Abstract
Triarylmethyl (trityl, TAM) based spin labels represent promising alternative to nitroxides for EPR distance measurements in biomolecules. Herewith, we report synthesis and comparative study of series of model DNA duplexes, 5′-spin-labeled with TAMs and nitroxides. We have found that the accuracy (width) of distance distributions obtained by Double Electron-Electron Resonance (DEER/PELDOR) strongly depends on the type of radical. Replacement of both nitroxides by TAMs in the same spin-labeled duplex allows narrowing of the distance distributions by a factor of three. Replacement of one nitroxide by TAM (orthogonal labeling) leads to a less pronounced narrowing, but at the same time gains sensitivity in DEER experiment due to efficient pumping on narrow EPR line of TAM. Distance distributions in nitroxide/nitroxide pairs are influenced by the structure of linker: the use of a short amine-based linker improves the accuracy by a factor of two. At the same time, negligible dependence on the linker length is found for distribution width in TAM/TAM pairs. Molecular dynamics calculations indicate greater conformational disorder of nitroxide labels compared to TAM ones, thus rationalizing the experimentally observed trends. Thereby, we conclude that double spin-labeling using TAMs allows obtaining narrower spin-spin distance distributions and potentially more precise distances between labeling sites compared to traditional nitroxides.
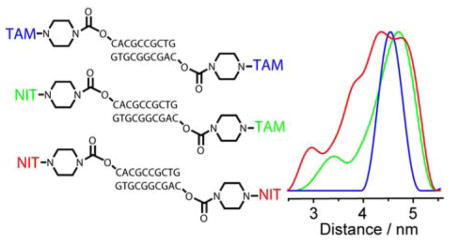
Graphical Abstract
Introduction
Pulsed dipolar EPR spectroscopy is nowadays routinely used in structural studies of biological systems, especially those that cannot be crystallized and investigated by X-ray diffraction methods.1–6 Pulsed Double Electron-Electron Resonance7–10 (DEER/PELDOR) or Double Quantum Coherence11,12 (DQC) techniques are used for obtaining distance distributions between paramagnetic sites in biomolecules. Although in some cases naturally present radical or metal centers can be used as EPR-active reporters, in most situations site-directed introduction of spin labels is required.13–15 Stable nitroxides with various linkers have been widely employed as spin labels in proteins and nucleic acids, with methanethiosulfonate label (MTSSL) being probably the most used. Recently, Cu2+, Gd3+ and Mn2+ based labels have appeared as alternatives to traditional nitroxides, proven to have advantageous properties in certain cases.16–24 Even more recently, triarylmethyl (trityl, TAM) radicals began to be used as spin labels for distance measurements in model systems and biomolecules.25–28 Although synthetic strategies for spin-labeling using TAMs are by far not enough elaborated compared to nitroxides up to date, the advantages of TAMs are already evident. First, much narrower EPR line of TAM compared to nitroxide allows sensitivity improvement and straightforward use of DQC method (requiring excitation of the whole spectrum) on commercial EPR spectrometers.25–27 Second, superior relaxation properties of TAMs, namely, much longer phase memory time, allow distance measurements even at room or physiological temperatures, thus avoiding the necessity to freeze samples and potentially alter naturally-occurring structures.26,28
In this work we address an important aspect of using TAM labels as compared to traditional nitroxides – the accuracy (width) of distance distributions obtained. Although distance distributions obtained using TAMs and nitroxides (NITs) have been previously compared for model biradicals,25 more detailed study is required on spin-labeled biomolecules. For this sake, we have synthesized identical model DNA duplexes and spin-labeled them using pairs TAM/TAM, TAM/NIT, NIT/NIT. To discriminate between the effect of radical vs. effect of linker used to attach the radical to DNA, we have also studied duplexes with two types of linkers. Since a comparison with nitroxides was a major point of the work, we have performed all measurements using DEER at 80 K, analyzed the distributions obtained and draw corresponding conclusions, supported by molecular dynamics (MD) calculations.
Experimental
Synthetic procedures
The TAM spin label used in this work is TAM-Cl – the tris-acyl chloride derivative of the Finland trityl radical.28 Contrary to apprehensions, the multi-functional nature of the reagent did not cause problems in selective labeling of the oligonucleotide targets with the only acyl chloride function of the TAM participated in attachment. Two remaining acyl chloride functions were efficiently hydrolyzed in the course of labeling and further work-up. The details of reaction procedure, labeling efficiency and spectroscopy data compiled for the title products are given in Supporting Information (SI).
To prepare samples I–IV (see Scheme 3) for pulsed dipolar EPR distance measurements, we have synthesized two 10-mer complementary oligonucleotides ON1 (5′–CACGCCGCTG–3′) and ON2 (5′–CAGCGGCGTG–3′) which formed duplex 1 (D1). Each of oligonucleotides was obtained by the phosphoramidite chemistry on CPG (Controlled Pore Glass) support. The CPG-attached 5′-detritylated oligonucleotides were treated by N,N′-carbonyldiimidazole (CDI) in 1,4-dioxane (see Scheme 1). After washing the support was treated with 1,4-piperazine solution in anhydrous 1,4-dioxane. Derivatives tethered 5′-piperazine residue (Pip-ONn, n=1, 2) was fully deprotected in concentrated aqueous ammonia and purified by HPLC. Purified oligonucleotide was transferred to a water-insoluble cetyltrimethylammonium (CTAB) salt and dried. CTAB salt of oligonucleotide derivative was dissolved in anhydrous dimethylsulfoxide (DMSO)/N,N-diisopropylethylamine (DIPEA) mixture (DMSO/DIPEA, 20/1, v/v) and treated with 10-fold excess of TAM-Cl or NIT-OSU (N-hydroxysuccinimidic ether). The latter operation afforded the required derivative of TAM to be attached to the only oligonucleotide molecule.
Scheme 3.
Structures of synthesized spin-labeled DNA duplexes D1 (I–V) and D2 (VI–VIII).
Scheme 1.
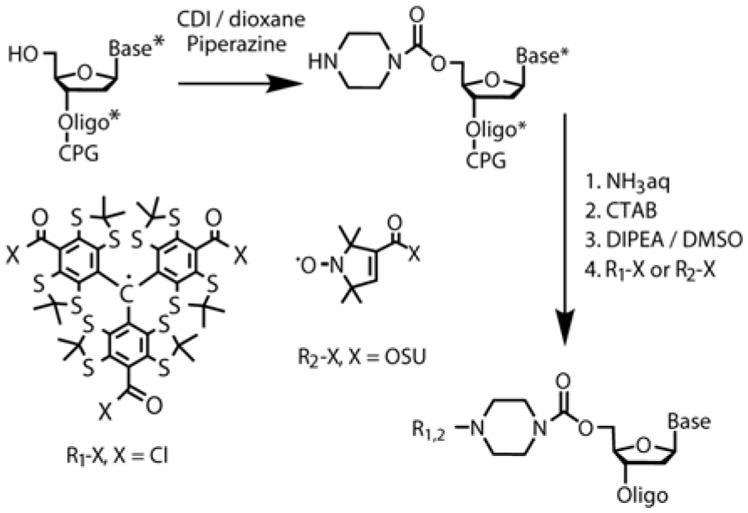
Synthesis of TAM and Nitroxide labeled oligonucleotide with piperazine linker. * means the presence of protecting groups at the synthetic step.
To prepare samples V, VI, VIII containing short amine-based –NH– linker we purchased commercially available phosphoramidite («Nanotech-C», Russia), containing 5′-monimetoxytrityl protected amino group (see SI). Modified phosphoramidite has been used in protocols of automated oligonucleotide synthesis on CPG support with extended condensation time (10 minutes instead of standard 30 seconds) and extended amounts of phosproramidite solution (see SI for details). We have synthesized single self-complementary oligonucleotide ON3 (5′–CGCGATCGCG–3′) which formed duplex 2 (D2). CPG-attached 5′-detritilated oligonucleotide was washed by absolute acetonitrile and treated with 10-fold excess of TAM-Cl in the presence of DMAP in toluene solution. After reaction TAM-containing oligonucleotide with two remaining acyl chloride functions was hydrolyzed by 0.01 M alkaline solution for a few minutes and then fully deprotected in concentrated aqueous ammonia and purified by HPLC. In the case of labeling of NH-contained oligonucleotide ON3 with nitroxide label CPG-attached 5′-detritilated oligonucleotide ON3 was fully deprotected in concentrated aqueous ammonia, purified by HPLC and then transferred to a water-insoluble cetyltrimethylammonium (CTAB) salt and dried (see Scheme 2). CTAB salt of oligonucleotide derivative was dissolved in anhydrous mixture DMSO/DIPEA (20/1, v/v) and treated with 10-fold excess of Nitr-OSU.
Scheme 2.
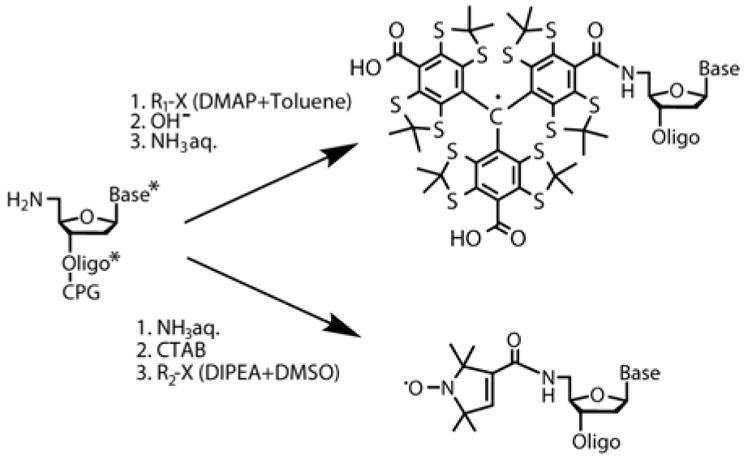
Synthesis of TAM and NIT labeled oligonucleotide with short (–NH–) linker. * means the presence of protecting groups at the synthetic step.
Synthetic procedures for nitroxide labeling of piperazine-based linker oligonucleotide ON3 (sample VII) were similar to those used for TAM labeling of piperazine-contained oligonucleotides ON1 and ON2.28 The details of reaction procedure, labeling efficiency and spectroscopy data compiled for the title product are given in SI.
EPR measurements
Samples for DEER measurements were prepared at room temperature in glass capillary tubes (OD 1.5 mm, ID 0.9 mm, with the sample volume being ca. 10 μl), shock-frozen in liquid nitrogen and investigated at T=80 K. The data were collected at the Q-band (34 GHz) using a Bruker Elexsys E580 pulse/cw EPR spectrometer equipped with an EN5107D2 resonator and Oxford Instruments temperature control system. Maximum available power was limited to 1 W, therefore resonator was critically coupled with the bandwidth (at the power level 0.5) not exceeding 30 MHz. A standard four-pulse DEER sequence has been used with pulse lengths of 20/40 ns for probe and 44 ns for pump frequency. In addition to the traditional two-step phase cycle, we implemented cycling of the second pulse at the probe frequency, which gave slightly better results for our experimental setup. All experimental results have been processed using DeerAnalysis2013.29
Note that the effects of orientation selection, known for doubly-labeled rigid systems, are not anticipated for TAM/TAM labeling. The reason for that is relatively narrow linewidth of TAM in frozen solution (<0.7 mT), so that efficient excitation of the whole spectrum is achieved. For TAM/NIT, and especially NIT/NIT pairs30 the orientation selection at Q-band might be significant. However, as is shown in Supporting Information, in our experimental conditions it can safely be neglected, and therefore will not be discussed any further.
Molecular dynamics simulations
The molecular dynamics (MD) simulations were performed using Amber 12 software.31 Structures and library files of spin labels with linker and internal cytosine nucleotide without phosphate residue connected to linker were generated using AmberTools 12. Particular atoms charges were calculated using Gaussian 09 software. Semiempirical PM3 parameterization was used for calculation of TAM labels with the linker. Nitroxide labels and internal cytosine nucleotide without phosphate residue connected to the spin labels were calculated using Hartree-Fock method and 6-31G* basis set. MD library files were used to build spin-labeled DNA duplex in the B-form. Nine series of MD simulation with various initial atom’s speed distribution in explicit solvent were performed. Each series included 12 stages (see SI). We used ff99bsc0 force field. The final stage of MD simulation was performed in explicit TIP3 water model with radius 12 Å. To neutralize net charge 18–22 Na+ ions were added to the system. Periodic boundary conditions, NTP ensemble with isotropic position scaling at 300 K and Andersen temperature coupling scheme were used. The particle mesh Ewald (PME) method with cutoff 8 Å was applied. SHAKE algorithm for bonds involving hydrogen was used. A time step 2 fs was used and the translational and rotation motion of the center of mass were removed every 1 ps.
In addition, to prevent unwinding of terminal base pairs, the H-bonds were restrained. The distances between atoms with unpaired electron were measured using ptraj program (AmberTools 12). These results were analyzed via superposition of a number of normal distribution functions.
Results and Discussion
Synthesis of model DNA duplexes
We have synthesized a series of 10-mer DNA duplexes 5′-spin-labeled with TAM or NIT radicals (Scheme 3). First, to investigate the effect of radical type on distance distributions obtained we have prepared all combinations of labels, namely TAM/TAM, TAM/NIT, NIT/TAM and NIT/NIT for the same double-stranded DNA sequence D1 (duplexes I–IV, respectively). Second, in order to investigate the effect of linker on the distributions, we used piperazine-type linker and newly developed short amine-based –NH– linker for symmetric pairs of labels (Scheme 3, duplexes IV and V). We did not succeed in spin-labeling of D1 sequence with two TAMs via –NH– linker, therefore another 10-mer duplex D2 was used for this task (VI). Then, for completeness of comparison, we synthesized D2 duplex with two TAM labels and piperazine linker as well (VII) and D2 duplex with two nitroxide labels and –NH– linker (VIII).
We have demonstrated previously that TAM label does not noticeably perturb the B-form conformation of DNA duplex.28 We also assume that nitroxide labels have insignificant influence on the structure and thermodynamic properties of DNA complexes. Melting temperature analysis additionally confirmed this observation: only insignificant increase of thermal stability of the duplex arose from introduction of terminal TAM and nitroxide labels with piperazine as well as short –NH– linkers (see SI).
Effect of radical type on distance distributions
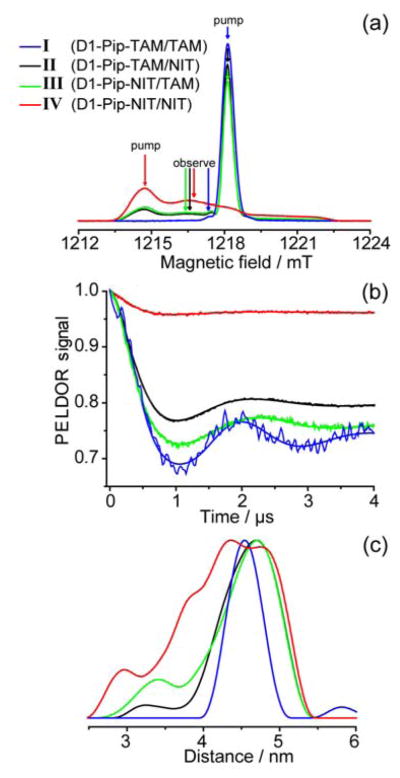
Figure 1 shows the obtained echo-detected (ED) EPR spectra, DEER time traces and distance distributions for duplexes I–IV with piperazine linkers. Here and below we number duplexes according to Scheme 3 and, in addition, use the following convention: “duplex type – linker type – spin label type” (e.g. “D1-Pip-TAM/TAM” refers to D1 duplex with two TAM labels attached via piperazine linker). The obtained mean distances and distribution widths (defined as standard deviation parameter σ) are summarized in Table 1.
Fig. 1.
(a) ED EPR spectra of duplexes I–IV. Spectral positions of pump and observe pulses are indicated for each duplex. (b) DEER time traces for each duplex after removal of relaxation background. (c) Distance distributions obtained for each duplex. Regularization parameter 100.
Table 1.
Mean distances <rDEER> obtained by EPR and calculated distances <rMD> obtained in MD experiments for all studied duplexes. Distances <rMD> which most closely correspond to <rDEER> are shown in bold.
| No. | Complex type | <rDEER> ± σ/nm | <rMD> ± σ/nm state 1 – 4 |
|---|---|---|---|
| I | D1-Pip-TAM/TAM | 4.54 ± 0.20 | 4.57 ± 0.12 |
| 4.45 ± 0.27 | |||
| 4.79 ± 0.18 | |||
|
| |||
| II | D1-Pip-TAM/NIT | 4.54 ± 0.42 | 4.34 ± 0.22 |
| 4.92 ± 0.29 | |||
| 3.36 ± 0.36 | |||
| 2.31 ± 0.40 | |||
|
| |||
| III | D1-Pip-NIT/TAM | 4.42 ± 0.53 | 4.35 ± 0.16 |
| 4.76 ± 0.24 | |||
| 3.40 ± 0.52 | |||
|
| |||
| IV | D1-Pip-NIT/NIT | 4.24 ± 0.62 | 4.20 ± 0.14 |
| 4.64 ±0.38 | |||
| 3.47 ± 0.40 | |||
| 2.11 ± 0.58 | |||
|
| |||
| V | D1-NH-NIT/NIT | 3.91 ± 0.37 | 3.90 ± 0.34 |
| 3.40 ± 0.48 | |||
|
| |||
| VI | D2-NH-TAM/TAM | 4.49 ± 0.25 | 4.53 ± 0.16 |
| 4.31 ± 0.28 | |||
|
| |||
| VII | D2-Pip-TAM/TAM | 4.50 ± 0.18 | 4.57 ± 0.11 |
| 5.55 ± 0.19 | |||
| 5.01 ± 0.19 | |||
| 4.30 ± 0.41 | |||
|
| |||
| VIII | D2-NH-NIT/NIT | 4.05±0.36 | 4.20 ± 0.29 |
| 3.56 ± 0.28 | |||
| 3.01 ± 0.38 | |||
In case of NIT/NIT pairs the position of pump and probe pulses in DEER experiment were traditional, corresponding roughly to MS=−1 for pump and MS=0 for probe frequencies (Fig. 1a). For TAM/TAM pairs the spectrum is dominated by single narrow EPR line, therefore we selected the 13C satellite for detection and the main line for pumping.25 Of course, this is not an optimum condition for distance measurements, and DQC would be best suitable for TAM/TAM pairs; however, in this work we focus on distance distributions rather than signal intensities, and thus to make clear comparison between nitroxides and TAMs we prefer to use the same method (DEER at 80 K). For TAM/NIT and NIT/TAM pairs we chose main TAM line for pumping and position close to MS=0 of the nitroxide spectrum for probing (Fig. 1a).
DEER time traces demonstrate quite deep dipolar modulation for all duplexes I–IV. We do not compare modulation depths quantitatively for technical reasons (see Experimental section, low available mw power is disadvantageous for nitroxides in comparison with TAMs). However, at least it is evident that the use of TAM labels provides reasonable modulation depth and high-quality distance distributions.
The most narrow distance distribution is obtained for pair TAM/TAM (σ≈0.20 nm). Substitution of one TAM by nitroxide results in noticeably broader distribution (σ≈0.42–0.53 nm). Finally, substitution of both TAMs by nitroxides results in the largest distribution width (σ=0.62 nm). The trend is absolutely clear, and the apparent advantage of using TAM/TAM pair is undoubted. However, the reason for this improvement is not immediately obvious. Possibly, it can be ascribed to the different delocalization of electron density in TAMs vs. nitroxides, to different conformational dynamics, different types of linkers used for attachment to DNA and accompanying effects (e.g. different stability of the duplex ends). Note also that previous study on model biradicals with TAM and nitroxide moieties (in different combinations) did not reveal such tremendous narrowing of distance distributions for TAM/TAM pair, and both TAM/TAM and NIT/NIT biradicals demonstrated much narrower distributions than those obtained by us here on DNA.25 Therefore, we reasonably assume that the conformational dynamics, which seems to be strongly restricted in biradicals compared to DNA duplexes, plays an important role in observed trends. To get deeper insight in this issue, below we compare identical duplexes with the same spin labels, but with linkers of different length and rigidity.
Effect of linker length on distance distributions
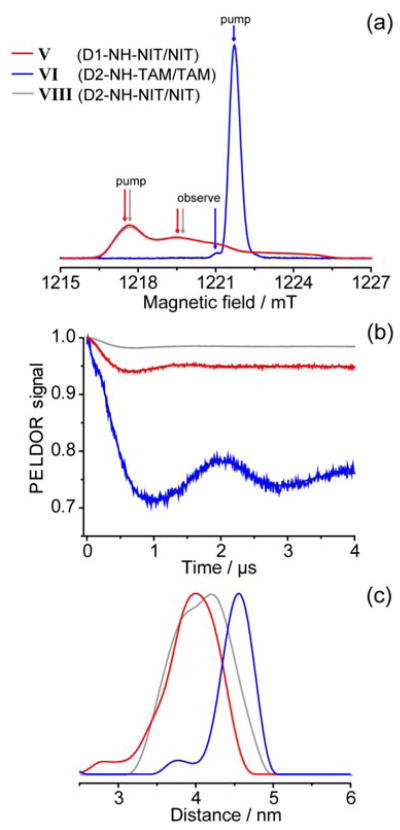
Figure 2 shows ED EPR spectra, DEER time traces and distance distributions for duplexes V, VI and VIII with new amine-based –NH– linker developed in this work. Such linker is much shorter compared to piperazine (Pip) one and, perhaps, represents the shortest possible linker for attachment of nitroxides or TAMs to the 5′-terminal positions of double-stranded DNA.
Fig. 2.
(a) ED EPR spectra of duplexes V, VI and VIII. Spectral positions of pump and observe pulses are indicated for each duplex. (b) DEER time traces for each duplex after removal of relaxation background. (c) Distance distributions obtained for each duplex. Regularization parameter 100.
Similar to the duplexes with piperazine linker, it is evident that the width of distance distribution is noticeably narrower for TAM/TAM pair (σ=0.25 nm) compared to NIT/NIT pair (σ=0.36 nm). Note, that the distribution width for TAM/TAM pair is very similar for piperazine and amine-based linkers, whereas for NIT/NIT pair short amine-based linker gains significant improvement in the accuracy of the measurement. This implies that the conformational dynamics of nitroxide label becomes more restricted by using a short linker, whereas in case of TAM the linker is not a “bottleneck” limiting the distribution width.
Another interesting difference between TAM and nitroxide labels is the dependence of mean distance <rDEER> on the linker length. Reasonably enough, <rDEER> for NIT/NIT pair is notably smaller when short amine linker is used. At the same time, for TAM/TAM pair the <rDEER> value is roughly the same for both piperazine and amine linkers used. The reason for this can be different DNA sequence used for TAM/TAM labeling via –NH– linker (D2 duplex), because the corresponding labeling of D1 duplex was not synthetically feasible. To avoid this ambiguity, we investigated in addition the D2 duplex with TAM/TAM labeling via piperazine linker (Table 1 and Supporting Information). Again, mean distances were roughly the same for two types of linker; at the same time, remarkably, distance distribution width remained closely the same (σ≈0.20 nm). To clarify structural grounds for these observations, we performed a series of molecular dynamics (MD) calculations.
Molecular dynamics simulation
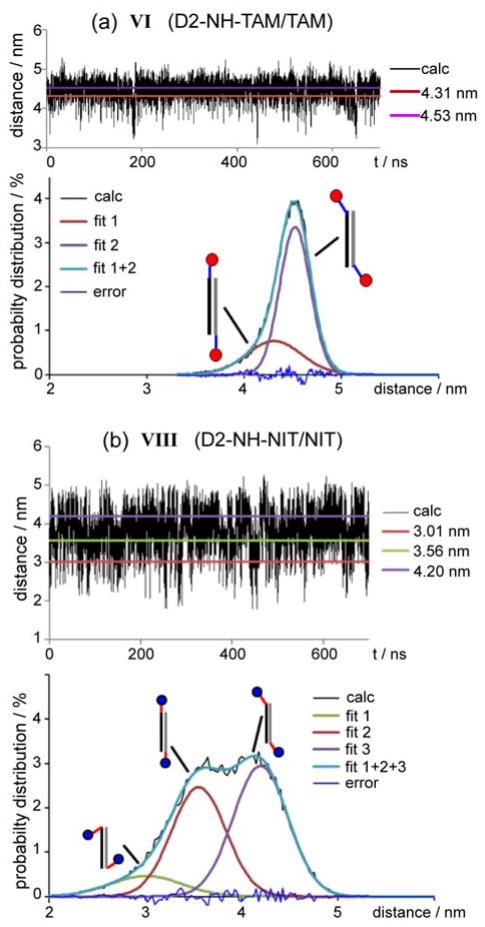
Molecular dynamics simulations were performed in explicit water with total length of trajectory ~0.7 μs for each duplex. The structures of all labeled DNA duplexes were analyzed. During the simulations the structures of labeled double helix were close to unmodified B-form of DNA. The distances between TAM and/or nitroxide labels along the trajectories were measured by analyzing each of ~70000 snapshots. Typical dependences of the interspin distances on time of MD simulation are shown in Figure 3.
Figure 3.
Interspin distance calculated from MD trajectory for duplexes: (a) VI (D2-NH-TAM/TAM) and (b) VIII (D2-NH-NIT/NIT). Horizontal lines on upper plots correspond to the maxima of fitted distributions fit 1, fit 2 or fit 3 (Supporting information). Distance distributions corresponding to MD trajectories are given on the bottom plots. The sketches illustrate the conformation of labels corresponding to the maximum of each normal distribution.
Based on this data the distance probability distributions were built and fitted by superposition of 2 to 4 normal distributions, each of which corresponded to the certain conformational state of spin-labeled duplex (Supporting Information). The maxima and widths of the obtained distributions are given in Table 1, whereas several preferential orientations of labels for piperazine and –NH– linkers are illustrated in Figure 4.
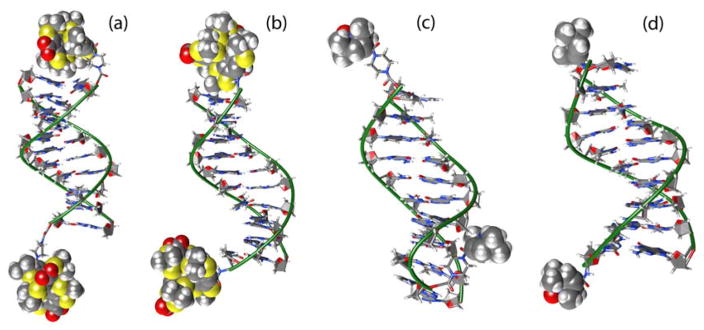
Figure 4.
Typical positions of spin labels relative to DNA duplexes: (a) I (D1-Pip-TAM/TAM); (b) VI (D1-NH-TAM/TAM); (c) IV (D1-Pip-NIT/NIT); (d) V (D1-NH-NIT/NIT).
It was found that TAM labels may or may not interact with terminal base pairs (Figure 4a upper and lower labels, respectively). However, it is reasonable to expect that the preferred conformation should correspond to the “capped” structure. This is consistent with the observation that the presence of TAM labels slightly (by ~1 °C) increases the duplex melting temperature, owing to the hydrophobic interactions of TAMs with terminal base pairs and protection of these base pairs from water molecules. Remarkably, average spin-spin distances obtained by MD simulations for such “capped” conformations of TAM-labels are in good agreement with mean DEER distances (bold values in Table 1). Therefore, we reasonably conclude that, in general, TAM labels tend to occupy “capping” positions with respect to the terminal base pairs of the duplex.
Contrary to TAMs, our MD data shows that nitroxide labels do not occupy any well-defined conformations, perhaps because this label is noticeably smaller compared to TAM and the hydrophobic interactions with terminal base pairs are also weaker. As a result, nitroxide does not “cap” terminal base pair and freely moves relative to DNA, being restricted only by steric interactions (Figures 4c and d). That is why the obtained distance distribution is much broader in case of NIT labels in comparison with TAM ones, as is shown in Figure 3. This is in perfect agreement with the EPR data that clearly demonstrate NIT labels to result in much broader distance distributions compared to TAMs.
In case of NIT/NIT pairs there is a clear dependence of the mean distance and the distribution width on the linker type. The replacement of piperazine linker by –NH– linker leads to a decrease of interspin distance by ~0.33 nm (Table 1). This agrees well with the analysis of MD trajectories that gives the distance C5′-O* (between terminal C5′ and oxygen of NO-group) equal to 1.1 nm for D1-NH-NIT/NIT and 0.8 nm for D1-pip-NIT/NIT, i.e. ~0.3 nm shorter for the latter duplex. Since the nitroxide radical can move freely relative to DNA, the length of linker strongly influences both the mean value and the width of distance distribution observed.
In contrast to nitroxides, in the case of TAM labels there is no significant effect of linker length on the distance distribution width. This is mainly ascribed, again, to the interaction between TAM label and terminal base pair. Qualitatively, the piperazine linker is long and flexible enough to let TAM interact with the terminal base pair and “cap” the duplex. At the same time, even though –NH– linker is noticeably shorter, it still allows the realization of similar “capping” conformation, as is revealed by MD calculations. As a result, TAM/TAM distances can be very close for piperazine and –NH– linkers. Note that EPR gives very close mean distances for VII (D2-Pip-TAM/TAM) and VI (D2-NH-TAM/TAM), in agreement with MD calculations yielding the same mean distances within the accuracy of modeling.
Overall, in most cases calculated MD distributions agree with the distributions obtained by EPR rather well, especially for duplexes doubly-labeled with TAMs (Supporting Information). For TAM/NIT and NIT/NIT duplexes the agreement is worse, but despite the differences in distribution shapes the experimental trends are reproduced: calculated distributions for TAM/TAM duplexes are narrower than those for NIT/TAM with the same linker, and NIT/NIT duplexes have the broadest distributions.
Conclusions
Recent burst of interest to TAMs as a new class of spin labels for dipolar EPR spectroscopy raised a lot of topical questions, one of which is the accuracy of distance measurements in biomolecules. In this paper we have addressed this question using model DNA duplexes doubly spin-labeled with TAMs and/or nitroxides. We have demonstrated that TAMs provide sensible advancement (up to three-fold) in the width of the distance distribution compared to nitroxides attached to the same sites via similar linkers. This occurs because the conformational disorder is noticeably smaller for TAM compared to nitroxide due to specific interaction of TAM with terminal base pair, as has been evidenced by experiments with different linkers and by molecular dynamics calculations. Such specific interactions may be dependent on the labeling method and site, thus complicating the translation of obtained distance distributions into desired structural information. Therefore, further methodological studies are required to address other typical situations for SDSL of DNAs using TAMs and to facilitate data interpretation in arbitrary case. Note that in some situations labeling of DNAs using rigid nitroxides also allows very narrow distance distributions to be obtained.32–35 Current temporary disadvantages of using TAMs include their less availability and more complicated spin-labeling procedures compared to nitroxides. However, we emphasize that superior spectral characteristics and relaxation properties of TAMs (evidenced earlier), as well as better accuracy of obtained distance distributions (demonstrated here), may boost the broad use of these new labels in various fields of biochemistry and structural biology in the nearest future.
Acknowledgments
Synthesis and spin-labeling of DNA duplexes, EPR measurements and interpretation were supported by Russian Science Foundation (no. 14-14-00922). Synthesis of TAM labels was supported by Russian Foundation for Basic Research (grant No. 13-04-00680A) and the National Institute of Biomedical Imaging and Bioengineering (grant No. 5P41EB002034). MD simulations were supported by Russian Foundation for Basic Research (no. 13-04-01176) and MES RF (Agreement No 14. B25.31.0028). We thank Marat Kassakin for MS Characterization and Dr. Igor A. Kirilyuk for kindly providing us with the NIT-OSU. We thank Siberian Supercomputer Center for providing computing resources.
Footnotes
NOTES
The authors declare no competing financial interests.
Supporting Information available: Synthesis of 5′-piperazine tethered derivatives of oligonucleotides; Synthesis of TAM- and Nitroxyl-labeled oligonucleotide with piperazine linker; Synthesis of 5′-NH2 tethered derivatives of oligonucleotides; Synthesis of TAM-labeled oligonucleotide with short –NH– linker; Synthesis of Nitroxyl-labeled oligonucleotide with short –NH– linker; Characterization of synthesized spin-labeled oligonucleotides; Molecular Dynamics Simulations; CW EPR settings; DEER results for duplexes I, VI and VII. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jeschke G. DEER Distance Measurements on Proteins. Annu Rev Phys Chem. 2012;63:419. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 2.Borbat PP, Freed JH. Pulse Dipolar Electron Spin Resonance: Distance Measurements. Struct Bond. 2013:1. [Google Scholar]
- 3.Borbat PP, Costa-Filho AJ, Earle KA, Moscicki JK, Freed JH. Electron spin resonance in studies of membranes and proteins. Science. 2001;291:266. doi: 10.1126/science.291.5502.266. [DOI] [PubMed] [Google Scholar]
- 4.Schiemann O, Prisner TF. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Quart Rev Biophys. 2007;40:1. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 5.Tsvetkov YD, Grishin YA. Techniques for EPR Spectroscopy of Pulsed Electron Double Resonance (PELDOR): A Review. Instrum Exp Tech. 2009;52:615. [Google Scholar]
- 6.Duss O, Michel E, Yulikov M, Schubert M, Jeschke G, Allain FHT. Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature. 2014;509:588. doi: 10.1038/nature13271. [DOI] [PubMed] [Google Scholar]
- 7.Milov AD, Salikhov KM, Shirov MD. Application of the double resonance method to electron spin echo in a study of the spatial distribution of paramagnetic centers in solids. Fiz Tverd Tela. 1981;23:975. [Google Scholar]
- 8.Milov AD, Ponomarev AB, Tsvetkov YD. Electron electron double-resonance in electron-spin echo - model biradical systems and the sensitized photolysis of decalin. Chem Phys Lett. 1984;110:67. [Google Scholar]
- 9.Larsen RG, Singel DJ. Double electron-electron resonance spin-echo modulation - spectroscopic measurement of electron-spin pair separations in orientationally disordered solids. J Chem Phys. 1993;98:5134. [Google Scholar]
- 10.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 11.Borbat PP, Mchaourab HS, Freed JH. Protein structure determination using long-distance constraints from double-quantum coherence ESR: Study of T4 lysozyme. J Am Chem Soc. 2002;124:5304. doi: 10.1021/ja020040y. [DOI] [PubMed] [Google Scholar]
- 12.Saxena S, Freed JH. Theory of double quantum two-dimensional electron spin resonance with application to distance measurements. J Chem Phys. 1997;107:1317. [Google Scholar]
- 13.Hubbell WL, Lopez CJ, Altenbach C, Yang Z. Technological advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 2013;23:725. doi: 10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:644. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Bordignon E. Site-Directed Spin Labeling of Membrane Proteins. Top Curr Chem. 2012;321:121. doi: 10.1007/128_2011_243. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Becker J, Saxena S. On Cu(II)-Cu(II) distance measurements using pulsed electron electron double resonance. J Magn Reson. 2007;188:337. doi: 10.1016/j.jmr.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Raitsimring AM, Gunanathan C, Potapov A, Efremenko I, Martin JML, Milstein D, Goldfarb D. Gd3+ complexes as potential spin labels for high field pulsed EPR distance measurements. J Am Chem Soc. 2007;129:14138. doi: 10.1021/ja075544g. [DOI] [PubMed] [Google Scholar]
- 18.Potapov A, Yagi H, Huber T, Jergic S, Dixon NE, Otting G, Goldfarb D. Nanometer-Scale Distance Measurements in Proteins Using Gd3+ Spin Labeling. J Amer Chem Soc. 2010;132:9040. doi: 10.1021/ja1015662. [DOI] [PubMed] [Google Scholar]
- 19.Martorana A, Bellapadrona G, Feintuch A, Di Gregorio E, Aime S, Goldfarb D. Probing Protein Conformation in Cells by EPR Distance Measurements using Gd3+ Spin Labeling. J Amer Chem Soc. 2014;136:13458. doi: 10.1021/ja5079392. [DOI] [PubMed] [Google Scholar]
- 20.Goldfarb D. Gd3+ spin labeling for distance measurements by pulse EPR spectroscopy. Phys Chem Chem Phys. 2014;16:9685. doi: 10.1039/c3cp53822b. [DOI] [PubMed] [Google Scholar]
- 21.Lueders P, Jeschke G, Yulikov M. Double Electron-Electron Resonance Measured Between Gd3+ Ions and Nitroxide Radicals. J Phys Chem Lett. 2011;2:604. [Google Scholar]
- 22.Lueders P, Jager H, Hemminga MA, Jeschke G, Yulikov M. Distance Measurements on Orthogonally Spin-Labeled Membrane Spanning WALP23 Polypeptides. J Phys Chem B. 2013;117:2061. doi: 10.1021/jp311287t. [DOI] [PubMed] [Google Scholar]
- 23.Kisseleva N, Khvorova A, Westhof E, Schiemann O. Binding of manganese(II) to a tertiary stabilized hammerhead ribozyme as studied by electron paramagnetic resionance spectroscopy. RNA. 2005;11:1. doi: 10.1261/rna.7127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee D, Yagi H, Huber T, Otting G, Goldfarb D. Nanometer-Range Distance Measurement in a Protein Using Mn2+ Tags. J Phys Chem Lett. 2012;3:157. [Google Scholar]
- 25.Reginsson GW, Kunjir NC, Sigurdsson ST, Schiemann O. Trityl Radicals: Spin Labels for Nanometer-Distance Measurements. Chem Eur J. 2012;18:13580. doi: 10.1002/chem.201203014. [DOI] [PubMed] [Google Scholar]
- 26.Yang ZY, Liu YP, Borbat P, Zweier JL, Freed JH, Hubbell WL. Pulsed ESR Dipolar Spectroscopy for Distance Measurements in Immobilized Spin Labeled Proteins in Liquid Solution. J Am Chem Soc. 2012;134:9950. doi: 10.1021/ja303791p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunjir NC, Reginsson GW, Schiemann O, Sigurdsson ST. Measurements of short distances between trityl spin labels with CW EPR, DQC and PELDOR. Phys Chem Chem Phys. 2013;15:19673. doi: 10.1039/c3cp52789a. [DOI] [PubMed] [Google Scholar]
- 28.Yu Shevelev G, Krumkacheva OA, Kuzhelev AA, Lomzov AA, Rogozhnikova Yu O, Trukhin DV, Troitskaya TI, Tormyshev VM, Fedin MV, Pyshnyi DV, Bagryanskaya EG. Physiological-Temperature Distance Measurement in Nucleic Acid using Triarylmethyl-Based Spin Labels and Pulsed Dipolar EPR Spectroscopy. J Amer Chem Soc. 2014;136:9874. doi: 10.1021/ja505122n. [DOI] [PubMed] [Google Scholar]
- 29.Jeschke G, Chechik V, Ionita G, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. DeerAnalysis2006 - a comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30:473. [Google Scholar]
- 30.Polyhach Y, Bordignon E, Tschaggelar R, Gandra S, Godt A, Jeschke G. High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Phys Chem Chem Phys. 2012;14:10762. doi: 10.1039/c2cp41520h. [DOI] [PubMed] [Google Scholar]
- 31.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Götz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Wu JL, Brozell X, Steinbrecher SR, Gohlke T, Cai H, Ye Q, Wang X, Hsieh JM-J, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 12. University of California; San Francisco: San Francisco, CA: 2012. [Google Scholar]
- 32.Prisner TF, Marko A, Sigurdsson STh. Conformational dynamics of nucleic acid molecules studied by PELDOR spectroscopy with rigid spin labels. J Magn Reson. 2015;252:187. doi: 10.1016/j.jmr.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Schiemann O, Cekan P, Margraf D, Prisner TF, Sigurdsson S. Th. Relative Orientation of Rigid Nitroxides by PELDOR: Beyond Distance Measurements in Nucleic Acids. Angew Chem Int Ed. 2009;48:3292. doi: 10.1002/anie.200805152. [DOI] [PubMed] [Google Scholar]
- 34.Krstic I, Hansel R, Romainczyk O, Engels JW, Dotsch V, Prisner TF. Long-Range Distance Measurements on Nucleic Acids in Cells by Pulsed EPR Spectroscopy. Angew Chem Int Ed. 2011;50:5070. doi: 10.1002/anie.201100886. [DOI] [PubMed] [Google Scholar]
- 35.Gophane DB, Endeward B, Prisner TF, Sigurdsson S. Th. Conformationally Restricted Isoindoline-Derived Spin Labels in Duplex DNA: Distances and Rotational Flexibility by Pulsed Electron–Electron Double Resonance Spectroscopy. Chem Eur J. 2014;20:15913. doi: 10.1002/chem.201403726. [DOI] [PubMed] [Google Scholar]