Abstract
Programmable transcription factors can enable precise control of gene expression triggered by a chemical inducer or light. To obtain versatile transgene system with combined benefits of a chemical inducer and light inducer, we created various chimeric promoters through the assembly of different copies of the tet operator and Gal4 operator module, which simultaneously responded to a tetracycline-responsive transcription factor and a light-switchable transactivator. The activities of these chimeric promoters can be regulated by tetracycline and blue light synergistically or antagonistically. Further studies of the antagonistic genetic circuit exhibited high spatiotemporal resolution and extremely low leaky expression, which therefore could be used to spatially and stringently control the expression of highly toxic protein Diphtheria toxin A for light regulated gene therapy. When transferring plasmids engineered for the gene switch-driven expression of a firefly luciferase (Fluc) into mice, the Fluc expression levels of the treated animals directly correlated with the tetracycline and light input program. We suggest that dual-input genetic circuits using TET and light that serve as triggers to achieve expression profiles may enable the design of robust therapeutic gene circuits for gene- and cell-based therapies.
INTRODUCTION
The development and progress in synthetic biology have been facilitated by trigger-controlled genetic circuits capable of precisely fine-tuning the expression of transgenes, enabling the rational and predictable reprogramming of cells for complex physiological activities or to interface with complex metabolic pathways and potentially leading to new strategies for cell-based therapies and diagnostics (1,2). Similar to electronic circuits, cells operate as information-processing systems that dynamically integrate and respond to distinct input signals. Both intracellular information and extracellular information are collected by sensors, which communicate the input signal states into a network. This network processes the data according to logic and arithmetic operations, resulting in decisions that are finally executed by output signals. However, intrinsic noise is always unavoidable and unpredictable for living cells, which enables stochastic decision-making even if cells exposed to the same inputs, leading to phenotypic diversification of completely identical cells (3).
Simple one-input one-output gene switches feature robust and predictable performances and have various biotechnological and therapeutic applications; the triggers include small chemicals (4–18), heat (19) or even light (20–28). However, in certain cases, a single endogenous or environmental input does not provide sufficient information to determine whether a transition should be observed in a circuit state. In a pioneering example of the catabolic sugar pathways of Escherichia coli, the activation of particular alternative sugar-metabolizing enzymes depends not only on whether an alternative sugar is present but also on the absence of glucose (29). Therefore, multi-input cellular biosensors are essential to recognize complex conditions under which an enhanced sensing specificity and accuracy of the output response are necessary. However, the design of the multi-input gene circuits is always time-consuming and is characterized by trial and error mainly because of the unpredictability resulting from cross talk, switchability and diverse dynamics of each gene circuit. Several novel bipartite transcription factors (30,31) or artificial regulatory networks (32–38) have been developed in mammalian cells, whose functions were regulated by two or more different trigger molecules that provided novel control topologies and regulation dynamics of gene expression. Nevertheless, most of the developed multi-input controllers in mammalian cells are triggered by the same type of signals—e.g. chemical and chemical (30–32,34–38). In recent years, several light-switchable genetic circuits have been developed to control diverse processes in living cells, and can serve as a transient, non-invasive and reversible means of control over gene expression with superb spatiotemporal resolution (20–28). However, a multi-input genetic circuit simultaneously responding to a chemical inducer and light inducer, which may have the combined benefits of chemical- and light-induced transgene systems and can provide a novel control topology that improves the regulation performance and regulatory expression windows of the transgenes, has not been developed yet.
Until now, most artificial transcription controllers are based on a binary design concept by fusing prokaryotic response regulators with mammalian transactivation or transrepression domains, which bind modulator-specific operator-containing promoters in a small molecule-adjustable manner. In the past 20 years, the most widely used tetracycline (Tc)-responsive expression systems, frequently referred to as the ‘Tet-On’ and ‘Tet-Off’ systems, consist of a bacterial repressor protein (TetR) and a transactivation domain. Both systems oppositely respond to the presence of TETs (e.g. doxycycline, Dox), by either activating (Tet-On) or inactivating gene expression (Tet-Off) (17,39). We previously developed a light-switchable transgene expression system termed LightOn, which consists of only one photoactive transactivator GAVPO. GAVPO can homodimerize and bind to UASG sequence upstream of a minimal promoter and subsequently activate the transcription of target genes upon blue light exposure. The LightOn system allows good spatial, temporal and quantitative control of transgene expression in cultured cells and in mice (26). Previous studies have shown that both the TET and light inducers can be easily and safely introduced into living organisms; therefore, the combination of TET and light have enormous potential for the development of novel control topology for future gene- and cell-based therapies in vivo.
In this study, we have pioneered synthetic versatile two-input genetic circuits based on the synergistic or antagonistic effect in a TET and blue light trigger-adjustable manner, leading to a multitude of regulatory systems where regulation properties, background noise and maximal gene expression levels were highly diverse in combination with different chimeric promoters. The maximum induction ratios could reach more than 1500-fold for the synergistic genetic circuit and more than 1800-fold for the antagonistic genetic circuit in transiently transfected cells, which can be used to program the transgene expression level over a wide range. The gene expression levels from both of the genetic circuits can be quantitatively defined by modulating the Dox concentration and light irradiance. Further studies of the antagonistic genetic circuit revealed high spatiotemporal resolution and extremely low leakage, allowing spatial and stringent control of toxin gene expression for safe and effective gene therapy. When transferring the plasmids engineered for the gene switch-driven expression of a firefly luciferase (Fluc) into mice, the Fluc expression levels of the treated animals directly correlated with the TET and light input. Furthermore, gene expression in the antagonistic dual-input genetic circuit can be locked in ‘OFF’ state even when cells were illuminated with light. Thus, cells or animals being investigated do not have to be always kept in darkness before or after gene induction, which is convenient and avoids resulting in disrupted circadian behavior. On the other hand, the synergistic genetic circuit offers the flexibility to switch on gene expression with either Dox or light, which may be very useful for animal studies. Taken together, such versatile genetic circuits with the combined benefits of a chemical inducer and light inducer would contribute well to synthetic biology approaches and are expected to be of great interest for gene- and cell-based therapies, tissue engineering and biopharmaceutical manufacturing communities.
MATERIALS AND METHODS
DNA cloning
The construction of expression vectors is given in detail in Table 1.
Table 1. Plasmids designed and used in this study.
| Plasmid | Description | Reference or source |
|---|---|---|
| pTetR-VP16 | Constitutive transactivator tTA expression vector | Clontech, CA |
| prTetR-VP16 | Constitutive transactivator rtTA expression vector | Clontech, CA |
| pTRE2 | A Tc-response plasmid containing 7xtetO sequence and CMV minimal promoter | Clontech, CA |
| pTRIPz | Entiviral vector containing 6xtetO sequence | Open Biosystems |
| pGAVPO | Constitutive light-switchable transactivation factor GAVPO expression vector. | (26) |
| pU5-Gluc | A reporter vector for LigthOn system containing 5xUASG and E1b minimal promoter driven expression of Gluc. | (26) |
| pU5-Fluc | A reporter vector for LigthOn system containing 5xUASG and E1b minimal promoter driven expression of Fluc. | (26) |
| pU5-hrGFP | A reporter vector for LigthOn system containing 5xUASG and E1b minimal promoter driven expression of hrGFP. | (26) |
| pCDNA3.1-hrGFP | Constitutive hrGFP expression vector. | (26) |
| pU5-mCherry | A reporter vector for LigthOn system containing 5xUASG and E1b minimal promoter driven expression of mCherry. | (26) |
| pU3-Gluc | A reporter vector for LigthOn system containing 3xUASG and E1b minimal promoter. | (40) |
| pU2-Gluc | A reporter vector for LigthOn system containing 2xUASG and E1b minimal promoter. | (40) |
| pU1-Gluc | A reporter vector for LigthOn system containing 1xUASG and E1b minimal promoter. | (40) |
| pTetR-KRAB | Constitutive transsilencer TetR-KRAB (tTS) expression vector. TetR was PCR-amplified from pTetR-VP16 using oligonucleotides TetR/rTetR-F (5′- cccgaattcaccatgtctagattagataaa -3′) and TetR/rTetR-R (5′-cactgacactgctagggacccactttcacattt-3′), and then was fused to KRAB commercially synthesized by Shanghai generay Biotech Co.,Ltd using overlap PCR. The fused fragment was restricted with EcoRI/SalI and cloned into the corresponding sites (EcoRI/SalI) of pTetR-VP16. | This work |
| pT6U1-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 6xTetO and 1xUASG adjacent to E1b minimal promoter. 6xTetO was PCR-amplified from pTRIPz using oligonucleotides 6xTetO-F (5′-gccctcgaggtccgaggttctagacgag-3′) and 6xTetO-R(5′-gggagcgctcaccatgtctagactggacaagag-3′), restricted with XhoI/Eco47III and cloned into the corresponding sites (XhoI/Eco47III) of pU1-Gluc. | This work |
| pT6U2-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 6xTetO and 2xUASG adjacent to E1b minimal promoter. Similar to pT6U1-Gluc, 6xTetO was inserted into the XhoI/Eco47III sites of pU2-Gluc. | This work |
| pT6U3-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 6xTetO and 3xUASG adjacent to E1b minimal promoter. Similar to pT6U1-Gluc, 6xTetO was inserted into the XhoI/Eco47III sites of pU3-Gluc. | This work |
| pT6U5-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 6xTetO and 5xUASG adjacent to E1b minimal promoter. 5xUASG was cut off from pU5-Gluc by KpnI/NheI digestion and inserted into the corresponding sites (EcoRI/SalI) of pT6U3-Gluc. | This work |
| pU5T1-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 1xTetO and 5xUASG adjacent to E1b minimal promoter. The oligonucleotides 1xTet-F (5′-ctagcggctcgagtttactccctatcagtgatagagaacgtatgagct-3′) and 1xTet-R(5′-catacgttctctatcactgatagggagtaaactcgagccg-3′) were annealed, phosphorylated and then inserted into the NheI/SacI sites of pU5-Gluc. | This work |
| pU5T2-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 2xTetO and 5xUASG adjacent to E1b minimal promoter. The oligonucleotides 2xTet-F (5′- ctagcggctcgagtttactccctatcagtgatagagaacgtatgtcgagtttactccctatcagtgatagagaacgatgtcgaccgagct-3′) and 2xTet-R (5′- cggtcgacatcgttctctatcactgatagggagtaaactcgacatacgttctctatcactgatagggagtaaactcgagccg-3′) were annealed, phosphatized and then inserted into the NheI/SacI sites of pU5-Gluc. | This work |
| pU5T4-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 4xTetO and 5xUASG adjacent to E1b minimal promoter. 2xTetO was cut off from pU5T2-Gluc by XhoI/SacI double digestion and inserted into the SalI/SacI sites of pU5T2-Gluc to generate pU5T4-Gluc. | This work |
| pU5T6-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 6xTetO and 5xUASG adjacent to E1b minimal promoter. 2xTetO was cut off from pU5T2-Gluc by XhoI/SacI double digestion and inserted into the SalI/SacI sites of pU5T4-Gluc to generate pU5T6-Gluc. | This work |
| p(TU)1-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 1xTetO and 1xUASG (1xTU) adjacent to E1b minimal promoter. The oligonucleotide(5′-ggtaccctgagctggatgagccgcgctcgagtttactccctatcagtgatagagaacgtatgtccggagtactgtcctccggtcgactatcgtagtccagcgctacgagctc-3′) was commercially synthesized by Shanghai generay Biotech Co.,Ltd and was inserted into the KpnI/SacI sites of pU5T2-Gluc. | This work |
| p(TU)2-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 2xTU adjacent to E1b minimal promoter. 1xTU was cut off from p(TU)1-Gluc by XhoI/SacI double digestion and inserted into the SalI/SacI sites of p(TU)1-Gluc to generate p(TU)2-Gluc. | This work |
| p(TU)3-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 3xTU adjacent to E1b minimal promoter. 1xTU was cut off from p(TU)1-Gluc by XhoI/SacI double digestion and inserted into the SalI/SacI sites of p(TU)2-Gluc to generate p(TU)3-Gluc. | This work |
| p(TU)4-Gluc | Tetracycline and blue light-responsive Gluc expression vector containing 3xTU adjacent to E1b minimal promoter. 1xTU was cut off from p(TU)1-Gluc by XhoI/SacI double digestion and inserted into the SalI/SacI sites of p(TU)3-Gluc to generate p(TU)4-Gluc. | This work |
| pTRE2-Gluc | Gluc gene amplified from pU5-Gluc was cloned into BamHI/HindIII sites of pTRE2. | This work |
| pT6U3-DTA | Tetracycline and blue light-responsive Gluc expression vector containing 6xTetO and 3xUASG adjacent to E1b minimal promoter, the reporter gene is DTA. DTA gene was commercially synthesized by Shanghai generay Biotech Co.,Ltd and cloned into HindIII/BamHI sites of pT6U3-Gluc to generate pT6U3-DTA. | This work |
| pU5-DTA | A reporter vector for LigthOn system containing 5xUASG and E1b minimal promoter driven expression of DTA. Gluc gene in pU5-Gluc was replaced by DTA by HindIII/ BamHI digestion. | This work |
| pT6U3-Fluc | Tetracycline and blue light-responsive Gluc expression vector containing 6xTetO and 3xUASG adjacent to E1b minimal promoter, the reporter gene is Fluc. Fluc gene was PCR-amplified from pU5-Fluc using oligonucleotides Fluc-F (5′- cccaagcttcaccatggaagacgccaaaaacat-3′) and Fluc-R (5′- cccggatccttacacggcgatctttccgc-3′), restricted with HindIII/BamHI and cloned into the corresponding sites (HindIII/BamHI) of pT6U3-Gluc. | This work |
| pT6U3-mCherry | Tetracycline and blue light-responsive Gluc expression vector containing 6xTetO and 3xUASG adjacent to E1b minimal promoter. Gluc gene of pT6U3-Gluc was replaced by mCherry by HindIII/BamHI digestion. | This work |
TetR, E.coli Tn10-derived repressor of the TET resistance gene; VP16, Herpes simplexvirus-derived transactivation domain; rTetR, amino acid exchanges in the TetR, reversing the response of the presence of the allosteric effector Dox; GAVPO, a light-switchable transcription factor consisting of DNA binding domain of Gal4, a light-oxygen-voltage (LOV) domain–containing protein VIVID and p65 activation domain from NF-κB. GLuc, Gaussia princeps luciferase; Fluc, Firefly luciferase. mCherry, a red fluorescent protein; KRAB, Krueppel-associated box domain of the human kox-1 gene; tTA, TET-dependent transactivator; rtTA, TET-dependent transactivator shows reverse response to the presence of Dox relative to tTA; UASG, upstream active sequence, an enhancer to which GAL4 specifically binds to activate gene transcription. TetO, operator sequence can be recognized and bond by tTA and rtTA to activate gene transcription; DTA, a segment of the diphtheria toxin (tox), inhibits protein synthesis in cells; hrGFP, humanized Renilla reniformis green fluorescent protein.
Reagents and materials
All restriction enzymes, T4 ligase and T4 polynucleotide kinase were purchased from Fermentas (Thermo Scientific). Fetal bovine serum (FBS), Opti-MEM and 0.25% trypsin-EDTA were obtained from GIBCO. Tetracycline-screened FBS, antibiotic-free high-glucose Dulbecco's modified Eagle's medium (DMEM) and 10 000 U/ml penicillin and 10 000 μg/ml streptomycin were purchased from HyClone. Lipofectamine 2000 was purchased from Invitrogen. The Gaussia luciferase assay kit was purchased from NEB. D-Luciferin was purchased from Sigma-Aldrich. Human embryonic kidney 293 cells (HEK293), human cervical carcinoma cells (HeLa) and MDA-MB-468 cells were obtained from ATCC. Four-week-old male ICR mice (≈20 g body weight) were purchased from SLRC Laboratory Animals.
Cell culture and transfection
Human embryonic kidney 293 cells (HEK293), human cervical carcinoma cells (HeLa) and MDA-MB-468 cells were cultivated in high-glucose DMEM (HyClone) supplemented with 10% fetal bovine serum (FBS) (GIBCO) and 1% of 10 000 U/ml penicillin and 10 000 μg/ml streptomycin (Hyclone) at 37°C and 5% CO2. Before transfection, the cells were plated in high-glucose DMEM supplemented with 10% tetracycline-screened FBS (Hyclone).
All cells were transfected using Lipofectamine 2000 (Invitrogen). In detail, the cells were seeded onto a 24-well plate and cultivated overnight to be 70%–90% confluent at the time of transfection. For each well, 0.6 μg of vector DNA for the synergistic and antagonistic dual-input genetic circuits (0.2 μg of pGAVPO, 0.2 μg of reporter plasmid and 0.2 μg of pTetR-VP16 or prTetR-VP16 or pTetR-KRAB) or 0.4 μg of vector DNA for the LightOn system (0.2 μg of pGAVPO and 0.2 μg of reporter plasmid) or 0.4 μg of vector DNA for the Tet-On and Tet-Off system (0.005 μg of reporter plasmid and 0.2 μg of prTetR-VP16 or pTetR-VP16 supplemented with 0.195 μg of pCDNA3.1 empty vector to 0.4 μg of total vector DNA, which could result in higher on/off ratios in our experiment) and 1.2 μl of Lipofectamine 2000 reagent were diluted in 50 μl of Opti-MEM (GIBCO) and then mixed after a 5-min incubation. After a 20-min incubation at room temperature, the DNA-lipid complex was added to the cells. For other plate formats, the cell number and amount of reagents were scaled up according to the growth area.
Light irradiation and doxycycline addition
Unless indicated, the transfected cells were kept in the dark for 8 h, and then were illuminated by 1.4 W·m−2 (average irradiance) blue light from an LED lamp (460-nm peak) from below or remained in the dark for 24 h before characterization. The LED lamps were controlled with a timer to adjust the overall dose of blue light illumination during the specified period. Neutral density filters were used to adjust the light irradiance. Doxycycline (Sigma) was dissolved in water at the concentration of 1.5 mg·ml−1 and added into the culture 8 h after transfection. To spatially control gene expression in cultured mammalian cells, single layers of the engineered HEK293 cells cultured on glass-bottomed dishes were illuminated with a spatial pattern using a photomask printed with a specific image for 48 h.
Chemiluminescence assays
The Synergy 2 multi-mode microplate reader (BioTek) was used to measure the chemiluminescence and fluorescence of samples. A Gaussia luciferase assay kit was used to assay secreted Gluc activity in cell culture supernatants according to the manufacturer's protocol (NEB). Ten-microliter cell culture supernatants were transferred to each well of a white 384-well plate (Greiner), and 10 μl of coelenterazine solution (1 μM coelenterazine, 0.1 M Tris-HCl buffer at pH 7.4, and 0.3 M sodium ascorbate) was added. Light emission was recorded as relative light units.
Imaging
For live-cell fluorescence microscopy, images were acquired using an S Plan Fluor ELWD 20×, 0.45 numerical aperture (NA) objective and a digital sight camera on an Eclipse Ti inverted microscope system (Nikon), using an FITC filter for hrGFP. For mCherry and hrGFP imaging of cultured cells with a spatial pattern, a Kodak In-Vivo Multispectral System FX (Carestream Health) was used with 550-nm excitation and 600-nm emission filters for mCherry and 480-nm excitation and 535-nm emission filters for hrGFP.
Mice
All procedures involving animals were approved by the Institutional Animal Care and Use Committee of Shanghai and were conducted in accordance with the National Research Council Guide for Care and Use of Laboratory Animals. Four-week-old male ICR mice (≈20 g body weight) purchased from SLRC Laboratory Animals, were used for some experiments. Unless otherwise mentioned, mice were subjected to intravenous co-injection of 100 μg of pT6U3-Fluc, 10 μg of pGAVPO and 10 μg prTetR-VP16 or 100 μg pTetR-KRAB in 2–3 ml (12% of the body weight in grams) of Ringer's solution (147 mM NaCl, 4 mM KCl, 1.13 mM CaCl2) within 5–7 seconds. We removed the abdominal fur of the injected mice using a shaver and 8% sodium sulfide. Mice were then rested in cages with glass bottoms and illuminated from below using a blue LED lamp (90 mW·cm−2) or in the dark. Next, 50 mg/kg Dox dissolved in saline was administered i.p. (41). Imaging of firefly luminescence was carried out 12 h after intravenous injection of plasmids by tail intravenous injection of D-luciferin (150 μg/g of body weight, i.p.) under ether anesthesia (42). Images were acquired using the In-Vivo Multispectral System FX system (Kodak) 10 min after injection of D-luciferin.
Measurement of DTA-mediated protein synthesis inhibition and cell death
The HEK293 cells were transfected with 1.4 μg of plasmids (0.2 μg of pCDNA3.1-hrGFP, 0.073 μg of pGAVPO, 0.73 μg of pTetR-KRAB and 0.4 μg of pT6U3-DTA or pT6U3 empty plasmid) per well in a 12-well plate, and cultured under blue light irradiance with Dox or in the dark without Dox. For the LightOn system-controlled DTA expression, 1 μg of the plasmids (0.2 μg of pCDNA3.1-hrGFP, 0.4 μg of pGAVPO and 0.4 μg of pU5-DTA or pU5 empty plasmid) was used for the transfection, and the cells were then grown in the dark or under blue light irradiance. The fluorescence intensity of hrGFP was detected 48 h after transfection using fluorescence microscopy with an FITC channel. After imaging, the cells were harvested by Trypsin-EDTA treatment and washed with phosphate-buffered saline (PBS). Fluorescence of the cells was measured using a Synergy 2 Multi-Mode Microplate Reader (BioTek) with a 485/20 excitation filter and a 528/20 emission filter. To detect the cell death caused by DTA, HEK293 cells were transfected with 3 μg of plasmids (1.82 μg of pGAVPO, 0.18 μg of pTetR-KRAB and 1 μg of pT6U3-DTA for the antagonistic dual-input genetic circuit) and 2 μg of plasmids (1 μg pGAVPO and 1 μg pU5-DTA for the LightOn system) per well in a 6-well plate and were cultured under blue light irradiance with Dox or in the dark without Dox for 24 h. The cells were then plated at a density of 104 cells in each well of a 96-well plate and were cultured for another 72 h under the previous conditions. Phase imaging was performed by fluorescence microscopy, and quantitative analysis of cell viability was conducted using Cell Counting Kit-8 (Dojindo Laboratories, Gaithersburg, MD) according to the manufacturer's protocol. The absorbance at 450 nm was determined using a microplate reader (BioTek).
RESULTS AND DISCUSSION
Synergistic control of gene circuits by TET and light
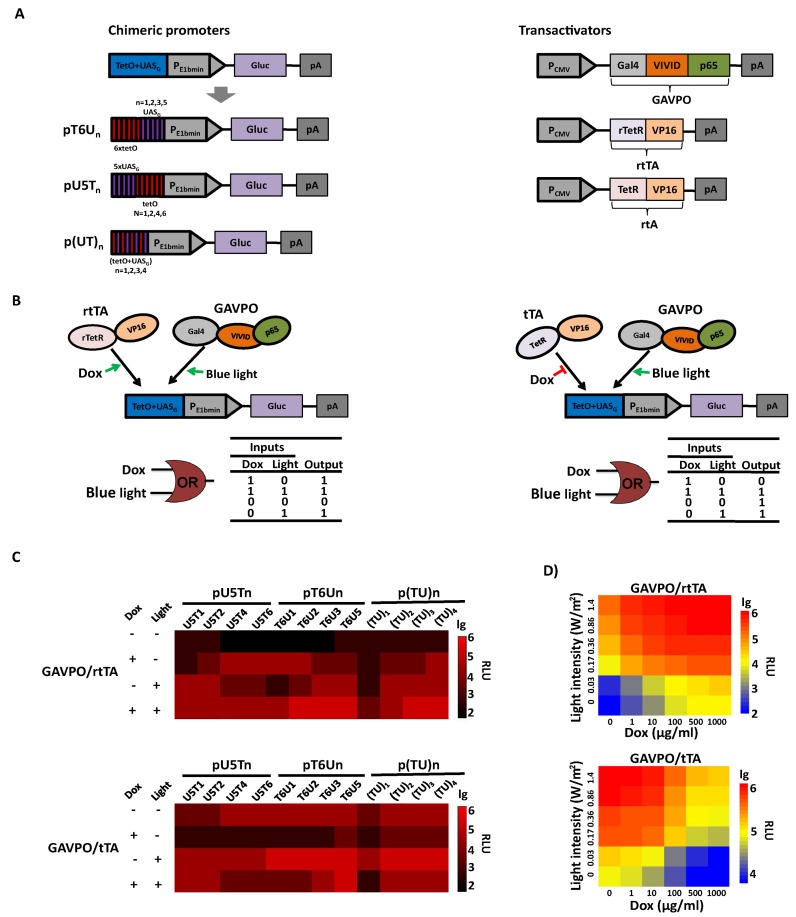
In the TET systems, Tc responsive transactivators rtTA or tTA bind to tetO with or without TET, respectively, resulting in activation of transcription (17,39). Similarly, in the LightOn system, the light-switchable transactivator GAVPO bound to the UASG element and activated transcription under blue light (26). To make promoters that respond simultaneously to TET and light, we assembled different copies of the tet operator (tetO) and UASG element upstream of the E1b minimal promoter, using Gaussia luciferase (Gluc) as a reporter. In these chimeric promoters, the tetO sequence was upstream of UASG (U5Tn), or vice versa (T6Un), or these sequences were alternated with each other (Figure 1A). Under such a configuration, the Tc-responsive transactivator and GAVPO can be activated by chemical and light, respectively, bind to their cognate sequences, and demonstrate a synergistic dual-input control of transgene expression. An ‘OR’ like gate is predicted for the synergistic effect based on rtTA and GAVPO when TET or light is present as two inputs. For tTA and GAVPO based synergistic control, illumination with blue light or the absence of TET would initiate the transcription of downstream genes (Figure 1B). We next tested the effects of doxycycline (Dox, a synthetic tetracycline derivative) and blue light on transgene activation in HEK293 cells with transient co-transfection of a Tc-responsive transactivator plasmid (encoding tTA or rtTA), a light-switchable transactivator plasmid (encoding GAVPO) and a reporter plasmid (encoding Gluc under the control of different chimeric promoters). In the ‘OR’ like gate configuration, the data showed that most combinations of chimeric promoters and activators showed robust synergistic dual-input control of transgene expression, with maximum induction occurring when the cells were activated by Dox and light simultaneously; however, leaky expression and maximal induction levels varied markedly (Figure 1C). For pU5Tn reporters, with increasing copy numbers of tetO inserted downstream of UASG copies, Dox-triggered Gluc expression increased, whereas light-induced Gluc expression decreased, likely due to the increasing spacer length between the UASG and minimal promoter (Figure 1C). Similar results were obtained for pT6Un, showing that Dox-triggered Gluc expression decreased, while light-induced Gluc expression increased, with increasing copies of UASG in pT6Un. For p(TU)n, both Dox- and blue light-induced transcription increased with increasing ‘TU’ units. Among them, the T6U3 chimeric promoter showed the maximum range of transcription control and balanced activation by Dox and light. The maximum induction ratio could reach more than 1500-fold (Figure 1C, Supplementary Figure S1), which can be used to program the transgene expression level over a wide range. The tTA- and GAVPO-based synergistic dual-input gene circuit exhibited similar results for most of the chimeric promoters; however, the T6U2 chimeric promoter had the maximum induction ratio, ≈300-fold, and balanced activation by the two activators instead of T6U3 (Figure 1C, Supplementary Figure S1). The synergistic control of gene expression by TET and light was also observed in other cell lines (Supplementary Figure S2). Therefore, such synergistic control of the dual-input gene circuits enable improved ranges of transcription control compared with the one-input genetic circuit (Supplementary Figure S3), and offer the flexibility to switch on gene expression with either Dox or light.
Figure 1.
Synergistic dual-input genetic circuits triggered by TET and light in mammalian cells. (A) Design of the synergistic controllers by GAVPO and tTA or rtTA triggered by TET and blue light. Three types of promoter configurations were constructed: (i) pU5Tn (n = 1, 2, 4, 6), six copies of tetO were placed at the 5′ end of different copies of the UASG element adjacent to the E1b minimal promoter; (ii) pT6Un (n = 1, 2, 3, 5), where five copies of the UASG element are placed at the 5′ end of different copies tetO adjacent to the minimal promoter and (iii) p(TU)n (n = 1, 2, 3, 4), the operator region comprises tetO alternating with UASG, and the copy number differs from one to four. The Gluc gene was used as the reporter gene and placed downstream of the E1b minimal promoter. Expression of the light-switchable transactivator GAVPO, Tc-responsive transactivators tTA and rtTA were all driven by the strong human cytomegalovirus (CMV) promoter. (B) Schematic representation of synergistic dual-input genetic circuits. Binding of the transactivator tTA and rtTA to the tetO in the absence and presence of Dox, respectively, or the binding of GAVPO to the UASG upon blue light exposure would result in gene activation. (C) Validation of the synergistic dual-input genetic circuits with different chimeric promoters. HEK293 cells were transiently co-transfected with a Tc-responsive transactivator plasmid (encoding tTA or rtTA), a light-switchable transactivator plasmid (encoding GAVPO), and a reporter plasmid (encoding Gluc under the control of different chimeric promoters). Gluc expression at different conditions was scored in the culture supernatant after 24 h. (D) Qualitative expression by modulating light irradiance and Dox concentration. The engineered cells were cultured at different light irradiances (0–1.4 W/m2) and Dox concentrations (0–1000 ng/ml). Gluc activity was determined after 24 h. The data in (C) and (D) were collected from three independent experiments.
Previous studies have shown that the expression levels of a transgene can be modulated by altering the Dox concentration and light irradiance in the TET and LightOn systems. To determine the combined regulation characteristics of the dual-input gene switch, transiently transfected HEK293 cells were cultured under different light irradiance and Dox concentrations. The result indicated that the engineered cells produced and secreted Gluc at levels in a light irradiance- and Dox concentration-dependent manner, allowing highly precise control of the transgene expression by combining the Dox concentration and light irradiance (Figure 1D). Taken together, our data suggest that the synergistic dual-input genetic circuit allows tunable, flexible and qualitative control of transgene expression with improved ranges of transcription control.
Antagonistic dual-input expression of the genetic circuit by TET and light
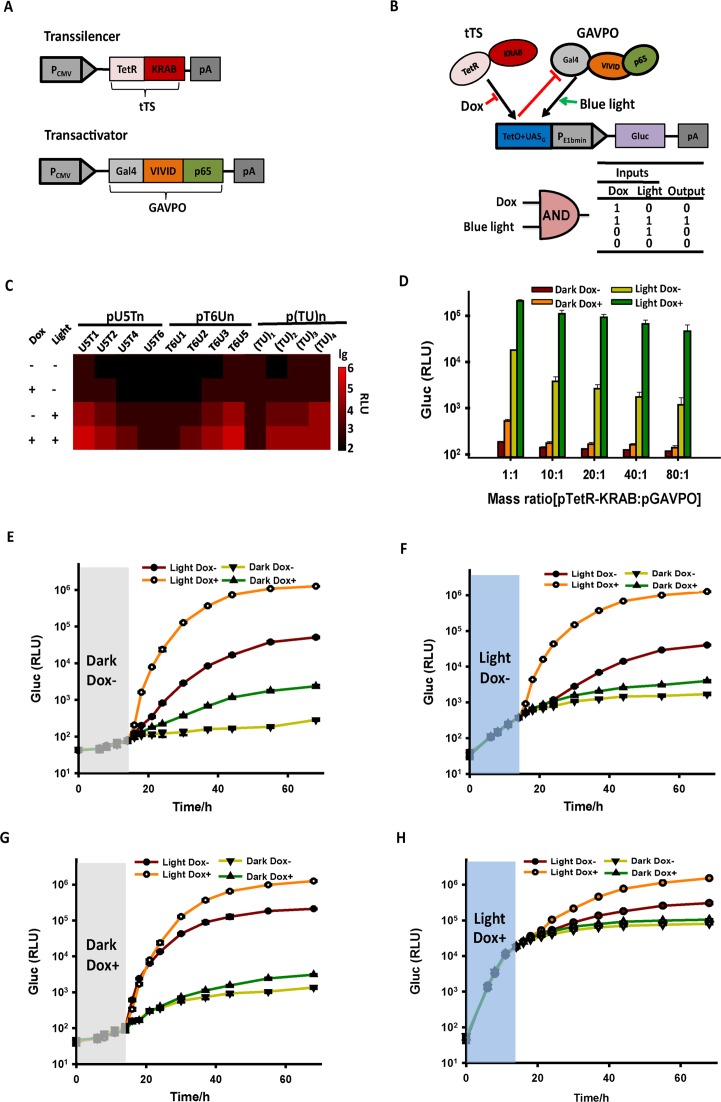
In the antagonistic dual-input genetic circuit by TET and light, the Tc-responsive activator was replaced by the Tc-responsive transsilencer tTS (Figure 2A). tTS was obtained by fusing the KRAB domain of the human kox-1 gene to TetR, which can actively suppress background expression from tetO-containing minimal promoters in the absence of Dox (43). We hypothesized that the light-switchable transactivator GAVPO binds to UASG to activate transcription under blue light illumination, whereas the binding of tTS to tetO would counteract the activation of the chimeric promoters by GAVPO in the absence of Dox. Thus, gene expression would only occur upon blue light illumination when tTS was released from tetO, demonstrating an antagonistic dual-input control by tTS and GAVPO. An ‘AND’ like gate is predicted for the antagonistic effect when Dox and light were used as the two inputs (Figure 2B). The antagonistic efficiencies were measured for different chimeric promoters in HEK293 cell transiently co-transfected with the Tc-responsive transsilencer plasmid (encoding tTS) and light-switchable transactivator plasmid (encoding GAVPO). The results showed that, in most of the chimeric promoters, the binding of tTS to tetO could efficiently repress the leakage under the darkness, or gene activation when cells were illuminated by blue light. Among these chimeric promoters, T6U3 exhibited the most significant antagonistic effect by tTS, as shown by the more than 10-fold reduction of Gluc expression in the absence of Dox upon blue light exposure (Figure 2C, Supplementary Figure S4), compared with cells kept in the presence of Dox upon light exposure. Such antagonistic effects were also observed in other cell lines as well when using pT6U3-Gluc as the reporter (Supplementary Figure S5). Further studies showed that the antagonistic efficiencies of tTS increased with higher mass ratios of the tTS-encoding plasmid to GAVPO-encoding plasmid following transfection, probably due to the high levels of transsilencer expression. The antagonistic efficiency could reach ≈30-fold when the mass ratio increased from 1:1 to 10:1 (Figure 2D, Supplementary Figure S6). The increased mass ratios further reduced the leakage under the non-induced conditions (Dark Dox-), enabling stringently controlled transgene expression (Figure 2D).
Figure 2.
Design and validation of the antagonistic dual-input genetic circuits in mammalian cells. (A) The VP16 transactivation domain of tTA was replaced with the Krueppel-associated box domain (KRAB) of human kox-1, resulting in trans-silencer TetR-KRAB (tTS). (B) Schematic representation of the antagonistic control triggered by TET and light. GAVPO bound to UASG to activate transgene expression upon light illumination, whereas the binding of tTS to tetO could counteract the activation by GAVPO in the absence of Dox. Therefore, only when tTS was released from TetO in the presence of Dox, gene expression occurred upon light illumination. (C) Validation of the antagonistic dual-input genetic circuit with different chimeric promoters. HEK293 cells were transiently co-transfected with a reporter plasmid (encoding Gluc under the control of different chimeric promoters), the Tc-responsive transsilencer plasmid (encoding tTS) and light-switchable transactivator plasmid (encoding GAVPO). Gluc expression at different conditions was determined in the culture supernatant after 24 h. (D) The antagonistic efficiencies of different mass ratios of the tTS-encoding plasmid pTetR-KRAB to the GAVPO-encoding plasmid pGAVPO. HEK293 cells were transiently co-transfected with different ratios (w/w; 0.8 μg of total DNA) of pTetR-KRAB to pGAVPO, and Gluc activity was scored in the culture supernatant after 24 h. The data are shown as the mean ± SD (n = 3). (E–G) ON kinetics of the gene expression from the antagonistic controller. The engineered HEK293 cells were cultured in the non-inducing conditions (Dark Dox+, Dark Dox- or Light Dox-) for the first 14 h, and then were transferred to inducing conditions (Light Dox+) or kept in non-inducing conditions as the controls. Gluc activities were determined at the indicated time points. (H) OFF kinetics of the gene expression from the antagonistic controller. The engineered HEK293 cells were cultured in inducing conditions (Light Dox+) for the first 14 h, and then were transferred to non-inducing conditions or kept in inducing conditions as the control. Gluc activities were determined at the indicated time points. The data in (E–H) are shown as the mean ± SD (n = 3). (I) Quantitative expression by modulating the light irradiance and Dox concentration. The engineered cells were cultured at different light irradiances (0–1.4 W/m2) and Dox concentrations (0–1000 ng/ml). The data in (C) and (I) were collected from three independent experiments. (J) Spatial control of gene expression by the antagonistic dual-input genetic circuit. Ten hours after transfection, the engineered cells were cultured at different conditions with a spatial pattern using a printed mask with a specific image (left panel) for 48 h before the image of mCherry fluorescence was taken (middle and right panel). The orange circle indicated the glass bottom of the dish, where the cells were attached. Scale bar, 1 cm.
The kinetics of the antagonistic dual-input genetic circuit was also investigated. The engineered HEK293 cells were cultured in the non-inducing conditions (Dark Dox+, Dark Dox- or Light Dox-) for the first 14 h, and then were transferred to inducing conditions (Light Dox+) or kept in non-inducing conditions as the controls. The results showed that Gluc activities maintained at low levels in the first 14 h and increased rapidly after transferring cells to blue light illumination with Dox addition (Figure 2E, F and G), while much lower Gluc production was observed in the cells kept in non-inducing conditions. We further tested the OFF kinetics of the antagonistic dual-input genetic circuit. Gluc expression increased significantly in the first 14 h when cells were cultured in inducing conditions (Light Dox+). The cells were then transferred to non-inducing conditions or kept in inducing conditions as the control. The results demonstrated that removal of light or Dox led to a plateau in the amount of Gluc protein (Figure 2H). In contrast, cells always kept in inducing conditions maintained rapid Gluc production during the time course (Figure 2H). For the cells transferred to darkness without Dox, tTS bound to tetO to repress the continuous mRNA synthesis by the activated GAVPO (half-life for the activated GAVPO was ≈2 h (26)), which further accelerated the shutdown of mRNA synthesis and protein production. The data showed that Gluc release significantly dropped 2 h after the system was turned OFF (Supplemental Figure S7A and B), however the small increase of Gluc level continued for another 20 h (Figure 2H, Supplemental Figure S7A). Such a delay of OFF kinetics is due to slow degradation of the Gluc mRNA transcript and the time for Gluc protein to secrete out of the cells. We previously showed that OFF kinetics can be accelerated significantly by modifying the 3′ untranslated region of the Gluc reporter gene by inserting the conserved AU-rich element (ARE) from the gene encoding GM-CSF, which mediates selective degradation of mRNA (26,44). Such approach can also be applied to the dual-input genetic circuits to accelerate the OFF kinetics.
Further studies showed that for the antagonistic dual-input genetic circuit, short-term exposure of the cells to blue light resulted in little changes of Gluc expression comparing with the cells kept in the dark without Dox addition. By contrast, the LightOn system controlling Gluc expression exhibited significant activation even under short-term exposure to ambient light (Supplementary Figure S8).
We next tested the dose-dependent induction of Gluc production by controlling the light irradiance and Dox concentration. Our results showed that Gluc production increased along with increased light irradiance and Dox concentration (Figure 2I). To spatially control the gene expression in cultured cells, we illuminated the engineered HEK293 cells using the red fluorescent protein mCherry as the reporter under a specific pattern. Only cells exposed to Dox and light illumination exhibited the pattern of the original image used as a mask (Figure 2J). Collectively, these data suggest that the antagonistic dual-input genetic circuit provides a versatile tool allowing convenient, tunable, quantitative and spatial control of transgene expression.
In vivo study of the synergistic and antagonistic dual-input expression
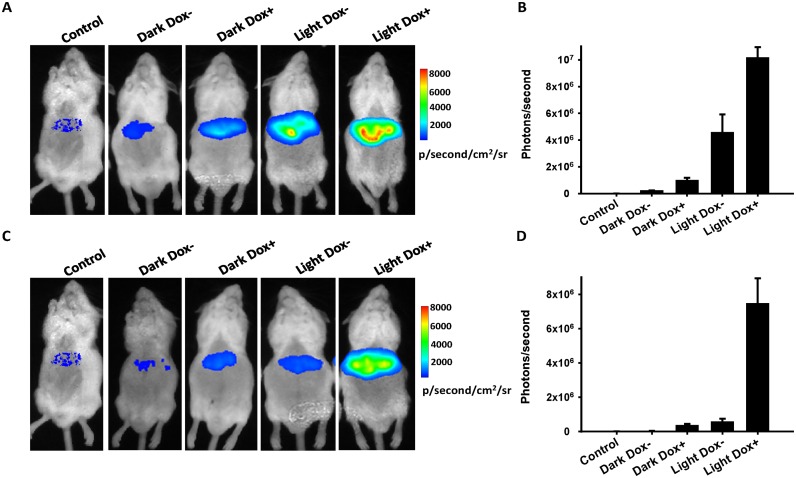
To assess the synergistic and antagonistic dual-input genetic circuits in mice, the reporter plasmid (encoding firefly luciferase (Fluc) under the control of T6U3 chimeric promoter), pGAVPO and Tc-responsive transactivator or transsilencer plasmid (encoding rtTA or tTS) were transferred into mice livers through a hydrodynamic procedure. The mice were exposed to blue light illumination or kept in darkness, with or without Dox intake, and the correlating Fluc levels in the livers were measured. Similar to the results from the cultured cells, the synergistic effects by the rtTA and GAVPO transactivators were observed using in situ imaging of Fluc expression. The intake of Dox or illumination with blue light could result in notable expression of the Fluc reporter, while maximum Fluc expression was achieved when mice were fed under blue light illumination with Dox intake (Figure 3A and B). Fluc expression in mice injected with tTS- and GAVPO-encoding plasmids showed that the trans-silencer tTS could also efficiently counteract the activation of GAVPO under blue light irradiance by more than 10-fold in mice. The intake of Dox enabled the release of tTS from tetO, mitigating the antagonistic effect and resulting in Fluc expression under blue light exposure (Figure 3C and D). Notably, the antagonistic dual-input genetic circuit still exhibited extremely low leakage in mice kept under non-inducing conditions, which showed no significant differences with that of the control mice injected with Ringer's solution without any plasmids (Figure 3C and D, Supplementary Figure S9), indicating stringent control of transgene expression in mice. Taken together, these data suggest that the synergistic and antagonistic dual-input genetic circuits can also efficiently and stringently regulate transgene expression in mice, providing potential application for in vivo gene therapy.
Figure 3.
In vivo study of the synergistic and antagonistic dual-input genetic circuits. Mice were co-transfected with 100 μg of pT6U3-Fluc, 10 μg of pGAVPO and 10 μg of prTetR-VP16 (A) or 100 μg of pTetR-KRAB (C) via a hydrodynamic procedure. The mice were then fed in cages with glass bottoms in darkness without (Dark Dox-) or with (Dark Dox+) Dox, or in 90 mW·cm−2 blue light irradiance without (Light Dox-) or with (Light Dox+) Dox. Mice got injection of only Ringer's solution without any plasmids and kept in darkness without Dox addition were used as the control. In situ imaging of firefly luminescence was carried out 12 h after intravenous injection of plasmids by tail intravenous injection of D-luciferin (150 μg/g of body weight, i.p.) under ether anesthesia. (B) and (D), a region of interest (ROI) was drawn around each liver location, and the number of photons/second was calculated. The data are shown as the mean ± SD (n = 3–4).
Stringent control of toxin expression by the antagonistic dual-input genetic circuit
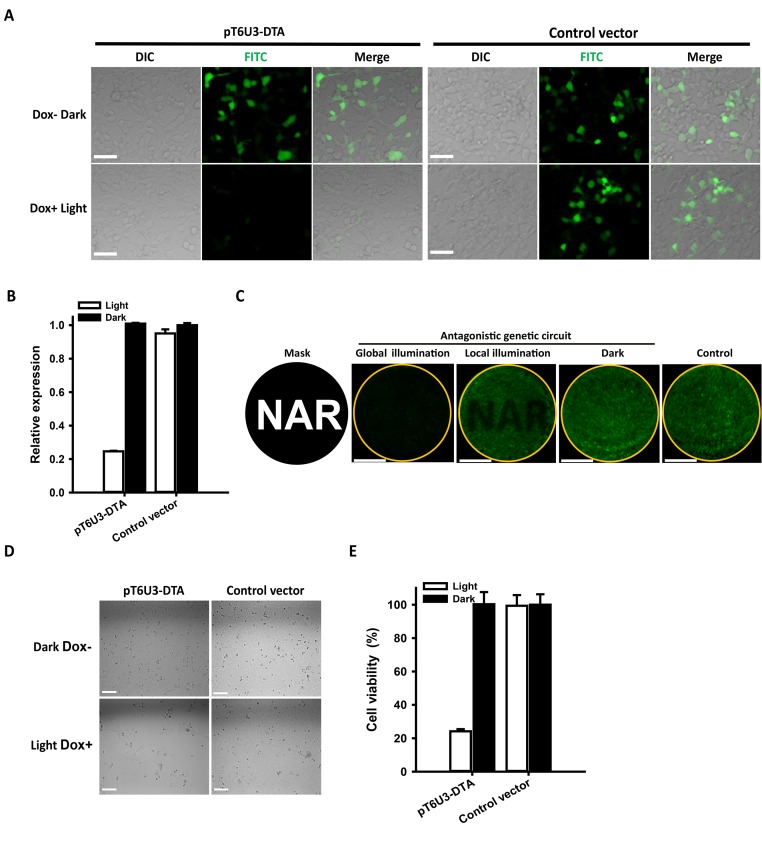
Diphtheria toxin A (DTA), a segment of the diphtheria toxin (tox), catalyzes ADP-ribosylation and inactivation of the elongation factor 2 (eEF2), resulting in protein synthesis inhibition and cell death even at extraordinary low concentrations (45,46). DTA has been used in gene therapy studies (47). As a proof-of-principle demonstration of the potential of the antagonistic dual-input genetic circuit for the stringent regulation of toxin gene expression for gene therapy, the regulation and induction of the DTA gene were examined in transient transfection assays involving co-transfection of a reporter plasmid (encoding DTA under the control of T6U3 chimeric promoter), pGAVPO, pTetR-KRAB and an ‘indicator’ plasmid (encoding green fluorescent protein hrGFP under the control of the CMV promoter). The rationale for the indicator plasmid is that the degree of expression of the hrGFP gene provides a highly sensitive means of measuring the DTA protein effectiveness to abolish protein synthesis and cause cell death. The empty pT6U3 vector was used to replace pT6U3-DTA as the negative control. The results of fluorescence microscopic analysis showed a significantly reduced hrGFP expression when cells were cultured under blue light irradiance with Dox addition, whereas the hrGFP fluorescence of the cells in the dark without Dox was almost the same with control cells (Figure 4A and B). By contrast, the LightOn system-controlled DTA expression exhibited more marked light-induced inhibition of protein synthesis; however, significant background inhibition was also observed when cells were kept in the dark (Supplementary Figure S10A and B). Spatial inhibition of protein synthesis was also achieved with local illumination using a spatial pattern by the antagonistic dual-input genetic circuit (Figure 4C). In contrast, we found LightOn system had significant leak expression of DTA in the non-inducing conditions, which resulted in marked inhibition of GFP expression even in the dark regions of the culture (Supplementary Figure S10C). These data indicated that the antagonistic dual-input genetic circuit has much lower leakage of DTA expression under non-inducing conditions compared with the LightOn system. Similar results were obtained in the cell viability analysis, indicating that, for the antagonistic dual-input genetic circuit, marked cell death occurred when DTA was induced by Dox and light, whereas cells remained viable under darkness without Dox (Figure 4D and E). For the LightOn system, leaked expression of DTA in cells under darkness led to 40% reduction of cell viability, while induction of DTA when the cells were illuminated further reduced cell viability (Supplementary Figure S10D and E). Taken together, these data demonstrate that the antagonistic dual-input genetic circuit can be used for extremely stringent control of therapeutic gene expression for gene- and cell-based therapies with precise spatiotemporal resolution.
Figure 4.
Stringent control of toxin DTA expression by the antagonistic dual-input genetic circuit. (A) Fluorescence imaging of hrGFP expression. HEK293 cells were co-transfected with pT6U3-DTA, pGAVPO, pTetR-KRAB and pCDNA3.1-hrGFP, and cultured in the darkness without Dox, or upon blue light exposure with Dox. An empty plasmid pT6U3 was used to replace pT6U3-DTA as the negative control. Imaging of hrGFP was conducted 48 h after transfection by fluorescence microscopy using the FITC channel. Scale bar, 50 μm. (B) Quantitative analysis of hrGFP expression. The transfected cells after imaging were digested with trypsin-EDTA and washed with phosphate-buffered saline (PBS). Fluorescence was determined using a Microplate Reader (BioTek) with 485/20 excitation filter and a 528/20 emission filter. The data were normalized to the cells transfected with control vector and kept in darkness without Dox. The data are shown as the mean ± SD (n = 3). (C) Spatial control of DTA expression for local inhibition of hrGFP expression. The engineered cells, transfected with pT6U3-DTA, pGAVPO, pTetR-KRAB and pCDNA3.1-hrGFP, were illuminated by blue light with a spatial pattern using a printed mask with a specific image (local illumination) or with no pattern (global illumination) in the presence of Dox, or were kept in darkness without Dox addition (Dark). The image of hrGFP fluorescence was taken 48 h after transfection. The orange circle indicated the glass bottom of the dish, where the cells were attached. An empty plasmid pT6U3 was used to replace pT6U3-DTA as the negative control. Scale bar, 1 cm. (D) Phase imaging of cells transfected with the pGAVPO, pTetR-KRAB and pT6U3-DTA or pT6U3 empty vector. The engineered cells were cultured in blue light irradiance with Dox or in darkness without Dox, and then were plated with a density of 104 cells in each well of a 96-well plate after 24 h. The cells were cultured for another 72 h, and phase imaging was performed. Scale bar, 100 μm. (E) Quantitative analysis of cell viability by CCK-8. Briefly, 10 μl of CCK-8 solution was added to each well and incubated for 2 h at 37°C. The absorbance at 450 nm was determined using a microplate reader (BioTek). The data were normalized to the cells transfected with control vector and kept in darkness without Dox. The data are shown as the mean ± SD, n = 3.
DISCUSSION
Continuous expansion of the gene circuits is required for the rational design of highly complex devices that can program and probe diverse mammalian cell behaviors. Cells receive various cellular and environmental signals, which are often combinatorially processed to generate specific genetic responses. To date, available gene switches typically use a single type of control input that either induces or represses the gene switch and modulates the expression of a single or set of target genes. Several bipartite transcription factors have been developed to sense two separate small-molecular chemical input signals and switch genes on or off. Pioneering examples include the following: (i) synthetic mammalian trigger-controlled bipartite transcription factors, which are based on artificial TetR and GntR family-derived bipartite transcription factors, and exhibit control dynamics reminiscent of double-pole double-throw relay switches by TET and c-butyrolactones, phloretin, or vanillic acid (30); and (ii) an antagonistic dual-input expression device based on the CbaR-derived transactivator or trans-silencer triggered by the food additives benzoate and vanillate (31). In contrast to the development of artificial regulators with novel regulatory characteristics, the combination of heterologous transcription controls in mammalian cells may improve the regulation performance and regulatory expression window of the transgenes. Several investigations aimed to construct artificial regulatory networks containing multiple small-molecular chemical inputs are based on developed heterologous transcription factors. However, most of the developed multi-input controllers in mammalian cells are triggered by the same type of signals, while only a few utilized two different types of triggers. Chemical- and light-induced genetic devices have been widely used to switch transgene expression for the study of diverse life processes; however, a multi-input genetic circuit simultaneously regulated by a chemical and light has not been developed yet, which may have the combined benefits of a chemical inducer and light inducer and can provide novel control topologies with improved regulation performance and regulatory expression windows.
In the present study, several versatile dual-input genetic circuits triggered by tetracycline and light were constructed. In the first synergistic dual-input genetic circuit, the binding of either GAVPO or tTA or rtTA to the operator region of chimeric promoters could efficiently induce gene expression, which offers flexibility to switch on gene expression with either Dox or light and may be very useful for animal studies. The maximum induction ratio reached more than 1500-fold. The second antagonistic control impact exhibits a more than 1800-fold maximum induction ratio and high spatiotemporal resolution. Gene expression levels from both of the genetic circuits can be quantitatively defined by modulating the Dox concentration and light irradiance. The LightOn system has been well studied previously and shows relatively low leakage, and has been used in regulating transgene expression both in cultured cells and in mice. However, the developed antagonistic dual-input genetic circuit exhibits much lower leakage than the LightOn system in the non-inducing conditions, allowing extremely stringent control of transgene expression, even with the highly toxic DTA gene, which is difficult for other gene circuits in mammalian cells. Furthermore, for almost all of the developed light-regulated gene expression systems, cells or animals being investigated have to be always kept in darkness or at least avoid the exposure to light of specific color, which is inconvenient and may result in disrupted circadian behavior. However, gene expression in the antagonistic dual-input genetic circuit can be locked in ‘OFF’ state even when cells were illuminated with light. Therefore, it is not necessary to keep the samples always in darkness before or after gene induction, which offers great convenience for manipulating the samples and avoids disrupted circadian behavior.
Both fluorescent protein and luciferase were used in our studies to report the level of gene expression. Though the former allows single-cell analysis, the latter is more suitable for in vivo studies. Usually, mammalian cells are transiently transfected with plasmids, which leads to a broad range of transgene expression (48). In contrast, stable chromosomal integration of genetic payloads can help achieve long-term, defined and reproducible expression of transgene. On this end, genomic integration of synthetic circuits is still one of the key challenges for mammalian synthetic biology. Conventional integrative gene transfer methods are limited by the consequences of random and multi-copy integration that contribute to unstable or silenced transgene expression (49). Though Several approaches allows loci-specific integration of exogenous DNA sequences have been developed (50–52), the choice of a suitable genomic acceptor site for the optimal design of the transgene cassette to ensure robust expression without perturbing nearby endogenous transcription remains to be investigated (53). Moreover, it is hard for stable cells to quantitative express the transcription factors, which is directly relevant to the induction characteristics, e.g. balanced regulation, induction ratio or leak expression in the currently developed genetic circuits, as well as in many previously reported circuits. Therefore, though construction of a stably-integrated cell line with appropriate expression of the transcription factors is attractive, it is quite challenging and labor-intensive. To date, the majority of synthetic gene networks in mammalian cells are done in transfected cells, but not stable cell lines (54).
The developed dual-input controllers offer additional advantages when used in vivo. First, tetracycline as an effector molecule has well-established pharmacological features and is broadly distributed in various tissues following ingestion. Regulation of gene expression in the brain has also been achieved by replacing tetracycline with its derivatives, such as minocycline, which has a higher ability to penetrate the blood-brain barrier. Second, light is an ideal inducer of gene expression because it is easy to obtain, is highly tunable, is non-toxic and, most importantly, has high spatiotemporal resolution. Light-based photodynamic therapy (PDT) has been a promising new clinical treatment of cancers. Laser light can be directed through fiber optic cables (thin fibers that transmit light) to deliver light to areas inside the body. Thus, the developed dual-input controllers combining the TET and light can also be easily and safely applied in gene therapy in vivo.
Overall, as a new addition to the synthetic biology toolbox, we have shown that the TET- and light-triggered dual-input gene circuits combining the benefits of a chemical inducer and light inducer have different regulation properties compared with the existing regulatory systems. Accordingly, these versatile transgene systems have a significant potential in the design of novel and extremely compact gene network topologies that may foster advances in gene- and cell-based therapies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Yuzheng Zhao, Rongkun Tao and Xiaopei Xu for technical assistance.
FUNDING
Funding for open access charge: 973 Program [2013CB531200]; NSFC [31225008, 91313301, 31170815, 31470833]; Shanghai Science and Technology Commission [12JC1402900, 11DZ2260600, 15YF1402600]; Dawn Program of the Shanghai Education Commission [11SG31]; State Key Laboratory of Bioreactor Engineering; 111 Project [B07023]; Fundamental Research Funds for the Central Universities.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bugaj L.J., Schaffer D.V. Bringing next-generation therapeutics to the clinic through synthetic biology. Curr. Opin. Chem. Biol. 2012;16:355–361. doi: 10.1016/j.cbpa.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Auslander S., Fussenegger M. From gene switches to mammalian designer cells: present and future prospects. Trends Biotechnol. 2013;31:155–168. doi: 10.1016/j.tibtech.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Balazsi G., van Oudenaarden A., Collins J.J. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gossen M., Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fussenegger M., Morris R.P., Fux C., Rimann M., von Stockar B., Thompson C.J., Bailey J.E. Streptogramin-based gene regulation systems for mammalian cells. Nat. Biotechnol. 2000;18:1203–1208. doi: 10.1038/81208. [DOI] [PubMed] [Google Scholar]
- 6.Roman G., Davis R.L. Conditional expression of UAS-transgenes in the adult eye with a new gene-switch vector system. Genesis. 2002;34:127–131. doi: 10.1002/gene.10133. [DOI] [PubMed] [Google Scholar]
- 7.Weber W., Fux C., Daoud-el Baba M., Keller B., Weber C.C., Kramer B.P., Heinzen C., Aubel D., Bailey J.E., Fussenegger M. Macrolide-based transgene control in mammalian cells and mice. Nat. Biotechnol. 2002;20:901–907. doi: 10.1038/nbt731. [DOI] [PubMed] [Google Scholar]
- 8.Weber W., Schoenmakers R., Spielmann M., El-Baba M.D., Folcher M., Keller B., Weber C.C., Link N., van de Wetering P., Heinzen C., et al. Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice. Nucleic Acids Res. 2003;31:e71. doi: 10.1093/nar/gng071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber W., Bacchus W., Daoud-El Baba M., Fussenegger M. Vitamin H-regulated transgene expression in mammalian cells. Nucleic Acids Res. 2007;35:e116. doi: 10.1093/nar/gkm466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartenbach S., Daoud-El Baba M., Weber W., Fussenegger M. An engineered L-arginine sensor of Chlamydia pneumoniae enables arginine-adjustable transcription control in mammalian cells and mice. Nucleic Acids Res. 2007;35:e136. doi: 10.1093/nar/gkm652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gitzinger M., Kemmer C., El-Baba M.D., Weber W., Fussenegger M. Controlling transgene expression in subcutaneous implants using a skin lotion containing the apple metabolite phloretin. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10638–10643. doi: 10.1073/pnas.0901501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber W., Lienhart C., Baba M.D., Fussenegger M. A biotin-triggered genetic switch in mammalian cells and mice. Metab. Eng. 2009;11:117–124. doi: 10.1016/j.ymben.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Kemmer C., Gitzinger M., Daoud-El Baba M., Djonov V., Stelling J., Fussenegger M. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat. Biotechnol. 2010;28:355–360. doi: 10.1038/nbt.1617. [DOI] [PubMed] [Google Scholar]
- 14.Gitzinger M., Kemmer C., Fluri D.A., El-Baba M.D., Weber W., Fussenegger M. The food additive vanillic acid controls transgene expression in mammalian cells and mice. Nucleic Acids Res. 2012;40:e37. doi: 10.1093/nar/gkr1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim T., Folcher M., Charpin-El Hamri G., Fussenegger M. A synthetic cGMP-sensitive gene switch providing Viagra((R))-controlled gene expression in mammalian cells and mice. Metab. Eng. 2015;29:169–179. doi: 10.1016/j.ymben.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 16.Wang H., Ye H., Xie M., Daoud El-Baba M., Fussenegger M. Cosmetics-triggered percutaneous remote control of transgene expression in mice. Nucleic Acids Res. 2015;43:e91. doi: 10.1093/nar/gkv326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossger K., Charpin-El-Hamri G., Fussenegger M. Bile acid-controlled transgene expression in mammalian cells and mice. Metab. Eng. 2014;21:81–90. doi: 10.1016/j.ymben.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Nevozhay D., Zal T., Balazsi G. Transferring a synthetic gene circuit from yeast to mammalian cells. Nat. Commun. 2013;4:1451. doi: 10.1038/ncomms2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanley S.A., Gagner J.E., Damanpour S., Yoshida M., Dordick J.S., Friedman J.M. Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science. 2012;336:604–608. doi: 10.1126/science.1216753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller K., Engesser R., Schulz S., Steinberg T., Tomakidi P., Weber C.C., Ulm R., Timmer J., Zurbriggen M.D., Weber W. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 2013;41:e124. doi: 10.1093/nar/gkt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller K., Engesser R., Metzger S., Schulz S., Kampf M.M., Busacker M., Steinberg T., Tomakidi P., Ehrbar M., Nagy F., et al. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res. 2013;41:e77. doi: 10.1093/nar/gkt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kennedy M.J., Hughes R.M., Peteya L.A., Schwartz J.W., Ehlers M.D., Tucker C.L. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yazawa M., Sadaghiani A.M., Hsueh B., Dolmetsch R.E. Induction of protein-protein interactions in live cells using light. Nat. Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 24.Ye H., Daoud-El Baba M., Peng R.W., Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–1568. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 25.Motta-Mena L.B., Reade A., Mallory M.J., Glantz S., Weiner O.D., Lynch K.W., Gardner K.H. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., Chen X., Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 27.Polstein L.R., Gersbach C.A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nihongaki Y., Yamamoto S., Kawano F., Suzuki H., Sato M. CRISPR-Cas9-based photoactivatable transcription system. Chem. Biol. 2015;22:169–174. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan S., Bren A., Zaslaver A., Dekel E., Alon U. Diverse two-dimensional input functions control bacterial sugar genes. Mol. Cell. 2008;29:786–792. doi: 10.1016/j.molcel.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Folcher M., Xie M., Spinnler A., Fussenegger M. Synthetic mammalian trigger-controlled bipartite transcription factors. Nucleic Acids Res. 2013;41:e134. doi: 10.1093/nar/gkt405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie M., Ye H., Hamri G.C., Fussenegger M. Antagonistic control of a dual-input mammalian gene switch by food additives. Nucleic Acids Res. 2014;42:e116. doi: 10.1093/nar/gku545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nissim L., Bar-Ziv R.H. A tunable dual-promoter integrator for targeting of cancer cells. Mol. Syst. Biol. 2010;6:444. doi: 10.1038/msb.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auslander D., Auslander S., Charpin-El Hamri G., Sedlmayer F., Muller M., Frey O., Hierlemann A., Stelling J., Fussenegger M. A synthetic multifunctional mammalian pH sensor and CO2 transgene-control device. Mol. Cell. 2014;55:397–408. doi: 10.1016/j.molcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Xie Z., Wroblewska L., Prochazka L., Weiss R., Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 35.Lienert F., Torella J.P., Chen J.H., Norsworthy M., Richardson R.R., Silver P.A. Two- and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic Acids Res. 2013;41:9967–9975. doi: 10.1093/nar/gkt758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aubrecht J., Manivasakam P., Schiestl R.H. Controlled gene expression in mammalian cells via a regulatory cascade involving the tetracycline transactivator and lac repressor. Gene. 1996;172:227–231. doi: 10.1016/0378-1119(96)00216-8. [DOI] [PubMed] [Google Scholar]
- 37.Kramer B.P., Fischer C., Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol. Bioeng. 2004;87:478–484. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- 38.Kramer B.P., Weber W., Fussenegger M. Artificial regulatory networks and cascades for discrete multilevel transgene control in mammalian cells. Biotechnol. Bioeng. 2003;83:810–820. doi: 10.1002/bit.10731. [DOI] [PubMed] [Google Scholar]
- 39.Gossen M., Freundlieb S., Bender G., Muller G., Hillen W., Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 40.Ma Z., Du Z., Chen X., Wang X., Yang Y. Fine tuning the LightOn light-switchable transgene expression system. Biochem. Biophys. Res. Commun. 2013;440:419–423. doi: 10.1016/j.bbrc.2013.09.092. [DOI] [PubMed] [Google Scholar]
- 41.Zabala M., Wang L., Hernandez-Alcoceba R., Hillen W., Qian C., Prieto J., Kramer M.G. Optimization of the Tet-on system to regulate interleukin 12 expression in the liver for the treatment of hepatic tumors. Cancer Res. 2004;64:2799–2804. doi: 10.1158/0008-5472.can-03-3061. [DOI] [PubMed] [Google Scholar]
- 42.Wendt M.K., Molter J., Flask C.A., Schiemann W.P. In vivo dual substrate bioluminescent imaging. J. Vis. Exp. 2011;56:e3245. doi: 10.3791/3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deuschle U., Meyer W.K., Thiesen H.J. Tetracycline-reversible silencing of eukaryotic promoters. Mol. Cell. Biol. 1995;15:1907–1914. doi: 10.1128/mcb.15.4.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaw G., Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 45.Collier R.J. Diphtheria toxin: mode of action and structure. Bacteriol. Rev. 1975;39:54–85. doi: 10.1128/br.39.1.54-85.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaizumi M., Mekada E., Uchida T., Okada Y. One molecule of diphtheria toxin fragment A introduced into a cell can kill the cell. Cell. 1978;15:245–250. doi: 10.1016/0092-8674(78)90099-5. [DOI] [PubMed] [Google Scholar]
- 47.Zheng J.Y., Chen D.L., Chan J., Yu D., Ko E., Pang S. Regression of prostate cancer xenografts by a lentiviral vector specifically expressing diphtheria toxin A. Cancer Gene Therapy. 2003;10:764–770. doi: 10.1038/sj.cgt.7700629. [DOI] [PubMed] [Google Scholar]
- 48.Gopal T.V. Gene transfer method for transient gene expression, stable transformation, and cotransformation of suspension cell cultures. Mol. Cell. Biol. 1985;5:1188–1190. doi: 10.1128/mcb.5.5.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duportet X., Wroblewska L., Guye P., Li Y., Eyquem J., Rieders J., Rimchala T., Batt G., Weiss R. A platform for rapid prototyping of synthetic gene networks in mammalian cells. Nucleic Acids Res. 2014;42:13440–13451. doi: 10.1093/nar/gku1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lombardo A., Genovese P., Beausejour C.M., Colleoni S., Lee Y.L., Kim K.A., Ando D., Urnov F.D., Galli C., Gregory P.D., et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 51.Boch J. TALEs of genome targeting. Nat. Biotechnol. 2011;29:135–136. doi: 10.1038/nbt.1767. [DOI] [PubMed] [Google Scholar]
- 52.Ledford H. CRISPR, the disruptor. Nature. 2015;522:20–24. doi: 10.1038/522020a. [DOI] [PubMed] [Google Scholar]
- 53.Lombardo A., Cesana D., Genovese P., Stefano B., Provasi E., Colombo D.F., Neri M., Magnani Z., Cantore A., Lo Riso P., et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 54.Kis Z., Pereira H.S., Homma T., Pedrigi R.M., Krams R. Mammalian synthetic biology: emerging medical applications. J. R. Soc. Interface. 2015;12:20141000. doi: 10.1098/rsif.2014.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.