Abstract
The nitrogen-fixing symbiosis between Sinorhizobium meliloti and its leguminous host plant Medicago truncatula occurs in a specialized root organ called the nodule. Bacteria that are released into plant cells are surrounded by a unique plant membrane compartment termed a symbiosome. We found that in the symbiosis-defective dnf1 mutant of M. truncatula, bacteroid and symbiosome development are blocked. We identified the DNF1 gene as encoding a subunit of a signal peptidase complex that is highly expressed in nodules. By analyzing data from whole-genome expression analysis, we propose that correct symbiosome development in M. truncatula requires the orderly secretion of protein constituents through coordinated up-regulation of a nodule-specific pathway exemplified by DNF1.
To study the mechanisms governing bacterial infection and nodule organogenesis, we previously isolated a number of Medicago truncatula fast neutron bombardment mutants that were defective in nitrogen fixation (dnf mutants) (1). Among these, dnf1 is blocked at an intermediate stage of nodule development, as indicated by abolished induction of a set of host and bacterial genes and by microscopic analyses of thin longitudinal sections of nodules (Fig. 1, A and B) (2). After release from infection threads into the host cytoplasm, rhizobial bacteria in wild-type (WT) nodule cells divide and quickly differentiate into very large, elongated bacteroids that are dedicated to nitrogen fixation (Fig. 1, A and E, and fig. S1). Bacteria in dnf1 nodules are released from infection threads, are surrounded by symbiosome membrane, and divide, as do bacteria in WT plants. However, the bacteroids in dnf1 mutant hosts remain small and resemble freshly released rhizobia (Fig. 1B and fig. S1) or stage 1 bacteroids (3) (Fig. 1, C and D) that are present in about 10 cell layers of plant tissue. This is in contrast to WT nodules, which contain the stage 1 bacteroids only in the first one or two cell layers of the infection zone, beyond which they rapidly enlarge (fig. S1) to become about 5 to 10 times longer than freshly released rhizobia. Light and electron microscopic analyses show that bacteroids in dnf1 nodules are arrested at a very early stage of development. Further, numerous (pre-)autophagic bodies/autophagosomes (4) were observed in dnf1 nodule cells (Fig. 1C), whereas they are rare in WT cells. Whether the accumulation of autophagosomes is a primary or more indirect consequence of the dnf1 mutation remains to be determined. This accumulation of autophagosomes may reflect delayed vesicle maturation in the dnf1 mutant, or may relate to overall impaired vacuole development seen in young nodule cells of dnf1 (Fig. 1B) (4).
Fig. 1.
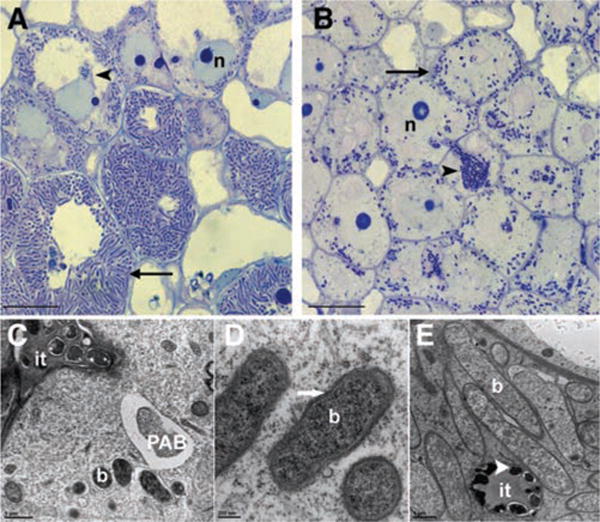
Cytological characterization of dnf1 root nodules. (A and B) High-magnification light microscopic image of longitudinal sections of a WT (A) and dnf1 (B) nodule 12 dpi, showing that, in all layers of the central tissue, the bacteroids in dnf1 are arrested at stage 1 (20). In WT, after release from the infection thread (it, arrowhead), the bacteria soon differentiate into elongated bacteroids (arrow). In dnf1, although many bacteroids are visible in the central part of the nodule, they have a size similar to that of the bacteria that are present in the infection threads. n, nucleus. (C) Transmission electron microscope (TEM) image of a freshly infected dnf1 nodule cell showing the infection thread (it), several bacteroids surrounded by a symbiosome membrane (b), and a pre-autophagic body (PAB) containing cytoplasm. (D) TEM image of an infected dnf1 nodule cell containing symbiosomes that remain arrested at stage 1. Arrows indicate symbiosome membrane. (E) Mature bacteroids in a WT nodule. These bacteroids are markedly bigger than the bacteria that are present in the it (arrowhead). Scale bars in (A) and (B), 10 μm; (C), 1 μm; (D), 200 nm; (E), 1 μm.
Recently, we showed that the symbiosome membrane has a “mosaic” identity (5). The plasma membrane SNARE SYP132 is present directly after the release of the bacteria from the infection threads (5, 6) and is maintained on the symbiosome membrane through later stages of development. The late endosome marker Rab7 appears only when the symbiosomes stop dividing, whereas the vacuolar SNAREs occur on the symbiosome membrane only at the onset of senescence. To test whether the arrest of symbiosome development in dnf1 correlates with an altered symbiosome membrane identity, dnf1 roots were transformed with constructs, each encoding a fusion of green fluorescent protein (GFP) to one of these membrane identity markers (5). Our data showed that dnf1 symbiosomes obtain Rab7 (fig. S2A) as well as the vacuolar SNARE VTI11 before senescence (fig. S2B). Because the occurrence of membrane identity markers on dnf1 symbiosome membranes is very similar to that of WT symbiosomes, a changed membrane identity cannot explain the block in symbiosome development.
To identify candidate genes for DNF1, we compared the transcriptomes of WT versus dnf1 plants. Such an analysis has previously been used to isolate the M. truncatula DMI3 calcium- and calmodulin-dependent (CCaM) kinase gene (7), because fast neutron-bombardment mutagenesis frequently causes deletions in the genome, and the consequent abolition of transcript can be detected by array hybridization. In plants that are homozygous for the dnf1-1 mutant allele, a set of sequences showed drastically reduced expression, including the sequence of TC121074. Further investigation identified a large deletion of at least 20 kb that removed the entire gene for TC121074. A second allele, dnf1-2, displayed an independent disruption of the TC121074 locus as a result of chromosome rearrangement. The deletion of TC121074 in dnf1-1 cosegregated with the mutant phenotype (32 F2 individuals), implying a causal relationship.
By using the dnf1-1 and dnf1-2 mutants, we transformed roots with a genomic copy of the predicted coding region (Fig. 2A) for TC121074 under its inferred native promoter (3 kb upstream of the translational start site). At 14 days post inoculation (dpi) with Sinorhizobium meliloti Rm1021, transgenic roots produced pink nodules that were indistinguishable from those of WT (Fig. 2B). We concluded that DNF1 corresponds to TC121074.
Fig. 2.
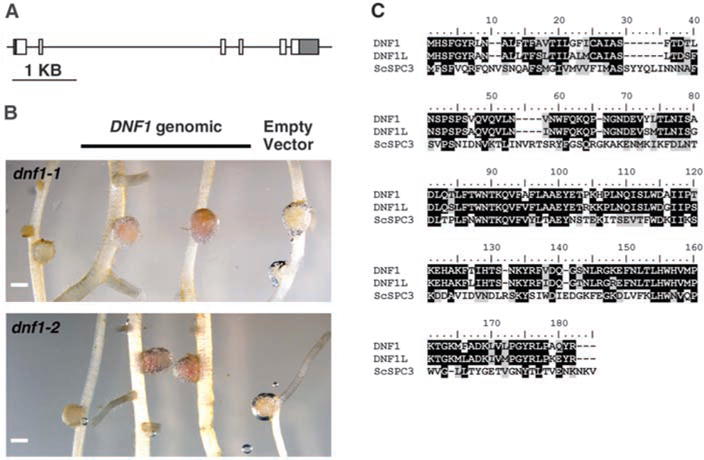
DNF1 encodes a subunit of the SPC. (A) Structure of the candidate DNF1 gene. Boxes represent exons, and the lines between them are introns. 5′ and 3′ untranslated regions are shaded. (B) Complementation of mutant nodule phenotype by introducing the genomic fragment of the predicted DNF1 gene. Pink color indicates the presence of leghemoglobin. Scale bars, 1 mm. (C) Alignment of protein sequences between DNF1, DNF1L of M. truncatula, and the Sa. cerevisiae homolog ScSPC3.
The DNF1 protein is annotated as the 22-kD subunit (SPC22) of the signal peptidase complex (SPC). Being an early component of the protein secretory pathway in the endoplasmic reticulum (ER), the SPC cleaves signal peptides off nascent polypeptides that are destined for intracellular compartments and the extracellular matrix (8). Although it is not the catalytic domain, SPC22 is nonetheless essential for viability in species where it has been tested (9, 10). In the available genome and expressed sequence tag (EST) databases, we found another SPC22 gene (DNF1L), whose protein had a sequence that is almost identical to that of DNF1. Both proteins display substantial sequence similarity to the Saccharomyces cerevisiae SPC3 (8–11) (Fig. 2C). We hypothesize that housekeeping activities important for viability can be performed by DNF1L alone, while DNF1 has evolved to specialize in symbiosis.
To test this hypothesis, we queried the M. truncatula Gene Expression Atlas (12) for the expression profiles of DNF1 and DNF1L. Their profiles are essentially uncorrelated (R2 = 0.024). DNF1L is expressed evenly in all organs at a low level, with modestly higher levels being expressed in pods and nodules (fig. S3). In contrast, DNF1 expression is highly elevated in the nodule compared with other source tissues (Fig. 3A). The nodule expression of DNF1 reaches the highest level at 4 dpi, which is the earliest available time point in the dataset (Fig. 3B). This early induction of DNF1 expression is consistent with the mutant phenotype. Previous transcriptional profiling studies have also noted the up-regulation of DNF1 during nodule formation (13, 14).
Fig. 3.
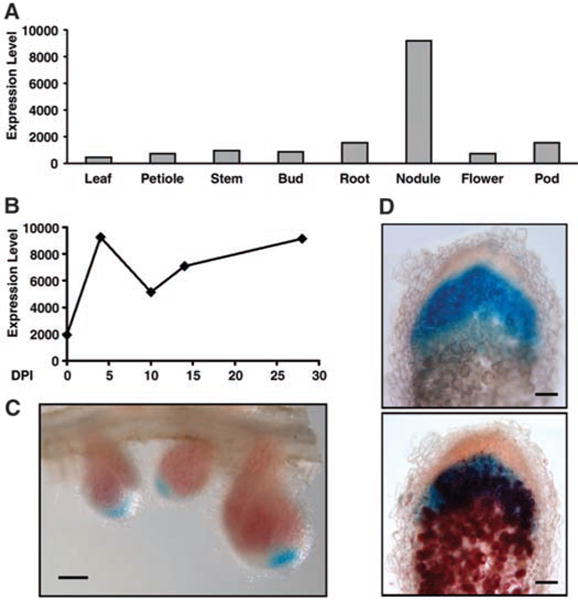
DNF1 expression pattern. (A) Expression level of DNF1 in various tissues. (B) Temporal profile of DNF1 expression in nodule. (C) Spatial pattern of DNF1 expression revealed by a promoter::GUS construct in 14-day-old nodules. Nodule samples were stained briefly to preserve the color of leghemoglobin. (D) DNF1 promoter activity in nodules inoculated with Rm1021 carrying hemA::lacZ. A DNF1 promoter::GUS transgenic nodule was stained for GUS activity, photographed (top), briefly fixed, sectioned manually, and then stained with Salmon-gal (bottom). Values in (A) and (B) are levels of Affymetrix probe signal based on microarray data from the M. truncatula Gene Expression Atlas. DPI: days post inoculation. Scale bars in (C), 1 mm; (D), 100 mm.
We further investigated the tissue specificity of DNF1 expression in the nodule. The DNF1 promoter was fused to β-glucuronidase (GUS) and transformed into roots. In roots before inoculation, the promoter is active in the vasculature. GUS activity becomes visible inside very young nodules (fig. S4). By 14 dpi, the nodule has differentiated into distinct zones. GUS activity appears the highest in the growing tip of the nodule (Fig. 3C). By visualizing bacteria in transgenic nodules, we found that the DNF1 gene is expressed in the infection zone, where bacteria invade the host cell and symbiosomes subsequently differentiate (Fig. 3D). Its expression decreases but is still detectable in differentiated cells of the fixation zone (fig. S5). This expression pattern is consistent with a function of DNF1 in the differentiation of symbiosomes.
The ER is a dynamic, multifunctional structure with roles in protein targeting and secretion (15, 16). We made a translational DNF1-GFP fusion under its own promoter, transformed the construct into dnf1-2 mutant roots, and then inoculated them with mCherry-labeled S. meliloti. Transgenic nodules recovered the pink color of leghemoglobin, which is an indication of normal nitrogen-fixing activity. The bacteria were able to differentiate into elongated bacteroids similar to WT, proving that the GFP moiety does not interfere with DNF1 function (Fig. 4). DNF1-GFP exhibits a robust signal in cells containing bacteria, and shows ER-like distributions (Fig. 4B). In an accompanying study, Van de Velde et al. demonstrate that M. truncatula produces nodule-specific cysteine rich proteins (NCRs) governing bacteroid differentiation, which require DNF1 to be processed from the preprotein (11). Failure to process the propeptides in dnf1 results in the accumulation of these propeptides in the ER.
Fig. 4.
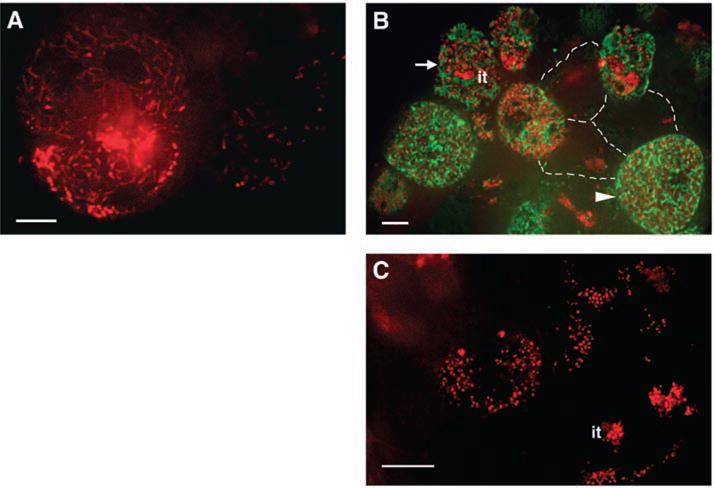
Localization of DNF1. Roots were inoculated with Rm1021 carrying hemA::mCherry, nodules were hand sectioned at 14 dpi, and the infection zones were imaged under a confocal microscope. (A) In WT nodule cells, bacteroids quickly become highly elongated. (B) DNF1 was fused to GFP, expressed from its own promoter, and introduced into dnf1-2 roots. In uninvaded cells, three of which are outlined with dotted lines, no robust GFP signals were observed. In cells being invaded by bacteria (arrow), where the end of the it is discernible, DNF1-GFP fluorescence first appears. In cells where bacteroids are fully mature (arrowhead), DNF1-containing structures are numerous and appear elongated and closely associated with symbiosomes. (C) In dnf1-2 mutant nodule cells, bacteroids remain small and undifferentiated after release from the it. Green, DNF1-GFP; red, Rm1021 hemA::mCherry. Scale bars, 10 μm.
In plants, the SPC complex is composed of four subunits. In the M. truncatula Gene Expression Atlas, the three genes with the highest expression correlation to DNF1 encode SPC25, SPC12, and SPC18 (the catalytic subunit) (17). The correlation at the transcript level suggests that their protein products probably form the SPC complex in nodule cells. Therefore, we named them DAS12 (for DNF1-associated SPC12), DAS18, and DAS25 (Fig. 5A). Following the DAS genes on the correlation list is a gene that encodes a signal peptide peptidase (18), which degrades the signal peptides after cleavage (Fig. 5A). Further strengthening the notion of a common regulatory mechanism of the protein secretory pathway during symbiosome biogenesis, the expression of genes coding for the small GTPase Rab11b and an isoform of SYP132 is also highly correlated to that of DNF1 (Fig. 5A). In Nicotiana tabacum, Rab11b regulates vesicle trafficking for protein secretion in the pollen tube (19). The comparatively lower correlation for Rab11b may reflect additional mechanisms to regulate Rab protein activity beyond transcriptional control.
Fig. 5.
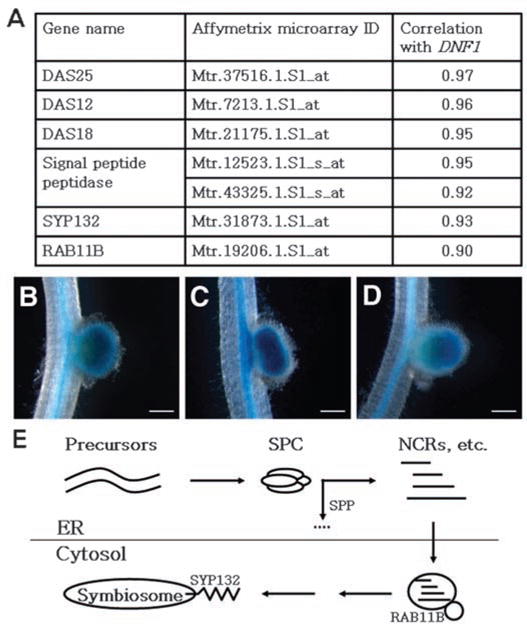
Coordinated up-regulation of a nodule-specific protein secretory pathway. (A) Correlation in gene expression between DNF1 and additional components of the protein secretory pathway. Correlation values are provided by the M. truncatula Gene Expression Atlas (12) based on microarray data on vegetative and reproductive tissues, seed development, nodulation, mycorrhization, and chemical treatments. The signal peptide peptidase gene is represented by two probe sets. (B to D) Promoter::GUS activity in the nodule of DNF1 and its associated SPC genes DAS12 and DAS25. Transgenic roots were inoculated with Rm1021, and nodules were photographed at 14 dpi. (B) pDNF1::GUS; (C) pDAS12::GUS; and (D) pDAS25::GUS. Background GUS activity in the vasculature confirmed the presence of the transgene. In (C), a nodule with double primodia is being formed. (E) A model for a nodule-specific pathway dedicated to secrete protein constitutes toward the developing symbiosome. SPC denotes the DNF1-containing SPC, SPP refers to the signal peptide peptidase listed in (A), and NCRs are mature peptides from precursor proteins processed by SPC and SPP. Scale bar, 1 μm.
We produced promoter::GUS fusions for two genes, DAS12 and DAS25, to both verify the transcriptome data and probe their spatial expression. Reporter fusion data indicate that both genes are expressed in the infection zone of the nodule in a pattern that is indistinguishable from that of DNF1 (Fig. 5, B to D). Overall, the high correlation among the expression profiles of multiple functionally related genes strongly supports the hypothesis that upon rhizobial infection, the plant machinery for processing secretory proteins is mobilized coordinately (Fig. 5E).
In dnf1 mutants, bacterial release into the host cell is apparently normal, but the subsequent differentiation of the bacteria is blocked. Such a phenotype suggests that among the substrates of the DNF1 complex are important host determinants of symbiosome development, such as the processed NCR peptides described by Van de Velde et al. (11). The NCR proteins are found only in legumes such as Medicago spp. that subject their microbial partners to terminal differentiation; however, it is possible that the DNF1 apparatus is present even in legume species that lack NCR type proteins, such as Lotus or bean (18). If so, it will be interesting to test whether disrupting function of a DNF1 homolog affects symbiosome function in these legumes. Such a result would imply that there are other substrates for the DNF1 SPC, a possibility that can be empirically tested. Potential substrates include the A1b/leginsulin family proteins ENOD8, ENOD16, and nodulin-25, all of which are proteins with a signal peptide, are up-regulated during nodulation, and in some cases are shown to localize to the symbiosome (13, 20, 21). Some of these proteins are conserved in Lotus japonicas and soybean, suggesting that they may be processed by a common mechanism in a variety of legume species. Alternatively, the DNF1 and co-expressed signal peptidase genes may have co-evolved with the NCR genes within the clade of legumes that show terminal bacteroid differentiation as a specialized means to control bacterial proliferation and function within the host cells.
Supplementary Material
Acknowledgments
We thank D. Ehrhardt for assistance with confocal microscope and E. Kondorosi, P. Mergaert, and W. Van de Velde for critical reviews of the manuscript and for sharing unpublished results. This work was supported by the Helen Hay Whitney Foundation (to J.G.), a National Institutes of Health training grant (to C.S.), The Netherlands Organisation for Scientific Research (NWO, to E.L., S.I., E.F., and T.B.), the Hoover Circle fund, and prior support from the Howard Hughes Medical Institute and the U.S. Department of Energy grant no. DE–FG03–90ER20010 (to S.R.L.).
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/327/5969/1126/DC1
Materials and Methods
References
References and Notes
- 1.Starker CG, Parra-Colmenares AL, Smith L, Mitra RM, Long SR. Plant Physiol. 2006;140:671. doi: 10.1104/pp.105.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitra RM, Long SR. Plant Physiol. 2004;134:595. doi: 10.1104/pp.103.031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasse J, de Billy F, Camut S, Truchet G. J Bacteriol. 1990;172:4295. doi: 10.1128/jb.172.8.4295-4306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson AR, Vierstra RD. Curr Opin Plant Biol. 2005;8:165. doi: 10.1016/j.pbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Limpens E, et al. Plant Cell. 2009;21:2811. doi: 10.1105/tpc.108.064410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catalano CM, Czymmek KJ, Gann JG, Sherrier DJ. Planta. 2007;225:541. doi: 10.1007/s00425-006-0369-y. [DOI] [PubMed] [Google Scholar]
- 7.Mitra RM, et al. Proc Natl Acad Sci USA. 2004;101:4701. doi: 10.1073/pnas.0400595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paetzel M, Karla A, Strynadka NC, Dalbey RE. Chem Rev. 2002;102:4549. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 9.Fang H, Mullins C, Green N. J Biol Chem. 1997;272:13152. doi: 10.1074/jbc.272.20.13152. [DOI] [PubMed] [Google Scholar]
- 10.Meyer HA, Hartmann E. J Biol Chem. 1997;272:13159. doi: 10.1074/jbc.272.20.13159. [DOI] [PubMed] [Google Scholar]
- 11.Van de Velde W, et al. Science. 2010;327 [THIS ISSUE] [Google Scholar]
- 12.Benedito VA, et al. Plant J. 2008;55:504. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- 13.El Yahyaoui F, et al. Plant Physiol. 2004;136:3159. doi: 10.1104/pp.104.043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manthey K, et al. Mol Plant Microbe Interact. 2004;17:1063. doi: 10.1094/MPMI.2004.17.10.1063. [DOI] [PubMed] [Google Scholar]
- 15.Raikhel N, Chrispeels MJ. In: Biochemistry and Molecular Biology of Plants. Buchanan BB, Gruissem W, Jones RL, editors. American Society of Plant Biologists; Rockville, MD: 2000. pp. 160–201. [Google Scholar]
- 16.Du Y, Ferro-Novick S, Novick P. J Cell Sci. 2004;117:2871. doi: 10.1242/jcs.01286. [DOI] [PubMed] [Google Scholar]
- 17.Dalbey RE, Lively MO, Bron S, van Dijl JM. Protein Sci. 1997;6:1129. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mergaert P, et al. Plant Physiol. 2003;132:161. doi: 10.1104/pp.102.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Graaf BH, et al. Plant Cell. 2005;17:2564. doi: 10.1105/tpc.105.033183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Catalano CM, Lane WS, Sherrier DJ. Electrophoresis. 2004;25:519. doi: 10.1002/elps.200305711. [DOI] [PubMed] [Google Scholar]
- 21.Coque L, et al. Mol Plant Microbe Interact. 2008;21:404. doi: 10.1094/MPMI-21-4-0404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


