Abstract
Loss of ‘Survival of Motor Neurons’ (SMN) leads to spinal muscular atrophy (SMA), a disease characterized by degeneration of spinal cord alpha motor neurons, resulting in muscle weakness, paralysis and death during early childhood. SMN is required for assembly of the core splicing machinery, and splicing defects were documented in SMA. We previously uncovered that Coactivator-Associated Methyltransferase-1 (CARM1) is abnormally up-regulated in SMA, leading to mis-regulation of a number of transcriptional and alternative splicing events. We report here that CARM1 can promote decay of a premature terminating codon (PTC)-containing mRNA reporter, suggesting it can act as a mediator of nonsense-mediated mRNA decay (NMD). Interestingly, this pathway, while originally perceived as solely a surveillance mechanism preventing expression of potentially detrimental proteins, is now emerging as a highly regulated RNA decay pathway also acting on a subset of normal mRNAs. We further show that CARM1 associates with major NMD factor UPF1 and promotes its occupancy on PTC-containing transcripts. Finally, we identify a specific subset of NMD targets that are dependent on CARM1 for degradation and that are also misregulated in SMA, potentially adding exacerbated targeting of PTC-containing mRNAs to the already complex array of molecular defects associated with this disease.
INTRODUCTION
Autosomal-recessive proximal spinal muscular atrophy (SMA) is a progressive neuromuscular disorder characterized by the selective loss or dysfunction of α-motoneurons in the anterior horn of the spinal cord (1). With a prevalence of at least 1 in 10 000 live births and a carrier frequency of ∼1 in 40, SMA is amongst the leading genetic cause of infant mortality (2,3). Based on the time of onset of the disease and its severity, SMA can be divided into five types, with Type 0 and Type I (Werdnig–Hoffman syndrome) being the most severe forms (4). Patients with severe Type I SMA will usually develop weakness of the proximal muscles of the trunk and body, ultimately leading to muscle atrophy and death from respiratory distress within ∼2 years of age, depending on the choice of palliative care (5). SMA is caused by disruption of the survival of motor neuron (SMN1) gene (6). In humans, a second copy of the SMN gene exists but naturally harbours a non-polymorphic C→T transition that interferes with the normal splicing of exon 7, resulting in the expression of a truncated and unstable form of the protein (7–10). The low level of full-length functional SMN protein produced in human SMA patients is sufficient to sustain embryonic development and survival of all cells, except lower motoneurons, which seem to have a lower ‘tolerance threshold’ for SMN levels, a phenomenon which still remains one of foremost questions in the field. SMN exists in cells as part of a stable ∼50S macromolecular complex consisting of at least eight tightly associated components that include Gemins 2–8 and unrip (11–13). The best understood function for the core SMN complex is its essential role in promoting the efficiency and specificity of the cytoplasmic assembly of Sm proteins and U snRNAs into small nuclear ribonucleoprotein particles (snRNPs), the core components of the pre-mRNA splicing machinery (11–14). Accordingly, a number of studies have now reported lower levels of specific U snRNAs and widespread splicing defects in SMA tissues, including a recently uncovered feedback loop affecting the splicing of SMN2 exon 7 and misregulation of a subset of U12-dependent introns (15–24).
Parallel work also supports additional and distinct roles for SMN in motoneurons, where it localizes to so-called RNA granules along axonal processes and interacts with a number of RNA binding proteins such as FUS/TLS, IMP1/ZBP, TDP-43, hnRNP R/Q, FMRP, KSRP and HuD (25–36). RNA granules are responsible for the transport, along microtubules, of specific mRNAs in dendrites and axons, and contribute to the regulation of mRNA stability and local translation at synapses and growth cones, which in turn is crucial for neuronal differentiation, axon outgrowth and synaptic function (37–39). Recent studies from our group and others have provided evidence suggesting SMN is somehow required for the proper assembly of RNA granules (25–27). Therefore, defects in RNA granules assembly and/or function may account for the various neurite outgrowth and synaptic maturation, stability and functional phenotypes documented in SMA (30,32,40–49). It was also demonstrated for the first time recently that local translation of β-actin mRNA was deregulated in motorneurons from a severe mouse model of SMA (50), although a direct involvement of SMN in this process was not investigated.
In a recent study (51), we demonstrated that SMN co-fractionates with polyribosomes and represses translation in vitro. In this work, we further identified the protein arginine methyltransferase CARM1 as a target that is repressed by SMN at the translational level in motoneuron-derived MN-1 cells. Accordingly, we documented that CARM1 is abnormally up-regulated at the protein level in spinal cord tissue from SMA mice and in severe Type I SMA patient cells. CARM1 is best-known as a transcriptional regulator through its methylation of histones and transcription factors/co-regulators (52–55), but has also emerged as a factor able to influence post-transcriptional processes, including alternative pre-mRNA splicing and mRNA stability, through methylation of specific splicing factors and RNA-binding proteins (27,56–61). Based on these observations, we used genome-wide exon array technology to identify specific transcriptional and splicing targets of CARM1 in motoneuron-derived MN-1 cells and found that a number of these targets were also misregulated in a SMA cell culture model (51), strongly suggesting that CARM1 up-regulation contributes to altered gene expression profiles observed in the pathology.
Nonsense-mediated mRNA decay (NMD) is a surveillance mechanism that in addition to preventing production of truncated protein isoforms with potentially noxious consequences for the cell (62), has also been shown to act on numerous mRNAs thereby contributing to general post-transcriptional gene regulation (63). In the present study, we report that CARM1 interacts with the essential NMD factor UPF1 in an RNA-dependent fashion and can promote NMD on a generic β-globin pre-mRNA reporter. Additionally, CARM1 was required for interaction of UPF1 with a premature termination codon (PTC)-containing mRNA. We identified a number of well established endogenous NMD targets that were misregulated in the face of modulated CARM1 levels and in SMA conditions. Since there is an aberrant upregulation of CARM1 in SMA, we believe that this (and potentially other mechanisms) induce NMD misregulation of several key mRNAs that could contribute to the SMA pathophysiology.
MATERIALS AND METHODS
Cell culture, treatments and transfection experiments
Motoneuron-derived Ctrl MN-1 cells, SMN stable knockdown MN-1 cells (shSMN) and CARM1 stable knockdown MN-1 cells (shCARM1) are described previously (27). Cells were cultured in DMEM (GIBCO) supplemented with 10% fetal bovine serum and maintained in 2 μg/ml of puromycin. For transient transfection, Lipofectamine and Plus Reagents from Invitrogen were used. Cells were harvested for biochemical analyses 48 h after transfection. For the USPL1 depletion experiment, the pGIPZ (OpenBiosystems) control vector and the pGIPZ-shRNA-UPF1 (RHS4430–101031100, V3LHS-352951, OpenBiosystems) were transiently transfected. For cycloheximide (CHX) treatment, cells were treated 4 h with 10 μg/ml. For actinomycin D treatment, cells were incubated for the times indicated in the Figure with 5 μg/ml of the drug. For wortmannin treatment, cells were treated 6 h with 5 μM. The β-Globin WT and MT (NMD) reporters (64) were a kind gift from Dr Adrian R Krainer (Cold Spring Harbor Laboratory, USA) and the CARM1-WT and CARM1-E266Q plasmids (57) used for the rescue experiments were a generous gift from Dr Mark T. Bedford (University of Texas M.D. Anderson Cancer Center, USA).
Animals
Spinal cord tissue was harvested from 1 to 3 month old C57 Black 6 SMN+/− mice (obtained from Drs. Alex MacKenzie and Rashmi Kothary), flash frozen in liquid nitrogen and stored at –80°C.
RNA purification, RT-PCR and RT-qPCR
RNA was extracted by using Trizol (Invitrogen) and treated with DNase I (DNAfree, Ambion). Reverse-transcription (RT) was done by using AMV reverse transcriptase (Promega) and random primers. After completion of the RT reaction, cDNA samples were diluted to a final concentration of 2.5 ng/μl. 5 μl of cDNA samples were used per 25 μl PCR or qPCR reactions. PCR was performed by using GoTaq Flexi DNA Polymerase (Promega) and Quantitative PCR was performed by using iQ SYBR Green Supermix (BioRad) on a Chromo4 Real-Time Detector (BioRad). The relative amounts of cDNA targets in samples were determined on the basis of the threshold cycle for each PCR product (Ct). All semi-quantitative RT- and RT-qPCR primers used in this study and resulting PCR amplicons are shown in Supplementary Table S2.
Co- and RNA-immunoprecipitation experiments
Proteins were extracted from cells collected from 100 mm plates. Cells were first washed in 1× PBS and then incubated in lysis buffer (50 mM Tris-HCl pH 7.5; 150 mM NaCl; 1% NP40; 0.5% Sodium Deoxycholate; and complete protease inhibitor cocktail from Roche Applied Sciences). Immunoprecipitations were done by using Protein A/G PLUS-Agarose beads (sc-2003, Santa Cruz) and 3 μg of specific antibody or 3 μg of control IgG (sc-2027, Santa Cruz). After an overnight incubation, beads were washed four times in lysis buffer. Subsequently, samples were subjected to SDS-PAGE and western blotting analysis. For RNase A treatment (Qiagen, 19101), the cell pellets were first incubated with lysis buffer. Then, RNase A (1 μg/ml) was added to supernatants and the samples were incubated for 1 h at 37°C prior to realize an immunoprecipitation with an UPF1 antibody. RNA-immunoprecipations were performed as described previously (27). RNA was immunoprecipitated using 4 μg of UPF1 antibody and 4 μg of control rabbit IgG (sc-2027, Santa Cruz).
Immunoblotting
The following antibodies were used: CARM1 (A300–421A, Bethyl Laboratories); GAPDH (MMS-580S, Covance); β-Actin (sc-47778, Santa Cruz Biotechnology); Tubulin (T6199, Sigma-Aldrich); UPF1 (07–1014, Millipore); UPF2 (D3B10, Cell Signaling); UPF3b (sc-48800, Santa Cruz Biotechnology); Magoh (Ab10686, Abcam); eIF4A3 (Ab32485, Abcam); RBM8a (NB100–55326, Novus Biologicals); Casc3 (LS-C100827, Lifespan Biosciences); Asparagine Synthetase (sc365809, Santa Cruz Biotechnology); Arc (sc15325, Santa Cruz Biotechnology); GADD45a (sc-796, Santa Cruz Biotechnology). Quantitative analyses were done with the ImageJ software.
Statistical analysis
For statistical analysis, unpaired t-test was used. However, when three or more groups are compared, a one-way ANOVA and post hoc Tukey analysis was used. The minimum α-level of significance was set at 0.05. Data are presented as means ± SEM. In the figures, a single asterisk shows P < 0.05, a double asterisk shows P < 0.01, a triple asterisk shows P < 0.001. Analyses were realized with Graphpad Prism 6 software.
RESULTS
Aberrant splicing of USPL1 pre-mRNA is observed in human SMA patients cells
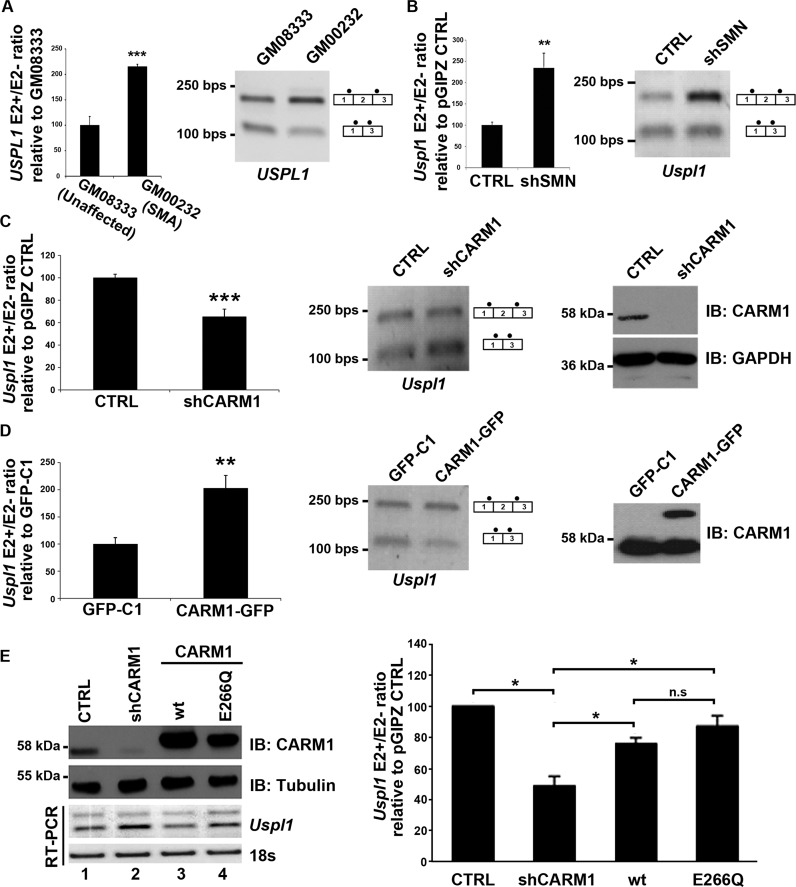
A number of studies have assessed genome-wide gene expression profiles in various models of SMA (15,20,22,65–66), but intriguingly, very little overlap has been found between these studies. Amongst the handful of genes identified as misregulated in the pathology is the ubiquitin-specific protease-like 1 (USPL1) gene, which was recently found to code for a SUMO isopeptidase shown to localize to Cajal bodies (67). The USPL1 pre-mRNA is alternatively spliced within its 5′ end to produce isoforms including or skipping 169 bp cassette-type exon 2. It has been reported that the isoform including exon 2 (E2+) is selectively over-represented in spinal cord tissues from mouse models of severe SMA, relative to the E2- transcript (15,20,68). To determine if this aberrant splicing event is also observed in SMA patients cells, we assessed the inclusion of exon 2 in primary fibroblasts derived from a severe type I SMA patient. As shown in Figure 1A, increased inclusion of USPL1 exon 2 was observed, indicating for the first time that this alteration is present in human SMA patient cells. We next quantified by RT-qPCR the USPL1 E2+/E2- ratio in a stable motoneuron MN-1 cell line expressing either a shRNA against SMN (shSMN) or a control shRNA (27,51). As expected, depletion of SMN resulted in a relative increase in Uspl1 E2+ mRNA levels relative to E2- isoform, as described in the pathology (Figure 1B), indicating that we can recapitulate this molecular defect in a motoneuron-like cell culture model of SMA.
Figure 1.
The ratio of USPL1 splicing variants is altered in SMA and by CARM1. (A) USPL1 E2+/E2- ratio obtained with the GM00232 primary fibroblasts from a SMA patient was expressed relative to GM08333 primary fibroblasts from an unaffected control. Data are means +/− SEM (n = 4). (B) Quantification of the Uspl1 E2+/E2- ratio in the MN-1 pGIPZ CTRL cell line and MN-1 shSMN cell line which stably expressed a shRNA against SMN (shSMN). Results obtained in the shSMN condition were expressed relative to the CTRL cell line. Data are means +/−SEM (n = 5). (C) CARM1 affects the USPL1 E2+/E2- ratio. Total RNA was isolated from the MN-1 pGIPZ CTRL cell line and the MN-1 shCARM1 cell line which stably expressed a shRNA against CARM1. Uspl1 E2+/E2- ratio levels obtained in the shCARM1 cells were expressed relative to CTRL. Data are means +/− SEM (n = 6). (D) Quantification of the Uspl1 E2+/E2- ratio in the MN-1 stable cell lines expressing either the pEGFP-C1 empty vector (GFP-C1) or a CARM1-GFP construct. Data obtained in the CARM1-GFP condition were expressed relative to GFP-C1. Data are means +/− SEM (n = 6). CARM1 protein levels were assessed in the MN-1 GFP-C1 and MN-1 CARM1-GFP cell lines (upper band, CARM1-GFP). (E) MN-1 cell lines were transiently transfected with either a pGIPZ CTRL or shCARM1 plasmid for 24 h. A rescue was performed in the MN-1 shCARM1 cell line by transfecting with either the WT-CARM1 or the CARM1-E266Q mutant expression vectors for an additional 24 h. Protein lysate from transfected MN-1 lines confirm the knockdown and overexpression of CARM1 or the E266Q mutant, normalized to Tubulin. Total RNA was extracted and RT-PCR performed using Uspl1 primers to amplify both the E2+ and E2- isoforms of the mRNA. 18s RNA was also shown as a loading control. Quantification of the Uspl1 E2+/E2- ratios in the CARM1 rescue MN-1 cell lines. Uspl1 mRNA levels are shown as relative to CTRL. Data are means +/− SEM (n = 3).
CARM1 regulates the relative expression of Uspl1 alternatively spliced mRNA isoforms
Our previous work has uncovered that protein levels of the protein arginine methyltransferase CARM1 are up-regulated in spinal cord motor neurons of SMA mouse models and in severe Type I SMA patient cells (51). This study also identified aberrant gene expression and alternative splicing profiles due to CARM1 up-regulation (51). To assess if Uspl1 alternative splicing was also sensitive to CARM1 levels, the E2+/E2- ratio was quantified by RT-qPCR in a motoneuron MN-1 cell line stably expressing an shRNA against CARM1 (27,51). Strikingly, CARM1 down-regulation resulted in a decrease of the E2+/E2- ratio (Figure 1C), thus shifting the ratio opposite to what is observed in the pathology. The E2+/E2- ratio was then quantified in a stable motoneuron MN-1 cell line overexpressing a CARM1-GFP construct. Accordingly, overexpression of CARM1 led to an increased E2+/E2- ratio and thus, mirrors what is seen in the SMA pathology (Figure 1D). To insure that the observed changes were not due to non-specific and/or off-target effects, a CARM1 expression rescue experiment was performed, using either wild type (wt) CARM1 or a mutant allele (E266Q) lacking methyltransferase activity (57) (Figure 1E). First, CARM1 levels were knockdown using an shRNA and confirmed by western blot in MN-1 cell lines. RT-PCR was used to confirm the decrease in the Uspl1 E2+/E2- ratio as previously observed (Figure 1E, lanes 1 and 2), which accompanies the decrease in CARM1. As expected, re-introduction of wt CARM1 into the shCARM1 MN-1 cells rescued the Uspl1 E2+/E2- ratios, returning them towards levels observed in the control condition (Figure 1E, lane 3). Intriguingly, re-introduction of the CARM1 E266Q mutant allele rescued E2+/E2- ratios almost as efficiently as the wt allele (Figure 1E, lane 4 and quantification from n=3 experiments). Altogether, these results indicate that CARM1 can regulate the expression of Uspl1 E2+/E2- ratios, through a mechanism that is seemingly independent of its methyltransferase activity.
Skipping of Uspl1 alternative exon 2 creates a premature termination codon that elicits NMD
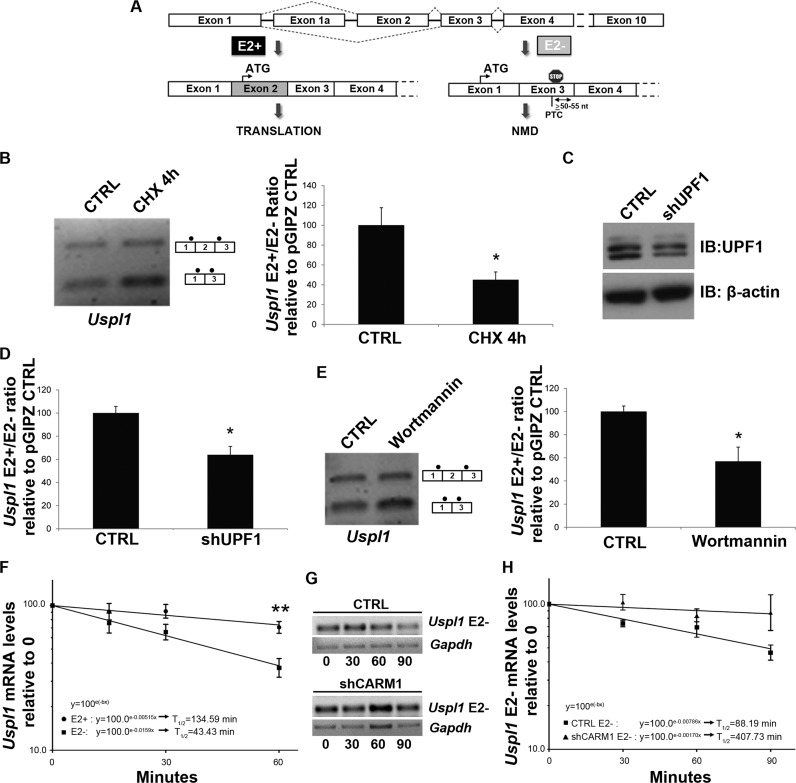
Upon closer inspection of the nucleotide sequence surrounding Uspl1 alternative exon 2, we noted that skipping of exon 2 causes a shift in the coding reading frame resulting in the creation of a premature termination codon (PTC; Figure 2A), which, based on its position >50–55 nt upstream of the exon junction is predicted to elicit the NMD pathway (69,70). We refer to this transcript as Uspl1 E2- (Uspl1–006 - ENSMUST00000121416) and the WT transcript which includes exon 2 we denote Uspl1 E2+ (Uspl1–001 - ENSMUST00000050472). We also observed an additional Uspl1 isoform containing a ‘cryptic’ exon upstream of exon 2 which we denoted as exon 1α (Uspl1–002 - ENSMUST00000122160). This isoform did not appear to be regulated by CARM1 and was not included in future analysis and specific qRT-PCR primers were designed to avoid amplification of transcripts containing this exon (Primer list - Supplementary Table S2).
Figure 2.
Uspl1 is a NMD-regulated target. (A) Schematic representation of Uspl1 E2+ and Uspl1 E2- transcripts. As depicted in this illustration, the skipping of Uspl1 exon 2 introduces a Premature Termination Codon (PTC) in exon 2 of the Uspl1 E2- transcript. (B) Impact of the translational inhibitor cycloheximide on the Uspl1 E2+/E2- ratio. Cells were treated 4 h with cycloheximide (CHX). Then, results obtained in response to CHX were expressed relative to a carrier CTRL. Data are means +/− SEM (n = 3). (C) UPF1 protein levels were assessed from cell transiently transfected with a control shRNA vector CTRL or a shRNA against UPF1. β-actin was used as a loading control. (D) Effect of UPF1 down-regulation on the Uspl1 E2+/E2- ratio. Data are means +/− SEM (n = 3). (E) Impact of the SMG-1 inhibitor wortmannin on the Uspl1 E2+/E2- ratio. Cells were treated 6 h with wortmannin. Subsequently, results obtained in response to wortmannin were expressed relative to DMSO carrier CTRL. Data are means +/− SEM (n = 3). (F) Quantification of the Uspl1 E2+ and Uspl1 E2- mRNA half-lives. MN-1 wt cell lines were treated with actinomycin D (5 μg/ml) for 15, 30 and 60 min. Semi-logarithmic graph shows the decay rate of mRNAs calculated using formula: (x) = ln(0.5)/b, where b = slope obtained from the trendline formula: y = ne−bx. Quantification shows the mean +/− SEM (n = 6), of Uspl1 E2+/18S or Uspl1 E2-/18S. (G) Representative RT-PCR gels showing time course of Uspl1 E2- and Gapdh mRNA levels in DMSO carrier CTRL or shCARM1 conditions, treated with actinomycin D. (H) Quantification of the Uspl1 E2- mRNA half-lives in MN-1 pGIPZ CTRL or shCARM1 cell lines. Quantification shows the mean +/− SEM (n = 3), of Uspl1 E2-/Gapdh.
Since NMD is a process coupled to translation (70–73), MN-1 cells were treated with an inhibitor of translation (cycloheximide, CHX) followed by RT-PCR analysis as above to determine the ratio of Uspl1 isoforms. In response to cycloheximide treatment, the E2+/E2- ratio was decreased relative to carrier treatment (Figure 2B), consistent with stabilization of the E2- mRNA in the absence of translation-dependent NMD activity. To further confirm that the E2- transcript is regulated by NMD, a shRNA against UPF1 (Figure 2C) was transiently transfected into MN-1 cells. As a result of the down-regulation of the main trans-effector of the NMD process, the Uspl1 E2+/E2- ratio, as determined using RT-qPCR, was again decreased as compared to an shRNA CTRL (Figure 2D). Finally, to ascertain that the Uspl1 E2- transcript is a NMD target, MN-1 cells were treated with wortmannin, a drug known to inhibit NMD by preventing the phosphorylation of UPF1 by SMG-1 (74,75). Again, this resulted in a decrease in the E2+/E2- ratio relative to carrier treatment (Figure 2E). To further confirm that the E2- transcript is indeed subjected to NMD and regulated at the level of mRNA stability, we determined the half-life of each Uspl1 alternatively spliced isoform (Figure 2F). MN-1 cells were treated with the transcription inhibitor actinomycin D for 15-90 min. Quantification of Uspl1 E2+ and E2- RNA variants at each time points indicated that the putative E2- NMD transcripts has a considerably shorter half-life of approximately 43 min, relative to the E2+ mRNA at approximately 135 min (Figure 2F). Thus, together, these results strongly support the notion that the Uspl1 E2- isoform is target of the NMD pathway. Since we determined that CARM1 levels influenced the relative ratio of Uspl1 E2+/E2- isoforms, we next assessed whether this effect was due to an effect on mRNA stability of the E2- mRNA isoform. Thus, the same experiment as in Figure 2F was performed using MN1 cells stably transfected with a control shRNA vector or with a shRNA expression plasmid targeting CARM1. Coherent with our findings, the reduction of CARM1 levels resulted in a ∼4.6-fold increase in the Uspl1 E2- mRNA half-life compared to CTRL conditions (Figure 2G,H), although these results did not quite reach statistical significance. These results thus suggest that CARM1 can regulate Uspl1 expression at the level of alternative splicing, and potentially to some extent also at the level of mRNA stability through regulation of the NMD pathway acting on the E2- mRNA isoform.
CARM1 can affect the fate of a generic NMD reporter
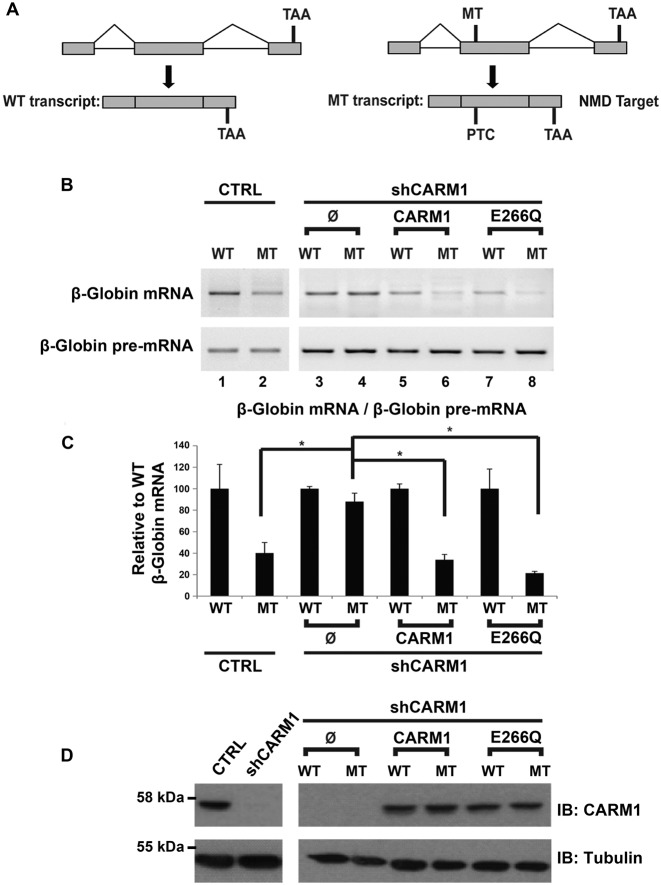
These results prompted us to investigate whether CARM1 has a general impact on NMD. To test this hypothesis, we used a well-characterized NMD reporter system (76) consisting of the full-length β-globin pre-mRNA as well as a mutant version containing in exon 2, a nonsense mutation which introduces a PTC following splicing (Figure 3A). The PTC-containing β-globin mRNA has been thoroughly demonstrated to be a bone fide NMD-regulated target (64,76). Wild type (WT) or mutant (MT) β-globin reporters were transiently transfected into MN-1 pGIPZ CTRL and MN-1 shCARM1 cell lines (Figure 3B, lanes 1–4). In order to quantify the effect of the CARM1 knockdown on the NMD process, levels of the WT and MT reporters were first normalized to their respective pre-mRNA levels (thus accounting for potential differences in transfection efficiency and/or expression levels). Then, the results obtained for the MT reporter were expressed relative to the WT reporter (Figure 3C). As shown in Figure 3B, the relative abundance of the PTC-containing MT reporter mRNA in the MN-1 pGIPZ CTRL cell line is lower than in its WT counterpart (Figure 3B lanes 1–2, and 3C). Remarkably, in the MN-1 CARM1 hypomorph cell line, the amount of the WT minigene is no longer significantly different from the WT (Figure 3B lanes 3–4, and 3C), suggesting CARM1 is required for recognition and/or degradation of the PTC-containing mRNA through NMD.
Figure 3.
CARM1 can affect the fate of a generic NMD reporter. (A) Schematic representation of the wild-type (WT) β-Globin plasmid and the mutant (MT) β-Globin reporter containing a PTC which elicit degradation by the NMD pathway. (B) The WT and MT reporters were transiently transfected either into the MN-1 pGIPZ CTRL or the MN-1 shCARM1 cell line. Furthermore, the WT and MT reporters were also transiently co-transfected with a pcDNA3.1 empty vector, CARM1 or CARM1 E266Q expression vectors into the MN-1 shCARM1 cell line. Representative RT-PCRs of the β-Globin mRNA and β-Globin pre-mRNA are shown. (C) After normalization of the β-Globin mRNA to the β-Globin pre-mRNA levels in each condition, the data were expressed relative to the WT. Data are means +/− SEM, (MN-1 CTRL cell line, n = 5 and MN-1 shCARM1 cell line, n = 3). (D) CARM1 protein levels were assessed by western blotting in MN-1 pGIPZ CTRL and MN-1 shCARM1 cell lines. Tubulin was used as loading control.
Since CARM1 is a well-known regulator of gene expression through various mechanisms (52–55,77–79), we wanted to rule out the possibility that CARM1 might be mediating its effect on NMD by regulating the expression of one or more of the core NMD factors. Consequently, protein levels of the tetrameric EJC core complex components (80) as well as the NMD factors UPF1, UPF2 and UPF3b (aka. UPF3X) (81) were compared in the MN-1 pGIPZ CTRL and MN-1 shCARM1 cell lines (Supplementary Figure S1A,B). Additionally, the mRNA expression patterns measured by RT-qPCR, of the NMD factors Smg1, Smg6, Smg7 and Upf2 were also compared in the MN-1 pGIPZ CTRL and shCARM1 cell lines (Supplementary Figure S1C). From these analyses, no significant difference in expression was observed for these key NMD factors upon depletion of CARM1. Altogether, these results strongly suggest that CARM1 can promote NMD, through a mechanism other than through transcriptional regulation of major known NMD factors. To ensure that the observed changes in mRNA levels are not due to off-target effects, a rescue experiment was also performed as above with either wt or E226Q CARM1 alleles. Consistent with our previous observation, re-expression of either WT or mutant CARM1 alleles was able to rescue the efficiency of the NMD process (Figure 3B lanes 5–8, and 3C,D). This observation suggests that CARM1 can promote NMD through a mechanism that is somehow independent of its methyltransferase activity.
CARM1 interacts with main NMD effector UPF1
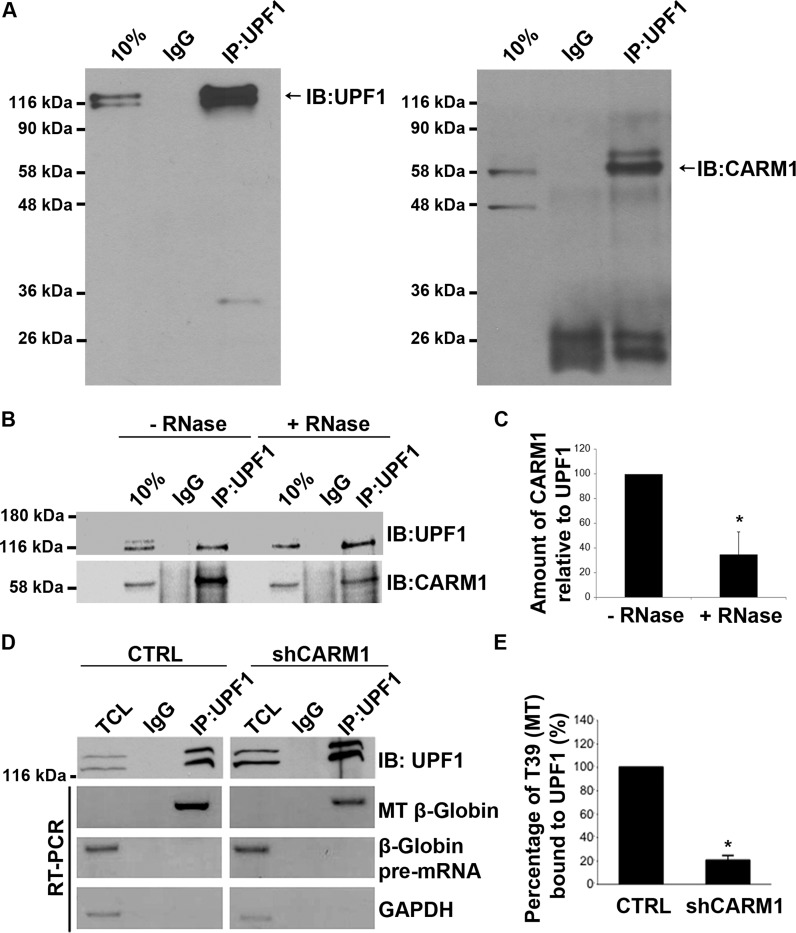
In order to get some insights into the mechanism through which CARM1 regulates NMD, we assessed its potential interaction with core NMD factors. Since UPF1 is central to the NMD process, an endogenous UPF1 immunoprecipitation was performed in extracts from MN-1 cells and analysed by western blotting for the presence of CARM1. As shown in Figure 4A, CARM1 was detected in the endogenous UPF1 immunoprecipitate. Immunoprecipitations with a UPF3 antibody, which detects UPF3b (aka. UPF3X) the first protein to associate with the EJC and recruit UPF2 to the EJC (74), were also performed, but failed to pull-down CARM1 (Supplementary Figure S2). We also wanted to assess whether CARM1 may interact with UPF1 through a mechanism independent of NMD, like perhaps Staufen1 (Stau1)-mediated decay (SMD). SMD is thought to be a process in which the double-stranded RNA binding protein, Stau1, binds to sites within the 3′ UTR of a subset of transcripts and through interaction with UPF1, has been shown to promote the decay of those transcripts (82–84). However, we have not been able to detect any interaction between CARM1 and Stau1, and reducing Stau1 expression using shRNAs did not affect the relative ratio of Uspl1 E2+/E2- isoforms (data not shown), strongly suggesting that CARM1 is not affecting mRNA fate through an impact on SMD. Because our attempts at demonstrating a potential direct interaction between CARM1 and UPF1 purified from Escherichia coli have so far failed (F. Fiorini and H. Le Hir, personal communication), it appears that post-translational modifications might be required for the interaction to occur or, alternatively, that it may be mediated through an intermediate (e.g. protein or RNA). To test the idea that RNA may be necessary for the interaction between UPF1 and CARM1, we next performed co-immunoprecipitations with MN-1 extracts pre-incubated or not with RNase A and assessed the presence of CARM1 in the UPF1 immunoprecipitate by western blotting (Figure 4B). Whereas an equivalent amount of UPF1 was immunoprecipitated in both conditions, less CARM1 protein was detected following RNase A treatment, thereby indicating that the interaction is likely mediated via a RNA moiety (Figure 4B,C).
Figure 4.
CARM1 can be co-immunoprecipitated with UPF1 and the interaction between UPF1 and the β-Globin T39 mutant (MT) decreases with CARM1 knockdown. (A) Total cell lysates were prepared from MN-1 cells and subjected to immunoprecipitation with an IgG CTRL or UPF1 antibodies. Immunoprecipitated proteins were then analysed by western blot using antibodies against UPF1 and CARM1. (B) UPF1 immunoprecipitation experiments were performed with or without (w/o) pretreatment of the cell lysate with RNase A (1 μg/ml) for 30 min at 37°C. (C) Then, the CARM1/UPF1 ratios in response to the RNase A treatments were assessed. Values shown in the bar graph are means +/− SEM (n = 3). (D) The MT reporter was transiently transfected either into the MN-1 pGIPZ CTRL or the MN-1 shCARM1 cell line. RT-PCR analysis was performed using primers specific for the MT mRNA or pre-mRNA, on total RNA extracted from the CTRL (left panel) or shCARM1 (right panel). (E) β-Globin mRNA levels, shown here as percent bound to UPF1, were normalized to overall immunoprecipated UPF1 levels and Gapdh mRNA was used as a loading control. Data are means +/− SEM (n = 3).
UPF1 occupancy on a PTC-containing transcript is reduced in the absence of CARM1
Since CARM1 interacts with UPF1, and based on our observations indicating it plays more of a scaffolding role (i.e. independent of its methyltransferase activity), we sought to assess whether CARM1 may influence steady-state levels of UPF1 on PTC-containing mRNAs. Using RNA-immunoprecipations, we investigated the effect of CARM1 depletion on the ability of UPF1 to bind to a generic NMD target. MN-1 cells were co-transfected with the PTC-containing β-globin MT reporter pre-mRNA and either a CTRL plasmid or shCARM1. Forty-eight hours after transfection, UPF1 was immunoprecipitated and the amount of the MT reporter mRNAs associated with UPF1 was measured by RT-PCR. After normalization to the amount of UPF1 protein immunoprecipitated in each condition (Figure 4D; top panel), the amount of the β-globin MT reporter mRNAs bound to UPF1 was significantly less in cells with reduced CARM1 levels, as compared with CTRL (Figure 4D,E). It is worth noting that although some level of UPF1 occupancy was also detected on the WT β-globin reporter, consistent with previous reports (85,86) CARM1 had no impact in this context (Supplementary Figure S3A). Moreover, no detectable occupancy of UPF1 was observed on an unrelated, non PTC-containing pre-mRNA minigene, whether CARM1 was present or not (Supplementary Figure S3B). Together, these experiments strongly suggest that CARM1 promotes NMD, at least in part, by favouring UPF1 association with PTC-containing mRNA transcripts.
CARM1 regulates a diverse subset of known endogenous NMD targets
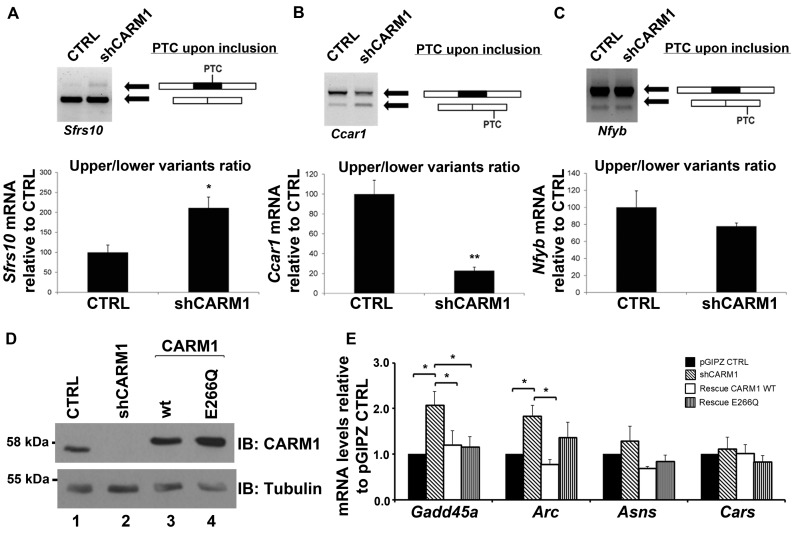
The impact of CARM1 on synthetic NMD reporters led us to speculate that it may have a broad impact on NMD-regulated transcripts. We therefore assessed the expression levels of several previously validated endogenous NMD targets (87–94) in the face of reduced CARM1 levels in MN-1 cells. These transcripts are targets of the NMD machinery because they either: (i) harbour a PTC following inclusion or exclusion of an exon through alternative splicing; (ii) contain an upstream open reading frame (uORF); or (iii) contain introns within their 3′ UTRs (95). Amongst eight Alternative Splicing-Coupled NMD transcripts that we tested, three (Sfrs10, hnrnpa2b1 and Ccar1) were sensitive to CARM1 levels (Figure 5A,B and Supplementary Table S1), while for example, the Nfyb mRNA was not dependent on CARM1 (Figure 5C). As shown by semi-quantitative RT-PCR, the Sfrs10 isoform containing a PTC upon exon inclusion is increased relative to the skipped isoform in response to CARM1 depletion (Figure 5A). However, for Ccar1, while the relative ratio of spliced isoform is affected in the face of reduced CARM1 levels (Figure 5B), total amount of transcripts appear to stay constant, suggesting that CARM1 likely influences this pre-mRNA at the alternative splicing level rather than through an effect on NMD. In any case, more experiments would be required in order to determine precisely whether CARM1 affects the expression of each of these genes at the level of splicing and/or NMD.
Figure 5.
CARM1 regulates a diverse subset of known NMD targets. (A) Total RNA was extracted from MN-1 pGIPZ CTRL and MN-1 shCARM1 cell lines. PCR primer sets were designed to flank the PTC resulting either by exon skipping or exon inclusion. For Sfrs10, PTC occurs upon exon inclusion. Values shown in the bar graph are means +/− SEM, n = 4. (B) For Ccar1, PTC is generated upon exon skipping. Values shown in the bar graph are means +/− SEM. (C) In the case of Nfyb gene, PTC is generated upon exon skipping. After optical quantification of the upper/lower splicing variants ratio, results are expressed relative to the MN-1 pGIPZ CTRL cell line. Values shown in the bar graph are means +/− SEM, n = 3 (D) A rescue was performed in two of the MN-1 shCARM1 cell lines by then transfecting with either the WT-CARM1 or the CARM1-E266Q mutant expression vectors for an additional 24 h. Protein lysate from transfected MN-1 lines confirm the knockdown and overexpression of CARM1 or the E266Q mutant, normalized to Tubulin. (E) Total RNA was extracted and RT-qPCR was used to measure the mRNA levels of targets which were normalized to 18s RNA levels and presented as relative to pGIPZ CTRL target mRNA levels. Data are means +/− SEM (n = 3).
We next assessed eight transcripts with distinct NMD-inducing features, that all have been documented elsewhere to be subjected to NMD (Supplementary Table S1), in order to determine whether their steady-state levels was influenced by CARM1. As before, rescue experiments were conducted to ensure that the observed changes were not due to off-target effects. Of the seven transcripts assessed, Gadd45a and Arc, showed significant dependence on CARM1 levels (Figure 5D,E). A general trend towards CARM1-sensitivity was also observed for Asns, although statistical significance was not quite attained using CARM1 shRNA knock-downs (see below). For Gadd45a, both wt and methyltransferase-dead CARM1 alleles were as efficient at rescuing mRNA levels to that of CTRL (Figure 5D,E). However, for Arc, the E266Q mutant did not fully rescue mRNA levels to that of CTRL, suggesting that methyltransferase activity could be involved in the mechanism by which CARM1 regulates this specific target (Figure 5D,E and see Discussion).
To further and independently confirm that CARM1 is involved in regulating those specific endogenous NMD targets at the level of mRNA stability, we made use of mouse embryonic fibroblasts (MEFs) derived from Carm1−/− mice (96) (Figure 6). Using Actinomycin D treatments as described above, we measured the half-lives of the endogenous NMD targets Arc, Gadd45a and Asns (90,94), by RT-qPCR in wt and Carm1−/− MEFs. As predicted, all three NMD target mRNAs, this time including Asns, showed stabilization in Carm1 −/− MEFs (Figure 6B,C and Supplementary Figure S4). Specifically, for Gadd45a, the calculated half-life was of ∼3.2 h in wt MEFs, compare with ∼5 h in Carm1 -/- cells (Figure 6B). Interestingly, this was comparable to the level of stabilization observed upon inhibition of NMD using Wortmannin in wt MEFs (see gel in Figure 6B). A similar phenomenon was also observed with Asns, with the half-life of the mRNA increasing by > 2-fold between wt and Carm1 -/- MEFs (Figure 6C), and this difference also being reflected at the protein level (Figure 6A). However, for Arc, even if an ∼2-fold increase in mRNA steady-state (Figure 5E) and protein (Figure 6A) levels was seen in CARM1-depleted cells, the measured difference in actual mRNA stability did not quite reach statistical significance (Supplementary Figure S4), suggesting CARM1 may affect this particular target mostly at the level of transcription. Altogether, these experiments strongly suggest that, in addition to its well-known roles in regulating transcription and alternative splicing, CARM1 can also promote degradation of a specific subset of mRNAs through the NMD pathway.
Figure 6.
NMD target stability and protein expression in Carm1 +/+ and Carm1 -/- MEFs. (A) Western blotting analysis and quantification shows levels of CARM1, UPF1 and NMD targets ASNS and ARC protein levels in Carm1 +/+ (wt) and Carm1 -/- MEFs. Tubulin was used as loading control. Data are means +/− SEM (n = 3). (B,C) Quantification of NMD transcripts Gadd45A and Asns mRNA half-lives normalized to Gapdh. Semi-logarithmic graph shows the decay rate of mRNAs calculated using formula: (x) = ln(0.5)/b, where b = slope obtained from the trendline formula: y = ne−bx. Carm1 +/+ and Carm1 -/- MEFs were pretreated with either a vehicle or Wortmannin (5 μM) and followed by treatment with actinomycin D (5 μg/ml) and RNA extraction at 0, 2, 4, 6 and 8 h. Quantification shows the means +/− SEM (n = 3) and data are presented as relative to zero time point.
CARM1-dependent NMD targets misregulated in SMA
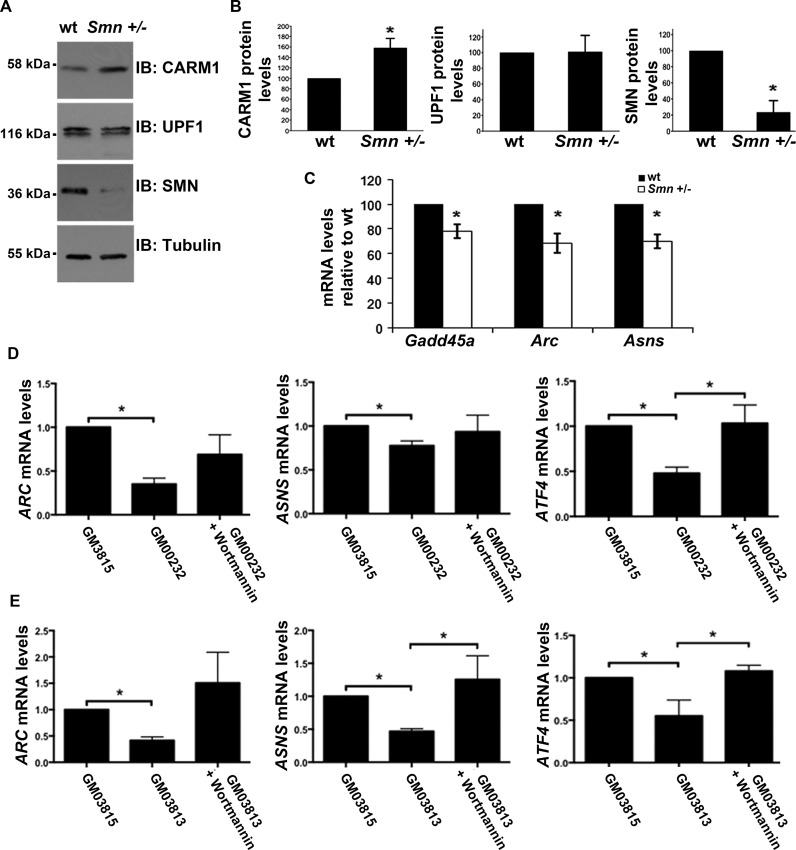
Since we previously reported that CARM1 levels are up-regulated in SMA (51), we reasoned that NMD targets found here to be CARM1-sensitive should be down-regulated in SMA-like settings. We first extended our findings to an in vivo setting by investigating the expression levels of the known endogenous NMD targets which we found to be CARM1 sensitive, in Smn+/− mice. Mice heterozygous for Smn show ∼50% motor neuron attrition by ∼6 months of age and are used as a model for mild SMA in humans (97). Western blot analysis confirmed upregulation of CARM1 in these mice at pre-symptomatic time-points, and we observed no significant changes in UPF1 protein levels (Figure 7A,B). RT-qPCR analysis of CARM1-sensitive NMD targets Gadd45a, Arc and Asns, revealed a significant decrease in mRNA expression levels in Smn+/− mouse spinal cord tissue as compared to wt (Figure 7C). These data, in conjunction with the increased CARM1 levels seen in the Smn+/− mice and the steady UPF1 levels in both conditions, are consistent with these targets being misregulated in SMA, at least in part, through a CARM1-dependent decay mechanism, although further experiments would be warranted to clearly demonstrate this link. Finally, we observed misregulation of ARC, ASNS and ATF4 (another known endogenous NMD target) mRNAs in two independent type I SMA patients fibroblast cell lines, when compared to a fibroblast line derived from an unaffected carrier (Figure 7D,E). Interestingly, treatment of SMA fibroblasts with Wortmannin to inhibit the NMD pathway led to an increase in the levels of these mRNAs (Figure 7D,E), consistent with these mRNAs being misregulated in SMA due to a defect in NMD. Altogether, these results suggest that at least a subset of CARM1-sensitive NMD targets are decreased in SMA-like settings, supporting the notion that the NMD pathway may somehow be exacerbated in SMA.
Figure 7.
Endogenous NMD targets are mis-regulated in Smn +/− mice and SMA type I fibroblasts. (A) Protein lysate was collected from the spinal cord tissue of wt and Smn +/− mice. CARM1, UPF1 and SMN protein levels were assessed by western blotting. (B) Quantification of protein levels were gathered by normalizing to tubulin levels. Data are means +/− SEM (n = 3) (C) Total RNA was extracted from spinal cord tissue of wt and Smn +/− mice. RT-qPCR was used to measure the mRNA levels of targets which were normalized to Gapdh mRNA levels and presented as relative to wt target mRNA levels. Data are means +/− SEM (n = 3). (D,E) Fibroblast cell lines from two independent human SMA type I patients (GM00232, GM03813) were treated with Wortmannin (5 μM) for 6 h. Untreated fibroblast cell lines from a carrier (GM03815) were used as CTRL. Total RNA was isolated from the cell lines and three non-ASC-NMD target mRNA expression levels were measured by RT-qPCR: ARC, ASNS and ATF4 using GAPDH as a normalization control. Data are means +/− SEM (ARC, ATF4n = 6; ASNSn = 5).
DISCUSSION
We report in the present study that CARM1 promotes down-regulation of a specific subset of NMD substrates. Specifically, we found that CARM1 is required to elicit NMD on a generic NMD reporter, through a mechanism that is independent of its methyltransferase activity. We also demonstrate that CARM1 interacts with major NMD factor UPF1, in an RNA-dependent fashion, and that occupancy of UPF1 on a PTC-containing transcript is significantly decreased in CARM1-depleted cells. Finally, consistent with the fact that CARM1 is upregulated in SMA-like settings, we provide evidence that the NMD pathway may somehow be exacerbated in SMA.
CARM1 promotes down-regulation of mRNAs with different NMD-inducing features
We have initially focussed our efforts on Uspl1 in the present study and uncovered that an alternatively spliced isoform resulting from the skipping of exon 2 was a target for NMD. This finding was also corroborated by the fact that Uspl1 was identified in a recent unbiased, genome-wide screen for novel NMD targets (98). While we clearly demonstrated that reduced levels of CARM1 affected the relative ratio of Uspl1 alternatively spliced isoforms, our mRNA stability assessments suggested that this effect was likely mediated principally at the splicing level, although we can not rule out some contribution at the level of NMD. We also observed an effect of CARM1 on the relative ratio of spliced isoforms for three other genes, Sfrs10, HnRNPa2b1 and Ccar1 (Supplementary Table S1), although additional experiments would be required to determine precisely whether CARM1 affects splicing and/or NMD for those targets. Because it is often more difficult for splicing-coupled NMD targets to discern between an effect on splicing and/or NMD, we next decided to focus on well-established endogenous NMD targets with distinct NMD-inducing features. We documented three such endogenous transcripts (Gadd45a, Arc and Asns) which are targets of NMD due to features unrelated to alternative splicing (e.g. uORFs or introns in the 3′ UTR), that are CARM1-sensitive. This suggest that CARM1's influence on NMD targets is not dependent on the mechanism through which NMD is elicited. Nevertheless, not all previously validated endogenous NMD targets we assayed were CARM1-sensitive (see Supplementary Table S1), suggestive of a significant degree of target specificity for CARM1's regulation of NMD targets. For example, we have found that CARM1 did not influence mRNA or protein levels of major NMD factors (Supplementary Figure S1), many of which are known to be regulated through the NMD pathway in a negative feedback regulatory loop involved in establishing and maintain NMD homeostasis (63,99). There is mounting evidence in the literature that several NMD branches exist (100–102), each with distinct substrate specificity features, so it would be interesting to determine whether this may explain CARM1's NMD target discrimination. Identification of additional CARM1-dependent NMD targets in future studies, ideally through unbiased genome-wide approaches, should allow to determine the precise mechanism(s) through which CARM1 target specificity is arising.
CARM1 interacts with major NMD factor UPF1
We have observed that CARM1 interacts with UPF1, in an RNA-dependent fashion, using co-immunoprecipitation experiments, with endogenous proteins. However, we have not been able to detect a signal for CARM1 upon UPF3b or UFP2 immunoprecipitations (Supplementary Figure S2 and data not shown, respectively). Nevertheless, both these antibodies were not very efficient at immunoprecipitating their respective target, so we can not rule out at this point that a lack of signal for CARM1 in these experiments could be due to technical issues, like e.g. insufficient sensitivity or masking of an interaction domain by the antibodies. In an attempt to address the functional relevance of the CARM1/UPF1 interaction, we used RIP assays to measure the occupancy of UPF1 on a PTC-containing transcript, and found that occupancy was reduced upon CARM1 depletion by RNA interference. This provides further evidence that CARM1 promotes NMD, and suggest that it may play a scaffolding role at some point along the NMD pathway. Nevertheless, it was recently shown that target discrimination is likely determined by the ATPase-dependent duration of UPF1 occupancy (off-rate) on mRNAs (85), so it will be interesting to investigate whether CARM1 affects this process. Such a scaffolding role for CARM1 would be consistent with our observation that CARM1's methyltransferase activity is dispensable for its function in promoting down-regulation of NMD substrates. This is also in line with the fact that we were unable to detect any methylation mark in UPF1 using mass spectrometry (data not shown). Interestingly, the methyltransferase-DEAD E266Q CARM1 mutant was as efficient as wt CARM1 in rescue experiments, for all targets tested except for Arc, which we in fact showed is likely regulated by CARM1 mostly at the level of transcription. There are other documented cases of activity-independent roles for PRMTs. For example, Jayne et al. have reported that CARM1 methyltransferase activity was dispensable for the expression of a subset of NF-κB target genes in response to either TNFα or PMA/ionomycin stimuli (103). In the case of Rmt3, the homolog of human PRMT3 in Schizosaccharomyces pombe, Perrault et al. (104) demonstrated that enzymatic inactive mutants of Rmt3 were able to rescue the ribosomal 40S formation defect in Rmt3-null cells. Interestingly, they also observed that an Rps2 mutant in which the methylated arginines were substituted by lysines (which are not substrates for PRMTs but still have a positive charge) was still able to rescue the defects whereas Rmt3 mutants failing to interact with Rps2 did not. Altogether, these data argue that in addition to their enzymatic activity, PRMTs can also have methyltransferase-independent roles in scaffolding and/or stabilization of complexes through protein-protein and protein-RNA interactions. Nevertheless, it is known that transcriptional events can impact on the efficiency of NMD (105–107), so considering that CARM1 is a well-known transcriptional and splicing regulator, further investigations will be required in order to determine the precise mechanism by which it can promote NMD.
Potential contribution of misregulated NMD targets to the pathophysiology of SMA?
In the current study we have used the Smn+/− mice to confirm our results in an in vivo setting that would allow us to assess protein expression profiles at a pre-phenotypic or disease onset time-point. Using this model, we reproduced our previous observation that CARM1 was upregulated (∼60%) in spinal cord tissues, with no significant difference in main NMD mediator UPF1 levels, thus providing an ideal in vivo model to validate CARM1-sensitive NMD targets identified in our cell culture models. We were also able to corroborate some of our findings in two SMA Type I patient fibroblast cell lines, importantly showing that treatment with NMD inhibitor Wortmannin was able to restore steady-state expression levels of a subset of NMD substrates found to be misregulated in these cells. However, these experiments were performed with a very limited number of lines and lacked a proper, genetically matched, control to allow for a stringent comparative analysis. It is interesting to note that amongst the NMD targets that we have found to be misregulated in SMA, both ATF4 and GADD45a, are well known mediators of cellular stress response (108,109). More specifically, for example the ATF4/GADD45a axis was recently shown to contribute to skeletal muscle atrophy (109,110). Nevertheless, the fact that we observe a decrease in ATF4 and GADD45a levels may seem counter-intuitive, but it will be important to investigate this further in relevant tissues (i.e. motorneurons, skeletal muscles, glial cells) and at various developmental stages in more severe SMA models. Another known NMD target that we have found to be downregulated in SMA, and which might have important implications for the pathophysiology of the disease, is the immediate-early gene Arc. It was elegantly shown by Melissa Moore's group in 2007 that Arc mRNA are subjected to ‘translation-dependent decay (TDD)’ in dentrites of neuronal cells, as part of the mechanisms involved in maintaining its restricted protein expression at synapses (94). The Arc protein, through its dynamic expression, is a crucial mediator of homeostatic synaptic scaling of AMPA type glutamate in order to exert minute control over individual synaptic strength and overall cellular excitability (111,112). Strikingly, the existence of sensory-motor circuitry defects was recently demonstrated in SMA model mice, which among other things involved intrinsic hyperexcitability of SMA motoneurons (47). This hyperexcitability phenotype is in fact consistent with the reduced Arc levels that we have documented in the current study, suggesting misregulated expression of Arc in motoneuron dentrites may contribute, at least in part, to defects in the SMA sensory-motor circuit. Altogether, our results are consistent with the notion that the NMD pathway may somehow be exarcebated in SMA, although obviously, additional experiments, in more relevant models of the disease, will be necessary to clearly establish whether this contributes to SMA pathophysiology.
Supplementary Material
Acknowledgments
We thank Dr Adrian Krainer (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA) for generously providing the WT and MT β-Globin reporters. We also thank Dr Mark Bedford (The University of Texas MD Anderson Cancer Center, Smithville, TX, USA) for the generous gift of the CARM1 and CARM1 E266Q methyl dead expression vectors and CARM1−/− MEFs, and Dr Jens Lykke-Andersen for reagents to perform tethering experiments. Finally, we thank members of the Côté and Jasmin labs for helpful discussions and technical advice, as well as Dr Jean-Claude Béïque for his insights into the function of Arc in synaptic plasticity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (CIHR) [MOP-86746 to J.C.) and Families of SMA Canada (FSMA; now known as CureSMA); PDF fellowship from the Association Française contre les Myopathies (AFM) [to G.S.]; STaR Awards from the University of Ottawa Centre for Neuromuscular Disease (CNMD) and the University of Ottawa Brain and Mind Research Institute (uOBMRI) [E.B.-C. and A.D.]; Canada Research Chair (Tier II) in RNA Metabolism [to J.C.]. Funding for open access charge: CIHR [MOP-86746] and FSMA Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.Markowitz J.A., Singh P., Darras B.T. Spinal muscular atrophy: a clinical and research update. Pediatr. Neurol. 2012;46:1–12. doi: 10.1016/j.pediatrneurol.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Ogino S., Wilson R.B. Spinal muscular atrophy: molecular genetics and diagnostics. Expert Rev. Mol. Diagn. 2004;1:15–29. doi: 10.1586/14737159.4.1.15. [DOI] [PubMed] [Google Scholar]
- 3.Swoboda K.J. Of SMN in mice and men: a therapeutic opportunity. J. Clin. Invest. 2011;121:2978. doi: 10.1172/JCI58752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostova F.V., Williams V.C., Heemskerk J., Iannaccone S., DiDonato C., Swoboda K., Maria B.L. Spinal muscular atrophy: classification, diagnosis, management, pathogenesis, and future research directions. J. Child Neurol. 2007;22:926–945. doi: 10.1177/0883073807305662. [DOI] [PubMed] [Google Scholar]
- 5.Gregoretti C., Ottonello G., Testa M.B.C., Mastella C., Ravà L., Bignamini E., Veljkovic A., Cutrera R. Survival of patients with spinal muscular atrophy type 1. Pediatrics. 2013;131:e1509–e1514. doi: 10.1542/peds.2012-2278. [DOI] [PubMed] [Google Scholar]
- 6.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 7.Cho S., Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24:438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 10.Burnett B.G., Muñoz E., Tandon A., Kwon D.Y., Sumner C.J., Fischbeck K.H. Regulation of SMN protein stability. Mol. Cell. Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eggert C., Chari A., Laggerbauer B., Fischer U. Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 2006;12:113–121. doi: 10.1016/j.molmed.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Kolb S.J., Battle D.J., Dreyfuss G. Molecular functions of the SMN complex. J. Child Neurol. 2007;22:990–994. doi: 10.1177/0883073807305666. [DOI] [PubMed] [Google Scholar]
- 13.Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer U., Englbrecht C., Chari A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdis. Rev. RNA. 2011;2:718–731. doi: 10.1002/wrna.87. [DOI] [PubMed] [Google Scholar]
- 15.Baumer D., Lee S., Nicholson G., Davies J.L., Parkinson N.J., Murray L.M., Gillingwater T.H., Ansorge O., Davies K.E., Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabanella F., Butchbach M., Saieva L., Carissimi C., Burghes A., Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PloS One. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jodelka F.M., Ebert A.D., Duelli D.M., Hastings M.L. A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum. Mol. Genet. 2010;19:4906–4917. doi: 10.1093/hmg/ddq425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lotti F., Imlach W.L., Saieva L., Beck E.S., Hao L.T., Li D.K., Jiao W., Mentis G.Z., Beattie C.E., McCabe B.D. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151:440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruggiu M., McGovern V.L., Lotti F., Saieva L., Li D.K., Kariya S., Monani U.R., Burghes A.H., Pellizzoni L. A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol. Cell. Biol. 2012;32:126–138. doi: 10.1128/MCB.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulisfane N., Choleza M., Rage F., Neel H., Soret J., Bordonné R. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum. Mol. Genet. 2011;20:641–648. doi: 10.1093/hmg/ddq508. [DOI] [PubMed] [Google Scholar]
- 22.Campion Y., Neel H., Gostan T., Soret J., Bordonné R. Specific splicing defects in S. pombe carrying a degron allele of the Survival of Motor Neuron gene. EMBO J. 2010;29:1817–1829. doi: 10.1038/emboj.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.See K., Yadav P., Giegerich M., Cheong P.S., Graf M., Vyas H., Lee S.G., Mathavan S., Fischer U., Sendtner M., et al. SMN deficiency alters Nrxn2 expression and splicing in zebrafish and mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2014;23:1754–1770. doi: 10.1093/hmg/ddt567. [DOI] [PubMed] [Google Scholar]
- 24.Huo Q., Kayikci M., Odermatt P., Meyer K., Michels O., Saxena S., Ule J., Schumperli D. Splicing changes in SMA mouse motoneurons and SMN-depleted neuroblastoma cells: evidence for involvement of splicing regulatory proteins. RNA Biol. 2014;11:1430–1446. doi: 10.1080/15476286.2014.996494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akten B., Kye M.J., Hao le T., Wertz M.H., Singh S., Nie D., Huang J., Merianda T.T., Twiss J.L., Beattie C.E., et al. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fallini C., Zhang H., Su Y., Silani V., Singer R.H., Rossoll W., Bassell G.J. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly (A) mRNA in primary motor neuron axons. J. Neurosci. 2011;31:3914–3925. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubers L., Valderrama-Carvajal H., Laframboise J., Timbers J., Sanchez G., Cote J. HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum. Mol. Genet. 2011;20:553–579. doi: 10.1093/hmg/ddq500. [DOI] [PubMed] [Google Scholar]
- 28.Peter C.J., Evans M., Thayanithy V., Taniguchi-Ishigaki N., Bach I., Kolpak A., Bassell G.J., Rossoll W., Lorson C.L., Bao Z.-Z. The COPI vesicle complex binds and moves with survival motor neuron within axons. Hum. Mol. Genet. 2011;20:1701–1711. doi: 10.1093/hmg/ddr046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piazzon N., Rage F., Schlotter F., Moine H., Branlant C., Massenet S. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J. Biol. Chem. 2008;283:5598–5610. doi: 10.1074/jbc.M707304200. [DOI] [PubMed] [Google Scholar]
- 30.Rossoll W., Jablonka S., Andreassi C., Kröning A.-K., Karle K., Monani U.R., Sendtner M. Smn, the spinal muscular atrophy–determining gene product, modulates axon growth and localization of β-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossoll W., Kröning A.-K., Ohndorf U.-M., Steegborn C., Jablonka S., Sendtner M. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum. Mol. Genet. 2002;11:93–105. doi: 10.1093/hmg/11.1.93. [DOI] [PubMed] [Google Scholar]
- 32.Tadesse H., Deschenes-Furry J., Boisvenue S., Cote J. KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum. Mol. Genet. 2008;17:506–524. doi: 10.1093/hmg/ddm327. [DOI] [PubMed] [Google Scholar]
- 33.Sun M., Yamashita T., Shang J., Liu N., Deguchi K., Liu W., Ikeda Y., Feng J., Abe K. Acceleration of TDP43 and FUS/TLS protein expressions in the preconditioned hippocampus following repeated transient ischemia. J. Neurosci. Res. 2014;92:54–63. doi: 10.1002/jnr.23301. [DOI] [PubMed] [Google Scholar]
- 34.Fallini C., Rouanet J.P., Donlin-Asp P.G., Guo P., Zhang H., Singer R.H., Rossoll W., Bassell G.J. Dynamics of survival of motor neuron (SMN) protein interaction with the mRNA-binding protein IMP1 facilitates its trafficking into motor neuron axons. Develop. Neurobiol. 2014;74:319–332. doi: 10.1002/dneu.22111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freibaum B.D., Chitta R.K., High A.A., Taylor J.P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dombert B., Sivadasan R., Simon C.M., Jablonka S., Sendtner M. Presynaptic localization of Smn and hnRNP R in axon terminals of embryonic and postnatal mouse motoneurons. PLoS One. 2014;9:e110846. doi: 10.1371/journal.pone.0110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiebler M.A., Bassell G.J. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Liu-Yesucevitz L., Bassell G.J., Gitler A.D., Hart A.C., Klann E., Richter J.D., Warren S.T., Wolozin B. Local RNA translation at the synapse and in disease. J. Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas M.G., Loschi M., Desbats M.A., Boccaccio G.L. RNA granules: the good, the bad and the ugly. Cell. Signal. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biondi O., Grondard C., Lécolle S., Deforges S., Pariset C., Lopes P., Cifuentes-Diaz C., Li H., Della Gaspera B., Chanoine C. Exercise-induced activation of NMDA receptor promotes motor unit development and survival in a type 2 spinal muscular atrophy model mouse. J. Neurosci. 2008;28:953–962. doi: 10.1523/JNEUROSCI.3237-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowerman M., Shafey D., Kothary R. Smn depletion alters profilin II expression and leads to upregulation of the RhoA/ROCK pathway and defects in neuronal integrity. J. Mol. Neurosci. 2007;32:120–131. doi: 10.1007/s12031-007-0024-5. [DOI] [PubMed] [Google Scholar]
- 42.Cifuentes-Diaz C., Nicole S., Velasco M.E., Borra-Cebrian C., Panozzo C., Frugier T., Millet G., Roblot N., Joshi V., Melki J. Neurofilament accumulation at the motor endplate and lack of axonal sprouting in a spinal muscular atrophy mouse model. Hum. Mol. Genet. 2002;11:1439–1447. doi: 10.1093/hmg/11.12.1439. [DOI] [PubMed] [Google Scholar]
- 43.Kariya S., Park G.-H., Maeno-Hikichi Y., Leykekhman O., Lutz C., Arkovitz M.S., Landmesser L.T., Monani U.R. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kong L., Wang X., Choe D.W., Polley M., Burnett B.G., Bosch-Marcé M., Griffin J.W., Rich M.M., Sumner C.J. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J. Neurosci. 2009;29:842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H., Beauvais A., Baker A.N., Tsilfidis C., Kothary R. Smn deficiency causes neuritogenesis and neurogenesis defects in the retinal neurons of a mouse model of spinal muscular atrophy. Develop. Neurobiol. 2011;71:153–169. doi: 10.1002/dneu.20840. [DOI] [PubMed] [Google Scholar]
- 46.McGovern V.L., Gavrilina T.O., Beattie C.E., Burghes A.H. Embryonic motor axon development in the severe SMA mouse. Hum. Mol. Genet. 2008;17:2900–2909. doi: 10.1093/hmg/ddn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mentis G.Z., Blivis D., Liu W., Drobac E., Crowder M.E., Kong L., Alvarez F.J., Sumner C.J., O'Donovan M.J. Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron. 2011;69:453–467. doi: 10.1016/j.neuron.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray L.M., Comley L.H., Thomson D., Parkinson N., Talbot K., Gillingwater T.H. Selective vulnerability of motor neurons and dissociation of pre-and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 49.Shafey D., MacKenzie A.E., Kothary R. Neurodevelopmental abnormalities in neurosphere-derived neural stem cells from SMN-depleted mice. J. Neurosci. Res. 2008;86:2839–2847. doi: 10.1002/jnr.21743. [DOI] [PubMed] [Google Scholar]
- 50.Rathod R., Havlicek S., Frank N., Blum R., Sendtner M. Laminin induced local axonal translation of β-actin mRNA is impaired in SMN-deficient motoneurons. Histochem. Cell Biol. 2012;138:737–748. doi: 10.1007/s00418-012-0989-1. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez G., Dury A.Y., Murray L.M., Biondi O., Tadesse H., El Fatimy R., Kothary R., Charbonnier F., Khandjian E.W., Côté J. A novel function for the survival motoneuron protein as a translational regulator. Hum. Mol. Genet. 2012;22:668–684. doi: 10.1093/hmg/dds474. [DOI] [PubMed] [Google Scholar]
- 52.Di Lorenzo A., Bedford M.T. Histone arginine methylation. FEBS Lett. 2011;585:2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawabe Y.-i., Wang Y.X., McKinnell I.W., Bedford M.T., Rudnicki M.A. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012;11:333–345. doi: 10.1016/j.stem.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J., Xu W. Histone H3R17me2a mark recruits human RNA polymerase-associated factor 1 complex to activate transcription. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5675–5680. doi: 10.1073/pnas.1114905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee Y.-H., Stallcup M.R. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol. Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Calvanese V., Lara E., Suárez-Álvarez B., Dawud R.A., Vázquez-Chantada M., Martínez-Chantar M.L., Embade N., López-Nieva P., Horrillo A., Hmadcha A. Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:13736–13741. doi: 10.1073/pnas.1001399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng D., Cote J., Shaaban S., Bedford M.T. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 58.Fauquier L., Duboé C., Joré C., Trouche D., Vandel L. Dual role of the arginine methyltransferase CARM1 in the regulation of c-Fos target genes. FASEB J. 2008;22:3337–3347. doi: 10.1096/fj.07-104604. [DOI] [PubMed] [Google Scholar]
- 59.Fujiwara T., Mori Y., Chu D.L., Koyama Y., Miyata S., Tanaka H., Yachi K., Kubo T., Yoshikawa H., Tohyama M. CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol. Cell. Biol. 2006;26:2273–2285. doi: 10.1128/MCB.26.6.2273-2285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng Q., Yi P., Wong J., O'malley B.W. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol. Cell. Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Y., Bedford M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 62.Wagner E., Lykke-Andersen J. mRNA surveillance: the perfect persist. J. Cell Sci. 2002;115:3033–3038. doi: 10.1242/jcs.115.15.3033. [DOI] [PubMed] [Google Scholar]
- 63.Yepiskoposyan H., Aeschimann F., Nilsson D., Okoniewski M., Muhlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–2118. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sinha R., Allemand E., Zhang Z., Karni R., Myers M.P., Krainer A.R. Arginine methylation controls the subcellular localization and functions of the oncoprotein splicing factor SF2/ASF. Mol. Cell. Biol. 2010;30:2762–2774. doi: 10.1128/MCB.01270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olaso R., Joshi V., Fernandez J., Roblot N., Courageot S., Bonnefont J.P., Melki J. Activation of RNA metabolism-related genes in mouse but not human tissues deficient in SMN. Physiol. Genomics. 2006;24:97–104. doi: 10.1152/physiolgenomics.00134.2005. [DOI] [PubMed] [Google Scholar]
- 66.Maeda M., Harris A.W., Kingham B.F., Lumpkin C.J., Opdenaker L.M., McCahan S.M., Wang W., Butchbach M.E. Transcriptome profiling of spinal muscular atrophy motor neurons derived from mouse embryonic stem cells. PLoS One. 2014;9:e106818. doi: 10.1371/journal.pone.0106818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schulz S., Chachami G., Kozaczkiewicz L., Winter U., Stankovic-Valentin N., Haas P., Hofmann K., Urlaub H., Ovaa H., Wittbrodt J., et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012;13:930–938. doi: 10.1038/embor.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu H., Shafey D., Moores J.N., Kothary R. Neurodevelopmental consequences of Smn depletion in a mouse model of spinal muscular atrophy. J. Neurosci. Res. 2010;88:111–122. doi: 10.1002/jnr.22189. [DOI] [PubMed] [Google Scholar]
- 69.Maquat L.E. Nonsense-mediated mRNA decay. Curr. Biol. 2002;12:R196–R197. doi: 10.1016/s0960-9822(02)00747-9. [DOI] [PubMed] [Google Scholar]
- 70.Ishigaki Y., Li X., Serin G., Maquat L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 71.Lejeune F., Ishigaki Y., Li X., Maquat L.E. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lejeune F., Ranganathan A.C., Maquat L.E. eIF4G is required for the pioneer round of translation in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:992–1000. doi: 10.1038/nsmb824. [DOI] [PubMed] [Google Scholar]
- 73.Maquat L.E. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 74.Isken O., Maquat L.E. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pal M., Ishigaki Y., Nagy E., Maquat L.E. Evidence that phosphorylation of human Upf1 protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001;7:5–15. doi: 10.1017/s1355838201000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J., Sun X., Qian Y., Maquat L. Intron function in the non-sense mediated decay of b-globin mRNA: Indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Covic M., Hassa P.O., Saccani S., Buerki C., Meier N.I., Lombardi C., Imhof R., Bedford M.T., Natoli G., Hottiger M.O. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-κB-dependent gene expression. EMBO J. 2005;24:85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stallcup M.R. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene. 2001;20:3014–3020. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- 79.Bedford M.T., Richard S. Arginine methylation: an emerging regulatorof protein function. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 80.Tange T.Ø., Shibuya T., Jurica M.S., Moore M.J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Serin G., Gersappe A., Black J.D., Aronoff R., Maquat L.E. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol. Cell. Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park E., Gleghorn M.L., Maquat L.E. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc. Natl. Acad. Sci. U.S.A. 2013;110:405–412. doi: 10.1073/pnas.1213508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim Y.K., Furic L., Desgroseillers L., Maquat L.E. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′ UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 84.Kim Y.K., Furic L., Parisien M., Major F., DesGroseillers L., Maquat L.E. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee S.R., Pratt G.A., Martinez F.J., Yeo G.W., Lykke-Andersen J. Target Discrimination in Nonsense-Mediated mRNA Decay Requires Upf1 ATPase Activity. Mol. Cell. 2015;59:413–425. doi: 10.1016/j.molcel.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Silva A.L., Ribeiro P., Inácio Â., Liebhaber S.A., Romão L. Proximity of the poly (A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA. 2008;14:563–576. doi: 10.1261/rna.815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saltzman A.L., Kim Y.K., Pan Q., Fagnani M.M., Maquat L.E., Blencowe B.J. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McIlwain D.R., Pan Q., Reilly P.T., Elia A.J., McCracken S., Wakeham A.C., Itie-Youten A., Blencowe B.J., Mak T.W. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tani H., Torimura M., Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS One. 2013;8:e55684. doi: 10.1371/journal.pone.0055684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morris C., Wittmann J., Jack H.M., Jalinot P. Human INT6/eIF3e is required for nonsense-mediated mRNA decay. EMBO Rep. 2007;8:596–602. doi: 10.1038/sj.embor.7400955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sharova L.V., Sharov A.A., Nedorezov T., Piao Y., Shaik N., Ko M.S. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Linde L., Boelz S., Neu-Yilik G., Kulozik A.E., Kerem B. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur. J. Hum. Genet. 2007;15:1156–1162. doi: 10.1038/sj.ejhg.5201889. [DOI] [PubMed] [Google Scholar]
- 93.Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 94.Giorgi C., Yeo G.W., Stone M.E., Katz D.B., Burge C., Turrigiano G., Moore M.J. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 95.Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 96.Yadav N., Lee J., Kim J., Shen J., Hu M.C.-T., Aldaz C.M., Bedford M.T. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Balabanian S., Gendron N.H., MacKenzie A.E. Histologic and transcriptional assessment of a mild SMA model. Neurol. Res. 2007;29:413–424. doi: 10.1179/016164107X159243. [DOI] [PubMed] [Google Scholar]
- 98.Hurt J.A., Robertson A.D., Burge C.B. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23:1636–1650. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang L., Lou C.H., Chan W., Shum E.Y., Shao A., Stone E., Karam R., Song H.W., Wilkinson M.F. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol. Cell. 2011;43:950–961. doi: 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gehring N.H., Kunz J.B., Neu-Yilik G., Breit S., Viegas M.H., Hentze M.W., Kulozik A.E. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 101.Chan W.K., Huang L., Gudikote J.P., Chang Y.F., Imam J.S., MacLean J.A., Wilkinson M.F. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Metze S., Herzog V.A., Ruepp M.-D., Mühlemann O. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA. 2013;19:1432–1448. doi: 10.1261/rna.038893.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jayne S., Rothgiesser K.M., Hottiger M.O. CARM1 but not its enzymatic activity is required for transcriptional coactivation of NF-κB-dependent gene expression. J. Mol. Biol. 2009;394:485–495. doi: 10.1016/j.jmb.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 104.Perreault A., Gascon S., D'Amours A., Aletta J.M., Bachand F. A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis. J. Biol. Chem. 2009;284:15026–15037. doi: 10.1074/jbc.M109.004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Enssle J., Kugler W., Hentze M.W., Kulozik A.E. Determination of mRNA fate by different RNA polymerase II promoters. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10091–10095. doi: 10.1073/pnas.90.21.10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bühler M., Mohn F., Stalder L., Mühlemann O. Transcriptional silencing of nonsense codon-containing immunoglobulin minigenes. Mol. Cell. 2005;18:307–317. doi: 10.1016/j.molcel.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 107.Gudikote J.P., Imam J.S., Garcia R.F., Wilkinson M.F. RNA splicing promotes translation and RNA surveillance. Nat. Struct. Mol. Biol. 2005;12:801–809. doi: 10.1038/nsmb980. [DOI] [PubMed] [Google Scholar]
- 108.Jiang H.Y., Jiang L., Wek R.C. The eukaryotic initiation factor-2 kinase pathway facilitates differential GADD45a expression in response to environmental stress. J. Biol. Chem. 2007;282:3755–3765. doi: 10.1074/jbc.M606461200. [DOI] [PubMed] [Google Scholar]
- 109.Ebert S.M., Dyle M.C., Kunkel S.D., Bullard S.A., Bongers K.S., Fox D.K., Dierdorff J.M., Foster E.D., Adams C.M. Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. J. Biol. Chem. 2012;287:27290–27301. doi: 10.1074/jbc.M112.374777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bongers K.S., Fox D.K., Ebert S.M., Kunkel S.D., Dyle M.C., Bullard S.A., Dierdorff J.M., Adams C.M. Skeletal muscle denervation causes skeletal muscle atrophy through a pathway that involves both Gadd45a and HDAC4. Am. J. Physiol. Endocrinol. Metab. 2013;305:E907–E915. doi: 10.1152/ajpendo.00380.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shepherd J.D., Rumbaugh G., Wu J., Chowdhury S., Plath N., Kuhl D., Huganir R.L., Worley P.F. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Béïque J.C., Na Y., Kuhl D., Worley P.F., Huganir R.L. Arc-dependent synapse-specific homeostatic plasticity. PNAS. 2011;108:816–821. doi: 10.1073/pnas.1017914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.