Abstract
The adeno-associated virus (AAV)-based targeting vector has been one of the tools commonly used for genome modification in human cell lines. It allows for relatively efficient gene targeting associated with 1–4-log higher ratios of homologous-to-random integration of targeting vectors (H/R ratios) than plasmid-based targeting vectors, without actively introducing DNA double-strand breaks. In this study, we sought to improve the efficiency of AAV-mediated gene targeting by introducing a 2A-based promoter-trap system into targeting constructs. We generated three distinct AAV-based targeting vectors carrying 2A for promoter trapping, each targeting a GFP-based reporter module incorporated into the genome, PIGA exon 6 or PIGA intron 5. The absolute gene targeting efficiencies and H/R ratios attained using these vectors were assessed in multiple human cell lines and compared with those attained using targeting vectors carrying internal ribosome entry site (IRES) for promoter trapping. We found that the use of 2A for promoter trapping increased absolute gene targeting efficiencies by 3.4–28-fold and H/R ratios by 2–5-fold compared to values obtained with IRES. In CRISPR-Cas9-assisted gene targeting using plasmid-based targeting vectors, the use of 2A did not enhance the H/R ratios but did upregulate the absolute gene targeting efficiencies compared to the use of IRES.
INTRODUCTION
Gene targeting in human cell lines is a useful technology allowing for the biological analyses of human gene function under physiological conditions (1). This technology enables the establishment of isogenic sibling clones which differ from each other only by the presence or absence of a designed genetic alteration in a gene of interest. By comparing the properties of sibling clones, logical and definitive studies of human gene function can be conducted. Gene-targeted human cell lines also serve as a platform for developing and validating molecularly targeted therapies (2–4).
Although gene targeting is generally difficult to achieve in somatic cells partly because of the low homologous recombination efficiency (5), gene targeting in human somatic cell lines has been successfully achieved using adeno-associated virus (AAV)-based targeting vectors (6,7). AAV-based targeting vectors were shown to have 1–4-log higher efficiency of gene targeting (the ratio of homologous to random integration of targeting vectors into the genome; H/R ratio) compared to that using plasmid-based targeting vectors (6,8–11). AAV-based targeting vectors do not actively introduce DNA double-strand breaks into the genome and thus likely produce a relatively low frequency of off-target genetic alterations. Although several different technologies for genome editing have been developed (12–17), AAV-mediated gene targeting remains a useful alternative method for introducing designed genetic alterations in human somatic cell lines (18). AAV-mediated gene targeting has also been applied to gene targeting in human primary cells and stem cells ex vivo to develop new therapies against an inherited genetic disorder, epidermolysis bullosa (19,20). In addition, the AAV vector is thought to be relatively safe as a vehicle for in vivo gene delivery (21,22). AAV-based targeting vectors have been employed in a preclinical in vivo study to correct the deficiency of coagulation factor IX (F9) using the targeted knock-in approach in murine hepatocytes (23).
Significant improvements have been made in the technology used in AAV-mediated gene targeting to facilitate its procedures and improve its efficiency (7,24–26). One of these improvements was the introduction of a promoter-trap system exploiting an internal ribosome entry site (IRES) to express a promoterless antibiotic resistance gene in targeting vectors (24). The promoter-trap system reduces the number of antibiotic-resistant clones formed by random integration of targeting vectors into the genome, thus allowing for the enrichment of gene-targeted cell clones and leading to a higher H/R ratio of the targeting vector. IRES has proven useful to conduct promoter trapping; however, a more powerful promoter-trap system would further facilitate AAV-mediated gene targeting and enable the application of this technology to a wide range of human cell lines.
The 2A sequence, which has been characterized in foot and mouth disease virus and some other viruses, encodes a short peptide that mediates translational skip close to its 3′ end, resulting in multicistronic expression of polypeptides from a single mRNA (27,28). Equal amounts of proteins are yielded from the two genes connected via 2A, while IRES often induces lower protein production from the downstream gene compared with the upstream gene (29). Because of these properties, many studies have exploited 2A as a tool for genetic engineering in cells, such as to express multiple genes from a single vector (30,31). In addition, 2A has been used in plasmid-based targeting vectors to express a promoterless antibiotic resistance gene within a promoter-trap system for gene targeting assisted by designed endonucleases (32–34). The use of 2A should also be of particular advantage in AAV vectors as it is small in size and AAV vectors have a strict size limit of ∼5 kb in total. Indeed, 2A has been used in AAV vectors to express multiple genes from the same vector (35–37) or to place an exogenous gene downstream of an endogenous gene to allow for stable expression of a therapeutic gene (23). However, to our knowledge, few studies have reported the use of 2A within AAV-based targeting vectors for the enrichment of gene-targeted cell clones using the promoter-trap strategy.
In this study, we generated multiple AAV-based targeting vectors bearing a 2A-based promoter-trap system and systematically compared their capacity to enrich gene-targeted cell clones with the levels achieved using an IRES-based promoter-trap system. We first created a versatile AAV plasmid to serve as a platform for constructing AAV-based targeting vectors bearing 2A and a promoterless neomycin resistance (NeoR) gene for promoter trapping. Multiple targeting vectors were then constructed based on this plasmid, and gene targeting efficiencies achieved with these targeting vectors were determined in multiple human somatic cell lines. In addition, we also compared the capacities of these promoter-trap systems to enrich gene-targeted clones in gene targeting assisted by a clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated (Cas) system.
MATERIALS AND METHODS
General molecular biology techniques
Extraction of genomic DNA (gDNA) and total RNA as well as the synthesis of complementary DNA (cDNA) from total RNA were carried out using PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, Waltham, MA, USA), NucleoSpin RNA Kit (TaKaRa Bio, Otsu, Japan) and High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific), respectively, following the manufacturers’ instructions. gDNA and cDNA were amplified by polymerase chain reaction (PCR) performed using Platinum Taq (Thermo Fisher Scientific) or Pwo SuperYield (Roche, Basel, Switzerland) DNA polymerase with a Veriti Thermal Cycler (Thermo Fisher Scientific). Sequence analyses were carried out using the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific). Southern blotting was performed as described previously (10).
AAV vectors
To construct pAAV-2Aneo, pSEPT (a gift from Dr Fred Bunz, Johns Hopkins University; ref (9)) was initially cleaved with BstXI to remove a loxP site, synthetic intron (IVS) and IRES. The remaining part of the plasmid was ligated to annealed oligonucleotides containing Thosea asigna virus 2A, another set of annealed oligonucleotides containing a ‘frame adjuster’ and then a loxP site and IVS excised from pSEPT. The resulting plasmid carried an assembled 2A-based promoter-trap module between two loxP sites. To incorporate a pair of inverted terminal repeats (ITRs) into this plasmid, the backbone of the plasmid was switched to that derived from pAAV-MCS (Agilent Technologies, Santa Clara, CA, USA).
For the gene targeting assay based on the hygromycin B phosphotransferase gene fused to a 3′-truncated enhanced GFP (HygR–5′ EGFP) reporter system, we utilized an AAV-based promoter-trap targeting vector described in our previous study (11), the ATG-less targeting vector (TV), as EGFP-TV-IRES in this study. To produce EGFP-TV-2A, pAAV-2Aneo was cleaved with BsrGI and then self-ligated to adjust the reading frame. The resultant plasmid was cleaved at multiple cloning site-1 (MCS-1) and ligated with a 5′ homology arm that had been PCR-amplified with the oligonucleotide primers shown in Supplementary Table S1 using pEGFP-C1 (TaKaRa Bio) as a template. A 3′ homology arm was transferred from EGFP-TV-IRES to the resulting plasmid at MCS-2.
To disrupt PIGA exon 6, the targeting vector PIGAex6-TV-2A was produced by transferring the 5′ and 3′ homology arms from PIGAex6-TV-IRES to pAAV-2Aneo, in which the frame adjuster was processed by BsrGI. PIGAex6-TV-IRES used in this study was identical to the AAV-based promoter-trap PIGA targeting vector without a negative selection cassette, which was described in our previous study (10).
To disrupt PIGA intron 5, we initially modified the frame adjuster in pAAV-2Aneo by double-digesting it with BspEI and BsrGI and ligating the resultant linearized plasmid with a pair of annealed oligonucleotides (Supplementary Table S1). The resultant plasmid was cut with BspEI and then self-religated to truncate the new frame adjuster. The 5′ and 3′ homology arms were then PCR-amplified using the oligonucleotide primers listed in Supplementary Table S1 and were inserted into MCS-1 and MCS-2, respectively, to create PIGAint5-TV-2A. For PIGAint5-TV-IRES, both 5′ and 3′ homology arms of PIGAex6-TV-IRES were replaced with those excised from PIGAint5-TV-2A.
AAV-based targeting vectors were designed so that the combination of 5′ and 3′ homology arms in the respective vectors were between 1738 and 1992 bp in length, because the AAV vector has limited packaging capacity. Supplementary Table S2 summarizes the length of the homology arms in the respective targeting vectors.
Upon production of the vectors, the PCR-amplified fragments were fully sequenced after incorporation into the plasmids to verify their integrity.
Cell culture
The human cell lines used in this study include HCT116, DLD-1, AsPC-1, PL5 and HEK293T (American Type Culture Collection, Manassas, VA, USA) and HuP-T3 (Sigma-Aldrich, St Louis, MO, USA). Cell culture was carried out in McCoy's 5A modified medium (Sigma-Aldrich; for HCT116), RPMI-1640 medium (Wako, Osaka, Japan; for DLD-1, AsPC-1 and PL5) and Dulbecco's modified Eagle's medium (Wako; for HuP-T3 and HEK293T). Media were supplemented with 5% fetal bovine serum (BioWest, Nuaillé, France) and 1% penicillin/streptomycin (Wako) prior to use for regular cell culture. The transfection of cells with the plasmids was performed using TransIT-LT1 transfection reagent (Mirus Bio, Madison, WI, USA) according to the manufacturer's instructions. Selection of cells with G418 (Life Technologies) was carried out at concentrations of 0.4 mg/ml (PL5 and HuP-T3), 0.8 mg/ml (HCT116 and AsPC-1) or 1 mg/mL (DLD-1) throughout the study.
AAV-based gene targeting assays
AAV-based gene targeting assays (an assay based on the HygR–5′ EGFP reporter system as well as assays targeting PIGA exon 6 and PIGA intron 5, respectively) were performed as previously described with some modifications (10,11,38). For all AAV-based gene targeting assays, AAV particles (serotype 2) were produced by co-transfection of HEK293T cells with AAV-based targeting plasmids along with the plasmids pAAV-RC and pHelper included in the AAV Helper-Free System (Agilent Technologies). Before infection, the cells were plated in 75-cm2 flasks in triplicate at a density of 5 × 105 cells/flask (HCT116, DLD-1 and their derivatives) or 1 × 106 cells/flask (AsPC-1, PL5 and HuP-T3), and infected with AAV vectors at a multiplicity of infection (MOI) of 1 × 104 on the following day. The MOIs of the AAV vectors were determined based on their copy numbers, which were quantified by real-time PCR using the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) as described previously (10,11). Infected cells were selected with G418 for ∼2 weeks, dissociated with Accutase (Innovative Cell Technologies, San Diego, CA, USA) and collected as bulk populations of cells. Collected cells were directly analyzed by flow cytometry (FCM) in the HygR–5′ EGFP reporter gene targeting assay; while, in PIGA gene targeting assays, dissociated cells were stained with Alexa 488-conjugated inactive aerolysin variant (FLAER; Pinewood Scientific Services, Victoria, Canada) as per the manufacturer's instructions and then subjected to FCM analyses. FCM analyses were performed using a FACSCanto II flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) based on an FL1-A (530 ± 15 nm) versus FL2-A (585 ± 21 nm) dot plot and the H/R ratios of the targeting vectors were determined as the ratios of GFP-positive and FLAER-negative cells in the HygR–5′ EGFP reporter and the PIGA gene targeting assays, respectively. To isolate GFP-positive and FLAER-negative single cell clones, the cells were sorted using FACSAria III (BD Biosciences). All AAV-based gene targeting assays were repeated twice and representative data are presented in the Figures and the ‘Results’ section.
Total integration frequency and plating efficiency in AAV-based gene targeting assays
Total integration frequency was represented as the efficiency of G418-resistant colony formation by the transduction of AAV vectors. To determine this efficiency, the cells were plated in 25-cm2 flasks in triplicate at a density of 2 × 104 cells/flask (HCT116, DLD-1 and their derivatives), 1 × 105 cells/flask (PL5), 3 × 105 cells/flask (HuP-T3) or 1 × 106 cells/flask (AsPC-1), and infected with the respective AAV vectors at a MOI of 1 × 104 on the following day. The cells were then selected with G418 for 1–2 weeks until visible colonies were formed, fixed/stained with 3.7% formaldehyde (Sigma-Aldrich) containing 0.2% (wt/vol) crystal violet (Sigma-Aldrich) and subjected to colony counting. To determine the plating efficiencies, the cells were inoculated into 25-cm2 flasks in triplicate at the same densities described above, and dissociated and counted on the following day. The average values of triplicate samples were used as the plating efficiencies of the respective cell lines.
Quantitative reverse transcription (qRT)-PCR
For qRT-PCR of the NeoR gene, HCT116, DLD-1 or their derivative clones were plated into 6-well plates at a density of 1 × 104 cells/well and infected with respective AAV-based targeting vectors at a MOI of 1 × 104 on the following day. Four days after infection, total RNA was extracted from the cells in each well and converted to cDNA. PCR was carried out for each cDNA template in triplicate using SYBR Green Dye (Takara Bio) and StepOnePlus. A standard curve was constructed for each round of qRT-PCR using serially diluted samples, and gene expression in each sample was determined in reference to the standard curve. For qRT-PCR analyses of the HygR–5′ EGFP fusion gene and the PIGA gene, the HygR–5′ EGFP reporter clones as well as five cell lines employed in the disruption of PIGA exon 6 were propagated in exponential growth conditions and then processed for total RNA extraction and cDNA synthesis. PCR using cDNA samples was carried out using the same procedure as that for qRT-PCR analysis of the NeoR gene. Oligonucleotide primers used to amplify 186-, 160-, 211- and 184-bp fragments in the NeoR, HygR–5′ EGFP, PIGA and the GAPDH genes, respectively, are listed in Supplementary Table S1.
Western blotting
Five micrograms of whole cell protein extracts solubilized in Laemmli's sample buffer were resolved by 15% (for NeoR) and 10% (for GAPDH) SDS-polyacrylamide gel electrophoresis, transferred onto Immobilon-P membranes (Millipore, Bedford, MA, USA) and probed with primary and secondary antibodies. Primary antibodies used in this study include anti-neomycin phosphotransferase rabbit polyclonal antibody (Millipore) and anti-GAPDH rabbit monoclonal antibody (Cell Signaling Technology, Danvers, MA, USA). A horseradish peroxidase-linked anti-rabbit antibody (Cell Signaling Technology) was used as a secondary antibody. Membranes were visualized using ImmunoStar LD (Wako) and signals were quantified using ImageQuant LAS-4000 (GE Healthcare, Little Chalfont, UK) according to the manufacturers’ instructions.
CRISPR-Cas9-assisted gene targeting assay
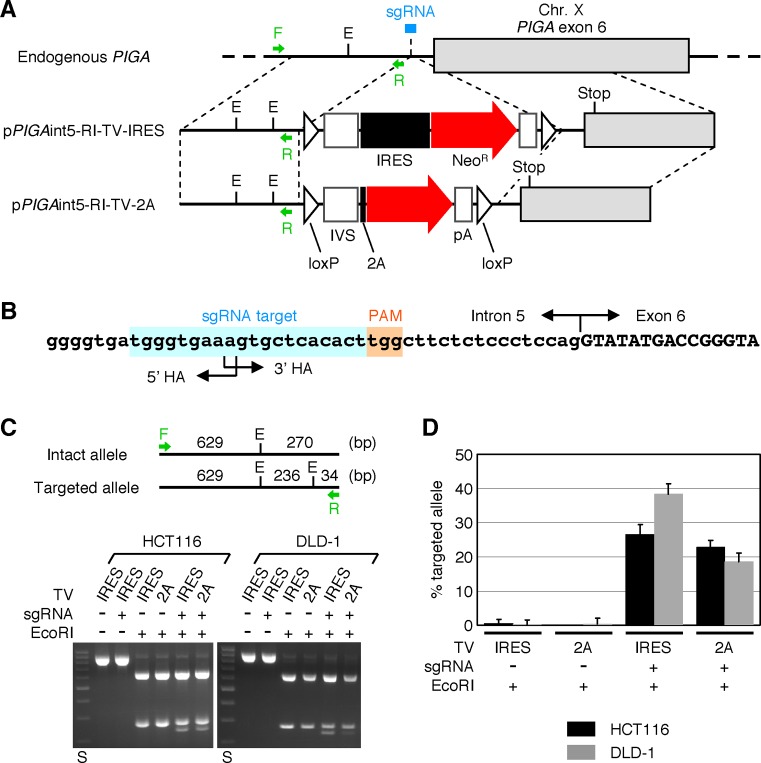
pSpCas9(BB)-2A-Puro (PX459) V2.0 (39), a versatile platform plasmid used for generating a CRISPR-Cas9 endonuclease, was a gift from Dr Feng Zhang (Broad Institute) obtained via Addgene (plasmid # 62988). To construct a CRISPR-Cas9 cleavase targeting the boundary of the 5′ and 3′ homology arms at PIGA intron 5 (PX459-PIGA), a pair of 25-mer oligonucleotides (Supplementary Table S1) were annealed with each other and cloned into BbsI-BbsI site in PX459. Unmodified PX459 was used as a vector control.
To construct the targeting vectors pPIGAint5-RI-TV-IRES and pPIGAint5-RI-TV-2A, a 5′ homology arm harboring an additional EcoRI site was generated by PCR using the pair of primers listed in Supplementary Table S1 and used to replace the 5′ homology arms in PIGAint5-TV-IRES and PIGAint5-TV-2A.
For gene targeting assays using these constructs, the HCT116 and DLD-1 cell lines were seeded onto 25-cm2 flasks in triplicate at a density of 2 × 104 cells/flask. On the following day, 5 μg/flask of PX459-PIGA (or PX459) and 11 μg/flask of either pPIGAint5-RI-TV-IRES or pPIGAint5-RI-TV-2A was mixed with 15 μl/flask of TransIT-LT1 and transfected into the cells as per the manufacturer's instruction. Transfected cells were selected with G418 for 1–2 weeks and processed for gDNA extraction. Restriction fragment length polymorphism (RFLP) analysis was then performed using the collected gDNA to determine the ratio of PIGA alleles in which an additional EcoRI site was introduced (representing targeted alleles) to the entire PIGA alleles (representing the sum of targeted and intact alleles). To this end, gDNA samples were first processed for PCR amplification using the primers shown in Supplementary Table S1, purified using the PureLink Quick PCR Purification Kit (Thermo Fisher Scientific), digested with EcoRI (or mock) and then separated by 2% agarose gel electrophoresis. Digital images of the gels were obtained using an AE-6932GXES Printgraph (ATTO, Tokyo, Japan) and analyzed to quantify the intensities of the 270- and 236-bp bands in each sample. The percentages of targeted alleles were calculated by the following equation: % targeted allele = 27000 × ‘236-bp band’/(236 × ‘270-bp band’ + 270 × ‘236-bp band’), where quotation marks represent the intensities of the indicated bands.
To determine the total integration frequencies in CRISPR-Cas9-assisted gene targeting assay, 3 × 104 cells/flask (HCT116) or 2 × 104 cells/flask (DLD-1) were seeded in 25-cm2 flasks in triplicate and, on the following day, co-transfected with PX459-PIGA (or PX459) and targeting vectors (pPIGAint5-RI-TV-IRES or pPIGAint5-RI-TV-2A) as described above. Cells were then selected with G418 until visible colonies were formed, fixed/stained and subjected to colony counting.
The CRISPR-Cas9-assisted gene targeting assay was repeated twice, and representative data are shown in Figure 6 and the ‘Results’ section.
Figure 6.
H/R ratios of plasmid-based targeting vectors in CRISPR-Cas9-assisted gene targeting. (A) A scheme describing the targeting of PIGA intron 5 with the assistance of a site-specific CRISPR-Cas9 endonuclease. Arrows associated with F or R indicate PCR primers used for RFLP analysis shown in (C). sgRNA: single guide RNA as a component of CRISPR-Cas9 system. E: EcoRI site. Other abbreviations are shown in the legends for Figures 1–3. (B) A sgRNA target sequence located at the junction of 5′ and 3′ homology arms. The genomic sequence of PIGA marked with a sgRNA target site, protospacer adjacent motif (PAM) and intron 5/exon 6 boundary is shown. (C) RFLP analysis. The HCT116 and DLD-1 cell lines were co-transfected with the above-described CRISPR-Cas9 cleavase along with either pPIGAint5-RI-TV-IRES (IRES) or pPIGAint5-RI-TV-2A (2A) and selected with G418. gDNA extracted from G418-resistant cells were processed for PCR amplification with the primer pair shown in (A), digested with EcoRI and separated by agarose gel electrophoresis. (Top) Deduced cleavage patterns of PCR products amplified from intact and targeted PIGA alleles. (Bottom) Representative gel images. A plasmid expressing Cas9 and a sgRNA without 20-nt guide RNA was used as a control for the CRISPR-Cas9 cleavase. DNA size standards (S) include 200–1000 bp DNA fragments with an interval of 100 bp. (D) The ratio of targeted versus entire PIGA alleles determined based on the intensities of 270 and 236-bp bands (mean ± s.e.m.; n = 3).
RESULTS
Platform plasmid pAAV-2Aneo to construct 2A-based promoter-trap gene targeting vectors with an AAV backbone
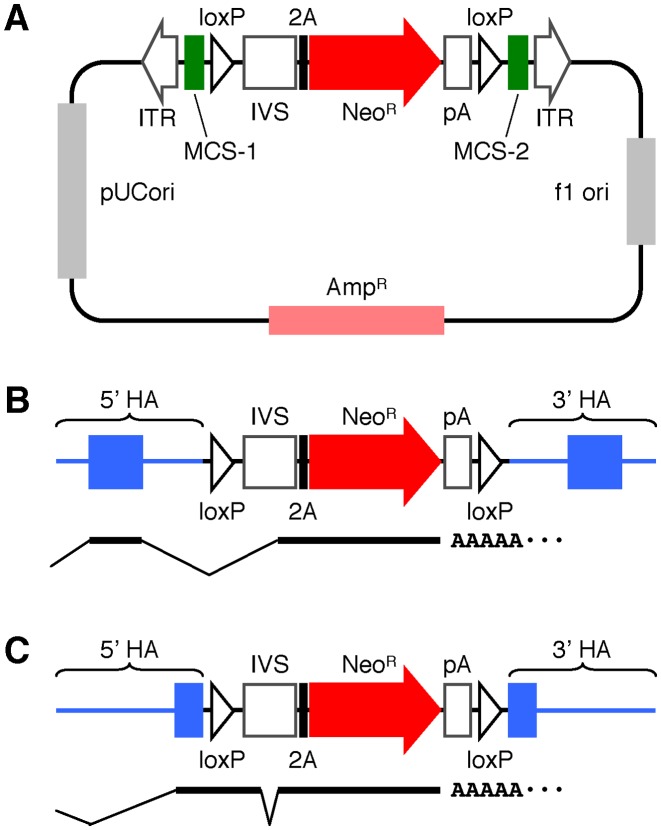
We first generated a platform plasmid, pAAV-2Aneo, which facilitates the construction of AAV-based targeting vectors using a 2A-based promoter-trap system. pAAV-2Aneo harbors a promoter-trap module consisting of an IVS, the insect Thosea asigna virus 2A, the NeoR gene and a polyadenylation site (Figure 1A). This promoter-trap module is encompassed by two loxP sites, which are flanked by two separate MCSs to allow for the ligation of 5′ and 3′ homology arms, respectively. In addition, a ‘frame adjuster’ consisting of two series of BspEI, MluI and BsrGI sites is located between IVS and 2A (Supplementary Figure S1A). Finally, all components described above are placed between two ITRs.
Figure 1.
Schematic representation of pAAV-2Aneo. (A) A plasmid map of pAAV-2Aneo. ITR: inverted terminal repeat, MCS: multiple cloning site, IVS: synthetic intron, 2A: Thosea asigna virus 2A, NeoR: neomycin resistance (neomycin phosphotransferase) gene, pA: polyadenylation site, f1 ori: fi single-strand DNA origin, AmpR: ampicillin resistance (β-lactamase) gene, pUCori: pUC plasmid replication origin. (B and C) Diagrams of targeting vectors constructed based on pAAV-2Aneo in which a promoter-trap module is inserted into an intron (B) or an exon (C). Predicted splicing of mRNAs derived from these targeting vectors is shown at the bottom of the diagrams. HA: homology arm.
When a targeting vector disrupting an intron of a gene upon homologous integration is constructed based on pAAV-2Aneo, the 5′ portion of the disrupted gene is transcribed as a fusion mRNA with 2A and NeoR (Figure 1B). By properly processing the frame adjuster during construction of the targeting vectors, 2A and NeoR are translated in the same reading frame with the disrupted gene (Supplementary Figure S1B). Meanwhile, if a targeting vector based on pAAV-2Aneo is designed such that it introduces the promoter-trap module into an exon, the 5′ portion of the disrupted gene is transcribed as a fusion mRNA with loxP, the exonic regions of IVS, 2A and NeoR (Figure 1C). By properly designing the junction of MCS-1 and the proximal end of the 5′ homology arm, the 5′ portion of the disrupted gene is translated as a fusion polypeptide with loxP and the exonic sequences of IVS (Supplementary Figure S1A). In addition, by cleaving the vector at two BsrGI sites within the frame adjuster followed by self-religation, the fusion polypeptide is translated in the same reading frame with 2A and NeoR. Collectively, production of the NeoR protein is permitted by proper processing of the frame adjuster during vector construction, regardless of whether the targeting vector is designed to disrupt an exon or an intron in a targeted gene.
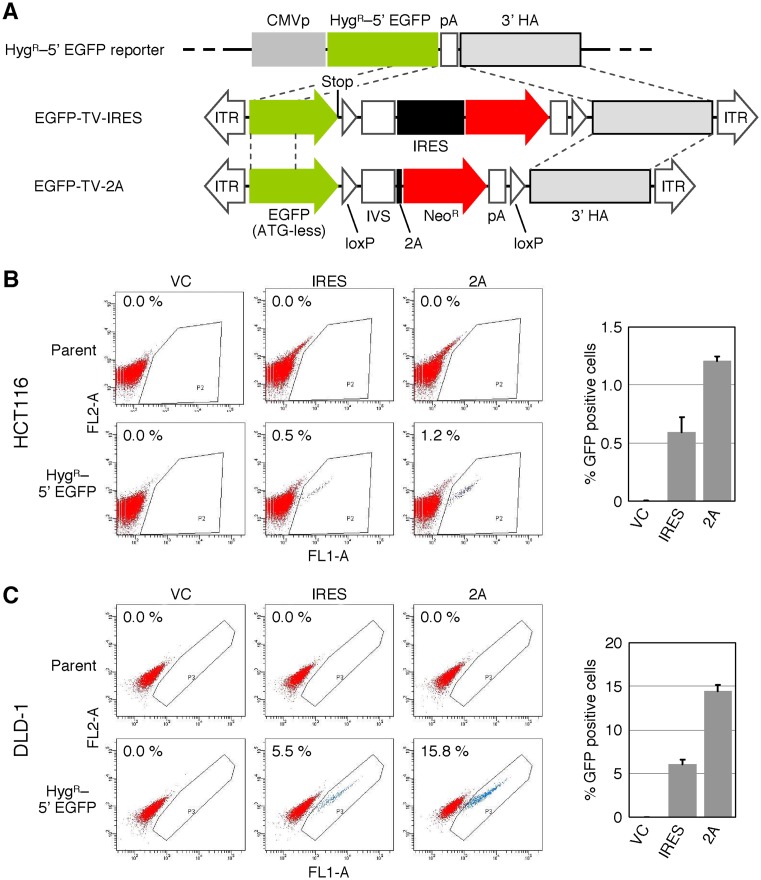
2A-based promoter trapping assessed by the HygR–5′ EGFP reporter gene targeting assay
We then sought to utilize pAAV-2Aneo to evaluate gene targeting efficiency achieved with 2A-based promoter trapping. To this end, we employed a molecular system which monitors the efficiency of homologous recombination using a reporter vector comprised of the HygR–5′ EGFP gene (Figure 2A) (11). An AAV-based targeting vector was created by inserting 5′ and 3′ arms with homology to this reporter vector into the multiple cloning sites of pAAV-2Aneo (EGFP-TV-2A). In addition, we employed an AAV-based targeting vector identical to EGFP-TV-2A except that IRES was placed instead of the frame adjuster and 2A (EGFP-TV-IRES). Homologous recombination between the HygR–5′ EGFP reporter and one of these targeting vectors leads to GFP signal emission from the cells. In our previous study, the HygR–5′ EGFP reporter vector was transfected into the HCT116 and the DLD-1 cell lines and a reporter clone in which a single reporter vector was stably integrated into an unknown locus of the genome was isolated from each cell line (11). We infected these reporter clones with the above-described AAV-based targeting vectors, selected the cells with G418 and performed FCM analysis. As a result, EGFP-TV-2A achieved 2.0–2.4-fold higher H/R ratios than EGFP-TV-IRES (Figure 2B and C). GFP-positive single-cell clones were then isolated from the cells infected with EGFP-TV-2A by cell sorting during FCM analysis, and the sequences of a fusion mRNA consisting of HygR, EGFP, loxP, IVS, 2A and NeoR were determined. As expected, the fusion mRNA was spliced as shown in the schematic Figure 1C (Supplementary Figure S2).
Figure 2.
H/R ratios of targeting vectors assessed with HygR–5′ EGFP reporter system. (A) A schematic description of the HygR–5′ EGFP reporter system. Thin dotted lines indicate homologous regions between reporter vector and two targeting vectors. CMVp: CMV promoter, HygR–5′ EGFP: fusion gene consisting of hygromycin resistance (hygromycin B phosphotransferase) gene and 3′-truncated enhanced GFP, TV: targeting vector, Stop: stop codon (TAA), IRES: internal ribosome entry site. For other abbreviations, see the legend for Figure 1. (B and C) H/R ratios of AAV-based targeting vectors determined using the HygR–5′ EGFP reporter system. The HCT116 (B) and DLD-1 (C) cell lines carrying a single HygR–5′ EGFP reporter vector integrated in the genome were infected with the AAV-based targeting vector EGFP-TV-IRES (IRES) or EGFP-TV-2A (2A), selected with G418, and then analyzed by FCM. An AAV vector targeting an unrelated gene created based on pAAV-2Aneo was used as a vector control (VC). For further controls, parental HCT116 and DLD-1 cells were infected with the same series of targeting vectors and analyzed in parallel. (Left) Representative dot plots obtained in FCM analyses. The percentage of GFP-positive cells is noted within each plot. (Right) Graphical representation of data obtained with cell clones carrying a HygR–5′ EGFP reporter vector (mean ± s.e.m.; n = 3).
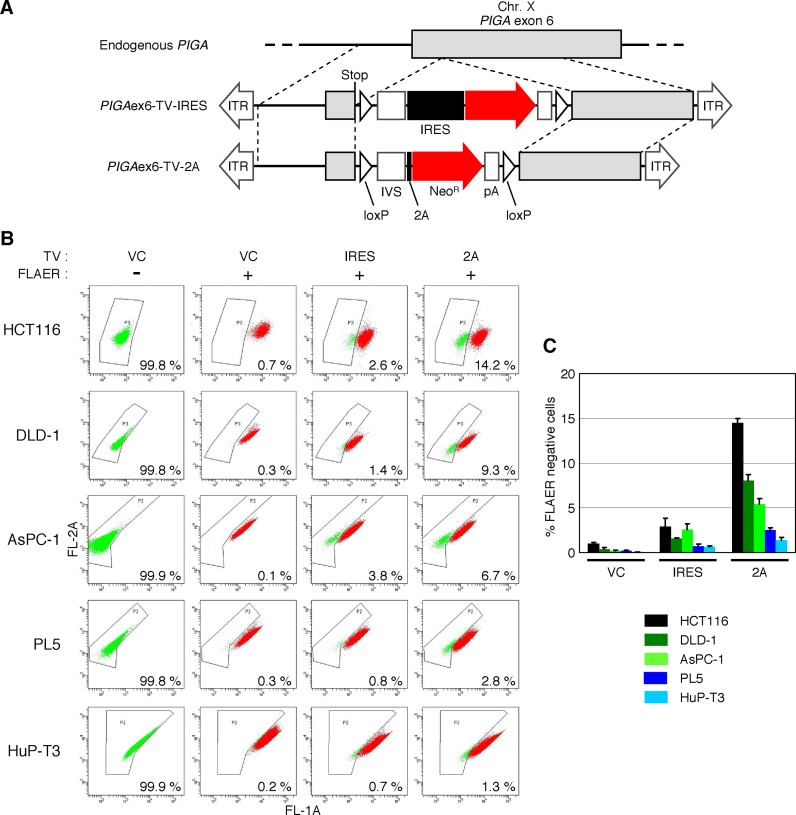
2A-based promoter trapping assessed by endogenous PIGA gene targeting assay
We also employed a reporter system exploiting an endogenous gene to evaluate the gene targeting efficiencies achieved with 2A-and IRES-based promoter-trap systems. The PIGA gene, which is indispensable for glycosylphosphatidylinositol (GPI)-anchor synthesis and is located on the X-chromosome (40,41), was chosen as the reporter gene, and a targeting vector disrupting PIGA exon 6 (PIGAex6-TV-2A) was constructed based on pAAV-2Aneo (Figure 3A). We also employed an AAV-based targeting vector that was mostly identical to PIGAex6-TV-2A but contained IRES in place of the frame adjuster and 2A (PIGAex6-TV-IRES). These targeting vectors were used to infect five cell lines of male origin carrying a single X-chromosome (Figure 3B). Infected cells were selected with G418 and then stained with FLAER, a reagent that specifically binds to GPI-anchors (42). Staining with FLAER followed by FCM analysis enabled identification of GPI-anchor-deficient cells emerging upon the disruption of PIGA as a FLAER-negative cell population. This assay demonstrated that ∼2–5-fold higher H/R ratios were achieved by the use of PIGAex6-TV-2A compared with the use of PIGAex6-TV-IRES (Figure 3B and C), which was largely concordant with the data acquired with the HygR–5′ EGFP reporter system. FLAER-negative single-cell clones were then isolated from the cells infected with PIGAex6-TV-2A by cell sorting during FCM analysis. We sequenced the fusion mRNA that included parts of PIGA, loxP, IVS, 2A and NeoR in these clones and confirmed that this mRNA was spliced as expected (Figure 1C, Supplementary Figure S3).
Figure 3.
H/R ratios of AAV-based targeting vectors assessed by disruption of PIGA exon 6. (A) A scheme describing the disruption of PIGA exon 6. Thin dotted lines indicate homologous regions between the wild-type PIGA locus and targeting vectors. Stop: artificially introduced stop codon (TAA). For other abbreviations, see legends for Figures 1 and 2. (B) H/R ratios of targeting vectors disrupting PIGA exon 6. Five cell lines denoted on the left were infected with the AAV-based targeting vector PIGAex6-TV-IRES (IRES), PIGAex6-TV-2A (2A) or an AAV vector targeting an unrelated gene created based on pAAV-2Aneo (VC). Infected cells were selected with G418, stained with FLAER, and then analyzed by FCM. Cells infected with VC but not stained with FLAER were included as further controls. Shown are representative dot plots with the ratio of FLAER-negative cells noted within respective plots. (C) Graphical representation of data obtained with cells subjected to FLAER-staining in the assay described in (B) (mean ± s.e.m.; n = 3).
2A-based promoter trapping assessed by targeting of a PIGA intron
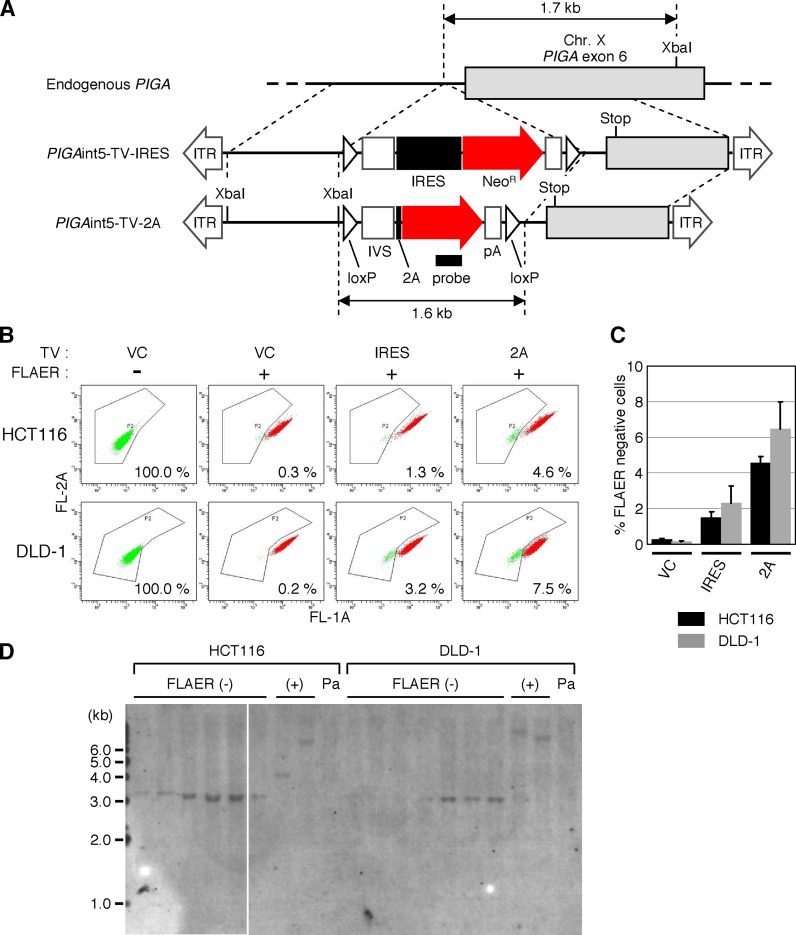
Targeting an intron of a gene (inserting an antibiotic resistance gene cassette into an intronic region) is a common strategy for targeted knock-in of a small mutation, insertion or deletion into an exon (3,43–46). We subsequently investigated whether the use of 2A for promoter trapping also enhances the efficiency of gene targeting when an intron of a gene is targeted. Using pAAV-2Aneo containing a modified frame adjuster (Supplementary Figure S4A) as a platform for construction, we created an AAV-based targeting vector to introduce a promoter-trap module into the PIGA intron 5 upon homologous integration into the genome (PIGAint5-TV-2A; Figure 4A). We also prepared another targeting vector that was very similar to PIGAint5-TV-2A but harbored IRES rather than the frame adjuster and 2A (PIGAint5-TV-IRES). These targeting vectors introduce a non-sense point mutation in exon 6 upon homologous integration into the PIGA locus, leading to the production of a non-functional PIGA protein. The assay was carried out using the HCT116 and the DLD-1 cell lines, as these cell lines showed higher H/R ratios than the other cell lines in the previous assay using PIGAex6-TV-2A. Cells were infected with the above-mentioned targeting vectors against PIGA intron 5, selected with G418, stained with FLAER and analyzed by FCM. We found that the use of 2A for promoter trapping led to ∼3-fold higher H/R ratios compared to the use of IRES in this assay (Figure 4B and C). FLAER-negative single-cell clones were then established and processed for Southern blot analysis, confirming homologous integration of the targeting vector PIGAint5-TV-2A into the PIGA gene locus (Figure 4D). In addition, sequence analysis of these clones indicated that splicing occurred as predicted (schematically shown in Figure 1B) in the fusion mRNA involving partial PIGA, 2A and NeoR (Supplementary Figure S4B).
Figure 4.
H/R ratios of AAV-based targeting vectors assessed by targeting of a PIGA intron. (A) A scheme describing the targeting of PIGA intron 5. A filled small rectangle indicates the location of a probe used in Southern blot analysis shown in (D). XbaI: restriction enzyme XbaI site. For other abbreviations, see legends for Figures 1–3. (B and C) H/R ratios of targeting vectors disrupting PIGA intron 5. The HCT116 and DLD-1 cell lines were infected with the AAV-based targeting vectors PIGAint5-TV-IRES (IRES), PIGAint5-TV-2A (2A) or an AAV vector targeting an unrelated gene created based on pAAV-2Aneo (VC). Infected cells were selected with G418, stained with FLAER and then analyzed by FCM. Cells infected with VC but not stained with FLAER were included as further controls. Shown are representative dot plots with the ratio of FLAER-negative cells noted in the respective plots (B) and a graphic representation of data obtained with cells subjected to FLAER-staining (mean ± s.e.m.; n = 3) (C). (D) Southern blot analysis of FLAER-negative and -positive single-cell clones. FLAER-negative and -positive cells were sorted from the HCT116 and DLD-1 cell lines infected with PIGAint5-TV-2A and subjected to single cell cloning. Genomic DNA was isolated from the resulting clones, digested with XbaI and processed for Southern blot analysis with a probe depicted in (A). A white line within the gel image indicates the border of two distinct gels. Pa: parental cell line.
Absolute gene targeting frequency achieved using 2A-based promoter trapping
We subsequently addressed how the use of 2A and IRES for promoter trapping affected the total integration frequencies and the absolute gene targeting frequencies of AAV-based targeting vectors in respective experiments (Table 1). Total integration frequency of a targeting vector can be defined as the ratio of ‘the cells in which a targeting vector is integrated in the genome’ versus ‘the cells exposed to AAV particles containing the targeting vector’, with the former represented by the number of G418-resistant colonies in our study. Absolute gene targeting frequencies can be determined by multiplying the total integration frequencies and H/R ratios. This analysis demonstrated that, although absolute gene targeting frequencies significantly varied depending on the genes targeted and the cell lines analyzed, higher absolute gene targeting frequencies (3.4–28-fold) were consistently achieved using 2A rather than IRES for promoter trapping. The total integration frequency was also higher by the use of 2A rather than IRES in all AAV-mediated gene targeting assays (1.6–5.4-fold).
Table 1. Total integration frequencies and absolute gene targeting frequencies achieved using the 2A-based or IRES-based promoter-trap system.
| Experiment | Cell line | Vector | Total integration frequencya | 2A to IRES ratio in total integration frequencyc | Absolute gene targeting frequencyb | 2A to IRES ratio in absolute gene targeting frequencyc | ||
|---|---|---|---|---|---|---|---|---|
| Mean | S.e.m. | Mean | S.e.m. | |||||
| HygR–5′ EGFP | HCT116 | IRES | 4.0 × 10−2 | 1.2 × 10−3 | 2.3 × 10−4 | 4.4 × 10−5 | ||
| 2A | 6.3 × 10−2 | 1.1 × 10−3 | 1.6 | 7.7 × 10−4 | 9.4 × 10−6 | 3.4 | ||
| DLD-1 | IRES | 1.7 × 10−2 | 1.5 × 10−4 | 1.0 × 10−3 | 8.4 × 10−5 | |||
| 2A | 3.7 × 10−2 | 4.3 × 10−4 | 2.2 | 5.3 × 10−3 | 2.1 × 10−4 | 5.1 | ||
| PIGA exon 6 | HCT116 | IRES | 4.5 × 10−3 | 2.6 × 10−4 | 1.2 × 10−4 | 3.7 × 10−5 | ||
| 2A | 8.3 × 10−3 | 3.1 × 10−4 | 1.8 | 1.2 × 10−3 | 1.4 × 10−5 | 9.6 | ||
| DLD-1 | IRES | 2.4 × 10−3 | 1.5 × 10−4 | 3.7 × 10−5 | 1.4 × 10−6 | |||
| 2A | 1.3 × 10−2 | 5.8 × 10−4 | 5.4 | 1.0 × 10−3 | 5.0 × 10−5 | 28 | ||
| AsPC-1 | IRES | 2.3 × 10−5 | 2.5 × 10−6 | 5.3 × 10−7 | 1.8 × 10−7 | |||
| 2A | 6.7 × 10−5 | 2.5 × 10−6 | 2.9 | 3.7 × 10−6 | 6.2 × 10−7 | 7.0 | ||
| PL5 | IRES | 2.8 × 10−4 | 1.4 × 10−5 | 1.9 × 10−6 | 6.6 × 10−7 | |||
| 2A | 4.4 × 10−4 | 2.1 × 10−5 | 1.6 | 1.2 × 10−5 | 7.1 × 10−7 | 6.1 | ||
| HuP-T3 | IRES | 2.3 × 10−4 | 1.1 × 10−5 | 1.2 × 10−6 | 4.4 × 10−7 | |||
| 2A | 4.0 × 10−4 | 1.9 × 10−5 | 1.8 | 5.3 × 10−6 | 8.8 × 10−8 | 4.6 | ||
| PIGA intron 5 | HCT116 | IRES | 3.5 × 10−3 | 5.1 × 10−5 | 4.5 × 10−5 | 1.7 × 10−5 | ||
| 2A | 1.3 × 10−2 | 3.6 × 10−4 | 3.6 | 6.0 × 10−4 | 6.9 × 10−5 | 13 | ||
| DLD-1 | IRES | 7.8 × 10−3 | 2.9 × 10−4 | 1.3 × 10−4 | 1.0 × 10−4 | |||
| 2A | 3.7 × 10−2 | 5.5 × 10−4 | 4.7 | 2.9 × 10−3 | 9.6 × 10−5 | 22 | ||
| + CRISPR-Cas9 | HCT116 | IRES | 1.6 × 10−2 | 2.8 × 10−4 | 4.2 × 10−3 | 3.8 × 10−4 | ||
| 2A | 7.9 × 10−2 | 6.4 × 10−4 | 5.0 | 1.8 × 10−2 | 1.4 × 10−3 | 4.3 | ||
| DLD-1 | IRES | 3.2 × 10−3 | 8.4 × 10−5 | 1.2 × 10−3 | 6.9 × 10−5 | |||
| 2A | 1.0 × 10−1 | 1.6 × 10−3 | 31 | 1.9 × 10−2 | 2.3 × 10−3 | 15 | ||
aTotal integration frequency was calculated by dividing ‘the number of G418-resistant colonies’ by ‘the number of cells inoculated for AAV infection (or plasmid transfection)’ and ‘plating efficiency’. Plating efficiencies empirically determined were 0.98 (HCT116 and its derivative), 0.87 (DLD-1 and its derivative), 0.79 (AsPC-1), 0.86 (PL5) and 0.95 (HuP-T3).
bAbsolute gene targeting frequency was calculated by multiplying total integration frequency and H/R ratio.
c‘2A to IRES ratio’ is the mean value for 2A divided by the mean value for IRES.
NeoR protein expression using the 2A-based promoter-trap system
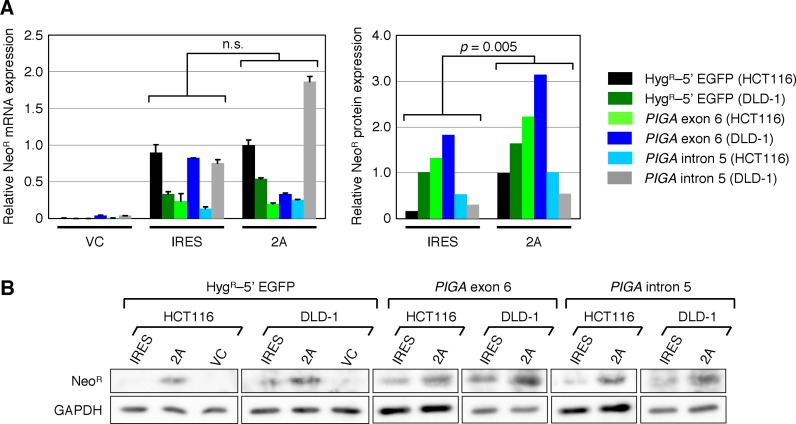
The higher absolute gene targeting frequencies and the larger number of G418-resistant colonies obtained using 2A in the promoter-trap system may be attributable to the higher yield of NeoR protein downstream of 2A rather than IRES. To examine whether the NeoR protein was indeed expressed in higher levels using 2A, we next determined the levels of NeoR expressed from targeting vectors at the mRNA and protein levels. As shown in Figure 5A, qRT-PCR analysis revealed no significant difference between the expression levels of NeoR mRNA using 2A and IRES. In contrast, western blot analyses demonstrated significantly higher levels of NeoR protein derived from targeting vectors with 2A rather than those with IRES (Figure 5A and B). These data may at least partially account for the higher absolute gene targeting frequencies and total integration frequencies attained using 2A rather than IRES for promoter trapping in AAV-based targeting vectors.
Figure 5.
Levels of NeoR expression from AAV-based targeting vectors carrying the IRES- or 2A-based promoter-trap systems. (A) Expression levels of the NeoR mRNA (left) and protein (right). HCT116 and DLD-1 clones carrying a HygR–5′ EGFP reporter were infected with EGFP-TV-IRES (IRES) or EGFP-TV-2A (2A). Parental HCT116 and DLD-1 cells were infected with AAV-based targeting vectors against PIGA exon 6 (PIGAex6-TV-IRES and PIGAex6-TV-2A) and intron 5 (PIGAint5-TV-IRES and PIGAint5-TV-2A). An AAV vector carrying no NeoR gene was included as a control (VC) in each set of infections. The expression levels of the NeoR mRNA and protein in the infected cells were determined by qRT-PCR (mean ± s.e.m.; n = 3) and western blotting, respectively, and shown after normalization to GAPDH. Two-tailed paired t-test was applied for statistical analyses of data. (B) Gel images of western blotting for the NeoR and GAPDH proteins.
Expression levels of the HygR–5′ EGFP and the PIGA genes in reporter cell lines (clones)
In the gene targeting assays described above, substantially distinct H/R ratios and absolute gene targeting frequencies were achieved depending on the cell line (clone) used for the assays. Because gene targeting in these assays was performed based on the promoter-trap strategy, the transcription levels of the reporter genes in each cell line (clone) may affect the overall gene targeting efficiency. In addition, transcriptional activity at the genomic loci may be correlated with the accessibility of the loci by targeting vectors. We thus assessed the expression levels of reporter genes, the HygR–5′ EGFP and the PIGA genes, in the reporter cell lines (clones) by quantitative RT-PCR. No direct correlation was observed between the expression levels of the reporter genes and either H/R ratios or absolute gene targeting frequencies (Supplementary Figure S5). These results suggest that the expression level of a target gene is not a primary factor determining gene targeting efficiency, and other factors, including the activities of homologous recombination and non-homologous end joining, may influence gene targeting efficiency in these assays.
Assessment of 2A-based promoter-trap system in plasmid-mediated gene targeting assisted by CRISPR-Cas9 system
Finally, we examined whether the advantage of using the 2A-based promoter-trap system in AAV-based targeting vectors could be applied to gene modification conducted using plasmid-based targeting vectors as well. An attempt to disrupt PIGA intron 5 by transfecting HCT116 and DLD-1 cell lines with the plasmid version of targeting vectors (pPIGAint5-RI-TV-IRES and pPIGAint5-RI-TV-2A) failed to exhibit appreciable FLAER-negative cell populations (Figure 6C and data not shown), which agrees with the results of previous reports demonstrating less efficient gene targeting by plasmid-based targeting vectors compared to AAV-based targeting vectors (6,8–11). We thus employed a site-specific endonuclease to introduce a DNA double-strand break and enhance homologous recombination between targeting vectors and the endogenous PIGA locus. A CRISPR-Cas9 cleavase was designed over the genomic position corresponding to the boundary of the 5′ and 3′ homology arms (Figure 6A and B), and transfected into the cell lines in combination with targeting vectors pPIGAint5-RI-TV-IRES or pPIGAint5-RI-TV-2A. gDNA was then extracted from the bulk populations of G418-resistant cells and PCR-amplified for subsequent RFLP analysis. This assay demonstrated that the use of 2A for promoter trapping lead to equivalent (HCT116) or lower (DLD-1) H/R ratios than the use of IRES in the context of plasmid-based gene targeting coupled with CRISPR-Cas9-mediated DNA double-strand break (Figure 6C and D).
To verify the integrity of targeted PIGA alleles, we stained subsets of G418-resistant HCT116 cells with FLAER, sorted FLAER-negative cell populations, isolated 24 single-cell clones and sequenced both homology arms and their flanking regions. All 24 clones were found to have undergone precise homologous integration of the targeting vector and harbored no additional CRISPR-Cas9-mediated genetic alterations in targeted PIGA alleles. The sequences of the primer-annealing sites for PCR amplification of RFLP fragments were intact in all 24 clones.
We also determined the number of G418-resistant colonies formed by each co-transfection, and then calculated the total integration frequencies and absolute gene targeting frequencies. Both of these frequencies were higher when 2A rather than IRES was used for promoter trapping (Table 1).These results collectively indicated that, in plasmid-based gene targeting assisted by CRISPR-Cas9 system, the use of 2A for promoter trapping did not increase H/R ratios of a targeting vector but did increase absolute gene targeting frequencies relative to the use of IRES.
DISCUSSION
In this study, we performed a comparative analysis of two distinct promoter-trap systems, those exploiting 2A and IRES respectively, within AAV-based targeting vectors and CRISPR-Cas9-assisted gene targeting. To our knowledge, this is the first systematic comparison of gene targeting efficiencies achieved using these promoter-trap systems. We found that, in AAV-mediated gene targeting, the absolute gene targeting frequencies achieved with 2A-based promoter trapping was 3.4–28-fold greater than those achieved with IRES-based promoter trapping. In addition, the number of cell clones obtained via G418 selection was 1.6–5.4-fold higher when 2A was used for promoter trapping. These results may be attributable to the distinct translation levels at the downstream of 2A and IRES; i.e. 2A allows the production of an equal amount of proteins from the genes connected by 2A (28), while a gene downstream of IRES is likely translated at a lower level than the gene upstream of IRES (29). Thus, if two distinct AAV-based targeting vectors, those employing 2A and IRES respectively for promoter trapping, are integrated into the same locus within the genome, the antibiotic resistance protein is likely translated at a higher level with the use of 2A. This agrees with our observations that the use of AAV-based targeting vectors carrying 2A rather than IRES resulted in the production of larger amounts of NeoR protein from the targeting vectors and the formation of larger numbers of gene-targeted cell clones and G418-resistant colonies.
In regard to the H/R ratios of targeting vectors, our data indicate that 2A-based promoter trapping permits 2–5-fold more efficient enrichment of gene-targeted cell clones compared to IRES-based promoter trapping in AAV-based targeting vectors. Unlike IRES, genes upstream and downstream of 2A must be connected in the same reading frame so that the downstream gene can be translated into a functional protein. Therefore, in theory, no more than one-third of the cell clones undergoing random integration of targeting vectors into intragenic genomic regions acquire antibiotic resistance when 2A is used for promoter trapping. In contrast, when promoter trapping is attained using IRES, all cell clones that undergo random integration of targeting vectors within intragenic genomic regions are eligible to acquire antibiotic resistance. Our data were largely in agreement with this argument as the H/R ratios of targeting vectors were consistently higher when promoter trapping was attained using 2A rather than IRES.
Another strategy to achieve promoter trapping that is as efficient as the 2A-based promoter-trap system may be to employ a targeting vector in which an antibiotic resistance gene is translated as a part of a fusion protein with the targeted endogenous gene (47–49). This type of targeting vector also secures the same amount of translation products from the antibiotic resistance gene in the vector and the endogenous gene at the targeted locus. In addition, when targeting vectors are randomly integrated into the genome and the antibiotic resistance gene is fused with an endogenous gene in a different reading frame, the antibiotic resistance gene is not translated into a functional protein, thus permitting the enrichment of gene-targeted cell clones similarly to 2A-based promoter trapping. However, an antibiotic resistance protein fused to the C-terminus of some polypeptides may fail to confer antibiotic resistance even when it is translated in-frame, depending on the function and/or structure of the polypeptide fused to the antibiotic resistance protein. In this regard, two genes connected via 2A are translated as proteins that are separate from each other, reducing the possibility that the targeting vectors homologously integrated into the genome fail to confer antibiotic resistance. Other types of promoter-trap targeting vectors that have been often used include those in which the coding region of a targeted gene was precisely replaced with the coding sequence of an antibiotic-resistance gene at the ATG start codon (50–52). However, although useful, this promoter-trap strategy can only be applied to a limited variety of gene modifications, typically gene knockout.
In this study, the use of 2A-based promoter trapping in targeting vectors led to better absolute gene targeting frequencies not only in AAV-mediated gene targeting but also in CRISPR-Cas9-assisted gene targeting. However, unlike in AAV-mediated gene targeting, the use of 2A-based promoter trapping did not result in higher H/R ratios in CRISPR-Cas9-assisted gene targeting. In this regard, we found that the number of NeoR colonies formed upon transfection of the targeting vector was greater when 2A rather than IRES was used for promoter trapping, and this upregulation was particularly remarkable (31-fold) in DLD-1 cells. Although the mechanism underlying enhanced random integration of the plasmid vector carrying 2A in comparison to IRES is unknown, this result might be related to the finding that AAV vectors are preferably integrated into gene-rich and transcriptionally active regions of the genome (53,54). Both 2A and IRES might be able to support the production of an amount of the NeoR protein sufficient for conferring G418-resistance from many randomly integrated AAV vectors, but many randomly integrated plasmids that are integrated into transcriptionally less active regions of the genome might confer G418 resistance only downstream of 2A.
In summary, our data indicate that the use of 2A compared to IRES for promoter trapping results in better gene targeting efficiencies in AAV-mediated gene targeting. In CRISPR-Cas9-assisted gene targeting, the utility of 2A-based promoter trapping may depend on the aim of the gene targeting experiments, as this method enhances the absolute gene targeting frequencies but not the H/R ratios of the targeting vectors. The findings in this study will help to improve the design of AAV-based targeting vectors and to develop an efficient technology of gene targeting readily applicable to various human cell lines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We would like to thank Drs Fred Bunz (Johns Hopkins University) and Feng Zhang (Broad Institute) for providing pSEPT and PX459 V2.0, respectively.
FUNDING
Grants-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science (JSPS) [25460395 to S.K., 15K19561 to A.O., 25640107 to H.K.]; Strategic Research Foundation Grant-aided Project for Private Universities from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan, 2011–2015 [S1101027 to S.K., Y.H., H.K.]; Takeda Science Foundation (to H.K.). Funding for open access charge: MEXT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sedivy J.M., Vogelstein B., Liber H.L., Hendrickson E.A., Rosmarin A. Gene targeting in human cells without isogenic DNA. Science. 1999;283:9a. [Google Scholar]
- 2.Di Nicolantonio F., Arena S., Gallicchio M., Zecchin D., Martini M., Flonta S.E., Stella G.M., Lamba S., Cancelliere C., Russo M., et al. Replacement of normal with mutant alleles in the genome of normal human cells unveils mutation-specific drug responses. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20864–20869. doi: 10.1073/pnas.0808757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sur S., Pagliarini R., Bunz F., Rago C., Diaz L.A., Jr, Kinzler K.W., Vogelstein B., Papadopoulos N. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3964–3969. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Nicolantonio F., Arena S., Tabernero J., Grosso S., Molinari F., Macarulla T., Russo M., Cancelliere C., Zecchin D., Mazzucchelli L., et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J. Clin. Invest. 2010;120:2858–2866. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arbonés M.L., Austin H.A., Capon D.J., Greenburg G. Gene targeting in normal somatic cells: inactivation of the interferon-gamma receptor in myoblasts. Nat. Genet. 1994;6:90–97. doi: 10.1038/ng0194-90. [DOI] [PubMed] [Google Scholar]
- 6.Russell D.W., Hirata R.K. Human gene targeting by viral vectors. Nat. Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan I.F., Hirata R.K., Russell D.W. AAV-mediated gene targeting methods for human cells. Nat. Protoc. 2011;6:482–501. doi: 10.1038/nprot.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porteus M.H., Cathomen T., Weitzman M.D., Baltimore D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol. Cell. Biol. 2003;23:3558–3565. doi: 10.1128/MCB.23.10.3558-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Topaloglu O., Hurley P.J., Yildirim O., Civin C.I., Bunz F. Improved methods for the generation of human gene knockout and knockin cell lines. Nucleic Acids Res. 2005;33:e158. doi: 10.1093/nar/gni160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karnan S., Konishi Y., Ota A., Takahashi M., Damdindorj L., Hosokawa Y., Konishi H. Simple monitoring of gene targeting efficiency in human somatic cell lines using the PIGA gene. PLoS One. 2012;7:e47389. doi: 10.1371/journal.pone.0047389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konishi Y., Karnan S., Takahashi M., Ota A., Damdindorj L., Hosokawa Y., Konishi H. A system for the measurement of gene targeting efficiency in human cell lines using an antibiotic resistance-GFP fusion gene. Biotechniques. 2012;53:141–152. doi: 10.2144/0000113911. [DOI] [PubMed] [Google Scholar]
- 12.Porteus M.H., Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 13.Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 14.Christian M., Cermak T., Doyle E.L., Schmidt C., Zhang F., Hummel A., Bogdanove A.J., Voytas D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F., Meng X., Paschon D.E., Leung E., Hinkley S.J., et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 16.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mali P., Esvelt K.M., Church G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howes R., Schofield C. Genome engineering using Adeno-Associated Virus (AAV) Methods Mol. Biol. 2015;1239:75–103. doi: 10.1007/978-1-4939-1862-1_5. [DOI] [PubMed] [Google Scholar]
- 19.Melo S.P., Lisowski L., Bashkirova E., Zhen H.H., Chu K., Keene D.R., Marinkovich M.P., Kay M.A., Oro A.E. Somatic correction of junctional epidermolysis bullosa by a highly recombinogenic AAV variant. Mol. Ther. 2014;22:725–733. doi: 10.1038/mt.2013.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sebastiano V., Zhen H.H., Haddad B., Bashkirova E., Melo S.P., Wang P., Leung T.L., Siprashvili Z., Tichy A., Li J., et al. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci. Transl. Med. 2014;6:264ra163. doi: 10.1126/scitranslmed.3009540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daya S., Berns K.I. Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 2008;21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller C., Flotte T.R. Clinical gene therapy using recombinant adeno-associated virus vectors. Gene Ther. 2008;15:858–863. doi: 10.1038/gt.2008.68. [DOI] [PubMed] [Google Scholar]
- 23.Barzel A., Paulk N.K., Shi Y., Huang Y., Chu K., Zhang F., Valdmanis P.N., Spector L.P., Porteus M.H., Gaensler K.M., et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517:360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirata R., Chamberlain J., Dong R., Russell D.W. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol. 2002;20:735–738. doi: 10.1038/nbt0702-735. [DOI] [PubMed] [Google Scholar]
- 25.Rago C., Vogelstein B., Bunz F. Genetic knockouts and knockins in human somatic cells. Nat. Protoc. 2007;2:2734–2746. doi: 10.1038/nprot.2007.408. [DOI] [PubMed] [Google Scholar]
- 26.Konishi H., Lauring J., Garay J.P., Karakas B., Abukhdeir A.M., Gustin J.P., Konishi Y., Park B.H. A PCR-based high-throughput screen with multiround sample pooling: application to somatic cell gene targeting. Nat. Protoc. 2007;2:2865–2874. doi: 10.1038/nprot.2007.409. [DOI] [PubMed] [Google Scholar]
- 27.Ryan M.D., King A.M., Thomas G.P. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol. 1991;72:2727–2732. doi: 10.1099/0022-1317-72-11-2727. [DOI] [PubMed] [Google Scholar]
- 28.Luke G., Escuin H., De Felipe P., Ryan M. 2A to the fore - research, technology and applications. Biotechnol. Genet. Eng. Rev. 2010;26:223–260. doi: 10.5661/bger-26-223. [DOI] [PubMed] [Google Scholar]
- 29.Hennecke M., Kwissa M., Metzger K., Oumard A., Kröger A., Schirmbeck R., Reimann J., Hauser H. Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res. 2001;29:3327–3334. doi: 10.1093/nar/29.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryan M.D., Drew J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 1994;13:928–933. doi: 10.1002/j.1460-2075.1994.tb06337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Percy N., Barclay W.S., Garcia-Sastre A., Palese P. Expression of a foreign protein by influenza A virus. J. Virol. 1994;68:4486–4492. doi: 10.1128/jvi.68.7.4486-4492.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B., et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeKelver R.C., Choi V.M., Moehle E.A., Paschon D.E., Hockemeyer D., Meijsing S.H., Sancak Y., Cui X., Steine E.J., Miller J.C., et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P., Cost G.J., Zhang L., Santiago Y., Miller J.C., et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellis B.L., Hirsch M.L., Porter S.N., Samulski R.J., Porteus M.H. Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by Food and Drug Administration-approved drugs. Gene Ther. 2013;20:35–42. doi: 10.1038/gt.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furler S., Paterna J.C., Weibel M., Büeler H. Recombinant AAV vectors containing the foot and mouth disease virus 2A sequence confer efficient bicistronic gene expression in cultured cells and rat substantia nigra neurons. Gene Ther. 2001;8:864–873. doi: 10.1038/sj.gt.3301469. [DOI] [PubMed] [Google Scholar]
- 37.Fang J., Qian J.J., Yi S., Harding T.C., Tu G.H., VanRoey M., Jooss K. Stable antibody expression at therapeutic levels using the 2A peptide. Nat. Biotechnol. 2005;23:584–590. doi: 10.1038/nbt1087. [DOI] [PubMed] [Google Scholar]
- 38.Damdindorj L., Karnan S., Ota A., Hossain E., Konishi Y., Hosokawa Y., Konishi H. A comparative analysis of constitutive promoters located in adeno-associated viral vectors. PLoS One. 2014;9:e106472. doi: 10.1371/journal.pone.0106472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyata T., Takeda J., Iida Y., Yamada N., Inoue N., Takahashi M., Maeda K., Kitani T., Kinoshita T. The cloning of PIG-A, a component in the early step of GPI-anchor biosynthesis. Science. 1993;259:1318–1320. doi: 10.1126/science.7680492. [DOI] [PubMed] [Google Scholar]
- 41.Takeda J., Miyata T., Kawagoe K., Iida Y., Endo Y., Fujita T., Takahashi M., Kitani T., Kinoshita T. Deficiency of the GPI anchor caused by a somatic mutation of the PIG-A gene in paroxysmal nocturnal hemoglobinuria. Cell. 1993;73:703–711. doi: 10.1016/0092-8674(93)90250-t. [DOI] [PubMed] [Google Scholar]
- 42.Brodsky R.A., Mukhina G.L., Li S., Nelson K.L., Chiurazzi P.L., Buckley J.T., Borowitz M.J. Improved detection and characterization of paroxysmal nocturnal hemoglobinuria using fluorescent aerolysin. Am. J. Clin. Pathol. 2000;114:459–466. doi: 10.1093/ajcp/114.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilsker D., Petermann E., Helleday T., Bunz F. Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20752–20757. doi: 10.1073/pnas.0806917106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gustin J.P., Karakas B., Weiss M.B., Abukhdeir A.M., Lauring J., Garay J.P., Cosgrove D., Tamaki A., Konishi H., Konishi Y., et al. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2835–2840. doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konishi H., Karakas B., Abukhdeir A.M., Lauring J., Gustin J.P., Garay J.P., Konishi Y., Gallmeier E., Bachman K.E., Park B.H. Knock-in of mutant K-ras in nontumorigenic human epithelial cells as a new model for studying K-ras mediated transformation. Cancer Res. 2007;67:8460–8467. doi: 10.1158/0008-5472.CAN-07-0108. [DOI] [PubMed] [Google Scholar]
- 46.Konishi H., Mohseni M., Tamaki A., Garay J.P., Croessmann S., Karnan S., Ota A., Wong H.Y., Konishi Y., Karakas B., et al. Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17773–17778. doi: 10.1073/pnas.1110969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshihara T., Ishida M., Kinomura A., Katsura M., Tsuruga T., Tashiro S., Asahara T., Miyagawa K. XRCC3 deficiency results in a defect in recombination and increased endoreduplication in human cells. EMBO J. 2004;23:670–680. doi: 10.1038/sj.emboj.7600087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou S., Buckhaults P., Zawel L., Bunz F., Riggins G., Dai J.L., Kern S.E., Kinzler K.W., Vogelstein B. Targeted deletion of Smad4 shows it is required for transforming growth factor beta and activin signaling in colorectal cancer cells. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2412–2416. doi: 10.1073/pnas.95.5.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhee I., Jair K.W., Yen R.W., Lengauer C., Herman J.G., Kinzler K.W., Vogelstein B., Baylin S.B., Schuebel K.E. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 50.Hanson K.D., Sedivy J.M. Analysis of biological selections for high-efficiency gene targeting. Mol. Cell. Biol. 1995;15:45–51. doi: 10.1128/mcb.15.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waldman T., Kinzler K.W., Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 52.Khan I.F., Hirata R.K., Wang P.R., Li Y., Kho J., Nelson A., Huo Y., Zavaljevski M., Ware C., Russell D.W. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol. Ther. 2010;18:1192–1199. doi: 10.1038/mt.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller D.G., Rutledge E.A., Russell D.W. Chromosomal effects of adeno-associated virus vector integration. Nat. Genet. 2002;30:147–148. doi: 10.1038/ng824. [DOI] [PubMed] [Google Scholar]
- 54.Nakai H., Montini E., Fuess S., Storm T.A., Grompe M., Kay M.A. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat. Genet. 2003;34:297–302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.