Abstract
The Hox genes encode transcription factors that determine embryonic pattern formation. In embryonic stem cells, the Hox genes are silenced by PRC2. Recent studies have reported a role for long noncoding RNAs in PRC2 recruitment in vertebrates. However, little is known about how PRC2 is recruited to the Hox genes in ESCs. Here, we used stable knockdown and knockout strategies to characterize the function of the long noncoding RNA Gm15055 in the regulation of Hoxa genes in mouse ESCs. We found that Gm15055 is highly expressed in mESCs and its expression is maintained by OCT4. Gm15055 represses Hoxa gene expression by recruiting PRC2 to the cluster and maintaining the H3K27me3 modification on Hoxa promoters. A chromosome conformation capture assay revealed the close physical association of the Gm15055 locus to multiple sites at the Hoxa gene cluster in mESCs, which may facilitate the in cis targeting of Gm15055 RNA to the Hoxa genes. Furthermore, an OCT4-responsive positive cis-regulatory element is found in the Gm15055 gene locus, which potentially regulates both Gm15055 itself and the Hoxa gene activation. This study suggests how PRC2 is recruited to the Hoxa locus in mESCs, and implies an elaborate mechanism for Hoxa gene regulation in mESCs.
INTRODUCTION
The pivotal roles of the Hox genes in metazoan embryonic development have long been recognized. However, the regulation of the Hox genes themselves is not well understood, especially in vertebrates. At early developmental stages, all of the Hox genes are repressed. In mouse embryonic stem cells (mESCs), all four repressed Hox gene clusters possess both the active histone modification H3K4me3 and the repressive histone modification H3K27me3, which is referred to as a bivalent chromatin state (1). Recent studies showed that each repressed Hox gene cluster is packaged into a small topologically associated domain (TAD) (2). During activation of the Hox genes, the repressive histone mark H3K27me3 is removed, accompanied by an increase in the activation mark H3K4me3 (3). The activated Hox genes loop out from the repressive chromatin compartment and constitute an activated chromatin center (4). Determination of cell fate during differentiation is spatially and temporally regulated by Hox gene activation. The maintenance of bivalent chromatin in ESCs is therefore essential for the appropriate activation of the Hox genes and early embryonic development.
Polycomb Repressive Complex 2 (PRC2) occupies large domains of Hox genes (5,6) and maintains the repressive histone mark H3K27me3. The mechanism by which PRC2 is recruited to specific chromatin loci in vertebrates is poorly understood. Recent studies demonstrate that long noncoding RNAs (lncRNA) play important roles in the recruitment of PRC2 (7). LncRNA RepA within the Xist locus directly interacts with PRC2 and recruits it to X-chromosome for the initiation of X-chromosome inactivation (8). Another lncRNA Kcnq1ot1 has been shown to interact with PRC2 and is involved in Kcnq1 gene silencing (9). For Hox gene regulation, it has been reported that lncRNA HOTAIR is essential for PRC2 recruitment to the HOXD cluster at different developmental stages and in a variety of tissues (10–13). However, until now, lncRNAs have not been identified in the recruitment of PRC2 for Hox genes repression in ESCs. While lncRNAs regulate gene expression through diverse mechanisms, the most prevalent function of the lncRNAs is to target genes that are adjacent to their transcription site (14). Abundant lncRNAs are transcribed around the Hox gene cluster (3,10). In mESCs, RIP-sequencing identified thousands of lncRNAs that interact with PRC2 (15). Among these lncRNAs, several are located near the Hox gene cluster and one lncRNA, Gm15055 (also known as Haunt, Halr1 and linc-HOXA1, referred to as Gm15055 hereafter), is approximately 50 kb downstream of the Hoxa1 gene. Gm15055 was first predicted by the ‘K4-K36 domain’ feature in mESCs (16) and then identified by RNA-sequencing in mESCs (17). Maamar et al. reported that there are three isoforms of Gm15055 (isoform 1, 2, and 3) and that the transient knockdown of this lncRNA in mESCs specifically increases the expression of Hoxa1 (18). The function of Gm15055 has also been investigated in mice by replacing the second and third exons with lacZ and a selection marker; however, no obvious developmental defects were reported (19). Considering that the four Hox gene clusters may have redundant functions in vertebrate development, function of Gm15055 in the regulation of Hoxa genes remains to be characterized.
In this study, we applied lentiviral-mediated stable knockdown of Gm15055 and CRISPR/Cas9-mediated Gm15055-knockout in mESCs to investigate the regulation of Hoxa gene expression and chromatin status in mESCs by the lncRNA Gm15055. We found that Gm15055 was positively regulated by OCT4 and highly expressed in mESCs. Gm15055 interacted with PRC2 and recruited PRC2 to Hoxa gene cluster for maintaining the repressive histone mark H3K27me3 on the Hoxa gene promoters in mESCs. We further showed that long-distance chromatin interactions exist between the Gm15055 locus and the Hoxa gene cluster, which may help in the targeting of Gm15055-recruited PRC2 to the Hoxa gene cluster. Finally, a DNA fragment located within the Gm15055 gene locus was identified as an OCT4-dependent cis-regulatory element that potentially upregulates both Gm15055 and Hoxa genes expression.
MATERIALS AND METHODS
Cell culture
The mouse embryonic cell line JM8A3 was grown and maintained according to the supplier's instructions. Briefly, the cells were cultured at 37°C in a 5% (v/v) CO2 incubator in Knock-Out DMEM medium (Invitrogen) supplemented with 15% KSR (Invitrogen), 2 mM L-glutmax (Invitrogen), 1× nonessential amino acids (Invitrogen), 103 U/ml LIF (Millipore), and 55 μM 2-mercaptoethanol. The dishes were coated with 0.1% gelatin [v/v], and a layer of MMC-treated DR4 cells (for selection) or MEF cells was grown on top as feeder cells. The cells were passaged as needed, and the growth medium was changed daily. Upon harvesting the JM8A3 cells, the feeder cells were removed by incubation at 37°C on gelatin-coated dishes for 1.5 h. For embryoid body (EB) differentiation, the JM8A3 cells were suspended in DMEM basic medium (Invitrogen) supplemented with 10% FBS (Invitrogen) and cultured on ultra-low attachment cell culture plate (Corning). For all-trans retinoid acid (ATRA) induced differentiation, JM8A3 cells were maintained in ESC culture medium without feeder cells, and induced by withdraw of LIF and the addition of 5 μM ATRA (Sigma).
Gene knockdown and overexpression
For stable knockdown of Gm15055 in mESCs, shRNA hairpins were cloned into the plko.1 lentiviral vector, which contains a puromycin marker gene that allows for stable integration as previously described (20). Briefly, two hairpins were designed to target Gm15055, and one hairpin targeting GFP was used as the control. The target sequences of the three shRNAs were as follows: shGFP: TGACCCTGAAGTTCATCTGCA; shGm15055-1: ATATGAGTGAAGTCACAA ATG; shGm15055-2: GCCTAGTTCAACGACACATCA.
Lentiviruses containing the shRNAs were then prepared as previously described (20). For each shRNA plasmid, 1 μg of the shRNA vector and 0.5 μg of pMD2.G, 0.5 μg of pMDLg/pRRE, and 0.5 μg of pRSV-Rev were co-transfect into 2 × 106 293T packaging cells using 5 μl Lipofectamine2000 (Invitrogen). The viral supernatant was harvested at 48 and 72 h after transfection and purified with a 0.45-μm filter (Millipore). Then, 3 × 105 JM8A3 mESCs depleted of MEF cells were infected with the viruses at 37°C for 30 min before being cultured on DR4 feeder cells for 48 h. For selection, 1.5 ng/μl puromycin was added to the cultures, and the medium was changed daily. The transfected cells were harvested after 8 days of selection.
For Oct4 knockdown, two siRNAs (Invitrogen) targeting Oct4 and one control siRNA (Invitrogen, 4404021) were synthesized. Then, 5 × 105 JM8A3 cells were transfected with 125 or 100 pM of each siRNA with 5 μl Lipofectamine2000 and cultured on MEF feeder cells for 2 days before harvesting. The siRNA sequences are as follows: SiOct4-1: CGGAAGAGAAAGCGAACTA; SiOct4-2: CGGAAGAGAAAGCGAACTA.
For Gm15055 overexpression, the three isoforms of Gm15055 were cloned into the pcDNA3.1 vector, and the empty pcDNA3.1 was used as a control. Two micrograms of each plasmid was electrotransfected into 5 × 105 JM8A3 cells using the Neon transfection system (Invitrogen) with the parameters of 1350 V, 10 ms and three pulses. The cells were harvested 48 h after transfection.
For Oct4, Sox2 and Nanog overexpression in MEF, the same amount of pMXs-GFP/Oct4/ Sox2/Nanog viral supernatant was incubated with MEF and the cells were harvested 36 h after infection.
Gene targeting, selection and removal of selection markers
The CRISPR/Cas9 system for Gm15055 gene targeting was performed as previously described (21). Briefly, two guide RNAs (gRNA3: TAGAGGAACAATTGCTGCAT and gRNA5: CTTTGAAAAGAAGTGAAGGA) were cloned into the PX330 vector and used in combination with two targeting vectors (DTA and PB) with different selective markers, allowing for two rounds of knockout experiments to achieve homogenous Gm15055 ablation. The targeting vectors replaced a 1894-bp DNA fragment (mm9 chr6:52 055 891–52 057 784) of the Gm15055 locus with a transcription termination signal and a neomycin or puromycin resistance gene. Then, 5 μg of the guide RNA plasmid, 5 μg of the CAG-Cas9 vector and 5 μg of the targeting vector were co-transfected into 5 × 106 JM8A3 cells using the Neon transfection system (Invitrogen) and selected with 1.5 ng/μl puromycin or 150 ng/μl G418 for 8 days with daily media changes. Single JM8A3 clones were identified by PCR and RT-qPCR to verify the successful knockout of Gm15055. The selection markers were then removed from the homozygous Gm15055 knockout clones by electrotransfection of the PBase and pCAG-cre vectors and negative selection with 200 nM 1-(2-deoxy-2-fluoro-beta-d-arabinofuranosyl)-5-iodouracil (FIAU) for 10 days. The resulting clones were then selected and identified by PCR before subsequent analysis. The primers used for PCR identification of the knockout clones, as well as for the detection of potential off-target effects, are listed in Supplementary Table S1.
BoxB tethering assay
A BoxB tethering assay was performed as previously described with some modifications (22). A DNA fragment containing five tandem GAL4 UAS sites was inserted into the DTA targeting vector mentioned above and stably integrated into the Gm15055 gene locus of JM8A3 to prepare the UAS-JM8A3 cells after G418 selection. The vector encoding GAL4-λN hybrid protein was subsequently tranfected into the cells and selected with puromycin to obtain the UAS-GAL4-λN-JM8A3 clones. Finally, the vector harboring the isoform 3 Gm15055 fused with 5BoxB sequence was transfected into the UAS-GAL4-λN-JM8A3 cells and selected with 160 ng/μl hygromycin. The LacZ-5BoxB plasmid (addgene 29729) was used as a negative control. All the resultant JM8A3 clones were identified by PCR and RT-qPCR to verify the UAS insertion, GAL4-λN expression and 5BoxB-fused RNA expression. The positive clones were then harvested for RT-qPCR and ChIP-qPCR analyses to detect the transcription and PRC2 recruitment at the Neo gene downstream to the UAS site.
Chromatin immunoprecipitation (ChIP)
A ChIP assay was performed as previously described with some modifications (23). Briefly, mESCs were crosslinked with 1% formaldehyde for 15 min and lysed with lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris–HCl, pH 8.0) supplemented with protease inhibitors. The lysed cells were sheared until the chromatin DNA fragments were 200–1000 bp in size. The sonicated chromatin was then diluted 10-fold with ChIP dilution buffer (0.01% SDS, 1% Triton X-100, 150 mM NaCl, 2 mM EDTA and 20 mM Tris–HCl, pH 8.0) and pre-cleared using protein A/G Dynabeads (Invitrogen) before incubation with anti-Oct4 (ab19857, abcam), anti-H3K27me3 (07-449, Millipore), anti-H3K4me3 (07-473, Millipore), anti-H3 (ab1791, abcam), anti-Suz12 (17-661, Millipore or ab12073, abcam) and anti-RNA polymerase II CTD repeat YSPTSPS (phospho S5) (ab5131, abcam) antibodies or normal rabbit/mouse IgG (Santa Cruz) at 4°C overnight. The antibody-chromatin complexes were recovered by incubation with protein A/G Dynabeads (Invitrogen) at 4°C for 2 h. After washing and reverse cross-linking, the precipitated DNA was purified and subsequently analyzed by real-time PCR. The PCR products of the ChIP samples were normalized to the input DNA or H3 density. The IgG-precipitated DNA was used as a negative control for each ChIP assay. The ChIP was then repeated in triplicate with the Gm15055-knockdown cells and was repeated with at least three Gm15055 knockout clones. All the primers used for the ChIP-qPCR assay are listed in Supplementary Table S3.
Chromosome conformation capture
Chromatin conformation capture (3C) was performed as previously described (23,24). Briefly, the crosslinked mESCs (1% formaldehyde, 15 min) or MEFs (1% formaldehyde, 10 min) were digested with HindIII (NEB, R0104M) overnight, and the digestion efficiency was calculated by real-time PCR amplification of uncut fragments with primer pairs spanning the designated HindIII sites. A digestion efficiency of >80% at each individual restriction site is considered feasible for further 3C analysis. The samples were then ligated with 2 U/μl T4 DNA ligase (NEB, M0202M) at 16°C for 4 h at DNA concentrations of less than 2.5 ng/μl. After reversing the cross-links, the 3C sample was purified by phenol extraction and ethanol precipitation and then washed with 70% cold ethanol six times before qPCR quantification. The mouse Hoxa BAC clone (RP23–20F21) for control library construction was obtained from the Children's Hospital Oakland Research Institute and was used to correct the primer efficiency. Seven primer pairs amplifying control regions were used to correct for the template amount. The relative interaction frequencies between the two designated DNA fragments were then calculated by normalizing the PCR efficiency of the 3C samples to their respective controls. All of the qPCR was repeated in triplicate using at least three independent cultures. The primers used for the 3C-qPCR analyses are listed in Supplementary Table S4.
RNA immunoprecipitation (RIP)
RIP was performed as previously described (10,18). JM8A3 cell lysates were prepared by suspending 5 × 107 cells in 4 ml RIP buffer (150 mM KCl, 25 mM Tris at pH 7.4, 5 mM EDTA, 0.5 mM DTT, 0.5% NP40, 1× protease inhibitor cocktail [Sigma]) followed by incubation at 4°C for 30 min with gentle rotation. The cell lysates were then sonicated and cleared by centrifugation at 13 000 rpm for 20 min. The protein concentration of the lysate was quantified with the BCA protein assay kit (Pierce). Antibodies recognizing Suz12 (17-661, Millipore) or IgG (Santa Cruz) were incubated with 0.4 mg cell lysate at 4°C for 2 h with gentle rotation. Then, 30 μl of protein G/A Dynabeads (Invitrogen) was added and incubated for another 1 h. The beads were collected and washed three times with RIP buffer containing SUPERaseIn (Ambion). The RNA was extracted with 1 ml Trizol (Invitrogen) according to the manufacturer's instructions before quantification by RT-qPCR.
Reporter assay
The enhancer activity of the knockout DNA fragment was analyzed using the dual-luciferase reporter assay (Promega). Briefly, the DNA fragments were inserted into the pGL3-promoter vector or pGL3 basic vector and transfected into 293T, JM8A3 or MEF cells with a Renilla luciferase-encoding vector (pRL-CMV) as the internal control. The pMXs-control/Oct4 vectors were co-transfected with the reporters to test the regulatory effects of these factors to the target DNA fragments. The luciferase activity (firefly/Renilla) was measured using a Modulus Microplate Multimode Reader. For OCT4 binding sites mutation, the wildtype reporters were amplified by primers containing the mutated nucleotides, followed by Dpn I digestion to remove the wildtype templates. All mutated reporters were confirmed by sequencing. For each reporter assay, three biological replicates were examined and each sample was analyzed by three technical replicates. The primer pairs used for reporter vector construction are listed in Supplementary Table S5.
Statistical analysis
Data analysis was performed with GraphPad Prism6 (GraphPad Software, Inc.). The data are presented as the mean ± SD. The differences among samples were compared using Student's t-test and the P-values were indicated: **P < 0.01, *P < 0.05.
RESULTS
OCT4 regulates the elevated expression of lncRNA Gm15055 in mESCs
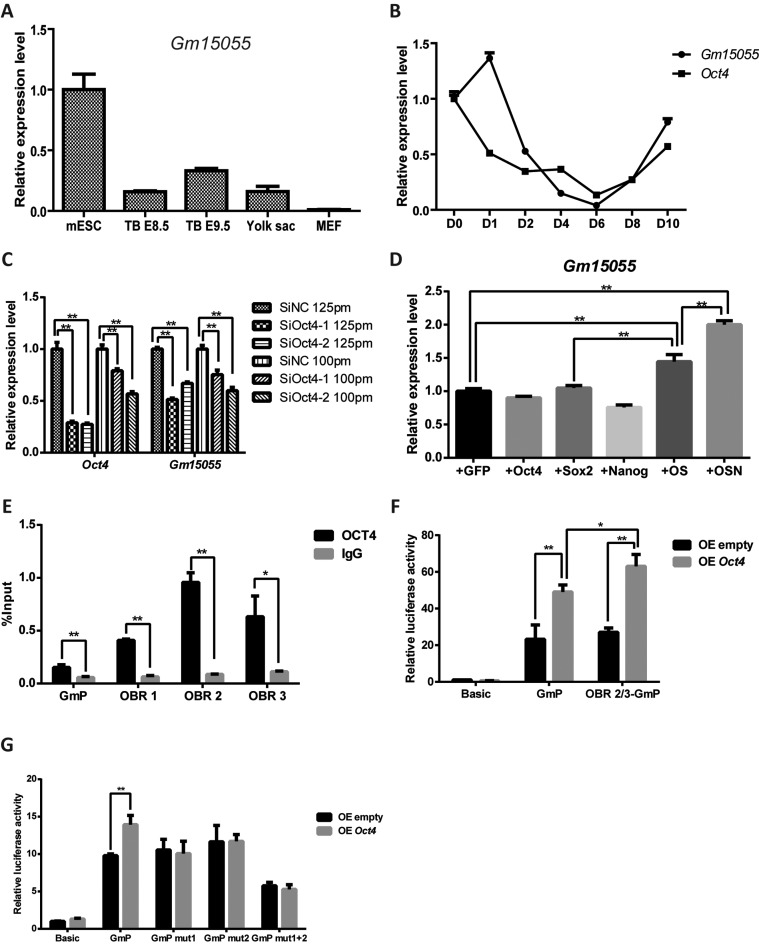
Gm15055 was first predicted and identified in mESC (16,17). To determine whether Gm15055 is expressed specifically in mESCs, we investigated its expression in mouse embryonic tissues at different developmental stages and during mESC differentiation. Tailbuds were collected from mouse embryos at 8.5 days and 9.5 days, and yolk sac cells and MEF cells were collected from mouse embryos at 14.5 days. The Gm15055 expression level was then detected by RT-qPCR and compared to its expression in mESCs. As expected, we observed that Gm15055 expression was higher in mESCs and obviously lower in the differentiated cells (Figure 1A). Similarly, we showed that the expression of Gm15055 is largely decreased during the first few days of embryoid body (EB) differentiation of mESCs (Figure 1B).
Figure 1.
LncRNA Gm15055 is maintained by OCT4 in mESCs. (A) Gm15055 expression in mESCs, E8.5 and E9.5 mouse tailbuds, E14.5 mouse yolk sac and MEF cells was quantified by RT-qPCR. The expression levels are shown relative to that of mESCs; Gapdh was used as an internal control. (B) The relative expression of Gm15055 and Oct4 during embryoid body differentiation of mESCs was detected by RT-qPCR at the indicated time points. (C) RT-qPCR detection of the Oct4 knockdown efficiency and the resulting Gm15055 expression in mESC. Two siRNAs targeting mOct4 (SiOct4-1 and SiOct4-2) and one control siRNA (SiNC) were used, with two different amounts of siRNA as indicated. (D) The pluripotency factors Oct4, Sox2, Nanog were overexpressed separately or in combination in MEF cells and the expression level of the endogenous Gm15055 was detected by RT-qPCR. GFP overexpression was used as a control. +OS: +Oct4+Sox2; +OSN: +Oct4+Sox2+Nanog. (E) Recruitment of OCT4 at the Gm15055 locus was analyzed by ChIP-qPCR. Three potential OCT4 binding regions at the Gm15055 locus (namely OBR 1, OBR 2 and OBR 3) and the Gm15055 promoter (GmP) were examined. (F) The activity of OCT4 on Gm15055 promoter with or without the downstream OCT4 binding regions were detected in reporter assay in 293T cells. The pGL3 basic (Basic), Gm15055 promoter-pGL3 (GmP) or OCT4 binding region 2/3-Gm15055 promoter-pGL3 (OBR 2/3-GmP) reporters, was co-transfected with empty or Oct4 overexpression vector, respectively. The relative luciferase activity was normalized to that of the Basic plus empty overexpression (OE empty) group. (G) Reporter assay in 293T cells showed that mutation to the OCT4 binding sites abolished the response of Gm15055 promoter to the OCT4 activity. The pGL3 basic (Basic), Gm15055 promoter-pGL3 (GmP) or that carrying single/double mutations at the Gm15055 promoter (GmP mut1, GmP mut2 and GmP mut1+2), was co-transfected with empty or Oct4 overexpression vector, respectively. The relative luciferase activity was normalized to that of the Basic plus empty overexpression (OE empty) group.
The expression pattern of Gm15055 was found highly correlated to that of Oct4 during EB differentiation, except at day 1 (Figure 1B). Given that the Oct4 gene is an important pluripotency marker of mESCs and that the previous ChIP-seq assay with OCT4 in mESCs showed a strong signal at the Gm15055 locus (25,26), it is possible that OCT4 participates in the maintenance of the high level of Gm15055 expression in mESCs. Knockdown Oct4 gene expression by two different siRNAs led to a decrease in Gm15055 expression (Figure 1C). Considering that decrease of Oct4 expression could lead to ESC differentiation, we repeated the Oct4 knockdown assay using a relatively lower amount of siRNA at 100pmol to avoid the potential ESC differentiation (27). RT-qPCR results showed that Gm15055 expression remained decreased upon the mild Oct4 knockdown (Figure 1C), meanwhile both immunofluorescence assays of the pluripotent factors OCT4, SOX2, NANOG, SSEA1 and the AP staining (Supplementary Figure S1) showed no traits of ESC differentiation with the mild knockdown of Oct4. Moreover, although overexpression of Oct4 alone showed no obvious effect on Gm15055 expression, the co-transfection with its co-factor Sox2 significantly promoted endogenous Gm15055 expression in the MEF cells (Figure 1D and Supplementary Figure S2A). Specifically, Recent OCT4 ChIP-seq data from Whyte et al. and Buecker et al. (25,26) showed two OCT4 binding regions (OBR) at Gm15055 locus, a minor one at the promoter of Gm15055 (GmP) and a major one around the Exon 2, which includes three peaks of OCT4 binding signals (OBR-1,-2,-3) (Supplementary Figure S2B). The binding of OCT4 at those sites in JM8A3 were then confirmed using conventional chromatin immunoprecipitation assay (Figure 1E). Further reporter assay in 293T cells showed that OCT4 stimulated transcription from Gm15055 promoter, and that the stimulation became a bit more obvious when a fragment of the OBR (containing OBR-2 and -3) was inserted into the reporter (Figure 1F). Two core OCT4 binding sites were then predicted from the Gm15055 promoter by FIMO (28), mutations to either of them abrogated the stimulative effect of OCT4 to the promoter (Figure 1G). Taken together, these results indicate that OCT4 maintains Gm15055 expression in mESCs.
Gm15055 represses Hoxa gene expression in mESC
Previous study by Maamar et al. found that Gm15055 functions as a repressor of the Hoxa1 gene (18). Using two separate lentiviruses carrying different Gm15055-targeting shRNA sequences, we stably knockdown Gm15055 expression in mESCs and observed a de-repression of all the Hoxa genes, except for Hoxa13, in the uninduced mESCs (Supplementary Figure S3A-B). The level of de-repression was comparable to that of Hoxa activation during the first few days of embryoid body differentiation (Supplementary Figure S3C). Alkaline phosphatase staining, as well as immunofluorescence and RT-qPCR analysis of the pluripotency markers SSEA1 and Oct4, confirmed that the pluripotency of the mESCs was not disrupted by Gm15055 knockdown, and excluded the possibility that the increased Hoxa gene expression is due to mESC differentiation (Supplementary Figure S4A–C). Overexpression of Gm15055 isoforms failed to cause obvious change in Hoxa expression (Supplementary Figure S5A-B), The observations were consistent to a recent paper published during the revision of this work, reporting that Gm15055 regulates the activation of Hoxa genes upon RA-induced ESC differentiation, and that Gm15055 acts on Hoxa gene in cis (29).
Gm15055 maintains the H3K27me3 modification of Hoxa genes by recruiting PRC2
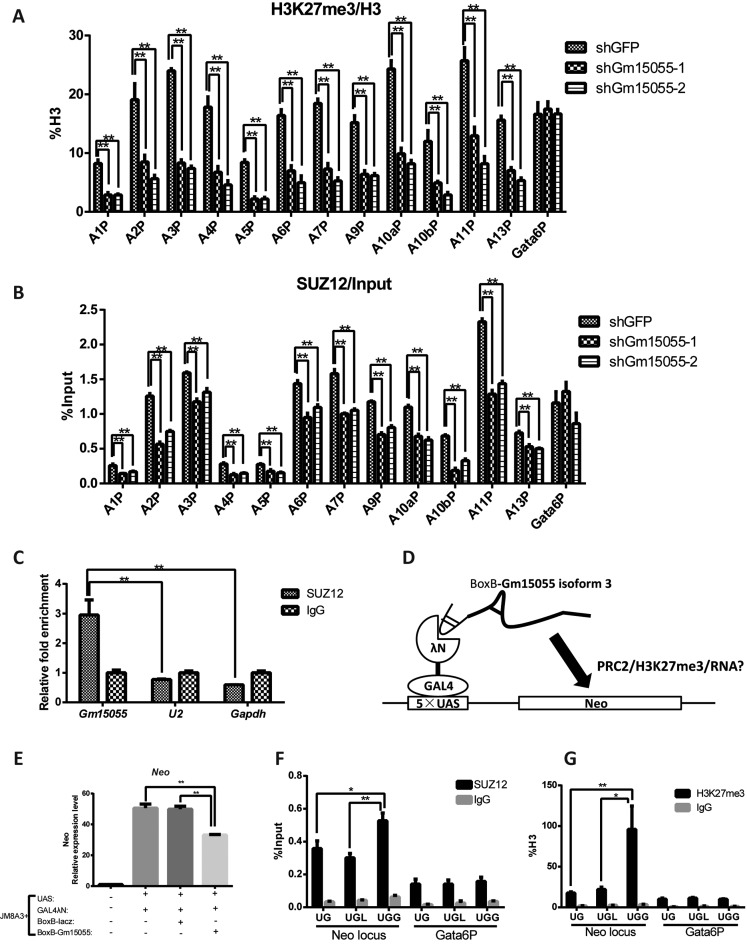
Bivalent chromatin is a prominent feature of the Hox genes in mESCs. We then examined whether Gm15055 represses the Hoxa genes by affecting chromatin modification. A ChIP assay showed that Gm15055 knockdown was accompanied by the decline of the H3K27me3 modification on all of the Hoxa gene promoters, the Gata-6 promoter was used as negative control to show that the effect was specific to the Hoxa locus (Figure 2A). H3K4me3 modification at the Hoxa gene promoters was largely unaffected (Supplementary Figure S6A). A similar but more 3′-proned elimination of H3K27me3 modification from the Hoxa locus was detected in RA-induced Gm15055 pKO ESC in the recent report (29).
Figure 2.
Gm15055 interacts with PRC2 and maintains the repressive H3K27me3 modification and PRC2 binding to the Hoxa gene promoters. (A) ChIP analysis of H3K27me3 modification at the Hoxa gene promoters in the control shGFP and the Gm15055-knockdown (shGm15055-1 and shGm15055-2) mESCs. All H3K27me3 signals were normalized to the H3 density at the same site, and the Gata6 promoter (Gata6P) was used as a control region. (B) ChIP analysis showing the recruitment of the PRC2 core component SUZ12 at the Hoxa gene promoters in the control shGFP and the Gm15055-knockdown mESCs. The Gata6 promoter (Gata6P) was used as a control region. (C) RNA immunoprecipitation (RIP) showed that PRC2 specifically interacted with Gm15055. An antibody against the PRC2 core component SUZ12 was used to pull down PRC2, and normal IgG was used as a negative control. Gm15055 and two other control RNAs (U2 and Gapdh) were then detected in both the anti-SUZ12 and IgG RIP samples. (D) Working model of the BoxB tethering assay. The GAL4-λN chimeric protein recognizes the BoxB-tethered RNA and targets the RNA to the 5×UAS locus that was integrated into the mESC genome. The cis-regulatory effect of the tethered Gm15055 isoform 3 RNA can then be evaluated by detecting the expression level and histone modification of the adjacent Neo gene. E. RT-qPCR analysis of the Neo gene adjacent to the UAS locus showing the repressive effect brought by the tethered Gm15055 (isoform 3) RNA. All the three constructs, the UAS-neo, GAL4-λN, BoxB-Gm15055 or BoxB-LacZ were stably integrated in the JM8A3 mESCs. (F–G) ChIP analysis showed the increased (F) recruitment of the PRC2 core component SUZ12 and (G) H3K27me3 modification at the Neo locus at the Gm15055 (isoform 3) group in the BoxB tethering assay. UG: JM8A3+UAS+GAL4λN, UGL: JM8A3+UAS+GAL4λN+BoxBLacZ, UGG: JM8A3+UAS+GAL4λN+BoxBGm15055. UG and UGL ESCs were used as the negative controls, and the Gata6 promoter (Gata6P) was used as an irrelevant control region in the ChIP assay.
PRC2 is the main protein complex responsible for H3K27me3 modification. We therefore investigated whether the recruitment of PRC2 to the Hoxa locus was affected by Gm15055 knockdown. A ChIP assay showed that the signal of SUZ12, a core component of PRC2, decreased at the Hoxa gene promoters, but not at the control Gata-6 promoter, in both Gm15055-knockdown mESCs (Figure 2B). The protein level of SUZ12 in the cells was shown unchanged (Supplementary Figure S6B). These results suggest that Gm15055 may repress the expression of the Hoxa genes by affecting PRC2 binding at the promoters of the Hoxa genes.
To clarify how Gm15055 may affect PRC2 recruitment to the Hoxa gene promoters, we first detected and confirmed the nuclear distribution of Gm15055 RNA (Supplementary Figure S6C). The nuclear RNA U2, and cytoplasm-enriched Gapdh and Oct4 mRNAs were using as the controls for the analysis. RNA immunoprecipitation (RIP) assay against SUZ12 was then performed to determine the ability of Gm15055 RNA in recruiting PRC2 complex. The results revealed that SUZ12 interacted specifically with Gm15055 in vivo. Interactions between SUZ12 and the two control RNAs U2 and Gapdh were not detected (Figure 2C). Next, a stable BoxB tethering assay was carried out to examine whether Gm15055 is able to target PRC2 to the neighboring genes (22). The longest isoform of Gm15055, isoform 3, was fused with the BoxB RNA and, through a mediating GAL4-λN chimeric protein, could be tethered to the UAS sequence upstream to a neo selection marker gene (Figure 2D, Supplementary Figure S7A). All the UAS-neo, GAL4-λN, and Gm15055-BoxB expressing vectors were stably integrated into the genome of mESCs. Blank, BoxB-lacZ RNA expressing vectors were used as the negative controls. The mESC clones that showed comparable expression of GAL4-λN and BoxB-tethered RNA were selected for further examination (Supplementary Figure S7B-C). RT-qPCR results showed that BoxB-Gm15055 overexpression led to obviously decreased neo gene expression, compared to that in the blank and BoxB-lacZ-overexpressing mESCs (Figure 2E). Moreover, the tethering of Gm15055 led to a significant increase of SUZ12 binding and H3K27me3 modification at the neo gene adjacent to the UAS sequence (Figure 2F-G), confirming the ability of Gm15055 to recruit PRC2 to a specific gene locus.
Long-range chromatin interaction may facilitate loading of PRC2 to the Hoxa locus
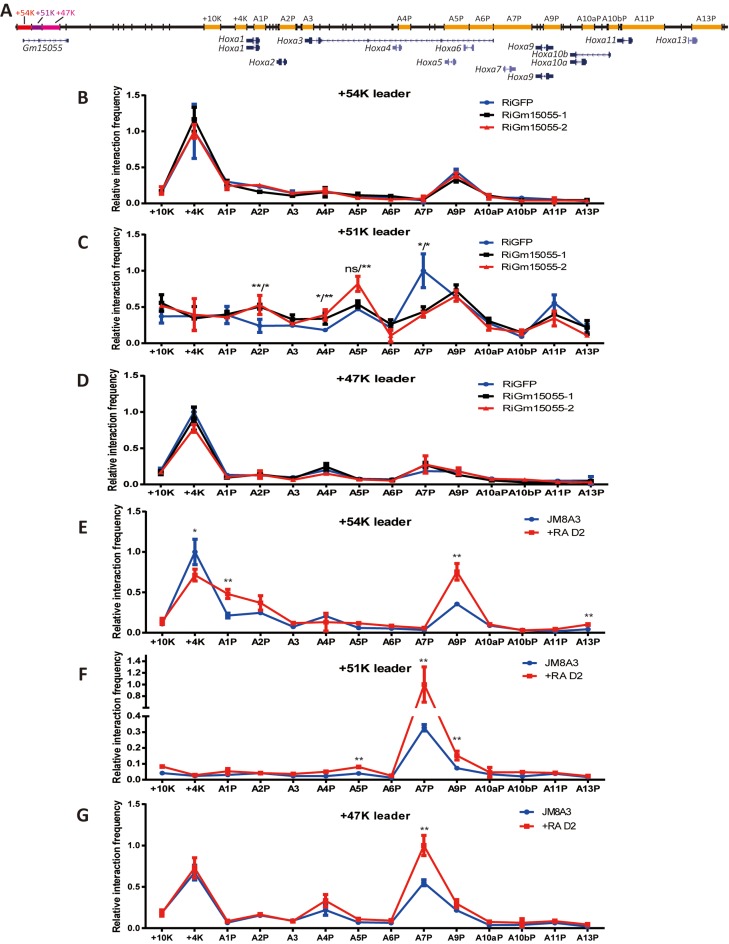
The Gm15055 gene locus is approximately 50 kb downstream of the Hoxa gene cluster. To understand how Gm15055 RNA may recruit PRC2 to the Hoxa gene cluster across such a long distance, we applied chromosome conformation capture (3C) to examine whether the Gm15055 locus interacts with the Hoxa gene cluster in wild type mESCs. The recent study showed a relatively weak chromatin interaction in between the two loci in uninduced ESCs, using the Hoxa1 and a5 promoters as the leader fragments (anchors) (29). Reversely, we used here the three HindIII restriction fragments from the Gm15055 locus as the 3C leaders: a ‘+54K’ fragment containing the Gm15055 promoter, a ‘+51K’ fragment containing exon 2 of Gm15055 and a CTCF binding site (CBS) (30), and a ‘+47K’ fragment, which covered the majority of the second intron of Gm15055 (Figure 3A). Restriction fragments covering the vicinity downstream of the Hoxa cluster and those containing the Hoxa promoters—the +10k, +4k, A1P, A2P, A3, A4P, A5P, A6P, A7P, A9P, A10aP, A10bP, A11P and A13P fragments—were analyzed by the 3C assay. The fragments are named according to their position relative to the Hoxa gene cluster or the Hoxa promoter. The 3C results showed that the +54K fragment was preferentially associated with the Hoxa9 promoter, and the +47K fragment selectively engaged with the Hoxa7 promoter. The +51K fragment, which contains the CBS, was extensively associated with most of the Hoxa promoter regions that have been detected (Figure 3B-D). Interestingly, the +4K fragment that was immediately downstream of the Hoxa cluster exhibited a high frequency of interaction with both the +54K and +47K leaders, bioinformatic analysis revealed enriched binding of the pluripotent factors OCT4, SOX2 and NANOG at the region (Supplementary Figure S8), implying a potential role of the fragment in mediating the interaction between Gm15055 locus and the Hoxa gene cluster.
Figure 3.
Long-range chromatin interactions exist between the Gm15055 gene locus and the Hoxa gene cluster. (A) A schematic representation of the Gm15055 and the Hoxa genes loci with the analyzed DNA fragments in the 3C experiment highlighted. The +54K, +51K and +47K leader fragments from the Gm15055 locus are marked as red, purple and pink, respectively. The other 3C fragments are marked as orange. Fragments outside of the Hoxa gene cluster are named according to their distance to the 3′-end of the cluster. Other fragments refer to the respective Hoxa gene promoters (e.g. A1P refers to promoter of Hoxa1). (B–D) Chromosome conformation capture assay of the interaction frequencies between the leaders (B: +54K; C:+51K; D: +47K) and the Hoxa gene cluster in control (shGFP) and two Gm15055 knockdown (shGm15055-1 and shGm15055-2) JM8A3 mESCs. All interaction frequencies were normalized to the highest interaction frequency detected for each respective leader fragment. The P-values are indicated in the form of shGm15055–1 vs shGFP/shGm15055–2 vs shGFP. (E–G) Chromosome conformation capture assay of the interaction frequencies between the leaders (E: +54K; F:+51K; G: +47K) and the Hoxa gene cluster in JM8A3 mESCs and the JM8A3 that was induced by all-trans retinoid acid for two days (+RA D2). All interaction frequencies were normalized to the highest interaction frequency detected for each respective leader fragment.
Upon knockdown of Gm15055, interactions of the +51K leader and the Hoxa cluster are specifically affected, with an increase observed at the 3′-part of the Hoxa cluster (+10K to A5P), and a decrease at the 5′ of the cluster, especially at the A7P fragment (Figure 3C). We further showed that the interactions between the two loci were strengthened after RA-induced mESC differentiation, with the only exception observed for the +4K fragment, where the 3C signal decreased (Figure 3E–G). The changes became even obvious in the more differentiated MEF cells (Supplementary Figure S9). Close physical associations have previous been shown to mediate the Xist-initiated X-chromosome inactivation (31). The physical interactions of Gm15055 and the Hoxa loci hence may supply a similar platform for the Gm15055-recruited PRC2 to target and spread on Hoxa gene cluster, and the Gm15055 RNA itself might participate in regulating this chromatin conformation to a more 5′-proned structure in mESC.
Gm15055 knockout decreases H3K27me3 modification and PRC2 binding on Hoxa genes
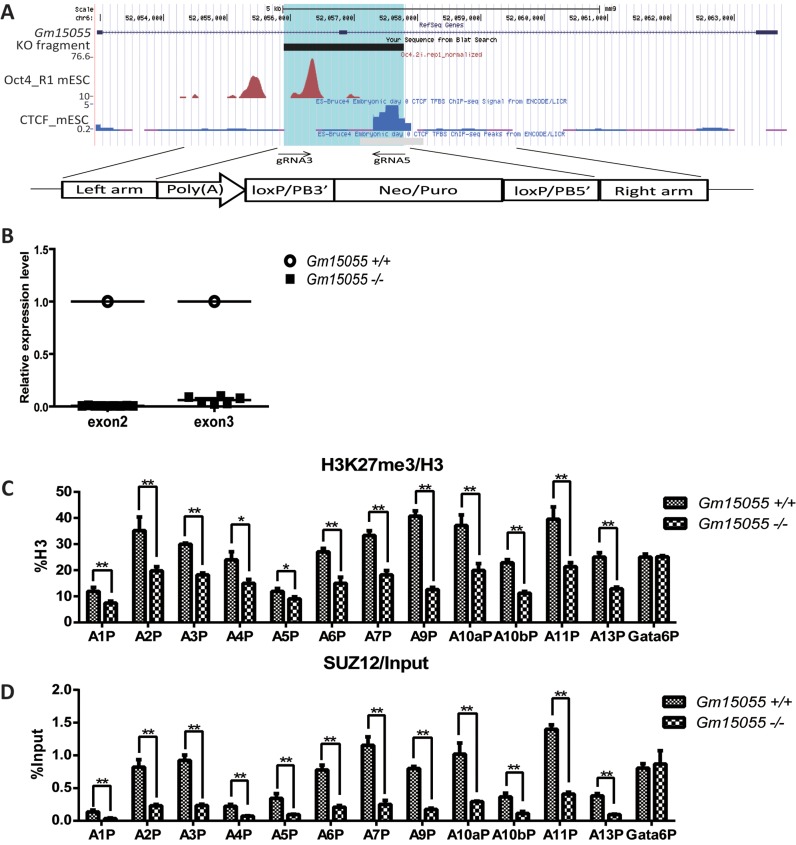
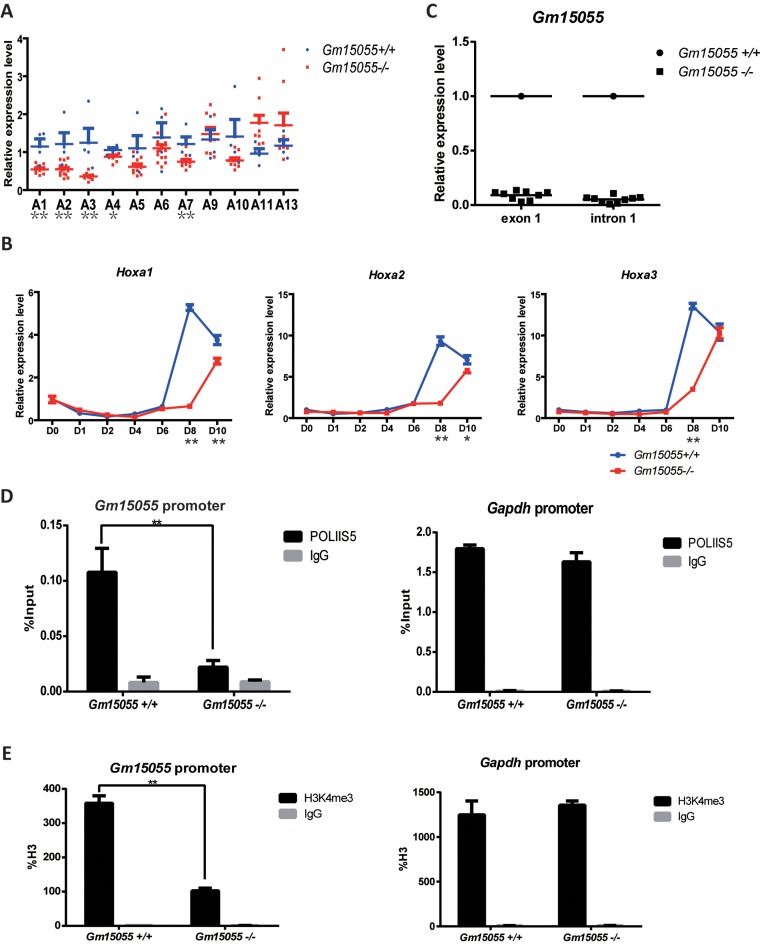
We went on to confirm the role of Gm15055 in PRC2 recruitment using CRISPR/Cas9- mediated knockout in JM8A3 mESCs. Two effective guide RNAs (gRNAs), gRNA3 or gRNA5, were used in combination with a gene targeting vector to homozygously delete a ∼1.9-kb DNA fragment of Gm15055 (mm9 chr6:52 055 891–52 057 784). The deleted region covers exon 2 of Gm15055 as well as the surrounding OCT4 and CTCF binding sites (26,30), and was replaced with a polyA transcription termination signal (Figure 4A). Homozygous Gm15055 knockout and the successful removal of the selective markers were validated by DNA sequencing. The sites that were predicted to have the highest off-target potential (32) were examined in both single-gRNA- and double-gRNA-targeted mESC clones and were confirmed to have no nucleotide mutations compared to the wild type sequence (Supplementary Figure S10). RT-PCR analysis of exons 2 and 3 in the homozygous Gm15055 knockout mESC clones confirmed the successful ablation of Gm15055 expression (Figure 4B). A pluripotency assay of the Gm15055 knockout clones with AP staining (Supplementary Figure S11A) and immunofluorescence analysis of SOX2, NANOG, OCT4, SSEA1 (Supplementary Figure S11B) was then carried out and indicated no significant changes in the Gm15055 knockout clones.
Figure 4.
Knockout of Gm15055 in mESCs decreases H3K27me3 modification and PRC2 binding on Hoxa gene promoters. (A) A schematic representation of the strategy and site of Gm15055 knockout. The highlighted blue region represents the knocked-out fragment of the Gm15055 locus. The DTA and PB targeting vector with a transcription termination poly(A) signal and a selection marker flanked by two homologous arms are shown at the bottom. Two CRISPR guide RNAs, the gRNA3 and gRNA5, were used in combination with the targeting vector to knockout each allele. The OCT4 ChIP-seq data are from Buecker, et al. 2014. Cell Stem Cell (26) and the CTCF ChIP-seq data are from ENCODE (30). (B) RT-qPCR confirmed the successful elimination of Gm15055 expression in Gm15055-knockout mESC clones. Two pairs of primers were used for the amplification of exon 2 and exon 3. All expression levels are shown relative to that of wild type JM8A3 cells. (C and D) Representative ChIP results of (C) H3K27me3 modification, (D) SUZ12 binding on Hoxa gene promoters before and after Gm15055 knockout in mESCs. All histone modification signals are normalized to H3 density and the Gata6 promoter (Gata6P) is used as a control region.
H3K27me3, H3K4me3 modifications and the binding of the PRC2 core component SUZ12 to the Hoxa gene locus were then examined in the Gm15055 knockout clones. Decreased H3K27me3 modification and SUZ12 binding were detected at the Hoxa gene promoters, the Gata-6 promoter was used as negative control (Figure 4C and D). The expression of Suz12 also remained unchanged, indicating that the defect involves the recruitment of SUZ12 (Supplementary Figure S12A). No significant changes in H3K4me3 modification were observed at the Hoxa gene promoters in the Gm15055 knockout clones, similar to the observations in the knockdown clones (Supplementary Figure S12B).
An OCT4-responsive cis-element within the Gm15055 locus potentially upregulates the transcription of both Gm15055 and the Hoxa genes
RT-qPCR examination of the Gm15055-knockout clones, however, revealed decreased expression of most Hoxa genes (Figure 5A). Moreover, embryoid body (EB) differentiation of the KO cells showed compromised activation of the Hoxa genes at day 8, when rapid Hoxa activation is started in wild-type mESC (Figure 5B and Supplementary Figure S13). Slight decrease in the activation of some Hoxa genes were also detected in all-trans retinoid acid (ATRA) induced differentiation of the KO cells, a model that is much faster than the EB differentiation model (Supplementary Figure S14). Our observation, together with the recent report about several other types of Gm15055-KO, support the existing of positive cis-regulatory element within the locus (29). Interestingly, severe decrease in the transcription of Gm15055 exon 1 and intron 1 was also observed in our Gm15055-knockout mESCs, although those regions were intact and protected by the polyA signal (Figure 5C). The decreases were accompanied with crippled Pol II binding and H3K4me3 modification at the Gm15055 promoter, the Gapdh promoter was used as a negative control (Figure 5D and E). These observations imply that the cis-element may regulate both the Gm15055 and the Hoxa genes expression.
Figure 5.
Knockout Gm15055 down-regulates most Hoxa genes and decreases transcriptional activity of Gm15055 promoter. (A) RT-qPCR detection of Hoxa genes expression in Gm15055-knockout and the wild type mESC clones. For each Hoxa gene, the expression level is shown relative to that of the corresponding Hoxa gene in wild-type JM8A3 cells. (B) RT-qPCR detection of Hoxa1-a3 expression in wild type and the Gm15055-knockout mESC during embryoid body differentiation. The time points (D0, D1, D2, D4, D6, D8, D10) indicated days after differentiation. See Supplementary Figure S13 for the analysis of the rest Hoxa genes. (C) RT-qPCR detection of Gm15055 exon 1 and intron 1 expression in the Gm15055-knockout ESC. The wild type ESC was used as a control. (D and E) ChIP-qPCR analysis of (D) RNA polymerase II phospho S5 (POLIIS5) binding and (E) H3K4me3 modification on Gm15055 promoter in the Gm15055-knockout and the wildtype ESCs. POLIIS5 binding and H3K4me3 modification at the Gapdh promoter in the two types of cells were shown as the control.
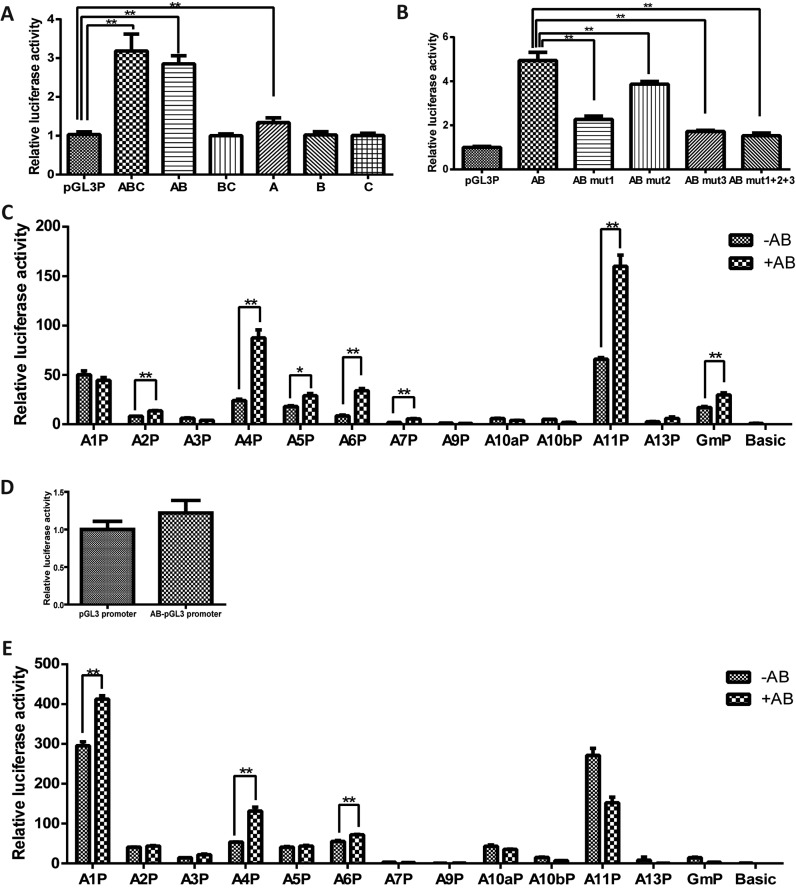
Reporter assays were then performed to confirm and dissect the knocked-out fragment for possible enhancer activity. The deleted region intersected the main OCT4 binding region of Gm15055 in the OBR-2 and -3, and was hence demarcated into three successive parts, part A, B and C, with the first two parts contain OBR-2 and -3 respectively (Supplementary Figure S15C). Enhancer activities of each part were then investigated individually and in combination (A, B, C, AB, BC, ABC) in mESCs by inserting these fragments downstream a basal SV40 promoter. The deleted region, as an entity, exhibited significantly increased luciferase activity in the reporter assay. Among the three individual sub-fragments, only fragment A exhibited a weak enhancer activity, which was notably facilitated by fragment B (Figure 6A). This result suggests that fragment A, which contains the OBR-2 of strong OCT4 binding, processes the core enhancer sequence for gene activation, and fragment B with the weak OBR-3 plays an auxiliary role in the activity.
Figure 6.
A DNA fragment within the Gm15055 gene locus possesses enhancer activity. (A) A reporter assay of the entire knockout fragment or the truncated fragment (named as ABC, AB, BC, A, B, C according to the contained sub-fragments) was performed in the JM8A3 ESC. The empty pGL3 promoter (pGL3P) vector was used as a control. The relative luciferase activity was normalized to that of the pGL3P. (B) Three OCT4 binding sites in the AB fragment were mutated separately (AB mut1, AB mut2, AB mut3) or in combination (AB mut1+2+3), and the reporter activities were detected in the JM8A3 ESC. The relative luciferase activity was normalized to that of the pGL3P. (C) Hoxa gene promoters with or without AB fragment were cloned into pGL3 basic vector and the reporter activities were detected in the JM8A3 ESC. The effect of AB fragment on the Gm15055 promoter (GmP) was also investigated. The relative luciferase activity was normalized to that of the pGL3 basic vector (Basic). (D) Reporter assay of the AB fragment cloned in the pGL3P vector was carried out in the MEF cells. The relative luciferase activity was normalized to that of the pGL3P vector. (E) Enhancer activity of the AB fragment to all the Hoxa gene promoters and the Gm15055 promoter (GmP) was detected in the MEFs. The relative luciferase activity was normalized to that of the pGL3 basic vector (Basic).
To further investigate how the enriched OCT4 binding contributes to the enhancer activity of the region, three OCT4 binding sites were predicted from the OBR-2 by JASPAR (33) and were mutated separately or together (Supplementary Figure S15C). The results showed that mutation to each OCT4 binding site significantly suppressed the enhancer activity of the AB fragment (mm9 chr6:52 055 891–52 057 264), and that simultaneous mutation to all the three sites impaired the enhancer activity to the most extent (Figure 6B), suggesting an important role of OCT4 binding to the enhancer activity of this region in mESCs.
We next examined the promoter-specificity (34) of the AB fragment. Each Hoxa gene promoter or the Gm15055 promoter was inserted into the reporter construct with or without the AB fragment. Reporter assay in mESCs showed that the AB fragment increased the activity of Gm15055 promoter and many Hoxa promoters, including the A2P, A4P, A5P, A6P, A7P and A11P (Figure 6C). Whereas in the mouse embryonic fibroblast (MEF) cells, where all Hoxa genes were activated (Supplementary Figure S16) and Gm15055 was inactivated, the AB fragment no longer increases the activity of SV40 and Gm15055 promoters (Figure 6D-E) and showed a limited enhancive effect to a few Hoxa promoters including the A1P, A4P and A6P (Figure 6E).
DISCUSSION
In the present study, we found that the long noncoding RNA Gm15055 is activated by the pluripotency factor OCT4 in mESCs and represses the adjacent Hoxa genes by recruiting PRC2 and maintaining the repressive histone mark H3K27me3 on the Hoxa gene cluster. Gm15055 loops to the Hoxa locus, which may facilitate PRC2 binding on the Hoxa cluster. In addition to producing the repressive lncRNA, the Gm15055 locus also contains an OCT4-dependent positive cis-regulatory element and may therefore contribute to both the high-level expression of Gm15055 itself and the maintenance of a low but steady level of Hoxa gene expression in mESC.
OCT4 activates Gm15055 expression in mESC
Oct4, together with Sox2 and Nanog are core transcriptional factors for maintaining pluripotency of stem cells (35–37). High-throughput analysis of OCT4 binding sites in mouse ESC has revealed thousands of target sites through which OCT4 activates the ESC-enriched genes or suppresses the differentiation genes (38). In the present study, we showed that OCT4 positively regulates Gm15055 expression without affecting cell differentiation, and demonstrated the direct targeting of OCT4 to the Gm15055 locus by conventional ChIP as well as point mutations in the reporter assays. Other than acting on the promoter of Gm15055, OCT4 also activates a cis-regulatory element at around exon 2 of the locus and could hence contribute to enriched Gm15055 expression in mESCs. Moreover, it was noticed that SOX2 and NANOG overlap with OCT4 in binding to the Gm15055 locus (Supplementary Figure S15B)(25,26). Considering the repressive effect of Gm15055 RNA to the downstream Hoxa genes, revealed by previous reports and the current data, and the essential roles of Hox genes in cell fate determination, we infer that mESC keeps abundant Gm15055 expression by OCT4 and other pluripotency factors, to help in maintaining a robust pluripotent state of the cells.
LncRNA Gm15055 recruits PRC2 to the Hoxa gene cluster in mESC
PRC2 is a well-recognized key regulator of developmental genes (5). The most well characterized recruiters of PRC2 are the polycomb repressive elements (PREs) found in Drosophila melanogaster, whereas only a few proteins or DNA elements have been identified to guide PRC2 complexes to target genes in mammals (39,40). LncRNAs have recently been discovered as a new group of mediators for the recruitment of PRC2 to a specific gene locus (8–10). Although a panel of lncRNAs have been shown to be capable of interacting with PRC2 in mESCs, only Gtl2 has been functionally characterized (15). Our study indicates that the long noncoding RNA Gm15055 is a new factor for PRC2 recruitment in mESCs. Both knockdown and knockout of Gm15055 in mESCs significantly decreased PRC2 binding and the H3K27me3 modification at the Hoxa gene promoters. Previous work showed that Gm15055 also recruited PURB (18). The recent study (29), however, failed to observe the effect of PUR proteins on Hoxa regulation, and indicated the involvement of Gm15055 in modulating the H3K27me3 at Hoxa gene locus in RA-induced ESC. Using a RIP assay, we observed in the present study that Gm15055 interacted with PRC2 in vivo and the BoxB-ChIP analysis further confirmed that Gm15055 could recruit PRC2 to the adjacent gene locus and facilitate H3K27me3 modification there, suggesting that Gm15055 recruits PRC2 to the Hoxa gene cluster in mESCs. Considering the promiscuous nature that PRC2 exhibited in the recent in vitro RNA binding studies (41–43), the direct Gm15055/PRC2 interaction remains to be determined in the future with more sophisticated approaches.
Our work consistent with the recent study (29) in finding that Gm15055 physically close associated with the Hoxa gene cluster, which may provide a molecular mechanism for the loading of Gm15055-recruited PRC2 to the Hoxa genes in mESCs. Instead of using the Hoxa1 and Hoxa5 promoters as the 3C leaders as in the previous study, we albeit used the Gm15055 fragments to screen the Hoxa locus, and have identified the active involvement of the +4K, and the Hoxa7, Hoxa9, Hoxa11 promoters in the association. The chromatin interaction appears relatively stable upon RA induction and after knockdown of the Gm15055 expression, implying multiple factors like OCT4, CTCF (44), which has binding sites at both loci, may participate in maintaining the association.
The Gm15055 locus coordinates the Hoxa genes expression
Interpretation of IncRNA function is complexed by the role of RNA per se, the existing of potential regulatory elements in the same locus and even the effect of RNA transcription event (45,46). By applying the knockdown strategy and establishing differentially knockout clones, recent study and the present work showed unexpected decrease in Hoxa genes expression in most of the Gm15055-knockout clones, which suggests potential positive cis-regulatory elements in the Gm15055 locus for maintaining a low but responsive Hoxa genes expression (29). Dissection and mutations of the knockout fragment here revealed enhancer activity mainly associated with OCT4 binding sites in the region and that disruption of the OCT4 sites largely impaired this activity. The cis-element selectively acted on Gm15055 and Hoxa2/4/5/6/7/11 promoters in reporter assay within mESC, and its activity diminished in differentiated MEF cells, emphasized the ESC specificity of the enhancer activity. In addition to this region, bioinformatics analysis using ChIP-seq data of MED12/NIPBL by Kagey et al. and that of OCT4/SOX2/NANOG showed enhancer characteristics at multiple sites in the rest part of the Gm15055 locus (Supplementary Figure S15A-B)(25,26,47,48). The recent study by Yin et al. indicated that knockout to different part of the locus variant in the range of Hoxa genes been affected (29). 3C assay in the present study further showed that different parts of Gm15055 locus diverse in their affinity to the respective Hoxa genes, implying a more differentiated and cooperative mechanism for the Gm15055 locus and its RNA product to elaborate Hoxa gene regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr Yue Huang for the generous gifts of the FIAU and the JM8A3 cells.
FUNDING
National Natural Science Foundation of China [31030026]; ‘973’ Program [2011CB965203, 2011CB964803 and 2011CB529903]; ‘863’ program [2011AA020116]; PUMC Youth funds [3332013138]. Funding for open access charge: National Natural Science Foundation of China [31030026]; ‘973’ Program [2011CB965203, 2011CB964803 and 2011CB529903]; ‘863’ program [2011AA020116]; PUMC Youth funds [3332013138].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 2.Noordermeer D., Leleu M., Schorderet P., Joye E., Chabaud F., Duboule D. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. Elife. 2014;3:e02557. doi: 10.7554/eLife.02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soshnikova N., Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324:1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- 4.Noordermeer D., Leleu M., Splinter E., Rougemont J., De Laat W., Duboule D. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 5.Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 6.Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-Dinardo D., Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 10.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L., Liu B., Wapinski O.L., Tsai M.C., Qu K., Zhang J., Carlson J.C., Lin M., Fang F., Gupta R.A., et al. Targeted Disruption of Hotair Leads to Homeotic Transformation and Gene Derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhan A., Mandal S.S. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttman M., Garber M., Levin J.Z., Donaghey J., Robinson J., Adiconis X., Fan L., Koziol M.J., Gnirke A., Nusbaum C., et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maamar H., Cabili M.N., Rinn J., Raj A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 2013;27:1260–1271. doi: 10.1101/gad.217018.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., Sanchez-Gomez D.B., Hacisuleyman E., Li E., Spence M., et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moffat J., Grueneberg D.A., Yang X., Kim S.Y., Kloepfer A.M., Hinkle G., Piqani B., Eisenhaure T.M., Luo B., Grenier J.K., et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 21.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A., et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu M., Zhao G.N., Lv X., Liu G., Wang L.Y., Hao D.L., Wang J., Liu D.P., Liang C.C. CTCF controls HOXA cluster silencing and mediates PRC2-repressive higher-order chromatin structure in NT2/D1 cells. Mol. Cell. Biol. 2014;34:3867–3879. doi: 10.1128/MCB.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naumova N., Smith E.M., Zhan Y., Dekker J. Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods. 2012;58:192–203. doi: 10.1016/j.ymeth.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buecker C., Srinivasan R., Wu Z., Calo E., Acampora D., Faial T., Simeone A., Tan M., Swigut T., Wysocka J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 28.Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin Y., Yan P., Lu J., Song G., Zhu Y., Li Z., Zhao Y., Shen B., Huang X., Zhu H., et al. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell. 2015;16:504–516. doi: 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Rosenbloom K.R., Sloan C.A., Malladi V.S., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R., Heitner S.G., et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engreitz J.M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C., Kadri S., Xing J., Goren A., Lander E.S., et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H., et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zabidi M.A., Arnold C.D., Schernhuber K., Pagani M., Rath M., Frank O., Stark A. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2014;518:556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng H.H., Surani M.A. The transcriptional and signalling networks of pluripotency. Nat. Cell Biol. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- 37.Orkin S.H., Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 39.Woo C.J., Kharchenko P.V., Daheron L., Park P.J., Kingston R.E. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jermann P., Hoerner L., Burger L., Schubeler D. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3415–E3421. doi: 10.1073/pnas.1400672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davidovich C., Zheng L., Goodrich K.J., Cech T.R. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cifuentes-Rojas C., Hernandez A.J., Sarma K., Lee J.T. Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell. 2014;55:171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davidovich C., Wang X., Cifuentes-Rojas C., Goodrich K.J., Gooding A.R., Lee J.T., Cech T.R. Toward a Consensus on the Binding Specificity and Promiscuity of PRC2 for RNA. Mol. Cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips J.E., Corces V.G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bassett A.R., Akhtar A., Barlow D.P., Bird A.P., Brockdorff N., Duboule D., Ephrussi A., Ferguson-Smith A.C., Gingeras T.R., Haerty W., et al. Considerations when investigating lncRNA function in vivo. Elife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L., Chang H.Y. Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol. 2014;24:594–602. doi: 10.1016/j.tcb.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C.Y., Morris Q., Mitchell J.A. Enhancer identification in mouse embryonic stem cells using integrative modeling of chromatin and genomic features. BMC Genomics. 2012;13:152. doi: 10.1186/1471-2164-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S., et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.