the genes trnL(uaa)_2 and trnX(uuaa), respectively. Nucleotides corresponding to the anticodon are in red. In case of tRNA-X, characterized by the anticodon loop expanded by 1 nt, it is unclear which of the 4 nt constitute the bona fide anticodon.
An even more interesting case of tRNA gene duplication in eustigmatophyte mitogenomes concerns a pair of related tRNA genes that appear to have been derived in the Vischeria lineage by two rounds of duplication and extensive modification of a canonical trnK(uuu) gene (supplementary fig. S6, Supplementary Material online). One of these novel tRNA genes is predicted to specify a tRNA molecule (referred to as tRNA-Leu2) possessing UAA as the anticodon (fig. 7), which would define it as cognate for the codons UUA and UUG encoding leucine. The biological significance of this putative tRNA is unclear, as the Vischeria mitogenome harbors a conventional trnL(uaa) gene that is theoretically sufficient to decode UUA and UUG codons, and it is also unknown whether this tRNA can be recognized by leucinyl-tRNA synthetase as a substrate.
The second related tRNA apparently derived from the trnK(uuu) gene in Vischeria, referred to as tRNA-X, is even more unusual, as its predicted anticodon loop is by one nucleotide longer than canonical anticodon loops, that is, it comprises eight rather than seven nucleotides (fig. 7). Although unusual, such a modification has been described before from various organisms. For example, an extended 8-nt anticodon loop occurs in one of two variants of tRNA-Thr specified by the mitogenome of the yeast S. cerevisiae (Su et al. 2011). This tRNA was proposed to have evolved from a duplication of the trnH(gug) gene and decodes CUN codons, underpinning the reassignment of these codons from leucine to threonine in the yeast mitochondria. Other tRNAs with extended 8-nt anticodon loops were described to suppress +1 frameshift mutations (Atkins and Björk 2009).
We did not notice any obvious frameshift mutation in the genes in the Vischeria mitogenome, but considering that the sequence that positionally corresponds to an anticodon in the unusual Vischeria tRNA-X is UUAA (fig. 7), one of the possible codons pairing with this tRNA might be the termination codon UAA. Interestingly, our inspection of protein-coding genes in the Vischeria mitogenome revealed that the coding sequence of the rps3 gene (encoding an essential ribosomal subunit) is interrupted by an in-frame TAA triplet. RT-PCR and sequencing of the resulting product ruled out modification of this triplet by RNA editing and confirmed that rps3 mRNA indeed includes an in-frame UAA codon and no putative nuclear genome-encoded mitochondrion-targeted Rps3 homolog could be found in our genomic and transcriptomic data for this species (data not shown). Therefore, we speculate that tRNA-X functions as a suppressor of this in-frame termination codon. The position of the codon corresponds to an amino acid residue in a poorly conserved region in the middle of the Rps3 protein (supplementary fig. S7, Supplementary Material online), so we cannot infer the amino acid specificity of the putative suppressor tRNA. In addition, we cannot exclude the possibility that the UAA codon in rps3 mRNA functions as a bona fide termination codon and that the Rps3 protein in Vischeria is assembled from two independently translated polypeptides corresponding to the N- and C-terminal conserved domains of the protein. Indeed, such a split of the rps3 genes has been observed in several distantly related eukaryotes (Swart et al. 2012; Fu et al. 2014). Nevertheless, the unique co-occurrence in Vischeria of the unusual tRNA with the expanded anticodon loop potentially pairing with the UAA codon and the rps3 gene interrupted by exactly this codon make the idea of the suppressor tRNA an interesting hypothesis.
Recurrent Simplification of Translation Termination in Eustigmatophyte Mitochondria
The standard genetic code includes three termination (stop) codons—UAA, UAG, and UGA. However, a number of exceptions have been described in various translation systems, one of which being the frequent abandoning of UGA as a termination codon in mitochondria (Duarte et al. 2012). We screened the predicted protein-coding genes in eustigmatophyte mitogenomes and found that no gene in Nannochloropsis/Microchloropsis spp. and Trachydiscus employs UGA as a termination codon, while two genes in the Monodopsis mitogenome—nad5 and rpl2, and one gene in the Vischeria mitogenome—tatC, do use it. In many mitochondria the UGA codon has been reassigned to code for the amino acid tryptophan (Lang et al. 2012); we investigated the eustigmatophyte mitogenomes for the possible occurrence of the reassigned UGA codons, but no candidate case could be discerned, even in the Nannochloropsis/Microchloropsis spp. and Trachydiscus mitogenomes.
Two release factors, mRF1 and mRF2, are typically needed by mitochondria utilizing all three canonical termination codons, as mRF1 recognizes UAA and UAG, whereas mRF2 recognizes UAA and UGA (Duarte et al. 2012). In accord with the lack of UGA-mediated translation termination inferred from the genome sequences, only mRF1 could be found in the nuclear genomes of Nannochloropsis/Microchloropsis spp. and our unpublished genomic and/or transcriptomic data generated for Trachydiscus, whereas both mRF1 and mRF2 orthologs exist in Monodopsis and Vischeria (supplementary fig. S8, Supplementary Material online). Given the phylogenetic relationships among eustigmatophytes, the absence of mitochondrial UGA termination codons and the associated protein machinery (mRF2) in the Trachydiscus and Nannochloropsis/Microchloropsis lineages must be due to two independent losses. Such reductive evolution of the mitochondrial translation termination mechanisms seen in eustigmatophytes is not without precedent among ochrophytes, as it was previously reported in diatoms (Duarte et al. 2012) and our inspection of available ochrophyte mitogenome sequences indicated the absence of UGA as a termination codon in the raphidophyte Heterosigma akashiwo (predicting that mRF2 is also absent, which can be confirmed only when the nuclear genome is sequenced).
Conclusions
By sequencing three new eustigmatophyte mitogenomes, we have considerably expanded the taxonomic sampling of this interesting, yet still poorly explored, algal group. Our comparative analyses using the previously analyzed mitogenomes of Nannochloropsis/Microchloropsis spp. and our new data enabled us to gain many interesting insights concerning the evolution of mitogenomes in eustigmatophyte (and even ochrophytes) in general as well as peculiarities of different eustigmatophyte lineages. One of the characteristic patterns is recurrent loss (or divergence beyond recognition) of genes (atp1, rpl10, rpl31, rps1) and other features (UGA as a termination codon) from different phylogenetic lineages. An opposite process—emergence of new mitochondrial genes (perhaps by various mechanisms including not only gene duplication, but potentially also creation de novo)—is also in place, as can particularly be documented by the case of eustigmatophytes as a whole (with their three potentially completely novel protein coding genes) or different eustigmatophyte sublineages (e.g., Vischeria with its two novel unusual tRNA genes). Figure 8 provides an overview of the major evolutionary events analyzed in detail in this study, mapped onto the ochrophyte phylogeny.
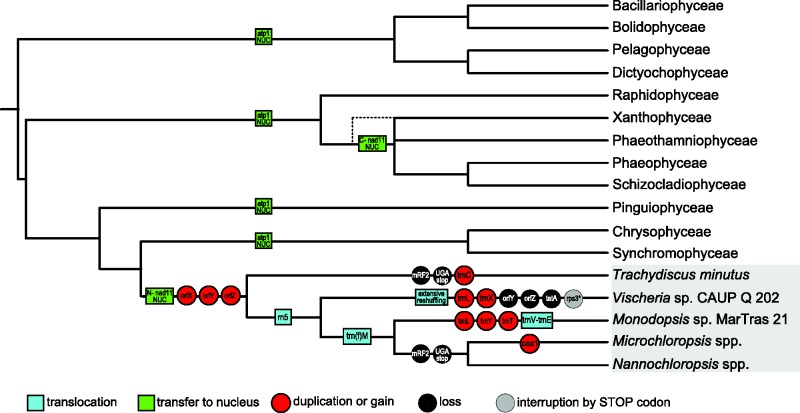
Fig. 8.—
Events in the evolution of mitogenomes in ochrophytes and in eustigmatophytes. The cartoon shows only those events affecting features of the mitogenomes (gene content, gene order, etc.) that were specifically addressed in this study. For simplicity, events specific for particular subgroups of classes other than eustigmatophytes (e.g., the split of the nad11 gene in some diatoms) and events that cannot be properly mapped due to insufficient sampling of ochrophyte mitogenomes (e.g., the presumed multiple losses of rpl10, rpl31, and rps1 genes) are omitted from the figure. The events were mapped onto a consensus ochrophyte phylogenetic tree based on recent multigene phylogenetic analyses (Yang et al. 2012; Ševčíková et al. 2015). The dashed line connected to the Xanthophyceae branch indicates the uncertainty in the phylogenetic position of this lineage with respect to other groups of the PX clade and the lack of direct evidence for relocation to the nuclear genome of the nad11 part encoding the C-terminal Nad11 domain.
An interesting observation that emerges from our analyses described above and which is also evident from figure 8 is that evolution of the mitogenome in the Vischeria lineage is exceptionally “active” when compared to lineages of other eustigmatophytes. This includes accelerated evolution of mitochondrial gene sequences, extensive reshuffling of the ancestral gene order, loss of three otherwise conserved genes, emergence of two novel tRNA genes, and interruption of the coding sequence of one of the genes by an in-frame termination codon. Interestingly, a correlation between accelerated substitution rates and increased rearrangements of organellar genomes has been observed also in some other groups, for example mitogenomes of arthropods (Xu et al. 2006) or plastid genomes of trebouxiophyte green algae (Turmel et al. 2015), suggesting possible common mechanistic causes.
Further studies of eustigmatophyte mitogenomes, which will fill-in important gaps in our current sampling (e.g., by adding first data from the hitherto uncovered deeply diverged Pseudellipsoidion group; Fawley et al. 2014), will enable us to further enrich our knowledge of their evolutionary history and more precisely pinpoint the different evolutionary events that have shaped the genomes. Our work also provides a basis for new ways of exploring the eustigmatophyte biology in general. For example, the availability of mitogenome sequences from a number eustigmatophytes now offers an opportunity for developing mitochondrial genes as useful markers for phylogenetic and taxonomic studies.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to Pavel Přibyl, Pavel Škaloud, and Martina Pichrtová for providing algal strains, to Jan Fousek for the help with the genome sequencing of T. minutus, to Matúš Valach for his help with the analyses of 5S rRNA, and to Vyacheslav Yurchenko for critical reading of the manuscript. This work was supported by the Czech Science Foundation (grant 13-33039S), the project CZ.1.05/2.1.00/03.0100 (IET) financed by the Structural Funds of the Europe Union, and the project LO1208 of the National Feasibility Program I of the Czech Republic.
Literature Cited
- Adl SM, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol. 59:429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen RA. 2004. Biology and systematics of heterokont and haptophyte algae. Am J Bot. 91:1508–1522. [DOI] [PubMed] [Google Scholar]
- Atkins JF, Björk GR. 2009. A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 73:178–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks BC. 2015. The twin-arginine protein translocation pathway. Annu Rev Biochem. 84:843–864. [DOI] [PubMed] [Google Scholar]
- Bernt M, et al. 2013. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol Phylogenet Evol. 69:352–364. [DOI] [PubMed] [Google Scholar]
- Bogsch EG, et al. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 273:18003–18006. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW, Forget L, Lang BF. 2013. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 5:418–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Jackson CJ, Waller RF. 2012. Unusual mitochondrial genomes and genes In: Bullerwell C, editor. Organelle genetics. New York: Springer; p. 41–77. [Google Scholar]
- Burger G, Nedelcu AM. 2012. Mitochondrial genomes of algae In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Advances in photosynthesis and respiration series. Vol. 35 London: Springer; p. 127–157. [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Scoble JM. 2013. Phylogeny of Heterokonta: Incisomonas marina, a uniciliate gliding opalozoan related to Solenicola (Nanomonadea), and evidence that Actinophryida evolved from raphidophytes. Eur J Protistol. 49:328–353. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. 2008. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics 24:861–862. [DOI] [PubMed] [Google Scholar]
- Corteggiani Carpinelli E, et al. 2014. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol Plant. 7:323–335. [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y. 2009. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. 1983. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1:19–21. [Google Scholar]
- Duarte I, Nabuurs SB, Magno R, Huynen M. 2012. Evolution and diversification of the organellar release factor family. Mol Biol Evol. 29:3497–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- Fawley KP, Eliáš M, Fawley MW. 2014. The diversity and phylogeny of the commercially important algal class Eustigmatophyceae, including the new clade Goniochloridales. J Appl Phycol. 26:1773–1782. [Google Scholar]
- Fawley MW, Jameson I, Fawley KP. 2015. The phylogeny of the genus Nannochloropsis (Monodopsidaceae, Eustigmatophyceae), with descriptions of N. australis sp. nov. and Microchloropsis gen. nov. Phycologia 54:545–552. [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu CJ, Sheikh S, Miao W, Andersson SG, Baldauf SL. 2014. Missing genes, multiple ORFs, and C-to-U type RNA editing in Acrasis kona (Heterolobosea, Excavata) mitochondrial DNA. Genome Biol Evol. 6:2240–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S, et al. 2015. Conservation of transit peptide-independent protein import into the mitochondrial and hydrogenosomal matrix. Genome Biol Evol. 7:2716–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobler CJ, et al. 2011. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc Natl Acad Sci U S A. 108:4352–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G. 2004. Mitochondria of protists. Annu Rev Genet. 38:477–524. [DOI] [PubMed] [Google Scholar]
- Hafez M, Burger G, Steinberg SV, Lang BF. 2013. A second eukaryotic group with mitochondrion-encoded tmRNA: in silico identification and experimental confirmation. RNA Biol. 10:1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd DJ, Leedale GF. 1970. Eustigmatophyceae—a new algal class with unique organization of the motile cell. Nature 225:758–760. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist FR. 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754–755. [DOI] [PubMed] [Google Scholar]
- Imanian B, Pombert JF, Dorrell RG, Burki F, Keeling PJ. 2012. Tertiary endosymbiosis in two dinotoms has generated little change in the mitochondrial genomes of their dinoflagellate hosts and diatom endosymbionts. PLoS One 7:e43763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Seif E, Paquet PO, Lang BF. 2004. Loss of the mRNA-like region in mitochondrial tmRNAs of jakobids. RNA 10:605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, et al. 2015. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci U S A. 112:10200–10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S, Rogozin IB, Koonin EV. 2014. MitoCOGs: clusters of orthologous genes from mitochondria and implications for the evolution of eukaryotes. BMC Evol Biol. 14:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Shapiro L, Williams KP. 2000. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter. Proc Natl Acad Sci U S A. 97:7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Kück P, Meusemann K. 2010. FASconCAT: Convenient handling of data matrices. Mol Phylogenet Evol. 56:1115–1118. [DOI] [PubMed] [Google Scholar]
- Lang BF, Burger G. 2012. Mitochondrial and eukaryotic origins: a critical review. Adv Bot Res. 63:1–20. [Google Scholar]
- Lang BF, Lavrov D, Beck N, Steinberg SV. 2012. Mitochondrial tRNA structure, identity, and evolution of the genetic code In: Bullerwell C, editor. Organelle genetics. New York: Springer; p. 431–474. [Google Scholar]
- Lang BF, O'Kelly C, Nerad T, Gray MW, Burger G. 2002. The closest unicellular relatives of animals. Curr Biol. 12:1773–1778. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Sedman J, Neupert W, Stuart RA. 1999. The DNA helicase, Hmi1p, is transported into mitochondria by a C-terminal cleavable targeting signal. J Biol Chem. 274:20937–20942. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiuchi T, Nozaki T. 2014. Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie. 100:3–17. [DOI] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 2010 Nov 14; New Orleans, LA, p. 1–8.
- Nelson DR, Tu ZJ, Lefebvre PA. 2013. Heterococcus sp. DN1 draft genome: focus on cold tolerance and lipid production. Microbiome Sci Med. 1:30–38. [Google Scholar]
- Nichols HW. 1973. Growth media—freshwater In: Stein J, editor. Handbook of phycological methods, culture methods and growth measurements. Cambridge: Cambridge University Press; p. 7–24. [Google Scholar]
- Oudot-Le Secq MP, Green BR. 2011. Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene 476:20–26. [DOI] [PubMed] [Google Scholar]
- Oudot-Le Secq MP, Loiseaux-de Goër S, Stam WT, Olsen JL. 2006. Complete mitochondrial genomes of the three brown algae (Heterokonta: Phaeophyceae) Dictyota dichotoma, Fucus vesiculosus and Desmarestia viridis. Curr Genet. 49:47–58. [DOI] [PubMed] [Google Scholar]
- Pombert JF, Otis C, Lemieux C, Turmel M. 2004. The complete mitochondrial DNA sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) highlights distinctive evolutionary trends in the chlorophyta and suggests a sister-group relationship between the Ulvophyceae and Chlorophyceae. Mol Biol Evol. 21:922–935. [DOI] [PubMed] [Google Scholar]
- Peabody DS. 1989. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 264:5031–5035. [PubMed] [Google Scholar]
- Přibyl P, Eliáš M, Cepák V, Lukavský J, Kaštánek P. 2012. Zoosporogenesis, morphology, ultrastructure, pigment composition, and phylogenetic position of Trachydiscus minutus (Eustigmatophyceae, Heterokontophyta). J Phycol. 48:231–242. [DOI] [PubMed] [Google Scholar]
- Radakovits R, et al. 2012. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat Commun. 3:686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riisberg I, et al. 2009. Seven gene phylogeny of heterokonts. Protist 160:191–204. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Salinas E, et al. 2012. Lineage-specific fragmentation and nuclear relocation of the mitochondrial cox2 gene in chlorophycean green algae (Chlorophyta). Mol Phylogenet Evol. 64:166–176. [DOI] [PubMed] [Google Scholar]
- Sargent F. 2007. The twin-arginine transport system: moving folded proteins across membranes. Biochem Soc Trans. 35:835–847. [DOI] [PubMed] [Google Scholar]
- Sazanov LA. 2007. Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry 46:2275–2288. [DOI] [PubMed] [Google Scholar]
- Ševčíková T, et al. 2015. Updating algal evolutionary relationships through plastid genome sequencing: did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci Rep. 5:10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. 2016. The past, present and future of mitochondrial genomics: have we sequenced enough mtDNAs? Brief Funct Genomics. 15:47–54. [DOI] [PMC free article] [PubMed]
- Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Starkenburg SR, et al. 2014. A pangenomic analysis of the Nannochloropsis organellar genomes reveals novel genetic variations in key metabolic genes. BMC Genomics 15:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, et al. 2011. An unusual tRNAThr derived from tRNAHis reassigns in yeast mitochondria the CUN codons to threonine. Nucleic Acids Res. 39:4866–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, et al. 2012. The Oxytricha trifallax mitochondrial genome. Genome Biol Evol. 4:136–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui CK, et al. 2009. Labyrinthulomycetes phylogeny and its implications for the evolutionary loss of chloroplasts and gain of ectoplasmic gliding. Mol Phylogenet Evol. 50:129–140. [DOI] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. 2015. Dynamic evolution of the chloroplast genome in the green algal classes Pedinophyceae and Trebouxiophyceae. Genome Biol Evol. 7:2062–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valach M, Burger G, Gray MW, Lang BF. 2014. Widespread occurrence of organelle genome-encoded 5S rRNAs including permuted molecules. Nucleic Acids Res. 42:13764–13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieler A, et al. 2012. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 8:e1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Keeling PJ. 2006. Alveolate and chlorophycean mitochondrial cox2 genes split twice independently. Gene 383:33–37. [DOI] [PubMed] [Google Scholar]
- Wang D, et al. 2014. Nannochloropsis genomes reveal evolution of microalgal oleaginous traits. PLoS Genet. 10:e1004094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, et al. 2013. Nannochloropsis plastid and mitochondrial phylogenomes reveal organelle diversification mechanism and intragenus phylotyping strategy in microalgae. BMC Genomics 14:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Jameson D, Tang B, Higgs PG. 2006. The relationship between the rate of molecular evolution and the rate of genome rearrangement in animal mitochondrial genomes. J Mol Evol. 63:375–392. [DOI] [PubMed] [Google Scholar]
- Yang EC, et al. 2012. Supermatrix data highlight the phylogenetic relationships of photosynthetic stramenopiles. Protist 163:217–231 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.