Abstract
The brain is built from a large number of cell types which have been historically classified using location, morphology and molecular markers. Recent research suggests an important role of epigenetics in shaping and maintaining cell identity in the brain. To elucidate the role of DNA methylation in neuronal differentiation, we developed a new protocol for separation of nuclei from the two major populations of human prefrontal cortex neurons—GABAergic interneurons and glutamatergic (GLU) projection neurons. Major differences between the neuronal subtypes were revealed in CpG, non-CpG and hydroxymethylation (hCpG). A dramatically greater number of undermethylated CpG sites in GLU versus GABA neurons were identified. These differences did not directly translate into differences in gene expression and did not stem from the differences in hCpG methylation, as more hCpG methylation was detected in GLU versus GABA neurons. Notably, a comparable number of undermethylated non-CpG sites were identified in GLU and GABA neurons, and non-CpG methylation was a better predictor of subtype-specific gene expression compared to CpG methylation. Regions that are differentially methylated in GABA and GLU neurons were significantly enriched for schizophrenia risk loci. Collectively, our findings suggest that functional differences between neuronal subtypes are linked to their epigenetic specification.
INTRODUCTION
The remarkable diversity of cell types that comprise complex nervous systems had been described more than a century ago by Ramon y Cajal (1), and it is universally agreed that the identification, characterization, and comparative analysis of these cell types is fundamentally important for understanding brain function. The mammalian neocortex contains two major classes of neurons, the excitatory glutamatergic projection neurons (hereinafter GLU neurons) and the inhibitory GABAergic interneurons (hereinafter GABA neurons) which constitute about 80% and 20% of all cortical neurons, respectively (2). Although less abundant than projection neurons, interneurons are extremely diverse in terms of morphology, connectivity, and physiological properties, and play crucial roles in the development and organization of cortical networks that underlie a wide range of brain functions (3). The majority of the GABAergic interneurons within the telencephalon originate from one of the two embryonic subcortical progenitor zones, the medial ganglionic eminence (MGE) and caudal ganglionic eminence (CGE) (4–6). Within the cortex, the MGE gives rise to the parvalbumin (PVALB)-expressing fast-spiking interneurons and the somatostatin (SST)-expressing neurons. The CGE produces relatively rarer subtypes, including neurogliaform, bipolar and vasointestinal peptide (VIP)-expressing multipolar interneurons.
Studies in mouse models and humans have shown that interneuron dysfunction is associated with neurological and psychiatric diseases such as schizophrenia (SCZ), autism and epilepsy (7–11). Moreover, the dysfunction of inhibitory interneurons in the prefrontal cortex (PFC) is the one of the most consistent findings in the SCZ research (12–16). The observed functional impairment is largely attributed to the MGE-derived interneurons which comprise 60–70% of all cortical interneurons. The mechanisms that lead to the molecular pathology of these interneurons have not been characterized in detail, but at least in SCZ, have been linked to an altered developmental pathway that involves ontogenetic transcription factors SRY (Sex Determining Region Y)-Box 6 (SOX6) and LIM Homeobox 6 (LHX6) (10). These transcription factors have been shown to play critical roles in the specification, migration, and maturation of the MGE-derived interneurons (17,18). Numerous studies have also implicated GLU signaling in SCZ (19). In particular, recent large scale genome-wide association study (GWAS) has identified highly significant associations for SNPs in genes involved in glutamatergic neurotransmission (20).
Studies in adult animals have shown that different subpopulations of GLU and GABA neurons express unique patterns of genes (21,22) which are likely to broadly determine their distinct functional capacities (5). Despite considerable effort, it remains poorly understood how the identity of each neuronal cell type is attained during development and how it is maintained throughout the life of a cell. DNA methylation in cytosine-guanine dinucleotides (mCpG) is believed to be a heritable epigenetic mark of cellular memory that maintains the unique gene expression pattern of a cell (23). However, it remains unclear how DNA methylation relates to cell type-specific gene expression.
Recent studies indicate that composition and dynamics of DNA methylation are highly distinct in the brain and/or neurons compared to other tissues or cells (24). Similar to stem cells, the adult human and mice brains show substantial cytosine methylation outside of the CpG context (non-CpG methylation or mCpH where H stands for A,T or C) as well as a relatively high level of hydroxymethylation (hmCpG), whereas in other somatic tissues, mCpH and hmCpG are nearly absent (24,25). Unlike mCpG that is mostly established during prenatal development (26), mCpH occurs de novo after birth and until the young adult age in neurons (but not in glia) (25). DNA hydroxymethylation is thought to be an intermediate of mC demethylation (27,28), but also could play a distinct role in gene regulation (29,30). Whereas mCpG in promoters and active enhancers is associated with transcriptional silencing (23,31–33), hmCpG is enriched throughout the bodies of highly expressed genes (34).
Although the majority of the studies on DNA methylation in the brain have been performed using bulk tissues, e.g. (26,35), several reports have clearly demonstrated robust differences in mCpG and mCpH patterns between neuronal and glial cell populations in human and rodent brains (24,36,37). A recent study in the adult mouse cortex, which used ‘nuclei tagged in specific cell types’ (INTACT) approach, has demonstrated widespread differences in DNA methylation among excitatory (glutamatergic), PVALB, and VIP neurons that reflect distinct mechanisms of gene regulation in these cellular populations (38). However, comparable genetic manipulations are not applicable in human studies, and differences in DNA methylation patterns among subpopulations of neurons in the human brain have never been explored. Such differences in epigenetic landscapes might ultimately contribute to the selective vulnerability of the MGE-derived GABA interneurons to neurodevelopmental or environmental insults that could culminate in diseases such as SCZ.
In the present study, we developed a novel method to separate neuronal nuclei from the autopsy specimens of the human PFC into two sub-populations containing GLU or MGE-derived GABA neurons, respectively, using fluorescence-activated cell sorting (FACS). We then performed genome-wide expression and DNA methylation analyses (including mCpG, mCpH and hmCpG) in these two distinct neuronal populations and found that, in addition to the expected differences in gene expression, these neuronal subtypes significantly differ in their DNA methylation landscapes. Comparison of the methylomes with the expression profiles revealed that non-CpG methylation is particularly pertinent in inferring neuron subtype-specific gene expression patterns. Finally, we detected a strong association between epigenetic specification of the neuronal subtypes and psychiatric disease.
MATERIALS AND METHODS
Brain specimens and tissue sample preparation
The brain specimens were obtained from the Brain Collection of Dr Yasmin Hurd (39,40). All specimens had been collected at autopsy within 24 h after death by the personnel of the Department of Forensic Medicine (Semmelweis University, Hungary) under approved local ethical guidelines. The subjects showed negative toxicology for common drugs of abuse (including alcohol) and for therapeutic agents. Nicotine toxicology was not conducted, but tobacco use is frequent in the general population from which the subjects were collected. The cause and manner of death were determined by a forensic pathologist after evaluating autopsy results, circumstances of death, toxicology data, and police reports, as well as family interviews and medical records. According to this assessment, all subjects used in our study died of non-suicide causes, such as cardiac failure or an accident. None of these individuals were diagnosed with any neurological or psychiatric condition at the time of death. Immediately after autopsy, brains were cut coronally in 1.5-cm slabs, frozen, and kept at −70°C. The dissections of the orbitofrontal cortex (OFC) were performed as described in (37). Specimens obtained from seven and two subjects were used in DNA methylation and gene expression studies, respectively (Supplementary Table S1). There were no significant differences in brain pH among all specimens, and post-mortem intervals for all subjects were ≤24 h. Age of death varied from 19 to 46 years for specimens used in DNA methylation studies; individuals used for RNA-seq studies were 37 and 44 years old.
Analysis of genetic background
Genome-wide genotyping of the study subjects' DNA was performed using Illumina HumanOmni2.5-8v1BeadChip. To test their genetic background, principal component analysis (PCA)-based population stratification was then conducted using these data and genotype data from the HapMap Project (http://hapmap.ncbi.nlm.nih.gov/) (Supplementary Figure S1).
Nuclei isolation and FACS-based nuclear sorting
The crude nuclear pellet was prepared as described (37) and was followed by incubation of nuclei in the blocking buffer [1% goat serum, 2 mM MgCl2, Tris-buffered saline, pH 7.6 (TBS)] in the presence of the mouse monoclonal Alexa488-conjugated anti-NeuN antibodies (1:1000 dilution, Millipore; MAB377X) and primary guinea pig polyclonal anti-Sox6 antibodies (1:1500 dilution) (41). After 1.5 h of incubation, nuclei were centrifuged for 15 min at 2800g through a layer of 1.1 M sucrose (37). Nuclear pellet was then resuspended in blocking buffer and incubated for 45 min with the secondary antibodies (Alexa647-conjugated goat anti-guinea pig, 1:1000 dilution, Jackson Immunoresearch, 106-606-003). Finally, nuclei were centrifuged through 1.1 M sucrose, resuspended in TBS containing 2 mM MgCl2, and mixed with a DNA dye 7-aminoactinomycin (7-AAD, Sigma, final concentration 2 μg/ml). FACS was performed using Vantage with DiVa. For DNA methylation studies, the sorted nuclear samples were pelleted by centrifugation at 4000 rpm for 20 min and stored at −80°C until the DNA isolation step.
Immunofluorescence and immunohistochemistry
For the immunofluorescence assay of non-sorted human brain nuclear samples, nuclei were isolated and stained with anti-NeuN and anti-Sox6 antibodies as described in the previous section. Nuclei were then stained with a DNA dye DAPI (0.1 μg/ml, 10 min), fixed with 2% formaldehyde for 10 min, and mounted on microscope slides. Confocal microscopy was performed on a LSM 700 microscope (Carl Zeiss), using 405, 488 and 639 nm lasers and a 63×/1.40 oil objective.
The procedure for double immunohistochemistry was similar to that employed previously with paraffin sections (42). Brain tissue was cut into 2-cm coronal slices and fixed in phosphate-buffered 10%’ formalin for 5 days, then transferred to phosphate-buffered saline (PBS) with 0.02% sodium azide and stored at 4°C until used. Sections of the PFC were cut at 40 μm on a vibratome into PBS, boiled in 10 mM citrate buffer, pH.6, incubated overnight at 4°C in guinea pig anti-SOX6 antibodies (41) diluted 1:8000 in PBS with 3% normal goat serum. Bound anti-SOX6 was detected by standard avidin-biotinylated peroxidase complex (ABC) methods with 0.05% 3,3′-diaminobenzidine (DAB), 0.02% nickel ammonium sulphate, 0.02% cobalt chloride, and 0.01% hydrogen peroxide, yielding a black reaction product. The sections were then incubated overnight at 4°C with anti-PVALB mouse monoclonal ascites (Swant, PV235) diluted 1:4000, followed by ABC with DAB as before, but without metals, yielding a brown reaction product, which cannot be seen in areas previously stained black.
RNA isolation, cDNA synthesis and qPCR
To isolate RNA from sorted GABA or GLU nuclei, the FACS-separated fractions (∼1 million nuclei for each neuronal subtype) were collected directly into tubes containing TRIzol LS reagent (Life Technologies). The final volume of TRIzol LS was then adjusted to be 3× the volume of the collected nuclear fractions, and the resulting samples were processed according to the manufacturer's protocol. RNA was precipitated in the presence of 1 μl of UltraPure Glycogen (20 μg/μl, Life Technologies), washed twice with 70% ethanol, and dissolved in RNAse-free water. cDNA synthesis was performed using iScript cDNA synthesis kit (BioRad) and quantitative real-time PCR (qPCR) was performed using specific TaqMan probes as described in (37).
Transcriptome profiling by RNA-seq
RNA-seq libraries were constructed using 0.7–1 μg of RNA and TruSeq Stranded Total RNA with Ribo-Zero Gold Kit (Illumina). The libraries were then sequenced using HiSeq 2000 with single-read (SR)-50 and paired-end (PE)-75 protocols for the first and the second pair of replicates, respectively. RNA-seq reads were trimmed for low quality ends (qphred scroe < 20) using Trimmomatic (v0.32) and aligned to human hg19 genome using the tophat aligner (v2.0.11) with the iGenomes UCSC hg19 gene annotation model under default alignment options. Python script HTSeq was used to count the number of reads aligned to the known gene features. Genes with at least 1 count-per-million reads in at least one of the libraries were considered ‘present’, and 17 884 such genes were identified in the data set, whereas the ‘absent’ genes were filtered from further analyses. In order to adjust for differences in library sizes, the scaling factors were estimated using the trimmed mean of M-values (TMM) normalization method.
Analysis of differential expression between GLU and GABA neurons was performed using the Bioconductor ‘limma’ package, with the sequencing batch incorporated as a covariate. Genes with false discovery rate (FDR)-adjusted P-value <0.05 and fold changes >2 were considered differentially expressed (DE) between the two neuronal subpopulations.
DNA methylation analysis by Infinium HM450K array and Enhanced Reduced Representation Bisulphite Sequencing (ERRBS)
Genomic DNA was extracted from the FACS-separated nuclei fractions (∼0.75–1 million nuclei for each neuronal subtype). Nuclear pellets were resuspended in DNA Prep Buffer (100 mM NaCl, 10 mM EDTA, 0.5% SDS, 10 mM Tris–HCl, pH 8.0), to which Proteinase K (Zymo Research) was added immediately before use to the final concentration 0.2 mg/ml. After incubation at 50°C for 4 hours, DNA was extracted using two rounds of phenol/chloroform extractions and ethanol-precipitated overnight in the presence of 20 μg glycogen.
HM450K (Illumina) assay was performed by Illumina Inc. as described in Supplementary Methods and in (37,43). The initial data processing was done using GenomeStudio (Illumina), generating β-values and detection P-values for each of the 482 421 CpG sites probed by the HM450K array. The CpG sites that overlapped with SNPs with MAF >5% (based on the 1000 Genomes Project database (www.1000genomes.org; N = 9462 sites), with missing β-values in at least one sample (additional N = 5382 sites) and with detection P-values >0.05 in at least one sample (additional N = 549 sites) were removed from all subsequent analyses, resulting in the final data set of 467 028 sites. Subsequent analyses of DNA methylation were done using R (www.r-project.org) or custom Perl scripts.
ERRBS was performed by the Epigenomics Core of the Weill Cornell Medical College (New York) as described in Supplementary Methods and in (37,44). Data analysis was done using the R package ‘methylKit’ (45).
Analysis of CpH methylation sequence context
A random sample of 10 000 CpH sites was selected among those CpH sites which displayed mean β-value >20% in the ERRBS data set. The human genomic sequence (±3 bp) surrounding the position of the methylated cytosine was then extracted using the tools available in Bioconductor/R. To generate the sequence logo profiles, the resulting multiple sequence alignments were submitted to the WebLogo online tool (http://weblogo.berkeley.edu).
Analysis of correlations between DNA methylation and gene expression
Spearman correlations between CpG or CpH methylation and RNA expression levels were analyzed separately for all expressed and for DE genes. For each gene, mCpG or mCpH sites were combined into 1kb bins as a function of the distance from TSS. Next, methylation levels for sites within each bin were averaged, and the resulting average value was correlated with gene expression using cor.test function in R. The significance was determined as P < 0.01 after Bonferroni-correction for the number of bins (N = 12).
Quantification of hydroxymethylation (hmC) and methylation (mC) by mass spectrometry
Global analysis of hmC and mC levels in GABA, GLU and GLIA cells was performed by Zymo Research Epigenetic Services (ZRES) using liquid chromatography/mass spectrometry (LC/MS). LC/MS enabled to determine the concentrations of 5-hydroxymethyl-2′-deoxycytidine (5hmdC) and 5-methyl-2′-deoxycytidine (5mdC) as a percentage of 2′-deoxyguanosine (dG) (i.e. [5hmdC]/[dG] and [5mdC]/[dG]). The calibration curves were constructed using internal standards, corresponding to the calibrated ranges of 0–2.5% for 5hmdC and 0–25% for 5mdC. The samples (100 ng of DNA isolated form each cell type) were run in technical replicates of 2 or 3.
Analysis of hmC by reduced representation hydroxymethylation profiling (RRHP)
The RRHP assay (Zymo Research) was performed by ZRES as described in Supplementary Methods and in (46). RRHP quantifies the level of hmCpG modification at the CpG sites within the MspI enzyme recognition sequence (CCGG; 2 297 198 sites in human genome) as a strand-specific number of sequencing reads corresponding to each such site. Only CpG sites that were flanked by 70–150 bp MspI digested fragments were considered in order to eliminate possible dependence of the number of reads on the fragment length, resulting in 465 487 sites. In this range of the MspI fragment lengths, the average number of reads was nearly independent (difference < 25%) from the length of the fragment and thus, enabled a reliable comparison of hmC levels between different loci within each sample. In order to normalize the data for the sequencing depth, the average levels of hmCpG for each CpG site in GABA or GLU cells were re-calculated as weighted averages between two replicates, using the total number of reads in each sample. For the CpG sites at which the hmC measures were available for both strands, the average number of reads was used in the strand-independent analyses. To compare hmC levels between the sense and the antisense strand, the RRHP read numbers for CpG sites that were flanked by 70–150 bp MspI digested fragments were averaged for each gene and for each strand orientation.
Analysis of the overlap between differential methylation and genetic associations
Summary statistics were downloaded from publically available GWAS datasets (http://www.med.unc.edu/pgc/downloads) for schizophrenia (SCZ) (20), autism spectrum disorder (ASD) (see AUT in PGC downloads), major depressive disorder (MDD) (47), Alzheimer's disease (AD) (48), rheumatoid arthritis (RA) (49), and the levels of total cholesterol (TC) and triglycerides (TG) (50). For each GWAS dataset, SNPs were ‘clumped’ using Plink 1.9 (https://www.cog-genomics.org/plink2) and samples of European ancestry from the 1000 genomes project phase 1 (51), using the following settings: two different thresholds of significance for disease-associated SNPs (P-values < 5 × 10−8 and < 1 × 10−6), r2 = 0.6, and a window of 500 kb.
Enrichment of the common disease-associated risk variants within differentially methylated regions (DMRs) was tested using Interval Enrichment Approach (INRICH) (52). We assessed if the GWAS variants significantly overlapped with DMRs within 250 bp, 500 bp or 1 kb upstream or downstream of each cell type-specific DM CpG site determined by HM450K assay in this study and in (37). Significance was determined using loci permuted within the genome, but matched to the associated loci by the number of SNPs, SNP density, and the number of overlapping DMRs by estimating empirical P-values based on 10 000 permutations. Multiple testing corrections were performed via a second, nested round of 10 000 permutations to assess the null distribution of the minimum empirical P-value across all tested gene sets.
RESULTS
Isolation of specific subpopulations of neuronal nuclei using FACS
Cell structure is not preserved in frozen autopsy brain specimens. However, the nuclei of different cell types remain intact and can be separated by FACS using antibodies against nuclear antigens, such as a RNA-binding protein RBFOX3 (also known as NeuN), which is expressed in neuronal nuclei and has been employed to separate neuronal and non-neuronal nuclei (53) and to compare DNA methylation between these two major classes of cells in the human brain (37). Here we expand this line of research by developing a multicolor FACS protocol that further separates neuronal [NeuN-positive, (+)] nuclei into the two major neuronal subtypes, GABA and GLU neurons. To this end, in addition to anti-NeuN antibodies, we used antibodies against SOX6, a transcription factor that regulates the ontogeny of the MGE-derived GABA neurons and is robustly expressed in these cells in the adult human and mouse cortex (10,18).
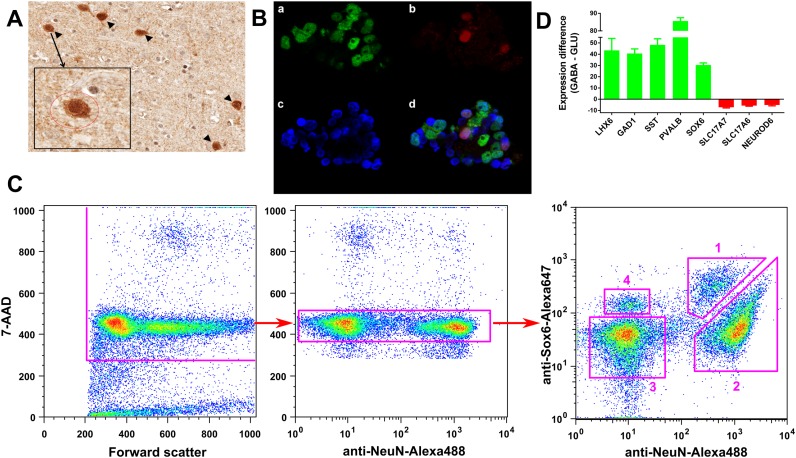
In order to validate the specificity of anti-SOX6 antibodies, we used immunohistochemistry on sections of the human PFC and immunofluorescence imaging of the unsorted nuclei that were isolated from the same PFC specimens and contained mixed population of nuclei from neuronal and non-neuronal cells (Figure 1). The immunohistochemical experiments confirmed that the anti-SOX6 staining significantly overlapped with staining of PVALB (Figure 1A)—a well-characterized marker of the MGE-derived GABA neurons (5). In our immunofluorescence experiments, the anti-SOX6 antibodies clearly distinguished a group of SOX6(+) nuclei over the background staining (Figure 1B). These SOX6(+) nuclei mostly belonged to the neuronal [NeuN(+)] population, and the double-positive SOX6(+)NeuN(+) population constituted ∼11% of the total NeuN(+) nuclei. In addition, a small fraction (<5%) of SOX6(+) nuclei was found within the NeuN(−) population (not shown). Expression of Sox6 has been described in oligodendrocyte precursor cells (18) and in Olig2-expressing cells in adult mice (41), suggesting that this SOX6(+)NeuN(−) population represents oligodendrocyte precursors and/or mature oligodendrocytes in the adult human PFC.
Figure 1.
Isolation of GABAergic (GABA) and glutamatergic (GLU) neuronal nuclei from the human PFC. (A) Immunohistochemical co-localization of PVALB positive (+) (brown) and SOX6(+) (dark grey) PFC cells. The majority of PVALB(+) cells are also SOX6(+) (arrow heads). Small round grey nucleus profile in the Insert is probably an oligodendrocyte. (B) Validation of anti-SOX6 antibodies by immunofluorescence confocal imaging of the PFC nuclei. The image is a representative example from several experiments. Nuclei were stained with anti-NeuN antibodies (a, green color), which label all neuronal nuclei, anti-SOX6 antibodies (b, red color), which label a subpopulation of GABA neuronal nuclei, and DNA stain DAPI (c, blue color). Panel (d) depicts the overlap of images (a–c). (C) Isolation of the PFC neuronal nuclei subpopulations by FACS. Figure shows sequential gating steps using anti-NeuN and anti-Sox6 antibodies, and 7-AAD DNA stain. Left: exclusion of debris and nuclear fragments. Middle: exclusion of doublets of nuclei and nuclei of dividing cells. Right: separation of neuronal and non-neuronal nuclei based on NeuN labelling intensity, and separation of SOX6(+) and SOX6(-) signals: (1) SOX6(+)NeuN(+)—GABA neurons; (2) SOX6(-)NeuN(+)—GLU neurons; (3) SOX6(-)NeuN(−)—GLIA; (4) SOX6(+)NeuN(−)—SOX6(+) GLIA. (D) Confirmation of enrichment of SOX6(+)NeuN(+) and SOX6(−)NeuN(+) fractions for known markers of GABA (green bars) and GLU (red bars) neurons, respectively. Shown are fold-differences that represent GABA to GLU (positive values) or GLU to GABA (negative values) gene expression ratios. The data were obtained using qPCR.
A typical flow cytometry experiment is schematically depicted in Figure 1C. During the first two gating steps, a DNA stain, 7-aminoactinomycin (7-AAD), was used as an additional marker to help eliminate the debris, doublets of nuclei and nuclei of dividing cells. At the final gating step, anti-NeuN and anti-SOX6 staining was combined in a dual-color protocol, resulting in efficient separation of four populations of nuclei: (1) SOX6(+)NeuN(+) (MGE-derived GABA neurons), (2) SOX6(-)NeuN(+) (largely GLU projection neurons), (3) SOX6(-)NeuN(−) (non-neuronal cells) and (4) SOX6(+)NeuN(−). The latter, non-neuronal SOX6(+) population was also detected in our immunofluorescence experiments. In agreement with the immunofluorescence experiments that were performed using non-sorted nuclear samples, the double-positive SOX6(+)NeuN(+) nuclei accounted for ∼12–14% of the total NeuN(+) fraction. This number is compatible with the published estimates according to which GABA interneurons constitute ∼20% of all neurons in human and monkey PFC, whereas the MGE-derived SOX6(+) cells comprise ∼60% of the cortical GABAergic interneurons (2).
As an initial validation of the cell-type identity of the collected nuclei, we isolated RNA from SOX6(+)NeuN(+) and SOX6(−)NeuN(+) populations and confirmed the enrichment of GABA cell-specific (LHX6, GAD1, SST, PVALB) or GLU cell-specific [SLC17A7 (VGLUT1), SLC17A6 (VGLUT2), NEUROD6] markers in the respective nuclear populations using qPCR (Figure 1D) (21,54). The SOX6(−)NeuN(−) population consists mostly of glial cells (such as oligodendrocytes, astrocytes and microglia) as well as a small population of endothelial cells (24,37). For the sake of brevity, we hereinafter refer to populations (1), (2) and (3) (Figure 1C) as ‘GABA’, ‘GLU’ and ‘GLIA’ cells, respectively.
RNA-seq analysis confirms the specificity of FACS-isolated neuronal subtypes
We next performed RNA-seq on the total RNA extracted from GLU and GABA FACS-separated nuclei. The nuclei were isolated from autopsy specimens dissected from a specific area of the PFC—OFC. The specimens were obtained from two male Caucasian individuals of similar age and similar (European) genetic background with no antemortem diagnoses of any neurological or psychiatric disorders and no discernible neuropathological lesions (Supplementary Table S1, Supplementary Figure S1). Compared to RNA extracted from brain tissue homogenate, RNA extracted from sorted nuclei showed significantly lower RIN numbers (4.0–4.8), suggestive of partial degradation (Supplementary Table S1, Supplementary Figure S2A). Therefore, high-throughput sequencing of ribosomal RNA-depleted total RNA was performed (see Methods). This approach has been shown to be compatible with quantification of partially degraded RNA (55). Approximately 100 million uniquely mapped sequence reads were generated for each of the cell types (Supplementary Table S2, Supplementary Figure S2B). We found that 66–70% of all mapped reads were located in introns and 12–17% of the reads mapped to exons, whereas the rest mapped to intergenic regions. This breakdown is in line with a previous analysis of the total brain RNA which revealed a higher proportion of reads originating from regions outside versus within known exons, whereas more exonic than intronic or intergenic RNA has been detected in polyA-selected RNA preparations (56). In addition, a higher coverage of introns could be expected in nuclear preparations used in our study compared with whole-cell RNA analysis. A substantial majority of human protein-coding genes (N = 15 460) and many genes for non-coding RNAs (N = 2424) were represented in at least one of the samples (see Materials and Methods). For each neuronal subtype, RNA-seq profiles were similar across replicate specimens (r = 0.97–0.98); the correlations were weaker when samples from different subtypes were compared (r = 0.91–0.92) (Supplementary Figure S2C).
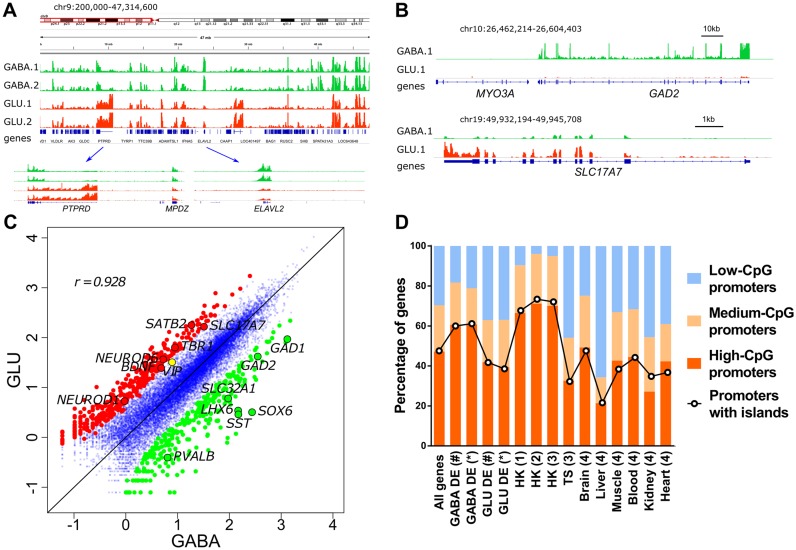
The GABA-specific and GLU-specific transcriptomes appeared concordant on a >10-Mb scale; nevertheless, the presence of cell-specific gene expression was immediately apparent as well (Figure 2A). We identified 1286 and 1366 genes that showed >2-fold differential expression (DE) in GABA and GLU cells, respectively (FDR-adjusted P < 0.05), and which we for convenience hereinafter denote GABA-DE and GLU-DE genes (Supplementary Table S3). Notably, whereas the numbers of protein-coding genes were similar among the GABA-DE and GLU-DE genes (1224 and 1084 genes, respectively), there were ∼4.5-fold more GLU-DE than GABA-DE non-coding RNAs (282 versus 62 genes). As expected, known markers of GABA (e.g. GAD1, GAD2, SOX6, SLC32A1, LHX6, PVALB, SST) or GLU (e.g. SLC17A7, NEUROD1, NEUROD6, BDNF, SATB2, TBR1) neurons (21,54) were enriched in the corresponding types of purified nuclei (Figure 2B and C), including the genes that we confirmed to be GABA- or GLU-specific using qPCR (Figure 1D). SOX6 is not expressed in non-MGE-derived GABA neurons which are, therefore, expected to sort together with SOX6(−)NeuN(+) population of mostly GLU neurons. Although the proportion of these non-MGE GABA neurons is relatively small (∼8% of the total sorted GLU population) (2), we indeed detected significantly higher expression of the specific marker of the CGE-derived GABA neurons (VIP) in the GLU- versus GABA neurons (Figure 2C). Collectively, these findings confirm the specificity of the SOX6(+)NeuN(+) fraction to MGE-derived GABA neurons. We, however, note that, in addition to GLU neurons, the SOX6(−)NeuN(+) fraction contains a small percentage of non-MGE GABA neurons.
Figure 2.
Cell-specific intranuclear transcriptomes of GABA and GLU neurons obtained by RNA-seq. (A) Integrative Genomics Viewer (IGV) (96) snapshot of intranuclear transcriptomes derived from GABA and GLU neuronal nuclei obtained from the OFC of 2 individuals. Tracks are colored by cell type; GABA neurons are shown in green, and GLU neurons in red; 1 and 2 denote individual subjects. Two regions that represent genes with predominant expression in GLU (PTPRD) or in GABA (ELAVL2) neurons are shown in the lower panel. (B) Profiles for archetypical GABA-specific (GAD2, encodes GABA-synthesizing enzyme glutamate decarboxylase 2; upper panel) and GLU-specific (SLC17A7, encodes vesicular glutamate transporter, VGLUT1; lower panel) genes. Note that MYO3A (neighboring GAD2), which is specifically expressed in cochlea and retina, does not show any significant number of reads mapped to its locus. (C) Pairwise comparisons of gene expression measured by RNA-seq in GABA and GLU neurons. The most differentially expressed genes (>5-fold change) are shown as colored circles, and selected neuron subtype-specific genes are labelled. r, Spearman correlation of log10-transformed RNA-seq read counts. (D) Percentage of GABA-DE and GLU-DE genes with low-, medium- or high-CpG content within promoter regions, and with promoters that overlap CpG islands. GABA-DE and GLU-DE genes were defined using criteria of (#) >2-fold enrichment and FDR < 0.05 or (*) >4-fold enrichment and FDR < 0.01. All genes, data for all RefSeq genes; (1) housekeeping (HK) genes identified in (63,64); (2) HK genes identified in (62); (3) HK genes identified in (61); TS, all tissue specific genes identified in (61); (4) TS genes identified in individual tissues in (65). Promoter regions were defined as regions proximal to TSS (−1000 bp to +100 bp). Assignment of promoters to low, medium or high-CpG groups is shown in Supplementary Figure S4.
To the best of our knowledge, the present work is the first to report GABA-specific and GLU-specific gene expression profiling in the human brain. Therefore, to further validate the results, we compared our data set with previously described neuron subtype-specific expression data for the mouse brain (38). We detected a highly significant overlap between human and mouse preferentially expressed genes from the similar neuronal subtypes (fold enrichments 4.0–4.4; P-values <1e−116), but no significant overlap between different subtypes (Supplementary Table S4).
Lastly, gene ontology analysis of GABA-DE and GLU-DE gene lists using WebGestalt (57) showed enrichment for multiple ontologies related to function or development of nervous system for both neuronal subtypes (e.g. ‘synaptic transmission’, ‘neuron differentiation’) as well as enrichment for categories that are specific to GABA (e.g. ‘glutamatergic amino acid catabolic process’, ‘clathrin-sculpted GABA transport vesicle membrane’) or GLU (e.g. ‘microtubule motor activity’, ‘dynein complex’) neurons and appeared to be biologically relevant for the respective neuronal subtypes (Supplementary Figure S3).
To summarize, we identified substantial differences between the expression profiles of the two isolated neuronal populations, and the nature of the observed differences is compatible with the assignment of these populations as GABA and GLU neurons.
GABA-specific genes are enriched for high-CpG promoters
CpG islands represent an important class of regulatory regions in mammalian genomes that are characterized by high local concentration of CpG sites (58). Previous studies have shown that whereas the promoters of somatic tissue-specific genes are often CpG-poor and lack CpG islands, the promoters of housekeeping genes are CpG-rich and predominantly contain CpG islands (59,60). We, therefore, asked if the promoters of the GABA- and GLU-specific genes show features similar to those of tissue-specific promoters. We first divided promoters of all genes in the RefSeq human genome annotation into three groups, with low, medium and high CpG content (33) (Supplementary Figure S4), and then identified promoters for each of the neuron subtype-specific genes. In line with previous findings, the proportion of genes with high-CpG promoters was lower for the GABA-DE and GLU-DE genes compared to housekeeping genes (61–64). Surprisingly, this proportion was higher for the GABA-DE compared to GLU-DE genes (60.6% versus 41.9%, respectively) (Figure 2D). We detected a comparable difference (60.8% versus 39.5%) when we applied more stringent criteria to define the DE genes (FDR-adjusted P-value < 0.01 and fold change > 4), and thus analyzed significantly fewer DE genes (261 and 277 DE genes in GABA or GLU neurons, respectively). Similarly, there were more GABA-DE than GLU-DE genes with promoters overlapping with a CpG island (Figure 2D). Compared to previously identified tissue-specific genes (65), the GABA-DE genes contained noticeably higher proportion of genes with high-CpG promoters, whereas the data for GLU-DE genes was comparable to those previously reported for brain-specific genes. These findings suggest that the promoters of genes that are preferentially expressed in GABA or GLU neurons have distinct sequence features that might translate into functionally important differences in epigenetic regulation (33).
DNA methylation landscapes significantly differ between GABA and GLU neurons in the human PFC
We next characterized cell type-specific total (mC+hmC) DNA methylation in GABA and GLU neuronal populations using OFC autopsy specimens obtained from five age-matched male Caucasian subjects without antemortem diagnoses of any neurological or psychiatric diseases and no visible neuropathological lesions (Supplementary Table S1). Following the FACS protocol described above, we isolated nuclei from 3 populations of cells: (1) GABA, (2) GLU, and (3) GLIA (see Figure 1C). We then measured genome-wide methylation of the DNA isolated from these sorted nuclei using the Infinium HM450K beadchip array which includes probes for ∼480 000 individual CpG positions across the genome, covering most of the predicted genes, CpG islands, as well as many predicted enhancers (66). A ‘β-value’ was obtained for each CpG site, corresponding to the level of the total DNA methylation (in the range between 0 and 1). We then filtered out the probes that could potentially produce unreliable data (see Materials and Methods).
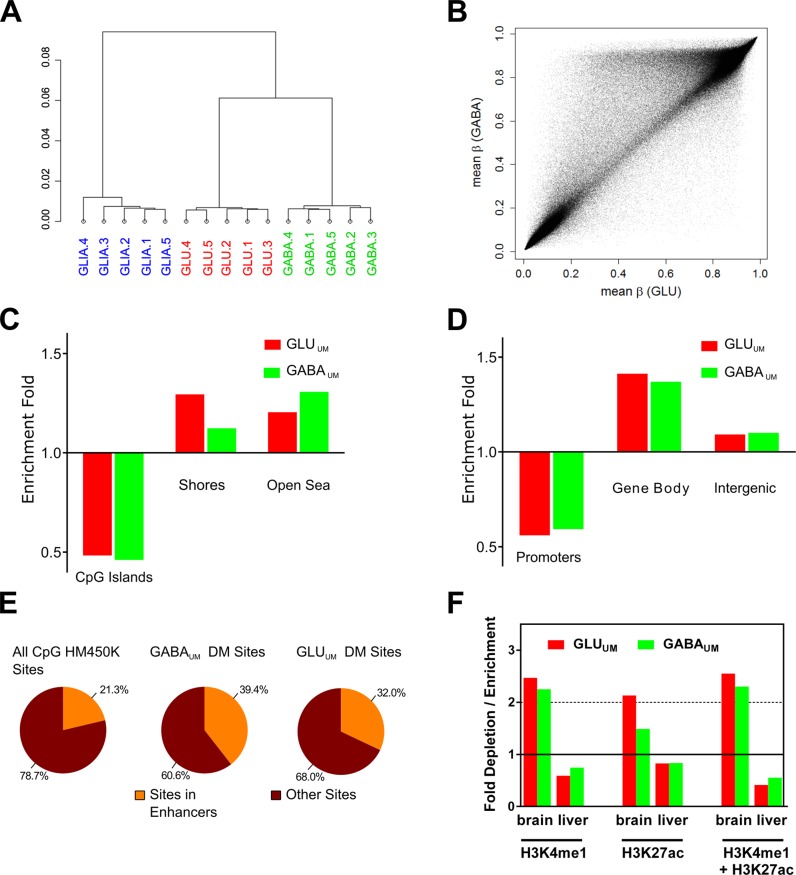
In all three types of cells (GABA, GLU and GLIA), the overall methylation frequency of individual sites showed the expected bimodal distribution (with the majority of sites showing β-values either <20% or >80%) (Supplementary Figure S5A) (67). Unsupervised hierarchical clustering demonstrated a clear separation of the samples according to the cell type, with the two neuronal (GABA and GLU) populations clustering together, to the exclusion of the GLIA population (Figure 3A). Correlation analysis showed that, in contrast to the readily detectable differences among the cell types, there was low variability in DNA methylation among individuals for the same cell type (Supplementary Table S5). The average level of DNA methylation across all CpG sites probed by the HM450K assay was ∼3.2% and 3.9% higher in GABA versus GLU neurons or GLIA, respectively (P-values <0.001 by paired t-test) (Supplementary Figure S5B).
Figure 3.
Characterization of DNA methylation in GABA and GLU neurons and in GLIA by Infinium HM450K array. (A) Unsupervised hierarchical clustering of CpG methylation data. 1, 2, 3, 4, 5 denote individual subjects. (B) Scatter plot comparing averaged (across all five subjects) CpG methylation between GABA and GLU samples. Each point represents an averaged β value for one CpG site from the array. The data demonstrate significant differences in total CpG methylation among neuronal subtypes. (C) Enrichment / depletion of the differentially methylated (DM) CpG sites in CpG islands and shores. ‘Open sea regions’ denote areas outside CpG islands and shores. (D) Enrichment / depletion of the DM CpG sites within promoters, gene bodies and intergenic regions. (E) Enrichment of the DM CpG sites within predicted enhancers defined according to HM450K array annotation. (F) Enrichment of the distal CpG DM sites within predicted enhancers defined based on ChIP-seq data from REMC for human brain (combined data sets for medial prefrontal cortex and hippocampus), liver, muscle and heart (74). Tissue-specific histone marks were defined as mark-enriched regions which are only present in one specific tissue but not in three other tissues.
Differences in DNA methylation between neuronal and glial cells have been recently described in detail (37). Here we focused on the comparison between the two neuronal subpopulations. Pairwise comparison of methylation across all HM450K CpG sites showed a substantially greater number of sites with higher methylation in GABA compared with GLU neurons (Figure 3B), indicative of pronounced differences in DNA methylation between the two neuronal subtypes. In subsequent analyses, a CpG site was considered to be differentially methylated (DM) between GABA and GLU neurons (i) if it showed a difference in total methylation between the neuronal subtypes above 0.1 [|delta (β)| > 0.1] (68–72) and (ii) if its methylation level was significantly different between these cell types by FDR-corrected (<5%) paired t-test. Under these criteria, we identified 59 941 DM sites (∼12.8% of all assayed CpG sites) with a significantly lower methylation in GLU versus GABA cells (GLU-undermethylated, or ‘GLUUM’ sites), and 10 433 DM sites (∼2.2% of all assayed CpG sites) with a significantly lower methylation in GABA versus GLU cells (‘GABAUM’ sites) (Supplementary Table S6). Thus, in line with data illustrated in Figure 3B, this analysis revealed significantly more GLUUM sites than GABAUM CpG sites.
Both the GLUUM and GABAUM sites were depleted in CpG islands but enriched within CpG island ‘shores’ (defined as 2-kb regions flanking CpG islands) and in the areas of genome distant from islands (‘open sea’ regions) (Figure 3C). The enrichment within shores was more pronounced for the GLUUM versus GABAUM sites. In line with our findings for CpG islands (which overlap with a high percentage of mammalian promoters (73)), both GABAUM and GLUUM sites were strongly depleted from promoters (Figure 3D). Notably, a strong enrichment of GABAUM and GLUUM sites was detected within gene bodies, whereas only marginal enrichment was detected in intergenic regions (Figure 3D). The GABAUM and GLUUM sites were much more likely to occur at positions distal (>1000 bp) to the nearest TSS (Supplementary Figure S5C). This distal DM was significantly enriched within gene bodies and shores located both upstream and downstream of the nearest TSS, whereas a markedly smaller enrichment was observed in the upstream and downstream intergenic regions (Supplementary Figure S5D, E). All enrichments and depletions mentioned above were statistically significant by Fisher's exact test (P-values <2.0e−9).
It has been recently reported that regions that are differentially methylated between neuronal and non-neuronal cells are enriched within putative enhancer elements (37). Similarly, both GLUUM and GABAUM sites were strongly enriched in putative enhancer elements defined according to the HM450K array annotation (P-values <2.2e−16) (Figure 3E). The majority of the DM sites overlapping with the HM450K-annotated enhancers were located at the regions distal to TSS (Supplementary Figure S5F). In addition, consistent with the epigenetic signatures of active and poised enhancers, we observed strong enrichment of both GABAUM and GLUUM sites within TSS-distal regions marked by human brain-specific histone H3 mono-methylation at lysine 4 (H3K4me1) and histone H3 acetylation at lysine 27 (H3K27ac) as well as by combination of these two marks that have been reported by the NIH Roadmap Epigenomics Mapping Consortium (REMC) (74) (Figure 3F). In contrast, depletion of the DM sites within tissue-specific predicted enhancers was observed in tissues that originate from different germ layers [e.g. liver (Figure 3F), heart or muscle (not shown)]. Although we identified the enrichment of the distal GABAUM and GLUUM sites within both predicted enhancers and CpG island shores, little overlap was observed between these two subsets of the DM sites (Supplementary Figure S5G). Thus, the neuronal subtype-specific DM sites in the shores could not be assigned to the predicted enhancers, and so remain to be functionally characterized.
To validate the DNA methylation results obtained by the HM450K assay using a different methodology, we measured the total DNA methylation in GABA and GLU neuronal subpopulations using ERRBS, a next generation sequencing-based method (44,75). The measurements were performed for the OFC specimens obtained from three Caucasian subjects (two males and one female) (Supplementary Table S1). To ensure reproducibility of measurements across the full range of values, the analysis was restricted to sites with an at least 10x coverage (35).
The ERRBS analysis enabled a greater coverage than the HM450K assay, yielding measurements for nearly 2 million CpG sites which were represented in all six ERRBS samples (two cell types × three subjects). Similar to the HM450K data, the total methylation frequency of individual ERRBS sites showed the expected bimodal distribution, and hierarchical clustering of methylation measurements clearly separated GABA from GLU samples (Supplementary Figure S6A, B). The level of DNA methylation across all CpG sites was on average 2.6% higher in GABA versus GLU neurons (P = 0.019 by paired t-test); similar results were obtained when only autosomal CpG sites were analyzed (Supplementary Figure S6C). Analysis of the 106 248 CpG sites that were represented in both data sets demonstrated a good agreement between the results obtained with HM450K and ERRBS (Supplementary Figure S6D and Supplementary Table S7A).
Similar to the HM450K data, a comparison of methylation levels across all ERRBS-measured CpG sites demonstrated considerable differential methylation between the two cell types, with many more GLUUM than GABAUM CpG sites (Supplementary Figure S6E). Using criteria for differential methylation similar to those employed in the analysis of the HM450K data ([|delta (β)| > 0.1 and FDR-corrected P-value < 0.05), we identified 168 491 GLUUM CpG sites (∼9.2% of all assayed CpG sites) and 45 162 GABAUM CpG sites (∼2.5% of all assayed CpG sites) when all three subjects (two males and one female were analyzed (Supplementary Table S8). A similar number of DM sites was detected when only two male subjects were considered (Supplementary Table S9). We also detected high correlation between DM values obtained from three and two subjects (Supplementary Figure S6F).
The ERRBS and HM450K data sets showed a good agreement between the DM sites (GABAUM or GLUUM) detected within the overlapping set of CpG sites (Supplementary Table S7B). The distribution of the ERRBS-detected DM sites within different genomic features was largely compatible with that obtained with the HM450K method. Both the GABAUM and GLUUM sites identified by ERRBS were significantly depleted in CpG islands and promoters but enriched within CpG island shores; furthermore, strong enrichment of DM sites was detected in gene bodies and moderate enrichment was detected in intergenic regions (Supplementary Figure S7A, B) (all P-values <2.2e−16). Also, both the GABAUM and GLUUM ERRBS sites were significantly depleted in the vicinity of TSS, but were more likely to reside distal to TSS (Supplementary Figure S7C). These distal DM sites were strongly enriched within brain-specific H3K4me1 and/or H3K27ac histone marks reported by REMC (74), which are indicative of enhancers, but depleted in predicted liver-specific enhancers defined by the same epigenetic signatures (Supplementary Figure S7D). The enrichment of the DM sites within brain-specific enhancers was more pronounced for the GLUUM than for the GABAUM sites; this difference was also detected by the HM450K assay (Figure 3F).
To summarize, using two independent methods, we identified significant differences in CpG methylation between GABA and GLU neurons in the human PFC that were mostly concentrated within gene bodies, CpG island shores and predicted enhancers. Strikingly, among the DM sites, there were significantly more GLUUM versus GABAUM CpG sites.
Non-CpG methylation is prominent in both GABA and GLU neurons and shows cell-specific differences in distribution
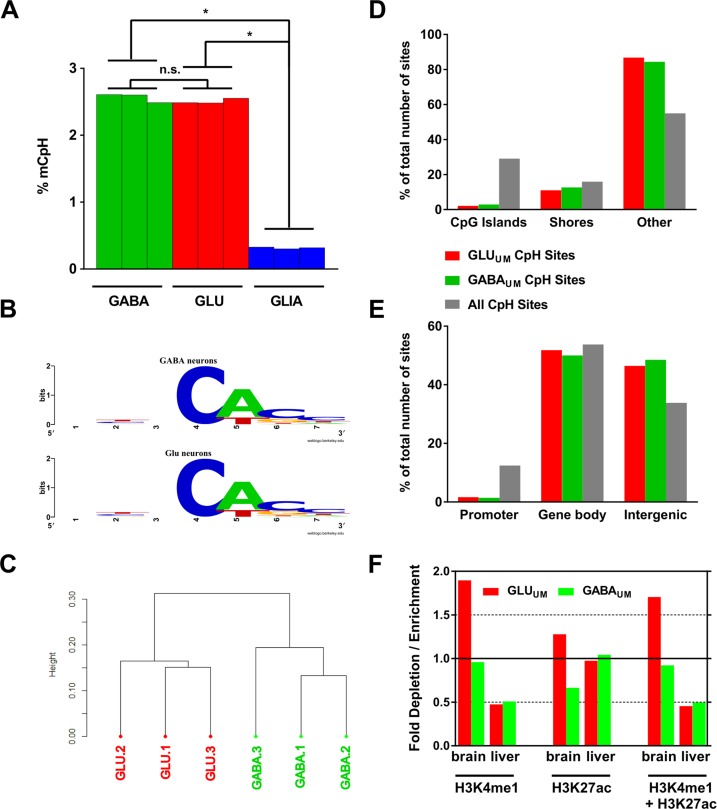
Whereas the HM450K assay probes included only 3000 individual cytosines in a non-CpG (CpH) sequence context, precluding a detailed analysis, ERRBS enabled us to assess methylation in ∼7.6 million CpH sites, considering only sites which were present in all 6 ERRBS samples with the coverage of ≥10 reads per site. Similar to the data recently reported for mouse brain (38), we detected substantial levels of CpH methylation in both GLU and GABA neuronal populations (Figure 4A). Because mCpH is negligible in the mouse and human fetal cortex and accumulates during early postnatal development long after the divergence of the excitatory and inhibitory neuronal lineages (24), these findings suggest that mCpH is a shared characteristic of the mature GABA and GLU neurons.
Figure 4.
Analysis of non-CpG (CpH) methylation measured by ERRBS. (A) Average CpH methylation in GABA (green) and GLU (red) neurons, and in GLIA (blue); N = 3 samples for each cell type. Data for GLIA are from (32); *P-values < 0.0001 for comparisons of GLIA with GABA or GLU (unpaired t-test). (B) Sequence context of CpH methylation in GABA and GLU neurons. In both neuronal subtypes CpH methylation occurs in mostly the same context—CACC. Methylated CpH cytosine is shown at position 4. (C) Unsupervised hierarchical clustering of CpH methylation data for GLU and GABA samples. 1, 2, 3 denote individual subjects. (D) Enrichment / depletion of CpH DM sites within CpG island-related genomic features. The data presented as% of total number of sites within each group (GLUUM, GABAUM and all CpH sites present in the ERRBS data set). (E) Enrichment/depletion of CpH DM sites within promoters, gene bodies and intergenic regions. (F) Enrichment of the distal CpH DM sites within predicted enhancers defined based on histone modification profiling data from REMC (74) (see Figure 3F for details).
In agreement with previous observations (24,37), mCpH was significantly higher in neurons versus non-neuronal cells from the same area of the PFC (Figure 4A). In contrast to our findings for mCpG and to recent findings in the mouse brain for mCpH (38), the average level of mCpH was not significantly different between the two neuronal subtypes (2.58% for GABA versus 2.52% for GLU neurons) when all CpH sites were considered. Similar results were obtained when CpH sites on sex chromosomes were excluded (Supplementary Figure S8A). We also detected a similar motif preference for mCpH in GABA and GLU neurons, with CAC having the highest methylation level (Figure 4B), which was in line with the data reported in the mouse brain (38). Unsupervised hierarchical clustering of GLU and GABA mCpH profiles showed a clear separation of the samples into two groups according to their neuronal subtype (Figure 4C), indicating a non-random cell-specific profile of this epigenetic modification. In both subtypes of neurons, the highest level of mCpH was detected in intergenic regions (∼4.0%) and the lowest in promoters (∼0.06%), whereas gene bodies showed an intermediate level of methylation (∼2.2%) (Supplementary Figure S8B). The levels of mCpH in these major genomic regions did not differ between GABA and GLU neurons.
Next, we identified CpH sites that were differentially methylated between the GABA and GLU neurons, using a threshold of [|delta (β)| > 0.1] and FDR-corrected P-value < 0.05. In contrast to the DM CpG sites, we detected comparable fractions of CpH sites that were undermethylated in GLU versus GABA cells (GLUUM; N = 96 071; 1.3% of all CpH sites) and in GABA versus GLU cells (GABAUM; N = 80 412; 1.1% of all CpH sites) (Supplementary Table S10). A comparable number of DM sites and high correlation between DM values was detected when only two male subjects were considered (Supplementary Table S9 and Supplementary Figure S6F).
Similar to the DM CpG sites, the DM CpH sites were strongly depleted in CpG islands and promoters; however, in contrast to the DM CpG sites, the DM CpH sites were slightly depleted also in shores and gene bodies, but were highly enriched in intergenic regions and outside CpG islands and shores (open sea regions) (Figure 4D and E) (all P-values <6.7e−33). In addition, only GLUUM but not GABAUM DM CpH sites were enriched within brain-specific distal enhancers predicted based on the REMC data (Figure 4F) (74). The latter finding suggests that a different subset of putative enhancer regions is marked by mCpG versus mCpH differential methylation. However, this result might also reflect the fact that the REMC data were obtained using homogenized cortical tissues. Because the majority of neurons in the cortical tissues are projection GLU neurons, the REMC histone modification profiling probably captures more GLU-specific than GABA-specific enhancers. Indeed, in the case of CpG DM sites, the REMC data showed a less pronounced enrichment for GABAUM versus GLUUM sites within putative brain-specific enhancers (see Figure 3F and Supplementary Figure S7D).
To summarize, we identified abundant mCpH in both GABA and GLU neurons, and detected cell-specific differences in the distribution of mCpH sites. In contrast to mCpG, the differences in mCpH methylation between the neuronal subtypes were more pronounced in intergenic regions, and there was a comparable number of GLUUM versus GABAUM CpH sites.
Correlation of DNA methylation with gene expression
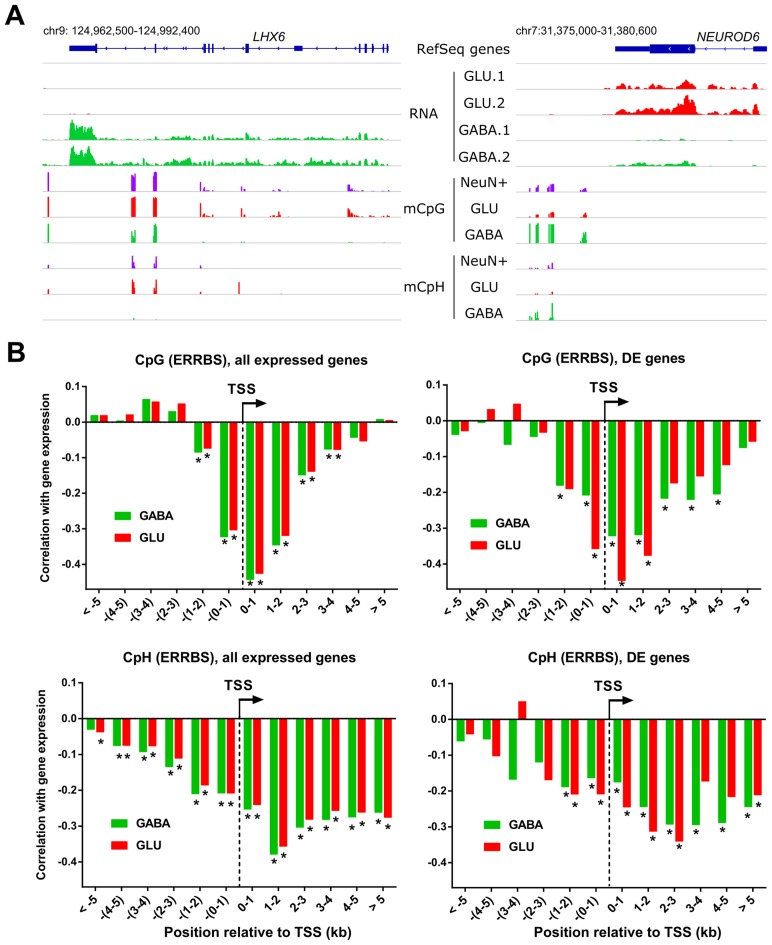
DNA methylation in promoters and gene bodies has been previously reported to inversely correlate with gene expression in the mixed NeuN(+) population of mammalian cortical neurons (24). Although these studies enabled the definitive characterization of the relationship between DNA methylation and gene expression for the projection GLU neurons, which constitute ∼80% of all NeuN(+) cells in the human and rodent PFC, the same could not be achieved for GABA neurons, which account for only ∼10–15% of all NeuN(+) cells. For example, NEUROD6 (a GLU specific transcription factor), which is highly expressed in GLU but not in GABA neurons, exhibits low promoter CpG and CpH methylation in the mixed NeuN(+) neuronal samples; however, LHX6 (a GABA-specific transcription factor), which is highly expressed in GABA but not in GLU neurons, appears to be highly methylated in this mixed specimen (Figure 5A). Our neuron subtype-specific data resolved this apparent discrepancy and showed that high level of DNA methylation and active gene expression do not typically co-occur in the same neuronal subtype. Indeed, NEUROD6 is significantly less methylated in GLU versus GABA neurons whereas the opposite is true for LHX6.
Figure 5.
Analysis of correlation between DNA methylation and gene expression. (A) RNA-seq read density and DNA methylation in CpG and non-CG contexts from ERRBS assay (CpG and CpH) at two genes, LHX6 and NEUROD6 that are specific for MGE-derived GABA and GLU neurons, respectively. Traces for GLU and GABA neurons are marked in green and red, respectively. NeuN(+) CpG and NeuN(+) CpH are mixed neuron ERRBS data from (37). 1 and 2 denote RNA-seq data for two different subjects. (B) Spearman correlations between mCpG or mCpH and RNA expression levels of all expressed genes (left panels) and differentially expressed genes (right panels). For each gene, mCpG or mCpH sites were combined into 1kb bins as a function of the distance from TSS. The significance (*) was determined as P < 0.01 after Bonferroni correction for the number of bins (N = 12).
When all expressed genes were analyzed (Figure 5B, left panels), we observed a strong inverse correlation between RNA expression and CpG methylation around the TSS region (±1 kb) for both GABA and GLU neurons (also see Supplementary Figure S9A). This inverse correlation extended into the gene body (but not into the intergenic regions) but rapidly subsided downstream from TSS. In contrast, the inverse correlation between RNA expression and mCpH stretched throughout the gene body for at least 5 kb reaching maximum at ∼1–2 kb downstream from the TSS and also extended into the intergenic region. These findings demonstrate that mCpH is not only abundant in different subpopulations of the PFC neurons but also correlates stronger with gene expression in the TSS-distal regions compared to mCpG. Similar patterns were observed in the mouse brain (38).
A similar TSS-distance-dependent pattern of correlations with DNA methylation was observed when DE genes were analyzed (Figure 5B right panels). However, stronger correlations for mCpG within the gene body and notable differences between GABA and GLU cells were detected for DE compared to all expressed genes. Specifically, the correlation between TSS-proximal mCpG or mCpH and DE gene expression was stronger for the GLU-DE genes in GLU cells versus the GABA-DE genes in GABA cells. At least for mCpG, this could stem from the higher proportion of CpG-rich promoters in GABA-DE versus GLU-DE genes (see Figure 2D). Because CpG-rich promoters are rarely marked by DNA methylation, a smaller proportion of GABA-DE versus GLU-DE genes should be associated with promoter methylation, resulting in a stronger correlation with mCpG in the latter. This is further supported by higher variability of promoter mCpG methylation for the GLU-DE genes compared to GABA-DE genes (Supplementary Figure S9B). Notably, the mCpG variability in gene bodies was comparable between GABA-DE and GLU-DE genes.
Hydroxymethylation is higher in GLU compared to GABA neurons
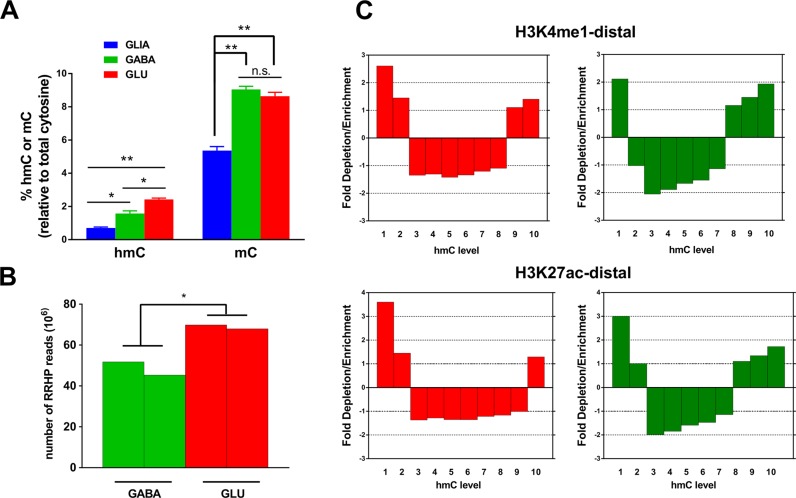
Because both assays that were used in this study (HM450K array and ERRBS) measured the total level of CpG methylation (mCpG plus hmCpG), we tested if the significantly greater number of GLUUM versus GABAUM CpG sites detected by these methods was due to higher hmCpG in GABA compared to GLU cells. To this end, we characterized the hmCpG modification in GABA and GLU cells using two independent methods. First, we measured the total levels of various cytosine modifications (mC, hmC or non-modified cytosine) in the genomic DNA samples from the FACS-separated GABA, GLU and GLIA nuclei obtained from 4 human autopsy OFC specimens (Supplementary Table S1) using LCMS/MS (Figure 6A and Supplementary Table S11). In agreement with previous reports (24), the level of mC was ∼2-fold higher in the neuronal compared with glial cells (p-values <0.0001). This difference is probably due to the substantially higher level of CpH methylation (which predominantly exists in the mC but not the hmC form) in neurons compared to glial cells. The levels of mC did not differ between GABA and GLU neurons; in both neuronal subpopulations, ∼9% of the cytosines were methylated. In contrast, the amounts of hmC differed significantly among the cell types: 2.4%, 1.6% and 0.7% of all cytosines were hydroxymethylated in GLU, GABA and in GLIA, respectively (P-values < 0.01 by ANOVA).
Figure 6.
Analysis of hydroxymethylation in GABA and GLU neurons. (A) Percentage of total cytosine residues containing hmC or mC modifications in human cortical non-neuronal cells (GLIA), GABA neurons or GLU neurons, as measured by the mass-spectrometry-based methodology (N = 4 samples for each cell type). *P <0.01, **P <0.0001 (ANOVA followed by Tukey's post hoc test). (B) Levels of hmC in GABA and GLU neurons measured by the RRHP method in two replicate samples. Total number of reads is the measure of hmC content; *P <0.01 (by t-test). (C) Enrichment or depletion of hmC within predicted distal enhancers. All CCGG regions for which the hmC data were obtained by the RRHP method were subdivided into 10 equally populated groups (from lowest to highest levels of hmC). Predicted enhancer positions were obtained from the REMC ChIP-seq data for distal H3K4me1 and H3K27ac histone marks (74). GLU neurons—red color, GABA neurons-green color.
Next, we used the recently introduced RRHP assay (46) to measure the level of hmC in GABA and GLU neurons from the OFC of two individuals. The RRHP method employs next generation sequencing and probes individual CpG sites within the CCGG sequence context. The output of RRHP is the number of sequencing reads corresponding to each specific CpG site, which serves as a strand-specific measure of its level of hmC. Initial data processing resulted in a data set of ∼465 000 CCGG sites (∼20% of all CCGG sequences in the human genome) (see Methods and Supplementary Tables S12A, B). In a good agreement with the results obtained by the mass-spectrometry, we detected significantly higher (1.4-fold) hmCpG in GLU versus GABA neurons (Figure 6B and Supplementary Table S13). Thus, using two independent methods, we detected a substantially higher level of CpG hydroxymethylation in GLU versus GABA cells, rejecting the hypothesis that the larger number of GLUUM versus GABAUM CpG sites stemmed from higher hmCpG content in GABA cells.
Previously reported data obtained in bulk (unsorted) human brain tissue samples suggest that hmC is enriched in enhancers (34). To test if this held true also for the GABA and GLU neurons, we subdivided the CpG sites within GABA and GLU neuronal subpopulations by their hmCpG level into 10 bins, from the lowest to highest number of RRHP reads. For both GLU and GABA samples, we observed enrichment within the predicted distal enhancers for sites with the lowest and the highest hmCpG levels, whereas sites with intermediate hmCpG were depleted from the enhancers (Figure 6C). This finding is in agreement with the previously published study (34) which suggested that enhancers with high and low hmCpG levels could correspond to ‘poised’ and ‘active’ enhancers, respectively. Another intriguing feature of hmCpG described in bulk brain tissue is its bias toward the sense compared with antisense strand within gene bodies (34). We analyzed 16 512 genes, for which the RRHP data were available, and detected a higher hmCpG level in the sense versus the antisense strand (differences of ∼3.0% for GLU and ∼4.6% for GABA neurons) (all P-values < 3e−14 by paired t-test, see Supplementary Table S12C). Thus, our results support the conclusions of previous studies performed with the human brain tissue, and extend these findings to the neuronal subpopulations.
Neuronal subtype-specific differentially methylated regions are enriched for schizophrenia risk loci
A previous study has demonstrated enrichment of schizophrenia (SCZ) risk variants within brain-specific regulatory sequences (76). Here we examined if common risk variants determined by the recent genome wide associated studies (GWAS) for several neuropsychiatric diseases—including SCZ, autism spectrum disorder (ASD), major depressive disorder (MDD), and Alzheimer's disease (AD) (20,47,48)—significantly overlap with regions that encompass GABAUM or GLUUM DM CpG sites from the HM450K data set (which we for convenience hereinafter denote differentially methylated regions, DMRs). We also performed a similar analysis to investigate if common risk variants for these diseases are enriched in the DMRs for NeuN(+)-specific and NeuN(−)-specific DM CpG sites using the data from our previous work (37). For comparison, we examined common variants associated with non-neuropsychiatric conditions, including rheumatoid arthritis (RA) (49) and the levels of total cholesterol (TC) and triglycerides (TG) (50).
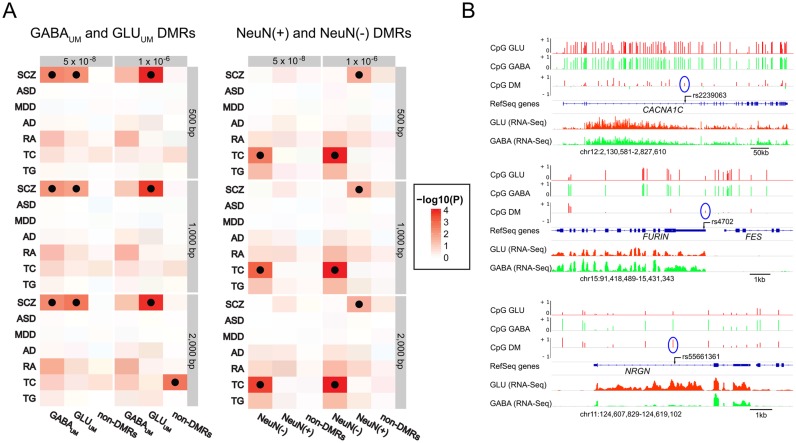
We analyzed the enrichment of SNPs in DMRs with windows of 3 different sizes (500 bp, 1 kb and 2 kb) encompassing 250 bp, 500 bp or 1kb regions upstream and downstream from each DM CpG site, and considered two different thresholds of genome-wide significance for disease-associated SNPs—P-values < 5 × 10−8 (stringent SNPs) and less stringent P-values < 1 × 10−6 (relaxed SNPs). We found that the SCZ risk variants were significantly enriched within the GABAUM DMRs (adjusted P-values < 0.05 for stringent SNPs) and within the GLUUM DMRs (adjusted P-values < 0.05 for both stringent and relaxed SNPs), but not within the non-DMRs (Figure 7A left panel). These enrichments were significant within each of the three windows analyzed. The SCZ GWAS identified 128 index SNPs, which are the most significant SNPs per a disease-associated locus (20). Among these 128 index SNPs, there were 13 and 3 SNPs that were found within 2 kb window GLUUM or GABAUM DMRs, respectively (Supplementary Table S14). There was no significant enrichment of risk variants for ASD, MDD or AD, even at nominal (uncorrected) P-value < 0.05, or for non-psychiatric data sets, except that the relaxed TC-associated SNPs were enriched within the 2 kb window non-DMRs (adjusted P-value = 0.011).
Figure 7.
Cell type-specific differentially methylated regions (DMRs) are enriched for schizophrenia risk loci. (A) Results of SNP enrichment analysis using GABA- and GLU-specific DMRs (left panel) and NeuN(+) and NeuN(−) DMRs (right panel) from (37). Heatmap plots visualize the −log10(P) value of disease-associated variants from recent GWAS of schizophrenia (SCZ), autism spectrum disorder (ASD), major depressive disorder (MDD), Alzheimer's disease (AD), rheumatoid arthritis (RA) and levels of total cholesterol (TC) and triglycerides (TG) within cell-specific DMRs. Black dots indicate enrichment that was significant at corrected P-value < 0.05. GWAS variants were selected based on two statistical thresholds: 5 × 10−8 and 1 × 10−6. Differentially methylated regions were defined as regions within 250 bp, 500 bp or 1kb upstream or downstream of the cell type-specific DM CpG sites. (B) CpG methylation profiles (measured by HM450K) and gene expression profiles (measured by RNA-seq) in the vicinity of 3 of the 16 SCZ risk SNPs which overlap with neuron subtype-specific DMRs. For each loci, top three traces depict β values in GLU (red) and GABA (green) cells and Δβ for CpG sites undermethylated in GLU (red) or GABA (green) neurons, respectively; bottom two traces show RNA-seq reads in GLU (red) or GABA (green) neurons, respectively. The position of the SCZ risk SNPs is shown as black arrow. The position of the nearest DM site in the HM450K array is encircled in blue.
We also detected a significant enrichment of the relaxed SCZ SNPs within NeuN(+) DMRs (adjusted P-values < 0.05), but not for the NeuN(−) DMRs or non-DMRs (Figure 7A right panel). In contrast, no significant enrichment of risk variants for ASD, MDD or AD was detected, even at nominal P-value < 0.05. Among non-psychiatric data sets, the TC-associated stringent and relaxed SNPs were significantly enriched within the NeuN(−) DMRs (adjusted P-value < 0.05). Because the NeuN(-) cell population comprises mostly of glial cells (24,37), this finding suggests a link between the level of TC and DNA methylation in glia.
DISCUSSION
To explore the potential role of DNA methylation in the functional differentiation of the two major neuronal subtypes in the human brain, we developed a FACS-based protocol to separate nuclei from the MGE-derived GABA interneurons and GLU projection neurons. Both neuronal subtypes have been specifically implicated in several neurological and psychiatric diseases, including SCZ (19,77).
In this study, we focused on characterizing GABA and GLU neurons in the PFC of healthy individuals. We confirmed the specificity of the two sorted nuclear populations using genome-wide transcriptome analysis (RNA-seq), which recovered the known GABA-specific and GLU-specific markers. This genome-wide approach enabled us to generate unique information-rich data sets that characterize neuron subtype-specific transcriptomes which, to the best of our knowledge, have not been so far available for the human brain. In addition to identifying novel GABA and GLU markers of the human PFC (e.g. ARPP21 for GLU and DOCK11 for GABA neurons; see Supplementary Table S3), we found that significantly more non-coding RNAs were preferentially expressed in GLU compared to GABA nuclei.
We identified a higher proportion of genes with CpG-rich promoters in the GABA-DE compared to GLU-DE genes. Our results suggest that this difference accounts for the weaker correlation between the promoter CpG methylation and the expression of the GABA-DE compared to GLU-DE genes. We speculate that the promoters of genes that are specifically expressed in GABA or GLU neurons differ in the preferential epigenetic mechanisms that affect their regulation. It has been previously reported that high CpG density promoters of low expressed genes tend to be enriched for H3K27me3, whereas promoters with a low CpG density are mostly marked by DNA methylation (33). Further studies of neuron subtype-specific histone modification profiling are necessary to test this hypothesis.
We compared mCpG and mCpH landscapes between GABA and GLU neurons using two independent methods (HM450K and ERRBS) and examined their hmCpG status using LCMS/MS and RRHP. We identified numerous CpG sites that were differentially methylated between these neuronal populations. These DM CpG sites were depleted in CpG-islands and promoters but concentrated within gene bodies as well as in CpG island shores and predicted enhancers although there was little overlap between the latter two sets of DM sites. These findings were in line with previous reports which mapped tissue-specific and cell-specific differential CpG methylation to shores and distal cis-regulatory elements (78,79), and in the case of neurons, to gene bodies versus intergenic regions (37). Unexpectedly, we identified a dramatically greater number of undermethylated CpG sites in GLU versus GABA neurons, suggesting the possibility that many more genes were preferentially expressed in GLU neurons compared to GABA neurons. This prediction, however, was not borne out by our RNA-Seq analysis which yielded comparable numbers of GLU- and GABA-DE genes. This is consistent with a recent study in the mouse cortex that reported about twice as many CpG hypomethylated regions in the excitatory (GLU) neurons compared to PVALB-expressing GABA interneurons, whereas there was no significant difference in the number of genes found to be differentially expressed between these neuronal subtypes (38).
In contrast to mCpG, hmCpG has been shown to be associated with active genes (30). Therefore, we tested the possibility that the lack of concordance between the differential CpG methylation and differential expression in GLU versus GABA neurons could be explained by higher hmCpG level in GABA cells. Should that be the case, the excess of hmCpG in GABA cells could mitigate the observed discrepancy between differential methylation and gene expression. However, we detected more hmC in GLU neurons compared to GABA neurons. The higher abundance of hmCpG sites in GLU versus GABA neurons appears indicative of a difference in transcriptional potential between the neuronal subtypes. The increased hydroxymethylation could enable certain genes (e.g. activity-dependent genes) to be more readily induced in GLU versus GABA neurons. Indeed, a recent study has examined activity-dependent transcriptional programs that were induced in the mouse brain-derived embryonic neuronal cultures 1hr (early-response genes) and 6 h (late-response genes) following membrane depolarization (80). In line with our hypothesis, it has been shown that significantly more early- (50 versus 28) and late- (808 versus 438) response transcripts were activated in cortical excitatory compared to MGE-derived inhibitory GABA neurons (80).
We observed a strong inverse correlation between gene expression and CpG methylation around the TSS in both GABA and GLU neurons. These correlations, however, were weaker within the gene body and rapidly subsided with the distance from the TSS. Similar results have been reported in the aforementioned mouse study (38) and appear surprising considering the depletion of the neuron-subtype specific GLUUM and GABAUM DM sites in the promoters and their enrichment in the regions distal to TSS and in putative distal enhancers (see Figure 3). These findings emphasize the importance of even subtle differences in the promoter CpG methylation for neuron subtype-specific gene expression. They also suggest that differences in CpG methylation within gene bodies and distal regulatory elements are not always directly reflected in differences in gene expression between neuronal subtypes.
We also detected abundant non-CpG methylation (mCpH) in both GABA and GLU neurons. In contrast to mCpG, mCpH showed no significant difference in the number of DM GABAUM versus GLUUM sites. It should be emphasized that methylation in the CpG and CpH sites was measured using the same assay (ERRBS). Thus, the CpH sites provide an internal control to the CpG sites, effectively eliminating the possibility of a cell-specific bias in methylation measurements. Despite the lack of bulk differences, the patterns of CpH methylation were clearly specific for the neuronal subtypes (see Figure 4C). This finding is compatible with the hypothesis that CpH methylation is an important determinant of the identity of mammalian neurons (24,25). Notably, and similar to the findings in the mouse brain (38), mCpH within gene bodies and intergenic regions was a better predictor of neuron subtype-specific gene expression compared to mCpG. This was again a surprising finding because, compared to the DM mCpG sites, we detected less (for GLUUM sites) or no (for GABAUM sites) enrichment of the DM mCpH sites within putative distal enhancers. Thus, the functional relevance of the association between gene expression and distal non-CpG methylation remains to be characterized. One possible mechanism is suggested by a recent work which has shown that methyl-DNA-binding protein MeCP2, previously proposed to function as a transcriptional regulator (81), represses expression of long genes by binding to mCA sites within gene bodies (82–84).
Finally, we investigated if the genetic risk for several neuropsychiatric diseases relates to the cell type-specific methylome. We found a significant overlap between neuron-specific NeuN(+) DMRs [but not glia-specific (NeuN–) DMRs] that were detected in our recent work (37) and SCZ-associated genetic loci that were reported in the recent comprehensive GWAS meta-analysis (20). We further observed that the SCZ-associated loci were enriched among both (GABA and GLU) neuron subtype-specific DMRs from this report. Importantly, we did not observe an enrichment of variants associated with non-neuropsychiatric conditions (RA, TC, TG) in neuron- or neuron subtype-specific DMRs. Collectively, these findings strongly suggest an association between the epigenetic specification of both GABA and GLU neurons and SCZ. Indeed, we found that 12.5% of SCZ index SNPs from (20) are situated in the vicinity of neuron subtype-specific DM CpG sites. Some of these SNPs and the neighbouring DMRs are located within genes which could contribute to cellular and molecular biology abnormalities underlying SCZ (see Supplementary Table S14 and Figure 7B). For example, CACNA1C encodes the subunit of the L-type voltage-gated calcium channel which is activated upon cellular depolarization and is involved in integration of dendritic information (85). Neurogranin, encoded by NRGN, is a calmodulin-binding protein that is an important component of the ionotropic glutamate receptor-signaling pathway that has been associated with synaptic plasticity and memory formation (86). Moreover, human brain imaging and behavioral studies have demonstrated morphological and functional alterations in individuals carrying the CACNA1C and NRGN schizophrenia risk alleles (87–91). Lastly, furin (encoded by FURIN) is a secretory proprotein convertase that converts the precursor of brain derived neurotrophic factor (BDNF) into the mature protein (92); BDNF has been implicated in SCZ by numerous studies (93).
Although we used the results of the largest GWAS analyses published so far for each disease, risk variants associates with ASD, MD, or AD were not enriched within neuron-specific or neuron subtype-specific DMRs, suggesting that this association is specific to SCZ. Notably, compared to the SCZ GWAS, the GWAS for other neuropsychiatric diseases resulted in a significantly smaller number of disease-associated loci. For example, even with the relaxed statistical threshold (P-values < 1 × 10−6), there are 344 SCZ-associated loci compared to 52 AD-associated, 5 MDD-associated and 2 ASD-associated ones. Therefore, the power of the present analysis could be insufficient to capture the link between the genetic risk and cell-specific methylation, especially for MDD and ASD. Subsequent, more extensive GWA studies might still reveal such associations. An alternative explanation of our negative results could be the involvement of different developmental stages and/or brain regions in different diseases.
To summarize, our results demonstrate striking differences in DNA methylation landscapes, including CpG, CpH and hCpG methylation, between GABA interneurons and projection GLU neurons in the human PFC. The apparent differences in the transcriptional potential that are reflected in DNA methylation differences are likely to be related to functional characteristics of the neuronal subtypes. Our data suggest that, compared to GABA interneurons, GLU projection neurons are characterized by more permissive chromatin state that is less constrained by repressive DNA methylation marks and is instead controlled by more dynamic means of transcription inhibition, such as non-coding RNAs and/or histone modifications (33,94). Indeed, we detected many more non-coding RNA genes that are specifically expressed in GLU compared with GABA neurons. Notably, a recent work, which integrated 111 reference human epigenomes for primary cells and tissues, emphasized the importance of chromatin state context in defining the relationship between DNA methylation and gene expression (95). Thus, mapping neuron subtype-specific histone modification marks in GABA and GLU neurons could further help to test this hypothesis and to better understand the relationship between cell-specific epigenome and vulnerability to disease.
Supplementary Material
Acknowledgments
The work was supported with resources and the use of facilities at the James J. Peters VA Medical Center, Bronx, New York.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Mental Health [R21MH103877 that is a part of the PsychENCODE consortium to S.D.]; U.S. Department of Veterans Affairs [Merit Review Award BX001829 to S.D.]; Intramural funds of the U.S. Department of Health and Human Services to National Library of Medicine (to E.V.K.). Funding for open access charge: National Institute of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ramon y Cajal S. Histology of the Nervous System. Wien: Springer; 1899. [Google Scholar]
- 2.Ma T., Wang C.M., Wang L., Zhou X., Tian M., Zhang Q.Q., Zhang Y., Li J.W., Liu Z.D., Cai Y.Q., et al. Subcortical origins of human and monkey neocortical interneurons. Nat. Neurosci. 2013;16:1588–1597. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- 3.Hu H., Gan J., Jonas P. Fast-spiking, parvalbumin(+) GABAergic interneurons: from cellular design to microcircuit function. Science. 2014;345:529. doi: 10.1126/science.1255263. [DOI] [PubMed] [Google Scholar]
- 4.Rudy B., Fishell G., Lee S., Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev. Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kepecs A., Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Magueresse C., Monyer H. GABAergic interneurons shape the functional maturation of the cortex. Neuron. 2013;77:388–405. doi: 10.1016/j.neuron.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Chao H.T., Chen H., Samaco R.C., Xue M., Chahrour M., Yoo J., Neul J.L., Gong S., Lu H.C., Heintz N., et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cobos I., Calcagnotto M.E., Vilaythong A.J., Thwin M.T., Noebels J.L., Baraban S.C., Rubenstein J.L. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat. Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- 9.Rubenstein J.L.R., Merzenich M.M. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volk D.W., Matsubara T., Li S., Sengupta E.J., Georgiev D., Minabe Y., Sampson A., Hashimoto T., Lewis D.A. Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. Am. J. Psychiatry. 2012;169:1082–1091. doi: 10.1176/appi.ajp.2012.12030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marin O. Interneuron dysfunction in psychiatric disorders. Nat. Rev. Neurosci. 2012;13:107–120. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- 12.Volk D.W., Austin M.C., Pierri J.N., Sampson A.R., Lewis D.A. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch. Gen. Psychiatry. 2000;57:237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- 13.Guidotti A., Auta J., Davis J.M., Di-Giorgi-Gerevini V., Dwivedi Y., Grayson D.R., Impagnatiello F., Pandey G., Pesold C., Sharma R., et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch. Gen. Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- 14.Straub R.E., Lipska B.K., Egan M.F., Goldberg T.E., Callicott J.H., Mayhew M.B., Vakkalanka R.K., Kolachana B.S., Kleinman J.E., Weinberger D.R. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Mol. Psychiatry. 2007;12:854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- 15.Fung S.J., Webster M.J., Sivagnanasundaram S., Duncan C., Elashoff M., Weickert C.S. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am. J. Psychiatry. 2010;167:1479–1488. doi: 10.1176/appi.ajp.2010.09060784. [DOI] [PubMed] [Google Scholar]
- 16.Curley A.A., Arion D., Volk D.W., Asafu-Adjei J.K., Sampson A.R., Fish K.N., Lewis D.A. Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: clinical, protein, and cell type-specific features. Am. J. Psychiatry. 2011;168:921–929. doi: 10.1176/appi.ajp.2011.11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y., Flandin P., Long J.E., Cuesta M.D., Westphal H., Rubenstein J.L. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J. Comp. Neurol. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batista-Brito R., Rossignol E., Hjerling-Leffler J., Denaxa M., Wegner M., Lefebvre V., Pachnis V., Fishell G. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron. 2009;63:466–481. doi: 10.1016/j.neuron.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu W., MacDonald M.L., Elswick D.E., Sweet R.A. The glutamate hypothesis of schizophrenia: evidence from human brain tissue studies. Ann. N. Y. Acad. Sci. 2015;1338:38–57. doi: 10.1111/nyas.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugino K., Hempel C.M., Miller M.N., Hattox A.M., Shapiro P., Wu C., Huang Z.J., Nelson S.B. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat. Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 22.Okaty B.W., Miller M.N., Sugino K., Hempel C.M., Nelson S.B. Transcriptional and electrophysiological maturation of neocortical fast-spiking GABAergic interneurons. J. Neurosci. 2009;29:7040–7052. doi: 10.1523/JNEUROSCI.0105-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 24.Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D., et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J.U., Su Y., Shin J.H., Shin J., Li H., Xie B., Zhong C., Hu S., Le T., Fan G., et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Numata S., Ye T., Hyde T.M., Guitart-Navarro X., Tao R., Wininger M., Colantuoni C., Weinberger D.R., Kleinman J.E., Lipska B.K. DNA methylation signatures in development and aging of the human prefrontal cortex. Am. J. Hum. Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito S., D'Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellen M., Ayata P., Dewell S., Kriaucionis S., Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stadler M.B., Murr R., Burger L., Ivanek R., Lienert F., Scholer A., van N.E., Wirbelauer C., Oakeley E.J., Gaidatzis D., et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 33.Xie W., Schultz M.D., Lister R., Hou Z., Rajagopal N., Ray P., Whitaker J.W., Tian S., Hawkins R.D., Leung D., et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen L., Li X.L., Yan L.Y., Tan Y.X., Li R., Zhao Y.Y., Wang Y., Xie J.C., Zhang Y., Song C.X., et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014;15:R49. doi: 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varley K.E., Gertz J., Bowling K.M., Parker S.L., Reddy T.E., Pauli-Behn F., Cross M.K., Williams B.A., Stamatoyannopoulos J.A., Crawford G.E., et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guintivano J., Aryee M.J., Kaminsky Z.A. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8:290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozlenkov A., Roussos P., Timashpolsky A., Barbu M., Rudchenko S., Bibikova M., Klotzle B., Byne W., Lyddon R., Di Narzo A.F., et al. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res. 2014;42:109–127. doi: 10.1093/nar/gkt838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mo A., Mukamel E.A., Davis F.P., Luo C., Henry G.L., Picard S., Urich M.A., Nery J.R., Sejnowski T.J., Lister R., et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drakenberg K., Nikoshkov A., Horvath M.C., Fagergren P., Gharibyan A., Saarelainen K., Rahman S., Nylander I., Bakalkin G., Rajs J., et al. Mu opioid receptor A118G polymorphism in association with striatal opioid neuropeptide gene expression in heroin abusers. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7883–7888. doi: 10.1073/pnas.0600871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikoshkov A., Drakenberg K., Wang X., Horvath M.C., Keller E., Hurd Y.L. Opioid neuropeptide genotypes in relation to heroin abuse: dopamine tone contributes to reversed mesolimbic proenkephalin expression. Proc. Natl. Acad. Sci. U.S.A. 2008;105:786–791. doi: 10.1073/pnas.0710902105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stolt C.C., Schlierf A., Lommes P., Hillgartner S., Werner T., Kosian T., Sock E., Kessaris N., Richardson W.D., Lefebvre V., et al. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev. Cell. 2006;11:697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Schnieder T.P., Trencevska I., Rosoklija G., Stankov A., Mann J.J., Smiley J., Dwork A.J. Microglia of prefrontal white matter in suicide. J. Neuropathol. Exp. Neurol. 2014;73:880–890. doi: 10.1097/NEN.0000000000000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bibikova M., Le J., Barnes B., Saedinia-Melnyk S., Zhou L., Shen R., Gunderson K.L. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 44.Akalin A., Garrett-Bakelman F.E., Kormaksson M., Busuttil J., Zhang L., Khrebtukova I., Milne T.A., Huang Y., Biswas D., Hess J.L., et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8:e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petterson A., Chung T.H., Tan D., Sun X.G., Jia X.Y. RRHP: a tag-based approach for 5-hydroxymethylcytosine mapping at single-site resolution. Genome Biol. 2014;15:456. doi: 10.1186/s13059-014-0456-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.CONVERGE consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Global Lipids Genetics, C. Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee P.H., O'Dushlaine C., Thomas B., Purcell S.M. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang Y., Matevossian A., Huang H.S., Straubhaar J., Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC.Neurosci. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeisel A., Manchado A.B., Codeluppi S., Lonnerberg P., La Manno G., Jureus A., Marques S., Munguba H., He L., Betsholtz C., et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 55.Zhao W., He X., Hoadley K.A., Parker J.S., Hayes D.N., Perou C.M. Comparison of RNA-Seq by poly (A) capture, ribosomal RNA depletion, and DNA microarray for expression profiling. BMC Genomics. 2014;15:419. doi: 10.1186/1471-2164-15-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ameur A., Zaghlool A., Halvardson J., Wetterbom A., Gyllensten U., Cavelier L., Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat. Struct. Mol. Biol. 2011;18:U1435–U1157. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 57.Wang J., Duncan D., Shi Z., Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deaton A.M., Bird A. CpG islands and the regulation of transcription. Gene Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schug J., Schuller W.P., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barrera L.O., Li Z., Smith A.D., Arden K.C., Cavenee W.K., Zhang M.Q., Green R.D., Ren B. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 2008;18:46–59. doi: 10.1101/gr.6654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang C.W., Cheng W.C., Chen C.R., Shu W.Y., Tsai M.L., Huang C.L., Hsu I.C. Identification of human housekeeping genes and tissue-selective genes by microarray meta-analysis. PLoS One. 2011;6:e22859. doi: 10.1371/journal.pone.0022859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisenberg E., Levanon E.Y. Human housekeeping genes, revisited. Trends Genet. 2013;29:569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 63.Zhu J., He F.H., Hu S.N., Yu J. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 64.Zhu J., He F.H., Song S.H., Wang J., Yu J. How many human genes can be defined as housekeeping with current expression data. BMC Genomics. 2008;9:172. doi: 10.1186/1471-2164-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu X., Yu X.P., Zack D.J., Zhu H., Qian J. TiGER:a database for tissue-specific gene expression and regulation. BMC Bioinformatics. 2008;9:271. doi: 10.1186/1471-2105-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L., et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 67.Cedar H., Bergman Y. Programming of DNA methylation patterns. Annu. Rev. Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 68.Bibikova M., Chudin E., Wu B., Zhou L., Garcia E.W., Liu Y., Shin S., Plaia T.W., Auerbach J.M., Arking D.E., et al. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hernando-Herraez I., Prado-Martinez J., Garg P., Fernandez-Callejo M., Heyn H., Hvilsom C., Navarro A., Esteller M., Sharp A.J., Marques-Bonet T. Dynamics of DNA methylation in recent human and great ape evolution. PLoS Genet. 2013;9:e1003763. doi: 10.1371/journal.pgen.1003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dedeurwaerder S., Defrance M., Bizet M., Calonne E., Bontempi G., Fuks F. A comprehensive overview of Infinium HumanMethylation450 data processing. Brief. Bioinformatics. 2014;15:929–941. doi: 10.1093/bib/bbt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gevaert O., Tibshirani R., Plevritis S.K. Pancancer analysis of DNA methylation-driven genes using MethylMix. Genome Biol. 2015;16:17. doi: 10.1186/s13059-014-0579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ziller M.J., Hansen K.D., Meissner A., Aryee M.J. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat. Methods. 2015;12:230–232. doi: 10.1038/nmeth.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saxonov S., Berg P., Brutlag D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu J., Adli M., Zou J.Y., Verstappen G., Coyne M., Zhang X., Durham T., Miri M., Deshpande V., De Jager P.L., et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meissner A., Gnirke A., Bell G.W., Ramsahoye B., Lander E.S., Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roussos P., Mitchell A.C., Voloudakis G., Fullard J.F., Pothula V.M., Tsang J., Stahl E.A., Georgakopoulos A., Ruderfer D.M., Charney A., et al. A role for noncoding variation in schizophrenia. Cell Rep. 2014;9:1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lewis D.A., Curley A.A., Glausier J.R., Volk D.W. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Irizarry R.A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P., Cui H., Gabo K., Rongione M., Webster M., et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hon G.C., Rajagopal N., Shen Y., McCleary D.F., Yue F., Dang M.D., Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013;45:U1198–U1340. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spiegel I., Mardinly A.R., Gabel H.W., Bazinet J.E., Couch C.H., Tzeng C.P., Harmin D.A., Greenberg M.E. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T., Qin J., Zoghbi H.Y. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sugino K., Hempel C.M., Okaty B.W., Arnson H.A., Kato S., Dani V.S., Nelson S.B. Cell-type-specific repression by methyl-CpG-binding protein 2 is biased toward long genes. J. Neurosci. 2014;34:12877–12883. doi: 10.1523/JNEUROSCI.2674-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen L., Chen K., Lavery L.A., Baker S.A., Shaw C.A., Li W., Zoghbi H.Y. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5509–5514. doi: 10.1073/pnas.1505909112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gabel H.W., Kinde B., Stroud H., Gilbert C.S., Harmin D.A., Kastan N.R., Hemberg M., Ebert D.H., Greenberg M.E. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522:89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Greer P.L., Greenberg M.E. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 86.Bliss T.V., Collingridge G.L. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 87.Franke B., Vasquez A.A., Veltman J.A., Brunner H.G., Rijpkema M., Fernandez G. Genetic variation in CACNA1C, a gene associated with bipolar disorder, influences brainstem rather than gray matter volume in healthy individuals. Biol. Psychiatry. 2010;68:586–588. doi: 10.1016/j.biopsych.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 88.Bigos K.L., Mattay V.S., Callicott J.H., Straub R.E., Vakkalanka R., Kolachana B., Hyde T.M., Lipska B.K., Kleinman J.E., Weinberger D.R. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch. Gen. Psychiatry. 2010;67:939–945. doi: 10.1001/archgenpsychiatry.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tesli M., Skatun K.C., Ousdal O.T., Brown A.A., Thoresen C., Agartz I., Melle I., Djurovic S., Jensen J., Andreassen O.A. CACNA1C Risk Variant and Amygdala Activity in Bipolar Disorder, Schizophrenia and Healthy Controls. PLoS One. 2013;8:e56970. doi: 10.1371/journal.pone.0056970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Walton E., Geisler D., Hass J., Liu J., Turner J., Yendiki A., Smolka M.N., Ho B.C., Manoach D.S., Gollub R.L., et al. The impact of genome-wide supported schizophrenia risk variants in the neurogranin gene on brain structure and function. PLoS One. 2013;8:e76815. doi: 10.1371/journal.pone.0076815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohi K., Hashimoto R., Yasuda Y., Fukumoto M., Yamamori H., Umeda-Yano S., Fujimoto M., Iwase M., Kazui H., Takeda M. Influence of the NRGN gene on intellectual ability in schizophrenia. J. Hum. Genet. 2013;58:700–705. doi: 10.1038/jhg.2013.82. [DOI] [PubMed] [Google Scholar]
- 92.Wetsel W.C., Rodriguiz R.M., Guillemot J., Rousselet E., Essalmani R., Kim I.H., Bryant J.C., Marcinkiewicz J., Desjardins R., Day R., et al. Disruption of the expression of the proprotein convertase PC7 reduces BDNF production and affects learning and memory in mice. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17362–17367. doi: 10.1073/pnas.1314698110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ahmed A.O., Mantini A.M., Fridberg D.J., Buckley P.F. Brain-derived neurotrophic factor (BDNF) and neurocognitive deficits in people with schizophrenia: a meta-analysis. Psychiatry Res. 2015;226:1–13. doi: 10.1016/j.psychres.2014.12.069. [DOI] [PubMed] [Google Scholar]
- 94.Gifford C.A., Ziller M.J., Gu H.C., Trapnell C., Donaghey J., Tsankov A., Shalek A.K., Kelley D.R., Shishkin A.A., Issner R., et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roadmap Epigenomics, C. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.