Abstract
Stable isotope labeling is central to NMR studies of nucleic acids. Development of methods that incorporate labels at specific atomic positions within each nucleotide promises to expand the size range of RNAs that can be studied by NMR. Using recombinantly expressed enzymes and chemically synthesized ribose and nucleobase, we have developed an inexpensive, rapid chemo-enzymatic method to label ATP and GTP site specifically and in high yields of up to 90%. We incorporated these nucleotides into RNAs with sizes ranging from 27 to 59 nucleotides using in vitro transcription: A-Site (27 nt), the iron responsive elements (29 nt), a fluoride riboswitch from Bacillus anthracis (48 nt), and a frame-shifting element from a human corona virus (59 nt). Finally, we showcase the improvement in spectral quality arising from reduced crowding and narrowed linewidths, and accurate analysis of NMR relaxation dispersion (CPMG) and TROSY-based CEST experiments to measure μs-ms time scale motions, and an improved NOESY strategy for resonance assignment. Applications of this selective labeling technology promises to reduce difficulties associated with chemical shift overlap and rapid signal decay that have made it challenging to study the structure and dynamics of large RNAs beyond the 50 nt median size found in the PDB.
INTRODUCTION
The tertiary architectures RNAs adopt are crucial for modulating gene expression across all domains of life, making them important targets of structural and dynamics studies. For instance, for riboswitches, the presence or absence of specific ligands drives the folding of one of two or more mutually exclusive, regulatory states (1–4). In viral RNA genomes, structured, untranslated regions commonly exercise direct control over viral gene expression (5,6). In the ribosome, the ability to distinguish between cognate and near-cognate tRNAs is governed in part by the extrahelical flipping of adenines A1492 and A1493 (7). Both the global architecture and the subtle motions of specific base and ribose moieties are thus demonstrably important and can profoundly modulate an RNA's function (8).
However, in spite of this importance, directly establishing how dynamics modulates the structure and function of RNAs has been difficult because X-ray crystallography and nuclear magnetic resonance spectroscopy are plagued by distinct but equally challenging problems. In crystallography, motions can only be observed in the ps–ns timescales and the strain imposed by crystal packing can obscure and distort structural data (9,10).
In contrast, NMR spectroscopy can probe dynamic fluctuations directly over a wide range of timescales. Unfortunately, NMR suffers from both narrow chemical shift dispersion and rapid signal decay, exacerbated by direct one-bond and multi-bond spin-spin couplings. The former leads to spectral crowding and the spin-spin couplings can lead to decreased spectral resolution and inaccurate measurements of 13C relaxation rates such as longitudinal relaxation rates (R1), transverse relaxation rates (R2), and heteronuclear Overhauser effect (hNOE) (11–13). Furthermore, these problems become more pronounced as the size of the RNA increases: the spectral quality deteriorates because of increased line broadening.
Addressing these problems requires the development of new technologies. In the past, spectral overlap has been addressed using heteronuclear multi-dimensional pulse sequences applied to uniformly 13C/15N labeled RNAs (14–16). By spreading the poorly dispersed proton resonances over the better resolved carbon and/or nitrogen dimensions, it is possible to resolve overlapped proton peaks in small RNAs. While these advances have greatly aided NMR structural studies of RNAs with a median size of 30 nt, they fail for RNAs larger than 60 nt. Out of 460 RNA structures in the PDB (Protein Data Bank), only seven RNA structures with sizes >60 nt have been solved by NMR (17–25). Of these seven, three RNA structures of 101, 132 and 155 nt have been solved using mostly homonuclear two-dimensional (2D) NOESY methods based on nucleotide-specific and fragmentation-based segmental 2H-labeling approaches (18,20,25). Thus, current uniform labeling approaches while valuable are quite limiting (26,27).
Also of great interest are the large couplings of adjacent 13C nuclei within the ribose and base ring systems which cause several complications in RNA relaxation measurements. The foremost concern is that uniform labeling introduces strong couplings that can render 13C R1, hNOE and CPMG (Carl-Purcell-Meiboom-Gill) relaxation measurements inaccurate. These couplings also complicate and limit the range of applicability of CEST (Chemical Exchange Saturation Transfer) and rotating-frame relaxation rate (R1ρ) measurements and analyses while also decreasing the attainable resolution and sensitivity of NMR experiments (11,13,28–34).
Numerous robust spectroscopic solutions have been proposed in the past to circumvent these coupling problems (33–41). Unwanted splittings can be removed using constant time (CT) evolution (35–38), adiabatic band selective decoupling (39–41), or a series of selective pulses. Constant time evolution limits the acquisition time that can be used to obtain adequate resolution. To improve resolution requires long constant-time delays that lead to significant signal loss for large RNA molecules (41). Additionally, obtaining accurate relaxation parameters are problematic for 13C-CPMG based relaxation dispersion rates for quantifying millisecond (ms) time-scale processes, as well as R1 and proton–carbon hNOE (28,42) important for quantifying ns-ps time-scale motions in RNA (43,44). Several precautions are needed to obtain accurate R1 and R1ρ measurements (28,45–46): provided R1 is derived from the initial slope of the relaxation decay curve, fairly accurate rates can be extracted for small RNAs; for R1ρ experiments, distortions can arise from transfer between adjacent 13C atoms with similar chemical shifts via a Hartmann–Hahn mechanism and these need to be minimized (28,45–46); to suppress the echo-modulation caused by the large scalar couplings during the 13C relaxation delay, R1ρ can be measured instead of CPMG (28,47–49). Still for nucleic acids high power spinlocks (>1 kHz) are needed to study isolated spin pairs such as C2 found in adenine and C8 in both adenine and guanine. For low spin lock power levels (<1 kHz), oscillations can be observed in the monoexponential decay of peak intensity, arising from residual scalar coupling interactions within neighbouring nuclei (48,50–51).
In addition, application of selective cross-polarization (52–55) using weak radio-frequency fields can effectively decouple homonuclear J-couplings. This elegant spectroscopic solution has been exploited to measure nitrogen R1ρ in proteins and both carbon and nitrogen R1ρ in uniformly labeled nucleic acids (56–59). While this scheme obviates the need for selective 13C isotopic enrichment, in uniformly labeled samples, the presence of large homonuclear scalar couplings again limits the range of applicability of these methods (56,51). Finally building upon schemes for protein 15N and 13C CEST measurements by Kay and co-workers, Zhang et al. developed a set of nucleic-acid-optimized 1D/2D 13C CEST experiments that use various shaped pulses to refocus carbon–carbon scalar coupling and showed that accurate exchange parameters can be obtained for all CEST profiles in uniformly labeled RNA samples for purines and ribose carbons(34,60–62). Nonetheless, they and others acknowledged the following limitations for both CEST and R1ρ in uniformly labeled RNA and protein samples (33–34,60–63). First, the lowest spinlock or saturating B1 field that can be used is limited (∼3× the scalar coupling) to ∼45 Hz for Ade C2, ∼45 Hz for purine C8, ∼150 Hz for C1’. For pyrimidine ring carbons with large carbon-carbon couplings of ∼66 Hz, it would require ∼200 Hz spinlock fields for C5 and C6, clearly intractable with uniformly labeled samples. Second, even though 13C–13C couplings do not introduce errors in extracted chemical shifts for purines, these homonuclear couplings decrease the resolution. Ultimately, the coupling effects need to be considered in the CEST data analyses for couplings greater than 15 Hz. Otherwise, exchange parameters (kex) are overestimated and population ratios are underestimated (33). Thus, uniformly labeled samples do limit the range of wide applicability of both CEST and R1ρ to biological problems.
These spectroscopic tools notwithstanding, an alternative, straightforward and effective solution for overcoming the problem of spectral crowding and J-coupling would complement existing methodologies. A promising method is to synthesize site-specific isotopically labeled nucleotides (11,64–65) as we recently demonstrated with our chemo-enzymatic production of pyrimidine nucleotides (30,31). Here, we extend that approach to improving the synthesis of purine nucleotides. Our synthesis offers improvements in speed, streamlined reaction conditions, and higher yields. By combining the newly developed purine nucleotides with our previous pyrimidine nucleotides we present an improvement to the traditional NOESY structural assignment protocol. Additionally, we show that the measurements of relaxation parameters using CPMG, R1ρ, and CEST are possible for both small and large RNAs. Furthermore, we demonstrate substantial improvements in signal-to-noise and line width for relaxation optimized spectroscopy (TROSY) experiments compared to the traditional heteronuclear single quantum coherence (HSQC) experiments for isolated two-spin systems approximated by our purine and pyrimidine labeling schemes (30–31,66–67).
MATERIALS AND METHODS
General procedures
Reagents and solvents were purchased from Sigma-Aldrich. 8-13C adenine and 8-13C guanine were either purchased from Cambridge Isotope Laboratories or synthesized as described in the supplementary materials. Similarly, preparative chemical synthesis of labeled adenine and guanine, chemo-enzymatic nucleotide synthesis, RNA preparation, and NMR experiments are detailed in the supplementary materials.
Data analysis
NMR spectral processing was done in Topspin (Bruker Biospin) and NVFx (One Moon Scientific). Peak intensities were selected using in-house software by David Fushman. CPMG data were fit to a two-state exchange model using the full Bloch-McConnell matrix. The time-dependent evolution of magnetization during the CPMG period was solved numerically by non-linear least squares fitting using in-house Matlab software. Errors in fits were calculated using Jacobian or 200 Monte Carlo simulations (33), and the larger of the two errors was reported for CPMG and CEST relaxation dispersion analysis.
NMRViewJ was used for peak assignments. Hydrogen and carbon chemical shifts are predicted based on the secondary structure of the input RNA molecule. Expected cross peaks for different experiment types and labeling patterns were then generated using the RNA Peak Generator tool. Expected cross-peaks were generated for HSQC spectra based on the covalent structure and for NOESY spectra using inter-atomic distances typically observed in RNA helices. For bacterial A-site RNA, of which there are no deposited chemical shifts in the BMRB database, the RNA Peak Generator accurately predicted 15 of the 18 expected C1′-H1′ resonances, 7 of the 9 C2′-H2′, and 20 of the 27 C6/8-H6/8 resonance within 0.1 ppm of their actual values in the HSQC spectra. Since the NOESY peak generator was used in a mode where it only predicts peaks in helical regions, peaks in bulge and tetraloop regions of the A-site RNA were not predicted. Further assignment of the NOESY spectra utilized the RNA peak slider tool within NMRViewJ. This links the predicted peaks into a network connected by atoms shared between the different peaks. Peaks are then interactively positioned in a way that utilizes the network of peaks typically connected within the NOESY ‘walk’. Overall, the combined tools of NMRViewJ allowed for relatively rapid assignment of the resonance in the specifically labeled A-site RNA model system and provides a powerful tool that, when combined with selective labeling, can streamline resonance assignment for RNA than previously reported (41).
RESULTS AND DISCUSSION
We have developed a protocol for synthesizing site-specific isotopically labeled purine nucleotides with varied ribose and base patterns (Supplementary Figure S1). In principle any combination of labeled base and ribose can be utilized, but here we demonstrate the use of both 1′,8-13C and 2′,8-13C purine and 1′,6-13C and 2′,6-13C pyrimidine nucleotides for assignment, structural, and dynamics measurements. These labeling patterns both remove the strong 13C–13C J-coupling found in uniformly labeled nucleotides and simultaneously reduce spectral crowding. The removal of 13C J-coupling creates isolated spin systems in both the base and ribose, and enables the use of TROSY pulse sequences for studying large RNAs, and these TROSY modules can be readily incorporated into CPMG, R1ρ and CEST pulse sequences for measuring μs-ms timescale dynamics (68–71).
In addition to the removal of strong J-couplings, the new labels help to greatly reduce spectral crowding and enable the creation of a new protocol for the NOESY assignment of RNAs. As a proof of this assignment concept, this protocol is demonstrated on the A-site RNA.
Chemo-enzymatic synthesis of GTP and ATP
We have created an improved method for the synthesis of site-selective isotopically labeled ATP and GTP with increased yields and speed of synthesis. Both purine reactions proceed to completion without the need to purify intermediate species. Final yields of >90% and >75% respectively for ATP and GTP were achieved relative to starting input adenine or guanine. Both yields are better than previously reported (72–77). In addition to improved yields, ATP synthesis is complete in 4–5 h while GTP synthesis is complete in 7–8 h. Previously, ATP synthesis was reported to take 29–48 h and GTP synthesis 48–70 h (72–77). These improvements allow reactions to be complete in a single day. Additionally we have taken advantage of the ability of creatine kinase to act on a variety of substrates to convert NDPs to NTPs and adapted the use of dATP as the energy source in the energy regeneration system (77–79). The use of dATP is ideal since the lack of a 2′-OH of the ribose in dATP prevents its interaction with the boronate column used to purify ATP and GTP. This offers a more robust synthesis, free of contaminants, and does not dilute the synthesized labels with unlabeled ATP. The effectiveness of these nucleotides is demonstrated by their incorporation into a number of interesting RNAs.
The production of GTP was achieved in a two-step, one pot reaction. Specifically-labeled ribose and guanine were combined in the presence of phosphoribosyl pyrophosphate synthetase (PRPPS), ribokinase (RK), and xanthine-guanine phosphoribosyl transferase (XGPRT), with a dATP regeneration system. The dATP regeneration system was composed of myokinase and creatine kinase, with creatine phosphate acting as the high energy phosphate donor. The formation of GMP was monitored by FPLC and NMR (Supplementary Figure S2). However, due to the low solubility of guanine (0.01 mM) FPLC was unsuitable to track its disappearance, thus making it difficult to monitor the progression of the reaction. However, by NMR spectroscopy, the resonance chemical shift between the labeled 13C-1′ position of unreacted ribose and newly formed GMP was used to determine the completion of the first step of the reaction (Supplementary Figure S2A). When the majority of guanine was converted to GMP, in approximately 4–5 h, guanylate kinase was added to the reaction. Guanylate kinase phosphorylates GMP to GDP. GDP is phosphorylated to GTP by creatine kinase which acts promiscuously to convert NDPs to NTPs. This was unexpected, as CK is said to be highly specific (78,79). GMP is completely converted to GTP in an additional 3 h. We confirmed by FPLC that conversion is complete and further validate this observation by 31P NMR (Supplementary Figure S2B).
The production of ATP and the progression of the reaction is monitored as reported for GTP. A notable difference is that adenine's greater solubility (8 mM) allowed the use of FPLC to monitor the disappearance of uncoupled base and the formation of product for all steps of the reaction. Labeled adenine and ribose were combined in the presence of PRPPS, RK, adenine phosphoribosyl transferase (APRT), and the dATP regeneration system. The dATP regeneration system acts on both AMP and ADP and takes the reaction to completion in ∼ 4 h. The reaction is similarly monitored by FPLC and NMR (Supplementary Figure S2C&D).
Synthesis of nucleotides with isolated base C6 or C8 offers large sensitivity improvements
When studying large RNAs (>40 nts) by NMR, slow molecular tumbling leads to broadened linewidths and losses in signal intensity. Careful selection of appropriate NMR experiments to address these losses are necessary for successful measurement of many NMR parameters. TROSY experiments take advantage of the interference between the dipolar coupling and chemical shift anisotropy (CSA) components of T2 relaxation (66). For the base C8 position of adenine and guanine, these contributions effectively cancel at ∼800 MHz field strength leading to reduction in the R2 relaxation rate (80,81). Thus, RNAs synthesized with our selective site-specifically labeled NTPs should benefit from TROSY based NMR experiments that reduce the problems of crowding, fast signal decay, low resolution, and decreased S/N ratios (12,34,31,66–67,80–81).
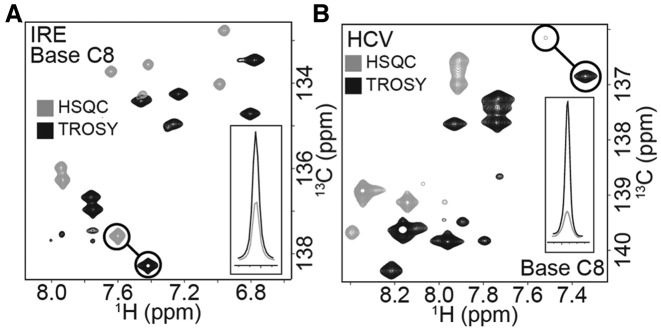
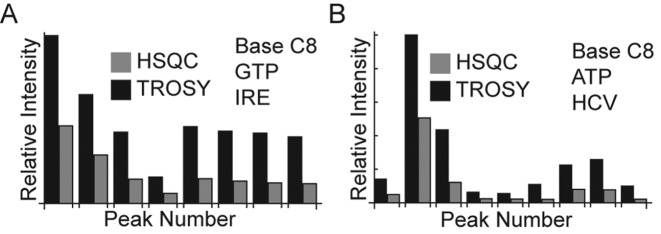
The benefits of TROSY increases with the size of the RNA. For small RNAs such as IRE (29 nt) we saw substantial, yet modest, improvements for the base region. These improvements in signal intensities ranged from 2.2- to 3.4-fold (average: 2.9 ± 0.5) when comparing TROSY with conventional HSQC sequences (Figures 1A and 2A). For the larger HCV SARS RNA (59 nt), the signal improvements are larger and ranged from 2.0- to 5.4-fold (average 3.3 ± 1.0) (Figures 1B and 2B). For the C1′ and C5′ peaks the improvements were more modest since these positions have lower CSA values (Supplementary Figure S3). Our labeled C8 approximates an isolated two spin system necessary for these gains in signal. Thus, the large improvements seen for these positions when using our site-specifically labeled nucleotides can be harnessed for assignment, structural, and dynamics measurements (82).
Figure 1.
TROSY improvements for IRE and HCV site-specifically labeled samples. (A) The base region of 1′,5′,8-13C3 GTP labeled IRE. Comparisons of the HSQC and TROSY pulse sequences show an average intensity improvement of 2.9 ± 0.5. The inset displays the overlaid slices of the highlighted residue. (B) The base region of 1′,5′,8-13C3 ATP labeled HCV. Again comparisons of the HSQC and TROSY pulse sequences show an average intensity improvement of 3.3 ± 1.0. The inset displays the overlaid slices of the highlighted residue.
Figure 2.
The improvement of HSQC versus TROSY pulse sequences for IRE (A) and HCV (B).
CEST measurements on RNAs >50 nucleotides
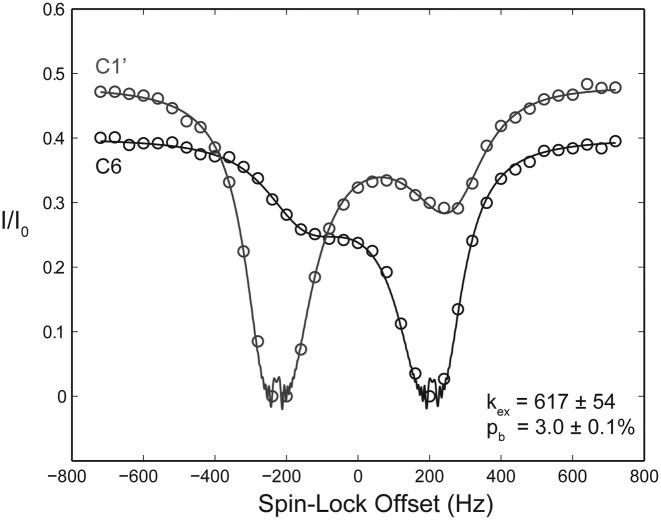
The above observations led us to run 13C-TROSY version of the 15N-TROSY experiment of Kay et al. (68). We decided that to validate this TROSY pulse sequence it would be appropriate to mirror measurements made previously on a fluoride riboswitch construct (34). Interestingly, our construct isolated from Bacillus anthracis showed similar behavior to the construct from Bacillus cereus studied by Zhang et al., albeit with slightly shifted values. While for B. cereus kex = 112 ± 4 s−1 and pb = 10.1 ± 0.1% at 30°C, the B. anthracis construct had exchange parameters of kex = 617 ± 54 s−1 and pb = 3.0 ± 0.1% at 35°C for a global fit to both C1′ and C6 (Figure 3). Further, when comparing TROSY CEST (68) to the traditional HSQC CEST (34), S/N improvements of ≈ 2:1 were obtained (Supplementary Figure S4). While the fits of both the HQSC and CEST data sets gave similar exchange parameters, comparing the χ2 of the fits showed significant improvements for the TROSY CEST experiment (0.6–42.5) when both experiments were run using the same parameters and experiment time. These measurements were made on the C1′ and C6 positions of a 1′,6-13C-1,3-15N-5-2H UTP labeled sample.
Figure 3.
The 13C-TROSY CEST profile for the fluoride riboswitch recorded at 35°C. Measurements were made on the C1′ (gray) and C6 (black) site of a 1′,6-13C-1,3-15N-5-2H UTP. Data were globally fit to a 2-site full Bloch–McConnell equation. The residue shown is proposed to be involved in an unstructured to base-paired transition involved in the formation of a pseudoknot. The B. anthracis construct had exchange parameters of kex = 617 ± 54 s-1 and pb = 3.0 ± 0.1%.
What then are some of the benefits of a selectively labeled sample when uniformly labeled samples have been shown to be adequate? Strong coupling eliminated between ribose carbons allowed a straightforward analysis of the CEST data without the need to account for and correct J-coupling (33,34). In particular obtaining CEST data for C6 pyrimidine is particularly problematic because of complications mentioned above in the introduction using uniformly labeled samples and that field strengths of >180 Hz needed preclude their use in uniformly labeled samples. With our selective labeled samples, we were able to obtain excellent CEST profiles readily for both purine and pyrimidines.
CPMG measurements on purine nucleotides and RNAs >50 nucleotides
CPMG relaxation dispersion measurements facilitate the extraction of information about exchange phenomenon occurring on the μs–ms timescale (83–89). Previously, others have used similar approaches to measure CPMG experiments for RNAs smaller than 50 nucleotides with specifically labeled pyrimidine bases (35,36). Here, we present data that illustrates the effect of creating an isolated, labeled C8 and C2′ positions in our nucleotides, and show that measurements of CPMG parameters are readily accessible without the problem of J-coupled induced oscillations (28,90).
We have transcribed a 59 nt viral RNA with 1′,8-13C labeling pattern as a proof of concept. The data indicate that while a majority of the nucleotides within the RNA do not experience exchange on the ms time-scale, a few residues sample a lowly populated state. Without data being fit at multiple static magnetic field strengths, the only meaningful parameter that can be extracted is a kex value (Figure 4A). The exchange rates extracted from the CPMG experiments on the viral RNA match well with those from CEST experiments (unpublished). Even though similar information, and perhaps more, can be derived from R1ρ data, we find that CPMG is straightforward to setup and analyse compared to R1ρ experiments. Thus having labeled RNA that facilitates CPMG measurements is important for the field.
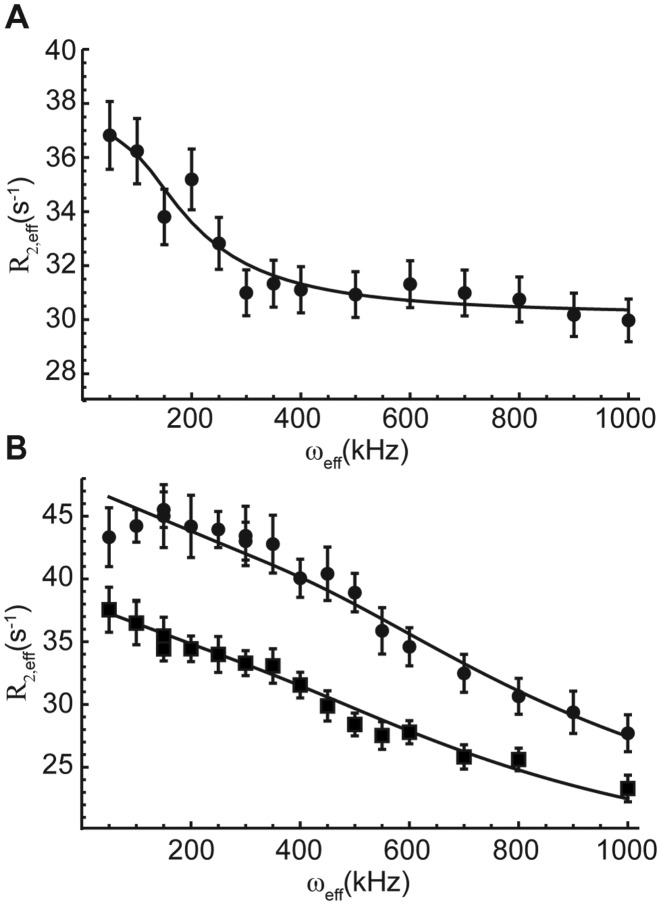
Figure 4.
CPMG fits for HCV SARS (A) and A-site (B). (A) The CPMG data revealed kex for select residues in the Human corona virus RNA element. Data were measured on the C1’ position of a 1′,8-13C labeled sample. (B) CPMG was run at 14.1 (▪) and 18.8 (•) T static field strengths for the A-site RNA. A global fit revealed kex = 3800 ± 200 s-1 and pb = 1.8 ± 0.1%. These results were in agreement with previously reported values (8). Measurements were made on the C2′ position of 2′,8-13C labeled sample. The A1493 position that these curves belong to has been implicated in the discrimination of cognate and near-cognate tRNAs in the ribosome.
Using in-house Matlab scripts, CPMG data were fit to a two-state exchange model using the Bloch–McConnell matrix as previously described by Kay et al. (90). Site-selective labels allow us to prepare isolated two spin systems without the carbon-carbon or carbon-nitrogen scalar couplings. In the past, such scalar couplings have hindered the interpretation of relaxation dispersion data (89,90). Using the bacterial A-site RNA as a model system, we were able to capture motions on the microsecond timescale using CPMG experiments to monitor exchange of the ribose C2′ residues. It is widely accepted that motions in residues A1492 and A1493 are involved in the discrimination between cognate and near-cognate tRNAs (7–8,91–93). Most notably, A1493, a residue that flips in and out of the bulge region of A-site showed characteristic dispersion profiles (Figure 4B). The extracted kex and pb values of 3800 ± 200 s−1 and 1.8 ± 0.1% match well the previously reported values of 4000 s−1 and 2.5% determined by relaxation dispersion measurements on the C1′ positions of the ribose moieties (8). Thus, our labels can be used to readily and straightforwardly capture lowly populated states in RNA.
Reduction of spectral crowding for large RNAs
The relatively narrow spectral width over which base and sugar carbons and protons resonate is a major limitation of RNA NMR that must be overcome (82). Overlap of signals is only partially alleviated by 2D and 3D NMR experiments in samples in which all 4 nucleotides are uniformly 13C- and 15N-labeled. We reasoned that what would be critical for de-cluttering spectra to manageable levels for large RNAs is not only the ability to choose which of the four nucleotides to label, but also which of the atomic sites to isotopically enrich. To demonstrate the power of this approach, we have examined RNAs ranging in size from 27 to 59 nucleotides in length.
For a large RNAs transcribed with only 1′,8-13C2 ATP, the resonances that belong to the adenine C8 can be identified rapidly when compared to a sample that has all four nucleotides fully-labeled (not shown). While it is possible to achieve a similar result using a fully labeled ATP only sample, one bond 13C–13C and 13C–15N couplings quickly degrade the quality of the spectrum. With a view to design a new NOESY assignment protocol, we synthesized RNA samples that maximize the information content of their spectra while simultaneously alleviating spectral overlap.
NOESY resonance assignments: an alternating 13C-1′ and 13C-2′ labeling scheme
The classic approach to assign resonances in a helical stretch of an RNA employs a NOESY walk methodology (41,94). Protons close in space (<5 Å) can produce cross peaks in a NOESY spectrum indicative of a through-space transfer of longitudinal magnetization between the adjacent nuclei. For nucleotides in a helix, the protons attached to the C8/C6 of the base and the C1′/C2′ of the sugar fulfill this distance requirement. By labeling all nucleotides at the C1′ and C8/C6, the base and ribose of adjacent nucleotides can be connected. However, as the size of the RNAs increases, spectral crowding becomes especially pronounced in the sugar resonances and may lead to incorrect peak assignments. In the past the solution to this problem might have been to remove these resonances by transcribing the RNA with unlabeled cytosine. While the spectra would then be simplified, the NOESY walk is broken in any helical stretches that contain cytosine. Here we propose an alternative approach. Instead of transcribing the RNA with unlabeled cytosine, a different labeling pattern such as 2′,8-13C could be used. In this way, the NOESY walk is preserved while removing the overlapping C1′ resonances.
Thus, by combining our previous work on pyrimidine synthesis with our current purine synthesis, we can make RNAs that provide labeling patterns that enable an important advance in NOESY assignment strategies (41,94). For the conventional uniformly labeled samples, the C2′ and C1′ resonances are both extremely crowded as discussed above. In a traditional NOESY walk all nucleotides or various permutations are fully-labeled. NOE crosspeaks between protons attached to the C1′ and C2′ and the C8/C6 of the same and previous nucleotides are observed for helical regions. As we have illustrated, spectral crowding can severely hinder this assignment process. However, by labeling the base of C6/C8 of each nucleotide and alternating the label on the ribose between C1′ and C2′ a sample is created that not only distinguishes the purines from the pyrimidines but also the A–U and the G–C pairs. We first made nucleotide specific labeled samples, and from the overlaid spectra, we could immediately tell that C/U and G/A showed more spectral overlap in their sugar resonances. Thus it was necessary to label C/G on their C1′ carbons and U/A on their C2′ carbons. As a proof of concept we have labeled the bacterial A-site RNA with 1′,6-13C-1,3-15N CTP, 2′,6-13C-1,3-15N UTP, 1′,8-13C GTP, and 2′,8-13C ATP (Figure 5). By combining this alternative labeling strategy with NOESY experiments that allow for filtering/editing of 1H cross-peaks based on the attached carbons (12C versus 13C), we can create a unique and powerful system to assign resonances without ambiguity (94–96). For ambiguous or overlapped cross-peaks, we utilized 3D 13C-NOESY-HSQC experiments. This alternating ribose pattern allowed us to unambiguously assign helical regions of RNA. In future work, we will streamline this methodology for use in larger RNAs.
Figure 5.
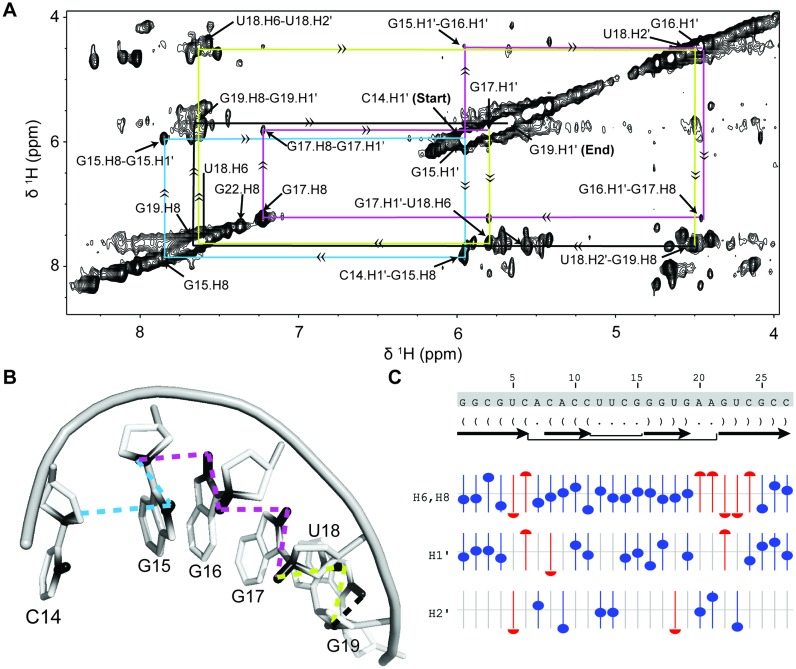
(A) NOESY walk of the bacterial A-site RNA using alternatively labeled nucleotides. Starting at the H1′ of C14, the connectivity from the sugar H1′/H2′ to C8/C6 of the N+1 base enables sequential assignment all the way to the H1′ of G19. This allowed the consecutive assignment of 6 residues present in the helical environment. Connectivities follow the placed arrows and move sequentially from cyan to magenta to yellow to black. (B) Pymol representation of the NOESY walk using the same coloring scheme as used in the NOESY walk (PDB: 1A3M (98)). (C) Predicted versus actual chemical shift values for the assigned residues as determined by NMRViewJ (One Moon Scientific). The offset from the central line represents how far the assigned resonance is from its predicted value. Blue circles represent resonances that are less than 0.1 ppm from predicted values, red circles have predicted ppm more than 0.1 ppm away from observed chemical shift.
The resulting assignment matched those previously determined (8). In situations where there is significant overlap in the base region, samples in which certain bases are unlabeled or even deuterated can be made allowing for the assignment bottleneck to be quickly circumvented.
CONCLUSIONS
This work extends our previous synthesis of pyrimidine (30,31) to purine nucleotides. We have shown that the ability to easily synthesize a variety of purine and pyrimidine nucleotides facilitates the study of large RNAs. These nucleotides are suitable for use in three key aspects of RNA NMR structural biology: assignment, structural and dynamics measurements.
The first advantage of these new site-specific labels is the potential for new assignment schemes. We have coupled alternate labeling of either C1′ or C2′ labeled ribose to C8 labeled purine bases. These combinations have allowed us to develop a new NOESY assignment strategy that benefits from reduced spectral crowding. This new strategy takes advantage of the large proton chemical shift differences between the C1′ and C2′ ribose carbons. By using an alternating C1′ and C2′ pattern with labeled bases, the NOESY spectrum is greatly simplified without compromising the information content present. Since all nucleotides are labeled, a complete NOESY walk is possible in helical regions. Additionally if the purines and pyrimidines labeled with C1′ and C2′ enrichment are reversed, orthogonal data is generated that can confirm the previous assignment.
The second advantage of these new labels is that the removal of the strong 13C J-coupling leads to substantial improvements in signal intensity in the protonated base C6 and C8 positions. Additionally, these isolated spin pairs have facilitated the measurement of μs-ms dynamics using CPMG and CEST pulse sequences without the complications of large carbon-carbon couplings. Finally with these isolated ‘two-spin’ labels, these couplings need not be explicitly taken into account in the data analysis of CEST profiles as required in previous studies using uniformly labeled RNA or protein (33,34), but also be able to probe more useful sites such as pyrimidine C5 and C6 sites. It is important to note that other dispersion experiments such as R1ρ will also benefit from using RNAs transcribed with isolated spin systems. The improvements we see from TROSY based pulse sequences scales with the size of the RNA.
A price to pay for not using spectroscopic tools to minimize the 13C-13C coupling problem is that the number of probe sites is now limited to the labeled sites. Nonetheless, it still remains useful because our method allows for very rapid accumulation of chemical shifts, a set of parameters that are easily and accurately measured and available at very early stages of NMR data analyses. Thus by measuring various chemical shifts (H1′/C1′, H2′/C2′, H5′,H5″/C5′, H2/C2, C4, H5/C5, H6/C6, H8/C8, N1, N3, N7, N9), we think the availability of such parameters will facilitate chemical shift based structure calculations of RNA, especially for constructing structural models for transiently and sparsely populated RNA states as has been done, so far, only for proteins (97).
We, therefore, anticipate that as the size of the RNAs under investigation becomes greater than 100 nucleotides the combined use of these selective labels with TROSY- and HMQC-based pulse elements will be critical for advancing NMR for the study of the structure and dynamics of a large number of new and interesting RNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Dr Q. Zhang for providing CEST fitting software that was adapted to analyze CPMG data.
FUNDING
National Institute of General Medical Sciences [P50GM103297 to B.A.J, T.K.D., and V.M.D.]; National Institute of General Medical Sciences [RO1AI104711 to V.M.D]; National Science Foundation for NMR instrumentation [DBI1040158 to T.K.D.]; Austrian Sciences Fund [I844 to C.K.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Winkler W., Nahvi A., Breaker R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 2.Baker J.L., Sudarsan N., Weinberg Z., Roth A., Stockbridge R.B., Breaker R.R. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen B., Zuo X., Wang Y.-X., Dayie T.K. Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy. Nucleic Acids Res. 2012;40:3117–3130. doi: 10.1093/nar/gkr1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reining A., Nozinovic S., Schlepckow K., Buhr F., Fürtig B., Schwalbe H. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature. 2013;499:355–359. doi: 10.1038/nature12378. [DOI] [PubMed] [Google Scholar]
- 5.Gao F., Kasprzak W.K., Szarko C., Shapiro B.A., Simon A.E. The 3′ untranslated region of pea enation mosaic virus contains two T-shaped, ribosome-binding, cap-independent translation enhancers. J. Virol. 2014;88:11696–11712. doi: 10.1128/JVI.01433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heng X., Kharytonchyk S., Garcia E.L., Lu K., Divakaruni S.S., LaCotti C., Edme K., Telesnitsky A., Summers M.F. Identification of a Minimal Region of the HIV-1 5-Leader Required for RNA Dimerization, NC Binding, and Packaging. J. Mol. Biol. 2012;417:224–239. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng X., Chugh J., Casiano-Negroni A., Al-Hashimi H.M., Brooks C.L. III. Flipping of the Ribosomal A-Site Adenines Provides a Basis for tRNA Selection. J. Mol. Biol. 2014;426:3201–3213. doi: 10.1016/j.jmb.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dethoff E.A., Petzold K., Chugh J., Casiano-Negroni A., Al-Hashimi H.M. Visualizing transient low-populated structures of RNA. Nature. 2012;491:724–728. doi: 10.1038/nature11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenwick R.B., van den Bedem H., Fraser J.S., Wright P.E. Integrated description of protein dynamics from room-temperature X-ray crystallography and NMR. Proc. Natl. Acad Sci. U.S.A. 2014;111:E445–E454. doi: 10.1073/pnas.1323440111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westenhoff S., Nazarenko E., Malmerberg E., Davidsson J., Katona G., Neutze R. Time-resolved structural studies of protein reaction dynamics: A smorgasbord of X-ray approaches. Acta Crystallogr. A. 2010;66:207–219. doi: 10.1107/S0108767309054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson J.E., Jr, Julien K.R., Hoogstraten C.G. Alternate-site isotopic labeling of ribonucleotides for NMR studies of ribose conformational dynamics in RNA. J. Biomol. NMR. 2006;35:261–274. doi: 10.1007/s10858-006-9041-x. [DOI] [PubMed] [Google Scholar]
- 12.Thakur C., Sama J., Jackson M., Chen B., Dayie T. Selective 13C labeling of nucleotides for large RNA NMR spectroscopy using an E. coli strain disabled in the TCA cycle. J. Biomol. NMR. 2010;48:179–192. doi: 10.1007/s10858-010-9454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thakur C., Sama J., Jackson M., Chen B., Dayie T.K. Selective 13C labeling of nucleotides for large RNA NMR spectroscopy using an E. coli strain disabled in the TCA cycle. J. Biomol. NMR. 2010;48:179–192. doi: 10.1007/s10858-010-9454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikonowicz E.P., Pardi A. Three-dimensional heteronuclear NMR studies of RNA. Nature. 1992;355:184–186. doi: 10.1038/355184a0. [DOI] [PubMed] [Google Scholar]
- 15.Nikonowicz E.P., Sirr A., Legault P., Jucker F.M., Baer L.M., Pardi A. Preparation of 13C and 15N labelled RNAs for heteronuclear multi-dimensional NMR studies. Nucleic Acids Res. 1992;20:4507–4513. doi: 10.1093/nar/20.17.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batey R. T., Inada M., Kujawinski E., Puglisi J. D., Williamson J. R. Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res. 1992;20:4515–4523. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukavsky P.J., Kim I., Otto G.A., Puglisi J.D. Structure of HCVIRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 18.D'Souza V., Dey A., Habib D., Summers M.F. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J. Mol. Biol. 2004;337:427–442. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 19.Grishaev A., Ying J., Canny M.D., Pardi A., Bax A. Solution structure of tRNA(Val) from refinement of homology model against residual dipolar coupling and SAXS data. J. Biomol. NMR. 2008;42:99–109. doi: 10.1007/s10858-008-9267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyazaki Y., Irobalieva R.N., Tolbert B.S., Smalls-Mantey A., Iyalla K., Loeliger K., D'Souza V., Khant H., Schmid M.F., Garcia E.L., et al. Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J. Mol. Biol. 2010;404:751–772. doi: 10.1016/j.jmb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo X., Wang J., Yu P., Eyler D., Xu H., Starich M.R., Tiede D.M., Simon A.E., Kasprzak W., Schwieters C.D., et al. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′ UTR of turnip crinkle virus. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1385–1390. doi: 10.1073/pnas.0908140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burke J.E., Sashital D.G., Zuo X., Wang Y.-X., Butcher S.E. Structure of the yeast U2/U6 snRNA complex. RNA. 2012;18:673–683. doi: 10.1261/rna.031138.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller S.B., Yildiz F.Z., Lo J.A., Wang B., D'Souza V.M. A structure-based mechanism for tRNA and retroviral RNA remodelling during primer annealing. Nature. 2014;515:591–595. doi: 10.1038/nature13709. [DOI] [PubMed] [Google Scholar]
- 24.Bonneau E., Girard N., Lemieux S., Legault P. The NMR structure of the II-III-VI three-way junction from the Neurospora VS ribozyme reveals a critical tertiary interaction and provides new insights into the global ribozyme structure. RNA. 2015;21:1621–1632. doi: 10.1261/rna.052076.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keane S.C., Heng X., Lu K., Kharytonchyk S., Ramakrishnan V., Carter G., Barton S., Hosic A., Florwick A., Santos J., et al. Structure of the HIV-1 RNA packaging signal. Science. 2015;348:917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dayie K.T. Key labeling technologies to tackle sizeable problems in RNA structural biology. Int. J. Mol. Sci. 2008;9:1214–1240. doi: 10.3390/ijms9071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu K., Miyazaki Y., Summers M.F. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR. 2010;46:113–125. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamazaki T., Muhandiram R., Kay L.E. NMR experiments for the measurement of carbon relaxation properties in highly enriched, uniformly 13C,15N-labeled proteins: application to 13Cα carbons. J. Am. Chem. Soc. 1994;116:8266–8278. [Google Scholar]
- 29.Dayie T.K., Thakur C. Site-specific labeling of nucleotides for making RNA for high resolution NMR studies using an E. coli strain disabled in the oxidative pentose phosphate pathway. J. Biomol. NMR. 2010;47:19–31. doi: 10.1007/s10858-010-9405-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarado L.J., LeBlanc R.M., Longhini A.P., Keane S.C., Jain N., Yildiz Z.F., Tolbert B.S., D'Souza V.M., Summers M.F., Kreutz C., et al. Regio-selective chemical-enzymatic synthesis of pyrimidine nucleotides facilitates RNA structure and dynamics studies. Chembiochem. 2014;15:1573–1577. doi: 10.1002/cbic.201402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alvarado L.J., Longhini A.P., LeBlanc R.M., Chen B., Kreutz C., Dayie T.K. In: Methods Enzymol. Burke-Aguero DH, editor. Vol. 549. Academic Press; 2014. pp. 133–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vallurupalli P., Bouvignies G., Kay L.E. A computational study of the effects of C-13-C-13 scalar couplings on C-13 CEST NMR spectra: towards studies on a uniformly C-13-labeled protein. Chembiochem. 2013;14:1709–1713. doi: 10.1002/cbic.201300230. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y., Yang D. Effects of J-couplings and unobservable minor states on kinetics parameters extracted from CEST data. J. Magn. Reson. 2014;249:118–125. doi: 10.1016/j.jmr.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Zhao B., Hansen A.L., Zhang Q. Characterizing slow chemical exchange in nucleic acids by carbon CEST and low spin-lock field R1ρ NMR spectroscopy. J. Am. Chem. Soc. 2014;136:20–23. doi: 10.1021/ja409835y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bax A., Mehlkopf A.F., Smidt J. Homonuclear broadband-decoupled absorption-spectra, with linewidths which are independent of the transverse relaxation rate. J. Magn. Reson. 1979;35:167–169. [Google Scholar]
- 36.Bax A., Freeman R. Investigation of complex networks of spin-spin coupling by two-dimensional NMR. J. Magn. Reson. 1981;44:542–561. [Google Scholar]
- 37.Grzesiek S., Bax A. Improved 3D triple-resonance NMR techniques applied to a 31-KDa protein. J. Magn. Reson. 1992;96:432–440. [Google Scholar]
- 38.Vandeven F.J.M., Philippens M.E.P. Optimization of constant-time evolution in multidimensional NMR experiments. J. Magn. Reson. 1992;97:637–644. [Google Scholar]
- 39.Kupce E., Wagner G. Multisite band-selective decoupling in proteins. J. Magn. Reson., Ser. B. 1996;110:309–312. doi: 10.1006/jmrb.1996.0048. [DOI] [PubMed] [Google Scholar]
- 40.Brutscher B., Boisbouvier J., Kupce E., Tisne C., Dardel F., Marion D., Simorre J.P. Base-type-selective high-resolution C-13 edited NOESY for sequential assignment of large RNAs. J. Biomol. NMR. 2001;19:141–151. doi: 10.1023/a:1008340210079. [DOI] [PubMed] [Google Scholar]
- 41.Dayie K.T. Resolution enhanced homonuclear carbon decoupled triple resonance experiments for unambiguous RNA structural characterization. J Biomol. NMR. 2005;32:129–139. doi: 10.1007/s10858-005-5093-6. [DOI] [PubMed] [Google Scholar]
- 42.Dayie K.T., Wagner G. Carbonyl carbon probe of local mobility in C-13,N-15-enriched proteins using high-resolution nuclear magnetic resonance. J. Am. Chem. Soc. 1997;119:7797–7806. [Google Scholar]
- 43.Johnson J.E., Jr, Julien K.R., Hoogstraten C.G. Alternate-site isotopic labeling of ribonucleotides for NMR studies of ribose conformational dynamics in RNA. J. Biomol. NMR. 2006;35:261–274. doi: 10.1007/s10858-006-9041-x. [DOI] [PubMed] [Google Scholar]
- 44.Johnson J.E., Hoogstraten C.G. Extensive backbone dynamics in the GCAA RNA tetraloop analyzed using 13C NMR spin relaxation and specific isotope labeling. J. Am. Chem. Soc. 2008;130:16757–16769. doi: 10.1021/ja805759z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boisbouvier J., Wu Z., Ono A., Kainosho M., Bax A. Rotational diffusion tensor of nucleic acids from 13C NMR relaxation. J. Biomol. NMR. 2003;27:133–142. doi: 10.1023/a:1024931619957. [DOI] [PubMed] [Google Scholar]
- 46.Hansen A.L., Al-Hashimi H.M. Dynamics of large elongated RNA by NMR carbon relaxation. J. Am. Chem. Soc. 2007;129:16072–16082. doi: 10.1021/ja0757982. [DOI] [PubMed] [Google Scholar]
- 47.Peng J.W., Thanabal V., Wagner G. 2D hetereonuclear NMR measurements of spin-lattice relaxation-times in the rotating frame of X nuclei in heteronuclear HX spin systems. J. Magn. Reson. 1991;94:82–100. [Google Scholar]
- 48.Hoogstraten C.G., Wank J.R., Pardi A. Active site dynamics in the lead-dependent ribozyme. Biochem. 2000;39:9951–9958. doi: 10.1021/bi0007627. [DOI] [PubMed] [Google Scholar]
- 49.Latham M.P., Brown D.J., McCallum S.A., Pardi A. NMR methods for studying the structure and dynamics of RNA. Chembiochem. 2005;6:1492–1505. doi: 10.1002/cbic.200500123. [DOI] [PubMed] [Google Scholar]
- 50.Teilum K., Brath U., Lundstrom P., Akke M. Biosynthetic C-13 labeling of aromatic side chains in proteins for NMR relaxation measurements. J. Am. Chem. Soc. 2006;128:2506–2507. doi: 10.1021/ja055660o. [DOI] [PubMed] [Google Scholar]
- 51.Xue Y., Kellogg D., Kimsey I.J., Sathyamoorthy B., Stein Z.W., McBrairty M., Al-Hashimi H.M. Characterizing RNA excited states using NMR relaxation dispersion. Methods Enzymol. 2015;558:39–73. doi: 10.1016/bs.mie.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiarparin E., Pelupessy P., Bodenhausen G. Selective cross-polarization in solution state NMR. Mol. Phys. 1998;95:759–767. [Google Scholar]
- 53.Pelupessy P., Chiarparin E., Bodenhausen G. Excitation of selected proton signals in NMR of isotopically labeled macromolecules. J. Magn. Reson. 1999;138:178–181. doi: 10.1006/jmre.1999.1715. [DOI] [PubMed] [Google Scholar]
- 54.Pelupessy P., Chiarparin E. Hartmann-Hahn polarization transfer in liquids: An ideal tool for selective experiments. Conc. Magn. Reson. 2000;12:103–124. [Google Scholar]
- 55.Ferrage F., Eykyn T.R., Bodenhausen G. Frequency-switched single-transition cross-polarization: A tool for selective experiments in biomolecular NMR. Chemphyschem. 2004;5:76–84. doi: 10.1002/cphc.200300905. [DOI] [PubMed] [Google Scholar]
- 56.Hansen A.L., Nikolova E.N., Casiano-Negroni A., Al-Hashimi H.M. Extending the range of microsecond-to-millisecond chemical exchange detected in labeled and unlabeled nucleic acids by selective carbon R-1 rho NMR spectroscopy. J. Am. Chem. Soc. 2009;131:3818–3819. doi: 10.1021/ja8091399. [DOI] [PubMed] [Google Scholar]
- 57.Massi F., Johnson E., Wang C.Y., Rance M., Palmer A.G. NMR R-1 rho rotating-frame relaxation with weak radio frequency fields. J. Am. Chem.Soc. 2004;126:2247–2256. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- 58.Korzhnev D.M., Orekhov V.Y., Kay L.E. Off-resonance R1(p) NMR studies of exchange dynamics in proteins with low spin-lock fields: An application to a fyn SH3 domain. J. Am. Chem. Soc. 2005;127:713–721. doi: 10.1021/ja0446855. [DOI] [PubMed] [Google Scholar]
- 59.Nikolova E.N., Gottardo F.L., Al-Hashimi H.M. Probing transient hoogsteen hydrogen bonds in canonical duplex DNA using NMR relaxation dispersion and single-atom substitution. J. Am. Chem. Soc. 2012;134:3667–3670. doi: 10.1021/ja2117816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen A.L., Bouvignies G., Kay L.E. Probing slowly exchanging protein systems via C-13α-CEST: monitoring folding of the Im7 protein. J. Biomol. NMR. 2013;55:279–289. doi: 10.1007/s10858-013-9711-4. [DOI] [PubMed] [Google Scholar]
- 61.Vallurupalli P., Kay L.E. Probing slow chemical exchange at carbonyl sites in proteins by chemical exchange saturation transfer NMR spectroscopy. Angew. Chem. Int. 2013;52:4156–4159. doi: 10.1002/anie.201209118. [DOI] [PubMed] [Google Scholar]
- 62.Bouvignies G., Vallurupalli P., Kay L.E. Visualizing side chains of invisible protein conformers by solution NMR. J. Mol. Biol. 2014;426:763–774. doi: 10.1016/j.jmb.2013.10.041. [DOI] [PubMed] [Google Scholar]
- 63.Zhou Y., Yang D. C-13α CEST experiment on uniformly C-13-labeled proteins. J. Biomol. NMR. 2015;61:89–94. doi: 10.1007/s10858-014-9888-1. [DOI] [PubMed] [Google Scholar]
- 64.Kloiber K., Spitzer R., Tollinger M., Konrat R., Kreutz C. Probing RNA dynamics via longitudinal exchange and CPMG relaxation dispersion NMR spectroscopy using a sensitive 13C-methyl label. Nucleic Acids Res. 2011;39:4340–4351. doi: 10.1093/nar/gkq1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wunderlich C.H., Spitzer R., Santner T., Fauster K., Tollinger M., Kreutz C. Synthesis of (6–13C)pyrimidine nucleotides as spin-labels for RNA dynamics. J. Am. Chem. Soc. 2012;134:7558–7569. doi: 10.1021/ja302148g. [DOI] [PubMed] [Google Scholar]
- 66.Pervushin K., Riek R., Wider G., Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miclet E., Williams D.C., Clore G.M., Bryce D.L., Boisbouvier J., Bax A. Relaxation-optimized NMR spectroscopy of methylene groups in proteins and nucleic acids. J. Am. Chem. Soc. 2004;126:10560–10570. doi: 10.1021/ja047904v. [DOI] [PubMed] [Google Scholar]
- 68.Long D., Bouvignies G., Kay L.E. Measuring hydrogen exchange rates in invisible protein excited states. Proc. Natl. Acad. Sci. U.S.A. 2014;111:8820–8825. doi: 10.1073/pnas.1405011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weininger U., Brath U., Modig K., Teilum K., Akke M. Off-resonance rotating-frame relaxation dispersion experiment for C-13 in aromatic side chains using L-optimized TROSY-selection. J. Biomol. NMR. 2014;59:23–29. doi: 10.1007/s10858-014-9826-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weininger U., Respondek M., Akke M. Conformational exchange of aromatic side chains characterized by L-optimized TROSY-selected C-13 CPMG relaxation dispersion. J. Biomol. NMR. 2012;54:9–14. doi: 10.1007/s10858-012-9656-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weininger U., Diehl C., Akke M. C-13 relaxation experiments for aromatic side chains employing longitudinal- and transverse-relaxation optimized NMR spectroscopy. J. Biomol. NMR. 2012;53:181–190. doi: 10.1007/s10858-012-9650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schultheisz H.L., Szymczyna B.R., Scott L.G., Williamson J.R. Pathway engineered enzymatic de novo purine nucleotide synthesis. ACS Chem. Biol. 2008;3:499–511. doi: 10.1021/cb800066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tolbert T.J., Williamson J.R. Preparation of specifically deuterated RNA for NMR studies using a combination of chemical and enzymatic synthesis. J. Am. Chem. Soc. 1996;118:7929–7940. [Google Scholar]
- 74.Tolbert T.J., Williamson J.R. Preparation of specifically deuterated and 13C-labeled RNA for NMR studies using enzymatic synthesis. J. Am. Chem. Soc. 1997;119:12100–12108. [Google Scholar]
- 75.Nikonowicz E.P., Michnicka M., Kalurachchi K., Dejong E. Preparation and characterization of a uniformly 2H/15N-labeled RNA oligonucleotide for NMR studies. Nucleic Acids Res. 1997;25:1390–1396. doi: 10.1093/nar/25.7.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dayie K.T., Tolbert T.J., Williamson J.R. 3D C(CC)H TOCSY experiment for assigning protons and carbons in uniformly 13C- and selectively 2H-labeled RNA. J. Mag. Reson. 1998;130:97–101. doi: 10.1006/jmre.1997.1286. [DOI] [PubMed] [Google Scholar]
- 77.Scott L.G., Geierstanger B.H., Williamson J.R., Hennig M. Enzymatic synthesis and 19F NMR studies of 2-fluoroadenine-substituted RNA. J. Am. Chem. Soc. 2004;126:11776–11777. doi: 10.1021/ja047556x. [DOI] [PubMed] [Google Scholar]
- 78.Wennefors C.K., Dobrikov M.I., Xu Z., Li P., Shaw B.R. Stereospecificity, substrate, and inhibitory properties of nucleoside diphosphate analogs for creatine and pyruvate kinases. Bioorg. Chem. 2008;36:169–177. doi: 10.1016/j.bioorg.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 79.McLeish M.J., Kenyon G.L. Relating structure to mechanism in creatine kinase. Crit. Rev. Biochem. Mol. 2005;40:1–20. doi: 10.1080/10409230590918577. [DOI] [PubMed] [Google Scholar]
- 80.Fiala R., Czernek J., Sklenar V. Transverse relaxation optimized triple-resonance NMR experiments for nucleic acids. J. Biomol. NMR. 2000;16:291–302. doi: 10.1023/a:1008388400601. [DOI] [PubMed] [Google Scholar]
- 81.Brutscher B., Boisbouvier J., Pardi A., Marion D., Simorre J.P. Improved sensitivity and resolution in 1H-13C NMR experiments of RNA. J. Am. Che. Soc. 1998;120:11845–11851. [Google Scholar]
- 82.Wijmenga S.S., van Buuren B.N.M. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Mag. Res. Sp. 1998;32:287–387. [Google Scholar]
- 83.Davis D.G., Perlman M.E., London R.E. Direct measurements of the dissociation-rate constant for inhibitor-enzyme complexes via the T1ρ and T2 (CPMG) methods. J. Magn. Res. Ser. B. 1994;104:266–275. doi: 10.1006/jmrb.1994.1084. [DOI] [PubMed] [Google Scholar]
- 84.Carr H.Y., Purcell E.M. Effect of diffusion on free precession in nuclear magnetic resonance experiments. Phys. Rev. 1954;94:630–638. [Google Scholar]
- 85.Meiboom S., Gill D. Modified spin-echo method for measuring nuclear relation times. Rev. Sci. Instrum. 1958;29:688–691. [Google Scholar]
- 86.Hahn E.L. Spin Echoes. Phys. Rev. 1950;80:580–594. [Google Scholar]
- 87.Carver J.P., Richards R.E. A general two-site solution for the chemical exchange produced dependence of T2 upon the carr-Purcell pulse separation. J. Magn. Reson. (1969) 1972;6:89–105. [Google Scholar]
- 88.Palmer A.G. III, Kroenke C.D., Patrick Loria J. Methods Enzymol. Vol. 339. Academic Press; 2001. pp. 204–238. [DOI] [PubMed] [Google Scholar]
- 89.Lundström P., Hansen D.F., Kay L. Measurement of carbonyl chemical shifts of excited protein states by relaxation dispersion NMR spectroscopy: comparison between uniformly and selectively 13C labeled samples. J. Biomol. NMR. 2008;42:35–47. doi: 10.1007/s10858-008-9260-4. [DOI] [PubMed] [Google Scholar]
- 90.Korzhnev D.M., Salvatella X., Vendruscolo M., Di Nardo A.A., Davidson A.R., Dobson C.M., Kay L.E. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 91.Kaul M., Barbieri C.M., Pilch D.S. Aminoglycoside-induced reduction in nucleotide mobility at the ribosomal RNA A-site as a potentially key determinant of antibacterial activity. J. Am. Chem. Soc. 2006;128:1261–1271. doi: 10.1021/ja056159z. [DOI] [PubMed] [Google Scholar]
- 92.Shandrick S., Zhao Q., Han Q., Ayida B.K., Takahashi M., Winters G.C., Simonsen K.B., Vourloumis D., Hermann T. Monitoring molecular recognition of the ribosomal decoding site. Angew. Chem. Int. 2004;116:3239–3244. doi: 10.1002/anie.200454217. [DOI] [PubMed] [Google Scholar]
- 93.Vaiana A.C., Sanbonmatsu K.Y. Stochastic gating and drug–ribosome interactions. J. Mol. Biol. 2009;386:648–661. doi: 10.1016/j.jmb.2008.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peterson R.D., Theimer C.A., Wu H.H., Feigon J. New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA-protein complexes. J. Biomol. NMR. 2004;28:59–67. doi: 10.1023/B:JNMR.0000012861.95939.05. [DOI] [PubMed] [Google Scholar]
- 95.SantaLucia J., Shen L.X., Cai Z., Lewis H., Tinoco I. Synthesis and NMR of RNA with Selective isotopic enrichment in the bases. Nucleic Acids Res. 1995;23:4913–4921. doi: 10.1093/nar/23.23.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Breeze A.L. Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog. Nucl. Mag. Res. Spec. 2000;36:323–372. [Google Scholar]
- 97.Sekhar A., Kay L.E. NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12867–12874. doi: 10.1073/pnas.1305688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fourmy D., Yoshizawa S., Puglisi J.D. Paromomycin binding induces a local conformational change in the A-site of 16 s rRNA1. J. Mol. Biol. 1998;277:333–345. doi: 10.1006/jmbi.1997.1551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.