Abstract
Genome engineering of human cells plays an important role in biotechnology and molecular medicine. In particular, insertions of functional multi-transgene cassettes into suitable endogenous sequences will lead to novel applications. Although several tools have been exploited in this context, safety issues such as cytotoxicity, insertional mutagenesis and off-target cleavage together with limitations in cargo size/expression often compromise utility. Phage λ integrase (Int) is a transgenesis tool that mediates conservative site-specific integration of 48 kb DNA into a safe harbor site of the bacterial genome. Here, we show that an Int variant precisely recombines large episomes into a sequence, termed attH4X, found in 1000 human Long INterspersed Elements-1 (LINE-1). We demonstrate single-copy transgenesis through attH4X-targeting in various cell lines including hESCs, with the flexibility of selecting clones according to transgene performance and downstream applications. This is exemplified with pluripotency reporter cassettes and constitutively expressed payloads that remain functional in LINE1-targeted hESCs and differentiated progenies. Furthermore, LINE-1 targeting does not induce DNA damage-response or chromosomal aberrations, and neither global nor localized endogenous gene expression is substantially affected. Hence, this simple transgene addition tool should become particularly useful for applications that require engineering of the human genome with multi-transgenes.
INTRODUCTION
Sustained multi-transgene expression from the human genome becomes increasingly important in applications involving stem cell engineering, gene therapy and synthetic biology (1,2). It can be accomplished by either site-specific or random genomic integration of foreign DNA. However, targeted integration at predetermined, so-called safe harbor sites is preferred over random insertions in order to prevent interference with transgene expression, insertional mutagenesis, activation of neighboring genes and cell toxicity (3,4). In this context, site-specific recombination systems have been developed using, for example, Flp recombinase from the 2 μm yeast plasmid and bacteriophage phiC31 integrase (Int), or custom recombinases that are derived from invertases/resolvases (5–7). However, their full potential in particular for safe harbor site transgenesis needs to be explored. The recent development of designer endonucleases such as ZFNs, TALENs and CRISPR/Cas9 has also led to more controlled and precise genome engineering, including the knock-in of transgenes at safe harbor sites such as AAVS1 on human chromosome 19 (8).
Designer nucleases introduce a double strand break (DSB) at the target sequence (9,10), and subsequent cellular DNA synthesis-dependent strand annealing and homology-directed repair synthesis involving a donor DNA template results in transgene insertion at DSBs (11). However, in the context of gene knock-in, some concerns and limitations still linger. These include off-target site cleavage which could lead to uncontrolled DNA damage response, cell death, chromosomal aberrations and unintended mutations due to induction of DSBs at sites apart from the targeted sequence (1,12). Furthermore, in case of linear donor DNA, illegitimate recombination frequently results in ‘bad’ or ‘ugly’ integrants at the target locus (3), in addition to true random integration events. Another limitation is the complete insertion of > 5 kb multi-gene constructs, in particular those containing repeat sequences (11,13).
We present here a novel transgenesis tool for the human genome on the basis of the well-studied integration system of phage λ Int which should help to address some of the above-mentioned concerns. The λ wild-type integration system requires Int as a recombinase, regulatory protein cofactors and two DNA attachment (att) sites: the target attB site (21 bp) on the bacterial chromosome and the more complex attP site (241 bp) located on the phage genome; the latter also requires negative DNA supercoiling to catalyze recombination (14). Integrative recombination between attB and attP leads to hybrid attL and attR sites that flank the prophage genome after integration into the bacterial chromosome.
Int shows exquisite target site specificity for the large (> 48 kb) circular transgenic phage λ genome (14). Unlike phage Ints of the serine type such as phiC31, the bacterial transposon-encoded resolvases/invertases or the above-mentioned designer nucleases, Int catalyzes conservative site-specific recombination via two successive rounds of DNA single strand exchanges, leading to a Holliday junction intermediate which is resolved into recombinants if partner recombination sequences are compatible (14). Int thus avoids generation of potentially dangerous DSBs at the genomic target site which may otherwise occur, for example, during aborted recombination attempts.
We previously generated a cofactor-independent Int variant, named Int-h/218, which recombines att sites in eukaryotic cells (15,16). Int-h/218 has been used for genome manipulation in mice, plants as well as for artificial chromosome engineering (17–19). In an attempt to improve Int-h/218 for human genome engineering, we recently applied a novel directed evolution strategy and selected variant Int-C3 which outperformed Int-h/218 both in vitro and ex vivo (20). Here, we used Int-C3 to develop a simple transgenesis tool for functional single-copy and multi-transgene cassette addition to the human genome by targeting a set of predetermined endogenous sequences that belong to Long INterspersed Elements-1 (LINE-1). At least some of these target sequences may be considered as genomic safe harbor sites.
MATERIALS AND METHODS
Cell lines
This study used human embryonic stem cell (hESC) line ‘Genea 047’ (Genea Biocells, Sydney, Australia) and cancer cell lines of Human origin A549 (lung epithelial carcinoma), HT1080 (fibrosarcoma), HeLa (Cervical epithelial adenocarcinoma) and NEB-1 (immortalized neonatal foreskin keratinocytes cell line).
Plasmids
Standard molecular cloning techniques were employed to generate plasmids used in this work. High fidelity Pfu polymerase (Thermo Scientific) was used for PCR amplifications and E. coli DH5α was used for plasmid DNA amplifications. The construction of Int expression vector (pCMVssInt-h/218) has been described (15). pCMVssInt-C3 was generated by PCR amplification of the Int-C3 coding sequence from pET-Int-C3 (20), using the primers Int_fwd_PstI and Int_rev_XbaI (all the primer sequences are listed in Supplementary Table S1). PCR products were cloned into pCMVssInt-h/218 between PstI and XbaI sites, thus replacing the Int-h/218 with the Int-C3 sequence.
pCMVssInt-C3CNLS was generated by inserting the SV40 nuclear localization signal (NLS) sequence at the 3′ end of Int-C3 coding sequence in pCMVssInt-C3. Int-C3CNLS sequence was PCR amplified from pCMVssInt-C3 using the primers CNLS_Xbal_Int (which provides the NLS sequence) and Int fwd PstI. Int-C3CNLS PCR products were cloned into pCMVssInt-C3 restricted with PstI and XbaI, thus replacing the Int-C3 sequence with Int-C3CNLS.
pCMVssInt-Ina (plasmid expressing Int with an inactivating mutation wherein the amino acid residue tyrosine at sequence position 342 is replaced by the amino acid alanine) was generated with a similar PCR based site-directed mutagenesis protocol as above using pCMVssInt-h/218 as template and primers SDM-Int-Y342A-F and SDM-Int-Y342A-R.
pCMVssInt-C3CNLS-H (for expression of HIS-tagged Int-C3 with C-terminal NLS) Int C3CNLS-6xHis coding sequence was generated by PCR amplification from pCMVssC3InthCNLS using the primers Int_fwd_PstI and NLS-HIS-XbaI Rev (providing the NLS signal and 6x Histidine coding sequences) and inserted into PstI and XbaI sites in pCMVssInt-h/218 (replacing the original Int-h/218 sequence).
pCMVssInt-Ina-H (for expression of HIS-tagged Int-Inactive) and pCMVssInt-C3-H (for expression of HIS-tagged Int-C3), was generated using the inactive Int coding sequence (from pCMVssInt-Ina) and that of Int-C3 (from pCMVssInt-C3) which were PCR amplified with primers Int_fwd_PstI and INT-HIS-XbaI Rev (providing the 6× Histidine coding sequence) and cloned into PstI and XbaI sites in pCMVssInt-h/218.
Int-C3 expression under the EF1α promoter in human embryonic stem cell (hESCs) were generated as follows (21). To construct pEF1α-Int-C3CNLS and pEF1α-Int-C3CNLS-H, the EF1α promoter sequence was PCR-amplified from pEF1α-eGFP using primers EF1Fw-EcoRI and EF1Rev-NsiI and cloned between EcoRI and NsiI sites in pCMVssInt-C3 and pCMVssInt-C3-H respectively, thus replacing the CMV promoter-ss hybrid intron sequences. To construct pEF1αss-Int-C3CNLS and pEF1αss-Int-C3CNLS-H, the EF1α promoter sequence was PCR-amplified from pEF1α-eGFP using the primers EF1Fw-EcoRI and EF1Rev-NheI and cloned between EcoRI and NheI sites in pCMVssInt-C3 and pCMVssInt-C3-H, respectively (replacing only the CMV promoter sequence but retaining the ‘ss’ hybrid intron (22,23).
For Int mRNA synthesis by in vitro transcription, pMRNAxp-based constructs (mRNA synthesis kit-mRNA Express, System Biosciences, CA, USA) were generated. To construct pMRNA-Int-C3 and pMRNA-Int-Ina, the Int-C3 and Int-Inactive sequences were PCR-amplified from pCMVssInt-C3 and pCMVssInt-Ina, respectively, using primers MRNA-Inth218Fw and MRNA-Inth218Rev and cloned into EcoRI and BamHI sites in pMRNAxp. To construct pMRNA-Int-C3CNLS the Int C3CNLS sequence was PCR-amplified from pCMVssInt-C3 using primers MRNA-Inth218Fw and C3Int-CNLS-BglII Rev and inserted between EcoRI and BamHI sites of pMRNAxp.
pattP4X-PGKssPuro was generated through PCR-based site-directed mutagenesis of the overlap sequence of attP present in pattP39-PGKssPuro (Tan and Droge, unpublished data) using primers attP(4X)sdm (For) and attP(4X)sdm (Rev). PCR-based site-directed mutagenesis using mutagenic primers was performed as described (24) with modifications. PCR was performed with the template plasmid and mutagenic primers using high fidelity Pfu polymerase (Thermo Scientific) in a 50 μl reaction. The thermal cycling parameters used for PCRs were: initial denaturation step at 95°C for 5 min, 25 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 1 min and extension at 68°C for 12 min and a final step of 68°C for 5 min. The PCR amplified product was purified using PCR purification kit (Qiagen, GmbH) and subjected to digestion with DpnI (New England Biolabs) to selectively digest the methylated parental DNA template. An aliquot of the reaction was transformed into E.coli DH5α. Plasmid DNA was isolated from transformants and analyzed by sequencing to verify the introduced mutation and integrity of the plasmid.
pTZ-attP4X-UN-EF1α-eGFP was generated by subcloning the attP4X sequence from pattP4X-PGKssPuro as an EcoRI fragment in the unique EcoRI site upstream of the human UTF1 promoter in pTZ-UN (25) to generate pTZ-attP4X-UN. The EF1α-eGFP sequence was PCR-amplified from pEF1α-EGFP using primers EcoRV_EF_fwd and ClaI_bgh_bpa_rev and cloned into pTZ-attP4X-UN digested with HindIII and XbaI and blunted by fill-in reaction using Klenow fragment (New England Biolabs).
pattP4X-PGKsspuro-UTF1-eGFP targeting vector was generated using the UTF1-eGFP cassette which was PCR amplified from pTZ-UTF1-EGFP (25) (using primers KpnI-UTF1-fwd and ClaI-UTF1enhancer-rev) and inserted into ClaI and KpnI sites of pattP4X-PGKssPuro in the opposite orientation with respect to the PGKssPuro cassette. Similarly, for the construction of pattP4X-PGKssPuro-EF-eGFP targeting vector, the EFα-eGFP cassette was PCR amplified from pEF1-eGFP (using primers EcoRV_EF_fwd and ClaI_bgh_bpa_rev) and inserted into pattP4X-PGKssPuro at ClaI and EcoRV sites in the opposite orientation with respect to the PGKssPuro cassette.
pTZ18R-attL/attR-PGKssPuro-UTF1-EGFP was constructed by cloning attL, attR and PGKssPuro-UTF1-EGFP cassette in the pTZ-18R vector backbone. The attL fragment was PCR amplified from pCMVssattL using primers attL(BOP′)Fwd-(KpnI) and attL(BOP′)Rev-(NotI). The attR site was PCR amplified from pCMVssattR as template using the primers attR(POB′)Fwd-(ClaI) and attR(POB′)Rev-(HindIII). pTZ18R along with PCR-amplified attL and attR were restricted with KpnI and HindIII enzymes and a three fragment ligation generated pTZ18R-attL/attR flanked by NotI and ClaI sites. The three fragment ligated product pTZ18R-attL/attR flanked by NotI and ClaI sites and the p(-attP4X) pgksspuro-UTFI-EGFP fragment were cleaved with NotI and ClaI enzymes and ligated to generate the pTZ18R-attL/attR-PGKssPuro-UTF1-EGFP target vector. All plasmids were confirmed by sequencing analysis.
Cell culture
HT1080, A549 and HeLa cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM) growth medium supplemented with 10% FBS, 1% L-glutamine and 100 Units/ml of Penicillin and Streptomycin each (Gibco, Life technologies) at 37°C under 5% CO2 in humidified condition. NEB-1 cells were cultured in RM+ medium [DME high glucose (326.25 ml), HAMS F12 (108.75 ml), Fetal Bovine Serum (10%), L-glutamine (1%), Penicillin/Streptomycin (100 units/ml each) and RM+ Supplement (1%). RM+ Supplement is composed of hydrocortisone (0.4 μg/ml), insulin (5 g/ml), Adenine (1.8 × 10−4 M), epidermal growth factor (10 ng/ml), cholera toxin (10−10 M), transferrin (5 μg/ml) and liothyronine (2 × 10−11 M)]. For selection of puromycin-resistant recombinants, puromycin (Gibco, Life technologies) was added in the growth medium (1 μg/ml). Trypsin-EDTA (Gibco, Life technologies) was used for detaching the adherent cells for passaging.
Human embryonic stem cells (GENEA 047) were cultured at 37°C under 5% CO2 and 5% O2 on Collagen I coated cell culture dishes (Biocoat, Corning) in Genea M2 Medium, (Genea Biocells, Sydney, Australia), supplemented with Penicillin and Streptomycin at 25 Units/ml each (Gibco, Life technologies). For selection of recombinants and maintenance of targeted clones, Neomycin (100–200 μg/ml) or Puromycin (300 ng/ml) (both from Gibco, Life technologies) was included in the growth medium. For passaging or preparing cell suspension for reverse transfections, adherent hESCs were rinsed with 1 × PBS, detached by incubating at 37°C for 3 min with passaging solution (Genea Biocells) (with a volume of 100 μl per well of a 6 well plate or 1 ml per 10 cm dish), dislodging cells by tapping and resuspending the cells with at least 3× volume of Neutralization solution (Genea Biocells). After counting the cells in a haemocytometer (Neubauer), they were pelleted by centrifuging at 300 × g for 4 min and resuspended in Genea M2 Medium to the required cell density and added drop-wise to Collagen I-coated dishes.
Differentiation of hESCs
Retinoic acid (RA) induced differentiation of the hESCs, was based on an established protocol (26). Briefly, hESCs were grown in 6-well plates to reach a confluence of 60–70% and M2 medium was replaced with DMEM (supplemented with 20% FBS, 1% L-glutamine and 100 Units/ml of penicillin and streptomycin each) containing RA (R 2625, Sigma) at a final concentration of 1 μM and cultured for 48 h at 37°C under 5% CO2 in humidified condition. Thereafter the cells were grown in DMEM (supplemented with 20% FBS, 1% L-glutamine and 100 Units/ml of penicillin and streptomycin each). Neomycin (at 100–200 μg/ml) was included in the growth medium after RA treatment in experiments testing functionality of the UTF1 reporter cassettes in hESCs clones and differentiated progenies. Microscopy data acquisition and analysis were done using OLYMPUS IX71 microscope with OLYMPUS DP70 camera and DP Controller.exe software tool (OLYMPUS, Japan) and CorrSight™ FEI microscope, Oregon, USA.
Cardiomyocytes were generated from hESCs as previously described (27). Briefly, cells were grown on Collagen I coated plates until they reached ∼85% confluency. The medium was changed to CDM3 (RPMI 1640 Life technologies), 500 μg/ml recombinant human albumin (A0237 Sigma-Aldrich), and 213 μg/ml L-ascorbic acid 2-phosphate (Sigma-Aldrich) with 6 μM CHIR99021 (Stemcell Technologies, 72052) for the first 48 h, then replaced by CDM3 plus 2 μM Wnt-C59 (Cellagen Technologies, C7641–2) for the following 48 h. Media were changed to CDM3 alone and continued to be replaced every other day. Cells began to beat between days 7 and 8.
Transfections
For transfections in HT1080, A549 and HeLa cell lines, 3 × 105 or 3 × 106 cells were seeded per well of 6-well plate (IWAKI, Japan) or per 10 cm tissue culture dishes (TPP, Switzerland), respectively, in DMEM growth medium a day before transfection to obtain 70–90% confluence at the time of transfection. Transfections were employed Lipofectamine 2000 (Invitrogen, Life technologies) with DNA/mRNA to Lipofectamine 2000 ratio of 1 μg : 2 μl. For every transfection per well, complexes were prepared by mixing DNA/mRNA and Lipofectamine 2000 reagent separately diluted in 100μl of Opti-MEM medium (Life technologies) and incubating for 20 min at room temperature. The transfection mix was added drop wise onto the cells (under DMEM growth medium without antibiotics) and transfection was allowed to proceed for 4–6 h before replacing with fresh growth medium.
For transfections in hESCs, FuGENE HD Transfection reagent (Promega) was used in a reverse transfection protocol. DNA to FuGENE ratio of 1 μg:3 μl was used. Transfection mixes were prepared by first diluting plasmid DNA in 100 μl of Opti-MEM and 5 min later the FuGENE HD reagent was added to the DNA dilution, mixed and incubated for 15 min at room temperature for the complexes to form. During the incubation period, hESCs were harvested (as described above) and resuspended in Genea M2 Medium (without antibiotics). The transfection complexes were added drop-wise to Collagen 1-coated plates and incubated at culturing conditions for 5 min after which the harvested cells were gently pipetted to the dishes at 5 × 105 cells per well of 6-well plate and 5 × 106 cells per 10 cm dish. Transfections were performed overnight under standard culture conditions for hESCs, and media containing transfection complex was replaced with fresh M2 media.
Antibiotic selection and screening for targeted cell clones
Forty-eight hours post transfection, selection with the respective antibiotic in growth medium at the concentrations indicated above was initiated. Selection medium was replaced once in 2 days until colonies expanded to about 0.3–0.4 cm in diameter. At this stage, the colonies were picked by carefully scraping patches of cells with a pipette tip and transferred to 96-well plates for clonal expansion. The clones were sequentially expanded from 96 wells to 24 wells and subsequently in 6-well plates. Genomic DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen, GmbH) as per manufacturer's protocol.
Identification of recombination events by PCR screening
PCR was performed using GoTaq Flexi DNA polymerase (Promega) to amplify attL or attR junctions using primers listed in the figure descriptions and 200 ng of genomic DNA from each recombinant clone or parental cells as template in 50 μl reactions. The thermal cycling parameters used for PCRs was as follows: initial denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 1 min, annealing at 57°C for 30 s and extension at 72°C for 1 min, and a final step of 72°C for 5 min. The PCR samples were analyzed by electrophoresis in 0.8% agarose (Seakem Agarose, Lonza, USA) gels in 1× TBE (Tris-Boric acid-EDTA buffer) containing 0.5 μg/ml ethidium bromide and PCR-amplified products were compared with DNA standard markers and digitally documented under UV illumination (Gel Doc 2K System, BioRad). PCR-amplified products were analyzed by sequencing.
Inverse PCR and corresponding nested PCRs were performed using Long Range PCR (Qiagen GmbH). Genomic DNA from pooled puromycin-resistant HT1080 colonies (obtained through co-transfection of pattP4X-PGKssPuro and pCMVssInt-h/218CNLS) or parental cell line was restricted with PstI restriction enzyme, followed by ligation (200 ng of DNA) favoring self-ligation of linearized DNA (as described by the NEB protocol) and purification of ligated DNA (PCR purification kit, Qiagen GmbH). DNA was subsequently used as template for inverse PCR (using primers Purorev303 and Purofw318). Around 1–2 μl of the purified inverse PCR sample was used as template for the subsequent nested PCR (using primers Purorev24 and Purofw509). The thermal cycling parameters were: an initial denaturation step at 93°C for 3 min, 35 cycles of denaturation at 93°C for 15 s, annealing at 60°C for 30 s and extension at 68°C for 5 min and a final extension step at 68°C for 5 min. PCR-amplified products were analyzed by sequencing.
Cell proliferation assays
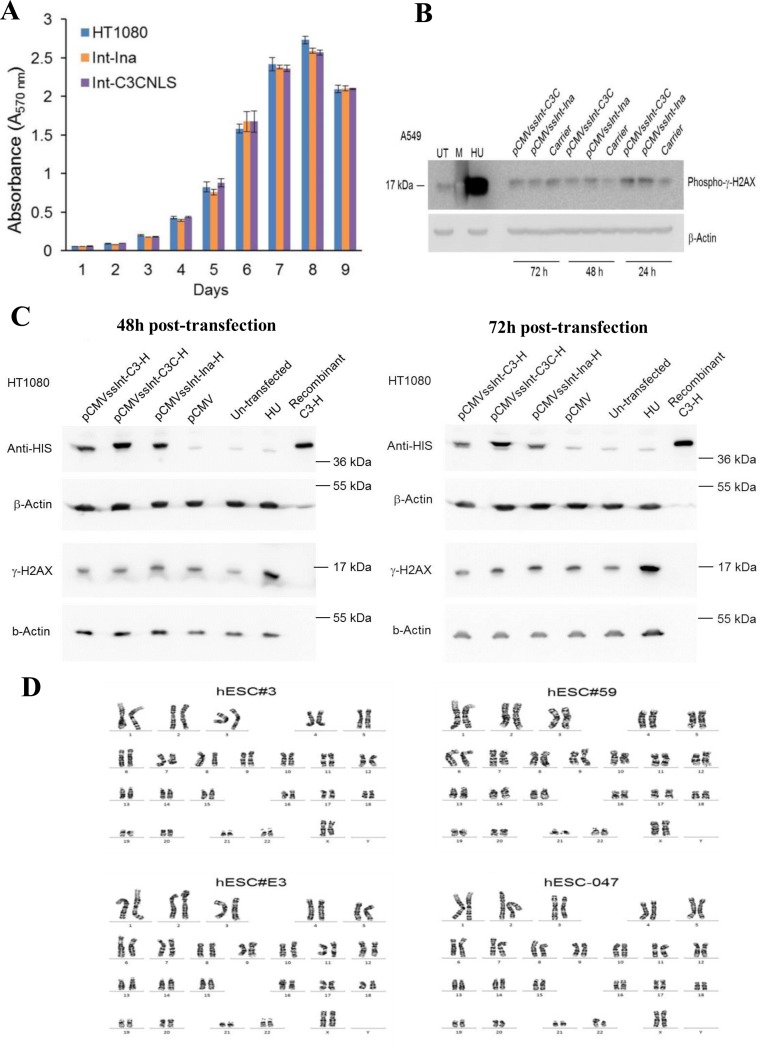
HT1080 cells were co-transfected with 1 μg each of pCMV-EGFP and pCMVssIna or pCMVss-C3CNLS in 10 cm plates. Forty eight hours post transfection, GFP+ cells from each sample were FACS sorted and plated in triplicates, at a density of 5 × 103 per well in 24-well plates and allowed to attach overnight. Untransfected HT1080 cells were included as control. Cell proliferation rates were measured every day for 9 days using MTT-based In Vitro Toxicology Assay Kit (Sigma-Aldrich) as per manufacturer's protocol. Briefly, 3 h post incubation with MTT dye-containing DMEM growth medium, cells were lysed with solubilization solution and 100 μl sample from each well was transferred to 96 well plate and absorbance measured at 570 nm in a microtiter plate reader (Infinite 200Pro, Tecan). The average values from triplicate readings were determined and values for the blanks subtracted from the average. Absorbance values were plotted against time points in days.
Flow cytometry
FACS Calibur Flow Cytometer (Becton Dickson) and CELL quest software (Becton Dickson) were used to analyze and quantify GFP+ cells. Cells were trypsinized, centrifuged and suspended in corresponding media. Dot plot of side scatter (SSC) versus forward scatter (FSC) was used to gate live cells in order to separate them from aggregated and dead cells. For gated cells, a dot plot of GFP versus FSC was constructed for further analyses. Data were analyzed with FlowJo software and GFP− and GFP+ cells for each sample were indicated ( in %) in the lower right and upper right quadrant, respectively.
Western analysis
Cell lysates were prepared as follows. At the indicated time points post transfection, cells were detached (with Passaging solution (GENEA) for hESCs and Trypsin-EDTA for HT1080 and A549 cells) and harvested with DMEM into Eppendorf tubes, pelleted by centrifugation (at 1000 rcf for 5 min at 4°C), washed once with 1× PBS and lysed in NP40 lysis buffer (NaCl (150 mM), NP-40 (1.0%), Tris-Cl (50 mM, pH 8.0), SDS (1%), protease inhibitor cocktail (11873580001, Roche) and phosphatase inhibitor (P0044, Sigma)) followed by incubation on ice for 20 min and sonication of lysates on ice (5 W, 10 × 3 s). Insoluble components were removed by centrifugation at 12 000 × g for 15 min and supernatant was collected. Protein concentrations were determined with DC protein assay reagent B (Bio-Rad). Proteins were separated by SDS-PAGE gels (10–15% acrylamide) and then transferred onto PVDF membranes with 0.2 μm pore size (Bio-Rad). Nonspecific binding was blocked by blocking buffer (5% nonfat milk (Bio-Rad) in 1 × TBS containing 0.1% Tween20) for 1 h at room temperature and incubated overnight at 4°C with primary antibodies against Histidine tag (1:1000 dilution; MA1-21315, Thermo Fisher Scientific), Phospho-H2AX (1:1000 dilution; MA1-2022, Thermo Fisher Scientific), Nanog (1:1000 dilution; MA1-017, Thermo Fisher Scientific), Sox2 (1:1000; MA1-014, Thermo Fisher Scientific) Oct-4 (1:1000 dilution; sc-365509, Santa Cruz Biotechnology) in blocking buffer. Amounts of β-actin protein were determined by monoclonal antibodies raised against human β-actin (1:10 000 dilution; A1978, Sigma). Blots were washed with 1 × TBS containing 0.1% Tween 20 and incubated for 1 h at room temperature with secondary antibody HRP-conjugated polyclonal goat anti-mouse (1:10 000; Dako, Denmark). After washing with 1 × TBS containing 0.1% Tween 20, immunoreactive bands were detected using the Western HRP substrate (Luminata Forte, Millipore) in an infrared Imager (LAS-4000, Fuji).
Southern analysis
Genomic DNA was purified using DNeasy Blood & Tissue Kit (Qiagen, GmbH). 15 μg of genomic DNA was subjected to restriction digestion using 50 U of the respective enzyme in 200 μl overnight at 37°C. DNA was ethanol precipitated and dissolved in 20 μl TE buffer (pH 8.0). Targeting vectors were linearized with single cutter restriction enzyme and diluted to 107, 108, 109 copies per μl. Digested genomic DNA samples were resolved overnight on a 1% agarose gel in 1× TAE (Tris-Acetate-Boric acid) buffer, with 1 kb DNA ladder (New England Biolabs) and 1 μl of positive control samples. Southern blotting employing the respective probes, as indicated, was performed using the DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche) as per the manufacturers’ protocol. The probe-target hybrids on the blots were detected by chemiluminescent assay followed by exposure to an X-ray film (Kodak MXG film, Kodak) and developed on a Kodak X-OMAT 2000 Processor.
Karyotyping
The targeted hESC lines hESC#3, hESC#59, hESC#E3 and the parental hESC-047 were karyotyped by G-banding of metaphase chromosomes as previously described (28). For each cell line 20 GTG-banded cells were scored and at least five GTG-banded cells were analyzed. Analysis and interpretation of data were in accordance to the International System for Human Cytogenetic Nomenclature (ISCN 2013). This analysis was carried out at the Cytogenetics Lab, Department of Pathology, Singapore General Hospital, Singapore.
Transcriptome analysis
Whole transcriptome analysis for three hESC clones (#3, #24 and E3) was carried out by AIT Biotech and Life Technologies, Singapore who provided the following protocol. Briefly, 100 ng of total RNA was used as input for library construction using the Ion AmpliSeq™ Transcriptome Human Gene Expression Kit, according to the manufacturer's protocol. Total RNA was reverse-transcribed using random priming, and target genes were amplified using the Human Gene Expression Core Panel with the Ion AmpliSeq™ Library Kit Plus. Following target amplification, the resulting amplicons were treated with FuPA reagent for partial digestion of the primers and phosphorylation of the amplicons. Amplicons were ligated to the Ion Xpress™ Barcode Adapters for barcoding. The eight barcoded libraries were then quantified by qPCR (Ion Library Quantitation Kit) normalized and combined into a single library prior to template preparation and enrichment on the Ion OneTouch™ 2 System. During template preparation, libraries were clonally amplified on Ion Sphere Particles™ by emulsion PCR using the Ion PI™ Template OT2 200 Kit v3 on the Ion OneTouch™ 2 Instrument. Enrichment was carried out on the Ion OneTouch™ ES (enrichment system) to isolate the template-positive Ion Sphere Particles™. The template-positive Ion Sphere Particles™ was loaded onto the Ion PI™ Chip for subsequent sequencing using the Ion PI™ Sequencing 200 Kit v3 chemistry. Raw data were then processed on the Ion Proton™ Sequencer and transferred to the Ion Proton™ Torrent Server for primary data analysis with gene-level transcript quantification from sequence read data performed using the associated Torrent Suite™ analysis plugin, ampliSeqRNA. Sequence reads were aligned to the Ion AmpliSeq™ Transcriptome reference file in Torrent Suite™ Software using the Ion Torrent Mapping Alignment Program (TMAP). The reference file contains the entire set of RefSeq transcripts from which all 20,802 Ion AmpliSeq™ Transcriptome panel primers were designed. Following alignment, the ampliSeqRNA plugin examines the number of reads mapping to the expected amplicon ranges and assigns counts per gene for reads which align to these regions as defined in the BED file. The Ion AmpliSeq™ Transcriptome BED is a formatted file containing the nucleotide positions of each amplicon per transcript in the mapping reference. Reads aligning to the expected amplicon locations are referred to as ‘on target’ reads and are reported as a percentage of total reads by the plugin.
RESULTS
Identification of functional λ Int target sequences in the human genome
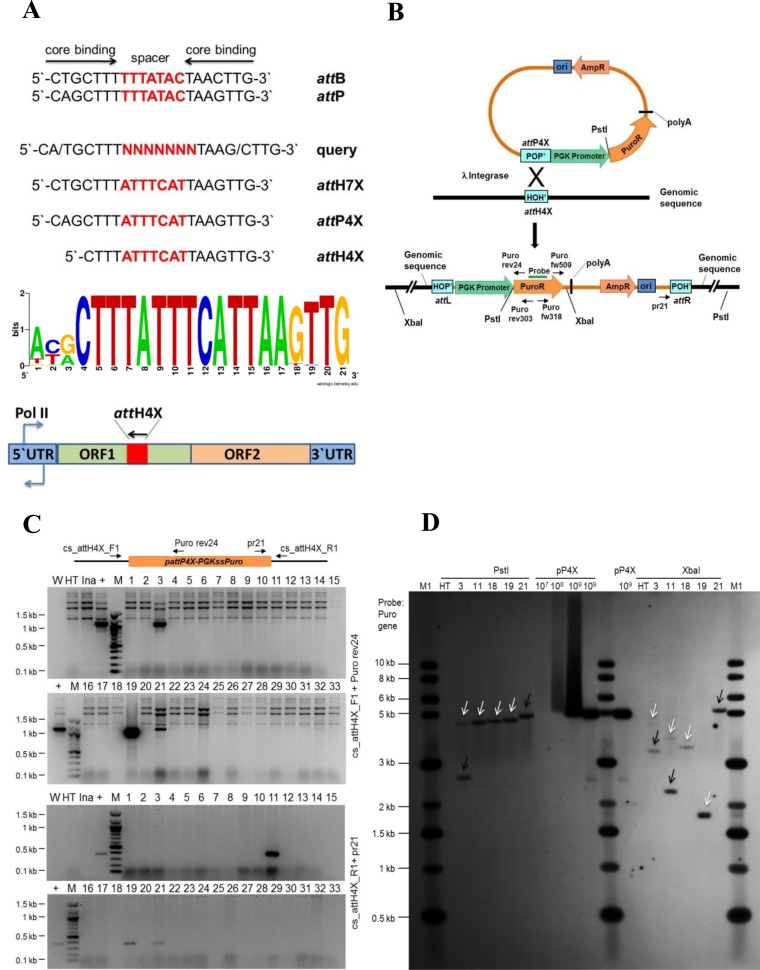
We set out to develop Int-C3 into a site-specific multi-transgene addition tool and used a PERL script to search the human genome sequence (hs_ref-GRCh37.p5) for targets resembling cores of att sites. Our query sequence encompassed λ Int core binding sites from attB and attP, and a degenerate 7 bp spacer (Figure 1A). Out of 147 hits, a sequence termed attH7X showed the highest occurrence in the human genome (Figure 1A; Supplementary Table S2). This attH7x sequence is found in some families of LINE-1 elements (Supplementary Table S3). In order to target attH7X with attP, the 7bp spacer in the attP core was adjusted to match the attH7X spacer. Outside the core, functionally important parts of attP, in particular Int arm type DNA binding sites, remained unchanged (not shown). This resulted in the core attP4X sequence as potential recombination partner for the genomic attH7X sequence (Figure 1A).
Figure 1.
Targeting endogenous attH4X in HT1080 cells. (A) Diagram showing the 21 nucleotide sequences comprising the core binding and spacer sequences of various att sites and features of the LINE-1 retrotransposon. Wild type attB and attP sequences aligned to show the respective core binding and spacer sequences. The query sequence was used in a bioinformatics search for targets resembling core att sites. Sequence logo analysis was performed for the 18 bp attH4X sequences in LINE-1 elements in the targeted cell lines including additional three nucleotides at the 5′ end in order to compare with the 21 bp attB sequence. A LINE-1 retrotransposon diagram with the position and orientation of attH4X is shown at the bottom. See text for details. (B) Schematic drawing showing pattP4X-PGKssPuro target vector and predicted recombination between attP4X and genomic attH4X. Positions of relevant primers (Puro rev24, Puro rev303, Puro fw318, Puro fw509 and pr21), restriction sites and the probe used for Southern blotting are indicated. (C) Screening for attH4X × attP4X recombination events in HT1080 clones. PCR was performed with genomic DNA and primers cs_attH4X_F1 and Puro rev24 (for attL junction) and cs_attH4X_R1 and pr21 (for attR junction). PCR amplified products of the expected size (1100 bp; for the left junction) were detected in clones 3, 19 and 21 (top two panels) and (∼375 bp; for the right junction) in clones 11, 19 and 21 (bottom two panels). W, no DNA template control; HT, negative control (genomic DNA from parental cells); Ina, genomic DNA from puromycin resistant clones obtained through co-transfection of pattP4X-PGKssPuro and pCMVssIna, the latter expressed inactive Int as negative control; +, positive control (genomic DNA from a HT1080 clone carrying an attH4X x attP4X integration event); M, 100 bp DNA ladder; 1 to 33, genomic DNA from puromycin resistant HT1080 clones obtained through co-transfection of pattP4X-PGKssPuro and pCMVssInt-C3CNLS. (D) Southern blot analysis. Genomic DNA purified from five targeted HT1080 clones, as indicated, and parental HT1080 cell line was subjected to digestion with PstI or XbaI. A PCR-derived digoxigenin-labeled probe complementary to the puromycin resistance gene was used. Lanes: M1, 1 kb DNA ladder; HT, genomic DNA from HT1080; 3, 11, 18*, 19 and 21, genomic DNA from targeted clones; pP4X (107, 108, 109), copies of linearized pattP4X-PGKssPuro loaded as positive control. The arrows indicate fragments of expected size for clones 3 and 11. *HT 1080 Clone 18 (carrying a single-copy transgene) was obtained from a screen of puromycin-resistant HT1080 clones that resulted from a previous co-transfection of pattP4X-PGKssPuro and pCMVssInt-h/218. White arrow heads indicate fragments of the expected size and black arrow heads indicate extra or unexpected fragments in the targeted clones.
Human HT1080 fibrosarcoma cells were co-transfected with target vector pattP4X-PGKssPuro (Figure 1B) and, in our initial experiment, Int-h/218 expression plasmid pCMVssInt-h/218. Self-ligated genomic restriction fragments from puromycin-resistant bulk cultures were subsequently analyzed by inverse PCR followed by sequencing of PCR products (primers rev303 and fw318 in Figure 1B; Supplementary Figure S1a). This revealed site-specific integration into a sequence which we named attH4X.
The attH4X sequence was found in open reading frame 1 (ORF 1) of a LINE-1 element on chromosome 3. LINE-1 ORF1 encodes an RNA-binding protein required for ribonucleoprotein particle assembly during retro-transposition (29). The ORF1-targeted attH4X matched attH7X, except for three nucleotides at the 5′-end (Figure 1A).
We next searched the human genome with the 18 bp attH4X sequence as query and identified 935 hits, all belonging to ORF1 of various LINE-1 subfamilies scattered throughout the human genome. Based on sequence homology between the 935 hits, consensus ORF1 primers flanking attH4X were designed to enable first round PCR-screening of integration events (Supplementary Table S4). In different genome searches, exact copies of attH4X were also found in LINE-1 sequences of other primates, notably in the Chimpanzee and Gorilla genome (data not shown).
In order to verify targeting of attH4X, 3 × 106 HT1080 human fibrosarcoma cells were co-transfected with pattP4X-PGKssPuro (Figure 1B) and Int-C3 expression vector pCMVssInt-C3CNLS. The latter expressed the more active Int-C3 variant (20) which carried a C-terminal NLS. The NLS was found to boost integrative recombination reactions by 2- to 3-fold in engineered HeLa test cells which harbored a single copy artificial genomic att site (data not shown).
Using ORF1 screening primers in combination with target vector primers, 4 out of 33 puromycin-resistant HT1080 clones identified via PCR/DNA sequencing revealed at least one correct recombination junction (Figure 1C). Based on the predicted genomic locations of the corresponding four LINE-1 elements, specific genomic primers were designed and subsequent sequencing of PCR products confirmed integration of full-length pattP4X-PGKssPuro in attH4X in all 4 clones (data not shown). Importantly, in each case both recombination junction sequences revealed precise vector integration as expected for Int-mediated catalysis, i.e. without nucleotide additions or deletions (Supplementary Table S5). Southern blotting showed that clones #19 and #21 carried a single-copy transgene whereas clones #3 and #11 either carried one additional integration event or were not entirely clonal (Figure 1D). The genomic restriction pattern of clone #19 matched the predicted intergenic location of the targeted LINE-1 element on human chromosome 2 (Supplementary Tables S5 and S6).
Int-mediated LINE-1 targeting in different cell types
Using PCR/DNA sequencing and Southern blotting, we confirmed attH4X-targeting in HeLa cells and human immortalized NEB-1 keratinocytes (30). With HeLa cells, five of the 44 analyzed clones showed attH4x targeting events, and single-copy LINE-1 integration was confirmed in four of them by Southern analysis (data not shown). Genomic restriction fragments from three clones matched the predicted locations (Supplementary Tables S5 and S6). One additional sequence that was targeted in LINE-1 was identified in the PCR screen but, although present in ORF1, substantially deviated from attH4x and will be described further below in the context of off-target events. With NEB-1 cells, two of the 26 clones showed attH4X-targeting, and single-copy integration was detected in both cases with genomic fragments matching the predicted loci (Supplementary Tables S5 and S6). Together, these data revealed that Int-C3CNLS-mediated site-specific transgene addition to LINE-1 elements can be achieved with different human cell types and occurred in about 10% of analyzed clones, with more than half carrying intact single-copy transgenes.
It is possible that clones carrying multiple transgene copies resulted from insertions into different LINE-1 elements rather than from a mixture of targeted and random integration events. However, we did not investigate this possibility further in the current study. The data also showed that even difficult-to-transfect human cells, such as keratinocytes, could be targeted by Int-C3CNLS. Importantly, we also achieved site-specific integration into attH4X in HT1080 cells using co-transfected mRNA to express Int-C3CNLS (Supplementary Table S5), thus eliminating the possibility of random background integration events with recombinase expression vectors.
LINE-1 targeting in hESCs
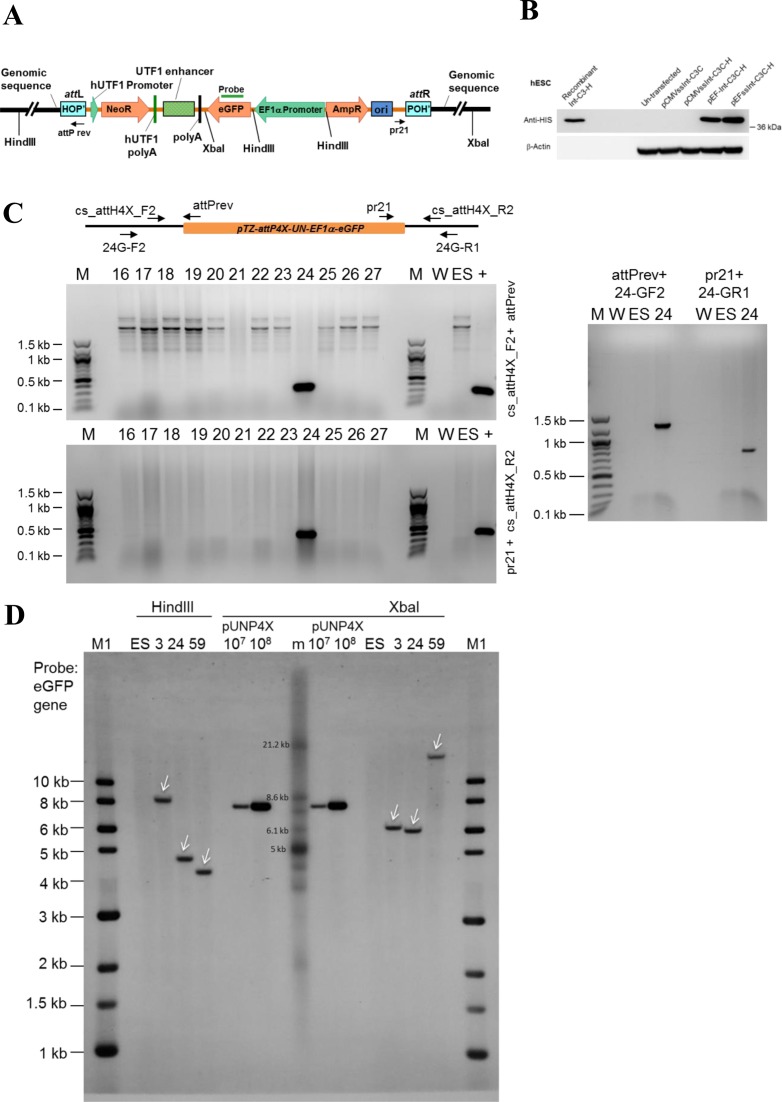
We next targeted attH4X in hESCs by employing our established human Undifferentiated Transcription Factor 1 (UTF1) gene-based pluripotency reporter cassette. Expression of this reporter is under the control of pluripotency factors OCT4, SOX2 and, most likely, NANOG (25,31). Expression of endogenous UTF1 is known as one of the most reliable indicators of human and mouse ESC pluripotency (32). Successful targeting attH4X yields a recombinant product comprised of the entire 8.2 kb vector, i.e. the neomycin resistance gene controlled by UTF1 promoter/enhancer elements, a reporter gene (EGFP) under the control of the constitutive EF1α promoter, as well as bacterial plasmid sequences. The inserted vector is flanked by hybrid attL/R sites in the genome (Figure 2A).
Figure 2.
attH4X targeting in human embryonic stem cell (hESCs). (A) Schematic diagram of pTZ-attP4X-UN-EF1α-eGFP targeting vector after integration into attH4X. Positions of relevant primers, the Southern probe targeting EGFP and HindIII and XbaI restriction sites are indicated. (B) Western blot showing Integrase expression in hESCs. Lysates from hESCs transfected with plasmids expressing Int-C3CNLS (pCMVssInt-C3C), 6xHIS-tagged Int-C3CNLS (pCMVssInt-C3C-H, pEF-Int-C3C-H, pEFssInt-C3C-H) and untransfected control cells were analyzed by western blotting with an anti-HIS tag antibody (top panel). Purified HIS-tagged Integrase C3 was employed as positive control. β-actin was used as loading control (bottom panel). (C) Example of screening for attH4X × attP4X recombination events in hESCs. PCR was performed with genomic DNA (extracted from neomycin-resistant, EGFP-positive hESC recombinants) and primers cs_attH4X_F2 and attP rev (for the left junction; top left panel) and cs_attH4X_R2 and pr21 (for the right junction; bottom left panel). PCR amplified products of the expected sizes (278 and 439 bp) were detected in clone #24. The right panel shows a PCR analysis to confirm site-specific recombination in clone #24 using different genomic locus-specific primers. PCR-amplified products of the expected sizes (∼1.25 kb with primers attP rev and 24G-F2, and ∼750 bp with primers pr21 and 24G-R1) were obtained and confirmed by sequencing. W, no DNA template control; ES, negative control (genomic DNA from parental hESCs); +, positive control (genomic DNA from HT1080 clone #19); M, 100 bp DNA ladder; M1, 1 kb DNA ladder; 16 to 27, genomic DNA from neomycin resistant hESC clones obtained through co-transfection of pTZ-attP4X-UN-EF1α-eGFP and pEF1α-ssInt-C3CNLS. (D) Southern blot analysis. Genomic DNA purified from three targeted hESC clones and parental hESC cell lines were digested with HindIII or XbaI. A probe complementary to EGFP was employed. Lanes: M1, 1 kb DNA ladder; m, DNA ladder (TeloTAGGG Telomere Length Assay kit, Roche); ES, parental DNA; 3, 24, 59, genomic DNA from targeted hESC clones; pUN4X (107, 108), copies of linearized targeting vector pTZ-attP4X-UN-EF1α-eGFP. White arrow heads indicate fragments of the expected size in the targeted clones.
First, we noted that transient Int-C3CNLS expression from pCMVssInt-C3CNLS is strongly attenuated in hESCs and therefore employed the EF1α promoter for recombinase expression (Figure 2B). Co-transfection of hESCs with pTZ-attP4X-UN-EF1α-eGFP and Int-C3NLS expression vector followed by selection with neomycin resulted in 68 colonies. PCR screening plus sequencing revealed successful attH4X-targeting in three clones (Figure 2C and data not shown). Southern analysis showed that each clone harbored a single-copy transgene and that their genomic fragment sizes matched the predicted locations, i.e. two introns and one intergenic region (Figure 2D; Supplementary Tables S5 and S6).
Transgene cassettes remain functional at targeted LINE-1 loci
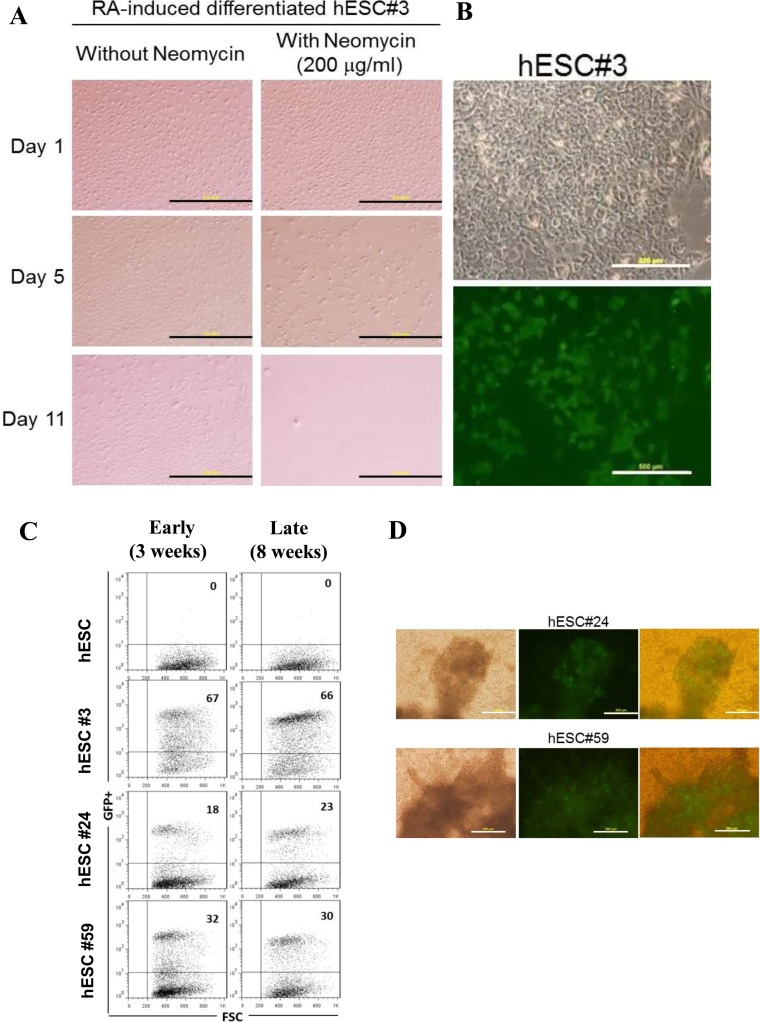
Our UTF1-based neomycin selection cassette has been used before to ablate differentiated cells from hESC cultures (25). We subjected the three single-copy transgenic hESC clones identified in the previous section to RA-induced differentiation and observed complete loss of cell viability within 11 days in the presence of neomycin (Figure 3A; data not shown). Western blotting of pluripotency markers confirmed induction of differentiation for all three hESC clones (Supplementary Figure S2; clones #3, #24 and #59). Hence, the UTF1-based selection cassette remained functional after single-copy integration into LINE-1 in hESCs and was turned off in differentiated progenies leading to complete cell ablation under selection pressure. In addition, robust expression of the constitutive EGFP reporter was detected in different clones, ranging from 18 to 67%, and the expression levels remained essentially unchanged over a period of at least two months of continued passaging (Figure 3B and C). Expression of the reporter was also largely maintained in fully differentiated, beating cardiomyocytes (Figure 3D; Supplementary Movies 1–3). The latter result indicated that transgenesis at the respective LINE-1 elements does not substantially interfere with the differentiation potential of these targeted cell lines.
Figure 3.
Functionality of transgenes in LINE-1-targeted hESCs. (A) Functional test for UTF1 promoter-driven neomycin resistance. hESC clone #3 was induced to differentiate by retinoic acid (RA) treatment and cultured in differentiation medium with or without neomycin for up to 12 days. In differentiated cells, cell death was evident from day 5, following neomycin treatment (bottom panels) while cells without neomycin treatment continued to grow (top panels). Magnification of 4×, Scale bars, 2 mm. (B) GFP expression in targeted hESC clones. A representative phase-contrast light micrograph (top panel) and fluorescence micrograph showing stable GFP expression (bottom panel) in hESC#3. Magnification of 10x, Scale bars, 500 μm. (C) FACS analysis of targeted clones. Dot plots representing GFP+ cells (upper right quadrant) and GFP− cells (lower right quadrant) for untargeted hESCs (parental cells) and targeted hESC clones #3, #24 and #59 after 3 weeks (early) and 8 weeks (late) of culturing the cells. (D) Cardiac differentiation of targeted hESC clones. Representative photomicrographs of cardiomyocytes generated from hESC clones #24 and #59. Light micrographs (left) and fluorescence micrographs showing EGFP expression in differentiated cells (middle). Overlay of light and fluorescence micrographs (right). Pulsating EGFP-positive cardiomyocytes could be seen at day 7 post induction. Movies of beating cardiomyocytes (Supplementary Movies 1–3).
It is perhaps interesting to note here that we isolated EGFP+ cells from individual undifferentiated hESC clones and monitored transgene expression over a period of time. Beginning with close to 100% EGFP+ cells, this fraction became smaller and eventually petered out to levels that were very similar to those seen before sorting (data not shown). The mechanism regulating this homeostatic behavior of transgene expression is, to our knowledge at least, completely unknown.
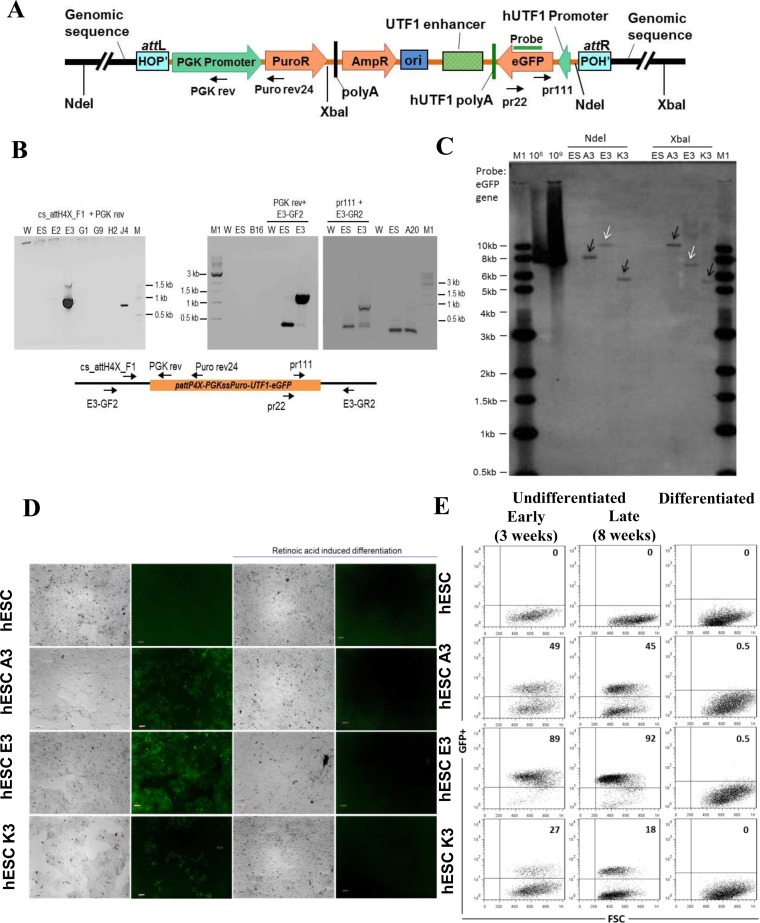
In another application of our UTF1 reporter cassette (31), we employed target vector pattP4X-PGKssPuro-UTF1-EGFP. Here, the UTF1 promoter/enhancer elements control EGFP expression, while the puromycin resistance cassette is constitutively expressed after targeted genomic integration (Figure 4A). Co-transfection of the target vector pattP4X-PGKssPuro-UTF1-EGFP and Int-C3NLS expression vector followed by selection with puromycin resulted in 120 colonies. Preliminary PCR screening using ORF1 forward primer followed by PCR product sequencing identified 17 clones containing the recombination junction attL, indicating successful attH4X targeting (data not shown). Both attL and attR junctions were confirmed in 5/17 clones by genomic PCR and sequencing, and clones A3, E3 and K3 were chosen because of the intergenic chromosomal location of transgenes and the level of EGFP expression. (Figure 4B; data not shown; Supplementary Table S5).
Figure 4.
Targeting attH4X in hESCs with pattP4X-PGKssPuro-UTF1-eGFP. (A) Schematic diagram of pattP4X-PGKssPuro-UTF1-eGFP targeting vector after integration. Locations of primers (Puro rev24, PGK rev, pr111 and pr22) and the Southern probe are indicated. (B) Screening for attH4X x attP4X recombination events in selected hESC clones. Semi-nested PCR with primers cs_attH4X_F1 and PGK rev (for the left junction) using templates obtained with primary PCR (primers cs_attH4X_F1 and Puro rev24). PCR products of the expected size (∼900 bp) were detected in hESC clone E3 (left panel). Confirmatory PCR with genomic locus specific primers were performed for clone E3. PCR products of expected size (∼1100 bp) were obtained in a semi-nested PCR with primer PGK rev and genomic locus-specific forward primer E3-GF2 using templates from a primary PCR (primers Puro rev24 and primer E3-GF2; middle panel). PCR products of expected size (∼1000 bp) were obtained in a semi-nested PCR with primer pr111 and genomic locus specific reverse primer E3-GR2 (for the right junction) using templates from a primary PCR (primers pr22 and E3-GR2; right panel). W, no DNA template control; ES, negative control (genomic DNA from parental cells); M, 100 bp DNA ladder; M1, 1 kb DNA ladder; E2, E3, G1, G9, H2, J4, B16, A20, genomic DNA from puromycin resistant and GFP-positive hESC clones obtained through co-transfection of pattP4X-PGKssPuro-UTF1-eGFP and pEF1α-ss-Int-C3CNLS. (C) Southern blot analysis. Genomic DNA from three hESC clones and parental hESC cells were digested with NdeI or XbaI. Digoxigenin-labeled probe to EGFP was employed. Lanes: M1, 1 kb DNA ladder; 108, 109, copies of linearized targeting vector as positive control; ES, parental DNA; A3, E3 and K3, genomic DNA from targeted hESC clones. White arrow heads indicate fragments of the expected size and black arrow heads indicate extra or unexpected fragments in the targeted clones. (D) Functional test for UTF1 promoter-driven EGFP expression in targeted hESC clones. Fluorescence microscopic analysis of undifferentiated and RA-induced, differentiated parental hES-047 cells and clones A3, E3 and K3. EGFP expression was detected with the undifferentiated hESC clones A3, E3 and K3 (column 2, panels 2, 3 and 4) but disappeared in differentiated progenies (column 4, panels 2, 3 and 4) respectively. Panels in columns 1 and 3 are phase-contrast light micrographs of undifferentiated and differentiated cells, respectively. Magnification 5×; Scale bars 100 μm. (E) FACS analysis for undifferentiated and differentiated hESCs clones Dot plots representing GFP+ cells (upper right quadrant) and GFP− cells (lower right quadrant) for the untargeted hESCs, undifferentiated targeted hESC clones (A3, E3, K3) after 3 weeks (early) and 8 weeks (late) of culturing the cells (left and middle panel) and their differentiated progenies (right panel).
Southern blotting for these three clones revealed that they carried single-copy transgenes, with clone E3 matching the predicted genomic fragment sizes (Figure 4C; Supplementary Table S6). The UTF1-driven EGFP expression pattern for each clone was determined by cell imaging and quantified by FACS. The undifferentiated clones A3, E3 and K3 showed fractions of 49, 89 and 27% EGFP+ cells, respectively, and this level remained largely unchanged during extended cell passaging (Figure 4D and E). Importantly, the fraction of EGFP+ cells was substantially reduced to 0.5% or less as a result of RA-induced differentiation (Figure 4D and E; Supplementary Figure S2). The nearly complete loss of detectable EGFP expression demonstrated again the sustained functionality of the UTF1 reporter cassette integrated at the respective LINE-1 loci during extended periods of cell culture.
Int-C3CNLS neither induces DNA damage response nor cytotoxicity
Phage λ Int is the prototypical tyrosine recombinase (14). Other members of this enzyme family, notably Cre, cause cytotoxicity when expressed at high levels (33). It was therefore important to determine whether Int-C3CNLS induces signs of cell toxicity and/or DNA damage at expression levels suitable for genome engineering.
We used a recombination-inactive Int-h/218 variant carrying an alanine substitution for the scissile bond-forming tyrosine 342 as negative control and observed no impact on cell viability during 9 days after transfection in (p53 wild-type) HT1080 fibrosarcoma cells (Figure 5A). We next employed human A549 lung cancer cells and, over a period of 3 days, determined the amount of phosphorylated γH2AX as marker for DNA damage. Compared to cells transfected with inactive Int and hydroxyurea (HU)-treated cells as negative and positive controls, respectively, Int-C3CNLS failed to notably trigger p-γH2AX expression (Figure 5B). This was repeated with HT1080 cells and we observed again no detectable p-γH2AX induction over a period of 48 and 72 h using HU-treated cells as positive control (Figure 5C). Lastly, we subjected three single-copy transgenic hESC clones as well as the parental hESCs to karyotype analysis and found no statistically significant chromosomal aberrations (Figure 5D). Together, these data indicated that, at least at this level of analysis, transient expression of Int-C3CNLS in human cells for attH4X-targeting is safe with respect to cell toxicity and host cell genome integrity.
Figure 5.
Int-C3CNLS does not induce DNA damage or cytotoxicity. (A) MTT-based cell proliferation assays were performed to assess effects on cell proliferation rates upon Int expression in human cell lines. HT1080 cells untransfected (HT1080), and FACS sorted GFP+ cells obtained after co-transfection of pCMV-EGFP with either pCMVssInt-Ina (INA; expressing inactive integrase) or pCMVssInt-C3CNLS (C3CNLS) were analyzed for the effect on cellular proliferation using the colorimetric MTT assays over the indicated time course. Data show the mean of triplicates and standard deviation of a representative experiment. n = 2. (B) Western blot analysis to determine phospho-γ-H2AX levels to assess DNA damage induced by expression of Int in A549 cells. Cell lysates prepared at time points of 24, 48 and 72 h (post transfection) from A549 cells transfected with pCMVssInt-Ina or pCMVssInt-C3C (expressing Int-C3CNLS) and from control cells treated with the carrier (Lipofectamine2000 Transfection reagent) were subjected to western blot analysis using antibodies against phospho-γ-H2AX (top panel). UT, untreated cells as negative control; HU, cells treated with hydroxy urea (10 mM for 24 h) as positive control; M, Marker lane. β-actin was used as loading control (bottom panel). (C) Western blot analysis to determine phospho-γ-H2AX levels to assess DNA damage induced by expression of Int in HT1080 cells. Forty-eight hours post transfection, top and 72 h post transfection, bottom. Lysates from HT1080 cells transfected with pCMV vector, plasmids expressing 6xHIS-tagged Inactive integrase (pCMVssInt-Ina-H), 6xHIS-tagged Int-C3 (pCMVssInt-C3-H), 6xHIS-tagged Int-C3-CNLS (pCMVssInt-C3C-H) were analyzed by western blotting with anti-HIS tag antibodies and phospho-γ-H2AX antibodies. UT, untreated cells; HU, cells treated with hydroxy urea (10 mM for 24 h) as positive control; C3-H, purified recombinant HIS-tagged Int-C3. HIS-tagged Int variants were detected at the expected size of 40 kDa in lysates from cells transfected with the integrase expression plasmids. There was no detectable induction of phospho-γ-H2AX upon expression of Int-C3-H or Int-C3CNLS-H compared to inactive Int-expressing cells and HU-treated cells. β-Actin protein levels were determined as loading controls. (D) Karyotyping to verify chromosomal stability. The targeted hESC lines hESC#3, hESC#59, hESC#E3 and the parental hESC-047 were karyotyped by G-banding of metaphase chromosomes. A representative karyotype (from 20 scored and five analyzed GTG-banded cells) for each cell line is shown. Results indicated no apparent chromosomal abnormalities in the tested cell lines.
A set of attH4X loci as putative safe harbor sites
One important criterion to classify a genomic site as safe harbor site is no or minimal interference of the transgene with the rest of the genome. To investigate this possibility for selected attH4X sites, we performed global transcriptome analysis on single-copy integrant hESC clones #3, #24 (Figure 2D) and E3 (Figure 4C) and determined expression of about 20 000 protein-coding genes through targeted next-generation sequencing (Ion AmpliSeq™ Transcriptome Human Gene Expression; Life Technologies) in combination with the Ion Proton System, which covers >95% of human RefSeq genes (see ‘Materials and Methods’ section). The subsequent bioinformatics analysis to determine differential gene expression was performed by Torrent Suite™ Software.
We isolated mRNA from the three single-copy integrant clones and determined comparative expression levels from cells harvested at early and late cell passages, and from their corresponding samples of parental cells. Initially, a comparison of gene expression data from parental cell lines at the different passages revealed that out of 20,000 genes, the majority showed up to 4-fold differential expression, which can be regarded as background variation. Hence, we used a 4-fold difference in expression levels as cut-off for genes consistently differentially expressed, irrespective of cell passage, and identified expression variations for 22, 2, and 45 out of 20 000 genes for clones #3, #24 and E3, respectively (Supplementary Table S7). This indicated that, at least for these three LINE-1 elements, targeting had a rather negligible effect on the global transcriptome.
A shorter attL variant recombines into LINE-1 sites
An interesting question that we addressed next was whether the shorter hybrid attL or attR sites, which differ from attP in the arrangement of Int arm and accessory binding sites, can functionally replace attP4X for LINE-1 targeting. The adjusted attL/R spacer variants, termed attL4X and attR4X, were cloned as direct repeats into target vector pTZ18R-attL4X/attR4X-pgksspuro-UTF1-EGFP (Supplementary Figure S3a) and co-transfected with Int-C3CNLS expression vector into HT1080 cells. ORF1 primer-based PCR screening of 60 puromycin-resistant clones revealed eight with targeted attH4X loci (Supplementary Figure S3b). Southern blotting showed that five clones carried single-copy transgenes (Supplementary Figure S3c). Sequence analyses for both junctions revealed that each clone resulted from attL4X recombination with attH4X, indicating that attL4X functionally replaced attP4X (Supplementary Tables S5 and S6). These data implied that intra-molecular attL4X x attR4X recombination by Int-C3CNLS appeared to be strongly disfavored over inter-molecular attL4X × attH4X recombination. One interpretation of this surprising finding is that the arrangement of Int arm binding sites on the target vector is critical for the recombinogenic potential of att sites in combination with Int-C3, at least in human cells.
Int-C3-mediated off-target events
Our data revealed that about 10% of analyzed cell clones obtained from different cell lines showed attH4x targeting in LINE-1 elements and, based on our Southern blot analysis, half of them carried a single copy of the respective transgene cassette (Supplementary Table S6). Transfection of any target vector for transgene insertion or donor DNA template for homology-directed DNA synthesis/integration with subsequent selection inevitably generates a fraction of cell clones that result from random integration due to illegitimate recombination events. This is irrespective of the system employed for attempted sequence-specific genome targeting, such as CRISPR/Cas9, designer nucleases or site-specific recombinases. In addition, every targeting system leads to off-target events where the transgenic DNA is integrated by the system into other genomic sequences than the intended target.
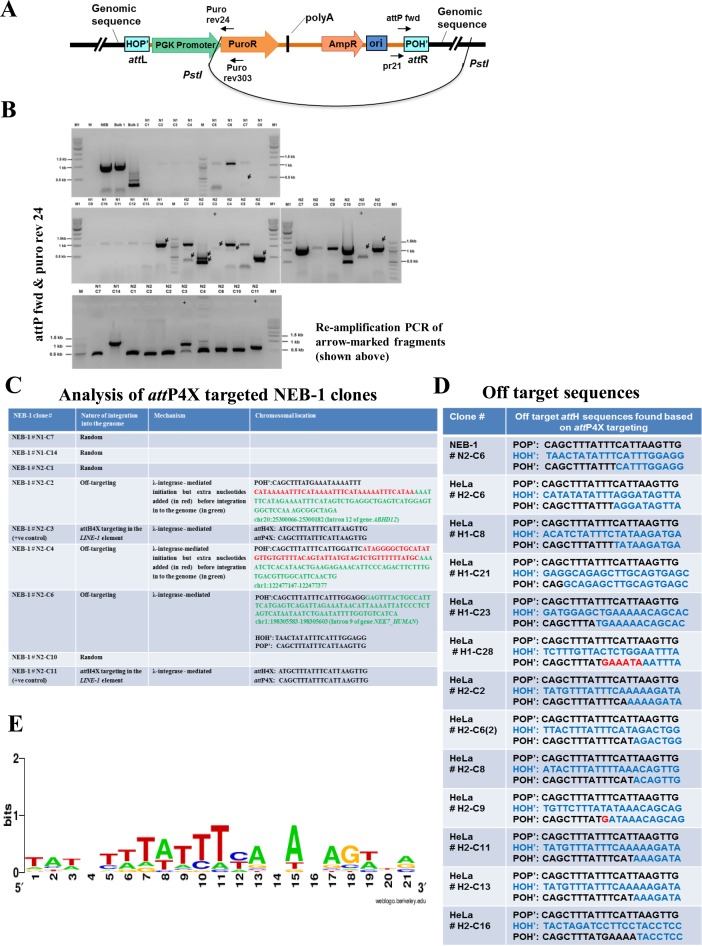
In order to analyze off-target events as a result of Int-C3-mediated recombination, we employed inverse PCR with DNA sequencing (Figure 6A) and analyzed restricted genomic DNA from the NEB-1 and HeLa cell clones described above. It is important to note that such an analysis depends on a number of parameters which are difficult to control, such as efficiency of both fragment self-ligation and subsequent PCR, and the position of restriction sites in the genome.
Figure 6.
Off-target analysis in NEB-1 and HeLa cell lines. (A) Schematic drawing showing self-ligation of PstI digested genomic DNA consisting of integrated pattP4X-PGKssPuro target vector. Positions of relevant primers (Puro rev24, Puro rev303, pr21, attP fwd) used for inverse nested PCR to determine the potential attR site are indicated. (B) Screening for off-targeting events in NEB-1 clones. Inverse nested PCR was performed using genomic DNA from 26 puromycin-resistant NEB-1 clones which were obtained from two independent transfections. PCR products that were not present in control PCRs (with genomic DNA of NEB-1 parental cells as template) and were larger than 200 bp (fragments marked with black arrows) were detected in clones N1-C7, N1-C14, N2-C1, N2-C2, N2-C3, N2-C4, N2-C6, N2-C10, N2-C11 (upper two panels). They were extracted and re-amplified (bottom panel) for sequencing. W, no DNA template control; NEB, negative control (genomic DNA from parental cells); Bulk 1 & bulk 2, genomic DNA template from all puromycin resistant colonies obtained through two independent co-transfections of pattP4X-PGKssPuro and pCMVssInt-h/218CNL; N1(C1-C14) and N2 (C1-C12), genomic DNA from puromycin resistant NEB-1 clones obtained through two independent co-transfections of pattP4X-PGKssPuro and pCMVssInt-C3CNLS; +, positive control (genomic DNA from a NEB-1 clone carrying an attH4X × attP4X integration event); M, 100 bp DNA ladder; M1, 1 kb DNA ladder. (C) Table showing sequence analysis of NEB-1 targeted clones (nature, possible mechanism and chromosomal location of genomic integration of target vector). (D) Table showing the observed off-target sequences based on attP4X targeting in NEB-1 and HeLa cell lines. (E) Sequence logo analysis for the 21bp off-target attH (HOH‘) sequences found based on attP4X targeting in NEB-1 and HeLa cell lines.
Inverse PCR with DNA from the 26 NEB-1 clones as template resulted in ten products which were re-amplified and sequenced (Figure 6B). Sequence analysis confirmed attH4x targeting in clones N2-C3 and N2-C11, as described above (Figure 6C; Supplementary Table S5). Sequencing of four PCR products returned only vector sequences, which most likely indicated random integration events. Four sequences revealed off-target integration into three different genomic loci, whereby the two sequences from clone N2-C2 produced the same hit. Two off-target events apparently involved the addition of a few extra nucleotides; one of these short sequences (N2-C4) matched attP and could indicate a PCR artifact, while the other sequence (N2-C2) could not be identified (Figure 6C).
Our initial PCR-based screen for attH4x targeting in HeLa cells revealed five events and, as mentioned above, an additional event which occurred at a different sequence but also within LINE-1 and hence qualified as an off-target event (#H2-C6; Figure 6D). To identify other off-target events, the same inverse PCR strategy was employed to analyze genomic DNA from 44 HeLa clones (data not shown). This confirmed two of the five attH4x targeting events reported above (H1-C7 and H2-C15; Supplementary Table S5) and revealed 12 off-target events which appeared to be Int-C3-mediated (Figure 6D). A sequence logo analysis (34) did not return a consensus target sequence for these events (Figure 6E). Finally, it should be noted that exactly the same off-target event which included the addition of few nucleotides and occurred on chromosome 20 in NEB-1 clone # N2-C2 (Figure 6C) was identified by inverse PCR in two of the 44 HeLa clones (data not shown). This indicated that at least some off-target events occurred at preferred genomic sequences. It could be interesting to investigate in the future the mechanistic basis for these off-target preferences, which is briefly discussed below.
DISCUSSION
Over the past two decades, efforts have been made to harness the potential of genome engineering and to establish proof of concept for functional and safe human genome engineering, and subsequent translation for gene therapy and other clinically relevant applications. Most efforts in this direction are compromised due to the critical thresholds of safety and efficacy issues. This necessitates expansion and refinement of the current editing toolbox.
Apart from safety issues such as genotoxicity, chromosomal aberrations, off-target effects associated with random (viral and plasmid-mediated) as well as site-specific integration (e.g. via ZFNs, TALENs, CRISPR/Cas9), the difficulties of integrating functional multi-transgene cassettes is yet another limitation. For instance, in applications such as stem cell engineering wherein faithful expression of large transgenes from a single locus is required to monitor various developmental lineages, cargo size limitations become critical factors. Therefore, gene addition tools which provide the flexibility of inserting large multi-transgenes that remain functional in a simple and efficient manner would make an impact.
In this context, we have developed a λ Int-mediated site-specific transgenesis tool for the human genome. Our tool faithfully inserts multi-transgene cassettes into an endogenous human sequence, attH4X. The attH4X sequence is found in LINE-1 elements and occurs at 935 different loci scattered throughout the genome. Integration events at attH4X can initially be screened by PCR using our consensus primers in the conserved regions flanking attH4X sequences. Our prototype is simple: it can yield the desired outcome within 5–6 weeks, i.e. from cell transfection until derivation of a panel of fully characterized single-copy transgenic clones. The technology has been validated in various cell lines (HeLa, fibrosarcoma HT1080, NEB-1 keratinocytes and hESCs), demonstrating that recombination between genomic attH4X and vector-borne attP4X occurred as predicted, i.e. without generating indels.
We identified a total of 42 Int-targeted LINE-1 sequences in this study, which were identical or nearly identical to attH4x (Supplementary Table S5). In order to compare with the 21 bp attB sequence, we included three additional 5′ nucleotides obtained from the respective genomic loci for sequence logo analysis. This revealed an attH4x consensus sequence shown in Figure 1A. It is clear that the spacer sequence, the 3′ Int core binding site and the right half of the 5′core site in attH4x are critical for Int-C3-mediated recombination. The first three nucleotides of the 5′ core site, however, do not seem to be functionally important (Figure 1A). The reason for this functional asymmetry within attH4x, when compared to the attB site, is not clear. Similar recombination site promiscuities have recently been described for the wild-type HK022 Int, but extend in this case also into the 3′ core site (35).
The overall targeting efficiency of attH4X was about 10%; however, its real value might be higher because sequence polymorphism in the 935 LINE-1 ORF1 likely compromises consensus primer binding. Hence, some successful targeting events may have been missed in our first round screens. Furthermore, fine-tuning of the ratios of target vectors and Int expression levels may increase the proportion of LINE-1-targeted to random integration events. Adjusting Int expression levels will also help to reduce the fraction of observed off-target events. Nevertheless, each attH4X targeting attempt, starting with about 106 human cells, resulted in tractable single-copy transgene addition to LINE-1. Further upscaling of transfected cells should readily lead to a sizeable panel of a dozen or so single-copy integrant clones from a single targeting attempt, which could be used for evaluation of transgene performances.
We observed that our transgene cassettes remained functional and receptive to cellular signaling at targeted attH4X loci in hESCs. It is possible that Int-C3-mediated recombination preferentially occurs at loci with a more accessible or recombination-permissive chromatin structure which also promotes sustained transgene functionality. This idea is supported by the fact that integrants still contained bacterial plasmid sequences, which are prone to trigger genomic silencing in hESCs (36). In this context, it is worth noting that full length yet inactive LINE-1 elements, particularly those belonging to the L1PA7 subfamily, seem to be overrepresented in the list of targeted attH4X sequences (Supplementary Table S5). This may also indicate that a particularly permissive chromatin state could be present at these loci, especially in hESCs.
It is also clear, however, that the pattern of transgene expression from these targeted loci can vary considerably between individual clones. Because there is no a priori size limit for Int target vectors as long as they can be generated in circular form and introduced into human cells, it should be possible in the future to include flanking regulatory insulator sequences in order to minimize heterogeneity in transgene expression pattern. Another interesting future improvement of our prototype may include elimination of remaining bacterial sequences from target vectors through in vitro attP4X x attH4X intra-molecular recombination before transfection, resulting in seamless attL4X vectors which can recombine with the genomic attH4X site. This may also contribute to increased homogeneity in transgene expression from LINE-1 loci.
About 500 000 LINE-1 elements sculpted the human genome during mammalian evolution (37) and their integration sites are generally considered benign with respect to human genome function (4,38). The attH4X target sequence resides in ORF1 of the LINE-1 encoded RNA-binding protein, which is required for ribonucleoprotein particle assembly during retro-transposition (29). The vast majority of ORF1 sequences are non-functional due to mutations and truncations acquired throughout human evolution (38). Disruption of ORF1 through transgenesis may thus pose little or no risk to human genome function. In particular, attH4X loci located in long intergenic regions may fulfill this criterion of safe harbor sites (4).
Our global transcriptome analyses obtained for three hESC clones at different passages in comparison with parental cells revealed that transgenes at the respective LINE-1 loci can be considered safe since RNA expression profiles of these clones did not show substantial up/down regulation of proximal genes. Out of 20,000 genes analyzed for each clone, at most 40 genes showed more than differential 4-fold expression compared to parental cells (Supplementary Table S7). In addition, none of these differentially expressed genes fell in the category of oncogenes and most of them were located either on different chromosomes or at least 107 bp apart from the targeted attH4X locus (Supplementary Table S7). Taken together, at least for some attH4x loci, sequence-specific transgenesis leaves global transcriptomes largely undisturbed and allows functional and sustained expression of transgenes, albeit with variable efficiency, thus meeting criteria formulated for safe harbor sites (4). In this context, it is worth noting that LINE-1 elements are more prevalent in AT-rich, low-recombining and sparse gene regions of the genome (39), which also marks them as good safe harbor site candidates.
Based on sequence information obtained from genomic PCR products, we observed, in a few cases, a discrepancy between the predicted genomic location of the targeted LINE-1 element and the corresponding genomic fragment size obtained by Southern analysis. We consider two possible explanations for this discrepancy. First, the genome annotation of the corresponding LINE-1 elements may not be entirely correct perhaps due to the repetitive nature of these sequences. Second, sequence polymorphism surrounding these sequences could generate different restriction patterns in different genetic backgrounds.
Our safety profiling indicated that Int-mediated genomic targeting does not induce cytotoxicity. In addition, the karyotype of three transgenic clones compared to parental cells was normal. These data indicate that transient expression of Int-C3NLS in human cells in order to achieve attH4X targeting should be safe with respect to cell toxicity and genome integrity.
Based on current knowledge of the wild-type λ Int system (14), we consider the following scenario leading to off-target events: Int-C3, once expressed and bound to vector-borne attP inside human cells, captures a ‘naked’ attH sequence in the genome for synapsis, similar to the situation in the wild-type Int system (40) or with the target attH4x site. At this step, sequence identity between the two att site spacers is not tested by the system and strand cleavage at off-target attH and attP may occur if the synaptic complex is sufficiently stable and Int-C3 monomers in the complex are activated for catalysis. This is followed by a first round of single strand exchanges generating a Holliday junction intermediate (41).In the presence of non-homologous spacers at the off-target site and depending on the position of DNA strand cleavage, we think that Holliday junctions can be resolved by the variant Int-C3 into recombinant DNA molecules with or without generating mismatches. In the former case, mismatches can be repaired by host enzymes or, if they persist, resolved through the next round of replication. Possible examples of such events are represented by several junction sequences of off-target events (Figure 6D). In some instances of mismatch repair in recombinant products, a few nucleotides may be insertion at the site of repair. Two possible examples of such a scenario are presented by clones # H1-C28 and #H2-C9 (Figure 6D). A more detailed analysis of off-target events would require sequence information from both recombination junctions. Furthermore, biochemical analysis of in vitro recombination reactions could provide insight into the mechanism of off-target recombination catalyzed by Int-C3.
In summary, the novel λ Int-mediated sequence-specific transgenesis tool presented in this study is capable of inserting multi-transgene cassettes at putative human safe harbor sites. A distinct advantage of the system is that with a single targeting attempt, it is possible to choose the best performing clones from various single-copy integrants. Due to rapid progress at the intersection of systems/synthetic biology and gene therapy, with an increasing need for transgenesis using rather large multi-transgene cassettes for stem cell and tissue engineering (1), our prototype could become an important addition to the toolbox for human genome engineering.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Human ESCs were kindly provided by GENEA, Sydney, Australia. We thank B. Lane for a supply of NEB-1 cells and advice on cell culture, and R. Ivanyi-Nagy for comments on the manuscript.
Author Contribution: P.D. designed the study. S.H.V.C., H.M., C.M.M.W. and S.P. performed human cell-based targeting assays and analyzed data. H.M., J.Z., Z.R. and J.L. performed bioinformatic analyses. S.H.V.C. identified the targetable attH4X sequence, performed the Int mRNA based experiments and cell toxicity assays. S.H.V.C. and H.M. performed pluripotency assays. H.M. performed off-target event analyses. S.J.W., S.C. and F.J.G generated and characterized Int-C3. M.S. performed cardiomyocyte differentiation assays. All authors analyzed data. P.D., S.H.V.C. and H.M. wrote the paper.
Footnotes
Present address: Shree Harsha Vijaya Chandra, Institute of Medical Biology, Agency for Science Technology and Research, 8A Biomedical Grove, Immunos, Singapore 138648.
FUNDING
This work was supported by Exploit Pte Ltd [ETPL/11-R15COT-0006]; Nanyang Technological University PhD Scholarship [to H.M.]; Singapore Ministry of Education Academic Research Fund Tier 3 [MOE2012-T3-1-001]; National Natural Science Foundation of China [31371290 to J.L.]; Southern Medical University (to J.L.). Funding for open access charge: Singapore Ministry of Education Academic Research Fund Tier 3 [MOE2012-T3-1-001].
Conflict of interest statement. P.D., S.H.V.C., H.M., S.P., S.J.W., F.J.G. declare competing financial interests. P.D., S.H.V.C., H.M., S.P., S.J.W., F.J.G. are inventors on a related patent application. P.D. owns patent related to Int-h/218.
REFERENCES
- 1.Cheng J.K., Alper H.S. The genome editing toolbox: a spectrum of approaches for targeted modification. Curr. Opin. Biotechnol. 2014;30:87–94. doi: 10.1016/j.copbio.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Cheng A.A., Lu T.K. Synthetic biology: an emerging engineering discipline. Annu. Rev. Biomed. Eng. 2012;14:155–178. doi: 10.1146/annurev-bioeng-071811-150118. [DOI] [PubMed] [Google Scholar]
- 3.Holkers M., Maggio I., Henriques S.F., Janssen J.M., Cathomen T., Goncalves M.A. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat. Methods. 2014;11:1051–1057. doi: 10.1038/nmeth.3075. [DOI] [PubMed] [Google Scholar]
- 4.Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2012;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 5.Calos M.P. The phiC31 integrase system for gene therapy. Curr. Gene Ther. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- 6.Shah R., Li F., Voziyanova E., Voziyanov Y. Target-specific variants of Flp recombinase mediate genome engineering reactions in mammalian cells. FEBS J. 2015;282:3323–3333. doi: 10.1111/febs.13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gersbach C.A., Gaj T., Barbas C.F. 3rd. Synthetic zinc finger proteins: the advent of targeted gene regulation and genome modification technologies. Accounts Chem. Res. 2014;47:2309–2318. doi: 10.1021/ar500039w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 10.Carroll D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 11.Byrne S.M., Ortiz L., Mali P., Aach J., Church G.M. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 2015;43:e21. doi: 10.1093/nar/gku1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabriel R., von Kalle C., Schmidt M. Mapping the precision of genome editing. Nat. Biotechnol. 2015;33:150–152. doi: 10.1038/nbt.3142. [DOI] [PubMed] [Google Scholar]
- 13.Li K., Wang G., Andersen T., Zhou P., Pu W.T. Optimization of genome engineering approaches with the CRISPR/Cas9 system. PloS One. 2014;9:e105779. doi: 10.1371/journal.pone.0105779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landy A. The lambda integrase site-specific recombination pathway. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0051-2014. MDNA3-0051-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorbach E., Christ N., Schwikardi M., Droge P. Site-specific recombination in human cells catalyzed by phage lambda integrase mutants. J. Mol. Biol. 2000;296:1175–1181. doi: 10.1006/jmbi.2000.3532. [DOI] [PubMed] [Google Scholar]
- 16.Tan S.M., Droge P. Comparative analysis of sequence-specific DNA recombination systems in human embryonic stem cells. Stem Cells. 2005;23:868–873. doi: 10.1634/stemcells.2005-0044. [DOI] [PubMed] [Google Scholar]
- 17.Christ N., Droge P. Genetic manipulation of mouse embryonic stem cells by mutant lambda integrase. Genesis. 2002;32:203–208. doi: 10.1002/gene.10031. [DOI] [PubMed] [Google Scholar]
- 18.Suttie J.L., Chilton M-D., Que Q. Lambda integrase mediated recombination in plants. 2008. US Patent number 7351877 B2.
- 19.Lindenbaum M., Perkins E., Csonka E., Fleming E., Garcia L., Greene A., Gung L., Hadlaczky G., Lee E., Leung J., et al. A mammalian artificial chromosome engineering system (ACE System) applicable to biopharmaceutical protein production, transgenesis and gene-based cell therapy. Nucleic Acids Res. 2004;32:e172. doi: 10.1093/nar/gnh169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siau J.W., Chee S., Makhija H., Wai C.M., Chandra S.H., Peter S., Droge P., Ghadessy F.J. Directed evolution of lambda integrase activity and specificity by genetic derepression. Protein Eng. Des. Sel. 2015;28:211–220. doi: 10.1093/protein/gzv015. [DOI] [PubMed] [Google Scholar]
- 21.Wen S., Zhang H., Li Y., Wang N., Zhang W., Yang K., Wu N., Chen X., Deng F., Liao Z., et al. Characterization of constitutive promoters for piggyBac transposon-mediated stable transgene expression in mesenchymal stem cells (MSCs) PloS One. 2014;9:e94397. doi: 10.1371/journal.pone.0094397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petitclerc D., Attal J., Theron M.C., Bearzotti M., Bolifraud P., Kann G., Stinnakre M.G., Pointu H., Puissant C., Houdebine L.M. The effect of various introns and transcription terminators on the efficiency of expression vectors in various cultured cell lines and in the mammary gland of transgenic mice. J. Biotechnol. 1995;40:169–178. doi: 10.1016/0168-1656(95)00047-t. [DOI] [PubMed] [Google Scholar]
- 23.Choi T., Huang M., Gorman C., Jaenisch R. A generic intron increases gene expression in transgenic mice. Molecular and Cellular Biology. 1991;11:3070–3074. doi: 10.1128/mcb.11.6.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng L., Baumann U., Reymond J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan S.M., Wang S.T., Hentze H., Droge P. A UTF1-based selection system for stable homogeneously pluripotent human embryonic stem cell cultures. Nucleic Acids Res. 2007;35:e118. doi: 10.1093/nar/gkm704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savatier P., Lapillonne H., van Grunsven L.A., Rudkin B.B., Samarut J. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 1996;12:309–322. [PubMed] [Google Scholar]
- 27.Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M., et al. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim A.S., Lim T.H., See K.H., Ng Y.J., Tan Y.M., Choo N.S., Lim S.X., Yee Y., Lau L.C., Tien S.L., et al. Cytogenetic and molecular aberrations of multiple myeloma patients: a single-center study in Singapore. Chinese Med. J. 2013;126:1872–1877. [PubMed] [Google Scholar]
- 29.Erwin J.A., Marchetto M.C., Gage F.H. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat. Rev. Neurosci. 2014;15:497–506. doi: 10.1038/nrn3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morley S.M., D'Alessandro M., Sexton C., Rugg E.L., Navsaria H., Shemanko C.S., Huber M., Hohl D., Heagerty A.I., Leigh I.M., et al. Generation and characterization of epidermolysis bullosa simplex cell lines: scratch assays show faster migration with disruptive keratin mutations. Brit. J. Dermatol. 2003;149:46–58. doi: 10.1046/j.1365-2133.2003.05493.x. [DOI] [PubMed] [Google Scholar]
- 31.Morshedi A., Soroush Noghabi M., Droge P. Use of UTF1 genetic control elements as iPSC reporter. Stem Cell Rev. 2013;9:523–530. doi: 10.1007/s12015-011-9342-7. [DOI] [PubMed] [Google Scholar]
- 32.Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K., Klemm S.L., van Oudenaarden A., Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silver D.P., Livingston D.M. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 34.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolot M., Malchin N., Elias A., Gritsenko N., Yagil E. Site promiscuity of coliphage HK022 integrase as tool for gene therapy. Gene Ther. 2015;22:602. doi: 10.1038/gt.2015.37. [DOI] [PubMed] [Google Scholar]
- 36.Fontes A., Lakshmipathy U. Advances in genetic modification of pluripotent stem cells. Biotechnol. Adv. 2013;31:994–1001. doi: 10.1016/j.biotechadv.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Burns K.H., Boeke J.D. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babatz T.D., Burns K.H. Functional impact of the human mobilome. Curr. Opin. Genet. Dev. 2013;23:264–270. doi: 10.1016/j.gde.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graham T., Boissinot S. The genomic distribution of L1 elements: the role of insertion bias and natural selection. J. Biomed. Biotechnol. 2006;2006:75327. doi: 10.1155/JBB/2006/75327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richet E., Abcarian P., Nash H.A. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein-DNA complex. Cell. 1988;52:9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- 41.Rajeev L., Malanowska K., Gardner J.F. Challenging a paradigm: the role of DNA homology in tyrosine recombinase reactions. Microbiol. Mol. Biol. Rev. 2009;73:300–309. doi: 10.1128/MMBR.00038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.