Abstract
A mechanism by which control DNA elements regulate transcription over large linear genomic distances is by achieving close physical proximity with genes, and looping of the intervening chromatin paths. Alterations of such regulatory ‘chromatin looping’ systems are likely to play a critical role in human genetic disease at large. Here, we studied the spatial organization of a ≈790 kb locus encompassing the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Dysregulation of CFTR is responsible for cystic fibrosis, which is the most common lethal genetic disorder in Caucasian populations. CFTR is a relatively large gene of 189 kb with a rather complex tissue-specific and temporal expression profile. We used chromatin conformation at the CFTR locus to identify new DNA sequences that regulate its transcription. By comparing 5C chromatin interaction maps of the CFTR locus in expressing and non-expressing human primary cells, we identified several new contact points between the CFTR promoter and its surroundings, in addition to regions featuring previously described regulatory elements. We demonstrate that two of these novel interacting regions cooperatively increase CFTR expression, and suggest that the new enhancer elements located on either side of the gene are brought together through chromatin looping via CTCF.
INTRODUCTION
Many different types of regulatory mechanisms can contribute to the correct spatial and temporal expression level of a gene. At the transcription stage, gene expression is regulated in part by control DNA elements that either promote or prevent transcription. These regulatory sequences are not necessarily located immediately next to the promoter they regulate but can be found downstream of the gene, within its introns, in other genes or non-coding regions (1,2). Control DNA elements can localize far away from their gene target—even megabases away—on the same or a different chromosome. DNA sequences can control gene expression over large distances by achieving close physical proximity with their target genes (3). Consequently, both changes in chromatin organization and variations in regulatory element sequences may affect gene expression. Alterations in this cis-acting regulatory mechanism has been linked to human genetic disease (4,5) like in the facioscapulohumeral muscular dystrophy (FSHD) disorder (6), the congenital eye malformation aniridia (7) or the blepharophimosis syndrome (BPES) (8). Therefore, identifying regulatory DNA sequences and investigating how they physically relate to each other and to genes are both essential to fully understand transcription.
Cystic fibrosis (CF) is the most frequent lethal inherited disorder in Caucasian populations. It is caused by mutation of both alleles of the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which was identified in 1989 (9–11). The CFTR gene is particularly expressed in the epithelial cells of the airway, the pancreas, the small intestine and the male genital ducts (12). Its expression is also strictly regulated both spatially and temporally (13). The molecular mechanisms underlying the strict transcription regulation of CFTR remain poorly described and understood. For instance, the CFTR promoter does not contain the regulatory elements responsible for complex cell-type specific and temporal regulation (14,15). In fact, it has many features of a ‘housekeeping gene’, as it does not possess a TATA box, is GC-rich, contains multiple transcriptional start sites and many putative Sp1 and AP-1 protein binding sites (16). Also, mutations within the coding regions of CFTR are not systematically found in individuals affected by the disease, suggesting that genetic variations in remote regulatory elements may change CFTR expression and induce the disease. CF-causing mutations have already been described in the large 5′ promoter region (17,18), in non-coding regions (19) and in the 3′ region (20) which could all contain remote regulatory elements.
Remote regulatory sequences can be identified by mapping DNAse I hypersensitive sites (DHS), which correspond to regions where DNA is more accessible, like the nucleosome-free regions often found at regulatory elements. This approach was used successfully to identify several CFTR enhancers. The first element was identified in 1996 within the gene in intron 1 (185 + 10 kb) (21). Its enhancer activity was next described in different studies particularly in intestinal cells (22–24) along with the binding of many transcription factors like HNF1α, CDX2, TCF4 and the histone acetyltransferases p300 (25–27). Other DHS regulatory elements were later identified upstream of the CFTR transcriptional start site (TSS) including the DHS -20,9 kb, which binds the CCCTC-binding factor (CTCF) (28), and one downstream of CFTR (DHS 4574 + 15,6 kb), which regulates CFTR expression through binding of CREB/ATF, AP-l and C/EBP and other factors like ARP-l and HNF-4 (29).
Another approach used successfully to identify CFTR enhancers is by mapping chromatin organization with the chromosome conformation capture (3C) technique developed by Dekker et al. in 2002 (30). 3C is a molecular approach that measures the frequency of pair-wise interactions between given chromosomal regions, and can therefore be used to identify DNA elements that regulate genes by physically interacting with them. 3C analysis of the CFTR locus revealed a physical proximity between the CFTR promoter and previously characterized DHS located upstream, downstream and intronically within the gene. A strong interaction was particularly shown between the CFTR promoter and a region located ≈80 kb upstream, which includes the DHS -79,5 kb (31). This contact was characterized in several cell lines (Caco-2, HT29 and HeLa) (32). A second study also described this proximity in 16HBE14o- cells (33). A region with the DHS 1811+0,8 kb within intron 11 (34), was further shown to interact specifically with the CFTR promoter in intestinal cells and act as a strong and specific enhancer by recruiting different transcriptional factors like HNF1, p300, FOXA1, FOXA2 and CDX2 (26,32,35). Additionally, a region with the DHS 4574 + 15,6 kb located downstream of the gene (36) is specifically associated with the CFTR promoter (26,32). Furthermore, a study in a 1.8-Mb region of human chromosome 7 encompassing the CFTR gene identified silencers and enhancer-blockers in this greater CFTR locus (37).
Despite having identified several regulatory elements, these do not fully explain CFTR expression or lack thereof in patients, perhaps in part because studies often make use of cell lines. Thus, several additional enhancers are likely involved in orchestrating the tissue-specific expression of CFTR, including the airway. Here, we use chromatin conformation as an approach to identify new regulatory elements functioning especially in CFTR-expressing cells. We analyse the spatial organization of a ≈790 kb region spanning the CFTR locus in expressing and non-expressing primary human cells using the 5C technique (see below). By comparing the interaction profile of the CFTR promoter in expressing human nasal epithelial cells (HNECs) and non-expressing skin fibroblasts (SFs), we identify several new contact points along the locus that are present only when the gene is active. Amongst these are regions corresponding to previously characterized regulatory elements, thereby validating our approach. We demonstrate that two new interacting regions cooperatively increase CFTR expression and suggest that these novel enhancer elements are brought together through chromatin looping via CTCF.
MATERIALS AND METHODS
Cell collection and culture
Human nasal epithelial cells (HNECs) from control individuals (carrying no CFTR mutation) were harvested using a sterile cytology brush (Gyneas, cat. no. 02–106). HNECs were collected by brushing the middle turbinate with backward-forward and rotatory movements and were placed in a collecting medium (Ham's F12 (Lonza) containing 1% Ultroser G (Pall, cat. no. 15950–17), and 1% antibiotic (PAA, cat. no. P11–002). Survival of the cells in this medium was tested until 96 h at room temperature. HNECs were detached from the brush by vortexing and were washed three times with Dulbecco's modified eagle medium (DMEM; Lonza) supplemented with decreasing antibiotic concentrations and a final wash with 1X phosphate buffered saline (PBS). Cell suspensions were centrifuged 5 min at 2200 g at 4°C. The cell pellets were resuspended in a selected and optimized cell culture media of human small airway epithelial cells (SAGM; Lonza) and cells were cultured following the manufacturer's instructions.
Abdomen SFs were isolated from adult women undergoing surgery. Samples were aseptically collected and washed three times with 70% ethanol and PBS. Skin samples were next cut into small pieces and treated with dispase (25 U/ml Gibco, USA) overnight in air-liquid interface at 4°C. The dermis was separated from the epidermis, and cells were dissociated with a trypsin-EDTA solution (LONZA) for 30 min. After filtration, cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS-PAA) and 1% antibiotic, at 37°C in 5% CO2 saturated humid air. For HNECs as for SFs, cell culture homogeneity was monitored by morphology under the microscope, and collected samples appeared 100% epithelial (HNEC) or fibroblastic (skin) in nature.
16HBE14o- human bronchial epithelial cells (38) were grown in DMEM with 10% FBS. All cells were grown on plastic at liquid interface. For HNECs and SFs, any suspension cells such as white or red blood cells collected along with the original samples will be washed away when replenishing the media during cell expansion.
CFTR expression analysis in HNEC and SF samples by endpoint reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted using RNeasy Plus mini kit (Qiagen) according to the manufacturer's instructions. Extracted RNA was eluted in RNase-free water and the concentration was determined using a nano-photometer (Implen GmbH, München, Germany). Reverse transcription was performed with 2 μg of total RNA using the SuperScript™ II Reverse Transcriptase kit (Life Technologies). PCR was performed with the HotStarTaq Polymerase (Qiagen) using the following primer sequences (5′-3′): CFTR forward-TTTCGTGTGGATCGCTCCTT, CFTR reverse-TCCAGCAACCGCCAACAACT, beta-actin forward-GTTGCTATCCAGGCTGTG, beta-actin reverse-CACTGTGTTGGCGTACAG.
Cell isolation and fixation for 5C analysis
HNEC and SF cells were fixed with 2% formaldehyde for 10 min at room temperature. Crosslinking was stopped with glycine (125 mM final concentration), by incubating 5 min at room temperature followed with 15 min on ice. Cells were scraped and centrifuged at 400 g for 10 min at 4°C. Supernatants were removed and the cell pellets were quick-frozen on dry ice.
3C library preparation
The fixed HNEC and SF cell pellets were treated as previously described (39,40). Briefly, 10 million cells were incubated in lysis buffer (10 mM Tris (pH 8.0), 10 mM NaCl, 0.2% NP-40, supplemented with fresh protease inhibitor cocktail) 10 min on ice. Cells were then disrupted on ice with a Dounce homogenizer (pestle B; 2 × 20 strokes). Nuclei were recovered by centrifugation, washed twice with 1X EcoRI buffer (NEB) and resuspended in 100 μl of 1X EcoRI buffer. 1X EcoRI buffer (337 μl) was added to 50 μl of cell suspension, and the mixture was incubated 10 min at 65°C with 0.1% SDS final concentration (38 μl). Triton X-100 (44 μl of 10% Triton X-100) was added before overnight digestion with EcoRI (400 U). The restriction enzyme was inactivated the next day by adding 86 μl of 10% SDS, and incubating 30 min at 65°C. Samples were then individually diluted into 7.62 ml of ligation mix (750 μl of 10% Triton X-100, 750 μl of 10X ligation buffer, 80 μl of 10 mg/ml BSA, 80 μl of 100 mM ATP and 3000 Cohesive end Units of T4 DNA ligase). Ligation was carried out at 16°C for 2 h.
The 3C libraries were next incubated overnight with 45 μl of Proteinase K (10 mg/ml) at 55°C. The DNA was purified by phenol-chloroform extraction and precipitated with 3 M NaOAc pH 5.2 (800 μl) and cold ethanol. After at least 1 h at –80°C, the DNA was recovered by centrifugation, the pellets were washed with cold 70% ethanol and then resuspended in 400 μl of 1X TE pH 8.0. A second phenol-chloroform extraction and precipitation with 3 M NaOAc pH 5.2 (40 μl) and cold ethanol were performed. DNA was recovered by centrifugation and washed eight times with cold 70% ethanol. The pellets were finally dissolved in 100 μl of 1X TE pH 8.0, and incubated with RNAse A (1 μl at 10 mg/ml) for 15 min at 37°C.
5C primer and library design
5C primers covering the CFTR region (hg19, chr7:116,600,610–117,391,651), the ERCC3 region (hg19, chr2:128,014,866–128,051,752) and the ENr313 region (hg19, chr16:62,276,449–62,776,448) were designed using ‘my5C.primer’ (41) and the following parameters: optimal primer length of 30 nt, optimal TM of 60°C, default primer quality parameters (mer: 800, U-blast: 3, S-blasr: 50). Primers were not designed for large (>20 kb) restriction fragments. Low complexity and repetitive sequences were excluded from our experimental design such that not all fragments could be probed in our assays. Primers with several genomic targets were also removed.
The universal A-key (CCATCTCATCCCTGCGTGTCTCCGACTCAG-(5C-specific)) and the P1-key tails ((5C-specific)-ATCACCGACTGCCCATAGAGAGG) were added to the Forward and Reverse 5C primers, respectively. Reverse 5C primers were phosphorylated at their 5′ ends. An anchored 5C design was used for the CFTR locus, 2 Reverse 5C primers targeted the CFTR promoter while 140 Forward 5C primers covered the surrounding region.
For the control regions, an alternating 5C design was used: alternating Forward and Reverse 5C primers covering entire ERCC3 and ENr313 regions were used to generate the 5C libraries. This design used 31 primers (2 Forward / 3 Reverse for the ERCC3 region, 13 Forward / 13 Reverse ENr313 region). All 5C primer sequences are listed in Supplementary Table S1.
5C library preparation
5C libraries were prepared and amplified with the A-key and P1-key primers following a procedure described previously (42). Briefly, 3C libraries were first titrated by PCR for quality control (single band, absence of primer dimers, etc.), and to verify that contacts were amplified at frequencies similar to what is usually obtained from comparable libraries (same DNA amount from the same species and karyotype) (39,43,44).
5C primer stocks (20 μM) were diluted individually in water on ice, and mixed to a final concentration of 0.002 μM. Mixed primers were combined with annealing buffer (10X NEBuffer 4, New England Biolabs Inc.) on ice in reaction tubes. Salmon testis DNA was added to each 5C reaction, followed by the 3C libraries and water for a final volume of 10 μl. Samples were denatured at 95°C for 5 min, and annealed at 50°C for 16 h before ligation with Taq DNA ligase (10 U) for 1 h. Each ligation reaction was then PCR-amplified individually with primers against the A-key and P1-key primer tails. We used 35 cycles based on dilution series showing linear PCR amplification within that cycle range. The 5C PCR products of corresponding 3C libraries were pooled before purifying the DNA on MinElute columns (Qiagen).
5C libraries were quantified on agarose gel and diluted to 0.0018 ng/μl (for Ion OneTouch™ 200 Template Kit v2 DL). One microliter of diluted 5C library was used for sequencing with an Ion PGM™ sequencer. Samples were sequenced onto Ion 314™ Chips or Ion 314™ Chips v2 following the Ion OneTouch™ 200 Template Kit v2 DL and Ion PGM™ 200 Sequencing Kit protocols as recommended by the manufacturer's instructions (Life TechnologiesTM).
5C data analysis
Analysis of the 5C sequencing data was performed as described earlier (42,45). The sequencing data were processed through a Torrent 5C data transformation pipeline on our local instance of Galaxy as previously described (42,46). This analysis generates an excel sheet containing interaction frequency lists (IFL) as well as a text file, which was used to visualize results using ‘my5C-heatmap’ (41).
Normalization between different libraries was done first by read count and then using the compaction profiles for the ERCC3 region and the gene desert region ENr313, setting one sample (HNEC) as a reference.
The ERCC3 and ENr313 5C data of each sample were normalized by dividing the number of reads of each 5C contact by the total number of reads from the corresponding sequence run. A ratio, calculated by dividing these normalized data by normalized data from a reference sample (HNEC), was applied to the corresponding raw data of the study region for each sample. The normalized 5C data for all the SF and HNEC samples are found in Supplementary Tables S3–S5 and Supplementary Tables S6–S8, respectively.
Databases and URLs
The Hi-C data from human ESC-H1 were downloaded from Gene Expression Omnibus (GEO) using accession number GSE35156 and processed as previously described through the hiclib software presented in (47), using the default parameters and mapped to hg19. The Hi-C data shown in heatmap format were binned at 30 kb. Topological Associating Domains (TAD) boundaries were calculated with the directionality index as previously described in (48), using a 500 kb window. Sub-TADs were outlined manually based on clustering of interaction frequencies.
DHS and CTCF data sets for the SAEC (Acc. No. GSE26328 and GSE30263) were downloaded from the GEO website at http://www.ncbi.nlm.nih.gov/geo/query/acc. The ‘my5C-primer’ and ‘my5C-heatmap’ bioinformatics tools are found at http://3dg.umassmed.edu/my5Cheatmap/heatmap.php.
The published 5C data were obtained and visualized using the UCSC genome browser (GM12878; wgEncodeEH001665, H1-hESC; wgEncodeEH001666, HeLa-S3; wgEncodeEH001667, K562; wgEncodeEH001668) (http://genome.ucsc.edu)(49). The corresponding CTCF ChIP-seq data were also found and visualized using the UCSC browser (GM12878; wgEncodeEH000029, H1-hESC; wgEncodeEH000085, HeLa-S3; wgEncodeEH001012, K562; wgEncodeEH000042). The DNAse I hypersensitivity cluster data from 125 cell types and the transcription factor ChIP-seq data of 161 factors from 91 cell types was uploaded from the Integrated Regulation from ENCODE tracks in the UCSC browser.
Plasmid constructs
All the cloning steps were done using the ‘In fusion®’ strategy from Clontech. Using the pGL3-Basic Vector (Promega), the 5′-flanking region of the CFTR gene (1662 bp, ‘PCFTR’) was cloned upstream the firefly luciferase cDNA, at the Hind III site. Candidate enhancer elements (A to G) were amplified and inserted downstream in the PCFTR construct.
Positive and negative controls, containing respectively a fragment of intron 23 and intron 11 of the CFTR gene were generated. These elements were previously described to have or not a significant enhancer activity (33). All the inserted fragments were verified by sequencing. The PCR primers used to amplify the CFTR promoter, candidate enhancer sequences and control sequences are shown in Supplementary Table S2.
Luciferase assay
2.5 × 105 cells were seeded in 6-well plates. Transfections were done 24 h later with the transit 2020 reagent (Mirus). 800 ng of the PCFTR constructs and 200 ng of a pCMV-LacZ construct, as internal control, were used for each condition. Every condition was done in triplicate.
48 h post-transfection the cells were washed once with 1X PBS and lysed with Passive lysis buffer (Promega). Cells lysates were clarified by centrifugation at 15 000 g for 5 min at 4°C. 20 μl of each protein extract were used to assay the luciferase activity and 25 μl for beta-galactosidase activity. We used Promega reagents and a multiwell plate reader Varioscan (Thermo Fisher). Results were presented as relative luciferase activity, with the PCFTR construct activity equal to 1. Significance of the increased luciferase activity was performed using unpaired t-tests using GraphPad Prism® software.
Chromatin immunoprecipitation (ChIP) and PCR
Formaldehyde (Sigma) was added to 10 million HNEC cells to a final concentration of 1.5%. Crosslinking was allowed to proceed for 10 min at room temperature and stopped by addition of glycine at a final concentration of 0.125M. Fixed cells were washed and harvested with cold PBS. Chromatin was prepared following the SimpleChIP® Plus Enzymatic Chromatin IP protocol (Cell Signaling Technology) with minor modifications. The Adaptive Focused Acoustics™ (AFA) Technology from Covaris was used in addition to enzymatic digestion with microccocal nuclease in order to produce DNA fragments of 150 to 900 bp. Chromatin was precleared with protein G magnetic beads (Cell Signaling Technology) for 2 h at 4°C and immunoprecipitation with the CTCF specific antibodies, a negative control IgG antibody or a positive control Histone H3 antibody (Cell Signaling Technology) were carried out overnight at 4°C. Immune complexes were recovered by adding protein G magnetic beads and incubated for 2 h at 4°C. Beads were washed, DNA was eluted and cross-links were reversed following the manufacturer's instructions. PCR were performed using HotStarTaq Mastermix (Qiagen) and specific primers, as outlined in Supplementary Table S9. Details are available upon request.
RESULTS
Analysing the human CFTR locus with the 5C technology
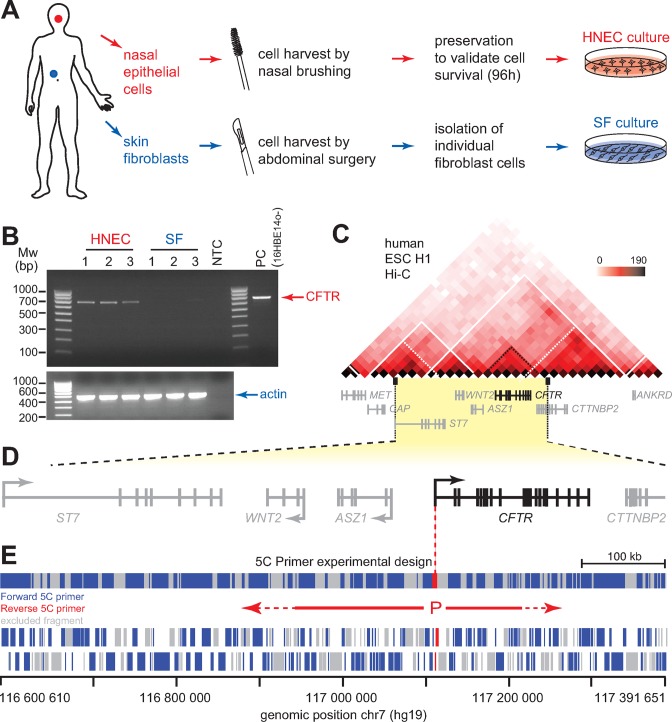
To identify new regulatory elements involved in the control of CFTR expression, we chose to conduct our analysis in primary human cells from healthy individuals. Primary cells offer physiological CFTR expression levels and a normal karyotype in contrast to cell lines and tissues where CFTR expression is highly variable. We used primary epithelial cells and primary SFs, which express high and very low levels of CFTR, respectively. Since the most easily recoverable primary epithelial cells are those from the middle turbinate of the nose, we developed a self-nasal epithelial cells sampling and culturing protocol for this study (Figure 1A). The procedure consists in nasal brushing for 1 min, followed by cell collection and conservation in Ham's F12/Ultroser G, and culturing in Small Airway Epithelial Growth Media (see MATERIAL AND METHODS). The primary SFs were collected from adult women undergoing surgery as described in MATERIAL AND METHODS. We collected several samples for both cell types to avoid variation due to genetic differences, and verified that CFTR is preferentially expressed in HNECs compared to SFs (Figure 1B).
Figure 1.
5C analysis of the human CFTR locus in primary human cells. (A) Schematic representation of the procedures used to collect SFs and nasal epithelial cells (HNECs) from control (wt CFTR) individuals. (B) Endpoint RT-PCR analysis of CFTR expression in collected SF and HNEC samples, and in control 16HBE14o- human bronchial epithelial cells. Actin is shown as control for RNA integrity. (C) Hi-C data from the H1 human ES cell line (H1-hESC) reveal that CFTR lies in a large TAD that contains several other genes (48). CFTR is further contained within a sub-TAD. The Hi-C data are shown in heatmap form where increasing colour intensity reflects higher interaction frequencies between genomic regions in 30 kb-resolution bins. White lines outline the TADs while the white dotted lines represent the sub-TADs. The genes contained within the area are shown under the heatmap. (D) Linear schematic representation of the ≈790 kb genomic region characterized in this study. Arrows indicate transcription orientation. The CFTR gene is shown in black and the neighbouring genes in grey. (E) Anchored 5C primers design scheme used to map the genomic environment of the CFTR promoter. The two reverse ‘bait’ 5C primers covering the CFTR promoter (P) are shown in red. 140 Forward 5C primers shown in blue represented the rest of the locus. Grey areas indicate regions not probed by 5C. Region coverage is summarized at the top, and individual fragments are shown at the bottom. 5C primer sequences are found in Supplementary Table S1.
Previous search for CFTR enhancers focused mostly on the 3′ region up to 80 kb from the end of the gene (50). However, Zhang et al. showed that the CFTR promoter also interacts with the 5′ region (33). We therefore chose to search for CFTR promoter-interacting partners in a larger area extending the analysis mainly towards the 5′ end. To delineate more precisely the domain where the CFTR promoter interactions are more likely to occur, we benefited from the Hi-C data generated from a human embryonic stem cell line (H1-hESC; Figure 1C)(48). Hi-C is a molecular technique used to quantify the frequency of chromatin interactions genome-wide in cell populations (51). This method was used to demonstrate that chromatin folds into TADs that correspond to chromosomal regions interacting more with themselves than with the rest of the chromosome (48,52). TADs represent intermediate units of chromosome folding, which tend to be conserved across cell types and species, and thought to play a role in the functional compartmentalization of genomes (53). Accordingly, genes within given TADs often display coordinated expression dynamics during differentiation, and at least in mouse, enhancer-promoter interactions are enriched within TADs (54,55).
Analysis of the H1-hESC Hi-C data revealed that CFTR lies within a TAD spreading from the ASZ1 to the ANKRD gene (Figure 1C). CFTR itself localizes within a TAD substructure (sub-TAD), which is also characterized as a chromatin region that preferentially interacts with itself but is smaller in size (56). A second TAD overlaps with the TAD containing CFTR, and extends up to the ST7 gene. As TADs are largely conserved between cell types, these data suggest that the chromosomal region between ST7 and ANKRD may potentially interact with the CFTR promoter in different cell lines (48). We therefore decided to focus our conformation analysis on a ≈790 kb domain (hg19, chr7:116,600,610–117,391,651) encompassing CFTR, but also its flanking genes: ASZ1, WNT2, ST7 and CTTNBP2 (Figure 1D).
We used 5C-seq to find and quantify chromosomal regions interacting with the CFTR promoter in cis (42,57). The chromosome conformation capture carbon copy (5C) has previously been used to map both chromatin organization and detect physical networks between promoters and regulatory elements (45,56,58). Like Hi-C, the 5C method measures interaction frequencies between chromatin segments in cell populations. 5C starts with the production of a 3C library where a population of cells is fixed with formaldehyde to capture chromatin interactions. The fixed chromatin is then digested into fragments with a restriction enzyme, and a ligation step follows to generate pair-wise ligation products at frequencies reflecting physical proximity or accessibility between chromatin segments in-vivo. The resulting 3C library is then converted into a 5C library by ligation-mediated amplification (LMA) where Forward and Reverse 5C primers are sequentially annealed and ligated at defined 3C junctions in a multiplex setting. The 5C ligation products are then amplified by PCR and processed for deep sequencing. We designed 5C primers within this region to measure interactions frequencies between the CFTR promoter (represented by 2 Reverse 5C primers or ‘baits’) and the rest of the ≈790 kb domain (Figure 1E, Supplementary Table S1). This 5C design therefore provides information about the genomic environment around the CFTR promoter in vivo, including which enhancer-containing regions have access to it.
CFTR expression associates with higher long-distance interaction frequencies
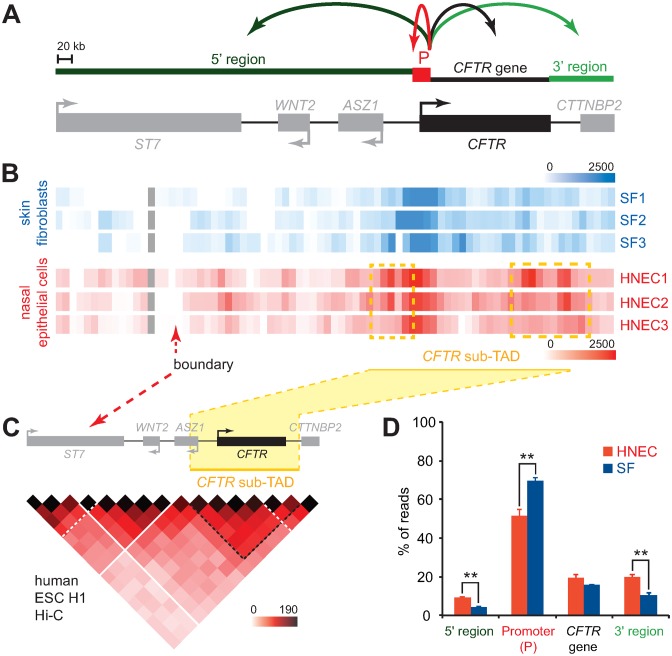
We mapped the interaction profile of the CFTR promoter within the ≈790 kb domain with 5C-seq in three SF and three HNEC samples isolated from different volunteers (Figure 2A, B). We first observed that the chromatin environment at the CFTR promoter captured by 5C-seq largely varies between individuals in both cell types (Figure 2B). For instance, the number and intensity of long-range interactions with upstream or downstream regions differed in SF samples. Similarly, not all long-range contacts with the CFTR promoter were the same in HNEC samples. Such variability is not unexpected and likely originates from many different sources including genetic variations amongst the donors, the overall transcription state of the cell populations (59), and the cell cycle distribution of the samples. Despite this variability however, we could detect long-range contacts preferentially found in HNECs in at least two individuals and by both promoter bait region. For example, one region located upstream and another downstream of the CFTR gene frequently and preferentially interacted with the promoter (highlighted in dashed yellow boxes). These contacts actually lie within and at opposite edges of the CFTR sub-TAD detected in the H1-hESC Hi-C data (Figure 2C). Moreover, we observed a dip in interaction frequency in both SF and HNEC, which corresponds to the upstream CFTR TAD boundary, suggesting that most CFTR promoter contacts lie within this domain (Figure 2B, C).
Figure 2.
CFTR expression corresponds to higher long-distance interaction frequencies. (A) Schematic linear representation of the ≈790 kb locus under study. The 4 regions quantified in (D) (5′ region, Promoter (P), CFTR gene and 3′ region) are shown above the diagram. Gene orientation is indicated with arrows. (B) 5C interaction maps of the CFTR promoter in 3 human SF and 3 human nasal epithelial cell (HNEC) samples. Interaction frequencies were measured from the CFTR promoter region using two Reverse 5C primer ‘baits’. The 5C data from the two anchor points were averaged, binned at 20 kb (2x step), and is represented in heatmap form where interaction frequencies are colour-code according to the respective scales. Averaged interaction frequencies are from the number of sequence reads after normalization. Heatmaps are aligned with the locus diagram shown in (A), and samples names are indicated on the right. (C) Regions interacting most frequently with the CFTR promoter are contained within the CFTR sub-TAD. The H1-hESC Hi-C data are from (48), and shown in heatmap form as in (A). (D) Graphical representation of the relative 5C signal distribution in SF and HNEC samples within each of the 4 regions shown in (A) (5′ region, CFTR promoter, CFTR gene body and 3′ region). The graph reveals higher long-range interaction frequencies are present when the CFTR promoter is active. Error bars indicate SEM, *P < 0.0002 and **P < 0.0001 using unpaired t-tests.
More general but different chromatin environment features could also be seen in SF and HNEC cells. In SF cells, which do not significantly express CFTR, the promoter was mostly engaged in short-range interactions with the CFTR gene body and the 3′ region (Figure 2B). Very high contact frequencies were found particularly immediately around the promoter, while the lowest interaction frequencies were measured with the 5′ region of the locus. This was confirmed by calculating the relative distribution of the sequence reads between the 5′ region, CFTR promoter (–16 to +0.5 kb from the TSS), CFTR gene body, and the 3′ region, where over 70% of the SF interactions mapped to the CFTR promoter region (Figure 2D). This CFTR promoter interaction pattern differs from the one seen in nasal epithelial cells where contacts with the CFTR promoter region is lower and redistributed to other regions along the domain, particularly at the 5′ and 3′ ends (Figure 2D). Considering that interaction frequency reflects relative in vivo distance and/or accessibility, these results suggest that the CFTR promoter is engaged in multiple long-range contacts with remote chromatin regions particularly in HNEC cells where CFTR is active. Previously uncharacterized regulatory elements controlling CFTR expression may therefore be present in either the 5′ or the 3′ region of the domain under investigation.
CFTR contact regions display characteristics of regulatory elements
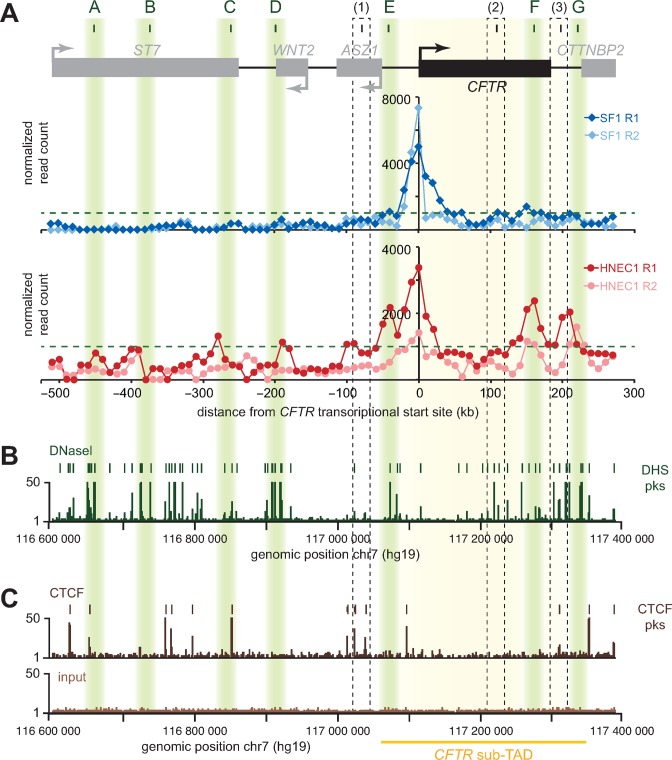
Since long-range contacts between the CFTR promoter and the 5′ and 3′ regions correlate with CFTR expression, some of them may correspond to DNA elements important for its regulation. To pinpoint candidate regions, we first compared the interaction profile of both CFTR promoter ‘baits’ in SF and HNEC samples (Figure 3A). As expected, both baits interacted more frequently around the promoter region in SF samples as compared to HNECs. Both baits also displayed similar interaction patterns in the respective cell lines, and as predicted, the one located further away from CFTR’s TSS (R2) interacted less frequently throughout the region than the R1 bait nearest to it, further supporting the specificity of these interactions with the CFTR promoter. In the nasal epithelial cell samples, we found several contact peaks bearing the canonical ‘interaction shoulders’ suggestive of looping contacts. Some of them correspond to previously characterized enhancer sequences, identified as DHS and shown to interact with the promoter by 3C in different cell lines, thereby validating our approach. These regions include one enhancer located ≈80 kb upstream of the promoter [1] (32), another within the gene body in intron 11 (26,32,60), and one downstream of the gene body (26,31–33,61).
Figure 3.
CFTR interacting regions display characteristics of regulatory elements. (A) Identification of candidate looping contacts. The 5C chromatin interaction profiles of two ‘bait’ regions upstream of CFTR's TSS for one of the skin fibroblast samples (SF; very low expressing) and one of the nasal epithelial cell samples (HNEC; expressing) are shown for comparison. Interaction frequency (y-axis) is correlated with the position from the transcriptional start of CFTR (x-axis). R1 is the bait closest to the TSS. The new contacting regions are highlighted in shaded green boxes (A to G). Previously reported looping contacts are indicated with black dashed boxes (1 to 3). (B) Alignment of DNAseI hypersensitivity data from SAEC over the region characterized. DHS peaks are indicated at the top and identify open chromatin sites. (C) Alignment of CTCF binding sites from SAEC over the area studied. The CFTR sub-TAD position is at the bottom and highlighted in yellow.
Several new ‘looping contacts’ were also captured preferentially in HNECs, and we examined whether some of them displayed enhancers features using marker data available in human Small Airway Epithelial Cells (SAECs). These cells originate from the lung and express CFTR, and we reasoned that regulatory elements controlling CFTR in this cell line would at least partially recapitulate the regulatory network in our nasal epithelial cells. We looked particularly for the presence of DHS and for binding of the CCCTC-binding factor (CTCF) because they often reflect the presence of regulatory elements and since CTCF is a major protein driving the formation of chromatin loops genome-wide (62). We found seven new contact regions in HNECs (Figure 3A, ‘A to G’) that coincide with DHS and/or CTCF sites in SAECs (Figure 3B,C). Five contacts involve sequences within the 5′ region (Figure 3, ‘A to E’), 1 is within the CFTR gene body (Figure 3, ‘F’) and 1 localizes to the 3′ region (Figure 3, ‘G’). Strong CTCF peaks can be observed in regions C and G, as well as in A although CTCF binding appeared less frequent in this region. E, F and G were within the very strong CFTR sub-TAD seen in the Hi-C data of H1-hESCs.
Several interacting DHS show enhancer activities
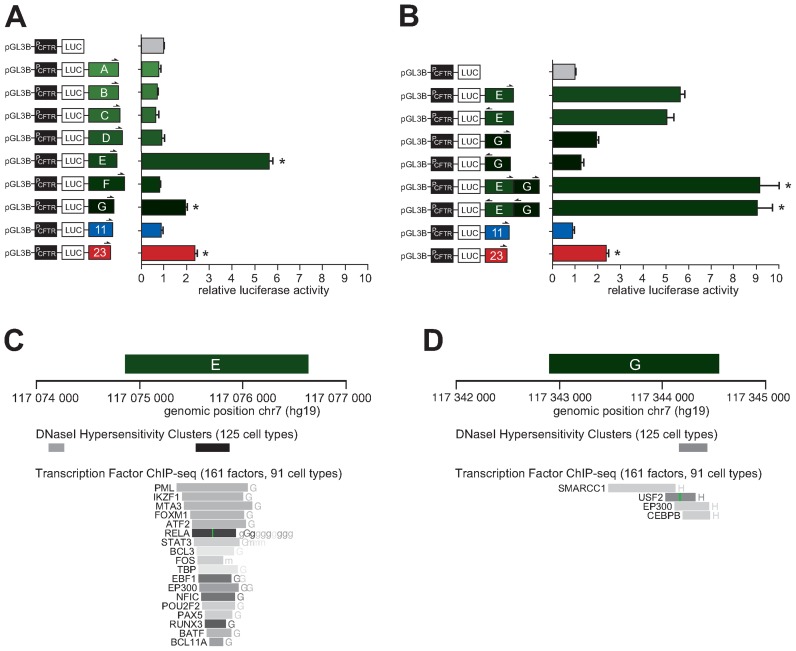
We tested whether any of the newly identified looping regions displayed enhancer activity in a reporter assay. For this analysis, we first prepared a ‘PCFTR’ construct by subcloning a 1662 bp CFTR promoter fragment into the pGL3-Basic vector, upstream of a modified firefly luciferase coding region optimized for analysis of transcription activity in eukaryotic cells. Regions A to G were then amplified by PCR and inserted into PCFTR (Supplementary Table S2). As positive control, we subcloned a fragment of the intron 23 (DHS 4374 + 1.3 kb) of CFTR. The intron 23 DHS was previously shown to display enhancer activity in airway epithelial cells and in intestinal cells at a lower extent (33). Our negative control was a PCFTR construct where the intron 11 (DHS 1811 + 0.8 kb) of CFTR was inserted downstream of the promoter. The intron 11 element was identified as an intestinal-specific enhancer active in Caco-2 cells that was later found inactive in airway epithelial cells (26,33).
We co-transfected these constructs individually into 16HBE14o- bronchial epithelial cells along with a LacZ plasmid as a control of transfection efficiency. Firefly luciferase expression was measured 48 h post-transfection and normalized against the CFTR promoter construct alone (PCFTR), which was set at 1. As shown in Figure 4A, two of our interacting regions significantly enhanced CFTR promoter activity. The fragment encompassing a DHS in the region E displayed a strong enhancer function with almost 6-fold effect on the CFTR promoter. A DHS in this region at −44 kb was reported previously in airway epithelial cells (33), and later shown to enhance CFTR expression (63), although whether it physically interacts with the CFTR promoter had not been shown. The G region containing a DHS and a CTCF binding site also acted as an enhancer, although more modestly at a level comparable to our positive (intron 23) control (≈2-fold). The other looping regions did not show enhancer activity.
Figure 4.
Two interacting DHS regions cooperatively increase CFTR expression in the airway. (A) 16HBE14o- cells were transfected with luciferase reporter constructs containing the CFTR basal promoter (PCFTR; 1662 bp), and fragments of DHS elements of the region A to G. An enhancer sequence inactive in the airway located in intron 11 and a known airway enhancer in intron 23 were used as negative and positive controls, respectively. (B) 16HBE14o- cells were transfected with luciferase reporter constructs containing the CFTR basal promoter (PCFTR; 1662 bp), and fragments at DHS-E or -G, individually or in combination in either orientation as indicated by half arrows. Luciferase data in (A) and (B) are shown relative to the CFTR basal promoter vector (set to 1). Error bars represent the standard error of the mean (SEM; n = 6), * P<0.0001 using unpaired t-tests. (C,D) Enhancer regions E and G overlap with DNaseI hypersensitivity cluster sites identified from 125 different cell lines, and with several transcription factor or chromatin remodeller binding sites with Factorbook motifs identified by ChIP-seq in 91 different cell lines.
Based on these results, we wondered whether these new enhancers might act supportively to activate the CFTR promoter. To address this question, we subcloned regions E and G together into the enhancer site of PCFTR construct in either orientation. Interestingly, the two elements increased luciferase transcription 9-fold when combined in either forward or reverse orientations (Figure 4B). These data suggest that regions E and G can act cooperatively to enhance CFTR promoter expression in airway epithelial cells. We examined the E and G enhancer regions against the DNAse I Hypersensitivity Clusters track in 125 cell lines from ENCODE and found that each overlap with a DHS (Figure 4C, D). Regions E and G also span sequences found to bind different types of transcription factors and chromatin remodellers in an ENCODE compilation of 161 factors with Factorbook motifs in 91 cell types (see MATERIAL AND METHODS).
The CFTR sub-TAD as an active chromatin hub
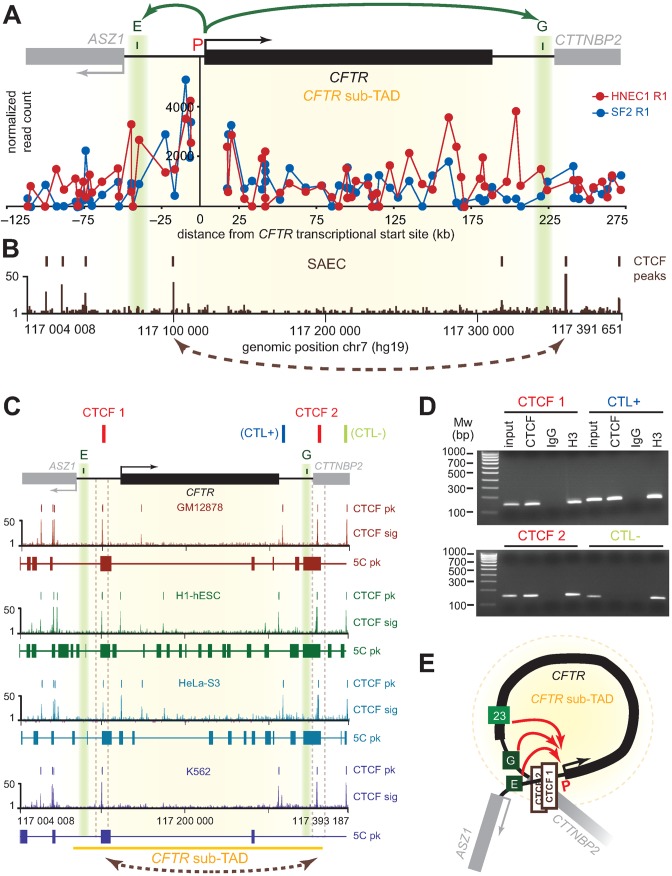
Enhancers E and G are located on opposite ends of the CFTR gene and at the sub-TAD boundaries it is contained within (Figure 5A). These elements are localized immediately upstream of strong CTCF peaks in SAECs (Figure 5B) as well as in H1-hESCs, GM12878, HeLa-S3 and K562 (Figure 5C). Interestingly, the regions containing these CTCF sites were previously found to interact in H1-hESCs, GM12878 and HeLa-S3 as measured by 5C (Figure 5C). CTCF binding is largely invariant across cell types in mammals (64), and we verified that it also binds at sites neighbouring the E and G enhancers in HNECs using ChIP-PCR (Figure 5D and Supplementary Figure S1). We found that CTCF binds at predicted sites close to these two regulatory elements in HNEC cells. We suggest that enhancers E and G cooperatively activate the CFTR promoter by achieving physical proximity via CTCF-CTCF binding and looping of the intervening chromatin (Figure 5E). The formation of such chromatin ‘hub’ might be more specific to certain tissues than dependent on transcription since low expression levels of CFTR are expected in both GM12878 and K562 (65), whereas long-range contacts between CTCF sites are absent only in K562.
Figure 5.
Formation of a CFTR ‘active chromatin hub’ explains the cooperative behaviour of the two newly identified CFTR enhancer sequences. (A) Comparison of the CFTR promoter interaction profile in SF and HNEC samples at the restriction fragment level within the CFTR sub-TAD. The CTCF interaction profile mapped by ChIP-seq in SAEC is aligned in (B) to highlight the position of enhancers E and G relative to CTCF peaks. (C) CTCF binding is conserved across cell lines and overlap previously reported 5C contacts in GM12878, H1-hESc and Hela-S3 cells. (D) CTCF binds at key locus sites adjacent to elements E and G in HNECs. CTCF binding was measured by ChIP-PCR with the primers listed in Supplementary Table S9. Regions examined include the two test sites (CTCF 1 and CTCF 2), a positive control (CTL+) already described (69), and a negative control (CTL–). The results of one biological replicate are shown. Full gel pictures and data from two additional replicates can be found in Supplementary Figure S1. (E) Three-dimensional regulation model of the CFTR gene. We suggest that enhancers E and G cooperatively activate the CFTR promoter by achieving physical proximity via CTCF-CTCF binding and looping of the intervening chromatin.
DISCUSSION
Investigating the mechanisms that regulate CFTR transcription is important because these remain poorly characterized. Also, cases of cystic fibrosis and CFTR-related disorders without genotypic explanation or mutation in the CFTR coding regions have been reported, which suggest that features of regulation remain uncharacterized, particularly long-distance regulation. Other non-coding regions have been associated with CFTR regulatory mutations in CFTR-related disease (66,67). Thus, we used 5C technology to identify distant regulatory elements that specifically interact with the active CFTR promoter. We chose to conduct our analysis in primary cells to avoid working with the altered expression patterns of cell lines. We used nasal epithelial cells as CFTR-expressing cellular type and skin fibroblastic cells as non-expressing cells. Although nasal epithelial cells do not show the highest expression, compared to intestinal or genital duct cells for instance, we chose them due to its ease of accessibility.
We observed two different chromatin interaction profiles according to the state of CFTR expression (Figure 2). Fibroblastic cell samples, where CFTR is expressed at very low levels, exhibited a profile where most contacts were located within the CFTR promoter region, and interaction frequencies rapidly decreased with more remote regions. This contact pattern is consistent with an unfolded organization of the region. The promoter region appeared more open in the nasal epithelial cells, which express much higher CFTR levels as compared to SFs. Indeed previous studies reported less frequent contact frequencies around active promoters due to local chromatin decondensation (68). Additionally, the whole locus is engaged in numerous interactions with the CFTR promoter through seven distal regions.
The CFTR interacting partners lie in the extragenic regions, upstream and downstream of the gene, as well as intronically. Some of them have already been identified in previous 3C analyses (Figure 3, elements 1 to 3), validating our 5C approach for regulatory element mapping. For example, we detected contacts between the CFTR promoter and a region located ≈80 kb upstream (Figure 3, region 1), which was originally observed in HeLa and Caco-2 cells, and later detected in Caco-2 and 16HBE14o- cells by a second group (32,33). We also found interactions between the promoter and an element located within intron 11 (109 kb downstream of the TSS) (Figure 3, region 2), which has been detected previously in the intestinal cell lines Caco-2 and HT29, in the bronchial epithelial cell line NHBE, and in primary epididymis cells (26). This contact was observed by Gheldof et al. and noted as element III (32). We detected interactions with a third regulatory element located 203 kb downstream of the promoter (Figure 3, region 3), which corresponds to a DHS previously referred as DHS 4574 + 15.6 kb (26,36).
Our 5C data revealed several new interacting regions in nasal epithelial cells (Figure 3, regions A to G) that contain DHS sites, which either overlap with or are next to a CTCF binding site. Two regions were found to have enhancer activity in a reporter assay. Region E, located around at −44 kb upstream of the gene showed the strongest enhancer activity with about 6-fold increase of the CFTR expression in bronchial epithelial cells (16HBE14o-) (Figure 4A). The Harris group had previously described this DHS at −44 kb in airway epithelial cells without pointing to any role (33), but have just recently published new data about this element (63). They also showed that this region is an airway-specific enhancer with a 6.4-fold increase of CFTR expression in 16HBE14o-. In addition, they identified a functional ARE (antioxidant response element) in this DHS, which could explain, in association with the DHS -35 kb, the regulation of CFTR expression in the airway based on environmental conditions and stress. Our reporter assays suggest that regions E and G, located downstream of the gene, cooperatively increase the CFTR promoter activity in airway epithelial cells (Figure 4B). Interestingly, region G contains a CTCF binding site (Figure 4D), which suggests that interaction of region E with the CFTR promoter could depend of a chromatin looping driven by this insulator factor. Although the five other contacting regions we identified in HNECs did not display any enhancer activity (Figure 3, regions A to D and H), these may still function in the regulation of CFTR through other mechanisms or by acting combinatorially.
In conclusion, we identified new cooperative CFTR regulatory elements flanking the gene in primary human nasal epithelial cells. As Blackledge et al. and Gheldof et al. had respectively proposed looping models of the CFTR locus organization in epididymis cells (69) and intestinal cells (32), we propose a similar conformation in airway cells via the formation of chromatin loop with CTCF-CTCF binding around the CFTR gene (Figure 5E).
Although the primary cell isolation protocol introduced here was used to generate samples only from healthy volunteers, it could easily be adapted to perform analyses with cells isolated from patients affected by cystic fibrosis or by CFTR-related disorders. Contacts profiles from patients with single mutations or none at all in the CFTR coding sequence could be compared to those found in healthy individuals in order to identify mechanisms contributing to the disease, involving our new contact regions and/or other sites. As synonymous substitutions may be significant factors in CF (70), genetic alterations within remote regions could lead to CFTR dysregulation, which might explain discrepancy in phenotypic features of CF and CFTR-related disorders.
Supplementary Material
Acknowledgments
We thank Pr Laurent Misery and its team for skin fibroblasts. We are grateful to the members of our laboratories for insightful discussions, and to D. Paquette for excellent technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French foundation ‘Vaincre la Mucoviscidose’; Gaëtan Salaün to C.F.; Canadian Institutes of Health Research to J.D. (CIHR; MOP-86716, MOP-142451). J.D. is an FRSQ Research Scholar (Fonds de la Recherche en Santé du Québec). Funding for open access charge: Université Bretagne Occidentale to C.F.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kleinjan D.A., van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsman J., Horsfield J.A. Long distance relationships: enhancer-promoter communication and dynamic gene transcription. Biochim. Biophys. Acta. 2012;1819:1217–1227. doi: 10.1016/j.bbagrm.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Crutchley J.L., Wang X.Q., Ferraiuolo M.A., Dostie J. Chromatin conformation signatures: ideal human disease biomarkers? Biomarkers Med. 2010;4:611–629. doi: 10.2217/bmm.10.68. [DOI] [PubMed] [Google Scholar]
- 5.Kleinjan D.J., Coutinho P. Cis-ruption mechanisms: disruption of cis-regulatory control as a cause of human genetic disease. Brief. Funct. Genomic. Proteomic. 2009;8:317–332. doi: 10.1093/bfgp/elp022. [DOI] [PubMed] [Google Scholar]
- 6.Bodega B., Ramirez G.D., Grasser F., Cheli S., Brunelli S., Mora M., Meneveri R., Marozzi A., Mueller S., Battaglioli E., et al. Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol. 2009;7:41. doi: 10.1186/1741-7007-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleinjan D.A., Seawright A., Schedl A., Quinlan R.A., Danes S., van Heyningen V. Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum. Mol. Genet. 2001;10:2049–2059. doi: 10.1093/hmg/10.19.2049. [DOI] [PubMed] [Google Scholar]
- 8.Beysen D., Raes J., Leroy B.P., Lucassen A., Yates J.R., Clayton-Smith J., Ilyina H., Brooks S.S., Christin-Maitre S., Fellous M., et al. Deletions involving long-range conserved nongenic sequences upstream and downstream of FOXL2 as a novel disease-causing mechanism in blepharophimosis syndrome. Am. J. Hum. Genet. 2005;77:205–218. doi: 10.1086/432083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerem B., Rommens J.M., Buchanan J.A., Markiewicz D., Cox T.K., Chakravarti A., Buchwald M., Tsui L.C. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 10.Riordan J.R., Rommens J.M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J.L., et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 11.Rommens J.M., Iannuzzi M.C., Kerem B., Drumm M.L., Melmer G., Dean M., Rozmahel R., Cole J.L., Kennedy D., Hidaka N., et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 12.Crawford I., Maloney P.C., Zeitlin P.L., Guggino W.B., Hyde S.C., Turley H., Gatter K.C., Harris A., Higgins C.F. Immunocytochemical localization of the cystic fibrosis gene product CFTR. Proc. Natl. Acad. Sci. U.S.A. 1991;88:9262–9266. doi: 10.1073/pnas.88.20.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris A., Chalkley G., Goodman S., Coleman L. Expression of the cystic fibrosis gene in human development. Development. 1991;113:305–310. doi: 10.1242/dev.113.1.305. [DOI] [PubMed] [Google Scholar]
- 14.Chou J.L., Rozmahel R., Tsui L.C. Characterization of the promoter region of the cystic fibrosis transmembrane conductance regulator gene. J. Biol. Chem. 1991;266:24471–24476. [PubMed] [Google Scholar]
- 15.Koh J., Sferra T.J., Collins F.S. Characterization of the cystic fibrosis transmembrane conductance regulator promoter region. Chromatin context and tissue-specificity. J. Biol. Chem. 1993;268:15912–15921. [PubMed] [Google Scholar]
- 16.Yoshimura K., Nakamura H., Trapnell B.C., Dalemans W., Pavirani A., Lecocq J.P., Crystal R.G. The cystic fibrosis gene has a ‘housekeeping’-type promoter and is expressed at low levels in cells of epithelial origin. J. Biol. Chem. 1991;266:9140–9144. [PubMed] [Google Scholar]
- 17.Giordano S., Amato F., Elce A., Monti M., Iannone C., Pucci P., Seia M., Angioni A., Zarrilli F., Castaldo G., et al. Molecular and functional analysis of the large 5′ promoter region of CFTR gene revealed pathogenic mutations in CF and CFTR-related disorders. J. Mol. Diagn. 2013;15:331–340. doi: 10.1016/j.jmoldx.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Viart V., Des Georges M., Claustres M., Taulan M. Functional analysis of a promoter variant identified in the CFTR gene in cis of a frameshift mutation. Eur. J. Hum. Genet. 2012;20:180–184. doi: 10.1038/ejhg.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonini J., Varilh J., Raynal C., Theze C., Beyne E., Audrezet M.P., Ferec C., Bienvenu T., Girodon E., Tuffery-Giraud S., et al. Small-scale high-throughput sequencing-based identification of new therapeutic tools in cystic fibrosis. Genet. Med. 2015;17:796–806. doi: 10.1038/gim.2014.194. [DOI] [PubMed] [Google Scholar]
- 20.Amato F., Seia M., Giordano S., Elce A., Zarrilli F., Castaldo G., Tomaiuolo R. Gene mutation in microRNA target sites of CFTR gene: a novel pathogenetic mechanism in cystic fibrosis. PloS One. 2013;8:e60448. doi: 10.1371/journal.pone.0060448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith A.N., Barth M.L., McDowell T.L., Moulin D.S., Nuthall H.N., Hollingsworth M.A., Harris A. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J. Biol. Chem. 1996;271:9947–9954. doi: 10.1074/jbc.271.17.9947. [DOI] [PubMed] [Google Scholar]
- 22.Mogayzel P.J., Jr, Ashlock M.A. CFTR intron 1 increases luciferase expression driven by CFTR 5′-flanking DNA in a yeast artificial chromosome. Genomics. 2000;64:211–215. doi: 10.1006/geno.2000.6119. [DOI] [PubMed] [Google Scholar]
- 23.Moulin D.S., Manson A.L., Nuthall H.N., Smith D.J., Huxley C., Harris A. In vivo analysis of DNase I hypersensitive sites in the human CFTR gene. Mol. Med. 1999;5:211–223. [PMC free article] [PubMed] [Google Scholar]
- 24.Rowntree R.K., Vassaux G., McDowell T.L., Howe S., McGuigan A., Phylactides M., Huxley C., Harris A. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum. Mol. Genet. 2001;10:1455–1464. doi: 10.1093/hmg/10.14.1455. [DOI] [PubMed] [Google Scholar]
- 25.Paul T., Li S., Khurana S., Leleiko N.S., Walsh M.J. The epigenetic signature of CFTR expression is co-ordinated via chromatin acetylation through a complex intronic element. Biochem. J. 2007;408:317–326. doi: 10.1042/BJ20070282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ott C.J., Blackledge N.P., Kerschner J.L., Leir S.H., Crawford G.E., Cotton C.U., Harris A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott C.J., Suszko M., Blackledge N.P., Wright J.E., Crawford G.E., Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J. Cell. Mol. Med. 2009;13:680–692. doi: 10.1111/j.1582-4934.2008.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackledge N.P., Carter E.J., Evans J.R., Lawson V., Rowntree R.K., Harris A. CTCF mediates insulator function at the CFTR locus. Biochem. J. 2007;408:267–275. doi: 10.1042/BJ20070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowntree R., Harris A. DNA polymorphisms in potential regulatory elements of the CFTR gene alter transcription factor binding. Hum. Genet. 2002;111:66–74. doi: 10.1007/s00439-002-0737-z. [DOI] [PubMed] [Google Scholar]
- 30.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 31.Smith A.N., Wardle C.J., Harris A. Characterization of DNASE I hypersensitive sites in the 120kb 5′ to the CFTR gene. Biochem. Biophys. Res. Commun. 1995;211:274–281. doi: 10.1006/bbrc.1995.1807. [DOI] [PubMed] [Google Scholar]
- 32.Gheldof N., Smith E.M., Tabuchi T.M., Koch C.M., Dunham I., Stamatoyannopoulos J.A., Dekker J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38:4325–4336. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z., Ott C.J., Lewandowska M.A., Leir S.H., Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J. Cell. Mol. Med. 2012;16:1321–1330. doi: 10.1111/j.1582-4934.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith D.J., Nuthall H.N., Majetti M.E., Harris A. Multiple potential intragenic regulatory elements in the CFTR gene. Genomics. 2000;64:90–96. doi: 10.1006/geno.1999.6086. [DOI] [PubMed] [Google Scholar]
- 35.Kerschner J.L., Gosalia N., Leir S.H., Harris A. Chromatin remodeling mediated by the FOXA1/A2 transcription factors activates CFTR expression in intestinal epithelial cells. Epigenetics. 2014;9:557–565. doi: 10.4161/epi.27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nuthall H.N., Moulin D.S., Huxley C., Harris A. Analysis of DNase-I-hypersensitive sites at the 3′ end of the cystic fibrosis transmembrane conductance regulator gene (CFTR) Biochem. J. 1999;341:601–611. [PMC free article] [PubMed] [Google Scholar]
- 37.Petrykowska H.M., Vockley C.M., Elnitski L. Detection and characterization of silencers and enhancer-blockers in the greater CFTR locus. Genome Res. 2008;18:1238–1246. doi: 10.1101/gr.073817.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cozens A.L., Yezzi M.J., Kunzelmann K., Ohrui T., Chin L., Eng K., Finkbeiner W.E., Widdicombe J.H., Gruenert D.C. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1994;10:38–47. doi: 10.1165/ajrcmb.10.1.7507342. [DOI] [PubMed] [Google Scholar]
- 39.Dostie J., Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat. Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- 40.Ferraiuolo M.A., Rousseau M., Miyamoto C., Shenker S., Wang X.Q., Nadler M., Blanchette M., Dostie J. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res. 2010;38:7472–7484. doi: 10.1093/nar/gkq644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lajoie B.R., van Berkum N.L., Sanyal A., Dekker J. My5C: web tools for chromosome conformation capture studies. Nat. Methods. 2009;6:690–691. doi: 10.1038/nmeth1009-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fraser J., Ethier S.D., Miura H., Dostie J. A Torrent of data: mapping chromatin organization using 5C and high-throughput sequencing. Methods Enzymol. 2012;513:113–141. doi: 10.1016/B978-0-12-391938-0.00005-7. [DOI] [PubMed] [Google Scholar]
- 43.Fraser J., Rousseau M., Blanchette M., Dostie J. Computing chromosome conformation. Methods Mol. Biol. 2010;674:251–268. doi: 10.1007/978-1-60761-854-6_16. [DOI] [PubMed] [Google Scholar]
- 44.Dostie J., Zhan Y., Dekker J. Chromosome conformation capture carbon copy technology. Curr Protoc Mol Biol. 2007 doi: 10.1002/0471142727.mb2114s80. doi:10.1002/0471142727.mb2114s80. [DOI] [PubMed] [Google Scholar]
- 45.Berlivet S., Paquette D., Dumouchel A., Langlais D., Dostie J., Kmita M. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 2013;9:e1004018. doi: 10.1371/journal.pgen.1004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rousseau M., Ferraiuolo M.A., Crutchley J.L., Wang X.Q., Miura H., Blanchette M., Dostie J. Classifying leukemia types with chromatin conformation data. Genome Biol. 2014;15:R60. doi: 10.1186/gb-2014-15-4-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imakaev M., Fudenberg G., McCord R.P., Naumova N., Goloborodko A., Lajoie B.R., Dekker J., Mirny L.A. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsui L.C., Dorfman R. The cystic fibrosis gene: a molecular genetic perspective. Cold Spring Harb. Perspect. Med. 2013;3:a009472. doi: 10.1101/cshperspect.a009472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J., et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dekker J., Marti-Renom M.A., Mirny L.A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V., et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraser J., Williamson I., Bickmore W.A., Dostie J. An Overview of Genome Organization and How We Got There: from FISH to Hi-C. Microbiol. Mol. Biol. Rev. 2015;79:347–372. doi: 10.1128/MMBR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phillips-Cremins J.E., Sauria M.E., Sanyal A., Gerasimova T.I., Lajoie B.R., Bell J.S., Ong C.T., Hookway T.A., Guo C., Sun Y., et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanyal A., Lajoie B.R., Jain G., Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rousseau M., Crutchley J.L., Miura H., Suderman M., Blanchette M., Dostie J. Hox in motion: tracking HoxA cluster conformation during differentiation. Nucleic Acids Res. 2014;42:1524–1540. doi: 10.1093/nar/gkt998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsu J.Y., Juven-Gershon T., Marr M.T. 2nd, Wright K.J., Tjian R., Kadonaga J.T. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 2008;22:2353–2358. doi: 10.1101/gad.1681808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelleher R.J. 3rd, Flanagan P.M., Kornberg R.D. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 62.Phillips J.E., Corces V.G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Z., Leir S.H., Harris A. Oxidative stress regulates CFTR gene expression in human airway epithelial cells through a distal antioxidant response element. Am. J. Respir. Cell Mol. Biol. 2015;52:387–396. doi: 10.1165/rcmb.2014-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lukowski S.W., Bombieri C., Trezise A.E. Disrupted post-transcriptional regulation of the cystic fibrosis transmembrane conductance regulator (CFTR) by a 5′ UTR mutation is associated with a CFTR-related disease. Hum. Mutat. 2011;32:E2266–E2282. doi: 10.1002/humu.21545. [DOI] [PubMed] [Google Scholar]
- 67.Lopez E., Viart V., Guittard C., Templin C., Rene C., Mechin D., Des Georges M., Claustres M., Romey-Chatelain M.C., Taulan M. Variants in CFTR untranslated regions are associated with congenital bilateral absence of the vas deferens. J. Med. Genet. 2011;48:152–159. doi: 10.1136/jmg.2010.081851. [DOI] [PubMed] [Google Scholar]
- 68.Gheldof N., Tabuchi T.M., Dekker J. The active FMR1 promoter is associated with a large domain of altered chromatin conformation with embedded local histone modifications. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12463–12468. doi: 10.1073/pnas.0605343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blackledge N.P., Ott C.J., Gillen A.E., Harris A. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 2009;37:1086–1094. doi: 10.1093/nar/gkn1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scott A., Petrykowska H.M., Hefferon T., Gotea V., Elnitski L. Functional analysis of synonymous substitutions predicted to affect splicing of the CFTR gene. J. Cyst. Fibros. 2012;11:511–517. doi: 10.1016/j.jcf.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.